/
+ - In this demo we would look in https://chtc.github.io/web-preview/preview-helloworld/
+
+**You can continue to push commits to this branch and have them populate on the preview at this point!**
+
+- When you are satisfied with these changes you can create a PR to merge into master
+- Delete the preview branch and Github will take care of the garbage collection!
+
+## Testing Changes Locally
+
+### Quickstart (Unix Only)
+
+1. Install Docker if you don't already have it on your computer.
+2. Open a terminal and `cd` to your local copy of the `chtc-website-source` repository
+3. Run the `./edit.sh` script.
+4. The website should appear at [http://localhost:8080](http://localhost:8080). Note that this system is missing the secret sauce of our setup that converts
+the pages to an `.shtml` file ending, so links won't work but just typing in the name of a page into the address bar (with no
+extension) will.
+
+### Run via Ruby
+
+```shell
+bundle install
+bundle exec jekyll serve --watch -p
+```
+
+### Run Docker Manually
+
+At the website root:
+
+```
+docker run -it -p 8001:8000 -v $PWD:/app -w /app ruby:2.7 /bin/bash
+```
+
+This will utilize the latest Jekyll version and map port `8000` to your host. Within the container, a small HTTP server can be started with the following command:
+
+```
+bundle install
+bundle exec jekyll serve --watch --config _config.yml -H 0.0.0.0 -P 8000
+```
+
+## Formatting
+
+### Markdown Reference and Style
+
+This is a useful reference for most common markdown features: https://daringfireball.net/projects/markdown/
+
+To format code blocks, we have the following special formatting tags:
+
+ ```
+ Pre-formatted text / code goes here
+ ```
+ {:.sub}
+
+`.sub` will generate a "submit file" styled block; `.term` will create a terminal style, and `.file` can
+be used for any generic text file.
+
+We will be using the pound sign for headers, not the `==` or `--` notation.
+
+For internal links (to a header inside the document), use this syntax:
+* header is written as
+ ```
+ ## A. Sample Header
+ ```
+* the internal link will look like this:
+ ```
+ [link to header A](#a-sample-header)
+ ```
+
+### Converting HTML to Markdown
+
+Right now, most of our pages are written in html and have a `.shtml` extension. We are
+gradually converting them to be formatted with markdown. To easily convert a page, you
+can install and use the `pandoc` converter:
+
+ pandoc hello.shtml --from html --to markdown > hello.md
+
+You'll still want to go through and double check / clean up the text, but that's a good starting point. Once the
+document is converted from markdown to html, the file extension should be `.md` instead. If you use the
+command above, this means you can just delete the `.shtml` version of the file and commit the new `.md` one.
+
+
+### Adding "Copy Code" Button to code blocks in guides
+
+Add .copy to the class and you will have a small button in the top right corner of your code blocks that
+when clicked, will copy all of the code inside of the block.
+
+### Adding Software Overview Guide
+
+When creating a new Software Guide format the frontmatter like this:
+
+software_icon: /uw-research-computing/guide-icons/miniconda-icon.png
+software: Miniconda
+excerpt_separator: <!--more-->
+
+Software Icon and software are how the guides are connected to the Software Overview page. The
+excerpt_seperator must be <!--more--> and can be placed anywhere in a document and all text
+above it will be put in the excerpt.
\ No newline at end of file
diff --git a/preview-fall2024-info/Record.html b/preview-fall2024-info/Record.html
new file mode 100644
index 000000000..6843c9727
--- /dev/null
+++ b/preview-fall2024-info/Record.html
@@ -0,0 +1,357 @@
+
+
+
+
+
+
+OSPool Hits Record Number of Jobs
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ OSPool Hits Record Number of Jobs
+
+
The OSPool processed over 2.6 million jobs during the week of April 14th - 17th this year and ran over half a million jobs on two separate days that week.
+
+
OSPool users and collaborators are smashing records. In April, researchers submitted a record-breaking number of jobs during the week of April 14th – 2.6 million, to be exact. The OSPool also processed over 500k jobs on two separate days during that same week, another record!
+
+
Nearly 60 projects from different fields contributed to the number of jobs processed during this record-breaking week, including these with substantial usage:
+
+ - BioMedInfo: University of Pittsburgh PI Erik Wright of the Wright Lab, develops and applies software tools to perform large-scale biomedical informatics on microbial genome sequence data.
+ - Michigan_Riles: University of Michigan PI Keith Riles leads the Michigan Gravitational Wave Group, researching continuous gravitational waves.
+ - chemml: PI Olexandr Isayev from Carnegie-Mellon University, whose group develops machine learning (ML) models for molecular simulations.
+ - CompBinFormMod: Researcher PI Geoffrey Hutchison from the University of Pittsburgh, looking at data-driven ML as surrogates for quantum chemical methods to improve existing processes and next-generation atomistic force fields.
+
+
+
Any researcher tackling a problem that can run as many self-contained jobs can harness the capacity of the OSPool. If you have any questions about the Open Science Pool or how to create an account, please visit the FAQ page on the OSG Help Desk website. Descriptions of active OSG projects can be found here.
+
+
…
+
+
Learn more about the Open Science Pool.
+
+
+
+
+
+
+
+ Resilience: How COVID-19 challenged the scientific world
+
+
In the face of the pandemic, scientists needed to adapt.
+The article below by the Morgridge Institute for Research provides a thoughtful look into how researchers have pivoted in these challenging times to come together and contribute meaningfully in the global fight against COVID-19.
+One of these pivots occurred in the spring of 2020, when Morgridge and the CHTC issued a call for projects investigating COVID-19, resulting in five major collaborations that leveraged the power of HTC.
+
+
For a closer look into how the CHTC and researchers have learned, grown, and adapted during the pandemic, read the full Morgridge article:
+
+
Resilience: How COVID-19 challenged the scientific world
+
+
+
+
+
+
+
+
+ OSG fuels a student-developed computing platform to advance RNA nanomachines
+
+
How undergraduates at the University of Nebraska-Lincoln developed a science gateway that enables researchers to build RNA nanomachines for therapeutic, engineering, and basic science applications.
+
+

+
+
The UNL students involved in the capstone project, on graduation day. Order from left to right: Evan, Josh, Dan, Daniel, and Conner.
+
+
When a science gateway built by a group of undergraduate students is deployed this fall, it will open the door for researchers to leverage the capabilities of advanced software and the capacity of the Open Science Pool (OSPool). Working under the guidance of researcher Joe Yesselman and longtime OSG contributor Derek Weitzel, the students united advanced simulation technology and a national, open source of high throughput computing capacity –– all within an intuitive, web-accessible science gateway.
+
+
Joe, a biochemist, has been fascinated by computers and mathematical languages for as long as he can remember. Reminiscing to when he first adopted computer programming and coding as a hobby back in high school, he reflects: “English was difficult for me to learn, but for some reason mathematical languages make a lot of sense to me.”
+
+
Today, he is an Assistant Professor of Chemistry at the University of Nebraska-Lincoln (UNL), and his affinity for computer science hasn’t waned. Leading the Yesselman Lab, he relies on the interplay between computation and experimentation to study the unique structural properties of RNA.
+
+
In September of 2020, Joe began collaborating with UNL’s Holland Computing Center (HCC) and the OSG to accelerate RNA nanostructure research everywhere by making his lab’s RNAMake software suite accessible to other scientists through a web portal. RNAMake enables researchers to build nanomachines for therapeutic, engineering, and basic science applications by simulating the 3D design of RNA structures.
+
+
Five UNL undergraduate students undertook this project as part of a year-long computer science capstone experience. By the end of the academic year, the students developed a science gateway –– an intuitive web-accessible interface that makes RNAMake easier and faster to use. Once it’s deployed this fall, the science gateway will put the Yesselman Lab’s advanced software and the shared computing resources of the OSPool into the hands of researchers, all through a mouse and keyboard.
+
+
The gateway’s workflow is efficient and simple. Researchers upload their input files, set a few parameters, and click the submit button –– no command lines necessary. Short simulations will take merely a few seconds, while complex simulations can last up to an hour. Once the job is completed, an email appears in their inbox, prompting them to analyze and download the resulting RNA nanostructures through the gateway.
+
+
This was no small feat. Collaboration among several organizations brought this seemingly simple final product to fruition.
+
+
To begin the process, the students received a number of startup allocations from the Extreme Science and Engineering Discovery Environment (XSEDE). When it was time to build the application, they used Apache Airavata to power the science gateway and they extended this underlying software in some notable ways. In order to provide researchers with more intuitive results, they implemented a table viewer and a 3D molecule visualization tool. Additionally, they added the ability for Airavata to submit directly to HTCondor, making it possible for simulations to be distributed across the resources offered by the OSPool.
+
+
The simulations themselves are small, short, and can be run independently. Furthermore, many of these simulations are needed in order to discover the right RNA nanostructures for each researcher’s purpose. Combined, these qualities make the jobs a perfect candidate for the OSPool’s distributed high throughput computing capabilities, enabled by computing capacity from campuses across the country.
+
+
Commenting on the incorporation of OSG resources, project sponsor Derek Weitzel explains how the gateway “not only makes it easier to use RNAMake, but it also distributes the work on the OSPool so that researchers can run more RNAMake simulations at the same time.” If the scientific process is like a long road trip, using high throughput computing isn’t even like taking the highway –– it’s like skipping the road entirely and taking to the skies in a high-speed jet.
+
+
The science gateway has immense potential to transform the way in which RNA nanostructure research is conducted, and the collaboration required to build it has already made lasting impacts on those involved. The group of undergraduate students are, in fact, no longer undergraduates. The team’s student development manager, Daniel Shchur, is now a software design engineer at Communication System Solutions in Lincoln, Nebraska. Reflecting on the capstone project, he remarks, “I think the most useful thing that my teammates and I learned was just being able to collaborate with outside people. It was definitely something that wasn’t taught in any of our classes and I think that was the most invaluable thing we learned.”
+
+
But learning isn’t just exclusive to students. Joe notes that he gained some unexpected knowledge from the students and Derek. “I learned a ton about software development, which I’m actually using in my lab,” he explains. “It’s very interesting how people can be so siloed. Something that’s so obvious, almost trivial for Derek is something that I don’t even know about because I don’t have that expertise. I loved that collaboration and I loved hearing his advice.”
+
+
In the end, this collaboration vastly improved the accessibility of RNAMake, Joe’s software suite and the focus of the science gateway. Perhaps he explains it best with an analogy: ”RNAMake is basically a set of 500 different LEGO® pieces. Using enthusiastic gestures, Joe continues by offering an example: “Suppose you want to build something from this palm to this palm, in three-dimensional space. It [RNAMake] will find a set of LEGO® pieces that will fit there.”
+
+
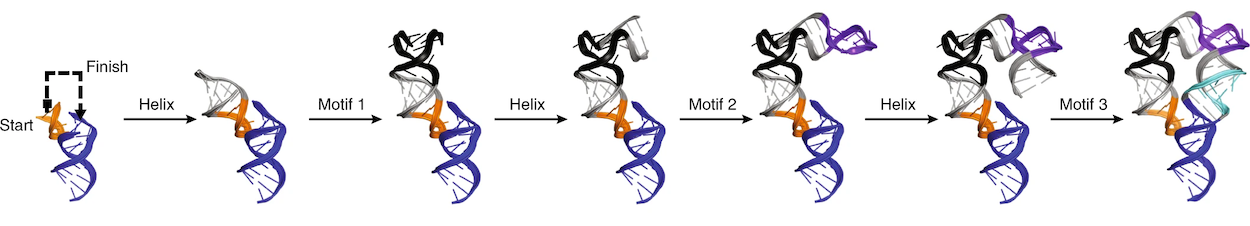
+
+
A demonstration of how RNAMake’s design algorithm works. Credit: Yesselman, J.D., Eiler, D., Carlson, E.D. et al. Computational design of three-dimensional RNA structure and function. Nat. Nanotechnol. 14, 866–873 (2019). https://doi.org/10.1038/s41565-019-0517-8
+
+
Since the possible combinations of these LEGO® pieces of RNA are endless, this tool saves users the painstaking work of predicting the structures manually. However, the installation and use of RNAMake requires researchers to have a large amount of command line knowledge –– something that the average biochemist might not have.
+
+
Ultimately, the science gateway makes this previously complicated software suddenly more accessible, allowing researchers to easily, quickly, and accurately design RNA nanostructures.
+
+
These structures are the basis for RNA nanomachines, which have a vast range of applications in society. Whether it be silencing RNAs that are used in clinical trials to cut cancer genes, or RNA biosensors that effectively bind to small molecules in order to detect contaminants even at low concentrations –– the RNAMake science gateway can help researchers design and build these structures.
+
+
Perhaps the most relevant and pressing applications are RNA-based vaccines like Moderna and Pfizer. These vaccines continue to be shipped across cities, countries, and continents to reach people in need, and it’s crucial that they remain in a stable form throughout their journey. Insight from RNA nanostructures can help ensure that these long strands of mRNA maintain stability so that they can eventually make their way into our cells.
+
+
Looking to the future, a second science gateway capstone project is already being planned for next year at UNL. Although it’s currently unclear what field of research it will serve, there’s no doubt that this project will foster collaboration, empower students and researchers, and impact society –– all through a few strokes on a keyboard.
+
+
+
+
+
+
+
+ Transforming research with high throughput computing
+
+
During the OSG Virtual School Showcase, three different researchers shared how high throughput computing has made lasting impacts on their work.
+
+

+
+
Over 40 researchers and campus research computing staff were selected to attend this year’s OSG Virtual School, all united by a shared desire to learn how high throughput computing can advance their work. During the first two weeks of August, school participants were busy attending lectures, watching demonstrations, and completing hands-on exercises; but on Wednesday, August 11, participants had the chance to hear from researchers who have successfully used high throughput computing (HTC) to transform their work. Year after year, this event –– the HTC Showcase –– is one highlight of the experience for many User School participants. This year, three different researchers in the fields of structural biology, psychology, and particle physics shared how HTC impacted their work. Read the articles below to learn about their stories.
+
+
Scaling virtual screening to ultra-large virtual chemical libraries – Spencer Ericksen, Carbone Cancer Center, University of Wisconsin-Madison
+
+
Using HTC for a simulation study on cross-validation for model evaluation in psychological science – Hannah Moshontz, Department of Psychology, University of Wisconsin-Madison
+
+
Antimatter: Using HTC to study very rare processes – Anirvan Shukla, Department of Physics, University of Hawai’i Mānoa
+
+
Collectively, these testimonies demonstrate how high throughput computing can transform research. In a few years, the students of this year’s User School might be the next Spencer, Hannah, and Anirvan, representing the new generation of researchers empowered by high throughput computing.
+
+
…
+
+
Visit the materials page to browse slide decks, exercises, and recordings of public lectures from OSG Virtual School 2021.
+
+
Established in 2010, OSG School, typically held each summer at the University of Wisconsin–Madison, is an annual education event for researchers who want to learn how to use distributed high throughput computing methods and tools. We hope to return to an in-person User School in 2022.
+
+
+
+
+
+
+
+
+ Scaling virtual screening to ultra-large virtual chemical libraries
+
+
+  + Image by the National Cancer Institute on Unsplash
+ Image by the National Cancer Institute on Unsplash
+
+
+
Kicking off the OSG User School Showcase, Spencer Ericksen, a researcher at the University of Wisconsin-Madison’s Carbone Cancer Center, described how high throughput computing (HTC) has made his work in early-stage drug discovery infinitely more scalable. Spencer works within the Small Molecule Screening Facility, where he partners with researchers across campus to search for small molecules that might bind to and affect the behavior of proteins they study. By using a computational approach, Spencer can help a researcher inexpensively screen many more candidates than possible through traditional laboratory approaches. With as many as 1033 possible molecules, the best binders from computational ‘docking’ might even be investigated as potential drug candidates.
+
+
With traditional laboratory approaches, researchers might test just 100,000 individual compounds using liquid handlers like the one pictured above. However, this approach is expensive, imposing limits both on the number of molecules tested and the number of researchers able to pursue potential binders of the proteins they study.
+
+
Spencer’s use of HTC allows him to take a different approach with virtual screening. By using computational models and machine learning techniques, he can inexpensively filter the masses of molecules and predict which ones will have the highest potential to interfere with a certain biological process. This reduces the time and money spent in the lab by selecting a subset of binding candidates that would be best to study experimentally.
+
+
“HTC is a fabulous resource for virtual screening,” Spencer attests. “We can now effectively validate, develop, and test virtual screening models, and scale to ever-increasing ultra-large virtual chemical libraries.” Today, Spencer is able to screen approximately 3.5 million molecules each day thanks to HTC.
+
+
There are a variety of virtual screening programs, but none of them are all that reliable individually. Instead of opting for a single program, Spencer runs several programs on the Open Science Pool (OSPool) and calculates a consensus score for each potential binder. “It’s a pretty old idea, basically like garnering wisdom from a council of fools,” Spencer explains. “Each program is a weak discriminator, but they do it in different ways. When we combine them, we get a positive effect that’s much better than the individual programs. Since we have the throughput, why not run them all?”
+
+
And there’s nothing stopping the Small Molecule Screening Facility from doing just that. Spencer’s jobs are independent from each other, making them “pleasantly parallelizable” on the OSPool’s distributed resources. To maximize throughput, Spencer splits the compound libraries that he’s analyzing into small increments that will run in approximately 2 hours, reducing the chances of a job being evicted and using the OSPool more efficiently.
+
+
…
+
+
This article is part of a series of articles from the 2021 OSG Virtual School Showcase. OSG School is an annual education event for researchers who want to learn how to use distributed high throughput computing methods and tools. The Showcase, which features researchers sharing how HTC has impacted their work, is a highlight of the school each year.
+
+
+
+
+
+
+
+ Technology Refresh
+
+
Thanks to the generous support of the Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation, CHTC has been able to execute a major refresh of hardware. This provided 207 new servers for our systems, representing over 40,000 batch slots of computing capacity. Most of this hardware arrived over the summer and we have started adding them to CHTC systems.
+
+
Continue reading to learn more about the types of servers we are adding and how to access them.
+
+
HTC System
+
+
On the HTC system, we are adding 167 servers of new capacity, representing 36,352 job slots and 40 high-end GPU cards.
+
+
The new servers will be running CentOS Linux 8 – CHTC users should see our website page about how to test your jobs and
+take advantage of servers running CentOS Stream 8. Details on user actions needed for this change can be found on the
+OS transition page.
+
+
New Server specs
+
+
PowerEdge R6525
+
+
+ - 157 servers with 128 cores / 256 job slots using the AMD Epyc 7763 processor
+ - 512 GB RAM per server
+
+
+
PowerEdge XE8545
+
+
+ - 10 servers, each with four A100 SXM4 80GB GPU cards
+ - 128 cores per server
+ - 512GB RAM per server
+
+
+
HPC Cluster
+
+
For the HPC cluster, we are adding 40 servers representing 5,120 cores. These servers have arrived but have not yet been added to the HPC cluster. In most cases, when we add them, they will form a new partition and displace some of our oldest servers, currently in the “univ2” partition.
+
+
New server specs:
+
+
Dell Poweredge R6525
+
+
+ - 128 cores using the AMD Epyc 7763 processor
+ - 512GB of memory
+
+
+
Users interested in early access to AMD processors before all 40 servers are installed should contact CHTC at chtc@cs.wisc.edu.
+
+
We have also obtained hardware and network infrastructure to completely replace the HPC cluster’s underlying file system and infiniband network fabric. We will be sending more updates to the chtc-users mailing list as we schedule specific transition dates for these major cluster components.
+
+
+
+
+
+
+
+ Save the dates for Throughput Computing 2023 - a joint HTCondor/OSG event
+
+
Don't miss these in-person learning opportunities in beautiful Madison, Wisconsin!
+
+
Save the dates for Throughput Computing 2023! For the first time, HTCondor Week and the OSG All-Hands Meeting will join together as a single, integrated event from July 10–14 to be held at the University of Wisconsin–Madison’s Fluno Center. Throughput Computing 2023 is sponsored by the OSG Consortium, the HTCondor team, and the UW-Madison Center for High Throughput Computing.
+
+
This will primarily be an in-person event, but remote participation (via Zoom) for the many plenary events will also be offered. Required registration for both components will open in March 2023.
+
+
If you register for the in-person event at the University of Wisconsin–Madison, you can attend plenary and non-plenary sessions, mingle with colleagues, and have planned or ad hoc meetings. Evening events are also planned throughout the week.
+
+
All the topics typically covered by HTCondor Week and the OSG All-Hands Meeting will be included:
+
+
+ - Science Enabled by the OSPool and the HTCondor Software Suite (HTCSS)
+ - OSG Technology
+ - HTCSS Technology
+ - HTCSS and OSG Tutorials
+ - State of the OSG
+ - Campus Services and Perspectives
+
+
+
The U.S. ATLAS and U.S. CMS high-energy physics projects are also planning parallel OSG-related topics during the event on Wednesday, July 12. (For other attendees, Wedneday’s schedule will also include parallel HTCondor and OSG tutorials and OSG Collaborations sessions.)
+
+
For questions, please contact us at events@osg-htc.org or htcondor-week@cs.wisc.edu.
+
+
View last year’s schedules for
+
+
+
+
+
+
+
+
+
+
+ How to Transfer 460 Terabytes? A File Transfer Case Study
+
+
When Greg Daues at the National Center for Supercomputing Applications (NCSA) needed to transfer 460 Terabytes of NCSA files from the National Institute of Nuclear and Particle Physics (IN2P3) in Lyon, France to Urbana, Illinois, for a project they were working with FNAL, CC-IN2P3 and the Rubin Data Production team, he turned to the HTCondor High Throughput system, not to run computationally intensive jobs, as many do, but to manage the hundreds of thousands of I/O bound transfers.
+
+
The Data
+
+
IN2P3 made the data available via https, but the number of files and their total size made the management of the transfer an engineering challenge. There were two kinds of files to be transferred, with 3.5 million files with a median size of roughly 100 Mb, and another 3.5 million smaller files, with a median size of about 10 megabytes. Total transfer size is roughly 460 Terabytes.
+
+
The Requirements
+
+
The requirement for this transfer was to reliably transfer all the files in a reasonably performant way, minimizing the human time to set up, run, and manage the transfer. Note the noni-goal of optimizing for the fastest possible transfer time – reliability and minimizing the human effort take priority here. Reliability, in this context implies:
+
+
Failed transfers are identified and re-run (with millions of files, a failed transfer is almost inevitable)
+Every file will get transferred
+The operation will not overload the sender, the receiver, or any network in between
+
+
The Inspiration
+
+
Daues presented unrelated work at the 2017 HTCondor Week workshop. At this workshop, he heard about the work of Phillip Papodopolous at UCSD, and his international Data Placement Lab (iDPL). iDPL used HTCondor jobs solely for transferring data between international sites. Daues re-used and adapted some of these ideas for NCSA’s needs.
+
+
The Solution
+
First, Daues installed a “mini-condor”, an HTCondor pool entirely on one machine, with an access point and eight execution slots on that same machine. Then, given a single large file containing the names of all the files to transfer, he ran the Unix split command to create separate files with either 50 of the larger files, or 200 of the smaller files. Finally, using the HTCondor submit file command
+
+
Queue filename matching files *.txt
+
+
the condor_submit command creates one job per split file, which runs the wget2 command and passes the list of filenames to wget2. The HTCondor access point can handle tens of thousands of idle jobs, and will schedule these jobs on the eight execution slots. While more slots would yield more overlapped i/o, eight slots were chosen to throttle the total network bandwidth used. Over the course of days, this machine with eight slots maintained roughly 600 MB/seconds.
+
+
(Note that the machine running HTCondor did not crash during this run, but if it had, all the jobs, after submission, were stored reliably on the local disk, and at such time as the crashed machine restarted, and the init program restarted the HTCondor system, all interrupted jobs would be restarted, and the process would continue without human intervention.)
+
+
+
+
+
+
+
+ The Future of Radio Astronomy Using High Throughput Computing
+
+
Eric Wilcots, UW-Madison dean of the College of Letters & Science and the Mary C. Jacoby Professor of Astronomy, dazzles the HTCondor Week 2022 audience.
+
+
+ 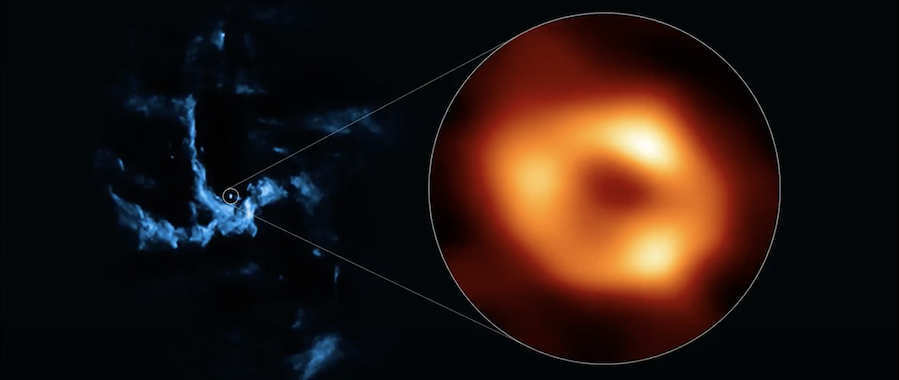 + Image of the black hole in the center of our Milky Way galaxy.
+ Image of the black hole in the center of our Milky Way galaxy.
+
+
+
+  + Eric Wilcots
+ Eric Wilcots
+
+
+
“My job here is to…inspire you all with a sense of the discoveries to come that will need to be enabled by” high throughput computing (HTC), Eric Wilcots opened his keynote for HTCondor Week 2022. Wilcots is the UW-Madison dean of the College of Letters & Science and the Mary C. Jacoby Professor of Astronomy.
+
+
Wilcots points out that the black hole image (shown above) is a remarkable feat in the world of astronomy. “Only the third such black hole imaged in this way by the Event Horizon Telescope,” and it was made possible with the help of the HTCondor Software Suite (HTCSS).
+
+
Beginning to build the future
+
+
Wilcots described how in the 1940s, a group of universities recognized that no single university could build a radio telescope necessary to advance science. To access these kinds of telescopes, the universities would need to have the national government involved, as it was the only one with this capability at that time. In 1946, these universities created Associated Universities Incorporated (AUI), which eventually became the management agency for the National Radio Astronomy Observatory (NRAO).
+
+
Advances in radio astronomy rely on current technology available to experts in this field. Wilcots explained that “the science demands more sensitivity, more resolution, and the ability to map large chunks of the sky simultaneously.” New and emerging technologies must continue pushing forward to discover the next big thing in radio astronomy.
+
+
This next generation of science requires more sensitive technology with higher spectra resolution than the Karl G. Jansky Very Large Array (JVLA) can provide. It also requires sensitivity in a particular chunk of the spectrum that neither the JVLA nor Atacama Large Millimeter/submillimeter Array (ALMA) can achieve. Wilcots described just what piece of technology astronomers and engineers need to create to reach this level of sensitivity. “We’re looking to build the Next Generation Very Large Array (ngVLA)…an instrument that will cover a huge chunk of spectrum from 1 GHz to 116 GHz.”
+
+
The fundamentals of the ngVLA
+
+
“The unique and wonderful thing about interferometry, or the basis of radio astronomy,” Wilcots discussed, “is the ability to have many individual detectors or dishes to form a telescope.” Each dish collects signals, creating an image or spectrum of the sky when combined. Because of this capability, engineers working on these detectors can begin to collect signals right away, and as more dishes get added, the telescope grows larger and larger.
+
+
Many individual detectors also mean lots of flexibility in the telescope arrays built, Wilcots explained. Here, the idea is to do several different arrays to make up one telescope. A particular scientific case drives each of these arrays:
+
+ - Main Array: a dish that you can control and point accurately but is also robust; it’ll be the workhorse of the ngVLA, simultaneously capable of high sensitivity and high-resolution observations.
+ - Short Baseline Array: dishes that are very close together, which allows you to have a large field of view of the sky.
+ - Long Baseline Array: spread out across the continental United States. The idea here is the longer the baseline, the higher the resolution. Dishes that are well separated allow the user to get spectacular spatial resolution of the sky. For example, the Event Horizon Telescope that took the image of the black hole is a telescope that spans the globe, which is the longest baseline we can get without putting it into orbit.
+
+
+
+ 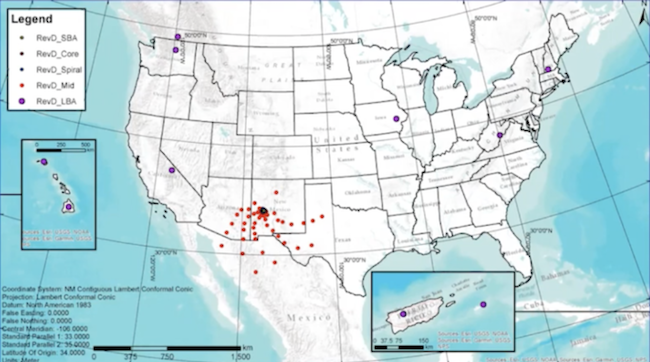 + The ngVLA will be spread out over the southwest United States and Mexico.
+ The ngVLA will be spread out over the southwest United States and Mexico.
+
+
+
A consensus study report called Pathways to Discovery in Astronomy and Astrophysics for the 2020s (Astro2020) identified the ngVLA as a high priority. The construction of this telescope should begin this decade and be completed by the middle of the 2020s.
+
+
Future of radio astronomy: planet formation
+
+
An area of research that radio astronomers are interested in examining in the future is imaging the formation of planets, Wilcot notes. Right now, astronomers can detect a planet’s presence and deduce specific characteristics, but being able to detect a planet directly is the next huge priority.
+
+
+ 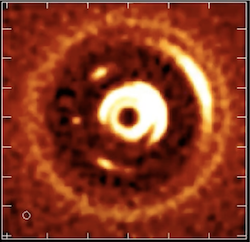 + A planetary system forming
+ A planetary system forming
+
+
+
One place astronomers might be able to do this with something like the ngVLA is in the early phases of planet formation within a planetary system. The thermal emissions from this process are bright enough to be detected by a telescope like the ngVLA. So the idea is to use this telescope to map an image of nearby planetary systems and begin to image the early stages of planet formation directly. A catalog of these planets forming will allow astronomers to understand what happens when planetary systems, like our own, form.
+
+
Future of radio astronomy: molecular systems
+
+
Wilcots explains that radio astronomers have discovered the spectral signature of innumerable molecules within the past fifty years. The ngVLA is being designed to probe, detect, catalog, and understand the origin of complex molecules and what they might tell us about star and planet formation. Wilcots comments in his talk that “this type of work is spawning a new type of science…a remarkable new discipline of astrobiology is emerging from our ability to identify and trace complex organic molecules.”
+
+
Future of radio astronomy: galaxy completion
+
+
Next, Wilcots discusses that radio astronomers want to understand how stars form in the first place and the processes that drive the collapse of clouds of gas into regions of star formations.
+
+
+ 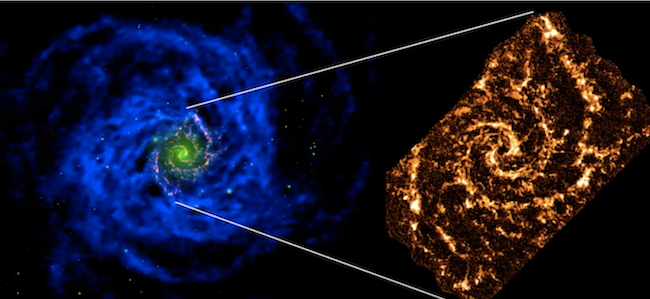 + An image of a blue spiral from the VLA of a nearby spiral galaxy is on the left. On the right an optical extent of the galaxy.
+ An image of a blue spiral from the VLA of a nearby spiral galaxy is on the left. On the right an optical extent of the galaxy.
+
+
+
The gas in a galaxy tends to extend well beyond the visible part of the galaxy, and this enormous gas reservoir is how the galaxy can make stars.
+
+
Astronomers like Wilcots want to know where the gas is, what drives that process of converting the gas into stars, what role the environment might play, and finally, what makes a galaxy stop creating stars.
+
+
ngVLA will be able to answer these questions as it combines the sensitivity and spatial resolution needed to take images of gas clouds in nearby galaxies while also capturing the full extent of that gas.
+
+
Future of radio astronomy: black holes
+
+
Wilcots’ look into the future of radio astronomy finishes with the idea and understanding of black holes.
+
+
Multi-messenger astrophysics helps experts recognize that information about the universe is not simply electromagnetic, as it is known best; there is more than one way astronomers can look at the universe.
+
+
More recently, astronomers have been looking at gravitational waves. In particular, they’ve been looking at how they can find a way to detect the gravitational waves produced by two black holes orbiting around one another to determine each black hole’s mass and learn something about them. As the recent EHT images show, we need radio telescopes’ high resolution and sensitivity to understand the nature of black holes fully.
+
+
A look toward the future
+
+
The next step is for the NRAO to create a prototype of the dishes they want to install for the telescope. Then, it’s just a question of whether or not they can build and install enough dishes to deliver this instrument to its full capacity. Wilcots elaborates, “we hope to transition to full scientific operations by the middle of next decade (the 2030s).”
+
+
The distinguished administrator expressed that “something that’s haunted radio astronomy for a while is that to do the imaging, you have to ‘be in the club,’ ” meaning that not just anyone can access the science coming out of these telescopes. The goal of the NRAO moving forward is to create science-ready data products so that this information can be more widely available to anyone, not just those with intimate knowledge of the subject.
+
+
This effort to make this science more accessible has been part of a budding collaboration between UW-Madison, the NRAO, and a consortium of Historically Black Colleges and Universities and other Minority Serving Institutions in what is called Project RADIAL.
+
+
“The idea behind RADIAL is to broaden the community; not just of individuals engaged in radio astronomy, but also of individuals engaged in the computing that goes into doing the great kind of science we have,” Wilcots explains.
+
+
On the UW-Madison campus in the Summer of 2022, half a dozen undergraduate students from the RADIAL consortium will be on campus doing summer research. The goal is to broaden awareness and increase the participation of communities not typically involved in these discussions in the kind of research in the radial astronomy field.
+
+
“We laid the groundwork for a partnership with a number of these institutions, and that partnership is alive and well,” Wilcots remarks, “so stay tuned for more of that, and we will be advancing that in the upcoming years.”
+
+
…
+
+
Watch a video recording of Eric Wilcots’ talk at HTCondor Week 2022.
+
+
+
+
+
+
+
+
+
+ European HTCondor Workshop: Abstract Submission Open
+
+
Share your experiences with HTCSS at the European HTCondor Workshop in Amsterdam!
+
+
See this recent post in htcondor-users for details.
+
+
+
+
+
+
+
+ Using HTC expanded scale of research using noninvasive measurements of tendons and ligaments
+
+
With this technique and the computing power of high throughput computing (HTC) combined, researchers can obtain thousands of simulations to study the pathology of tendons
+and ligaments.
+
+
A recent paper published in the Journal of the Mechanical Behavior of Biomedical Materials by former Ph.D.
+student in the Department of Mechanical Engineering (and current post-doctoral researcher at the University of Pennsylvania)
+Jonathon Blank and John Bollinger Chair of Mechanical Engineering
+Darryl Thelen used the Center for High Throughput Computing (CHTC) to obtain their results.
+Results that, Blank says, would not have been obtained at the same scale without HTC. “[This project], and a number of other projects, would have had a very small snapshot of the
+problem at hand, which would not have allowed me to obtain the understanding of shear waves that I did. Throughout my time at UW, I ran tens of thousands of simulations — probably
+even hundreds of thousands.”
+
+
+  + Post-doctoral researcher at the University of Pennsylvania Jonathon Blank.
+ Post-doctoral researcher at the University of Pennsylvania Jonathon Blank.
+
+
+
Using noninvasive sensors called shear wave tensiometers, researchers on this project applied HTC to study tendon structure and function. Currently, research in this field is hard
+to translate because most assessments of tendon and ligament structure-function relationships are performed on the benchtop in a lab, Blank explains. To translate the benchtop
+experiments into studying tendons in humans, the researchers use tensiometers as a measurement tool, and this study developed from trying to better understand these measurements
+and how they can be applied to humans. “Tendons are very complex materials from an engineering perspective. When stretched, they can bear loads far exceeding your body weight, and
+interestingly, even though they serve their roles in transmitting force from muscle to bone really well, the mechanisms that give rise to injury and pathology in these tissues aren’t
+well understood.”
+
+
+  + John Bollinger Chair of Mechanical Engineering Darryl Thelen.
+ John Bollinger Chair of Mechanical Engineering Darryl Thelen.
+
+
+
In living organisms, researchers have used tensiometers to study the loading of muscles and tendons, including the triceps surae, which connects to the Achilles tendon, Blank notes.
+Since humans are variable regarding the size, stiffness, composition, and length of their tendons or ligaments, it’s “challenging to use a model to accurately represent a parameter
+space of human biomechanics in the real world. High throughput computing is particularly useful for our field just because we can readily express that variability at a large scale”
+through HTC. With Thelen and Orthopedics and Rehabilitation assistant professor Josh Roth, Blank developed a pipeline for
+simulating shear wave propagation in tendons and ligaments with HTC, which Blank and Thelen used in the paper.
+
+
With HTC, the researchers of this paper were able to further explore the mechanistic causes of changes in wave speed. “The advantage of this technique is being able to fully explore
+an input space of different stiffnesses, geometries, microstructures, and applied forces. The advantage of the capabilities offered by the CHTC is that we can fill the entire input
+space, not just between two data points, and thereby study changes in shear wave speed due to physiological factors and the mechanical underpinning driving those changes,” Blank
+elaborates.
+
+
It wasn’t challenging to implement, Blank states, since facilitators were readily available to help and meet with him. When he first started using HTC, Blank attended the CHTC
+office hours to get answers to his questions, even during COVID-19; during this time, there were also numerous one-on-one meetings. Having this backbone of support from the CHTC
+research facilitators propelled Blank’s research and made it much easier. “For a lot of modeling studies, you’ll have this sparse input space where you change a couple of parameters
+and investigate the sensitivity of your model that way. But it’s hard to interpret what goes on in between, so the CHTC quite literally saved me a lot of time. There were some
+1,000 simulations in the paper, and HTC by scaling out the workload turned a couple thousand hours of simulation time into two or three hours of wall clock time. It’s a unique tool
+for this kind of research.”
+
+
The next step from this paper’s findings, Blank describes, is providing subject-specific measurements of wave speeds. This involves “understanding if when we use a tensiometer on
+someone’s Achilles tendon, for example, can we account for the tendon’s shape, size, injury status, etcetera — all of these variables matter when measuring shear wave speeds.”
+Researchers from the lab can then use wearable tensiometers to measure tension in the Achilles and other tendons to study human movement in the real world.
+
+
From his CHTC-supported studies, Blank learned how to design computational research, diagnose different parameter spaces, and manage data. “For my field, it [HTC] is very important
+because people are extremely variable — so our models should be too. The automation and capacity enabled by HTC makes it easy to understand whether our models are useful, and if
+they are, how best to tune them to inform human biomechanics,” Blank says.
+
+
+
+
+
+
+
+ About Our Approach
+
+
CHTC’s specialty is High
+Throughput Computing (HTC), which involves breaking up a single large
+computational task into many smaller tasks for the fastest overall
+turnaround. Most of our users find HTC to be invaluable
+in accelerating their computational work and thus their research.
+We support thousands of multi-core computers and use the task
+scheduling software called HTCondor, developed right here in Madison, to
+run thousands of independent jobs on as many total processors as
+possible. These computers, or “machines”, are distributed across several
+collections that we call pools (similar to “clusters”). Because machines are
+assigned to individual jobs, many users can be running jobs on a pool at any
+given time, all managed by HTCondor.
+
+
The diagram below shows some of the largest pools on campus and also
+shows our connection to the US-wide OS Pool where UW computing
+work can “backfill” available computers all over the country. The number
+under each resource name shows an approximate number of computing hours
+available to campus researchers for a typical week in Fall 2013. As
+demonstrated in the diagram, we help users to submit their work not only
+to our CHTC-owned machines, but to improve their throughput even further
+by seamlessly accessing as many available computers as possible, all
+over campus AND all over the country.
+
+
The vast majority of the computational work that campus researcher have
+is HTC, though we are happy to support researchers with a variety of
+beyond-the-desktop needs, including tightly-coupled computations (e.g.
+MPI), high-memory work (e.g. metagenomics), and specialized
+hardware like GPUs.
+
+

+
+
What kinds of applications run best in the CHTC?
+
+
“Pleasantly parallel” tasks, where many jobs can run independently,
+is what works best in the CHTC, and is what we can offer the greatest
+computational capacity for.
+Analyzing thousands of images, inferring statistical significance of hundreds of
+thousands of samples, optimizing an electric motor design with millions
+of constraints, aligning genomes, and performing deep linguistic search
+on a 30 TB sample of the internet are a few of the applications that
+campus researchers run every day in the CHTC. If you are not sure if
+your application is a good fit for CHTC resources, get in
+touch and we will be happy to help you figure it out.
+
+
Within a single compute system, we also support GPUs, high-memory
+servers, and specialized hardware owned by individual research groups.
+For tightly-coupled computations (e.g. MPI and similar programmed
+parallelization), our resources include an HPC Cluster, with faster
+inter-node networking.
+
+
How to Get Access
+
+
While you may be excited at the prospect of harnessing 100,000 compute
+hours a day for your research, the most valuable thing we offer is,
+well, us. We have a small, yet dedicated team of professionals who eat,
+breathe and sleep distributed computing. If you are a UW-Madison Researcher, you can request an
+account, and one of our dedicated Research Computing
+Facilitators will follow up to provide specific recommendations to
+accelerate YOUR science.
+
+
+
+
+
+
`-`` all receive top and bottom margins. We nuke the top\n// margin for easier control within type scales as it avoids margin collapsing.\n\n%heading {\n margin-top: 0; // 1\n margin-bottom: $headings-margin-bottom;\n font-family: $headings-font-family;\n font-style: $headings-font-style;\n font-weight: $headings-font-weight;\n line-height: $headings-line-height;\n color: $headings-color;\n}\n\nh1 {\n @extend %heading;\n @include font-size($h1-font-size);\n}\n\nh2 {\n @extend %heading;\n @include font-size($h2-font-size);\n}\n\nh3 {\n @extend %heading;\n @include font-size($h3-font-size);\n}\n\nh4 {\n @extend %heading;\n @include font-size($h4-font-size);\n}\n\nh5 {\n @extend %heading;\n @include font-size($h5-font-size);\n}\n\nh6 {\n @extend %heading;\n @include font-size($h6-font-size);\n}\n\n\n// Reset margins on paragraphs\n//\n// Similarly, the top margin on `
`s get reset. However, we also reset the\n// bottom margin to use `rem` units instead of `em`.\n\np {\n margin-top: 0;\n margin-bottom: $paragraph-margin-bottom;\n}\n\n\n// Abbreviations\n//\n// 1. Duplicate behavior to the data-bs-* attribute for our tooltip plugin\n// 2. Add the correct text decoration in Chrome, Edge, Opera, and Safari.\n// 3. Add explicit cursor to indicate changed behavior.\n// 4. Prevent the text-decoration to be skipped.\n\nabbr[title],\nabbr[data-bs-original-title] { // 1\n text-decoration: underline dotted; // 2\n cursor: help; // 3\n text-decoration-skip-ink: none; // 4\n}\n\n\n// Address\n\naddress {\n margin-bottom: 1rem;\n font-style: normal;\n line-height: inherit;\n}\n\n\n// Lists\n\nol,\nul {\n padding-left: 2rem;\n}\n\nol,\nul,\ndl {\n margin-top: 0;\n margin-bottom: 1rem;\n}\n\nol ol,\nul ul,\nol ul,\nul ol {\n margin-bottom: 0;\n}\n\ndt {\n font-weight: $dt-font-weight;\n}\n\n// 1. Undo browser default\n\ndd {\n margin-bottom: .5rem;\n margin-left: 0; // 1\n}\n\n\n// Blockquote\n\nblockquote {\n margin: 0 0 1rem;\n}\n\n\n// Strong\n//\n// Add the correct font weight in Chrome, Edge, and Safari\n\nb,\nstrong {\n font-weight: $font-weight-bolder;\n}\n\n\n// Small\n//\n// Add the correct font size in all browsers\n\nsmall {\n @include font-size($small-font-size);\n}\n\n\n// Mark\n\nmark {\n padding: $mark-padding;\n background-color: $mark-bg;\n}\n\n\n// Sub and Sup\n//\n// Prevent `sub` and `sup` elements from affecting the line height in\n// all browsers.\n\nsub,\nsup {\n position: relative;\n @include font-size($sub-sup-font-size);\n line-height: 0;\n vertical-align: baseline;\n}\n\nsub { bottom: -.25em; }\nsup { top: -.5em; }\n\n\n// Links\n\na {\n color: $link-color;\n text-decoration: $link-decoration;\n\n &:hover {\n color: $link-hover-color;\n text-decoration: $link-hover-decoration;\n }\n}\n\n// And undo these styles for placeholder links/named anchors (without href).\n// It would be more straightforward to just use a[href] in previous block, but that\n// causes specificity issues in many other styles that are too complex to fix.\n// See https://github.com/twbs/bootstrap/issues/19402\n\na:not([href]):not([class]) {\n &,\n &:hover {\n color: inherit;\n text-decoration: none;\n }\n}\n\n\n// Code\n\npre,\ncode,\nkbd,\nsamp {\n font-family: $font-family-code;\n @include font-size(1em); // Correct the odd `em` font sizing in all browsers.\n direction: ltr #{\"/* rtl:ignore */\"};\n unicode-bidi: bidi-override;\n}\n\n// 1. Remove browser default top margin\n// 2. Reset browser default of `1em` to use `rem`s\n// 3. Don't allow content to break outside\n\npre {\n display: block;\n margin-top: 0; // 1\n margin-bottom: 1rem; // 2\n overflow: auto; // 3\n @include font-size($code-font-size);\n color: $pre-color;\n\n // Account for some code outputs that place code tags in pre tags\n code {\n @include font-size(inherit);\n color: inherit;\n word-break: normal;\n }\n}\n\ncode {\n @include font-size($code-font-size);\n color: $code-color;\n word-wrap: break-word;\n\n // Streamline the style when inside anchors to avoid broken underline and more\n a > & {\n color: inherit;\n }\n}\n\nkbd {\n padding: $kbd-padding-y $kbd-padding-x;\n @include font-size($kbd-font-size);\n color: $kbd-color;\n background-color: $kbd-bg;\n @include border-radius($border-radius-sm);\n\n kbd {\n padding: 0;\n @include font-size(1em);\n font-weight: $nested-kbd-font-weight;\n }\n}\n\n\n// Figures\n//\n// Apply a consistent margin strategy (matches our type styles).\n\nfigure {\n margin: 0 0 1rem;\n}\n\n\n// Images and content\n\nimg,\nsvg {\n vertical-align: middle;\n}\n\n\n// Tables\n//\n// Prevent double borders\n\ntable {\n caption-side: bottom;\n border-collapse: collapse;\n}\n\ncaption {\n padding-top: $table-cell-padding-y;\n padding-bottom: $table-cell-padding-y;\n color: $table-caption-color;\n text-align: left;\n}\n\n// 1. Removes font-weight bold by inheriting\n// 2. Matches default `
` alignment by inheriting `text-align`.\n// 3. Fix alignment for Safari\n\nth {\n font-weight: $table-th-font-weight; // 1\n text-align: inherit; // 2\n text-align: -webkit-match-parent; // 3\n}\n\nthead,\ntbody,\ntfoot,\ntr,\ntd,\nth {\n border-color: inherit;\n border-style: solid;\n border-width: 0;\n}\n\n\n// Forms\n//\n// 1. Allow labels to use `margin` for spacing.\n\nlabel {\n display: inline-block; // 1\n}\n\n// Remove the default `border-radius` that macOS Chrome adds.\n// See https://github.com/twbs/bootstrap/issues/24093\n\nbutton {\n // stylelint-disable-next-line property-disallowed-list\n border-radius: 0;\n}\n\n// Explicitly remove focus outline in Chromium when it shouldn't be\n// visible (e.g. as result of mouse click or touch tap). It already\n// should be doing this automatically, but seems to currently be\n// confused and applies its very visible two-tone outline anyway.\n\nbutton:focus:not(:focus-visible) {\n outline: 0;\n}\n\n// 1. Remove the margin in Firefox and Safari\n\ninput,\nbutton,\nselect,\noptgroup,\ntextarea {\n margin: 0; // 1\n font-family: inherit;\n @include font-size(inherit);\n line-height: inherit;\n}\n\n// Remove the inheritance of text transform in Firefox\nbutton,\nselect {\n text-transform: none;\n}\n// Set the cursor for non-` |
+
+
+
+
+ |
+
+
+| |
+ |
+ |
+ |
+
+
+
+
+
+|
+
+
+ Feedback or content questions:
+ send email to "condor-admin" at the cs.wisc.edu server
+
+ Technical or accessibility issues:
+ chtc@cs.wisc.edu
+
+ |
+
+
+
+
+
+
diff --git a/preview-fall2024-info/includes/chtc_on_campus.png b/preview-fall2024-info/includes/chtc_on_campus.png
new file mode 100644
index 000000000..b77707bb1
Binary files /dev/null and b/preview-fall2024-info/includes/chtc_on_campus.png differ
diff --git a/preview-fall2024-info/includes/chtcusers.jpg b/preview-fall2024-info/includes/chtcusers.jpg
new file mode 100644
index 000000000..f647dbcaa
Binary files /dev/null and b/preview-fall2024-info/includes/chtcusers.jpg differ
diff --git a/preview-fall2024-info/includes/chtcusers_400.jpg b/preview-fall2024-info/includes/chtcusers_400.jpg
new file mode 100644
index 000000000..efa7e6675
Binary files /dev/null and b/preview-fall2024-info/includes/chtcusers_400.jpg differ
diff --git a/preview-fall2024-info/includes/chtcusers_L.jpg b/preview-fall2024-info/includes/chtcusers_L.jpg
new file mode 100644
index 000000000..fac6f5084
Binary files /dev/null and b/preview-fall2024-info/includes/chtcusers_L.jpg differ
diff --git a/preview-fall2024-info/includes/cron-generated/dynamic-resources-noedit.html b/preview-fall2024-info/includes/cron-generated/dynamic-resources-noedit.html
new file mode 100644
index 000000000..f4161e936
--- /dev/null
+++ b/preview-fall2024-info/includes/cron-generated/dynamic-resources-noedit.html
@@ -0,0 +1,39 @@
+| Pool/Mem | ≥1GB | ≥2GB | ≥4GB | ≥8GB | ≥16GB | ≥32GB | ≥64GB |
|---|
+| cm.chtc.wisc.edu |
+0 |
+0 |
+0 |
+0 |
+0 |
+0 |
+0 |
+
+| condor.cs.wisc.edu |
+0 |
+0 |
+0 |
+0 |
+0 |
+0 |
+0 |
+
+| condor.cae.wisc.edu |
+0 |
+0 |
+0 |
+0 |
+0 |
+0 |
+0 |
+
+| Totals |
+0 |
+0 |
+0 |
+0 |
+0 |
+0 |
+0 |
+
|---|
+
+As of Mon Jun 12 07:30:02 CDT 2017
diff --git a/preview-fall2024-info/includes/jahns/Bundle Proximity Losses Paragraph.doc b/preview-fall2024-info/includes/jahns/Bundle Proximity Losses Paragraph.doc
new file mode 100644
index 000000000..2695061cf
Binary files /dev/null and b/preview-fall2024-info/includes/jahns/Bundle Proximity Losses Paragraph.doc differ
diff --git a/preview-fall2024-info/includes/jahns/Thumbs.db b/preview-fall2024-info/includes/jahns/Thumbs.db
new file mode 100644
index 000000000..dd3f84614
Binary files /dev/null and b/preview-fall2024-info/includes/jahns/Thumbs.db differ
diff --git a/preview-fall2024-info/includes/old.intro.html b/preview-fall2024-info/includes/old.intro.html
new file mode 100755
index 000000000..7b02b4a01
--- /dev/null
+++ b/preview-fall2024-info/includes/old.intro.html
@@ -0,0 +1,26 @@
+Research is a computationally expensive endeavor, demanding on any computing resources available. Quite often, a researcher will require resources for computations for short bursts of time, frequently leaving the computer idle. This often results in wasted potential computation time. This issue can be addressed by means of high-throughput computing.
+High-throughput computing allows for many computational tasks to be done over a long period of time. It is concerned largely with the number of compute resources that are available to people who wish to use the system. It is a very useful system for researchers, who are more concerned with the number of computations they can do over long spans of time than they are with short-burst computations. Because of its value to research computations, the Univeristy of Wisconsin set up the Center for High-Throughput Computing to bring researchers and compute resources together.
+The Center for High-Throughput Computing (CHTC), approved in August 2006, has numerous resources at its disposal to keep up with the computational needs of UW Madison. These resources are being funded by the National Institute of Health (NIH), the Department of Energy (DOE), the National Science Foundation (NSF), and various grants from the University itself. Email us to see what we can do to help automate your research project at chtc@cs.wisc.edu It aims to pull four different resources together into one operation:
+
+ HTC Technologies: The CHTC leans heavily on the HTCondor project to provide a framework where high-throughput computing can take place. The HTCondor project aims to make grid and high-throughput computing a reality in any number of environments.
Dedicated Resources: CHTC HTCondor pool
+ The CHTC cluster is now composed of 1900 cores for use by researches across our campus. These rack mounted blade systems run Linux. Each core is 2.8Ghz with 1.5GB RAM or better. CHTC has provided 10 million CPU hours of research computation between 05/17/2008 and 02/23/2010 prior to the additional 960 cores. With the recent server purchase, CHTC provides in excess of 37,000 CPU hours per day.
+
+
+ CHTC Policy Description here
+
+ Middleware: The GRIDS branch at UW Madison will be an essential part towards keeping the CHTC running efficiently. GRIDS is funded by the NSF Middleware Initiative (NMI). At the University of Wisconsin, the HTCondor project makes heavy use of this system with their NMI Build & Test facility. The NMI Build & Test facility provides a framework to build and test software on a wide variety of platform and hardware combinations.
Computing Laboratory: The University of Wisconsin has many compute clusters at its disposal. In 2004 the university won an award to build the Grid Laboratory of Wisconsin (GLOW). GLOW is an interdepartmental pool of HTCondor nodes, containing 3000 CPUs and about 1 PB of storage.
+
+The University of Wisconsin-Madison (UW-Madison) campus is an excellent match for meeting the computational needs of your project. Existing UW technology infrastructure that can be leveraged includes CPU capacity, network connectivity, storage availability, and middleware connectivity. But perhaps most important, the UW has significant staff experience and core competency in deploying, managing, and using computational technology.
+
+
+To reiterate: The UW launched and funded the Center for High Throughput Computing (CHTC), a campus-wide organization dedicated to supercharging research on campus by working side-by-side with you, the domain scientists on infusing high throughput computing and grid computing techniques into your routine. Between the CHTC and the aforementioned HTCondor Project, the UW is home to over 20 full-time staff with a proven track record of making compute middleware work for scientists. Far beyond just being familiar with deployment and use of such software, UW staff has been intimately involved in its design and implementation.
+
+
+Applications: Many researchers are already using these facilities. More information about a sampling of those using the CHTC can be found here.
+And less recent projects in CHTC Older projects.
+
+
diff --git a/preview-fall2024-info/includes/onerow b/preview-fall2024-info/includes/onerow
new file mode 100644
index 000000000..9d97cdfc9
--- /dev/null
+++ b/preview-fall2024-info/includes/onerow
@@ -0,0 +1,18 @@
+
+
+
+
+ |
+
+
+Professor
+
+
+1,401,361 CPU hours since 1/1/2009
+
+
+
+
+
+ |
+
diff --git a/preview-fall2024-info/includes/web.css b/preview-fall2024-info/includes/web.css
new file mode 100644
index 000000000..293031cab
--- /dev/null
+++ b/preview-fall2024-info/includes/web.css
@@ -0,0 +1,447 @@
+/* The following six styles set attibutes for heading tags H1-H6 */
+
+div.announcement {
+ border: 1px solid #787878;
+ background-color: #efefef;
+ color: #787878;
+ padding: .5em;
+ margin: 1em 2em 1em 2em;
+ }
+
+table.gtable {
+ background: #B70101;
+ padding: 5px 10px;
+ border-radius: 5px;
+ -moz-border-radius: 5px;
+ -webkit-border-radius: 5px;
+
+ color: #FFFFFF;
+ overflow: hidden;
+ margin-bottom: 20px;
+
+ background: #ddd;
+ color: #333;
+ border: 0;
+ border-bottom: 3px solid #bbb;
+
+ -moz-box-shadow: 0px 2px 7px 1px #bbb;
+ -webkit-box-shadow: 0px 2px 7px 1px #bbb;
+ box-shadow: 0px 2px 7px 1px #bbb;
+}
+
+table.gtable img{
+ border-radius: 5px;
+ -moz-border-radius: 5px;
+ -webkit-border-radius: 5px;
+}
+
+table.gtable td {
+ padding: 0.6em 0.8em 0.6em 0.8em;
+ background-color: #ddd;
+ border-bottom: 1px solid #bbb;
+ border-top: 0px;
+ overflow: visible;
+}
+
+table.gtable th {
+ padding: 0.6em 0.8em 0.6em 0.8em;
+ background-color: #b70101;
+ color: #FFFFFF;
+ border: 0px;
+ border-bottom: 3px solid #920000;
+}
+
+H1 {
+ margin-bottom: -5px;
+ text-align: left;
+ color: #000000;
+ font-size: 180%;
+ font-weight: bold;
+ font-family: verdana, geneva, arial, sans-serif;
+ line-height: 195%
+ }
+
+
+H2 {
+ margin-top: 20px;
+ margin-bottom: 0px;
+ text-align: left;
+ color: #000000;
+ font-size: 150%;
+ font-weight: bold;
+ font-family: verdana, geneva, arial, sans-serif;
+ line-height: 180%
+ }
+
+
+
+H3 {
+ margin-top: 20px;
+ margin-bottom: -10px;
+ text-align: left;
+ color: #000000;
+ font-size: 120%;
+ line-height: 150%;
+ font-weight: bold;
+ font-family: verdana, geneva, arial, sans-serif;
+ width: 100%
+ }
+
+
+H4 {
+ margin-top: 15px;
+ margin-bottom: -10px;
+ text-align: left;
+ color: #000000;
+ font-size: 100%;
+ font-weight: bold;
+ font-family: verdana, geneva, arial, sans-serif;
+ width: 100%
+ }
+
+
+
+H5 {
+ margin-top: 10px;
+ margin-bottom: -10px;
+ text-align: left;
+ color: #000000;
+ font-size: 95%;
+ font-weight: bold;
+ font-family: verdana, geneva, arial, sans-serif;
+ width: 100%
+ }
+
+
+H6 {
+ margin-top: 10px;
+ margin-bottom: -10px;
+ text-align: left;
+ color: #000000;
+ font-size: 95%;
+ font-weight: bold;
+ font-family: verdana, geneva, arial, sans-serif;
+ width: 100%
+ }
+
+body {
+ background-color: #eee;
+ font-family: Verdana, Arial, Helvetica,sans-serif;
+}
+
+#main {
+ background: #fff;
+ margin: 10px 5px;
+ padding: 20px;
+ min-height: 1300px;
+ border-bottom: 3px solid #bbb;
+ border-left: 1px solid #ddd;
+ border-right: 1px solid #ddd;
+ border-radius: 5px;
+ -moz-border-radius-right: 5px;
+ -webkit-border-radius-right: 5px;
+
+ -moz-box-shadow: 0px 2px 7px 1px #bbb;
+ -webkit-box-shadow: 0px 2px 7px 1px #bbb;
+ box-shadow: 0px 2px 7px 1px #bbb;
+
+}
+
+.bgred {
+ background-color: #B70101;
+ -moz-border-top-right-radius: 10px;
+ -webkit-border-top-right-radius: 10px;
+ border-bottom-right-radius: 10px;
+ -moz-border-bottom-right-radius: 10px;
+ -webkit-border-bottom-right-radius: 10px;
+ margin: 10px 0px;
+
+ -moz-box-shadow: 0px 2px 7px 1px #bbb;
+ -webkit-box-shadow: 0px 2px 7px 1px #bbb;
+ box-shadow: 0px 2px 7px 1px #bbb;
+}
+
+#copyright {
+font-family: Verdana, Arial, Helvetica, sans-serif;
+font-size: 75%;
+
+background: #ddd;
+color: #333;
+border: 1px solid #bbb;
+border-bottom: 3px solid #bbb;
+border-top: 0px;
+
+padding: 5px 10px;
+border-radius: 5px;
+-moz-border-radius: 5px;
+-webkit-border-radius: 5px;
+
+ -moz-box-shadow: 0px 2px 7px 1px #bbb;
+ -webkit-box-shadow: 0px 2px 7px 1px #bbb;
+ box-shadow: 0px 2px 7px 1px #bbb;
+
+
+margin-top: 40px;
+}
+
+#copyright a {
+ color: #66a;
+}
+
+
+.navbodyblack {
+ font-family: Verdana,Arial, Helvetica, sans-serif;
+ color: #ffffff; background-color: #B70101;
+ text-decoration: none; padding-left: 8px;
+ padding-top: 5px; padding-bottom: 5px;
+ padding-right: 2px;
+ font-weight: normal;
+ margin: 0px;
+}
+
+.navbodyblack a:link {
+ color:#ffffff;
+ text-decoration: none;
+}
+
+.navbodyblack a:visited {
+ color:#ffffff;
+ text-decoration: none;
+}
+
+.navbodyblack a:hover {
+ color:#cc9900;
+ text-decoration: none;
+}
+
+code {
+ font-size: 120%;
+}
+
+pre {
+ border-radius: 5px;
+ -moz-border-radius: 5px;
+ -webkit-border-radius: 5px;
+ font-size: 120%;
+ margin: 1em 2em;
+
+ background: #ddd;
+ color: #333;
+ border: 1px solid #bbb;
+ border-bottom: 3px solid #bbb;
+ border-top: 0px;
+ border-left: 5px solid #b70101;
+ padding: 0.5em 1.2em;
+
+ -moz-box-shadow: 0px 2px 7px 1px #bbb;
+ -webkit-box-shadow: 0px 2px 7px 1px #bbb;
+ box-shadow: 0px 2px 7px 1px #bbb;
+}
+ul.sidebar {
+ width = 20%;
+ font-size: 80%;
+ margin: 0.8em;
+}
+ul.sidebar,ul.sidebar ul {
+ padding: 0;
+ border: 0;
+ color: white;
+}
+ul.sidebar ul {
+ margin: 0 0.8em;
+}
+ul.sidebar a:link, ul.sidebar a:visited, ul.sidebar a:active {
+ color: white;
+ text-decoration: none;
+ display: block;
+}
+ul.sidebar a:hover, ul.sidebar a:active {
+ /*text-decoration: underline;
+ color:#cc9900; */
+ color: #b70101;
+ background-color: white;
+ transition: all linear 0.2s 0s;
+ position: relative;
+ left:5px;
+
+ border-left:5px solid #d41a1a;
+}
+ul.sidebar li {
+ font-size: 16px;
+ margin: 0;
+ margin-top: .5em;
+ padding: 2px 4px;
+ border: 0;
+ list-style-type: none;
+}
+ul.sidebar li a{
+ position: relative;
+ top: 0px;
+
+ transition: all linear 0.2s 0s;
+
+ padding: 5px 10px;
+ border-left:5px solid #d41a1a;
+ background: #d41a1a;
+
+ -moz-box-shadow: 0px 2px 4px 1px #9E0000;
+ -webkit-box-shadow: 0px 2px 4px 1px #9E0000;
+ box-shadow: 0px 2px 4px 1px #9E0000;
+
+ color: white;
+ border-radius: 5px;
+ -moz-border-radius: 5px;
+ -webkit-border-radius: 5px;
+}
+
+ul.sidebar ul li a{
+ font-size: 14px;
+
+}
+
+ul.sidebar li.spacer {
+}
+
+#tile-wrapper {
+ margin-left:auto;
+ margin-right:auto;
+ padding:20px;
+}
+
+a.tile {
+ display: inline-block;
+ position:relative;
+ width:800px;
+ text-decoration: none;
+
+ overflow:hidden;
+
+ margin-left:auto;
+ margin-right:auto;
+
+ font-size:75%;
+ margin: 10px;
+ background: #cdcdcd;
+ padding: 10px;
+ border-radius: 5px;
+ border-bottom: 5px solid #aaa;
+ border-top: 5px solid #ddd;
+ color: #555;
+
+ -moz-box-shadow: 0px 1px 8px 1px #bbb;
+ -webkit-box-shadow: 0px 1px 8px 1px #bbb;
+ box-shadow: 0px 1px 8px 1px #bbb;
+}
+
+a.tile:hover {
+ top:-5px;
+
+
+ -moz-box-shadow: 0px 6px 8px 1px #bbb;
+ -webkit-box-shadow: 0px 6px 8px 1px #bbb;
+ box-shadow: 0px 6px 8px 1px #bbb;
+
+}
+
+
+a.tile p{
+ display:inline-block;
+ width:84%;
+ height:65px;
+ float:right;
+
+ text-align: left;
+
+ background: #fff;
+ color: #000;
+ padding: 10px;
+
+ font-size: 10px;
+
+ border-color: #fff;
+ background-color: #fff;
+ padding: 10px;
+ margin:5px;
+ border-radius: 5px;
+}
+
+a.tile img{
+ width:75px;
+ float:left;
+
+ display:inline-block;
+
+ border-color: #fff;
+ background-color: #fff;
+ padding: 5px;
+ margin:5px;
+ border-radius: 5px;
+}
+
+a.tile h2{
+ text-decoration: none;
+ color:#555;
+ margin:0px 5px;
+ font-size: 140%;
+}
+
+#hours {
+ width: 120px;
+ height: 92px;
+ font-size:75%;
+ margin-left:10px;
+ float: right;
+ background: #B70101;
+ padding: 5px 10px;
+ border-radius: 5px;
+ border-bottom: 5px solid #920000;
+ border-top: 5px solid #d41a1a;
+ color: #fff;
+
+ -moz-box-shadow: 0px 1px 4px 1px #bbb;
+ -webkit-box-shadow: 0px 1px 4px 1px #bbb;
+ box-shadow: 0px 1px 4px 1px #bbb;
+}
+
+#osg_power {
+ height: 92px;
+ margin-left:10px;
+ float: right;
+ background: #F29B12;
+ padding: 5px 10px;
+ border-radius: 5px;
+ border-bottom: 5px solid #EF7821;
+ border-top: 5px solid #FDC10A;
+ color: #fff;
+
+ -moz-box-shadow: 0px 1px 4px 1px #bbb;
+ -webkit-box-shadow: 0px 1px 4px 1px #bbb;
+ box-shadow: 0px 1px 4px 1px #bbb;
+}
+
+#osg_power img {
+ border-radius: 5px;
+}
+
+p.underconstruction {
+ border: 1px solid #666;
+ background-color: #FFA;
+ padding: 0.1em 0.5em;
+ margin-left: 2em;
+ margin-right: 2em;
+ font-style: italic;
+
+ -moz-box-shadow: 0px 2px 7px 1px #bbb;
+ -webkit-box-shadow: 0px 2px 7px 1px #bbb;
+ box-shadow: 0px 2px 7px 1px #bbb;
+}
+.num {
+ text-align: right;
+}
+table {
+ border-collapse: collapse;
+}
+td,tr {
+ padding-left: 0.2em;
+ padding-right: 0.2em;
+}
diff --git a/preview-fall2024-info/index.html b/preview-fall2024-info/index.html
new file mode 100644
index 000000000..05f7609c5
--- /dev/null
+++ b/preview-fall2024-info/index.html
@@ -0,0 +1,746 @@
+
+
+
+
+
+
+Home
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
CHTC is hiring, view the new position on the jobs page and apply now!
+ View Job Posting
+
+
+
+
+  +
+
+
+
+
+
+
+
+ The Center for High Throughput Computing (CHTC), established in 2006, aims to bring the power
+ of High Throughput Computing to all fields of research, and to allow the future of HTC to be shaped
+ by insight from all fields.
+
+
+
+
+
+
+ Are you a UW-Madison researcher looking to expand your computing beyond your local resources? Request
+ an account now to take advantage of the open computing services offered by the CHTC!
+
+
+
+
+
+
+ High Throughput Computing is a collection of principles and techniques which maximize the effective throughput
+ of computing resources towards a given problem. When applied for scientific computing, HTC can result in
+ improved use of a computing resource, improved automation, and help drive the scientific problem forward.
+
+
+ The team at CHTC develops technologies and services for HTC. CHTC is the home of the HTCondor Software
+ Suite which has over 30 years of experience in tackling HTC problems;
+ it manages shared computing resources
+ for researchers on the UW-Madison campus; and it leads the OSG Consortium,
+ a national-scale environment for distributed HTC.
+
+
+
+
+
+
+ Last Year Serving UW-Madison
+
+
+
+
+
+
+
+
+
+ HTCSS
+
+
+
+
The HTCondor Software Suite (HTCSS) provides sites and users with the ability to manage and execute
+HTC workloads. Whether it’s managing a single laptop or 250,000 cores at CERN, HTCondor
+helps solve computational problems through the application of the HTC principles.
+
+
+
+
+
+

+
+
+
+
+
+
+ Services
+
+
+
+
+
CHTC manages over 20,000 cores and dozens of GPUs for the UW-Madison
+campus; this resource, which is free and shared, aims to advance the
+mission of the University of Wisconsin in support of the Wisconsin
+Idea. Researchers can place their workloads on an access point at
+CHTC and utilize the resources at CHTC, across the campus, and across
+the nation.
+
+
Research Facilitation
+
CHTC’s Research Facilitation team empowers researchers to utilize computing to achieve
+their goals. The Research Facilitation approach emphasizes teaching users skills and
+methodologies to manage & automate workloads on resources like those at CHTC, the campus,
+or across the world.
+
+
+
+
+
+

+
+
+
+
+
+
+
+ As part of its many services to UW-Madison and beyond,
+ the CHTC is home to or supports the following Research
+ Projects and Partners.
+
+
+
+
+
+ The OSG is a consortium of research collaborations, campuses, national laboratories and software
+ providers dedicated to the advancement of all of open science via the practice of distributed High Throughput
+ Computing (dHTC), and the advancement of its state of the art. The OSG operates a fabric of dHTC services
+ for the national science and engineering community and CHTC has been a major force in OSG since its inception
+ in 2005.
+
+
+
+  +
+
+
+
+
+
+
+ The Partnership to Advance Throughput Computing (PATh) is a partnership between
+ CHTC and OSG to advance throughput computing. Funded through a major investment
+ from NSF, PATh helps advance HTC at a national level through support for
+ HTCSS and provides a fabric of services for the NSF science and engineering community
+ to access resources across the nation.
+
+
+
+  +
+
+
+
+
+
+
+ The Morgridge Institute for Research is a private, biomedical research institute
+ located on the UW-Madison campus. Morgridge’s Research Computing Theme is a unique
+ partner with CHTC, investing in the vision of HTC and its ability to advance basic
+ research.
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
 +
+  +
+
+
+
+
+
+
+
+
+ The Path to Internship and Fellowship Opportunities
+
+
+
+ Want to make a difference and see your work directly
+ impact science across the globe?
+
+
+ The Center for High Throughput Computing offers internship and summer fellowship opportunities
+ for undergraduate and graduate students.
+
+
+
The Mission
+
CHTC is a research computing organization located within the University
+ of Wisconsin-Madison CS Department and at the Morgridge Institute for
+ Research. CHTC is an internationally recognized leader in high
+ throughput computing and provides access to free large-scale computing
+ capacity for research. CHTC advances the field of research computing
+ through innovative software and services, leading distributed computing
+ projects across the campus and the nation.
+
+
+
The CHTC Fellows Program
+
Our Fellows Program provides undergraduate and graduate students with
+ learning opportunities in research computing, system administration,
+ and facilitation. Working with engineers and
+ dedicated mentors, fellows will have the opportunity to learn
+ from leaders in their field and access state of the art computing
+ facilities. Fellows can also attend workshops, lectures and social
+ and recreational events with CHTC team members. Learn more about the CHTC Fellows Program.
+
+
+
We Value Diversity
+
CHTC and Morgridge are committed to increasing diversity among
+ interns and staff. We believe that advancing throughput computing
+ and scientific research is enhanced by a wide range of backgrounds
+ and perspectives.
+
+
+
+
+
+
+
+
Questions: chtc-jobs@g-groups.wisc.edu
+
+
 + Photo: Morgridge Institute for Research
+
+
+
+ Photo: Morgridge Institute for Research
+
+
+
+
+
+
+ Available Positions
+
+
+
+ If advancing the state of the art of distributed computing in an
+ academic environment interests you, the Center for High Throughput
+ Computing (CHTC) at the University of Wisconsin-Madison (UW) offers a
+ unique working environment. Our project’s home is in the UW Department
+ of Computer Sciences, an internationally recognized department
+ consistently ranked in the top ten across the USA.
+
+
+ A position with CHTC
+ will provide you the opportunity to interact with both department
+ faculty and students to translate novel ideas into real-world solutions.
+ The software and infrastructure you will be working on is used by
+ scientists and engineers at hundreds of institutions, from universities
+ to national laboratories and from large high tech corporations to small
+ animation teams.
+
+
+
Internships
+
+ Our internship program provides undergraduate and graduate students with learning opportunities in
+ research computing, system administration, web development, and communication.
+
+
Learn More
+
+
+
+
+
Full Time Positions
+
+ Details about our open full-time positions are typically provided below.
+ Positions pertaining to "HTCondor" and "CHTC" search terms can also
+ be found on the University's
+ Position Vacancy List
+ (PVL).
+
+
+
+
+
+
Research Software Engineer
+
+
+ Full Time - Morgridge Institute for Research
+
+
+
+
+
+
The Research Software Engineer (RSE) will work to bring modern software engineering techniques and approaches
+to research projects at the institute as part of long-running engagements and collaborations between scientists.
+
+
At Morgridge, the RSE will sit at the nexus of exciting research, large-scale computing, and national
+cyberinfrastructure projects. Whether its bringing a codebase up to production quality, designing
+programmatic interfaces, or making workloads run more effectively across thousands of cores, the
+RSE will have a diversity of challenges and help advance Morgridge’s goals of Fearless Science.
+
+
Potential projects will be diverse, including facilitating machine learning projects, developing
+tools for global distributed data management, and enabling large-scale protein simulation;
+projects will be tailored based on the candidate.
+
+
+
+
+
+
+
+
+
+
+
Benefits
+
+
+ The University of Wisconsin-Madison is a great place to work. You can
+ read about the benefits in detail
+ elsewhere. In short, we have
+ five weeks of vacation/personal time per year, very good health
+ insurance (and cost effective for entire families), and a good
+ retirement plan. Please note that the minimum salary in our job listings
+ are just that - the minimum. Compensation will increase with experience.
+
+
+
In addition to the official benefits, there are many side benefits:
+
+
+ - You will work with the CHTC team. We are world leaders in solving
+ interesting distributed computing problems!
+
+ - You can attend interesting talks in the department
+ - Relatively flexible working hours — we value work-life balance.
+ - A Discounted Bus Pass!
+ - You get staff access to the Union,
+ the UW athletic facilities, and
+ the UW library system.
+
+ - We're in a lively neighborhood with great restaurants in easy
+ walking distance.
+
+
+
+
+
+
+ If you are interested in a position with CHTC, explore the job listings
+ below! If you would like to apply, send your resume and cover letter to
+ chtc-jobs@g-groups.wisc.edu, and indicate
+ which job you would like to apply for.
+
+
+
Please note:
+
+
+ - A criminal background check will be conducted prior to hiring.
+ - A period of evaluation will be required.
+ - UW-Madison is an equal opportunity/affirmative action employer. We
+ promote excellence through diversity and encourage all qualified
+ individuals to apply.
+
+
+
+
+
+
+
+
+
+ Machine learning insights into molecular science using the Open Science Pool
+
+
Machine learning insights into molecular science using the Open Science Pool
+Computation has extended what researchers can investigate in chemistry, biology, and material science. Studying complex systems like proteins or nanocomposites can use similar techniques for common challenges. For example, computational power is expanding the horizons of protein research and opening up vast new possibilities for drug discovery and disease treatment.
+
+
Olexandr Isayev is an assistant professor at the School of Pharmacy, University of North Carolina (UNC) at Chapel Hill. Isayev is part of a group at UNC using machine learning for chemical problems and material science.
+
+
“Specifically, we apply machine learning to chemical and material science data to understand the data, find patterns in it, and make predictive models,” says Isayev. “We focus on three areas: computer-aided design of novel materials, computational drug discovery, and acceleration of quantum mechanical methods with GPUs (graphic processing units) and machine learning.”
+
+
For studying drug discovery, where small organic molecule binds to a protein receptor, Isayev uses machine learning to build predictive models based on historical collection of experimental data. “We want to challenge models and find a new molecule with better binding properties,” says Isayev.
+
+
+
+
+
+
+
Protein Model
+
Example of a protein model that Isayev and his group study. Courtesy image.
+
+
+
+
Similar to the human genome project, five years ago President Obama created a new Materials Genome Initiative to accelerate the design of new materials. Using machine learning methods based on the crystal structure of the material he is studying, Isayev can predict its physical properties.
+
+
“Looking at a molecule or material based on geometry and topology, we can get the energy, and predict critical physical properties,” says Isayev. “This machine learning allows us to avoid many expensive uses of numeric simulation to understand the material.”
+
+
The challenge for Isayev’s group is that initial data accumulation is extremely numerically time consuming. So, they use the Open Science Pool to run simulations. Based on the data, they train their machine learning model, so the next time, instead of a time-consuming simulation model, they can use the machine learning model on a desktop PC.
+
+
“Using machine learning to do the preliminary screening saves a lot of computing time,” says Isayev. “Since we performed the hard work, scientists can save a lot of time by prioritizing a few promising candidate materials instead of running everything.”
+
+
For studying something like a photovoltaic semiconductor, Isayev selects a candidate after running about a thousand of quantum mechanical calculations. He then uses machine learning to screen 50,000 materials. “You can do this on a laptop,” says Isayev. “We prioritize a few—like ten to fifty. We can predict what to run next instead of running all of them. This saves a lot of computing time and gives us a powerful tool for screening and prioritization.”
+
+
On the OSG, they run “small density function (DFT) calculations. We are interested in molecular properties,” says Isayev. “We run a program package called ORCA (Quantum Chemistry Program), a free chemistry package. It implements lots of QM methods for molecules and crystals. We use it and then we have our own scripts, run them on the OSG, collect the data, and then analyze the data.”
+
+
“I am privileged to work with extremely talented people like Roman Zubatyuk,” says Isayev. Zubatyuk works with Isayev on many different projects. “Roman has developed our software ecosystem container using Docker. These simulations run locally on our machines through the Docker virtual environment and eliminate many issues. With a central database and set of scripts, we could seamlessly run hundreds of thousands of simulations without any problems.”
+
+
Finding new materials and molecules are hard science problems. “There is no one answer when looking for a new molecule,” says Isayev. “We cannot just use brute force. We have to be creative because it is like looking for a needle in a hay stack.”
+
+
For something like a solar cell device, researchers might find a drawback in the performance of the material. “We are looking to improve current materials, improve their performance, or make them cheaper, so we can move them to mass production so everyone benefits,” says Isayev.
+
+
“For us, the OSG is a fantastic resource for which we are very grateful,” says Isayev. “It gives us access to computation that enables our simulations that we could not do otherwise. To run all our simulations requires lots of computing resources that we cannot run on a local cluster. To do our simulation screening, we have to perform lots of calculations. We can easily distribute these calculations because they don’t need to communicate to each other. The OSG is a perfect fit.”
+
+
+
+
+
+
+
+
+ CHTC User Map
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+
+
+
+
![]() ')
+ .addClass('mapster_el')
+ .hide();
+
+ index=me._add(image[0]);
+
+ image
+ .bind('load',function(e) {
+ me.imageLoaded.call(me,e);
+ })
+ .bind('error',function(e) {
+ me.imageLoadError.call(me,e);
+ });
+
+ image.attr('src', src);
+ } else {
+
+ // use attr because we want the actual source, not the resolved path the browser will return directly calling image.src
+
+ index=me._add($(image)[0]);
+ }
+ if (id) {
+ if (this[id]) {
+ throw(id+" is already used or is not available as an altImage alias.");
+ }
+ me.ids.push(id);
+ me[id]=me[index];
+ }
+ return index;
+ },
+
+ /**
+ * Bind the images in this object,
+ * @param {boolean} retry when true, indicates that the function is calling itself after failure
+ * @return {Promise} a promise that resolves when the images have finished loading
+ */
+
+ bind: function(retry) {
+ var me = this,
+ promise,
+ triesLeft = me.owner.options.configTimeout / 200,
+
+ /* A recursive function to continue checking that the images have been
+ loaded until a timeout has elapsed */
+
+ check=function() {
+ var i;
+
+ // refresh status of images
+
+ i=me.length;
+
+ while (i-->0) {
+ if (!me.isLoaded(i)) {
+ break;
+ }
+ }
+
+ // check to see if every image has already been loaded
+
+ if (me.complete()) {
+ me.resolve();
+ } else {
+ // to account for failure of onLoad to fire in rare situations
+ if (triesLeft-- > 0) {
+ me.imgTimeout=window.setTimeout(function() {
+ check.call(me,true);
+ }, 50);
+ } else {
+ me.imageLoadError.call(me);
+ }
+ }
+
+ };
+
+ promise = me.deferred=u.defer();
+
+ check();
+ return promise;
+ },
+
+ resolve: function() {
+ var me=this,
+ resolver=me.deferred;
+
+ if (resolver) {
+ // Make a copy of the resolver before calling & removing it to ensure
+ // it is not called twice
+ me.deferred=null;
+ resolver.resolve();
+ }
+ },
+
+ /**
+ * Event handler for image onload
+ * @param {object} e jQuery event data
+ */
+
+ imageLoaded: function(e) {
+ var me=this,
+ index = me.indexOf(e.target);
+
+ if (index>=0) {
+
+ me.status[index] = true;
+ if ($.inArray(false, me.status) < 0) {
+ me.resolve();
+ }
+ }
+ },
+
+ /**
+ * Event handler for onload error
+ * @param {object} e jQuery event data
+ */
+
+ imageLoadError: function(e) {
+ clearTimeout(this.imgTimeout);
+ this.triesLeft=0;
+ var err = e ? 'The image ' + e.target.src + ' failed to load.' :
+ 'The images never seemed to finish loading. You may just need to increase the configTimeout if images could take a long time to load.';
+ throw err;
+ },
+ /**
+ * Test if the image at specificed index has finished loading
+ * @param {int} index The image index
+ * @return {boolean} true if loaded, false if not
+ */
+
+ isLoaded: function(index) {
+ var img,
+ me=this,
+ status=me.status;
+
+ if (status[index]) { return true; }
+ img = me[index];
+
+ if (typeof img.complete !== 'undefined') {
+ status[index]=img.complete;
+ } else {
+ status[index]=!!u.imgWidth(img);
+ }
+ // if complete passes, the image is loaded, but may STILL not be available because of stuff like adblock.
+ // make sure it is.
+
+ return status[index];
+ }
+ };
+ } (jQuery));
+/* mapdata.js
+ the MapData object, repesents an instance of a single bound imagemap
+*/
+
+
+(function ($) {
+
+ var m = $.mapster,
+ u = m.utils;
+
+ /**
+ * Set default values for MapData object properties
+ * @param {MapData} me The MapData object
+ */
+
+ function initializeDefaults(me) {
+ $.extend(me,{
+ complete: false, // (bool) when configuration is complete
+ map: null, // ($) the image map
+ base_canvas: null, // (canvas|var) where selections are rendered
+ overlay_canvas: null, // (canvas|var) where highlights are rendered
+ commands: [], // {} commands that were run before configuration was completed (b/c images weren't loaded)
+ data: [], // MapData[] area groups
+ mapAreas: [], // MapArea[] list. AreaData entities contain refs to this array, so options are stored with each.
+ _xref: {}, // (int) xref of mapKeys to data[]
+ highlightId: -1, // (int) the currently highlighted element.
+ currentAreaId: -1,
+ _tooltip_events: [], // {} info on events we bound to a tooltip container, so we can properly unbind them
+ scaleInfo: null, // {} info about the image size, scaling, defaults
+ index: -1, // index of this in map_cache - so we have an ID to use for wraper div
+ activeAreaEvent: null
+ });
+ }
+
+ /**
+ * Return an array of all image-containing options from an options object;
+ * that is, containers that may have an "altImage" property
+ *
+ * @param {object} obj An options object
+ * @return {object[]} An array of objects
+ */
+ function getOptionImages(obj) {
+ return [obj, obj.render_highlight, obj.render_select];
+ }
+
+ /**
+ * Parse all the altImage references, adding them to the library so they can be preloaded
+ * and aliased.
+ *
+ * @param {MapData} me The MapData object on which to operate
+ */
+ function configureAltImages(me)
+ {
+ var opts = me.options,
+ mi = me.images;
+
+ // add alt images
+
+ if ($.mapster.hasCanvas) {
+ // map altImage library first
+
+ $.each(opts.altImages || {}, function(i,e) {
+ mi.add(e,i);
+ });
+
+ // now find everything else
+
+ $.each([opts].concat(opts.areas),function(i,e) {
+ $.each(getOptionImages(e),function(i2,e2) {
+ if (e2 && e2.altImage) {
+ e2.altImageId=mi.add(e2.altImage);
+ }
+ });
+ });
+ }
+
+ // set area_options
+ me.area_options = u.updateProps({}, // default options for any MapArea
+ m.area_defaults,
+ opts);
+ }
+
+ /**
+ * Queue a mouse move action based on current delay settings
+ * (helper for mouseover/mouseout handlers)
+ *
+ * @param {MapData} me The MapData context
+ * @param {number} delay The number of milliseconds to delay the action
+ * @param {AreaData} area AreaData affected
+ * @param {Deferred} deferred A deferred object to return (instead of a new one)
+ * @return {Promise} A promise that resolves when the action is completed
+ */
+ function queueMouseEvent(me,delay,area, deferred) {
+
+ deferred = deferred || u.when.defer();
+
+ function cbFinal(areaId) {
+ if (me.currentAreaId!==areaId && me.highlightId>=0) {
+ deferred.resolve();
+ }
+ }
+ if (me.activeAreaEvent) {
+ window.clearTimeout(me.activeAreaEvent);
+ me.activeAreaEvent=0;
+ }
+ if (delay<0) {
+ return;
+ }
+
+ if (area.owner.currentAction || delay) {
+ me.activeAreaEvent = window.setTimeout((function() {
+ return function() {
+ queueMouseEvent(me,0,area,deferred);
+ };
+ }(area)),
+ delay || 100);
+ } else {
+ cbFinal(area.areaId);
+ }
+ return deferred;
+ }
+
+ /**
+ * Mousedown event. This is captured only to prevent browser from drawing an outline around an
+ * area when it's clicked.
+ *
+ * @param {EventData} e jQuery event data
+ */
+
+ function mousedown(e) {
+ if (!$.mapster.hasCanvas) {
+ this.blur();
+ }
+ e.preventDefault();
+ }
+
+ /**
+ * Mouseover event. Handle highlight rendering and client callback on mouseover
+ *
+ * @param {MapData} me The MapData context
+ * @param {EventData} e jQuery event data
+ * @return {[type]} [description]
+ */
+
+ function mouseover(me,e) {
+ var arData = me.getAllDataForArea(this),
+ ar=arData.length ? arData[0] : null;
+
+ // mouseover events are ignored entirely while resizing, though we do care about mouseout events
+ // and must queue the action to keep things clean.
+
+ if (!ar || ar.isNotRendered() || ar.owner.currentAction) {
+ return;
+ }
+
+ if (me.currentAreaId === ar.areaId) {
+ return;
+ }
+ if (me.highlightId !== ar.areaId) {
+ me.clearEffects();
+
+ ar.highlight();
+
+ if (me.options.showToolTip) {
+ $.each(arData,function(i,e) {
+ if (e.effectiveOptions().toolTip) {
+ e.showToolTip();
+ }
+ });
+ }
+ }
+
+ me.currentAreaId = ar.areaId;
+
+ if ($.isFunction(me.options.onMouseover)) {
+ me.options.onMouseover.call(this,
+ {
+ e: e,
+ options:ar.effectiveOptions(),
+ key: ar.key,
+ selected: ar.isSelected()
+ });
+ }
+ }
+
+ /**
+ * Mouseout event.
+ *
+ * @param {MapData} me The MapData context
+ * @param {EventData} e jQuery event data
+ * @return {[type]} [description]
+ */
+
+ function mouseout(me,e) {
+ var newArea,
+ ar = me.getDataForArea(this),
+ opts = me.options;
+
+
+ if (me.currentAreaId<0 || !ar) {
+ return;
+ }
+
+ newArea=me.getDataForArea(e.relatedTarget);
+
+ if (newArea === ar) {
+ return;
+ }
+
+ me.currentAreaId = -1;
+ ar.area=null;
+
+ queueMouseEvent(me,opts.mouseoutDelay,ar)
+ .then(me.clearEffects);
+
+ if ($.isFunction(opts.onMouseout)) {
+ opts.onMouseout.call(this,
+ {
+ e: e,
+ options: opts,
+ key: ar.key,
+ selected: ar.isSelected()
+ });
+ }
+
+ }
+
+ /**
+ * Clear any active tooltip or highlight
+ *
+ * @param {MapData} me The MapData context
+ * @param {EventData} e jQuery event data
+ * @return {[type]} [description]
+ */
+
+ function clearEffects(me) {
+ var opts = me.options;
+
+ me.ensureNoHighlight();
+
+ if (opts.toolTipClose
+ && $.inArray('area-mouseout', opts.toolTipClose) >= 0
+ && me.activeToolTip)
+ {
+ me.clearToolTip();
+ }
+ }
+
+ /**
+ * Mouse click event handler
+ *
+ * @param {MapData} me The MapData context
+ * @param {EventData} e jQuery event data
+ * @return {[type]} [description]
+ */
+
+ function click(me,e) {
+ var selected, list, list_target, newSelectionState, canChangeState, cbResult,
+ that = this,
+ ar = me.getDataForArea(this),
+ opts = me.options;
+
+ function clickArea(ar) {
+ var areaOpts,target;
+ canChangeState = (ar.isSelectable() &&
+ (ar.isDeselectable() || !ar.isSelected()));
+
+ if (canChangeState) {
+ newSelectionState = !ar.isSelected();
+ } else {
+ newSelectionState = ar.isSelected();
+ }
+
+ list_target = m.getBoundList(opts, ar.key);
+
+ if ($.isFunction(opts.onClick))
+ {
+ cbResult= opts.onClick.call(that,
+ {
+ e: e,
+ listTarget: list_target,
+ key: ar.key,
+ selected: newSelectionState
+ });
+
+ if (u.isBool(cbResult)) {
+ if (!cbResult) {
+ return false;
+ }
+ target = $(ar.area).attr('href');
+ if (target!=='#') {
+ window.location.href=target;
+ return false;
+ }
+ }
+ }
+
+ if (canChangeState) {
+ selected = ar.toggle();
+ }
+
+ if (opts.boundList && opts.boundList.length > 0) {
+ m.setBoundListProperties(opts, list_target, ar.isSelected());
+ }
+
+ areaOpts = ar.effectiveOptions();
+ if (areaOpts.includeKeys) {
+ list = u.split(areaOpts.includeKeys);
+ $.each(list, function (i, e) {
+ var ar = me.getDataForKey(e.toString());
+ if (!ar.options.isMask) {
+ clickArea(ar);
+ }
+ });
+ }
+ }
+
+ mousedown.call(this,e);
+
+ if (opts.clickNavigate && ar.href) {
+ window.location.href=ar.href;
+ return;
+ }
+
+ if (ar && !ar.owner.currentAction) {
+ opts = me.options;
+ clickArea(ar);
+ }
+ }
+
+ /**
+ * Prototype for a MapData object, representing an ImageMapster bound object
+ * @param {Element} image an IMG element
+ * @param {object} options ImageMapster binding options
+ */
+ m.MapData = function (image, options)
+ {
+ var me = this;
+
+ // (Image) main map image
+
+ me.image = image;
+
+ me.images = new m.MapImages(me);
+ me.graphics = new m.Graphics(me);
+
+ // save the initial style of the image for unbinding. This is problematic, chrome
+ // duplicates styles when assigning, and cssText is apparently not universally supported.
+ // Need to do something more robust to make unbinding work universally.
+
+ me.imgCssText = image.style.cssText || null;
+
+ initializeDefaults(me);
+
+ me.configureOptions(options);
+
+ // create context-bound event handlers from our private functions
+
+ me.mouseover = function(e) { mouseover.call(this,me,e); };
+ me.mouseout = function(e) { mouseout.call(this,me,e); };
+ me.click = function(e) { click.call(this,me,e); };
+ me.clearEffects = function(e) { clearEffects.call(this,me,e); };
+ };
+
+ m.MapData.prototype = {
+ constructor: m.MapData,
+
+ /**
+ * Set target.options from defaults + options
+ * @param {[type]} target The target
+ * @param {[type]} options The options to merge
+ */
+
+ configureOptions: function(options) {
+ this.options= u.updateProps({}, m.defaults, options);
+ },
+
+ /**
+ * Ensure all images are loaded
+ * @return {Promise} A promise that resolves when the images have finished loading (or fail)
+ */
+
+ bindImages: function() {
+ var me=this,
+ mi = me.images;
+
+ // reset the images if this is a rebind
+
+ if (mi.length>2) {
+ mi.splice(2);
+ } else if (mi.length===0) {
+
+ // add the actual main image
+ mi.add(me.image);
+ // will create a duplicate of the main image, we need this to get raw size info
+ mi.add(me.image.src);
+ }
+
+ configureAltImages(me);
+
+ return me.images.bind();
+ },
+
+ /**
+ * Test whether an async action is currently in progress
+ * @return {Boolean} true or false indicating state
+ */
+
+ isActive: function() {
+ return !this.complete || this.currentAction;
+ },
+
+ /**
+ * Return an object indicating the various states. This isn't really used by
+ * production code.
+ *
+ * @return {object} An object with properties for various states
+ */
+
+ state: function () {
+ return {
+ complete: this.complete,
+ resizing: this.currentAction==='resizing',
+ zoomed: this.zoomed,
+ zoomedArea: this.zoomedArea,
+ scaleInfo: this.scaleInfo
+ };
+ },
+
+ /**
+ * Get a unique ID for the wrapper of this imagemapster
+ * @return {string} A string that is unique to this image
+ */
+
+ wrapId: function () {
+ return 'mapster_wrap_' + this.index;
+ },
+ _idFromKey: function (key) {
+ return typeof key === "string" && this._xref.hasOwnProperty(key) ?
+ this._xref[key] : -1;
+ },
+
+ /**
+ * Return a comma-separated string of all selected keys
+ * @return {string} CSV of all keys that are currently selected
+ */
+
+ getSelected: function () {
+ var result = '';
+ $.each(this.data, function (i,e) {
+ if (e.isSelected()) {
+ result += (result ? ',' : '') + this.key;
+ }
+ });
+ return result;
+ },
+
+ /**
+ * Get an array of MapAreas associated with a specific AREA based on the keys for that area
+ * @param {Element} area An HTML AREA
+ * @param {number} atMost A number limiting the number of areas to be returned (typically 1 or 0 for no limit)
+ * @return {MapArea[]} Array of MapArea objects
+ */
+
+ getAllDataForArea:function (area,atMost) {
+ var i,ar, result,
+ me=this,
+ key = $(area).filter('area').attr(me.options.mapKey);
+
+ if (key) {
+ result=[];
+ key = u.split(key);
+
+ for (i=0;i<(atMost || key.length);i++) {
+ ar = me.data[me._idFromKey(key[i])];
+ ar.area=area.length ? area[0]:area;
+ // set the actual area moused over/selected
+ // TODO: this is a brittle model for capturing which specific area - if this method was not used,
+ // ar.area could have old data. fix this.
+ result.push(ar);
+ }
+ }
+
+ return result;
+ },
+ getDataForArea: function(area) {
+ var ar=this.getAllDataForArea(area,1);
+ return ar ? ar[0] || null : null;
+ },
+ getDataForKey: function (key) {
+ return this.data[this._idFromKey(key)];
+ },
+
+ /**
+ * Get the primary keys associated with an area group.
+ * If this is a primary key, it will be returned.
+ *
+ * @param {string key An area key
+ * @return {string} A CSV of area keys
+ */
+
+ getKeysForGroup: function(key) {
+ var ar=this.getDataForKey(key);
+
+ return !ar ? '':
+ ar.isPrimary ?
+ ar.key :
+ this.getPrimaryKeysForMapAreas(ar.areas()).join(',');
+ },
+
+ /**
+ * given an array of MapArea object, return an array of its unique primary keys
+ * @param {MapArea[]} areas The areas to analyze
+ * @return {string[]} An array of unique primary keys
+ */
+
+ getPrimaryKeysForMapAreas: function(areas)
+ {
+ var keys=[];
+ $.each(areas,function(i,e) {
+ if ($.inArray(e.keys[0],keys)<0) {
+ keys.push(e.keys[0]);
+ }
+ });
+ return keys;
+ },
+ getData: function (obj) {
+ if (typeof obj === 'string') {
+ return this.getDataForKey(obj);
+ } else if (obj && obj.mapster || u.isElement(obj)) {
+ return this.getDataForArea(obj);
+ } else {
+ return null;
+ }
+ },
+ // remove highlight if present, raise event
+ ensureNoHighlight: function () {
+ var ar;
+ if (this.highlightId >= 0) {
+ this.graphics.clearHighlight();
+ ar = this.data[this.highlightId];
+ ar.changeState('highlight', false);
+ this.setHighlightId(-1);
+ }
+ },
+ setHighlightId: function(id) {
+ this.highlightId = id;
+ },
+
+ /**
+ * Clear all active selections on this map
+ */
+
+ clearSelections: function () {
+ $.each(this.data, function (i,e) {
+ if (e.selected) {
+ e.deselect(true);
+ }
+ });
+ this.removeSelectionFinish();
+
+ },
+
+ /**
+ * Set area options from an array of option data.
+ *
+ * @param {object[]} areas An array of objects containing area-specific options
+ */
+
+ setAreaOptions: function (areas) {
+ var i, area_options, ar;
+ areas = areas || [];
+
+ // refer by: map_data.options[map_data.data[x].area_option_id]
+
+ for (i = areas.length - 1; i >= 0; i--) {
+ area_options = areas[i];
+ if (area_options) {
+ ar = this.getDataForKey(area_options.key);
+ if (ar) {
+ u.updateProps(ar.options, area_options);
+
+ // TODO: will not deselect areas that were previously selected, so this only works
+ // for an initial bind.
+
+ if (u.isBool(area_options.selected)) {
+ ar.selected = area_options.selected;
+ }
+ }
+ }
+ }
+ },
+ // keys: a comma-separated list
+ drawSelections: function (keys) {
+ var i, key_arr = u.asArray(keys);
+
+ for (i = key_arr.length - 1; i >= 0; i--) {
+ this.data[key_arr[i]].drawSelection();
+ }
+ },
+ redrawSelections: function () {
+ $.each(this.data, function (i, e) {
+ if (e.isSelectedOrStatic()) {
+ e.drawSelection();
+ }
+ });
+
+ },
+ ///called when images are done loading
+ initialize: function () {
+ var imgCopy, base_canvas, overlay_canvas, wrap, parentId, css, i,size,
+ img,sort_func, sorted_list, scale,
+ me = this,
+ opts = me.options;
+
+ if (me.complete) {
+ return;
+ }
+
+ img = $(me.image);
+
+ parentId = img.parent().attr('id');
+
+ // create a div wrapper only if there's not already a wrapper, otherwise, own it
+
+ if (parentId && parentId.length >= 12 && parentId.substring(0, 12) === "mapster_wrap") {
+ wrap = img.parent();
+ wrap.attr('id', me.wrapId());
+ } else {
+ wrap = $('');
+
+ if (opts.wrapClass) {
+ if (opts.wrapClass === true) {
+ wrap.addClass(img[0].className);
+ }
+ else {
+ wrap.addClass(opts.wrapClass);
+ }
+ }
+ }
+ me.wrapper = wrap;
+
+ // me.images[1] is the copy of the original image. It should be loaded & at its native size now so we can obtain the true
+ // width & height. This is needed to scale the imagemap if not being shown at its native size. It is also needed purely
+ // to finish binding in case the original image was not visible. It can be impossible in some browsers to obtain the
+ // native size of a hidden image.
+
+ me.scaleInfo = scale = u.scaleMap(me.images[0],me.images[1], opts.scaleMap);
+
+ me.base_canvas = base_canvas = me.graphics.createVisibleCanvas(me);
+ me.overlay_canvas = overlay_canvas = me.graphics.createVisibleCanvas(me);
+
+ // Now we got what we needed from the copy -clone from the original image again to make sure any other attributes are copied
+ imgCopy = $(me.images[1])
+ .addClass('mapster_el '+ me.images[0].className)
+ .attr({id:null, usemap: null});
+
+ size=u.size(me.images[0]);
+
+ if (size.complete) {
+ imgCopy.css({
+ width: size.width,
+ height: size.height
+ });
+ }
+
+ me.buildDataset();
+
+ // now that we have processed all the areas, set css for wrapper, scale map if needed
+
+ css = {
+ display: 'block',
+ position: 'relative',
+ padding: 0,
+ width: scale.width,
+ height: scale.height
+ };
+
+ if (opts.wrapCss) {
+ $.extend(css, opts.wrapCss);
+ }
+ // if we were rebinding with an existing wrapper, the image will aready be in it
+ if (img.parent()[0] !== me.wrapper[0]) {
+
+ img.before(me.wrapper);
+ }
+
+ wrap.css(css);
+
+ // move all generated images into the wrapper for easy removal later
+
+ $(me.images.slice(2)).hide();
+ for (i = 1; i < me.images.length; i++) {
+ wrap.append(me.images[i]);
+ }
+
+ //me.images[1].style.cssText = me.image.style.cssText;
+
+ wrap.append(base_canvas)
+ .append(overlay_canvas)
+ .append(img.css(m.canvas_style));
+
+ // images[0] is the original image with map, images[1] is the copy/background that is visible
+
+ u.setOpacity(me.images[0], 0);
+ $(me.images[1]).show();
+
+ u.setOpacity(me.images[1],1);
+
+ if (opts.isSelectable && opts.onGetList) {
+ sorted_list = me.data.slice(0);
+ if (opts.sortList) {
+ if (opts.sortList === "desc") {
+ sort_func = function (a, b) {
+ return a === b ? 0 : (a > b ? -1 : 1);
+ };
+ }
+ else {
+ sort_func = function (a, b) {
+ return a === b ? 0 : (a < b ? -1 : 1);
+ };
+ }
+
+ sorted_list.sort(function (a, b) {
+ a = a.value;
+ b = b.value;
+ return sort_func(a, b);
+ });
+ }
+
+ me.options.boundList = opts.onGetList.call(me.image, sorted_list);
+ }
+
+ me.complete=true;
+ me.processCommandQueue();
+
+ if (opts.onConfigured && typeof opts.onConfigured === 'function') {
+ opts.onConfigured.call(img, true);
+ }
+ },
+
+ // when rebind is true, the MapArea data will not be rebuilt.
+ buildDataset: function(rebind) {
+ var sel,areas,j,area_id,$area,area,curKey,mapArea,key,keys,mapAreaId,group_value,dataItem,href,
+ me=this,
+ opts=me.options,
+ default_group;
+
+ function addAreaData(key, value) {
+ var dataItem = new m.AreaData(me, key, value);
+ dataItem.areaId = me._xref[key] = me.data.push(dataItem) - 1;
+ return dataItem.areaId;
+ }
+
+ me._xref = {};
+ me.data = [];
+ if (!rebind) {
+ me.mapAreas=[];
+ }
+
+ default_group = !opts.mapKey;
+ if (default_group) {
+ opts.mapKey = 'data-mapster-key';
+ }
+ sel = ($.browser.msie && $.browser.version <= 7) ? 'area' :
+ (default_group ? 'area[coords]' : 'area[' + opts.mapKey + ']');
+ areas = $(me.map).find(sel).unbind('.mapster');
+
+ for (mapAreaId = 0;mapAreaId= 0; j--) {
+ key = keys[j];
+
+ if (opts.mapValue) {
+ group_value = $area.attr(opts.mapValue);
+ }
+ if (default_group) {
+ // set an attribute so we can refer to the area by index from the DOM object if no key
+ area_id = addAreaData(me.data.length, group_value);
+ dataItem = me.data[area_id];
+ dataItem.key = key = area_id.toString();
+ }
+ else {
+ area_id = me._xref[key];
+ if (area_id >= 0) {
+ dataItem = me.data[area_id];
+ if (group_value && !me.data[area_id].value) {
+ dataItem.value = group_value;
+ }
+ }
+ else {
+ area_id = addAreaData(key, group_value);
+ dataItem = me.data[area_id];
+ dataItem.isPrimary=j===0;
+ }
+ }
+ mapArea.areaDataXref.push(area_id);
+ dataItem.areasXref.push(mapAreaId);
+ }
+
+ href=$area.attr('href');
+ if (href && href!=='#' && !dataItem.href)
+ {
+ dataItem.href=href;
+ }
+
+ if (!mapArea.nohref) {
+ $area.bind('click.mapster', me.click);
+
+ if (!m.isTouch) {
+ $area.bind('mouseover.mapster', me.mouseover)
+ .bind('mouseout.mapster', me.mouseout)
+ .bind('mousedown.mapster', me.mousedown);
+
+ }
+
+ }
+
+ // store an ID with each area.
+ $area.data("mapster", mapAreaId+1);
+ }
+
+ // TODO listenToList
+ // if (opts.listenToList && opts.nitG) {
+ // opts.nitG.bind('click.mapster', event_hooks[map_data.hooks_index].listclick_hook);
+ // }
+
+ // populate areas from config options
+ me.setAreaOptions(opts.areas);
+ me.redrawSelections();
+
+ },
+ processCommandQueue: function() {
+
+ var cur,me=this;
+ while (!me.currentAction && me.commands.length) {
+ cur = me.commands[0];
+ me.commands.splice(0,1);
+ m.impl[cur.command].apply(cur.that, cur.args);
+ }
+ },
+ clearEvents: function () {
+ $(this.map).find('area')
+ .unbind('.mapster');
+ $(this.images)
+ .unbind('.mapster');
+ },
+ _clearCanvases: function (preserveState) {
+ // remove the canvas elements created
+ if (!preserveState) {
+ $(this.base_canvas).remove();
+ }
+ $(this.overlay_canvas).remove();
+ },
+ clearMapData: function (preserveState) {
+ var me = this;
+ this._clearCanvases(preserveState);
+
+ // release refs to DOM elements
+ $.each(this.data, function (i, e) {
+ e.reset();
+ });
+ this.data = null;
+ if (!preserveState) {
+ // get rid of everything except the original image
+ this.image.style.cssText = this.imgCssText;
+ $(this.wrapper).before(this.image).remove();
+ }
+
+ me.images.clear();
+
+ this.image = null;
+ u.ifFunction(this.clearTooltip, this);
+ },
+
+ // Compelete cleanup process for deslecting items. Called after a batch operation, or by AreaData for single
+ // operations not flagged as "partial"
+
+ removeSelectionFinish: function () {
+ var g = this.graphics;
+
+ g.refreshSelections();
+ // do not call ensure_no_highlight- we don't really want to unhilight it, just remove the effect
+ g.clearHighlight();
+ }
+ };
+} (jQuery));
+/* areadata.js
+ AreaData and MapArea protoypes
+*/
+
+(function ($) {
+ var m = $.mapster, u = m.utils;
+
+ /**
+ * Select this area
+ *
+ * @param {AreaData} me AreaData context
+ * @param {object} options Options for rendering the selection
+ */
+ function select(options) {
+ // need to add the new one first so that the double-opacity effect leaves the current one highlighted for singleSelect
+
+ var me=this, o = me.owner;
+ if (o.options.singleSelect) {
+ o.clearSelections();
+ }
+
+ // because areas can overlap - we can't depend on the selection state to tell us anything about the inner areas.
+ // don't check if it's already selected
+ if (!me.isSelected()) {
+ if (options) {
+
+ // cache the current options, and map the altImageId if an altimage
+ // was passed
+
+ me.optsCache = $.extend(me.effectiveRenderOptions('select'),
+ options,
+ {
+ altImageId: o.images.add(options.altImage)
+ });
+ }
+
+ me.drawSelection();
+
+ me.selected = true;
+ me.changeState('select', true);
+ }
+
+ if (o.options.singleSelect) {
+ o.graphics.refreshSelections();
+ }
+ }
+
+ /**
+ * Deselect this area, optionally deferring finalization so additional areas can be deselected
+ * in a single operation
+ *
+ * @param {boolean} partial when true, the caller must invoke "finishRemoveSelection" to render
+ */
+
+ function deselect(partial) {
+ var me=this;
+ me.selected = false;
+ me.changeState('select', false);
+
+ // release information about last area options when deselecting.
+
+ me.optsCache=null;
+ me.owner.graphics.removeSelections(me.areaId);
+
+ // Complete selection removal process. This is separated because it's very inefficient to perform the whole
+ // process for multiple removals, as the canvas must be totally redrawn at the end of the process.ar.remove
+
+ if (!partial) {
+ me.owner.removeSelectionFinish();
+ }
+ }
+
+ /**
+ * Toggle the selection state of this area
+ * @param {object} options Rendering options, if toggling on
+ * @return {bool} The new selection state
+ */
+ function toggle(options) {
+ var me=this;
+ if (!me.isSelected()) {
+ me.select(options);
+ }
+ else {
+ me.deselect();
+ }
+ return me.isSelected();
+ }
+
+ /**
+ * An AreaData object; represents a conceptual area that can be composed of
+ * one or more MapArea objects
+ *
+ * @param {MapData} owner The MapData object to which this belongs
+ * @param {string} key The key for this area
+ * @param {string} value The mapValue string for this area
+ */
+
+ m.AreaData = function (owner, key, value) {
+ $.extend(this,{
+ owner: owner,
+ key: key || '',
+ // means this represents the first key in a list of keys (it's the area group that gets highlighted on mouseover)
+ isPrimary: true,
+ areaId: -1,
+ href: '',
+ value: value || '',
+ options:{},
+ // "null" means unchanged. Use "isSelected" method to just test true/false
+ selected: null,
+ // xref to MapArea objects
+ areasXref: [],
+ // (temporary storage) - the actual area moused over
+ area: null,
+ // the last options used to render this. Cache so when re-drawing after a remove, changes in options won't
+ // break already selected things.
+ optsCache: null
+ });
+ };
+
+ /**
+ * The public API for AreaData object
+ */
+
+ m.AreaData.prototype = {
+ constuctor: m.AreaData,
+ select: select,
+ deselect: deselect,
+ toggle: toggle,
+ areas: function() {
+ var i,result=[];
+ for (i=0;i= 0; j -= 2) {
+ curX = coords[j];
+ curY = coords[j + 1];
+
+ if (curX < minX) {
+ minX = curX;
+ bestMaxY = curY;
+ }
+ if (curX > maxX) {
+ maxX = curX;
+ bestMinY = curY;
+ }
+ if (curY < minY) {
+ minY = curY;
+ bestMaxX = curX;
+ }
+ if (curY > maxY) {
+ maxY = curY;
+ bestMinX = curX;
+ }
+
+ }
+
+ // try to figure out the best place for the tooltip
+
+ if (width && height) {
+ found=false;
+ $.each([[bestMaxX - width, minY - height], [bestMinX, minY - height],
+ [minX - width, bestMaxY - height], [minX - width, bestMinY],
+ [maxX,bestMaxY - height], [ maxX,bestMinY],
+ [bestMaxX - width, maxY], [bestMinX, maxY]
+ ],function (i, e) {
+ if (!found && (e[0] > rootx && e[1] > rooty)) {
+ nest = e;
+ found=true;
+ return false;
+ }
+ });
+
+ // default to lower-right corner if nothing fit inside the boundaries of the image
+
+ if (!found) {
+ nest=[maxX,maxY];
+ }
+ }
+ return nest;
+ };
+} (jQuery));
+/* scale.js: resize and zoom functionality
+ requires areacorners.js, when.js
+*/
+
+
+(function ($) {
+ var m = $.mapster, u = m.utils, p = m.MapArea.prototype;
+
+ m.utils.getScaleInfo = function (eff, actual) {
+ var pct;
+ if (!actual) {
+ pct = 1;
+ actual=eff;
+ } else {
+ pct = eff.width / actual.width || eff.height / actual.height;
+ // make sure a float error doesn't muck us up
+ if (pct > 0.98 && pct < 1.02) { pct = 1; }
+ }
+ return {
+ scale: (pct !== 1),
+ scalePct: pct,
+ realWidth: actual.width,
+ realHeight: actual.height,
+ width: eff.width,
+ height: eff.height,
+ ratio: eff.width / eff.height
+ };
+ };
+ // Scale a set of AREAs, return old data as an array of objects
+ m.utils.scaleMap = function (image, imageRaw, scale) {
+
+ // stunningly, jQuery width can return zero even as width does not, seems to happen only
+ // with adBlock or maybe other plugins. These must interfere with onload events somehow.
+
+
+ var vis=u.size(image),
+ raw=u.size(imageRaw,true);
+
+ if (!raw.complete()) {
+ throw("Another script, such as an extension, appears to be interfering with image loading. Please let us know about this.");
+ }
+ if (!vis.complete()) {
+ vis=raw;
+ }
+ return this.getScaleInfo(vis, scale ? raw : null);
+ };
+
+ /**
+ * Resize the image map. Only one of newWidth and newHeight should be passed to preserve scale
+ *
+ * @param {int} width The new width OR an object containing named parameters matching this function sig
+ * @param {int} height The new height
+ * @param {int} effectDuration Time in ms for the resize animation, or zero for no animation
+ * @param {function} callback A function to invoke when the operation finishes
+ * @return {promise} NOT YET IMPLEMENTED
+ */
+
+ m.MapData.prototype.resize = function (width, height, duration, callback) {
+ var p,promises,newsize,els, highlightId, ratio,
+ me = this;
+
+ // allow omitting duration
+ callback = callback || duration;
+
+ function sizeCanvas(canvas, w, h) {
+ if ($.mapster.hasCanvas) {
+ canvas.width = w;
+ canvas.height = h;
+ } else {
+ $(canvas).width(w);
+ $(canvas).height(h);
+ }
+ }
+
+ // Finalize resize action, do callback, pass control to command queue
+
+ function cleanupAndNotify() {
+
+ me.currentAction = '';
+
+ if ($.isFunction(callback)) {
+ callback();
+ }
+
+ me.processCommandQueue();
+ }
+
+ // handle cleanup after the inner elements are resized
+
+ function finishResize() {
+ sizeCanvas(me.overlay_canvas, width, height);
+
+ // restore highlight state if it was highlighted before
+ if (highlightId >= 0) {
+ var areaData = me.data[highlightId];
+ areaData.tempOptions = { fade: false };
+ me.getDataForKey(areaData.key).highlight();
+ areaData.tempOptions = null;
+ }
+ sizeCanvas(me.base_canvas, width, height);
+ me.redrawSelections();
+ cleanupAndNotify();
+ }
+
+ function resizeMapData() {
+ $(me.image).css(newsize);
+ // start calculation at the same time as effect
+ me.scaleInfo = u.getScaleInfo({
+ width: width,
+ height: height
+ },
+ {
+ width: me.scaleInfo.realWidth,
+ height: me.scaleInfo.realHeight
+ });
+ $.each(me.data, function (i, e) {
+ $.each(e.areas(), function (i, e) {
+ e.resize();
+ });
+ });
+ }
+
+ if (me.scaleInfo.width === width && me.scaleInfo.height === height) {
+ return;
+ }
+
+ highlightId = me.highlightId;
+
+
+ if (!width) {
+ ratio = height / me.scaleInfo.realHeight;
+ width = Math.round(me.scaleInfo.realWidth * ratio);
+ }
+ if (!height) {
+ ratio = width / me.scaleInfo.realWidth;
+ height = Math.round(me.scaleInfo.realHeight * ratio);
+ }
+
+ newsize = { 'width': String(width) + 'px', 'height': String(height) + 'px' };
+ if (!$.mapster.hasCanvas) {
+ $(me.base_canvas).children().remove();
+ }
+
+ // resize all the elements that are part of the map except the image itself (which is not visible)
+ // but including the div wrapper
+ els = $(me.wrapper).find('.mapster_el').add(me.wrapper);
+
+ if (duration) {
+ promises = [];
+ me.currentAction = 'resizing';
+ els.each(function (i, e) {
+ p = u.defer();
+ promises.push(p);
+
+ $(e).animate(newsize, {
+ duration: duration,
+ complete: p.resolve,
+ easing: "linear"
+ });
+ });
+
+ p = u.defer();
+ promises.push(p);
+
+ // though resizeMapData is not async, it needs to be finished just the same as the animations,
+ // so add it to the "to do" list.
+
+ u.when.all(promises).then(finishResize);
+ resizeMapData();
+ p.resolve();
+ } else {
+ els.css(newsize);
+ resizeMapData();
+ finishResize();
+
+ }
+ };
+
+
+ m.MapArea = u.subclass(m.MapArea, function () {
+ //change the area tag data if needed
+ this.base.init();
+ if (this.owner.scaleInfo.scale) {
+ this.resize();
+ }
+ });
+
+ p.coords = function (percent, coordOffset) {
+ var j, newCoords = [],
+ pct = percent || this.owner.scaleInfo.scalePct,
+ offset = coordOffset || 0;
+
+ if (pct === 1 && coordOffset === 0) {
+ return this.originalCoords;
+ }
+
+ for (j = 0; j < this.length; j++) {
+ //amount = j % 2 === 0 ? xPct : yPct;
+ newCoords.push(Math.round(this.originalCoords[j] * pct) + offset);
+ }
+ return newCoords;
+ };
+ p.resize = function () {
+ this.area.coords = this.coords().join(',');
+ };
+
+ p.reset = function () {
+ this.area.coords = this.coords(1).join(',');
+ };
+
+ m.impl.resize = function (width, height, duration, callback) {
+ if (!width && !height) {
+ return false;
+ }
+ var x= (new m.Method(this,
+ function () {
+ this.resize(width, height, duration, callback);
+ },
+ null,
+ {
+ name: 'resize',
+ args: arguments
+ }
+ )).go();
+ return x;
+ };
+
+/*
+ m.impl.zoom = function (key, opts) {
+ var options = opts || {};
+
+ function zoom(areaData) {
+ // this will be MapData object returned by Method
+
+ var scroll, corners, height, width, ratio,
+ diffX, diffY, ratioX, ratioY, offsetX, offsetY, newWidth, newHeight, scrollLeft, scrollTop,
+ padding = options.padding || 0,
+ scrollBarSize = areaData ? 20 : 0,
+ me = this,
+ zoomOut = false;
+
+ if (areaData) {
+ // save original state on first zoom operation
+ if (!me.zoomed) {
+ me.zoomed = true;
+ me.preZoomWidth = me.scaleInfo.width;
+ me.preZoomHeight = me.scaleInfo.height;
+ me.zoomedArea = areaData;
+ if (options.scroll) {
+ me.wrapper.css({ overflow: 'auto' });
+ }
+ }
+ corners = $.mapster.utils.areaCorners(areaData.coords(1, 0));
+ width = me.wrapper.innerWidth() - scrollBarSize - padding * 2;
+ height = me.wrapper.innerHeight() - scrollBarSize - padding * 2;
+ diffX = corners.maxX - corners.minX;
+ diffY = corners.maxY - corners.minY;
+ ratioX = width / diffX;
+ ratioY = height / diffY;
+ ratio = Math.min(ratioX, ratioY);
+ offsetX = (width - diffX * ratio) / 2;
+ offsetY = (height - diffY * ratio) / 2;
+
+ newWidth = me.scaleInfo.realWidth * ratio;
+ newHeight = me.scaleInfo.realHeight * ratio;
+ scrollLeft = (corners.minX) * ratio - padding - offsetX;
+ scrollTop = (corners.minY) * ratio - padding - offsetY;
+ } else {
+ if (!me.zoomed) {
+ return;
+ }
+ zoomOut = true;
+ newWidth = me.preZoomWidth;
+ newHeight = me.preZoomHeight;
+ scrollLeft = null;
+ scrollTop = null;
+ }
+
+ this.resize({
+ width: newWidth,
+ height: newHeight,
+ duration: options.duration,
+ scroll: scroll,
+ scrollLeft: scrollLeft,
+ scrollTop: scrollTop,
+ // closure so we can be sure values are correct
+ callback: (function () {
+ var isZoomOut = zoomOut,
+ scroll = options.scroll,
+ areaD = areaData;
+ return function () {
+ if (isZoomOut) {
+ me.preZoomWidth = null;
+ me.preZoomHeight = null;
+ me.zoomed = false;
+ me.zoomedArea = false;
+ if (scroll) {
+ me.wrapper.css({ overflow: 'inherit' });
+ }
+ } else {
+ // just to be sure it wasn't canceled & restarted
+ me.zoomedArea = areaD;
+ }
+ };
+ } ())
+ });
+ }
+ return (new m.Method(this,
+ function (opts) {
+ zoom.call(this);
+ },
+ function () {
+ zoom.call(this.owner, this);
+ },
+ {
+ name: 'zoom',
+ args: arguments,
+ first: true,
+ key: key
+ }
+ )).go();
+
+
+ };
+ */
+} (jQuery));
+/* tooltip.js - tooltip functionality
+ requires areacorners.js
+*/
+
+(function ($) {
+
+ var m = $.mapster, u = m.utils;
+
+ $.extend(m.defaults, {
+ toolTipContainer: '
')
+ .addClass('mapster_el')
+ .hide();
+
+ index=me._add(image[0]);
+
+ image
+ .bind('load',function(e) {
+ me.imageLoaded.call(me,e);
+ })
+ .bind('error',function(e) {
+ me.imageLoadError.call(me,e);
+ });
+
+ image.attr('src', src);
+ } else {
+
+ // use attr because we want the actual source, not the resolved path the browser will return directly calling image.src
+
+ index=me._add($(image)[0]);
+ }
+ if (id) {
+ if (this[id]) {
+ throw(id+" is already used or is not available as an altImage alias.");
+ }
+ me.ids.push(id);
+ me[id]=me[index];
+ }
+ return index;
+ },
+
+ /**
+ * Bind the images in this object,
+ * @param {boolean} retry when true, indicates that the function is calling itself after failure
+ * @return {Promise} a promise that resolves when the images have finished loading
+ */
+
+ bind: function(retry) {
+ var me = this,
+ promise,
+ triesLeft = me.owner.options.configTimeout / 200,
+
+ /* A recursive function to continue checking that the images have been
+ loaded until a timeout has elapsed */
+
+ check=function() {
+ var i;
+
+ // refresh status of images
+
+ i=me.length;
+
+ while (i-->0) {
+ if (!me.isLoaded(i)) {
+ break;
+ }
+ }
+
+ // check to see if every image has already been loaded
+
+ if (me.complete()) {
+ me.resolve();
+ } else {
+ // to account for failure of onLoad to fire in rare situations
+ if (triesLeft-- > 0) {
+ me.imgTimeout=window.setTimeout(function() {
+ check.call(me,true);
+ }, 50);
+ } else {
+ me.imageLoadError.call(me);
+ }
+ }
+
+ };
+
+ promise = me.deferred=u.defer();
+
+ check();
+ return promise;
+ },
+
+ resolve: function() {
+ var me=this,
+ resolver=me.deferred;
+
+ if (resolver) {
+ // Make a copy of the resolver before calling & removing it to ensure
+ // it is not called twice
+ me.deferred=null;
+ resolver.resolve();
+ }
+ },
+
+ /**
+ * Event handler for image onload
+ * @param {object} e jQuery event data
+ */
+
+ imageLoaded: function(e) {
+ var me=this,
+ index = me.indexOf(e.target);
+
+ if (index>=0) {
+
+ me.status[index] = true;
+ if ($.inArray(false, me.status) < 0) {
+ me.resolve();
+ }
+ }
+ },
+
+ /**
+ * Event handler for onload error
+ * @param {object} e jQuery event data
+ */
+
+ imageLoadError: function(e) {
+ clearTimeout(this.imgTimeout);
+ this.triesLeft=0;
+ var err = e ? 'The image ' + e.target.src + ' failed to load.' :
+ 'The images never seemed to finish loading. You may just need to increase the configTimeout if images could take a long time to load.';
+ throw err;
+ },
+ /**
+ * Test if the image at specificed index has finished loading
+ * @param {int} index The image index
+ * @return {boolean} true if loaded, false if not
+ */
+
+ isLoaded: function(index) {
+ var img,
+ me=this,
+ status=me.status;
+
+ if (status[index]) { return true; }
+ img = me[index];
+
+ if (typeof img.complete !== 'undefined') {
+ status[index]=img.complete;
+ } else {
+ status[index]=!!u.imgWidth(img);
+ }
+ // if complete passes, the image is loaded, but may STILL not be available because of stuff like adblock.
+ // make sure it is.
+
+ return status[index];
+ }
+ };
+ } (jQuery));
+/* mapdata.js
+ the MapData object, repesents an instance of a single bound imagemap
+*/
+
+
+(function ($) {
+
+ var m = $.mapster,
+ u = m.utils;
+
+ /**
+ * Set default values for MapData object properties
+ * @param {MapData} me The MapData object
+ */
+
+ function initializeDefaults(me) {
+ $.extend(me,{
+ complete: false, // (bool) when configuration is complete
+ map: null, // ($) the image map
+ base_canvas: null, // (canvas|var) where selections are rendered
+ overlay_canvas: null, // (canvas|var) where highlights are rendered
+ commands: [], // {} commands that were run before configuration was completed (b/c images weren't loaded)
+ data: [], // MapData[] area groups
+ mapAreas: [], // MapArea[] list. AreaData entities contain refs to this array, so options are stored with each.
+ _xref: {}, // (int) xref of mapKeys to data[]
+ highlightId: -1, // (int) the currently highlighted element.
+ currentAreaId: -1,
+ _tooltip_events: [], // {} info on events we bound to a tooltip container, so we can properly unbind them
+ scaleInfo: null, // {} info about the image size, scaling, defaults
+ index: -1, // index of this in map_cache - so we have an ID to use for wraper div
+ activeAreaEvent: null
+ });
+ }
+
+ /**
+ * Return an array of all image-containing options from an options object;
+ * that is, containers that may have an "altImage" property
+ *
+ * @param {object} obj An options object
+ * @return {object[]} An array of objects
+ */
+ function getOptionImages(obj) {
+ return [obj, obj.render_highlight, obj.render_select];
+ }
+
+ /**
+ * Parse all the altImage references, adding them to the library so they can be preloaded
+ * and aliased.
+ *
+ * @param {MapData} me The MapData object on which to operate
+ */
+ function configureAltImages(me)
+ {
+ var opts = me.options,
+ mi = me.images;
+
+ // add alt images
+
+ if ($.mapster.hasCanvas) {
+ // map altImage library first
+
+ $.each(opts.altImages || {}, function(i,e) {
+ mi.add(e,i);
+ });
+
+ // now find everything else
+
+ $.each([opts].concat(opts.areas),function(i,e) {
+ $.each(getOptionImages(e),function(i2,e2) {
+ if (e2 && e2.altImage) {
+ e2.altImageId=mi.add(e2.altImage);
+ }
+ });
+ });
+ }
+
+ // set area_options
+ me.area_options = u.updateProps({}, // default options for any MapArea
+ m.area_defaults,
+ opts);
+ }
+
+ /**
+ * Queue a mouse move action based on current delay settings
+ * (helper for mouseover/mouseout handlers)
+ *
+ * @param {MapData} me The MapData context
+ * @param {number} delay The number of milliseconds to delay the action
+ * @param {AreaData} area AreaData affected
+ * @param {Deferred} deferred A deferred object to return (instead of a new one)
+ * @return {Promise} A promise that resolves when the action is completed
+ */
+ function queueMouseEvent(me,delay,area, deferred) {
+
+ deferred = deferred || u.when.defer();
+
+ function cbFinal(areaId) {
+ if (me.currentAreaId!==areaId && me.highlightId>=0) {
+ deferred.resolve();
+ }
+ }
+ if (me.activeAreaEvent) {
+ window.clearTimeout(me.activeAreaEvent);
+ me.activeAreaEvent=0;
+ }
+ if (delay<0) {
+ return;
+ }
+
+ if (area.owner.currentAction || delay) {
+ me.activeAreaEvent = window.setTimeout((function() {
+ return function() {
+ queueMouseEvent(me,0,area,deferred);
+ };
+ }(area)),
+ delay || 100);
+ } else {
+ cbFinal(area.areaId);
+ }
+ return deferred;
+ }
+
+ /**
+ * Mousedown event. This is captured only to prevent browser from drawing an outline around an
+ * area when it's clicked.
+ *
+ * @param {EventData} e jQuery event data
+ */
+
+ function mousedown(e) {
+ if (!$.mapster.hasCanvas) {
+ this.blur();
+ }
+ e.preventDefault();
+ }
+
+ /**
+ * Mouseover event. Handle highlight rendering and client callback on mouseover
+ *
+ * @param {MapData} me The MapData context
+ * @param {EventData} e jQuery event data
+ * @return {[type]} [description]
+ */
+
+ function mouseover(me,e) {
+ var arData = me.getAllDataForArea(this),
+ ar=arData.length ? arData[0] : null;
+
+ // mouseover events are ignored entirely while resizing, though we do care about mouseout events
+ // and must queue the action to keep things clean.
+
+ if (!ar || ar.isNotRendered() || ar.owner.currentAction) {
+ return;
+ }
+
+ if (me.currentAreaId === ar.areaId) {
+ return;
+ }
+ if (me.highlightId !== ar.areaId) {
+ me.clearEffects();
+
+ ar.highlight();
+
+ if (me.options.showToolTip) {
+ $.each(arData,function(i,e) {
+ if (e.effectiveOptions().toolTip) {
+ e.showToolTip();
+ }
+ });
+ }
+ }
+
+ me.currentAreaId = ar.areaId;
+
+ if ($.isFunction(me.options.onMouseover)) {
+ me.options.onMouseover.call(this,
+ {
+ e: e,
+ options:ar.effectiveOptions(),
+ key: ar.key,
+ selected: ar.isSelected()
+ });
+ }
+ }
+
+ /**
+ * Mouseout event.
+ *
+ * @param {MapData} me The MapData context
+ * @param {EventData} e jQuery event data
+ * @return {[type]} [description]
+ */
+
+ function mouseout(me,e) {
+ var newArea,
+ ar = me.getDataForArea(this),
+ opts = me.options;
+
+
+ if (me.currentAreaId<0 || !ar) {
+ return;
+ }
+
+ newArea=me.getDataForArea(e.relatedTarget);
+
+ if (newArea === ar) {
+ return;
+ }
+
+ me.currentAreaId = -1;
+ ar.area=null;
+
+ queueMouseEvent(me,opts.mouseoutDelay,ar)
+ .then(me.clearEffects);
+
+ if ($.isFunction(opts.onMouseout)) {
+ opts.onMouseout.call(this,
+ {
+ e: e,
+ options: opts,
+ key: ar.key,
+ selected: ar.isSelected()
+ });
+ }
+
+ }
+
+ /**
+ * Clear any active tooltip or highlight
+ *
+ * @param {MapData} me The MapData context
+ * @param {EventData} e jQuery event data
+ * @return {[type]} [description]
+ */
+
+ function clearEffects(me) {
+ var opts = me.options;
+
+ me.ensureNoHighlight();
+
+ if (opts.toolTipClose
+ && $.inArray('area-mouseout', opts.toolTipClose) >= 0
+ && me.activeToolTip)
+ {
+ me.clearToolTip();
+ }
+ }
+
+ /**
+ * Mouse click event handler
+ *
+ * @param {MapData} me The MapData context
+ * @param {EventData} e jQuery event data
+ * @return {[type]} [description]
+ */
+
+ function click(me,e) {
+ var selected, list, list_target, newSelectionState, canChangeState, cbResult,
+ that = this,
+ ar = me.getDataForArea(this),
+ opts = me.options;
+
+ function clickArea(ar) {
+ var areaOpts,target;
+ canChangeState = (ar.isSelectable() &&
+ (ar.isDeselectable() || !ar.isSelected()));
+
+ if (canChangeState) {
+ newSelectionState = !ar.isSelected();
+ } else {
+ newSelectionState = ar.isSelected();
+ }
+
+ list_target = m.getBoundList(opts, ar.key);
+
+ if ($.isFunction(opts.onClick))
+ {
+ cbResult= opts.onClick.call(that,
+ {
+ e: e,
+ listTarget: list_target,
+ key: ar.key,
+ selected: newSelectionState
+ });
+
+ if (u.isBool(cbResult)) {
+ if (!cbResult) {
+ return false;
+ }
+ target = $(ar.area).attr('href');
+ if (target!=='#') {
+ window.location.href=target;
+ return false;
+ }
+ }
+ }
+
+ if (canChangeState) {
+ selected = ar.toggle();
+ }
+
+ if (opts.boundList && opts.boundList.length > 0) {
+ m.setBoundListProperties(opts, list_target, ar.isSelected());
+ }
+
+ areaOpts = ar.effectiveOptions();
+ if (areaOpts.includeKeys) {
+ list = u.split(areaOpts.includeKeys);
+ $.each(list, function (i, e) {
+ var ar = me.getDataForKey(e.toString());
+ if (!ar.options.isMask) {
+ clickArea(ar);
+ }
+ });
+ }
+ }
+
+ mousedown.call(this,e);
+
+ if (opts.clickNavigate && ar.href) {
+ window.location.href=ar.href;
+ return;
+ }
+
+ if (ar && !ar.owner.currentAction) {
+ opts = me.options;
+ clickArea(ar);
+ }
+ }
+
+ /**
+ * Prototype for a MapData object, representing an ImageMapster bound object
+ * @param {Element} image an IMG element
+ * @param {object} options ImageMapster binding options
+ */
+ m.MapData = function (image, options)
+ {
+ var me = this;
+
+ // (Image) main map image
+
+ me.image = image;
+
+ me.images = new m.MapImages(me);
+ me.graphics = new m.Graphics(me);
+
+ // save the initial style of the image for unbinding. This is problematic, chrome
+ // duplicates styles when assigning, and cssText is apparently not universally supported.
+ // Need to do something more robust to make unbinding work universally.
+
+ me.imgCssText = image.style.cssText || null;
+
+ initializeDefaults(me);
+
+ me.configureOptions(options);
+
+ // create context-bound event handlers from our private functions
+
+ me.mouseover = function(e) { mouseover.call(this,me,e); };
+ me.mouseout = function(e) { mouseout.call(this,me,e); };
+ me.click = function(e) { click.call(this,me,e); };
+ me.clearEffects = function(e) { clearEffects.call(this,me,e); };
+ };
+
+ m.MapData.prototype = {
+ constructor: m.MapData,
+
+ /**
+ * Set target.options from defaults + options
+ * @param {[type]} target The target
+ * @param {[type]} options The options to merge
+ */
+
+ configureOptions: function(options) {
+ this.options= u.updateProps({}, m.defaults, options);
+ },
+
+ /**
+ * Ensure all images are loaded
+ * @return {Promise} A promise that resolves when the images have finished loading (or fail)
+ */
+
+ bindImages: function() {
+ var me=this,
+ mi = me.images;
+
+ // reset the images if this is a rebind
+
+ if (mi.length>2) {
+ mi.splice(2);
+ } else if (mi.length===0) {
+
+ // add the actual main image
+ mi.add(me.image);
+ // will create a duplicate of the main image, we need this to get raw size info
+ mi.add(me.image.src);
+ }
+
+ configureAltImages(me);
+
+ return me.images.bind();
+ },
+
+ /**
+ * Test whether an async action is currently in progress
+ * @return {Boolean} true or false indicating state
+ */
+
+ isActive: function() {
+ return !this.complete || this.currentAction;
+ },
+
+ /**
+ * Return an object indicating the various states. This isn't really used by
+ * production code.
+ *
+ * @return {object} An object with properties for various states
+ */
+
+ state: function () {
+ return {
+ complete: this.complete,
+ resizing: this.currentAction==='resizing',
+ zoomed: this.zoomed,
+ zoomedArea: this.zoomedArea,
+ scaleInfo: this.scaleInfo
+ };
+ },
+
+ /**
+ * Get a unique ID for the wrapper of this imagemapster
+ * @return {string} A string that is unique to this image
+ */
+
+ wrapId: function () {
+ return 'mapster_wrap_' + this.index;
+ },
+ _idFromKey: function (key) {
+ return typeof key === "string" && this._xref.hasOwnProperty(key) ?
+ this._xref[key] : -1;
+ },
+
+ /**
+ * Return a comma-separated string of all selected keys
+ * @return {string} CSV of all keys that are currently selected
+ */
+
+ getSelected: function () {
+ var result = '';
+ $.each(this.data, function (i,e) {
+ if (e.isSelected()) {
+ result += (result ? ',' : '') + this.key;
+ }
+ });
+ return result;
+ },
+
+ /**
+ * Get an array of MapAreas associated with a specific AREA based on the keys for that area
+ * @param {Element} area An HTML AREA
+ * @param {number} atMost A number limiting the number of areas to be returned (typically 1 or 0 for no limit)
+ * @return {MapArea[]} Array of MapArea objects
+ */
+
+ getAllDataForArea:function (area,atMost) {
+ var i,ar, result,
+ me=this,
+ key = $(area).filter('area').attr(me.options.mapKey);
+
+ if (key) {
+ result=[];
+ key = u.split(key);
+
+ for (i=0;i<(atMost || key.length);i++) {
+ ar = me.data[me._idFromKey(key[i])];
+ ar.area=area.length ? area[0]:area;
+ // set the actual area moused over/selected
+ // TODO: this is a brittle model for capturing which specific area - if this method was not used,
+ // ar.area could have old data. fix this.
+ result.push(ar);
+ }
+ }
+
+ return result;
+ },
+ getDataForArea: function(area) {
+ var ar=this.getAllDataForArea(area,1);
+ return ar ? ar[0] || null : null;
+ },
+ getDataForKey: function (key) {
+ return this.data[this._idFromKey(key)];
+ },
+
+ /**
+ * Get the primary keys associated with an area group.
+ * If this is a primary key, it will be returned.
+ *
+ * @param {string key An area key
+ * @return {string} A CSV of area keys
+ */
+
+ getKeysForGroup: function(key) {
+ var ar=this.getDataForKey(key);
+
+ return !ar ? '':
+ ar.isPrimary ?
+ ar.key :
+ this.getPrimaryKeysForMapAreas(ar.areas()).join(',');
+ },
+
+ /**
+ * given an array of MapArea object, return an array of its unique primary keys
+ * @param {MapArea[]} areas The areas to analyze
+ * @return {string[]} An array of unique primary keys
+ */
+
+ getPrimaryKeysForMapAreas: function(areas)
+ {
+ var keys=[];
+ $.each(areas,function(i,e) {
+ if ($.inArray(e.keys[0],keys)<0) {
+ keys.push(e.keys[0]);
+ }
+ });
+ return keys;
+ },
+ getData: function (obj) {
+ if (typeof obj === 'string') {
+ return this.getDataForKey(obj);
+ } else if (obj && obj.mapster || u.isElement(obj)) {
+ return this.getDataForArea(obj);
+ } else {
+ return null;
+ }
+ },
+ // remove highlight if present, raise event
+ ensureNoHighlight: function () {
+ var ar;
+ if (this.highlightId >= 0) {
+ this.graphics.clearHighlight();
+ ar = this.data[this.highlightId];
+ ar.changeState('highlight', false);
+ this.setHighlightId(-1);
+ }
+ },
+ setHighlightId: function(id) {
+ this.highlightId = id;
+ },
+
+ /**
+ * Clear all active selections on this map
+ */
+
+ clearSelections: function () {
+ $.each(this.data, function (i,e) {
+ if (e.selected) {
+ e.deselect(true);
+ }
+ });
+ this.removeSelectionFinish();
+
+ },
+
+ /**
+ * Set area options from an array of option data.
+ *
+ * @param {object[]} areas An array of objects containing area-specific options
+ */
+
+ setAreaOptions: function (areas) {
+ var i, area_options, ar;
+ areas = areas || [];
+
+ // refer by: map_data.options[map_data.data[x].area_option_id]
+
+ for (i = areas.length - 1; i >= 0; i--) {
+ area_options = areas[i];
+ if (area_options) {
+ ar = this.getDataForKey(area_options.key);
+ if (ar) {
+ u.updateProps(ar.options, area_options);
+
+ // TODO: will not deselect areas that were previously selected, so this only works
+ // for an initial bind.
+
+ if (u.isBool(area_options.selected)) {
+ ar.selected = area_options.selected;
+ }
+ }
+ }
+ }
+ },
+ // keys: a comma-separated list
+ drawSelections: function (keys) {
+ var i, key_arr = u.asArray(keys);
+
+ for (i = key_arr.length - 1; i >= 0; i--) {
+ this.data[key_arr[i]].drawSelection();
+ }
+ },
+ redrawSelections: function () {
+ $.each(this.data, function (i, e) {
+ if (e.isSelectedOrStatic()) {
+ e.drawSelection();
+ }
+ });
+
+ },
+ ///called when images are done loading
+ initialize: function () {
+ var imgCopy, base_canvas, overlay_canvas, wrap, parentId, css, i,size,
+ img,sort_func, sorted_list, scale,
+ me = this,
+ opts = me.options;
+
+ if (me.complete) {
+ return;
+ }
+
+ img = $(me.image);
+
+ parentId = img.parent().attr('id');
+
+ // create a div wrapper only if there's not already a wrapper, otherwise, own it
+
+ if (parentId && parentId.length >= 12 && parentId.substring(0, 12) === "mapster_wrap") {
+ wrap = img.parent();
+ wrap.attr('id', me.wrapId());
+ } else {
+ wrap = $('');
+
+ if (opts.wrapClass) {
+ if (opts.wrapClass === true) {
+ wrap.addClass(img[0].className);
+ }
+ else {
+ wrap.addClass(opts.wrapClass);
+ }
+ }
+ }
+ me.wrapper = wrap;
+
+ // me.images[1] is the copy of the original image. It should be loaded & at its native size now so we can obtain the true
+ // width & height. This is needed to scale the imagemap if not being shown at its native size. It is also needed purely
+ // to finish binding in case the original image was not visible. It can be impossible in some browsers to obtain the
+ // native size of a hidden image.
+
+ me.scaleInfo = scale = u.scaleMap(me.images[0],me.images[1], opts.scaleMap);
+
+ me.base_canvas = base_canvas = me.graphics.createVisibleCanvas(me);
+ me.overlay_canvas = overlay_canvas = me.graphics.createVisibleCanvas(me);
+
+ // Now we got what we needed from the copy -clone from the original image again to make sure any other attributes are copied
+ imgCopy = $(me.images[1])
+ .addClass('mapster_el '+ me.images[0].className)
+ .attr({id:null, usemap: null});
+
+ size=u.size(me.images[0]);
+
+ if (size.complete) {
+ imgCopy.css({
+ width: size.width,
+ height: size.height
+ });
+ }
+
+ me.buildDataset();
+
+ // now that we have processed all the areas, set css for wrapper, scale map if needed
+
+ css = {
+ display: 'block',
+ position: 'relative',
+ padding: 0,
+ width: scale.width,
+ height: scale.height
+ };
+
+ if (opts.wrapCss) {
+ $.extend(css, opts.wrapCss);
+ }
+ // if we were rebinding with an existing wrapper, the image will aready be in it
+ if (img.parent()[0] !== me.wrapper[0]) {
+
+ img.before(me.wrapper);
+ }
+
+ wrap.css(css);
+
+ // move all generated images into the wrapper for easy removal later
+
+ $(me.images.slice(2)).hide();
+ for (i = 1; i < me.images.length; i++) {
+ wrap.append(me.images[i]);
+ }
+
+ //me.images[1].style.cssText = me.image.style.cssText;
+
+ wrap.append(base_canvas)
+ .append(overlay_canvas)
+ .append(img.css(m.canvas_style));
+
+ // images[0] is the original image with map, images[1] is the copy/background that is visible
+
+ u.setOpacity(me.images[0], 0);
+ $(me.images[1]).show();
+
+ u.setOpacity(me.images[1],1);
+
+ if (opts.isSelectable && opts.onGetList) {
+ sorted_list = me.data.slice(0);
+ if (opts.sortList) {
+ if (opts.sortList === "desc") {
+ sort_func = function (a, b) {
+ return a === b ? 0 : (a > b ? -1 : 1);
+ };
+ }
+ else {
+ sort_func = function (a, b) {
+ return a === b ? 0 : (a < b ? -1 : 1);
+ };
+ }
+
+ sorted_list.sort(function (a, b) {
+ a = a.value;
+ b = b.value;
+ return sort_func(a, b);
+ });
+ }
+
+ me.options.boundList = opts.onGetList.call(me.image, sorted_list);
+ }
+
+ me.complete=true;
+ me.processCommandQueue();
+
+ if (opts.onConfigured && typeof opts.onConfigured === 'function') {
+ opts.onConfigured.call(img, true);
+ }
+ },
+
+ // when rebind is true, the MapArea data will not be rebuilt.
+ buildDataset: function(rebind) {
+ var sel,areas,j,area_id,$area,area,curKey,mapArea,key,keys,mapAreaId,group_value,dataItem,href,
+ me=this,
+ opts=me.options,
+ default_group;
+
+ function addAreaData(key, value) {
+ var dataItem = new m.AreaData(me, key, value);
+ dataItem.areaId = me._xref[key] = me.data.push(dataItem) - 1;
+ return dataItem.areaId;
+ }
+
+ me._xref = {};
+ me.data = [];
+ if (!rebind) {
+ me.mapAreas=[];
+ }
+
+ default_group = !opts.mapKey;
+ if (default_group) {
+ opts.mapKey = 'data-mapster-key';
+ }
+ sel = ($.browser.msie && $.browser.version <= 7) ? 'area' :
+ (default_group ? 'area[coords]' : 'area[' + opts.mapKey + ']');
+ areas = $(me.map).find(sel).unbind('.mapster');
+
+ for (mapAreaId = 0;mapAreaId= 0; j--) {
+ key = keys[j];
+
+ if (opts.mapValue) {
+ group_value = $area.attr(opts.mapValue);
+ }
+ if (default_group) {
+ // set an attribute so we can refer to the area by index from the DOM object if no key
+ area_id = addAreaData(me.data.length, group_value);
+ dataItem = me.data[area_id];
+ dataItem.key = key = area_id.toString();
+ }
+ else {
+ area_id = me._xref[key];
+ if (area_id >= 0) {
+ dataItem = me.data[area_id];
+ if (group_value && !me.data[area_id].value) {
+ dataItem.value = group_value;
+ }
+ }
+ else {
+ area_id = addAreaData(key, group_value);
+ dataItem = me.data[area_id];
+ dataItem.isPrimary=j===0;
+ }
+ }
+ mapArea.areaDataXref.push(area_id);
+ dataItem.areasXref.push(mapAreaId);
+ }
+
+ href=$area.attr('href');
+ if (href && href!=='#' && !dataItem.href)
+ {
+ dataItem.href=href;
+ }
+
+ if (!mapArea.nohref) {
+ $area.bind('click.mapster', me.click);
+
+ if (!m.isTouch) {
+ $area.bind('mouseover.mapster', me.mouseover)
+ .bind('mouseout.mapster', me.mouseout)
+ .bind('mousedown.mapster', me.mousedown);
+
+ }
+
+ }
+
+ // store an ID with each area.
+ $area.data("mapster", mapAreaId+1);
+ }
+
+ // TODO listenToList
+ // if (opts.listenToList && opts.nitG) {
+ // opts.nitG.bind('click.mapster', event_hooks[map_data.hooks_index].listclick_hook);
+ // }
+
+ // populate areas from config options
+ me.setAreaOptions(opts.areas);
+ me.redrawSelections();
+
+ },
+ processCommandQueue: function() {
+
+ var cur,me=this;
+ while (!me.currentAction && me.commands.length) {
+ cur = me.commands[0];
+ me.commands.splice(0,1);
+ m.impl[cur.command].apply(cur.that, cur.args);
+ }
+ },
+ clearEvents: function () {
+ $(this.map).find('area')
+ .unbind('.mapster');
+ $(this.images)
+ .unbind('.mapster');
+ },
+ _clearCanvases: function (preserveState) {
+ // remove the canvas elements created
+ if (!preserveState) {
+ $(this.base_canvas).remove();
+ }
+ $(this.overlay_canvas).remove();
+ },
+ clearMapData: function (preserveState) {
+ var me = this;
+ this._clearCanvases(preserveState);
+
+ // release refs to DOM elements
+ $.each(this.data, function (i, e) {
+ e.reset();
+ });
+ this.data = null;
+ if (!preserveState) {
+ // get rid of everything except the original image
+ this.image.style.cssText = this.imgCssText;
+ $(this.wrapper).before(this.image).remove();
+ }
+
+ me.images.clear();
+
+ this.image = null;
+ u.ifFunction(this.clearTooltip, this);
+ },
+
+ // Compelete cleanup process for deslecting items. Called after a batch operation, or by AreaData for single
+ // operations not flagged as "partial"
+
+ removeSelectionFinish: function () {
+ var g = this.graphics;
+
+ g.refreshSelections();
+ // do not call ensure_no_highlight- we don't really want to unhilight it, just remove the effect
+ g.clearHighlight();
+ }
+ };
+} (jQuery));
+/* areadata.js
+ AreaData and MapArea protoypes
+*/
+
+(function ($) {
+ var m = $.mapster, u = m.utils;
+
+ /**
+ * Select this area
+ *
+ * @param {AreaData} me AreaData context
+ * @param {object} options Options for rendering the selection
+ */
+ function select(options) {
+ // need to add the new one first so that the double-opacity effect leaves the current one highlighted for singleSelect
+
+ var me=this, o = me.owner;
+ if (o.options.singleSelect) {
+ o.clearSelections();
+ }
+
+ // because areas can overlap - we can't depend on the selection state to tell us anything about the inner areas.
+ // don't check if it's already selected
+ if (!me.isSelected()) {
+ if (options) {
+
+ // cache the current options, and map the altImageId if an altimage
+ // was passed
+
+ me.optsCache = $.extend(me.effectiveRenderOptions('select'),
+ options,
+ {
+ altImageId: o.images.add(options.altImage)
+ });
+ }
+
+ me.drawSelection();
+
+ me.selected = true;
+ me.changeState('select', true);
+ }
+
+ if (o.options.singleSelect) {
+ o.graphics.refreshSelections();
+ }
+ }
+
+ /**
+ * Deselect this area, optionally deferring finalization so additional areas can be deselected
+ * in a single operation
+ *
+ * @param {boolean} partial when true, the caller must invoke "finishRemoveSelection" to render
+ */
+
+ function deselect(partial) {
+ var me=this;
+ me.selected = false;
+ me.changeState('select', false);
+
+ // release information about last area options when deselecting.
+
+ me.optsCache=null;
+ me.owner.graphics.removeSelections(me.areaId);
+
+ // Complete selection removal process. This is separated because it's very inefficient to perform the whole
+ // process for multiple removals, as the canvas must be totally redrawn at the end of the process.ar.remove
+
+ if (!partial) {
+ me.owner.removeSelectionFinish();
+ }
+ }
+
+ /**
+ * Toggle the selection state of this area
+ * @param {object} options Rendering options, if toggling on
+ * @return {bool} The new selection state
+ */
+ function toggle(options) {
+ var me=this;
+ if (!me.isSelected()) {
+ me.select(options);
+ }
+ else {
+ me.deselect();
+ }
+ return me.isSelected();
+ }
+
+ /**
+ * An AreaData object; represents a conceptual area that can be composed of
+ * one or more MapArea objects
+ *
+ * @param {MapData} owner The MapData object to which this belongs
+ * @param {string} key The key for this area
+ * @param {string} value The mapValue string for this area
+ */
+
+ m.AreaData = function (owner, key, value) {
+ $.extend(this,{
+ owner: owner,
+ key: key || '',
+ // means this represents the first key in a list of keys (it's the area group that gets highlighted on mouseover)
+ isPrimary: true,
+ areaId: -1,
+ href: '',
+ value: value || '',
+ options:{},
+ // "null" means unchanged. Use "isSelected" method to just test true/false
+ selected: null,
+ // xref to MapArea objects
+ areasXref: [],
+ // (temporary storage) - the actual area moused over
+ area: null,
+ // the last options used to render this. Cache so when re-drawing after a remove, changes in options won't
+ // break already selected things.
+ optsCache: null
+ });
+ };
+
+ /**
+ * The public API for AreaData object
+ */
+
+ m.AreaData.prototype = {
+ constuctor: m.AreaData,
+ select: select,
+ deselect: deselect,
+ toggle: toggle,
+ areas: function() {
+ var i,result=[];
+ for (i=0;i= 0; j -= 2) {
+ curX = coords[j];
+ curY = coords[j + 1];
+
+ if (curX < minX) {
+ minX = curX;
+ bestMaxY = curY;
+ }
+ if (curX > maxX) {
+ maxX = curX;
+ bestMinY = curY;
+ }
+ if (curY < minY) {
+ minY = curY;
+ bestMaxX = curX;
+ }
+ if (curY > maxY) {
+ maxY = curY;
+ bestMinX = curX;
+ }
+
+ }
+
+ // try to figure out the best place for the tooltip
+
+ if (width && height) {
+ found=false;
+ $.each([[bestMaxX - width, minY - height], [bestMinX, minY - height],
+ [minX - width, bestMaxY - height], [minX - width, bestMinY],
+ [maxX,bestMaxY - height], [ maxX,bestMinY],
+ [bestMaxX - width, maxY], [bestMinX, maxY]
+ ],function (i, e) {
+ if (!found && (e[0] > rootx && e[1] > rooty)) {
+ nest = e;
+ found=true;
+ return false;
+ }
+ });
+
+ // default to lower-right corner if nothing fit inside the boundaries of the image
+
+ if (!found) {
+ nest=[maxX,maxY];
+ }
+ }
+ return nest;
+ };
+} (jQuery));
+/* scale.js: resize and zoom functionality
+ requires areacorners.js, when.js
+*/
+
+
+(function ($) {
+ var m = $.mapster, u = m.utils, p = m.MapArea.prototype;
+
+ m.utils.getScaleInfo = function (eff, actual) {
+ var pct;
+ if (!actual) {
+ pct = 1;
+ actual=eff;
+ } else {
+ pct = eff.width / actual.width || eff.height / actual.height;
+ // make sure a float error doesn't muck us up
+ if (pct > 0.98 && pct < 1.02) { pct = 1; }
+ }
+ return {
+ scale: (pct !== 1),
+ scalePct: pct,
+ realWidth: actual.width,
+ realHeight: actual.height,
+ width: eff.width,
+ height: eff.height,
+ ratio: eff.width / eff.height
+ };
+ };
+ // Scale a set of AREAs, return old data as an array of objects
+ m.utils.scaleMap = function (image, imageRaw, scale) {
+
+ // stunningly, jQuery width can return zero even as width does not, seems to happen only
+ // with adBlock or maybe other plugins. These must interfere with onload events somehow.
+
+
+ var vis=u.size(image),
+ raw=u.size(imageRaw,true);
+
+ if (!raw.complete()) {
+ throw("Another script, such as an extension, appears to be interfering with image loading. Please let us know about this.");
+ }
+ if (!vis.complete()) {
+ vis=raw;
+ }
+ return this.getScaleInfo(vis, scale ? raw : null);
+ };
+
+ /**
+ * Resize the image map. Only one of newWidth and newHeight should be passed to preserve scale
+ *
+ * @param {int} width The new width OR an object containing named parameters matching this function sig
+ * @param {int} height The new height
+ * @param {int} effectDuration Time in ms for the resize animation, or zero for no animation
+ * @param {function} callback A function to invoke when the operation finishes
+ * @return {promise} NOT YET IMPLEMENTED
+ */
+
+ m.MapData.prototype.resize = function (width, height, duration, callback) {
+ var p,promises,newsize,els, highlightId, ratio,
+ me = this;
+
+ // allow omitting duration
+ callback = callback || duration;
+
+ function sizeCanvas(canvas, w, h) {
+ if ($.mapster.hasCanvas) {
+ canvas.width = w;
+ canvas.height = h;
+ } else {
+ $(canvas).width(w);
+ $(canvas).height(h);
+ }
+ }
+
+ // Finalize resize action, do callback, pass control to command queue
+
+ function cleanupAndNotify() {
+
+ me.currentAction = '';
+
+ if ($.isFunction(callback)) {
+ callback();
+ }
+
+ me.processCommandQueue();
+ }
+
+ // handle cleanup after the inner elements are resized
+
+ function finishResize() {
+ sizeCanvas(me.overlay_canvas, width, height);
+
+ // restore highlight state if it was highlighted before
+ if (highlightId >= 0) {
+ var areaData = me.data[highlightId];
+ areaData.tempOptions = { fade: false };
+ me.getDataForKey(areaData.key).highlight();
+ areaData.tempOptions = null;
+ }
+ sizeCanvas(me.base_canvas, width, height);
+ me.redrawSelections();
+ cleanupAndNotify();
+ }
+
+ function resizeMapData() {
+ $(me.image).css(newsize);
+ // start calculation at the same time as effect
+ me.scaleInfo = u.getScaleInfo({
+ width: width,
+ height: height
+ },
+ {
+ width: me.scaleInfo.realWidth,
+ height: me.scaleInfo.realHeight
+ });
+ $.each(me.data, function (i, e) {
+ $.each(e.areas(), function (i, e) {
+ e.resize();
+ });
+ });
+ }
+
+ if (me.scaleInfo.width === width && me.scaleInfo.height === height) {
+ return;
+ }
+
+ highlightId = me.highlightId;
+
+
+ if (!width) {
+ ratio = height / me.scaleInfo.realHeight;
+ width = Math.round(me.scaleInfo.realWidth * ratio);
+ }
+ if (!height) {
+ ratio = width / me.scaleInfo.realWidth;
+ height = Math.round(me.scaleInfo.realHeight * ratio);
+ }
+
+ newsize = { 'width': String(width) + 'px', 'height': String(height) + 'px' };
+ if (!$.mapster.hasCanvas) {
+ $(me.base_canvas).children().remove();
+ }
+
+ // resize all the elements that are part of the map except the image itself (which is not visible)
+ // but including the div wrapper
+ els = $(me.wrapper).find('.mapster_el').add(me.wrapper);
+
+ if (duration) {
+ promises = [];
+ me.currentAction = 'resizing';
+ els.each(function (i, e) {
+ p = u.defer();
+ promises.push(p);
+
+ $(e).animate(newsize, {
+ duration: duration,
+ complete: p.resolve,
+ easing: "linear"
+ });
+ });
+
+ p = u.defer();
+ promises.push(p);
+
+ // though resizeMapData is not async, it needs to be finished just the same as the animations,
+ // so add it to the "to do" list.
+
+ u.when.all(promises).then(finishResize);
+ resizeMapData();
+ p.resolve();
+ } else {
+ els.css(newsize);
+ resizeMapData();
+ finishResize();
+
+ }
+ };
+
+
+ m.MapArea = u.subclass(m.MapArea, function () {
+ //change the area tag data if needed
+ this.base.init();
+ if (this.owner.scaleInfo.scale) {
+ this.resize();
+ }
+ });
+
+ p.coords = function (percent, coordOffset) {
+ var j, newCoords = [],
+ pct = percent || this.owner.scaleInfo.scalePct,
+ offset = coordOffset || 0;
+
+ if (pct === 1 && coordOffset === 0) {
+ return this.originalCoords;
+ }
+
+ for (j = 0; j < this.length; j++) {
+ //amount = j % 2 === 0 ? xPct : yPct;
+ newCoords.push(Math.round(this.originalCoords[j] * pct) + offset);
+ }
+ return newCoords;
+ };
+ p.resize = function () {
+ this.area.coords = this.coords().join(',');
+ };
+
+ p.reset = function () {
+ this.area.coords = this.coords(1).join(',');
+ };
+
+ m.impl.resize = function (width, height, duration, callback) {
+ if (!width && !height) {
+ return false;
+ }
+ var x= (new m.Method(this,
+ function () {
+ this.resize(width, height, duration, callback);
+ },
+ null,
+ {
+ name: 'resize',
+ args: arguments
+ }
+ )).go();
+ return x;
+ };
+
+/*
+ m.impl.zoom = function (key, opts) {
+ var options = opts || {};
+
+ function zoom(areaData) {
+ // this will be MapData object returned by Method
+
+ var scroll, corners, height, width, ratio,
+ diffX, diffY, ratioX, ratioY, offsetX, offsetY, newWidth, newHeight, scrollLeft, scrollTop,
+ padding = options.padding || 0,
+ scrollBarSize = areaData ? 20 : 0,
+ me = this,
+ zoomOut = false;
+
+ if (areaData) {
+ // save original state on first zoom operation
+ if (!me.zoomed) {
+ me.zoomed = true;
+ me.preZoomWidth = me.scaleInfo.width;
+ me.preZoomHeight = me.scaleInfo.height;
+ me.zoomedArea = areaData;
+ if (options.scroll) {
+ me.wrapper.css({ overflow: 'auto' });
+ }
+ }
+ corners = $.mapster.utils.areaCorners(areaData.coords(1, 0));
+ width = me.wrapper.innerWidth() - scrollBarSize - padding * 2;
+ height = me.wrapper.innerHeight() - scrollBarSize - padding * 2;
+ diffX = corners.maxX - corners.minX;
+ diffY = corners.maxY - corners.minY;
+ ratioX = width / diffX;
+ ratioY = height / diffY;
+ ratio = Math.min(ratioX, ratioY);
+ offsetX = (width - diffX * ratio) / 2;
+ offsetY = (height - diffY * ratio) / 2;
+
+ newWidth = me.scaleInfo.realWidth * ratio;
+ newHeight = me.scaleInfo.realHeight * ratio;
+ scrollLeft = (corners.minX) * ratio - padding - offsetX;
+ scrollTop = (corners.minY) * ratio - padding - offsetY;
+ } else {
+ if (!me.zoomed) {
+ return;
+ }
+ zoomOut = true;
+ newWidth = me.preZoomWidth;
+ newHeight = me.preZoomHeight;
+ scrollLeft = null;
+ scrollTop = null;
+ }
+
+ this.resize({
+ width: newWidth,
+ height: newHeight,
+ duration: options.duration,
+ scroll: scroll,
+ scrollLeft: scrollLeft,
+ scrollTop: scrollTop,
+ // closure so we can be sure values are correct
+ callback: (function () {
+ var isZoomOut = zoomOut,
+ scroll = options.scroll,
+ areaD = areaData;
+ return function () {
+ if (isZoomOut) {
+ me.preZoomWidth = null;
+ me.preZoomHeight = null;
+ me.zoomed = false;
+ me.zoomedArea = false;
+ if (scroll) {
+ me.wrapper.css({ overflow: 'inherit' });
+ }
+ } else {
+ // just to be sure it wasn't canceled & restarted
+ me.zoomedArea = areaD;
+ }
+ };
+ } ())
+ });
+ }
+ return (new m.Method(this,
+ function (opts) {
+ zoom.call(this);
+ },
+ function () {
+ zoom.call(this.owner, this);
+ },
+ {
+ name: 'zoom',
+ args: arguments,
+ first: true,
+ key: key
+ }
+ )).go();
+
+
+ };
+ */
+} (jQuery));
+/* tooltip.js - tooltip functionality
+ requires areacorners.js
+*/
+
+(function ($) {
+
+ var m = $.mapster, u = m.utils;
+
+ $.extend(m.defaults, {
+ toolTipContainer: '',
+ showToolTip: false,
+ toolTipFade: true,
+ toolTipClose: ['area-mouseout','image-mouseout'],
+ onShowToolTip: null,
+ onHideToolTip: null
+ });
+
+ $.extend(m.area_defaults, {
+ toolTip: null,
+ toolTipClose: null
+ });
+
+
+ /**
+ * Show a tooltip positioned near this area.
+ *
+ * @param {string|jquery} html A string of html or a jQuery object containing the tooltip content.
+ * @param {string|jquery} [template] The html template in which to wrap the content
+ * @param {string|object} [css] CSS to apply to the outermost element of the tooltip
+ * @return {jquery} The tooltip that was created
+ */
+
+ function createToolTip(html, template, css) {
+ var tooltip;
+
+ // wrap the template in a jQuery object, or clone the template if it's already one.
+ // This assumes that anything other than a string is a jQuery object; if it's not jQuery will
+ // probably throw an error.
+
+ if (template) {
+ tooltip = typeof template === 'string' ?
+ $(template) :
+ $(template).clone();
+
+ tooltip.append(html);
+ } else {
+ tooltip=$(html);
+ }
+
+ // always set display to block, or the positioning css won't work if the end user happened to
+ // use a non-block type element.
+
+ tooltip.css($.extend((css || {}),{
+ display:"block",
+ position:"absolute"
+ })).hide();
+
+ $('body').append(tooltip);
+
+ // we must actually add the tooltip to the DOM and "show" it in order to figure out how much space it
+ // consumes, and then reposition it with that knowledge.
+ // We also cache the actual opacity setting to restore finally.
+
+ tooltip.attr("data-opacity",tooltip.css("opacity"))
+ .css("opacity",0);
+
+ // doesn't really show it because opacity=0
+
+ return tooltip.show();
+ }
+
+
+ /**
+ * Show a tooltip positioned near this area.
+ *
+ * @param {jquery} tooltip The tooltip
+ * @param {object} [options] options for displaying the tooltip.
+ * @config {int} [left] The 0-based absolute x position for the tooltip
+ * @config {int} [top] The 0-based absolute y position for the tooltip
+ * @config {string|object} [css] CSS to apply to the outermost element of the tooltip
+ * @config {bool} [fadeDuration] When non-zero, the duration in milliseconds of a fade-in effect for the tooltip.
+ */
+
+ function showToolTipImpl(tooltip,options)
+ {
+ var tooltipCss = {
+ "left": options.left + "px",
+ "top": options.top + "px"
+ },
+ actalOpacity=tooltip.attr("data-opacity") || 0,
+ zindex = tooltip.css("z-index");
+
+ if (parseInt(zindex,10)===0
+ || zindex === "auto") {
+ tooltipCss["z-index"] = 9999;
+ }
+
+ tooltip.css(tooltipCss)
+ .addClass('mapster_tooltip');
+
+
+ if (options.fadeDuration && options.fadeDuration>0) {
+ u.fader(tooltip[0], 0, actalOpacity, options.fadeDuration);
+ } else {
+ u.setOpacity(tooltip[0], actalOpacity);
+ }
+ }
+
+ /**
+ * Hide and remove active tooltips
+ *
+ * @param {MapData} this The mapdata object to which the tooltips belong
+ */
+
+ m.MapData.prototype.clearToolTip = function() {
+ if (this.activeToolTip) {
+ this.activeToolTip.stop().remove();
+ this.activeToolTip = null;
+ this.activeToolTipID = null;
+ u.ifFunction(this.options.onHideToolTip, this);
+ }
+ };
+
+ /**
+ * Configure the binding between a named tooltip closing option, and a mouse event.
+ *
+ * If a callback is passed, it will be called when the activating event occurs, and the tooltip will
+ * only closed if it returns true.
+ *
+ * @param {MapData} [this] The MapData object to which this tooltip belongs.
+ * @param {String} option The name of the tooltip closing option
+ * @param {String} event UI event to bind to this option
+ * @param {Element} target The DOM element that is the target of the event
+ * @param {Function} [beforeClose] Callback when the tooltip is closed
+ * @param {Function} [onClose] Callback when the tooltip is closed
+ */
+ function bindToolTipClose(options, bindOption, event, target, beforeClose, onClose) {
+ var event_name = event + '.mapster-tooltip';
+
+ if ($.inArray(bindOption, options) >= 0) {
+ target.unbind(event_name)
+ .bind(event_name, function (e) {
+ if (!beforeClose || beforeClose.call(this,e)) {
+ target.unbind('.mapster-tooltip');
+ if (onClose) {
+ onClose.call(this);
+ }
+ }
+ });
+
+ return {
+ object: target,
+ event: event_name
+ };
+ }
+ }
+
+ /**
+ * Show a tooltip.
+ *
+ * @param {string|jquery} [tooltip] A string of html or a jQuery object containing the tooltip content.
+ *
+ * @param {string|jquery} [target] The target of the tooltip, to be used to determine positioning. If null,
+ * absolute position values must be passed with left and top.
+ *
+ * @param {string|jquery} [image] If target is an [area] the image that owns it
+ *
+ * @param {string|jquery} [container] An element within which the tooltip must be bounded
+ *
+ *
+ *
+ * @param {object|string|jQuery} [options] options to apply when creating this tooltip - OR -
+ * The markup, or a jquery object, containing the data for the tooltip
+ *
+ * @config {string} [closeEvents] A string with one or more comma-separated values that determine when the tooltip
+ * closes: 'area-click','tooltip-click','image-mouseout' are valid values
+ * then no template will be used.
+ * @config {int} [offsetx] the horizontal amount to offset the tooltip
+ * @config {int} [offsety] the vertical amount to offset the tooltip
+ * @config {string|object} [css] CSS to apply to the outermost element of the tooltip
+ */
+
+ function showToolTip(tooltip,target,image,container,options) {
+ var corners,
+ ttopts = {};
+
+ options = options || {};
+
+
+ if (target) {
+
+ corners = u.areaCorners(target,image,container,
+ tooltip.outerWidth(true),
+ tooltip.outerHeight(true));
+
+ // Try to upper-left align it first, if that doesn't work, change the parameters
+
+ ttopts.left = corners[0];
+ ttopts.top = corners[1];
+
+ } else {
+
+ ttopts.left = options.left;
+ ttopts.top = options.top;
+ }
+
+ ttopts.left += (options.offsetx || 0);
+ ttopts.top +=(options.offsety || 0);
+
+ ttopts.css= options.css;
+ ttopts.fadeDuration = options.fadeDuration;
+
+ showToolTipImpl(tooltip,ttopts);
+
+ return tooltip;
+ }
+
+ /**
+ * Show a tooltip positioned near this area.
+ *
+ * @param {string|jquery} [content] A string of html or a jQuery object containing the tooltip content.
+
+ * @param {object|string|jQuery} [options] options to apply when creating this tooltip - OR -
+ * The markup, or a jquery object, containing the data for the tooltip
+ * @config {string|jquery} [container] An element within which the tooltip must be bounded
+ * @config {bool} [template] a template to use instead of the default. If this property exists and is null,
+ * then no template will be used.
+ * @config {string} [closeEvents] A string with one or more comma-separated values that determine when the tooltip
+ * closes: 'area-click','tooltip-click','image-mouseout' are valid values
+ * then no template will be used.
+ * @config {int} [offsetx] the horizontal amount to offset the tooltip
+ * @config {int} [offsety] the vertical amount to offset the tooltip
+ * @config {string|object} [css] CSS to apply to the outermost element of the tooltip
+ */
+ m.AreaData.prototype.showToolTip= function(content,options) {
+ var tooltip, closeOpts, target, tipClosed, template,
+ ttopts = {},
+ ad=this,
+ md=ad.owner,
+ areaOpts = ad.effectiveOptions();
+
+ // copy the options object so we can update it
+ options = options ? $.extend({},options) : {};
+
+ content = content || areaOpts.toolTip;
+ closeOpts = options.closeEvents || areaOpts.toolTipClose || md.options.toolTipClose || 'tooltip-click';
+
+ template = typeof options.template !== 'undefined' ?
+ options.template :
+ md.options.toolTipContainer;
+
+ options.closeEvents = typeof closeOpts === 'string' ?
+ closeOpts = u.split(closeOpts) :
+ closeOpts;
+
+ options.fadeDuration = options.fadeDuration ||
+ (md.options.toolTipFade ?
+ (md.options.fadeDuration || areaOpts.fadeDuration) : 0);
+
+ target = ad.area ?
+ ad.area :
+ $.map(ad.areas(),
+ function(e) {
+ return e.area;
+ });
+
+ if (md.activeToolTipID===ad.areaId) {
+ return;
+ }
+
+ md.clearToolTip();
+
+ md.activeToolTip = tooltip = createToolTip(content,
+ template,
+ options.css);
+
+ md.activeToolTipID = ad.areaId;
+
+ tipClosed = function() {
+ md.clearToolTip();
+ };
+
+ bindToolTipClose(closeOpts,'area-click', 'click', $(md.map), null, tipClosed);
+ bindToolTipClose(closeOpts,'tooltip-click', 'click', tooltip,null, tipClosed);
+ bindToolTipClose(closeOpts,'image-mouseout', 'mouseout', $(md.image), function(e) {
+ return (e.relatedTarget && e.relatedTarget.nodeName!=='AREA' && e.relatedTarget!==ad.area);
+ }, tipClosed);
+
+
+ showToolTip(tooltip,
+ target,
+ md.image,
+ options.container,
+ template,
+ options);
+
+ u.ifFunction(md.options.onShowToolTip, ad.area,
+ {
+ toolTip: tooltip,
+ options: ttopts,
+ areaOptions: areaOpts,
+ key: ad.key,
+ selected: ad.isSelected()
+ });
+
+ return tooltip;
+ };
+
+
+ /**
+ * Parse an object that could be a string, a jquery object, or an object with a "contents" property
+ * containing html or a jQuery object.
+ *
+ * @param {object|string|jQuery} options The parameter to parse
+ * @return {string|jquery} A string or jquery object
+ */
+ function getHtmlFromOptions(options) {
+
+ // see if any html was passed as either the options object itself, or the content property
+
+ return (options ?
+ ((typeof options === 'string' || options.jquery) ?
+ options :
+ options.content) :
+ null);
+ }
+
+ /**
+ * Activate or remove a tooltip for an area. When this method is called on an area, the
+ * key parameter doesn't apply and "options" is the first parameter.
+ *
+ * When called with no parameters, or "key" is a falsy value, any active tooltip is cleared.
+ *
+ * When only a key is provided, the default tooltip for the area is used.
+ *
+ * When html is provided, this is used instead of the default tooltip.
+ *
+ * When "noTemplate" is true, the default tooltip template will not be used either, meaning only
+ * the actual html passed will be used.
+ *
+ * @param {string|AreaElement} key The area for which to activate a tooltip, or a DOM element.
+ *
+ * @param {object|string|jquery} [options] options to apply when creating this tooltip - OR -
+ * The markup, or a jquery object, containing the data for the tooltip
+ * @config {string|jQuery} [content] the inner content of the tooltip; the tooltip text or HTML
+ * @config {Element|jQuery} [container] the inner content of the tooltip; the tooltip text or HTML
+ * @config {bool} [template] a template to use instead of the default. If this property exists and is null,
+ * then no template will be used.
+ * @config {int} [offsetx] the horizontal amount to offset the tooltip.
+ * @config {int} [offsety] the vertical amount to offset the tooltip.
+ * @config {string|object} [css] CSS to apply to the outermost element of the tooltip
+ * @config {string|object} [css] CSS to apply to the outermost element of the tooltip
+ * @config {bool} [fadeDuration] When non-zero, the duration in milliseconds of a fade-in effect for the tooltip.
+ * @return {jQuery} The jQuery object
+ */
+
+ m.impl.tooltip = function (key,options) {
+ return (new m.Method(this,
+ function mapData() {
+ var tooltip, target, md=this;
+ if (!key) {
+ md.clearToolTip();
+ } else {
+ target=$(key);
+ if (md.activeToolTipID ===target[0]) {
+ return;
+ }
+ md.clearToolTip();
+
+ md.activeToolTip = tooltip = createToolTip(getHtmlFromOptions(options),
+ options.template || md.options.toolTipContainer,
+ options.css);
+ md.activeToolTipID = target[0];
+
+ bindToolTipClose(['tooltip-click'],'tooltip-click', 'click', tooltip, null, function() {
+ md.clearToolTip();
+ });
+
+ md.activeToolTip = tooltip = showToolTip(tooltip,
+ target,
+ md.image,
+ options.container,
+ options);
+ }
+ },
+ function areaData() {
+ if ($.isPlainObject(key) && !options) {
+ options = key;
+ }
+
+ this.showToolTip(getHtmlFromOptions(options),options);
+ },
+ {
+ name: 'tooltip',
+ args: arguments,
+ key: key
+ }
+ )).go();
+ };
+} (jQuery));
diff --git a/preview-fall2024-info/materials-science.html b/preview-fall2024-info/materials-science.html
new file mode 100644
index 000000000..1f8d76ee2
--- /dev/null
+++ b/preview-fall2024-info/materials-science.html
@@ -0,0 +1,362 @@
+
+
+
+
+
+
+
Empowering Computational Materials Science Research using HTC
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Empowering Computational Materials Science Research using HTC
+
+
Ajay Annamareddy, a research scientist at the University of Wisconsin-Madison, describes how he utilizes high-throughput computing in computational materials science.
+
+
+ .](https://raw.githubusercontent.com/CHTC/Articles/main/images/materials-science.jpg) + Computer screen with lines of code. Uploaded by AltumCode on Unsplash
+ Computer screen with lines of code. Uploaded by AltumCode on Unsplash
+
+
+
Groundbreaking research is in the works for the Computational Materials Group (CMG) at the University of Wisconsin-Madison (UW-Madison). Ajay Annamareddy, a research scientist within CMG, has been a leading user of GPU hours with the Center for High Throughput Computing (CHTC). He utilizes this capacity to run machine learning (ML) simulations as applied to material science problems that have gained tremendous interest in the past decade. CHTC resources have allowed him to study hugely data-driven problems that are practically impossible to deal with using regular resources.
+
+
Before coming to UW-Madison, Annamareddy received his Ph.D. in Nuclear Engineering from North Carolina State University. He was introduced to modeling and simulation work there, but he started using high-throughput computing (HTC) and CHTC services when he came to UW-Madison to work as a PostDoc with Prof. Dane Morgan in the Materials Science and Engineering department. He now works for CMG as a Research Scientist, where he’s been racking up GPU hours for over a year.
+
+
Working in the field of computational materials, Annamareddy and his group use computers to determine the properties of materials. So rather than preparing material and measuring it in experiments, they use a computer, which is less expensive and more time efficient. Annamareddy studies metallic glasses. These materials have many valuable properties and applications, but are not easy to make. Instead, he uses computer simulations of these materials to analyze and understand their fundamental properties.
+
+
Annamareddy’s group utilizes HTC and high-performance computing (HPC) for their work, so his project lead asked him to contact CHTC and set up an account. Christina Koch, the lead research computing facilitator, responded. “She helped me set up the account and determine how many resources we needed,” Annamareddy explained. “She was very generous in that whenever I exceeded my limits, she would increase them a bit more!”
+
+
CHTC resources have become critical for Annamareddy’s work. One of the projects involves running ML simulations, which he notes would be “difficult to complete” without the support of CHTC. Annamareddy uses graph neural networks (GNN), a powerful yet slightly inefficient deep learning technique. The upside to using GNN is that as long as there is some physics component in the underlying research problem, this technique can analyze just about anything. “The caveat is you need to provide lots of data for this technique to figure out a solution.”
+
+
Meeting this data challenge, Annamareddy put the input data he generates using high-performance computing (HPC) on the HTC staging location, which gets transferred to a local machine before the ML job starts running. “I use close to twenty gigabytes of data for my simulation, so this would be extremely inefficient to run without staging,” he explains. The CHTC provides Annamareddy with the storage and organization he needs to perform these potentially ground-breaking ML simulations.
+
+
Researchers often study materials in traditional atomistic simulations at different timescales, ranging from picoseconds to microseconds. Annamareddy’s goal with his work is to extend the time scales of these conventional simulations by using ML, which he found is well supported by HTC resources. “We have yet to reach it, but we hope we can use ML to extend the time scale of atomistic simulations by a few orders of magnitude. This would be extremely valuable when modeling systems like glass-forming materials where we should be able to obtain properties, like density and diffusion coefficients, much closer to experiments than currently possible with atomistic simulations,” Annamareddy elaborates. This is something that has never been done before in the field.
+
+
This project can potentially extend the time scales possible for conventional molecular dynamic simulations, allowing researchers in this field to predict how materials will behave over more extended periods of time. “It’s ambitious – but I’ve been working on it for more than a year, and we’ve made a lot of progress…I enjoy the challenge immensely and am happy I’m working on this problem!”
+
+
+
+
+
+
+
+
+
+
+
+
+
diff --git a/preview-fall2024-info/neuroscientist.html b/preview-fall2024-info/neuroscientist.html
new file mode 100644
index 000000000..d728d2ef5
--- /dev/null
+++ b/preview-fall2024-info/neuroscientist.html
@@ -0,0 +1,381 @@
+
+
+
+
+
+
+
For neuroscientist Chris Cox, the OSG helps process mountains of data
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ For neuroscientist Chris Cox, the OSG helps process mountains of data
+
+
Whether exploring how the brain is fooled by fake news or explaining the decline of knowledge in dementia, cognitive neuroscientists like Chris Cox are relying more on high-throughput computing resources like the Open Science Pool to understand how the brain makes sense of information.
+
+
Cognitive neuroscientist Chris Cox recently defended his dissertation at the University of Wisconsin Madison (UW-Madison). Unlike molecular or cellular study of neuroscience, cognitive neuroscience seeks a larger view of neural systems—of “how the brain supports cognition,” said Cox.
+
+
Cox and other neuroscience researchers seek to understand which parts of the brain support memory and decision making, and answer more nuanced questions like how objects are represented in the brain. For Cox, this has involved developing new techniques for studying the brain that rely heavily on high-throughput computing.
+
+
“Our research gets into the transformations that take place in the brain. We ask questions like ‘how is information from our senses combined to support abstract knowledge that seems to transcend our senses,’” said Cox. “For example, we can recognize a single object from different perspectives and scales as being the same thing, and when we read a word we can call to mind all kinds of meaning that have little if anything to do with the letters on the page.”
+
+
The brain is highly complex, so neural imaging methods like functional MRI yield thousands of individual data points for every two seconds of imaging. Cox first turned to high performance computing and finally to the Open Science Pool for high-throughput computing (HTC) to deal with the massive amounts of data. Because computing support at UW-Madison is so seamless, when he first started out on HTC, Cox wasn’t even aware that the OSG was powering the vast improvement in his research.
+
+
“The OSG at UW-Madison is like flipping a switch,” said Cox. “It cut my computing time in half and was totally painless. Our research was a good candidate for the OSG and the advantages of HTC. The OSG and the Center for High Throughput Computing at UW-Madison have empowered us to get results quickly that inform our next steps. This would be impossible without the extensive and robust HTC infrastructure provided by the OSG.”
+
+
A 45-minute experiment from many participants would produce enormous amounts of data. “From that, we can make inferences that generalize to humanity at large about how our brains work,” said Cox. “Our previous approach was to only look for activation that is located in the same place in the brain in different people and look for anatomical landmarks that we can line up across people. Then we ask whether they respond the same way (across people).”
+
+
“But now, we have expanded beyond that approach and look at how multiple parts of the brain are working together,” said Cox. “Even in one region of the brain, not every subcomponent might be working the same way, so when we start adding in all this extra diversity of the activation profile, we get very complicated models that have to be tuned to the data set.”
+
+
Cox’s major parameters now are how many data points to include when it’s time to build a model. “For cross-validation, that then increases the need for computing by an order of magnitude,” said Cox.
+
+
Each model can take 30 minutes to an hour to compute. Cox then runs hundreds of thousands of them to narrow in on the appropriate parameter values.
+
+
Further increasing the computational burden, this whole procedure has to be done multiple times, each time holding out a portion of the data for cross-validation. “By cross-validating and running simulations to determine what random performance looks like, we can test whether the models are revealing something meaningful about the brain,” said Cox.
+
+
Cox gains a particular advantage from high-throughput computing on the OSG by creating novel optimization procedures to probe MRI data that is more connected with cognitive theory.
+
+
“Saving a minute or two on each individual job is not important,” said Cox. “Our main priority can focus on the most conceptually sound algorithms and we can get to real work more quickly. We don’t need to optimize for a HPC cluster, we can just use the scale of HTC.”
+
+
Cox’s research is beginning to explore the neural dynamics involved when calling to mind a concept, with millisecond resolution. This requires looking at data collected with other methods like electroencephalography (EEG) and electrocortography (EcoG). Cox said that it takes about two full seconds for MRI to collect a single sample.
+
+
“The problem is that lots of cognitive activity is going on in those two seconds that is being missed,” said Cox. “When you gain resolution in the time domain you have a chance to notice qualitative shifts that may delimit different neural processes. Identifying when they occur has a lot of theoretical relevance, but also practical relevance in understanding when information is available to the person.”
+
+
“People think of the brain as a library—adding books to the stack and looking in a card catalog,” said Cox. “We are seeing knowledge more like Lego blocks than a library—no single block has meaning, but a collection can express meaning when properly composed. The brain puts those blocks together to give meaning. My research so far supports the Lego perspective over the library perspective.”
+
+
Cognitive neuroscience may offer clues to cognitive decline, which in turn could inform how we think about learning, instruction, and training. How we understand challenges like dementia can lead to better, more correct therapies by understanding the patterns of decline in the brain.
+
+
“Also, having a more accurate understanding of what it means to ‘know’ something can also help us understand how fake news and misinformation take hold in individuals and spread through social networks,” said Cox. “At the core of these issues are fundamental questions about how we process and assimilate information.
+
+
“We know it is hard to get someone to change their mind, so the question is what is happening in the brain. The answers depend on a better understanding of what knowledge is and how we acquire it. Our research is pointed to these higher level questions.”
+
+
“Once we had access to the computational resources of the OSG, we saw a paradigm shift in the way we think about research,” said Cox. “Previously, we might have jobs running for months. With HTC on the OSG, that job length became just a few days. It gave new legs to the whole research program and pushed us forward on new optimization techniques that we never would have tried otherwise.”
+
+
– Greg Moore
+
+
+
+
+
+
+
+
+
+
+
+
+
diff --git a/preview-fall2024-info/news-navigation.html b/preview-fall2024-info/news-navigation.html
new file mode 100644
index 000000000..0b632a710
--- /dev/null
+++ b/preview-fall2024-info/news-navigation.html
@@ -0,0 +1,357 @@
+
+
+
+
+
+
+
News
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
diff --git a/preview-fall2024-info/news.html b/preview-fall2024-info/news.html
new file mode 100644
index 000000000..1b4b0a360
--- /dev/null
+++ b/preview-fall2024-info/news.html
@@ -0,0 +1,2575 @@
+
+
+
+
+
+
+
News: How CHTC is Making An Impact
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ News: How CHTC is Making An Impact
+
+
+
+ CHTC’s computing pioneering continues to advance science and society in new ways. Located at the heart of
+ UW-Madison’s School for Computer, Data & Information Sciences (CDIS), CHTC offers
+ exceptional computing capabilities and experienced facilitation support to campus researchers and
+ international scientists alike. Working in collaboration with projects across all areas of study,
+ CHTC helps innovate solutions that otherwise might not have been possible, while at
+ the same time evolving the field of distributed computing.
+
+
+
+
+
+
+
+
Featured News
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
Read about some of CHTC’s latest news and projects:
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ 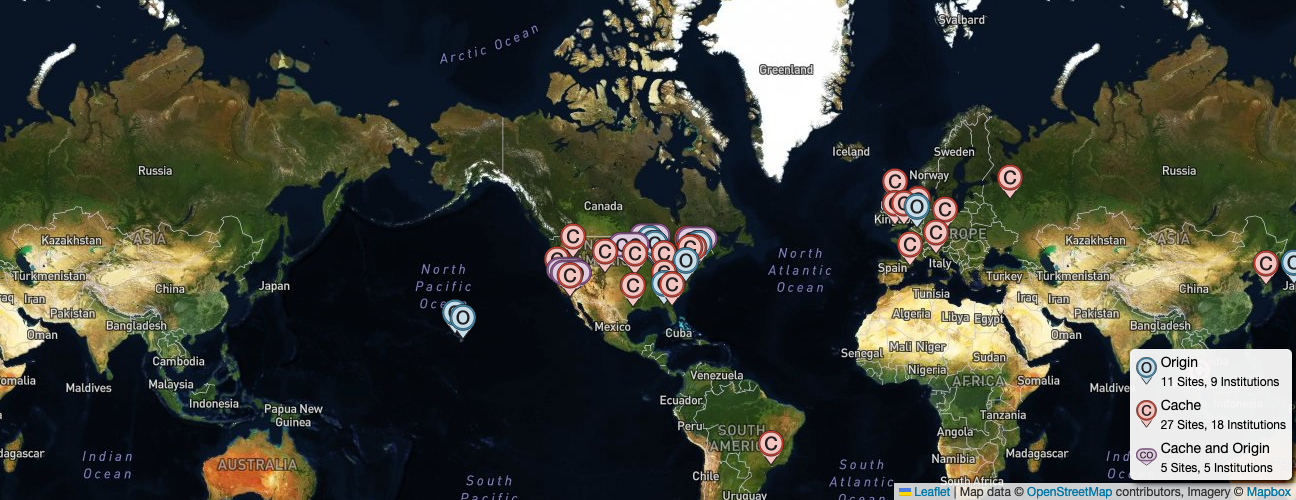 +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ 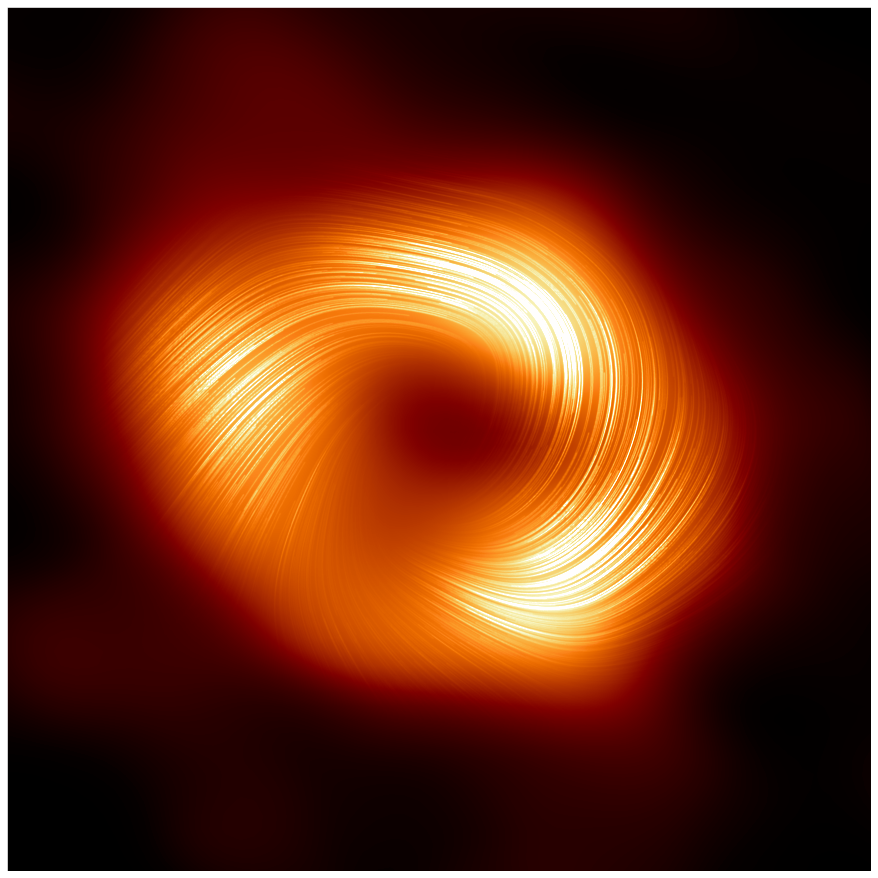 +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ 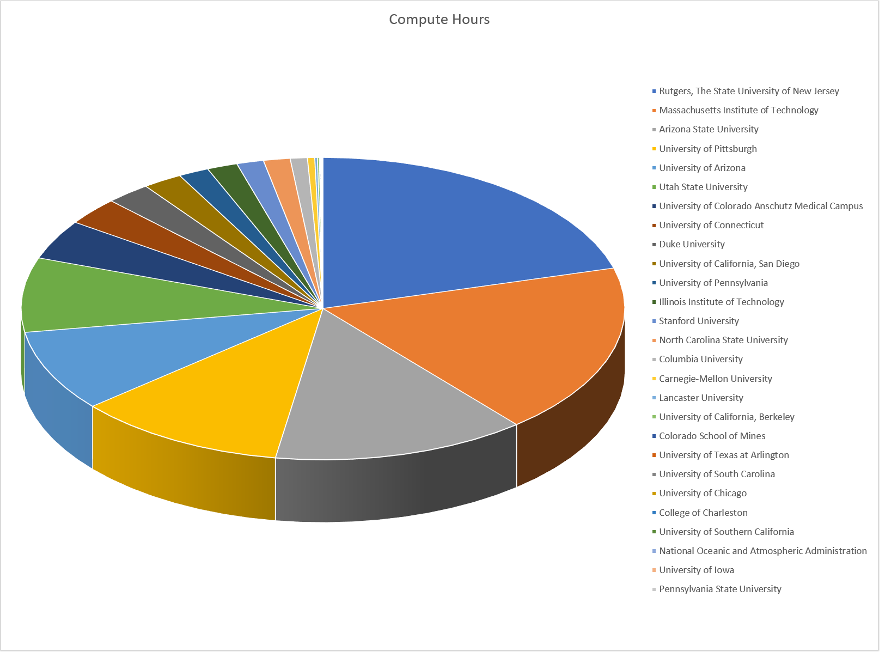 +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
diff --git a/preview-fall2024-info/news/PULL_REQUEST_TEMPLATE.md b/preview-fall2024-info/news/PULL_REQUEST_TEMPLATE.md
new file mode 100644
index 000000000..cde45773f
--- /dev/null
+++ b/preview-fall2024-info/news/PULL_REQUEST_TEMPLATE.md
@@ -0,0 +1,7 @@
+Article PR Template
+
+Article previews can be found [here](https://chtc.github.io/article-preview/)
+
+* [ ] Linked the preview url `https://chtc.github.io/article-preview/
`
+* [ ] Looked at the link in https://socialsharepreview.com/ to verify socials
+* [ ] Requested reviews from the correct parties
diff --git a/preview-fall2024-info/news/images/Ananya-headshot.jpeg b/preview-fall2024-info/news/images/Ananya-headshot.jpeg
new file mode 100644
index 000000000..80567facc
Binary files /dev/null and b/preview-fall2024-info/news/images/Ananya-headshot.jpeg differ
diff --git a/preview-fall2024-info/news/images/Anirvan-Showcase-1.png b/preview-fall2024-info/news/images/Anirvan-Showcase-1.png
new file mode 100644
index 000000000..6c5d44c6a
Binary files /dev/null and b/preview-fall2024-info/news/images/Anirvan-Showcase-1.png differ
diff --git a/preview-fall2024-info/news/images/Anirvan-Showcase-2.png b/preview-fall2024-info/news/images/Anirvan-Showcase-2.png
new file mode 100644
index 000000000..1bd6ba5b6
Binary files /dev/null and b/preview-fall2024-info/news/images/Anirvan-Showcase-2.png differ
diff --git a/preview-fall2024-info/news/images/Anirvan-Showcase-banner.png b/preview-fall2024-info/news/images/Anirvan-Showcase-banner.png
new file mode 100644
index 000000000..3f1a5cc95
Binary files /dev/null and b/preview-fall2024-info/news/images/Anirvan-Showcase-banner.png differ
diff --git a/preview-fall2024-info/news/images/Banner-Image.png b/preview-fall2024-info/news/images/Banner-Image.png
new file mode 100644
index 000000000..104a73aa8
Binary files /dev/null and b/preview-fall2024-info/news/images/Banner-Image.png differ
diff --git a/preview-fall2024-info/news/images/Bayly-work.png b/preview-fall2024-info/news/images/Bayly-work.png
new file mode 100644
index 000000000..7946597bc
Binary files /dev/null and b/preview-fall2024-info/news/images/Bayly-work.png differ
diff --git a/preview-fall2024-info/news/images/Black-hole-banner.jpg b/preview-fall2024-info/news/images/Black-hole-banner.jpg
new file mode 100644
index 000000000..478276c57
Binary files /dev/null and b/preview-fall2024-info/news/images/Black-hole-banner.jpg differ
diff --git a/preview-fall2024-info/news/images/BrianBockelman.jpeg b/preview-fall2024-info/news/images/BrianBockelman.jpeg
new file mode 100644
index 000000000..6890d9043
Binary files /dev/null and b/preview-fall2024-info/news/images/BrianBockelman.jpeg differ
diff --git a/preview-fall2024-info/news/images/CDIS_render.jpg b/preview-fall2024-info/news/images/CDIS_render.jpg
new file mode 100644
index 000000000..7f66984f2
Binary files /dev/null and b/preview-fall2024-info/news/images/CDIS_render.jpg differ
diff --git a/preview-fall2024-info/news/images/CHTC_Workshop.jpg b/preview-fall2024-info/news/images/CHTC_Workshop.jpg
new file mode 100644
index 000000000..6fca7382d
Binary files /dev/null and b/preview-fall2024-info/news/images/CHTC_Workshop.jpg differ
diff --git a/preview-fall2024-info/news/images/CLAS-Collaboration.jpg b/preview-fall2024-info/news/images/CLAS-Collaboration.jpg
new file mode 100644
index 000000000..059ab8694
Binary files /dev/null and b/preview-fall2024-info/news/images/CLAS-Collaboration.jpg differ
diff --git a/preview-fall2024-info/news/images/CLAS-Detector.jpg b/preview-fall2024-info/news/images/CLAS-Detector.jpg
new file mode 100644
index 000000000..1c7957786
Binary files /dev/null and b/preview-fall2024-info/news/images/CLAS-Detector.jpg differ
diff --git a/preview-fall2024-info/news/images/CLAS12-Banner.jpg b/preview-fall2024-info/news/images/CLAS12-Banner.jpg
new file mode 100644
index 000000000..45361cb26
Binary files /dev/null and b/preview-fall2024-info/news/images/CLAS12-Banner.jpg differ
diff --git a/preview-fall2024-info/news/images/California_wildfire.jpg b/preview-fall2024-info/news/images/California_wildfire.jpg
new file mode 100644
index 000000000..f3b588cea
Binary files /dev/null and b/preview-fall2024-info/news/images/California_wildfire.jpg differ
diff --git a/preview-fall2024-info/news/images/Central-Campus-Madison.jpg b/preview-fall2024-info/news/images/Central-Campus-Madison.jpg
new file mode 100644
index 000000000..c51e972a1
Binary files /dev/null and b/preview-fall2024-info/news/images/Central-Campus-Madison.jpg differ
diff --git a/preview-fall2024-info/news/images/Chan.jpg b/preview-fall2024-info/news/images/Chan.jpg
new file mode 100644
index 000000000..6e7e164fa
Binary files /dev/null and b/preview-fall2024-info/news/images/Chan.jpg differ
diff --git a/preview-fall2024-info/news/images/Connor-Natzke-Square-smaller.jpg b/preview-fall2024-info/news/images/Connor-Natzke-Square-smaller.jpg
new file mode 100644
index 000000000..55c179176
Binary files /dev/null and b/preview-fall2024-info/news/images/Connor-Natzke-Square-smaller.jpg differ
diff --git a/preview-fall2024-info/news/images/Dark-Matter-Map-II-e1521045294690.jpg b/preview-fall2024-info/news/images/Dark-Matter-Map-II-e1521045294690.jpg
new file mode 100644
index 000000000..393579eb2
Binary files /dev/null and b/preview-fall2024-info/news/images/Dark-Matter-Map-II-e1521045294690.jpg differ
diff --git a/preview-fall2024-info/news/images/DavidSwanson.png b/preview-fall2024-info/news/images/DavidSwanson.png
new file mode 100644
index 000000000..7f70da651
Binary files /dev/null and b/preview-fall2024-info/news/images/DavidSwanson.png differ
diff --git a/preview-fall2024-info/news/images/Devin-headshot.jpeg b/preview-fall2024-info/news/images/Devin-headshot.jpeg
new file mode 100644
index 000000000..295d73329
Binary files /dev/null and b/preview-fall2024-info/news/images/Devin-headshot.jpeg differ
diff --git a/preview-fall2024-info/news/images/Dorsey-headshot.jpg b/preview-fall2024-info/news/images/Dorsey-headshot.jpg
new file mode 100644
index 000000000..0b69815ce
Binary files /dev/null and b/preview-fall2024-info/news/images/Dorsey-headshot.jpg differ
diff --git a/preview-fall2024-info/news/images/East Side-Madison.jpg b/preview-fall2024-info/news/images/East Side-Madison.jpg
new file mode 100644
index 000000000..67ebd7152
Binary files /dev/null and b/preview-fall2024-info/news/images/East Side-Madison.jpg differ
diff --git a/preview-fall2024-info/news/images/Facilitation-cover.jpeg b/preview-fall2024-info/news/images/Facilitation-cover.jpeg
new file mode 100644
index 000000000..ff622665b
Binary files /dev/null and b/preview-fall2024-info/news/images/Facilitation-cover.jpeg differ
diff --git a/preview-fall2024-info/news/images/Fermion-Bag-300x300.jpg b/preview-fall2024-info/news/images/Fermion-Bag-300x300.jpg
new file mode 100644
index 000000000..962ac6e13
Binary files /dev/null and b/preview-fall2024-info/news/images/Fermion-Bag-300x300.jpg differ
diff --git a/preview-fall2024-info/news/images/Frank.jpg b/preview-fall2024-info/news/images/Frank.jpg
new file mode 100644
index 000000000..ae50ea2b8
Binary files /dev/null and b/preview-fall2024-info/news/images/Frank.jpg differ
diff --git a/preview-fall2024-info/news/images/Fulvio-card.jpeg b/preview-fall2024-info/news/images/Fulvio-card.jpeg
new file mode 100644
index 000000000..52710f382
Binary files /dev/null and b/preview-fall2024-info/news/images/Fulvio-card.jpeg differ
diff --git a/preview-fall2024-info/news/images/Fulvio-headshot.jpeg b/preview-fall2024-info/news/images/Fulvio-headshot.jpeg
new file mode 100644
index 000000000..bcca89ab3
Binary files /dev/null and b/preview-fall2024-info/news/images/Fulvio-headshot.jpeg differ
diff --git a/preview-fall2024-info/news/images/Fulvio-research.png b/preview-fall2024-info/news/images/Fulvio-research.png
new file mode 100644
index 000000000..cb1b59c05
Binary files /dev/null and b/preview-fall2024-info/news/images/Fulvio-research.png differ
diff --git a/preview-fall2024-info/news/images/Garcia-card.png b/preview-fall2024-info/news/images/Garcia-card.png
new file mode 100644
index 000000000..ad582e6d9
Binary files /dev/null and b/preview-fall2024-info/news/images/Garcia-card.png differ
diff --git a/preview-fall2024-info/news/images/Garcia-cycle.png b/preview-fall2024-info/news/images/Garcia-cycle.png
new file mode 100644
index 000000000..154da8ff1
Binary files /dev/null and b/preview-fall2024-info/news/images/Garcia-cycle.png differ
diff --git a/preview-fall2024-info/news/images/Garcia-workflow.png b/preview-fall2024-info/news/images/Garcia-workflow.png
new file mode 100644
index 000000000..473bdffc2
Binary files /dev/null and b/preview-fall2024-info/news/images/Garcia-workflow.png differ
diff --git a/preview-fall2024-info/news/images/Gaylen-Fronk-square.jpg b/preview-fall2024-info/news/images/Gaylen-Fronk-square.jpg
new file mode 100644
index 000000000..4290f44f7
Binary files /dev/null and b/preview-fall2024-info/news/images/Gaylen-Fronk-square.jpg differ
diff --git a/preview-fall2024-info/news/images/HTCondorWeek2022-Bike.jpg b/preview-fall2024-info/news/images/HTCondorWeek2022-Bike.jpg
new file mode 100644
index 000000000..845151635
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondorWeek2022-Bike.jpg differ
diff --git a/preview-fall2024-info/news/images/HTCondorWeek2022-Brian-Question.jpg b/preview-fall2024-info/news/images/HTCondorWeek2022-Brian-Question.jpg
new file mode 100644
index 000000000..95a4a0a7e
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondorWeek2022-Brian-Question.jpg differ
diff --git a/preview-fall2024-info/news/images/HTCondorWeek2022-Closing.jpg b/preview-fall2024-info/news/images/HTCondorWeek2022-Closing.jpg
new file mode 100644
index 000000000..0047a2cc9
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondorWeek2022-Closing.jpg differ
diff --git a/preview-fall2024-info/news/images/HTCondorWeek2022-Collage.jpg b/preview-fall2024-info/news/images/HTCondorWeek2022-Collage.jpg
new file mode 100644
index 000000000..76f7311f5
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondorWeek2022-Collage.jpg differ
diff --git a/preview-fall2024-info/news/images/HTCondorWeek2022-Conversation.jpg b/preview-fall2024-info/news/images/HTCondorWeek2022-Conversation.jpg
new file mode 100644
index 000000000..e4aa7d070
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondorWeek2022-Conversation.jpg differ
diff --git a/preview-fall2024-info/news/images/HTCondorWeek2022-Emile-Listening.jpg b/preview-fall2024-info/news/images/HTCondorWeek2022-Emile-Listening.jpg
new file mode 100644
index 000000000..ed2639df3
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondorWeek2022-Emile-Listening.jpg differ
diff --git a/preview-fall2024-info/news/images/HTCondorWeek2022-Justin-Presenting.jpg b/preview-fall2024-info/news/images/HTCondorWeek2022-Justin-Presenting.jpg
new file mode 100644
index 000000000..278d2e92c
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondorWeek2022-Justin-Presenting.jpg differ
diff --git a/preview-fall2024-info/news/images/HTCondorWeek2022-Lobby.jpg b/preview-fall2024-info/news/images/HTCondorWeek2022-Lobby.jpg
new file mode 100644
index 000000000..dacf03b49
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondorWeek2022-Lobby.jpg differ
diff --git a/preview-fall2024-info/news/images/HTCondorWeek2022-Outside.jpg b/preview-fall2024-info/news/images/HTCondorWeek2022-Outside.jpg
new file mode 100644
index 000000000..557294690
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondorWeek2022-Outside.jpg differ
diff --git a/preview-fall2024-info/news/images/HTCondorWeek2022-Terrace.jpg b/preview-fall2024-info/news/images/HTCondorWeek2022-Terrace.jpg
new file mode 100644
index 000000000..5184a8800
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondorWeek2022-Terrace.jpg differ
diff --git a/preview-fall2024-info/news/images/HTCondorWeek2022-Welcome.jpg b/preview-fall2024-info/news/images/HTCondorWeek2022-Welcome.jpg
new file mode 100644
index 000000000..f181610e5
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondorWeek2022-Welcome.jpg differ
diff --git a/preview-fall2024-info/news/images/HTCondorWeek2022-Wilcots.jpg b/preview-fall2024-info/news/images/HTCondorWeek2022-Wilcots.jpg
new file mode 100644
index 000000000..06d80a147
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondorWeek2022-Wilcots.jpg differ
diff --git a/preview-fall2024-info/news/images/HTCondorWeek2022-Yudhajit.jpg b/preview-fall2024-info/news/images/HTCondorWeek2022-Yudhajit.jpg
new file mode 100644
index 000000000..715140a42
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondorWeek2022-Yudhajit.jpg differ
diff --git a/preview-fall2024-info/news/images/HTCondor_Banner.jpeg b/preview-fall2024-info/news/images/HTCondor_Banner.jpeg
new file mode 100644
index 000000000..c6eca8bee
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondor_Banner.jpeg differ
diff --git a/preview-fall2024-info/news/images/HTCondor_Bird.png b/preview-fall2024-info/news/images/HTCondor_Bird.png
new file mode 100644
index 000000000..2d19edf9a
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondor_Bird.png differ
diff --git a/preview-fall2024-info/news/images/HTCondor_red_blk.png b/preview-fall2024-info/news/images/HTCondor_red_blk.png
new file mode 100644
index 000000000..667a492e8
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondor_red_blk.png differ
diff --git a/preview-fall2024-info/news/images/HTCondor_red_blk_notag.png b/preview-fall2024-info/news/images/HTCondor_red_blk_notag.png
new file mode 100644
index 000000000..f7d74b8c4
Binary files /dev/null and b/preview-fall2024-info/news/images/HTCondor_red_blk_notag.png differ
diff --git a/preview-fall2024-info/news/images/Hannah-Showcase-Banner.jpg b/preview-fall2024-info/news/images/Hannah-Showcase-Banner.jpg
new file mode 100644
index 000000000..f229dd844
Binary files /dev/null and b/preview-fall2024-info/news/images/Hannah-Showcase-Banner.jpg differ
diff --git a/preview-fall2024-info/news/images/Hannah-Showcase.jpg b/preview-fall2024-info/news/images/Hannah-Showcase.jpg
new file mode 100644
index 000000000..f999b4c5b
Binary files /dev/null and b/preview-fall2024-info/news/images/Hannah-Showcase.jpg differ
diff --git a/preview-fall2024-info/news/images/Hiemstra-card.png b/preview-fall2024-info/news/images/Hiemstra-card.png
new file mode 100644
index 000000000..72586d59e
Binary files /dev/null and b/preview-fall2024-info/news/images/Hiemstra-card.png differ
diff --git a/preview-fall2024-info/news/images/HimadriChakraborty2024.jpg b/preview-fall2024-info/news/images/HimadriChakraborty2024.jpg
new file mode 100644
index 000000000..72d2ed1fa
Binary files /dev/null and b/preview-fall2024-info/news/images/HimadriChakraborty2024.jpg differ
diff --git a/preview-fall2024-info/news/images/IMG_4838.JPG b/preview-fall2024-info/news/images/IMG_4838.JPG
new file mode 100644
index 000000000..1c25df2c9
Binary files /dev/null and b/preview-fall2024-info/news/images/IMG_4838.JPG differ
diff --git a/preview-fall2024-info/news/images/IMG_4839.JPG b/preview-fall2024-info/news/images/IMG_4839.JPG
new file mode 100644
index 000000000..abf88e3f3
Binary files /dev/null and b/preview-fall2024-info/news/images/IMG_4839.JPG differ
diff --git a/preview-fall2024-info/news/images/Icecube_Wins_Award.png b/preview-fall2024-info/news/images/Icecube_Wins_Award.png
new file mode 100644
index 000000000..05760b0d6
Binary files /dev/null and b/preview-fall2024-info/news/images/Icecube_Wins_Award.png differ
diff --git a/preview-fall2024-info/news/images/Jefferson-Lab.jpg b/preview-fall2024-info/news/images/Jefferson-Lab.jpg
new file mode 100644
index 000000000..a5becfb69
Binary files /dev/null and b/preview-fall2024-info/news/images/Jefferson-Lab.jpg differ
diff --git a/preview-fall2024-info/news/images/Jem-headshot.jpeg b/preview-fall2024-info/news/images/Jem-headshot.jpeg
new file mode 100644
index 000000000..148bcd8f8
Binary files /dev/null and b/preview-fall2024-info/news/images/Jem-headshot.jpeg differ
diff --git a/preview-fall2024-info/news/images/Joshi.png b/preview-fall2024-info/news/images/Joshi.png
new file mode 100644
index 000000000..a197e58e3
Binary files /dev/null and b/preview-fall2024-info/news/images/Joshi.png differ
diff --git a/preview-fall2024-info/news/images/Lacy-portrait.jpeg b/preview-fall2024-info/news/images/Lacy-portrait.jpeg
new file mode 100644
index 000000000..19218394d
Binary files /dev/null and b/preview-fall2024-info/news/images/Lacy-portrait.jpeg differ
diff --git a/preview-fall2024-info/news/images/Lacy-presentation-screenshot.png b/preview-fall2024-info/news/images/Lacy-presentation-screenshot.png
new file mode 100644
index 000000000..1ffa9053e
Binary files /dev/null and b/preview-fall2024-info/news/images/Lacy-presentation-screenshot.png differ
diff --git a/preview-fall2024-info/news/images/Lightning-Talks-card.jpeg b/preview-fall2024-info/news/images/Lightning-Talks-card.jpeg
new file mode 100644
index 000000000..0587fb1ef
Binary files /dev/null and b/preview-fall2024-info/news/images/Lightning-Talks-card.jpeg differ
diff --git a/preview-fall2024-info/news/images/ML_1.jpeg b/preview-fall2024-info/news/images/ML_1.jpeg
new file mode 100644
index 000000000..bccd59d57
Binary files /dev/null and b/preview-fall2024-info/news/images/ML_1.jpeg differ
diff --git a/preview-fall2024-info/news/images/Matthew-headshot.jpeg b/preview-fall2024-info/news/images/Matthew-headshot.jpeg
new file mode 100644
index 000000000..dd1b2270c
Binary files /dev/null and b/preview-fall2024-info/news/images/Matthew-headshot.jpeg differ
diff --git a/preview-fall2024-info/news/images/Maurizio.jpg b/preview-fall2024-info/news/images/Maurizio.jpg
new file mode 100644
index 000000000..187b3e98e
Binary files /dev/null and b/preview-fall2024-info/news/images/Maurizio.jpg differ
diff --git a/preview-fall2024-info/news/images/Max-headshot.jpeg b/preview-fall2024-info/news/images/Max-headshot.jpeg
new file mode 100644
index 000000000..08a87bf8c
Binary files /dev/null and b/preview-fall2024-info/news/images/Max-headshot.jpeg differ
diff --git a/preview-fall2024-info/news/images/Meng-headshot.jpeg b/preview-fall2024-info/news/images/Meng-headshot.jpeg
new file mode 100644
index 000000000..13fa73d1c
Binary files /dev/null and b/preview-fall2024-info/news/images/Meng-headshot.jpeg differ
diff --git a/preview-fall2024-info/news/images/Messick-card.png b/preview-fall2024-info/news/images/Messick-card.png
new file mode 100644
index 000000000..1062d3780
Binary files /dev/null and b/preview-fall2024-info/news/images/Messick-card.png differ
diff --git a/preview-fall2024-info/news/images/Messick-workflow.png b/preview-fall2024-info/news/images/Messick-workflow.png
new file mode 100644
index 000000000..615c50bfb
Binary files /dev/null and b/preview-fall2024-info/news/images/Messick-workflow.png differ
diff --git a/preview-fall2024-info/news/images/Mike-headshot.jpeg b/preview-fall2024-info/news/images/Mike-headshot.jpeg
new file mode 100644
index 000000000..02b65b992
Binary files /dev/null and b/preview-fall2024-info/news/images/Mike-headshot.jpeg differ
diff --git a/preview-fall2024-info/news/images/Miron.jpg b/preview-fall2024-info/news/images/Miron.jpg
new file mode 100644
index 000000000..47605b945
Binary files /dev/null and b/preview-fall2024-info/news/images/Miron.jpg differ
diff --git a/preview-fall2024-info/news/images/NIAID-banner.jpeg b/preview-fall2024-info/news/images/NIAID-banner.jpeg
new file mode 100644
index 000000000..4b8ab10f2
Binary files /dev/null and b/preview-fall2024-info/news/images/NIAID-banner.jpeg differ
diff --git a/preview-fall2024-info/news/images/NIAID-card.jpeg b/preview-fall2024-info/news/images/NIAID-card.jpeg
new file mode 100644
index 000000000..29755f70d
Binary files /dev/null and b/preview-fall2024-info/news/images/NIAID-card.jpeg differ
diff --git a/preview-fall2024-info/news/images/NIAID_students.jpeg b/preview-fall2024-info/news/images/NIAID_students.jpeg
new file mode 100644
index 000000000..f741f8462
Binary files /dev/null and b/preview-fall2024-info/news/images/NIAID_students.jpeg differ
diff --git a/preview-fall2024-info/news/images/Nathan.jpg b/preview-fall2024-info/news/images/Nathan.jpg
new file mode 100644
index 000000000..9d735ea50
Binary files /dev/null and b/preview-fall2024-info/news/images/Nathan.jpg differ
diff --git a/preview-fall2024-info/news/images/OSG-User-School.jpg b/preview-fall2024-info/news/images/OSG-User-School.jpg
new file mode 100644
index 000000000..4f795bae6
Binary files /dev/null and b/preview-fall2024-info/news/images/OSG-User-School.jpg differ
diff --git a/preview-fall2024-info/news/images/OSGVS21-Logo.png b/preview-fall2024-info/news/images/OSGVS21-Logo.png
new file mode 100644
index 000000000..888101cc5
Binary files /dev/null and b/preview-fall2024-info/news/images/OSGVS21-Logo.png differ
diff --git a/preview-fall2024-info/news/images/OSPool_Contributors.png b/preview-fall2024-info/news/images/OSPool_Contributors.png
new file mode 100644
index 000000000..07ccfb3ad
Binary files /dev/null and b/preview-fall2024-info/news/images/OSPool_Contributors.png differ
diff --git a/preview-fall2024-info/news/images/Olaitan-headshot.jpeg b/preview-fall2024-info/news/images/Olaitan-headshot.jpeg
new file mode 100644
index 000000000..f02d1fcd4
Binary files /dev/null and b/preview-fall2024-info/news/images/Olaitan-headshot.jpeg differ
diff --git a/preview-fall2024-info/news/images/Opotowsky-card.jpeg b/preview-fall2024-info/news/images/Opotowsky-card.jpeg
new file mode 100644
index 000000000..15f710fda
Binary files /dev/null and b/preview-fall2024-info/news/images/Opotowsky-card.jpeg differ
diff --git a/preview-fall2024-info/news/images/Opotowsky-headshot.png b/preview-fall2024-info/news/images/Opotowsky-headshot.png
new file mode 100644
index 000000000..2082adbe5
Binary files /dev/null and b/preview-fall2024-info/news/images/Opotowsky-headshot.png differ
diff --git a/preview-fall2024-info/news/images/PATh-Facility-Hardware.jpg b/preview-fall2024-info/news/images/PATh-Facility-Hardware.jpg
new file mode 100644
index 000000000..fe5e6ff85
Binary files /dev/null and b/preview-fall2024-info/news/images/PATh-Facility-Hardware.jpg differ
diff --git a/preview-fall2024-info/news/images/PATh-Facility-Map.jpg b/preview-fall2024-info/news/images/PATh-Facility-Map.jpg
new file mode 100644
index 000000000..a7da36c8c
Binary files /dev/null and b/preview-fall2024-info/news/images/PATh-Facility-Map.jpg differ
diff --git a/preview-fall2024-info/news/images/PATh_Facility_Preview.jpeg b/preview-fall2024-info/news/images/PATh_Facility_Preview.jpeg
new file mode 100644
index 000000000..b6a10d899
Binary files /dev/null and b/preview-fall2024-info/news/images/PATh_Facility_Preview.jpeg differ
diff --git a/preview-fall2024-info/news/images/Parul-Johri-headshot.jpg b/preview-fall2024-info/news/images/Parul-Johri-headshot.jpg
new file mode 100644
index 000000000..d18c7fc95
Binary files /dev/null and b/preview-fall2024-info/news/images/Parul-Johri-headshot.jpg differ
diff --git a/preview-fall2024-info/news/images/Pascal.jpg b/preview-fall2024-info/news/images/Pascal.jpg
new file mode 100644
index 000000000..eae5c53c6
Binary files /dev/null and b/preview-fall2024-info/news/images/Pascal.jpg differ
diff --git a/preview-fall2024-info/news/images/Peder-headshot.jpeg b/preview-fall2024-info/news/images/Peder-headshot.jpeg
new file mode 100644
index 000000000..9f6ffb1ff
Binary files /dev/null and b/preview-fall2024-info/news/images/Peder-headshot.jpeg differ
diff --git a/preview-fall2024-info/news/images/Pool-Record-Image.jpg b/preview-fall2024-info/news/images/Pool-Record-Image.jpg
new file mode 100644
index 000000000..943b7c885
Binary files /dev/null and b/preview-fall2024-info/news/images/Pool-Record-Image.jpg differ
diff --git a/preview-fall2024-info/news/images/RNAMake-Example.png b/preview-fall2024-info/news/images/RNAMake-Example.png
new file mode 100644
index 000000000..5fa43e2bb
Binary files /dev/null and b/preview-fall2024-info/news/images/RNAMake-Example.png differ
diff --git a/preview-fall2024-info/news/images/Raffaela.jpg b/preview-fall2024-info/news/images/Raffaela.jpg
new file mode 100644
index 000000000..317a1ce84
Binary files /dev/null and b/preview-fall2024-info/news/images/Raffaela.jpg differ
diff --git a/preview-fall2024-info/news/images/Ricarte.jpg b/preview-fall2024-info/news/images/Ricarte.jpg
new file mode 100644
index 000000000..4432ccfa1
Binary files /dev/null and b/preview-fall2024-info/news/images/Ricarte.jpg differ
diff --git a/preview-fall2024-info/news/images/SagA-black-hole.jpg b/preview-fall2024-info/news/images/SagA-black-hole.jpg
new file mode 100644
index 000000000..c44b0658a
Binary files /dev/null and b/preview-fall2024-info/news/images/SagA-black-hole.jpg differ
diff --git a/preview-fall2024-info/news/images/Science-Gateway-Students.jpeg b/preview-fall2024-info/news/images/Science-Gateway-Students.jpeg
new file mode 100644
index 000000000..748a2f3ba
Binary files /dev/null and b/preview-fall2024-info/news/images/Science-Gateway-Students.jpeg differ
diff --git a/preview-fall2024-info/news/images/Screen Shot 2022-12-19 at 12.34.31 PM.png b/preview-fall2024-info/news/images/Screen Shot 2022-12-19 at 12.34.31 PM.png
new file mode 100644
index 000000000..92dd004f6
Binary files /dev/null and b/preview-fall2024-info/news/images/Screen Shot 2022-12-19 at 12.34.31 PM.png differ
diff --git a/preview-fall2024-info/news/images/Shailesh-Chandrasekharan-150x150.jpg b/preview-fall2024-info/news/images/Shailesh-Chandrasekharan-150x150.jpg
new file mode 100644
index 000000000..4865839fc
Binary files /dev/null and b/preview-fall2024-info/news/images/Shailesh-Chandrasekharan-150x150.jpg differ
diff --git a/preview-fall2024-info/news/images/Side-by-Side-Madison-Aerial-Photography.jpg b/preview-fall2024-info/news/images/Side-by-Side-Madison-Aerial-Photography.jpg
new file mode 100644
index 000000000..9229b1d38
Binary files /dev/null and b/preview-fall2024-info/news/images/Side-by-Side-Madison-Aerial-Photography.jpg differ
diff --git a/preview-fall2024-info/news/images/Spectrometer.jpg b/preview-fall2024-info/news/images/Spectrometer.jpg
new file mode 100644
index 000000000..9069c7675
Binary files /dev/null and b/preview-fall2024-info/news/images/Spectrometer.jpg differ
diff --git a/preview-fall2024-info/news/images/Spencer-Showcase-Banner.jpg b/preview-fall2024-info/news/images/Spencer-Showcase-Banner.jpg
new file mode 100644
index 000000000..cf3f88729
Binary files /dev/null and b/preview-fall2024-info/news/images/Spencer-Showcase-Banner.jpg differ
diff --git a/preview-fall2024-info/news/images/Spencer-Showcase.jpg b/preview-fall2024-info/news/images/Spencer-Showcase.jpg
new file mode 100644
index 000000000..d75e9bc41
Binary files /dev/null and b/preview-fall2024-info/news/images/Spencer-Showcase.jpg differ
diff --git a/preview-fall2024-info/news/images/Submodule_Image.png b/preview-fall2024-info/news/images/Submodule_Image.png
new file mode 100644
index 000000000..b1d876d4e
Binary files /dev/null and b/preview-fall2024-info/news/images/Submodule_Image.png differ
diff --git a/preview-fall2024-info/news/images/USGS-collage.jpg b/preview-fall2024-info/news/images/USGS-collage.jpg
new file mode 100644
index 000000000..33bc74f42
Binary files /dev/null and b/preview-fall2024-info/news/images/USGS-collage.jpg differ
diff --git a/preview-fall2024-info/news/images/User_School_Collage.jpg b/preview-fall2024-info/news/images/User_School_Collage.jpg
new file mode 100644
index 000000000..e5eb84963
Binary files /dev/null and b/preview-fall2024-info/news/images/User_School_Collage.jpg differ
diff --git a/preview-fall2024-info/news/images/Venkitesh-150x150.jpg b/preview-fall2024-info/news/images/Venkitesh-150x150.jpg
new file mode 100644
index 000000000..1acaeb2f9
Binary files /dev/null and b/preview-fall2024-info/news/images/Venkitesh-150x150.jpg differ
diff --git a/preview-fall2024-info/news/images/WMS-hours-by-facility-comet.png b/preview-fall2024-info/news/images/WMS-hours-by-facility-comet.png
new file mode 100644
index 000000000..1f3521f32
Binary files /dev/null and b/preview-fall2024-info/news/images/WMS-hours-by-facility-comet.png differ
diff --git a/preview-fall2024-info/news/images/Wilcots-card.png b/preview-fall2024-info/news/images/Wilcots-card.png
new file mode 100644
index 000000000..bca604889
Binary files /dev/null and b/preview-fall2024-info/news/images/Wilcots-card.png differ
diff --git a/preview-fall2024-info/news/images/Wilcots-headshot.jpeg b/preview-fall2024-info/news/images/Wilcots-headshot.jpeg
new file mode 100644
index 000000000..2d73f6c46
Binary files /dev/null and b/preview-fall2024-info/news/images/Wilcots-headshot.jpeg differ
diff --git a/preview-fall2024-info/news/images/Wilcots-map.png b/preview-fall2024-info/news/images/Wilcots-map.png
new file mode 100644
index 000000000..efda3ef85
Binary files /dev/null and b/preview-fall2024-info/news/images/Wilcots-map.png differ
diff --git a/preview-fall2024-info/news/images/Wilcots-planet.png b/preview-fall2024-info/news/images/Wilcots-planet.png
new file mode 100644
index 000000000..973272b67
Binary files /dev/null and b/preview-fall2024-info/news/images/Wilcots-planet.png differ
diff --git a/preview-fall2024-info/news/images/Wilcots-spiral.png b/preview-fall2024-info/news/images/Wilcots-spiral.png
new file mode 100644
index 000000000..d5d4898f6
Binary files /dev/null and b/preview-fall2024-info/news/images/Wilcots-spiral.png differ
diff --git a/preview-fall2024-info/news/images/Zack-headshot.jpeg b/preview-fall2024-info/news/images/Zack-headshot.jpeg
new file mode 100644
index 000000000..42acfd935
Binary files /dev/null and b/preview-fall2024-info/news/images/Zack-headshot.jpeg differ
diff --git a/preview-fall2024-info/news/images/amnh.jpg b/preview-fall2024-info/news/images/amnh.jpg
new file mode 100644
index 000000000..bfb9eb17b
Binary files /dev/null and b/preview-fall2024-info/news/images/amnh.jpg differ
diff --git a/preview-fall2024-info/news/images/amnhgroup.jpeg b/preview-fall2024-info/news/images/amnhgroup.jpeg
new file mode 100644
index 000000000..8e83f0f03
Binary files /dev/null and b/preview-fall2024-info/news/images/amnhgroup.jpeg differ
diff --git a/preview-fall2024-info/news/images/asp-banner.jpeg b/preview-fall2024-info/news/images/asp-banner.jpeg
new file mode 100644
index 000000000..2de5da89c
Binary files /dev/null and b/preview-fall2024-info/news/images/asp-banner.jpeg differ
diff --git a/preview-fall2024-info/news/images/asp-horst.jpg b/preview-fall2024-info/news/images/asp-horst.jpg
new file mode 100644
index 000000000..b66ad50e5
Binary files /dev/null and b/preview-fall2024-info/news/images/asp-horst.jpg differ
diff --git a/preview-fall2024-info/news/images/asp-students.jpeg b/preview-fall2024-info/news/images/asp-students.jpeg
new file mode 100644
index 000000000..a3697875c
Binary files /dev/null and b/preview-fall2024-info/news/images/asp-students.jpeg differ
diff --git a/preview-fall2024-info/news/images/bannerimagedemo.png b/preview-fall2024-info/news/images/bannerimagedemo.png
new file mode 100644
index 000000000..ab261da4a
Binary files /dev/null and b/preview-fall2024-info/news/images/bannerimagedemo.png differ
diff --git a/preview-fall2024-info/news/images/banq-mw.jpg b/preview-fall2024-info/news/images/banq-mw.jpg
new file mode 100644
index 000000000..77dbbadea
Binary files /dev/null and b/preview-fall2024-info/news/images/banq-mw.jpg differ
diff --git a/preview-fall2024-info/news/images/banq-mw.png b/preview-fall2024-info/news/images/banq-mw.png
new file mode 100644
index 000000000..b2a97d94b
Binary files /dev/null and b/preview-fall2024-info/news/images/banq-mw.png differ
diff --git a/preview-fall2024-info/news/images/banq-patrie.jpg b/preview-fall2024-info/news/images/banq-patrie.jpg
new file mode 100644
index 000000000..b91b1d348
Binary files /dev/null and b/preview-fall2024-info/news/images/banq-patrie.jpg differ
diff --git a/preview-fall2024-info/news/images/banq-patrie.png b/preview-fall2024-info/news/images/banq-patrie.png
new file mode 100644
index 000000000..76aae24e7
Binary files /dev/null and b/preview-fall2024-info/news/images/banq-patrie.png differ
diff --git a/preview-fall2024-info/news/images/bat-carousel.jpg b/preview-fall2024-info/news/images/bat-carousel.jpg
new file mode 100644
index 000000000..034a58c6b
Binary files /dev/null and b/preview-fall2024-info/news/images/bat-carousel.jpg differ
diff --git a/preview-fall2024-info/news/images/bat-genomics-Ariadna.png b/preview-fall2024-info/news/images/bat-genomics-Ariadna.png
new file mode 100644
index 000000000..2c8b950e4
Binary files /dev/null and b/preview-fall2024-info/news/images/bat-genomics-Ariadna.png differ
diff --git a/preview-fall2024-info/news/images/beads.jpg b/preview-fall2024-info/news/images/beads.jpg
new file mode 100644
index 000000000..b7bda7b4a
Binary files /dev/null and b/preview-fall2024-info/news/images/beads.jpg differ
diff --git a/preview-fall2024-info/news/images/brian-speaking.png b/preview-fall2024-info/news/images/brian-speaking.png
new file mode 100644
index 000000000..f92873ed2
Binary files /dev/null and b/preview-fall2024-info/news/images/brian-speaking.png differ
diff --git a/preview-fall2024-info/news/images/brianbockelman.jpg b/preview-fall2024-info/news/images/brianbockelman.jpg
new file mode 100644
index 000000000..1ac0e21f1
Binary files /dev/null and b/preview-fall2024-info/news/images/brianbockelman.jpg differ
diff --git a/preview-fall2024-info/news/images/byregion.png b/preview-fall2024-info/news/images/byregion.png
new file mode 100644
index 000000000..0f2336f7a
Binary files /dev/null and b/preview-fall2024-info/news/images/byregion.png differ
diff --git a/preview-fall2024-info/news/images/cartwright-headshot.jpeg b/preview-fall2024-info/news/images/cartwright-headshot.jpeg
new file mode 100644
index 000000000..5a6ac61b8
Binary files /dev/null and b/preview-fall2024-info/news/images/cartwright-headshot.jpeg differ
diff --git a/preview-fall2024-info/news/images/cattle.png b/preview-fall2024-info/news/images/cattle.png
new file mode 100644
index 000000000..61e008495
Binary files /dev/null and b/preview-fall2024-info/news/images/cattle.png differ
diff --git a/preview-fall2024-info/news/images/chenimage.png b/preview-fall2024-info/news/images/chenimage.png
new file mode 100644
index 000000000..7ef14ac20
Binary files /dev/null and b/preview-fall2024-info/news/images/chenimage.png differ
diff --git a/preview-fall2024-info/news/images/christina-koch-chtc-featured.webp b/preview-fall2024-info/news/images/christina-koch-chtc-featured.webp
new file mode 100644
index 000000000..90914f9d8
Binary files /dev/null and b/preview-fall2024-info/news/images/christina-koch-chtc-featured.webp differ
diff --git a/preview-fall2024-info/news/images/christina-koch-square.jpg b/preview-fall2024-info/news/images/christina-koch-square.jpg
new file mode 100644
index 000000000..8b62fe376
Binary files /dev/null and b/preview-fall2024-info/news/images/christina-koch-square.jpg differ
diff --git a/preview-fall2024-info/news/images/chtc-facilitation.jpeg b/preview-fall2024-info/news/images/chtc-facilitation.jpeg
new file mode 100644
index 000000000..c65782a6c
Binary files /dev/null and b/preview-fall2024-info/news/images/chtc-facilitation.jpeg differ
diff --git a/preview-fall2024-info/news/images/chtc-map.jpg b/preview-fall2024-info/news/images/chtc-map.jpg
new file mode 100644
index 000000000..6183a371d
Binary files /dev/null and b/preview-fall2024-info/news/images/chtc-map.jpg differ
diff --git a/preview-fall2024-info/news/images/chtc-philosophy-banner.jpg b/preview-fall2024-info/news/images/chtc-philosophy-banner.jpg
new file mode 100644
index 000000000..98ba34bc2
Binary files /dev/null and b/preview-fall2024-info/news/images/chtc-philosophy-banner.jpg differ
diff --git a/preview-fall2024-info/news/images/chtc-philosophy-server.jpg b/preview-fall2024-info/news/images/chtc-philosophy-server.jpg
new file mode 100644
index 000000000..98ba34bc2
Binary files /dev/null and b/preview-fall2024-info/news/images/chtc-philosophy-server.jpg differ
diff --git a/preview-fall2024-info/news/images/clark_cropped-150x150.jpeg b/preview-fall2024-info/news/images/clark_cropped-150x150.jpeg
new file mode 100644
index 000000000..476d79464
Binary files /dev/null and b/preview-fall2024-info/news/images/clark_cropped-150x150.jpeg differ
diff --git a/preview-fall2024-info/news/images/classimage.png b/preview-fall2024-info/news/images/classimage.png
new file mode 100644
index 000000000..6f133b0f7
Binary files /dev/null and b/preview-fall2024-info/news/images/classimage.png differ
diff --git a/preview-fall2024-info/news/images/classroomimage.jpeg b/preview-fall2024-info/news/images/classroomimage.jpeg
new file mode 100644
index 000000000..192c5e891
Binary files /dev/null and b/preview-fall2024-info/news/images/classroomimage.jpeg differ
diff --git a/preview-fall2024-info/news/images/cole_beads.jpeg b/preview-fall2024-info/news/images/cole_beads.jpeg
new file mode 100644
index 000000000..2a4816e74
Binary files /dev/null and b/preview-fall2024-info/news/images/cole_beads.jpeg differ
diff --git a/preview-fall2024-info/news/images/cole_cage.jpg b/preview-fall2024-info/news/images/cole_cage.jpg
new file mode 100644
index 000000000..068b017ad
Binary files /dev/null and b/preview-fall2024-info/news/images/cole_cage.jpg differ
diff --git a/preview-fall2024-info/news/images/cole_math.jpg b/preview-fall2024-info/news/images/cole_math.jpg
new file mode 100644
index 000000000..93fc81718
Binary files /dev/null and b/preview-fall2024-info/news/images/cole_math.jpg differ
diff --git a/preview-fall2024-info/news/images/cole_pose.jpeg b/preview-fall2024-info/news/images/cole_pose.jpeg
new file mode 100644
index 000000000..28dadea37
Binary files /dev/null and b/preview-fall2024-info/news/images/cole_pose.jpeg differ
diff --git a/preview-fall2024-info/news/images/cole_terrace.jpg b/preview-fall2024-info/news/images/cole_terrace.jpg
new file mode 100644
index 000000000..aed60ab66
Binary files /dev/null and b/preview-fall2024-info/news/images/cole_terrace.jpg differ
diff --git a/preview-fall2024-info/news/images/cole_terrace1.jpg b/preview-fall2024-info/news/images/cole_terrace1.jpg
new file mode 100644
index 000000000..aed60ab66
Binary files /dev/null and b/preview-fall2024-info/news/images/cole_terrace1.jpg differ
diff --git a/preview-fall2024-info/news/images/cole_tree.jpeg b/preview-fall2024-info/news/images/cole_tree.jpeg
new file mode 100644
index 000000000..d947c5d0e
Binary files /dev/null and b/preview-fall2024-info/news/images/cole_tree.jpeg differ
diff --git a/preview-fall2024-info/news/images/collage-header.png b/preview-fall2024-info/news/images/collage-header.png
new file mode 100644
index 000000000..64f267c91
Binary files /dev/null and b/preview-fall2024-info/news/images/collage-header.png differ
diff --git a/preview-fall2024-info/news/images/core-comp-gpu.jpeg b/preview-fall2024-info/news/images/core-comp-gpu.jpeg
new file mode 100644
index 000000000..39f266964
Binary files /dev/null and b/preview-fall2024-info/news/images/core-comp-gpu.jpeg differ
diff --git a/preview-fall2024-info/news/images/costume.jpg b/preview-fall2024-info/news/images/costume.jpg
new file mode 100644
index 000000000..8ff22fcdf
Binary files /dev/null and b/preview-fall2024-info/news/images/costume.jpg differ
diff --git a/preview-fall2024-info/news/images/couvares_cropped-150x150.jpeg b/preview-fall2024-info/news/images/couvares_cropped-150x150.jpeg
new file mode 100644
index 000000000..2375b4f1f
Binary files /dev/null and b/preview-fall2024-info/news/images/couvares_cropped-150x150.jpeg differ
diff --git a/preview-fall2024-info/news/images/cow-ml-image-cropped-banner.jpg b/preview-fall2024-info/news/images/cow-ml-image-cropped-banner.jpg
new file mode 100644
index 000000000..998532c88
Binary files /dev/null and b/preview-fall2024-info/news/images/cow-ml-image-cropped-banner.jpg differ
diff --git a/preview-fall2024-info/news/images/cow-ml-image-cropped.jpg b/preview-fall2024-info/news/images/cow-ml-image-cropped.jpg
new file mode 100644
index 000000000..f823a76ff
Binary files /dev/null and b/preview-fall2024-info/news/images/cow-ml-image-cropped.jpg differ
diff --git a/preview-fall2024-info/news/images/darrylthelen.jpg b/preview-fall2024-info/news/images/darrylthelen.jpg
new file mode 100644
index 000000000..870a696a6
Binary files /dev/null and b/preview-fall2024-info/news/images/darrylthelen.jpg differ
diff --git a/preview-fall2024-info/news/images/davidswanson.jpg b/preview-fall2024-info/news/images/davidswanson.jpg
new file mode 100644
index 000000000..6d6a5b7d2
Binary files /dev/null and b/preview-fall2024-info/news/images/davidswanson.jpg differ
diff --git a/preview-fall2024-info/news/images/deforestation.png b/preview-fall2024-info/news/images/deforestation.png
new file mode 100644
index 000000000..dcaf103d9
Binary files /dev/null and b/preview-fall2024-info/news/images/deforestation.png differ
diff --git a/preview-fall2024-info/news/images/demobanner.png b/preview-fall2024-info/news/images/demobanner.png
new file mode 100644
index 000000000..57a133354
Binary files /dev/null and b/preview-fall2024-info/news/images/demobanner.png differ
diff --git a/preview-fall2024-info/news/images/demopic.png b/preview-fall2024-info/news/images/demopic.png
new file mode 100644
index 000000000..3a1086f41
Binary files /dev/null and b/preview-fall2024-info/news/images/demopic.png differ
diff --git a/preview-fall2024-info/news/images/derekweitzel.jpg b/preview-fall2024-info/news/images/derekweitzel.jpg
new file mode 100644
index 000000000..ad025794a
Binary files /dev/null and b/preview-fall2024-info/news/images/derekweitzel.jpg differ
diff --git a/preview-fall2024-info/news/images/dna.jpeg b/preview-fall2024-info/news/images/dna.jpeg
new file mode 100644
index 000000000..e025637fa
Binary files /dev/null and b/preview-fall2024-info/news/images/dna.jpeg differ
diff --git a/preview-fall2024-info/news/images/dog.jpg b/preview-fall2024-info/news/images/dog.jpg
new file mode 100644
index 000000000..7c8503930
Binary files /dev/null and b/preview-fall2024-info/news/images/dog.jpg differ
diff --git a/preview-fall2024-info/news/images/doit-summary-article.jpeg b/preview-fall2024-info/news/images/doit-summary-article.jpeg
new file mode 100644
index 000000000..713adc41c
Binary files /dev/null and b/preview-fall2024-info/news/images/doit-summary-article.jpeg differ
diff --git a/preview-fall2024-info/news/images/dr-wall.png b/preview-fall2024-info/news/images/dr-wall.png
new file mode 100644
index 000000000..fe0cee2bb
Binary files /dev/null and b/preview-fall2024-info/news/images/dr-wall.png differ
diff --git a/preview-fall2024-info/news/images/early-late-show.png b/preview-fall2024-info/news/images/early-late-show.png
new file mode 100644
index 000000000..55eaf3d78
Binary files /dev/null and b/preview-fall2024-info/news/images/early-late-show.png differ
diff --git a/preview-fall2024-info/news/images/elevator-picture.png b/preview-fall2024-info/news/images/elevator-picture.png
new file mode 100644
index 000000000..8cf911b2f
Binary files /dev/null and b/preview-fall2024-info/news/images/elevator-picture.png differ
diff --git a/preview-fall2024-info/news/images/eln.jpeg b/preview-fall2024-info/news/images/eln.jpeg
new file mode 100644
index 000000000..a689cce8e
Binary files /dev/null and b/preview-fall2024-info/news/images/eln.jpeg differ
diff --git a/preview-fall2024-info/news/images/epic-eic-collab.jpg b/preview-fall2024-info/news/images/epic-eic-collab.jpg
new file mode 100644
index 000000000..3122b5f8b
Binary files /dev/null and b/preview-fall2024-info/news/images/epic-eic-collab.jpg differ
diff --git a/preview-fall2024-info/news/images/ericjonasheadshot.png b/preview-fall2024-info/news/images/ericjonasheadshot.png
new file mode 100644
index 000000000..5c15efb2d
Binary files /dev/null and b/preview-fall2024-info/news/images/ericjonasheadshot.png differ
diff --git a/preview-fall2024-info/news/images/european-htcondor-week-2023.png b/preview-fall2024-info/news/images/european-htcondor-week-2023.png
new file mode 100644
index 000000000..6bf724e3a
Binary files /dev/null and b/preview-fall2024-info/news/images/european-htcondor-week-2023.png differ
diff --git a/preview-fall2024-info/news/images/fellow-speaking-1.png b/preview-fall2024-info/news/images/fellow-speaking-1.png
new file mode 100644
index 000000000..2597dd8f5
Binary files /dev/null and b/preview-fall2024-info/news/images/fellow-speaking-1.png differ
diff --git a/preview-fall2024-info/news/images/fellow-speaking-2.png b/preview-fall2024-info/news/images/fellow-speaking-2.png
new file mode 100644
index 000000000..286895040
Binary files /dev/null and b/preview-fall2024-info/news/images/fellow-speaking-2.png differ
diff --git a/preview-fall2024-info/news/images/fellows.jpeg b/preview-fall2024-info/news/images/fellows.jpeg
new file mode 100644
index 000000000..cc1d1a9e9
Binary files /dev/null and b/preview-fall2024-info/news/images/fellows.jpeg differ
diff --git a/preview-fall2024-info/news/images/finalosgschool.png b/preview-fall2024-info/news/images/finalosgschool.png
new file mode 100644
index 000000000..f5e0531b9
Binary files /dev/null and b/preview-fall2024-info/news/images/finalosgschool.png differ
diff --git a/preview-fall2024-info/news/images/firstmldemoimage-2022-12-19 at 12.34.31 PM.png b/preview-fall2024-info/news/images/firstmldemoimage-2022-12-19 at 12.34.31 PM.png
new file mode 100644
index 000000000..92dd004f6
Binary files /dev/null and b/preview-fall2024-info/news/images/firstmldemoimage-2022-12-19 at 12.34.31 PM.png differ
diff --git a/preview-fall2024-info/news/images/firstmldemoimage.png b/preview-fall2024-info/news/images/firstmldemoimage.png
new file mode 100644
index 000000000..92dd004f6
Binary files /dev/null and b/preview-fall2024-info/news/images/firstmldemoimage.png differ
diff --git a/preview-fall2024-info/news/images/firstmldemoimage2.png b/preview-fall2024-info/news/images/firstmldemoimage2.png
new file mode 100644
index 000000000..424954ade
Binary files /dev/null and b/preview-fall2024-info/news/images/firstmldemoimage2.png differ
diff --git a/preview-fall2024-info/news/images/gillett-card.jpeg b/preview-fall2024-info/news/images/gillett-card.jpeg
new file mode 100644
index 000000000..3d37411d3
Binary files /dev/null and b/preview-fall2024-info/news/images/gillett-card.jpeg differ
diff --git a/preview-fall2024-info/news/images/gillett-headshot.png b/preview-fall2024-info/news/images/gillett-headshot.png
new file mode 100644
index 000000000..cdb9937e2
Binary files /dev/null and b/preview-fall2024-info/news/images/gillett-headshot.png differ
diff --git a/preview-fall2024-info/news/images/gitter.jpeg b/preview-fall2024-info/news/images/gitter.jpeg
new file mode 100644
index 000000000..d02b7801d
Binary files /dev/null and b/preview-fall2024-info/news/images/gitter.jpeg differ
diff --git a/preview-fall2024-info/news/images/google-qvm.jpg b/preview-fall2024-info/news/images/google-qvm.jpg
new file mode 100644
index 000000000..1ecef9c2c
Binary files /dev/null and b/preview-fall2024-info/news/images/google-qvm.jpg differ
diff --git a/preview-fall2024-info/news/images/gpargo.png b/preview-fall2024-info/news/images/gpargo.png
new file mode 100644
index 000000000..3a4a122c8
Binary files /dev/null and b/preview-fall2024-info/news/images/gpargo.png differ
diff --git a/preview-fall2024-info/news/images/gpargonodes.png b/preview-fall2024-info/news/images/gpargonodes.png
new file mode 100644
index 000000000..8860fd951
Binary files /dev/null and b/preview-fall2024-info/news/images/gpargonodes.png differ
diff --git a/preview-fall2024-info/news/images/gpupflops.png b/preview-fall2024-info/news/images/gpupflops.png
new file mode 100644
index 000000000..9eceeb3f0
Binary files /dev/null and b/preview-fall2024-info/news/images/gpupflops.png differ
diff --git a/preview-fall2024-info/news/images/greenbay.jpg b/preview-fall2024-info/news/images/greenbay.jpg
new file mode 100644
index 000000000..379655784
Binary files /dev/null and b/preview-fall2024-info/news/images/greenbay.jpg differ
diff --git a/preview-fall2024-info/news/images/groom-2.jpg b/preview-fall2024-info/news/images/groom-2.jpg
new file mode 100644
index 000000000..adcfa1893
Binary files /dev/null and b/preview-fall2024-info/news/images/groom-2.jpg differ
diff --git a/preview-fall2024-info/news/images/groom-title.jpg b/preview-fall2024-info/news/images/groom-title.jpg
new file mode 100644
index 000000000..6d5f9879c
Binary files /dev/null and b/preview-fall2024-info/news/images/groom-title.jpg differ
diff --git a/preview-fall2024-info/news/images/groupphoto.png b/preview-fall2024-info/news/images/groupphoto.png
new file mode 100644
index 000000000..918fd7a03
Binary files /dev/null and b/preview-fall2024-info/news/images/groupphoto.png differ
diff --git a/preview-fall2024-info/news/images/hannahcard.png b/preview-fall2024-info/news/images/hannahcard.png
new file mode 100644
index 000000000..88ea89407
Binary files /dev/null and b/preview-fall2024-info/news/images/hannahcard.png differ
diff --git a/preview-fall2024-info/news/images/hannaheadshot.jpg b/preview-fall2024-info/news/images/hannaheadshot.jpg
new file mode 100644
index 000000000..9339d4fd2
Binary files /dev/null and b/preview-fall2024-info/news/images/hannaheadshot.jpg differ
diff --git a/preview-fall2024-info/news/images/hannahsisters.jpg b/preview-fall2024-info/news/images/hannahsisters.jpg
new file mode 100644
index 000000000..7f15c06ce
Binary files /dev/null and b/preview-fall2024-info/news/images/hannahsisters.jpg differ
diff --git a/preview-fall2024-info/news/images/hannalab.PNG b/preview-fall2024-info/news/images/hannalab.PNG
new file mode 100644
index 000000000..153b28649
Binary files /dev/null and b/preview-fall2024-info/news/images/hannalab.PNG differ
diff --git a/preview-fall2024-info/news/images/hannalab.jpg b/preview-fall2024-info/news/images/hannalab.jpg
new file mode 100644
index 000000000..72a4708f1
Binary files /dev/null and b/preview-fall2024-info/news/images/hannalab.jpg differ
diff --git a/preview-fall2024-info/news/images/header.jpeg b/preview-fall2024-info/news/images/header.jpeg
new file mode 100644
index 000000000..d2ed9376f
Binary files /dev/null and b/preview-fall2024-info/news/images/header.jpeg differ
diff --git a/preview-fall2024-info/news/images/headshot-use-of-osdf.png b/preview-fall2024-info/news/images/headshot-use-of-osdf.png
new file mode 100644
index 000000000..c64760721
Binary files /dev/null and b/preview-fall2024-info/news/images/headshot-use-of-osdf.png differ
diff --git a/preview-fall2024-info/news/images/high-resolution/User_School_Collage.png b/preview-fall2024-info/news/images/high-resolution/User_School_Collage.png
new file mode 100644
index 000000000..6a23fa618
Binary files /dev/null and b/preview-fall2024-info/news/images/high-resolution/User_School_Collage.png differ
diff --git a/preview-fall2024-info/news/images/high-resolution/bat-carousel.png b/preview-fall2024-info/news/images/high-resolution/bat-carousel.png
new file mode 100644
index 000000000..2cd406095
Binary files /dev/null and b/preview-fall2024-info/news/images/high-resolution/bat-carousel.png differ
diff --git a/preview-fall2024-info/news/images/high-resolution/cow-ml-image-cropped.png b/preview-fall2024-info/news/images/high-resolution/cow-ml-image-cropped.png
new file mode 100644
index 000000000..b04bf5820
Binary files /dev/null and b/preview-fall2024-info/news/images/high-resolution/cow-ml-image-cropped.png differ
diff --git a/preview-fall2024-info/news/images/high-resolution/cow-ml-image.png b/preview-fall2024-info/news/images/high-resolution/cow-ml-image.png
new file mode 100644
index 000000000..b04bf5820
Binary files /dev/null and b/preview-fall2024-info/news/images/high-resolution/cow-ml-image.png differ
diff --git a/preview-fall2024-info/news/images/high-resolution/dr-wall.png b/preview-fall2024-info/news/images/high-resolution/dr-wall.png
new file mode 100644
index 000000000..a7447269d
Binary files /dev/null and b/preview-fall2024-info/news/images/high-resolution/dr-wall.png differ
diff --git a/preview-fall2024-info/news/images/high-resolution/noaa-banner.png b/preview-fall2024-info/news/images/high-resolution/noaa-banner.png
new file mode 100644
index 000000000..5fee62d8a
Binary files /dev/null and b/preview-fall2024-info/news/images/high-resolution/noaa-banner.png differ
diff --git a/preview-fall2024-info/news/images/horus-logo.png b/preview-fall2024-info/news/images/horus-logo.png
new file mode 100644
index 000000000..bc7c41858
Binary files /dev/null and b/preview-fall2024-info/news/images/horus-logo.png differ
diff --git a/preview-fall2024-info/news/images/htc24-collage.png b/preview-fall2024-info/news/images/htc24-collage.png
new file mode 100644
index 000000000..6fea1f4d4
Binary files /dev/null and b/preview-fall2024-info/news/images/htc24-collage.png differ
diff --git a/preview-fall2024-info/news/images/intro-collage.jpeg b/preview-fall2024-info/news/images/intro-collage.jpeg
new file mode 100644
index 000000000..99033237a
Binary files /dev/null and b/preview-fall2024-info/news/images/intro-collage.jpeg differ
diff --git a/preview-fall2024-info/news/images/j_ayars_profile_pic.jpg b/preview-fall2024-info/news/images/j_ayars_profile_pic.jpg
new file mode 100644
index 000000000..0c9843d32
Binary files /dev/null and b/preview-fall2024-info/news/images/j_ayars_profile_pic.jpg differ
diff --git a/preview-fall2024-info/news/images/jobsubmission.png b/preview-fall2024-info/news/images/jobsubmission.png
new file mode 100644
index 000000000..a3d6366ae
Binary files /dev/null and b/preview-fall2024-info/news/images/jobsubmission.png differ
diff --git a/preview-fall2024-info/news/images/jobsubmitfile.png b/preview-fall2024-info/news/images/jobsubmitfile.png
new file mode 100644
index 000000000..5f9b298a9
Binary files /dev/null and b/preview-fall2024-info/news/images/jobsubmitfile.png differ
diff --git a/preview-fall2024-info/news/images/joes-cat.jpg b/preview-fall2024-info/news/images/joes-cat.jpg
new file mode 100644
index 000000000..216a2bd31
Binary files /dev/null and b/preview-fall2024-info/news/images/joes-cat.jpg differ
diff --git a/preview-fall2024-info/news/images/jonBlank3[5] copy.jpg b/preview-fall2024-info/news/images/jonBlank3[5] copy.jpg
new file mode 100644
index 000000000..4a2910321
Binary files /dev/null and b/preview-fall2024-info/news/images/jonBlank3[5] copy.jpg differ
diff --git a/preview-fall2024-info/news/images/jonblank.jpg b/preview-fall2024-info/news/images/jonblank.jpg
new file mode 100644
index 000000000..4a2910321
Binary files /dev/null and b/preview-fall2024-info/news/images/jonblank.jpg differ
diff --git a/preview-fall2024-info/news/images/kayak-picture.png b/preview-fall2024-info/news/images/kayak-picture.png
new file mode 100644
index 000000000..c18a298b6
Binary files /dev/null and b/preview-fall2024-info/news/images/kayak-picture.png differ
diff --git a/preview-fall2024-info/news/images/kayaking.jpeg b/preview-fall2024-info/news/images/kayaking.jpeg
new file mode 100644
index 000000000..c82361578
Binary files /dev/null and b/preview-fall2024-info/news/images/kayaking.jpeg differ
diff --git a/preview-fall2024-info/news/images/kevin.jpeg b/preview-fall2024-info/news/images/kevin.jpeg
new file mode 100644
index 000000000..af711297c
Binary files /dev/null and b/preview-fall2024-info/news/images/kevin.jpeg differ
diff --git a/preview-fall2024-info/news/images/keynote-speaking.png b/preview-fall2024-info/news/images/keynote-speaking.png
new file mode 100644
index 000000000..8a52599e1
Binary files /dev/null and b/preview-fall2024-info/news/images/keynote-speaking.png differ
diff --git a/preview-fall2024-info/news/images/lacysnapshot.jpg b/preview-fall2024-info/news/images/lacysnapshot.jpg
new file mode 100644
index 000000000..07af2e024
Binary files /dev/null and b/preview-fall2024-info/news/images/lacysnapshot.jpg differ
diff --git a/preview-fall2024-info/news/images/ligo20160211d-smaller-150x150.jpg b/preview-fall2024-info/news/images/ligo20160211d-smaller-150x150.jpg
new file mode 100644
index 000000000..914b2e619
Binary files /dev/null and b/preview-fall2024-info/news/images/ligo20160211d-smaller-150x150.jpg differ
diff --git a/preview-fall2024-info/news/images/lindleyimage.png b/preview-fall2024-info/news/images/lindleyimage.png
new file mode 100644
index 000000000..0fbedafeb
Binary files /dev/null and b/preview-fall2024-info/news/images/lindleyimage.png differ
diff --git a/preview-fall2024-info/news/images/lombardihiking.png b/preview-fall2024-info/news/images/lombardihiking.png
new file mode 100644
index 000000000..5a0e3c57b
Binary files /dev/null and b/preview-fall2024-info/news/images/lombardihiking.png differ
diff --git a/preview-fall2024-info/news/images/lombardiuganda.png b/preview-fall2024-info/news/images/lombardiuganda.png
new file mode 100644
index 000000000..3eb9dac1b
Binary files /dev/null and b/preview-fall2024-info/news/images/lombardiuganda.png differ
diff --git a/preview-fall2024-info/news/images/maizespalding.jpg b/preview-fall2024-info/news/images/maizespalding.jpg
new file mode 100644
index 000000000..85b0750fe
Binary files /dev/null and b/preview-fall2024-info/news/images/maizespalding.jpg differ
diff --git a/preview-fall2024-info/news/images/mars-image.jpg b/preview-fall2024-info/news/images/mars-image.jpg
new file mode 100644
index 000000000..e7e8232f5
Binary files /dev/null and b/preview-fall2024-info/news/images/mars-image.jpg differ
diff --git a/preview-fall2024-info/news/images/materials-science.jpg b/preview-fall2024-info/news/images/materials-science.jpg
new file mode 100644
index 000000000..22501de3b
Binary files /dev/null and b/preview-fall2024-info/news/images/materials-science.jpg differ
diff --git a/preview-fall2024-info/news/images/mattchristie.png b/preview-fall2024-info/news/images/mattchristie.png
new file mode 100644
index 000000000..5dda7d777
Binary files /dev/null and b/preview-fall2024-info/news/images/mattchristie.png differ
diff --git a/preview-fall2024-info/news/images/mental_health.jpeg b/preview-fall2024-info/news/images/mental_health.jpeg
new file mode 100644
index 000000000..0f05402a8
Binary files /dev/null and b/preview-fall2024-info/news/images/mental_health.jpeg differ
diff --git a/preview-fall2024-info/news/images/mental_health_banner.jpeg b/preview-fall2024-info/news/images/mental_health_banner.jpeg
new file mode 100644
index 000000000..610d7e910
Binary files /dev/null and b/preview-fall2024-info/news/images/mental_health_banner.jpeg differ
diff --git a/preview-fall2024-info/news/images/miron-livny-2021.jpeg b/preview-fall2024-info/news/images/miron-livny-2021.jpeg
new file mode 100644
index 000000000..2bc48838b
Binary files /dev/null and b/preview-fall2024-info/news/images/miron-livny-2021.jpeg differ
diff --git a/preview-fall2024-info/news/images/miron-speaking.png b/preview-fall2024-info/news/images/miron-speaking.png
new file mode 100644
index 000000000..3f86ad8db
Binary files /dev/null and b/preview-fall2024-info/news/images/miron-speaking.png differ
diff --git a/preview-fall2024-info/news/images/mironlivny.jpg b/preview-fall2024-info/news/images/mironlivny.jpg
new file mode 100644
index 000000000..bba9b716f
Binary files /dev/null and b/preview-fall2024-info/news/images/mironlivny.jpg differ
diff --git a/preview-fall2024-info/news/images/mldemowhatspossible at 1.05.07 PM.png b/preview-fall2024-info/news/images/mldemowhatspossible at 1.05.07 PM.png
new file mode 100644
index 000000000..e6cb99a60
Binary files /dev/null and b/preview-fall2024-info/news/images/mldemowhatspossible at 1.05.07 PM.png differ
diff --git a/preview-fall2024-info/news/images/mldemowhatspossible.png b/preview-fall2024-info/news/images/mldemowhatspossible.png
new file mode 100644
index 000000000..e6cb99a60
Binary files /dev/null and b/preview-fall2024-info/news/images/mldemowhatspossible.png differ
diff --git a/preview-fall2024-info/news/images/molcryst.png b/preview-fall2024-info/news/images/molcryst.png
new file mode 100644
index 000000000..a8b2ebfd2
Binary files /dev/null and b/preview-fall2024-info/news/images/molcryst.png differ
diff --git a/preview-fall2024-info/news/images/mrion-talking.jpeg b/preview-fall2024-info/news/images/mrion-talking.jpeg
new file mode 100644
index 000000000..638f5e518
Binary files /dev/null and b/preview-fall2024-info/news/images/mrion-talking.jpeg differ
diff --git a/preview-fall2024-info/news/images/neha.jpeg b/preview-fall2024-info/news/images/neha.jpeg
new file mode 100644
index 000000000..3579f0938
Binary files /dev/null and b/preview-fall2024-info/news/images/neha.jpeg differ
diff --git a/preview-fall2024-info/news/images/new-award.jpeg b/preview-fall2024-info/news/images/new-award.jpeg
new file mode 100644
index 000000000..76acd234d
Binary files /dev/null and b/preview-fall2024-info/news/images/new-award.jpeg differ
diff --git a/preview-fall2024-info/news/images/noaa-banner.png b/preview-fall2024-info/news/images/noaa-banner.png
new file mode 100644
index 000000000..5fee62d8a
Binary files /dev/null and b/preview-fall2024-info/news/images/noaa-banner.png differ
diff --git a/preview-fall2024-info/news/images/nrao-clean-image.png b/preview-fall2024-info/news/images/nrao-clean-image.png
new file mode 100644
index 000000000..70e233135
Binary files /dev/null and b/preview-fall2024-info/news/images/nrao-clean-image.png differ
diff --git a/preview-fall2024-info/news/images/nrao-dirty-image.png b/preview-fall2024-info/news/images/nrao-dirty-image.png
new file mode 100644
index 000000000..1c5195445
Binary files /dev/null and b/preview-fall2024-info/news/images/nrao-dirty-image.png differ
diff --git a/preview-fall2024-info/news/images/nrao-map-clear.jpeg b/preview-fall2024-info/news/images/nrao-map-clear.jpeg
new file mode 100644
index 000000000..1c67a4fd3
Binary files /dev/null and b/preview-fall2024-info/news/images/nrao-map-clear.jpeg differ
diff --git a/preview-fall2024-info/news/images/nrao-vla.png b/preview-fall2024-info/news/images/nrao-vla.png
new file mode 100644
index 000000000..1236fa431
Binary files /dev/null and b/preview-fall2024-info/news/images/nrao-vla.png differ
diff --git a/preview-fall2024-info/news/images/nrao_chtc_collab_map.jpeg b/preview-fall2024-info/news/images/nrao_chtc_collab_map.jpeg
new file mode 100644
index 000000000..47ad49f92
Binary files /dev/null and b/preview-fall2024-info/news/images/nrao_chtc_collab_map.jpeg differ
diff --git a/preview-fall2024-info/news/images/nsf-speaking.png b/preview-fall2024-info/news/images/nsf-speaking.png
new file mode 100644
index 000000000..9b7cdd549
Binary files /dev/null and b/preview-fall2024-info/news/images/nsf-speaking.png differ
diff --git a/preview-fall2024-info/news/images/nt.jpeg b/preview-fall2024-info/news/images/nt.jpeg
new file mode 100644
index 000000000..2c76d1473
Binary files /dev/null and b/preview-fall2024-info/news/images/nt.jpeg differ
diff --git a/preview-fall2024-info/news/images/numgpus.png b/preview-fall2024-info/news/images/numgpus.png
new file mode 100644
index 000000000..4e4523762
Binary files /dev/null and b/preview-fall2024-info/news/images/numgpus.png differ
diff --git a/preview-fall2024-info/news/images/osg-school-2023-01.jpg b/preview-fall2024-info/news/images/osg-school-2023-01.jpg
new file mode 100644
index 000000000..66f806ba2
Binary files /dev/null and b/preview-fall2024-info/news/images/osg-school-2023-01.jpg differ
diff --git a/preview-fall2024-info/news/images/osgmap.png b/preview-fall2024-info/news/images/osgmap.png
new file mode 100644
index 000000000..10a205cb2
Binary files /dev/null and b/preview-fall2024-info/news/images/osgmap.png differ
diff --git a/preview-fall2024-info/news/images/osgschool2023 b/preview-fall2024-info/news/images/osgschool2023
new file mode 100644
index 000000000..02db855a4
Binary files /dev/null and b/preview-fall2024-info/news/images/osgschool2023 differ
diff --git a/preview-fall2024-info/news/images/osgschool2023.png b/preview-fall2024-info/news/images/osgschool2023.png
new file mode 100644
index 000000000..cf5392979
Binary files /dev/null and b/preview-fall2024-info/news/images/osgschool2023.png differ
diff --git a/preview-fall2024-info/news/images/ospool-comp.jpg b/preview-fall2024-info/news/images/ospool-comp.jpg
new file mode 100644
index 000000000..842cb536e
Binary files /dev/null and b/preview-fall2024-info/news/images/ospool-comp.jpg differ
diff --git a/preview-fall2024-info/news/images/ospool-con-map.png b/preview-fall2024-info/news/images/ospool-con-map.png
new file mode 100644
index 000000000..7ef0923fd
Binary files /dev/null and b/preview-fall2024-info/news/images/ospool-con-map.png differ
diff --git a/preview-fall2024-info/news/images/pekowsy-150x150.jpeg b/preview-fall2024-info/news/images/pekowsy-150x150.jpeg
new file mode 100644
index 000000000..3208bc702
Binary files /dev/null and b/preview-fall2024-info/news/images/pekowsy-150x150.jpeg differ
diff --git a/preview-fall2024-info/news/images/pnas.2312909120fig01.jpg b/preview-fall2024-info/news/images/pnas.2312909120fig01.jpg
new file mode 100644
index 000000000..aca51b81a
Binary files /dev/null and b/preview-fall2024-info/news/images/pnas.2312909120fig01.jpg differ
diff --git a/preview-fall2024-info/news/images/pp.jpeg b/preview-fall2024-info/news/images/pp.jpeg
new file mode 100644
index 000000000..61ea60870
Binary files /dev/null and b/preview-fall2024-info/news/images/pp.jpeg differ
diff --git a/preview-fall2024-info/news/images/pratham.jpeg b/preview-fall2024-info/news/images/pratham.jpeg
new file mode 100644
index 000000000..7d9bb561a
Binary files /dev/null and b/preview-fall2024-info/news/images/pratham.jpeg differ
diff --git a/preview-fall2024-info/news/images/presentations.jpeg b/preview-fall2024-info/news/images/presentations.jpeg
new file mode 100644
index 000000000..698677d1a
Binary files /dev/null and b/preview-fall2024-info/news/images/presentations.jpeg differ
diff --git a/preview-fall2024-info/news/images/rachellombardi.jpg b/preview-fall2024-info/news/images/rachellombardi.jpg
new file mode 100644
index 000000000..acac1723d
Binary files /dev/null and b/preview-fall2024-info/news/images/rachellombardi.jpg differ
diff --git a/preview-fall2024-info/news/images/recordcores.png b/preview-fall2024-info/news/images/recordcores.png
new file mode 100644
index 000000000..2264cee59
Binary files /dev/null and b/preview-fall2024-info/news/images/recordcores.png differ
diff --git a/preview-fall2024-info/news/images/research-computing-partnership.jpeg b/preview-fall2024-info/news/images/research-computing-partnership.jpeg
new file mode 100644
index 000000000..a5fedcd96
Binary files /dev/null and b/preview-fall2024-info/news/images/research-computing-partnership.jpeg differ
diff --git a/preview-fall2024-info/news/images/researchcomp-pelican.jpg b/preview-fall2024-info/news/images/researchcomp-pelican.jpg
new file mode 100644
index 000000000..8e9539fbf
Binary files /dev/null and b/preview-fall2024-info/news/images/researchcomp-pelican.jpg differ
diff --git a/preview-fall2024-info/news/images/resilience-hero-large.jpeg b/preview-fall2024-info/news/images/resilience-hero-large.jpeg
new file mode 100644
index 000000000..394ebebf6
Binary files /dev/null and b/preview-fall2024-info/news/images/resilience-hero-large.jpeg differ
diff --git a/preview-fall2024-info/news/images/robotsoccer.jpeg b/preview-fall2024-info/news/images/robotsoccer.jpeg
new file mode 100644
index 000000000..874304c3b
Binary files /dev/null and b/preview-fall2024-info/news/images/robotsoccer.jpeg differ
diff --git a/preview-fall2024-info/news/images/school.png b/preview-fall2024-info/news/images/school.png
new file mode 100644
index 000000000..02db855a4
Binary files /dev/null and b/preview-fall2024-info/news/images/school.png differ
diff --git a/preview-fall2024-info/news/images/sidebyside.jpg b/preview-fall2024-info/news/images/sidebyside.jpg
new file mode 100644
index 000000000..9fefe67c4
Binary files /dev/null and b/preview-fall2024-info/news/images/sidebyside.jpg differ
diff --git a/preview-fall2024-info/news/images/sites-use-of-osdf.png b/preview-fall2024-info/news/images/sites-use-of-osdf.png
new file mode 100644
index 000000000..77daa15d2
Binary files /dev/null and b/preview-fall2024-info/news/images/sites-use-of-osdf.png differ
diff --git a/preview-fall2024-info/news/images/spaldinglab.jpg b/preview-fall2024-info/news/images/spaldinglab.jpg
new file mode 100644
index 000000000..d1b2a3216
Binary files /dev/null and b/preview-fall2024-info/news/images/spaldinglab.jpg differ
diff --git a/preview-fall2024-info/news/images/submittingjobs.png b/preview-fall2024-info/news/images/submittingjobs.png
new file mode 100644
index 000000000..96f7feeb5
Binary files /dev/null and b/preview-fall2024-info/news/images/submittingjobs.png differ
diff --git a/preview-fall2024-info/news/images/terawagner.jpg b/preview-fall2024-info/news/images/terawagner.jpg
new file mode 100644
index 000000000..156482911
Binary files /dev/null and b/preview-fall2024-info/news/images/terawagner.jpg differ
diff --git a/preview-fall2024-info/news/images/todd-t-article-0.jpg b/preview-fall2024-info/news/images/todd-t-article-0.jpg
new file mode 100644
index 000000000..f828a121f
Binary files /dev/null and b/preview-fall2024-info/news/images/todd-t-article-0.jpg differ
diff --git a/preview-fall2024-info/news/images/todd-t-article-1.png b/preview-fall2024-info/news/images/todd-t-article-1.png
new file mode 100644
index 000000000..bc7fd09d0
Binary files /dev/null and b/preview-fall2024-info/news/images/todd-t-article-1.png differ
diff --git a/preview-fall2024-info/news/images/todd-t-article-2.jpg b/preview-fall2024-info/news/images/todd-t-article-2.jpg
new file mode 100644
index 000000000..d41710a5d
Binary files /dev/null and b/preview-fall2024-info/news/images/todd-t-article-2.jpg differ
diff --git a/preview-fall2024-info/news/images/todd-t-article-6.jpg b/preview-fall2024-info/news/images/todd-t-article-6.jpg
new file mode 100644
index 000000000..d6760216b
Binary files /dev/null and b/preview-fall2024-info/news/images/todd-t-article-6.jpg differ
diff --git a/preview-fall2024-info/news/images/tree.jpg b/preview-fall2024-info/news/images/tree.jpg
new file mode 100644
index 000000000..2c0f8e391
Binary files /dev/null and b/preview-fall2024-info/news/images/tree.jpg differ
diff --git a/preview-fall2024-info/news/images/ucsd-public-relations.png b/preview-fall2024-info/news/images/ucsd-public-relations.png
new file mode 100644
index 000000000..e76d80ca2
Binary files /dev/null and b/preview-fall2024-info/news/images/ucsd-public-relations.png differ
diff --git a/preview-fall2024-info/news/images/unnamed_01.png b/preview-fall2024-info/news/images/unnamed_01.png
new file mode 100644
index 000000000..fa6d90b52
Binary files /dev/null and b/preview-fall2024-info/news/images/unnamed_01.png differ
diff --git a/preview-fall2024-info/news/images/veritas_1.png b/preview-fall2024-info/news/images/veritas_1.png
new file mode 100644
index 000000000..a9eda03a9
Binary files /dev/null and b/preview-fall2024-info/news/images/veritas_1.png differ
diff --git a/preview-fall2024-info/news/images/veritas_2.png b/preview-fall2024-info/news/images/veritas_2.png
new file mode 100644
index 000000000..857e73cc3
Binary files /dev/null and b/preview-fall2024-info/news/images/veritas_2.png differ
diff --git a/preview-fall2024-info/news/images/vla-hubble-ultra-deep.png b/preview-fall2024-info/news/images/vla-hubble-ultra-deep.png
new file mode 100644
index 000000000..1236fa431
Binary files /dev/null and b/preview-fall2024-info/news/images/vla-hubble-ultra-deep.png differ
diff --git a/preview-fall2024-info/news/images/wilcram.jpeg b/preview-fall2024-info/news/images/wilcram.jpeg
new file mode 100644
index 000000000..e9aed2abf
Binary files /dev/null and b/preview-fall2024-info/news/images/wilcram.jpeg differ
diff --git a/preview-fall2024-info/news/robotsoccer.jpeg b/preview-fall2024-info/news/robotsoccer.jpeg
new file mode 100644
index 000000000..874304c3b
Binary files /dev/null and b/preview-fall2024-info/news/robotsoccer.jpeg differ
diff --git a/preview-fall2024-info/newsletter.html b/preview-fall2024-info/newsletter.html
new file mode 100644
index 000000000..6a351693d
--- /dev/null
+++ b/preview-fall2024-info/newsletter.html
@@ -0,0 +1,471 @@
+
+
+
+
+
+
+CHTC Newsletter
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ CHTC Newsletter
+
+
+ The CHTC Newsletter is a quarterly email that includes information about upcoming events,
+ training opportunities, and other news from the CHTC. If you would like to receive
+ the CHTC Newsletter, please fill out the form below.
+
+
+
+
+
+
+
+
+
+
+
+ NOAA funded marine scientist uses OSPool access to high throughput computing to explode her boundaries of research
+
+
Dr. Carrie Wall, a research scientist at the University of Colorado Boulder, shares how access to OSPool resources has allowed her team to expand the scope of their research and to fail, unconstrained by the cost of computing in the cloud and the associated restraints that places on research.
+
+
+ +Water column sonar data collected on the NOAA Okeanos Explorer in the North Atlantic Ocean: One of NOAA’s research cruises.
+
+Water column sonar data collected on the NOAA Okeanos Explorer in the North Atlantic Ocean: One of NOAA’s research cruises.
+
+
+
A marine scientist faced the daunting challenge of processing sonar data from 65 research cruises spanning 20 years, totaling over 100,000 files. The researcher, Dr. Carrie Wall, braced herself for a grueling 30-week endeavor of single stream, desktop-based processing. However, good fortune intervened at a National Discovery Cloud for Climate (NDC-C) conference in January 2024 when she crossed paths with Brian Bockelman, the principal investigator (PI) of the Pelican Project and a Co-PI of the PATh Project.
+
+
Wall discussed with Bockelman the challenges of converting decades’ worth of sonar datasets into a format suitable for AI analysis —a crucial step for her NSF-funded project through the NDC-C. This initiative aimed to develop the cyberinfrastructure essential for implementing scalable self-supervised machine learning on the extensive water column sonar data accumulated over the years.“We all went around and did five-minute presentations explaining ‘here’s what I do, here’s what I work on,’ almost like speed dating,” recounted Bockelman. “Listening to her talk, it was like, ‘this is a great high throughput computing example.’” Recognizing the volume of Wall’s project, Bockelman introduced her to the OSPool, a shared computing resource freely available to researchers affiliated with US academic institutions. He observed that Wall’s computing style aligned seamlessly with OSPool’s capabilities and would address Wall’s sonar processing bottleneck.
+
+
With Bockelman’s encouragement, Wall and her team’s software developer, Rudy Klucik, easily created accounts and began modifying their computing workflow for high throughput computing.”The process was super easy and very accommodating. Rachel Lombardi, a Research Computing Facilitator for the Center for High Throughput Computing, walked me through all the details, answered my technical questions, and was very welcoming. It was a really nice onboarding,” enthused Klucik. What followed was nothing short of a paradigm shift.
+
+
+  + CIRES research scientist Dr. Carrie Wall
+
+ CIRES research scientist Dr. Carrie Wall
+
+
+
Within the walls of the University of Colorado Boulder lies CIRES: The Cooperative Institute for Research in Environmental Sciences, a partnership between the National Oceanic and Atmospheric Administration (NOAA) and the university itself. CIRES employs a workforce of over 800 scientists and staff, actively involved in various aspects of NOAA’s mission-critical endeavors. Among them are Wall and Klucik, both members of NOAA’s team. NOAA’s mission centers on supporting healthy oceans. Dr. Wall has dedicated the past 11 years to leading the development of national archives for water column sonar data, a task undertaken through the National Centers for Environmental Information (NCEI), NOAA’s archival arm.
+
+
Wall and her team have archived over 280TB of water column sonar data at NCEI, which serves not only NOAA’s scientists but also other agencies and academic institutions. However, there was a significant issue: it existed solely in its native, proprietary, and exceedingly complex industry format. Despite being hosted on Amazon Web Services (AWS) for accessibility, as Wall explained, “a lot of expert knowledge is needed to even open and read these files.”
+
+
“NOAA scientists, mostly from the National Marine Fisheries Service (NOAA Fisheries), have collected in all U.S. waters - from the Arctic Ocean to the Caribbean and off the entire U.S. coastline. In just the Gulf of Maine alone, NOAA Fisheries scientists have collected over 20 years of data going back to 1998. All of these data have been archived so not only do we have a very large volume of data, but also a very long time series covering critical habitats,” Wall explained. “The majority of these fascinating data have been used to support fishery stock assessments,” Wall emphasized. “There’s a lot of these data, and in collaboration with experts we want to find out more about them.”
+
+
With the help of the OSPool, Klucik has been able to successfully develop a workflow that he now executes smoothly. This involves reading files from an AWS bucket, processing and converting them into a cloud native Zarr format, and then writing that data out to a publicly accessible bucket, available under the NOAA Open Data Dissemination program. Wall added, “This will now serve as our input for the AI model.”
+
+
Before discovering the OSPool, the original plan was to “fully utilize a cloud native processing pipeline, mainly composed of AWS Lambdas to do all our data conversion” described Klucik. “One common misconception is that cloud computing is cheaper than traditional computing, if the technical elements are all aligned properly it can be cheap, but in a lot of situations it’s still extremely expensive; in the back of our minds we were afraid that it might even be cost prohibitive to process the archive as a whole.”
+
+
However, it is important to acknowledge that before being set up with the OSPool, there was a bit of a learning curve. “I was completely new to high throughput computing infrastructure and didn’t understand how the processing worked,” recalled Klucik. “So, a lot of my initial time was spent running ‘hello world’ examples to better understand the functionality. I started with one job, then scaled up to 100 and eventually 1,000 to get the concurrency we were looking to achieve. It involved a lot of trial and error to get everything right. It took about a month before I finally managed to run the full catalog of data properly.” Klucik noted that he was aware of the available resources, saying, “The OSPool documentation served as an invaluable resource for getting me oriented in a new computing environment.”
+
+
Although initially tasked with processing 100,000 files, their workflow using the OSPool has since surged beyond 400,000 files—an accomplishment that would have been financially daunting in a traditional cloud environment. Wall emphasized that “what OSPool has allowed us to do is fail, which is really, really good. Before [using the] OSPool, we started by processing a couple of cruises with a very small number of files in the cloud to be cost-effective; we didn’t want to make costly mistakes. Being able to use OSPool to iterate and strengthen our process, allowed us to then scale to the volume of data and number of files that we need to process. I don’t know where we would be without OSPool but it would’ve cost us tens of thousands of dollars. We didn’t have to sacrifice for a lesser workflow, one that we didn’t improve upon because it would have cost us more money. I’m really excited about where OSPool has allowed us to go, and now we can take that next step to say ‘okay, we have our foundation, which is our data and a great format, and we can build our models and additional workflows.’”
+
+
Wall’s testimony underscores OSPool’s role not just as computing capacity but as a catalyst for innovation, enabling teams to push boundaries and realize their full potential in research and model development.
+
+
+
+
+
+
+
+ Through the use of high throughput computing, NRAO delivers one of the deepest radio images of space
+
+
The National Radio Astronomy Observatory’s collaboration with the NSF-funded Partnership to Advance Throughput Computing
+(PATh; NSF grant #2030508) and the Pelican Project (NSF grant #2331480) leads to successfully imaged deep
+space and creates a first-of-its-kind nationally distributed workflow model for data-intensive scientific investigations.
+
+
Ten years ago, the National Radio Astronomy Observatory (NRAO) pointed its
+Very Large Array (VLA) telescopes toward a well-studied portion of the sky, searching for the
+oldest view of the universe. Deeper structures reflect older structures in space, as their light takes longer to travel through space
+and be picked up by telescopes. Radio astronomy can go even further, detecting structures beyond visible light. The VLA
+telescopes generated enough data that a single image of a portion of the sky resulted in two terabytes of data. Without the
+computing capacity to image the complete data set, it sat largely unprocessed — until now.
+
+
Researchers at NRAO knew that attempting to process this entire data set in-house was impractical. A previous computing run in 2016 using only a subset of this data took nearly two weeks of active processing. The high sensitivity of radio images requires a vast amount of computing to reach a final product, noted Felipe Madsen, an
+NRAO software engineer. The VLA telescopes are interferometers, meaning they point two antennas at the same portion of the
+sky; the differences in what these antennas provide eventually result in an image, Madsen explains. NRAO models and re-models
+the data to decrease the noise level until the noise is indistinguishable from structures in space. “This project is a lot
+more data-intensive than most other projects,” Madsen said.
+
+
Curious about how high-throughput computing (HTC) could enhance its capacity to process data from the VLA, NRAO joined
+forces with the Center for High Throughput Computing (CHTC) in 2018. After learning about
+what HTC could accomplish, NRAO began executing trial runs in 2019, experimenting with HTC. “Four years ago, we were
+beginning to use GPU software to process our data,” Madsen explained. “From the beginning, we understood that to be
+compatible with HTC we needed to make changes to our systems.”
+
+
Each team learned from and made improvements based on insights from each other. Greg Thain,
+an HTCondor Core Developer for the CHTC, met with NRAO weekly to discuss HTC and changes both parties
+could make. These weekly meetings resulted in the HTCondor team making changes to the software, eventually improving the
+experience of other users, he said. OSG Software Area Coordinator of CHTC Brian Lin
+helped NRAO manage their distributed infrastructure of resources across the country and transition workflows from CPUs to GPUs
+to make their workflows more compatible with HTC. Through distributed HTC, NRAO was able to run workflows across the country through the
+Open Science Pool (OSPool) and
+PATh Facility.
+
+
At NRAO, Madsen developed the software to interface the scientific software in the LibRA package
+developed by NRAO Algorithms Research & Development Group with the CHTC infrastructure software. This separation of software
+allowed the two teams to solve problems that arose in real-time as the data began to transfer across sites nationwide.
+
+
By December 2023, both parties were ready to tackle the VLA telescope deep sky data using HTC. Transitioning workflows to
+nationwide resources led to data movement issues, struggling to move efficiently from distributed resources. The December
+2023 image processing run relied upon resources from the Open Science Data Federation
+(OSDF) and the recently funded Pelican Project to speed up data
+transfers across sites. Brian Bockelman, PI of the
+Pelican Project, and his team helped NRAO improve data movement using the OSDF. “Both teams
+were working to solve problems as they were happening,” Madsen recounted. “That made for a very successful collaboration
+in this process.”
+
+
+  + The final product, looking into deep space.
+ The final product, looking into deep space.
+
+
+
Ultimately, the imaging process was 300 times faster than without using HTC, NRAO reported in
+a press release describing
+the project. What had previously taken two weeks now took only two hours to create the final result. The final image turned nine terabytes of data into a single
+product of one gigabyte. By
+the end, the collaboration resulted in one of the earliest radio images of the
+Hubble Ultra Deep Field.
+
+
The collaboration that led to this imaging is even bigger than NRAO and CHTC.
+The OSPool, which provided some of the computing capacity for the project,
+is supported by campuses and institutions across the country that share their excess capacity with the pool
+that NRAO utilized. For this project, 13 campuses contributed computing capacity, from small institutions
+like Emporia State University to larger ones like San Diego State University.
+
+
+ 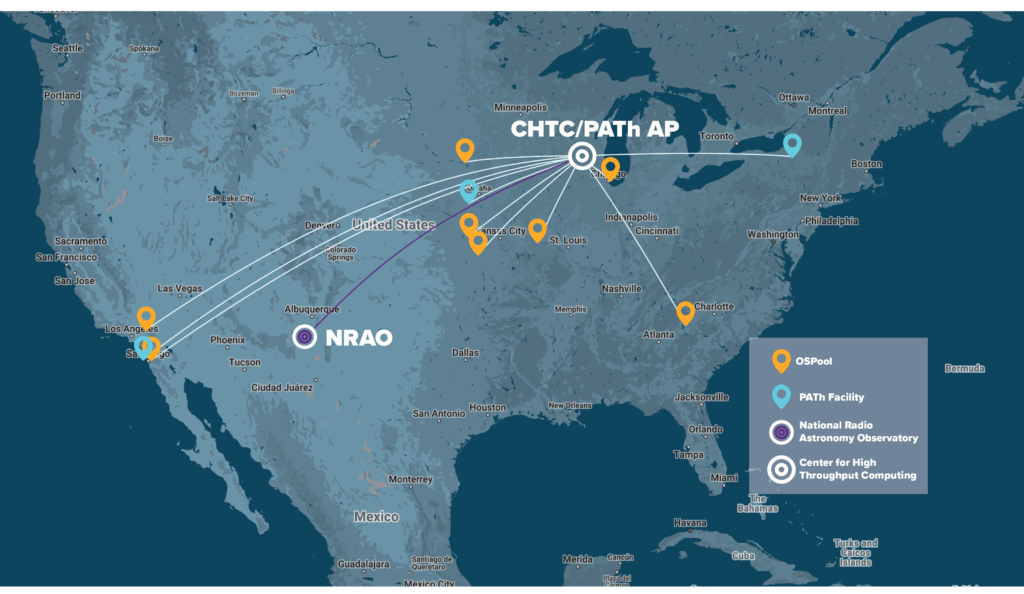 + A map of contributors across the OSPool and PATh Facility. Image courtesy of S. Dagnello, NRAO/AUI/NSF
+
+ A map of contributors across the OSPool and PATh Facility. Image courtesy of S. Dagnello, NRAO/AUI/NSF
+
+
+
+
+
The December 2023 run and the working relationship between CHTC and NRAO revolutionized information available to astronomers
+and proved that HTC is a viable option for the field. “It’s useful to do this run once. What’s exciting is doing it
+30,000 times for the entire sky,” Bockelman said. Although previous radio astronomy imaging workflows utilized HTC,
+this run was the first to image data on a distributed workflow nationwide from start to finish. Moving forward, NRAO
+and CHTC will continue covering the entire area of the sky seen by the VLA telescopes.
+
+
Madsen is enthusiastic about continuing this project, and how the use of HTC is revolutionizing astronomy, “I’ve always felt
+like, in this project, we are at the cutting edge of the current knowledge for making this kind of imaging.
+On the astronomy side, we can access a lot of new information with this image,” he said. “We have also imaged a data set that was
+previously impractical to image.”
+
+
+
+
+
+
+
+ OSG helps LIGO scientists confirm Einstein's unproven theory
+
+
Albert Einstein first posed the idea of gravitational waves in his general theory of relativity just over a century ago. But until now, they had never been observed directly. For the first time, scientists with the Laser Interferometer Gravitational-Wave Observatory (LIGO) Scientific Collaboration (LSC) have observed ripples in the fabric of spacetime called gravitational waves.
+
+
+
+
+

+
+
Image Courtesy Caltech/MIT/LIGO Laboratory
+
+
+
+
LIGO consists of two observatories within the United States—one in Hanford, Washington and the other in Livingston, Louisiana—separated by 1,865 miles. LIGO’s detectors search for gravitational waves from deep space. With two detectors, researchers can use differences in the wave’s arrival times to constrain the source location in the sky. LIGO’s first data run of its advanced gravitational wave detectors began in September 2015 and ran through January 12, 2016. The first gravitational waves were detected on September 14, 2015 by both detectors.
+
+
The LIGO project employs many concepts that the OSG promotes—resource sharing, aggregating opportunistic use across a variety of resources—and adds two twists: First, this experiment ran across LIGO Data Grid (LDG), OSPool and Extreme Science and Engineering Discovery Environment (XSEDE)-based resources, all managed from a single HTCondor-based system to take advantage of dedicated LDG, opportunistic OSG and NSF eXtreme Digital (XD) allocations. Second, workflows analyzing LIGO detector data proved more data-intensive than many opportunistic OSG workflows. Despite these challenges, LIGO scientists were able to manage workflows with the same tools they use to run on dedicated LDG systems—Pegasus and HTCondor.
+
+
Peter Couvares, data analysis computing manager for the Advanced LIGO project at Caltech, specializes in distributed computing problems. He and colleagues James Clark (Georgia Tech) and Larne Pekowsky (Syracuse University) explained LIGO’s computing needs and environment: The main focus is on optimization of data analysis codes, where optimization is broadly defined to encompass the overall performance and efficiency of their computing. While they use traditional optimization techniques to make things run faster, they also pursue more efficient resource management, and opportunistic resources—if there are computers available, they try to use them—thus the collaboration with OSG.
+
+
+
+
+

+
+
Peter Couvares, courtesy photo
+
+
+
+

+
+
James Clark, courtesy photo
+
+
+
+

+
+
Larne Pekowsky, courtesy photo
+
+
+
+
“When a workflow might consist of 600,000 jobs, we don’t want to rerun them if we make a mistake. So we use DAGMan (Directed Acyclic Graph Manager, a meta-scheduler for HTCondor) and Pegasus workflow manager to optimize changes,” added Couvares. “The combination of Pegasus, Condor, and OSG work great together.” Keeping track of what has run and how the workflow progresses, Pegasus translates the abstract layer of what needs to be done into actual jobs for Condor, which then puts them out on OSG.
+
+
The computing model
+
+
Since this work encompasses four types of computing – volunteer, dedicated, opportunistic (OSG), and allocated (XSEDE XD via OSG) – everything needs to be very efficient. Couvares helps with coordination, Pekowsky with optimization, and Clark with using OSG. In particular, OSG also enabled access to allocation-based resources from XSEDE. Allocations allow LIGO to get fixed amounts of time on dedicated NSF-funded supercomputers Comet and Stampede. While Stampede looks and behaves very much like a traditional supercomputer resource (batch, login node, shared file system), Comet has a new virtualization-based interface that eliminates the need to submit to a batch system. OSG provides this through a virtual machine (VM) image, then LIGO simply uses the OSG environment.
+
+
LIGO consumed 3,956,910 hours on OSG, out of which 628,602 hours were on the Comet and 430,960 on the Stampede XD resources. OSG’s Brian Bockelman (University of Nebraska-Lincoln) and Edgar Fajardo (UC San Diego/San Diego Supercomputer Center) used HTCondor to help LIGO implement their Pegasus workflow transparently across 16 clusters at universities and national labs across the US, including on the NSF-funded Comet and Stampede supercomputers.
+
+
“Normally our computing is done on dedicated clusters on the LIGO Data Grid,” said Couvares, “but we are moving toward also using outside and more elastic resources like OSG. OSG allows more flexibility as we add in systems that aren’t part of our traditional dedicated systems. The combination of OSG for unusual or dynamic workloads, and the LIGO Data Grid for regular workloads keeping up with new observational data is very powerful. In addition, Berkely Open Infrastructure for Network Computer (BOINC) allows us to use volunteers’ home computers when they are idle, running Pulsar searches around the world in the Einstein@Home project (E@H). The aggregated cycles from E@H are quite large but it is well-suited to only some kinds of searches where a computer must process a smaller amount of data for a longer amount of time.” We must rely on traditional HTC resources for our data-intensive analysis codes.
+
+
LIGO codes cannot all run as-is on OSG. The majority of codes are highly optimized for the LDG environment, so they identified the most compute-intensive and high science priority code to run on OSG. Of about 100 different data analysis codes, only a small handful are running on OSG so far. However, the research team started with the hardest code, their highest priority, which means they are now doing some of LIGO’s most important computing on OSG. Other low latency codes must run on dedicated local resources where they might need to be done in seconds or minutes.
+
+
“It is important that LIGO has a broad set of resources and an increasingly diverse set of resources. OSG is almost like a universal adapter for us,” said Couvares. “It is very powerful, users don’t need to care where a job runs, and it is another step toward that old promise of grid computing.
+
+
The importance of OSG and NSF support
+
+
Using data analysis run on the OSG, the LIGO team looked for a compact binary coalescence, that is, the merger of binary neutron stars or black holes. Couvares called it a modeled search—they have a signal that they believe is a strong indicator, they know what it’s going to look like, and they have optimal match filters to compare data with the signal they expect. But the search is computationally expensive because it’s not just one signal they are looking for: The parameters of the source may change or the objects may spin differently. The degree of match requires a search on the order of 100,000 different models/waveforms. This makes the OSG very valuable, because it can split up many of the match filters.
+
+
“The parallel nature of the OSG is what’s valuable,” said Couvares. “It is well suited to a high throughput environment. We would like to use more OSG resources because we could expand the parameter space of our searches beyond what is possible with dedicated resources. We need two things, really. We obviously need resources, but we also need people who can be a bridge between the data analysts/scientists and the computing resources. Resources alone are not enough. LIGO will always need dedicated in-house computing for low latency searches that need to be done quickly, and for our steady-state offline computing, but now we have the potential elasticity of OSG.”
+
+
“The nature of our collaboration with OSG has been absolutely great for a number of reasons,” said Couvares. “The OSG people have been extremely helpful. They are really unselfish and technical. That’s not always there in the open-source world. The basic idea of OSG has been good for LIGO—their willingness OSG services for LIGO, to reduce the barrier to entry, setting up computing elements, and so on. The barrier otherwise would have been too high. We couldn’t be happier with our partnership.”
+
+
Another big step has been the increase in network speed. The data was cached at the University of Nebraska and streamed to on-demand worker nodes that are able to read from a common location. This project benefited greatly from the NSF’s Campus Cyberinfrastructure – Network Infrastructure and Engineering (CC-NIE) program, which helped provide a hardware upgrade from 10Gbps to 100Gbps WAN connectivity. Receiving NSF support to upgrade to 100Gbps has enabled huge gains in workflow throughput.
+
+
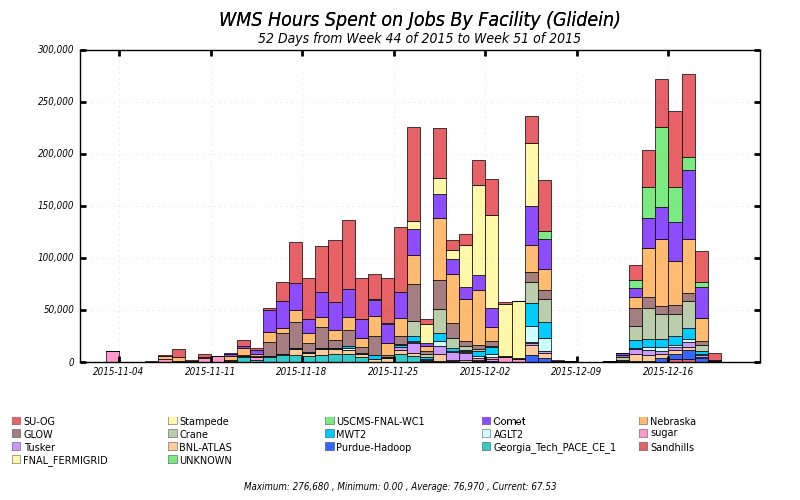
+
+
The LIGO analysis ran across 16 different OSG resources, for a total of 4M CPU hours:
+
+ - 1M CPU hour (25%) XSEDE contribution<
+ - 5 TB total input data, cached at the Holland Computing Center (HCC) at the University of Nebraska-Lincoln
+ - 1 PB total data volume distributed to jobs from Nebraska
+ - 10 Gbps sustained data rates from Nebraska storage to worker nodes
+
+
+
Couvares concluded, “What we are doing is pure science. We are trying to understand the universe, trying to do what people have wanted to do for 100 years. We are extending the reach of human understanding. It’s very exciting and the science is that much easier with the OSG.”
+
+
+
+
– Greg Moore
+
+
– Brian Bockelman (OSG, University of Nebraska at Lincoln) contributed to this story
+
+
+
+
+
+
+
+
+ OSPool As a Tool for Advancing Research in Computational Chemistry
+
+
Assistant Professor Eric Jonas uses OSG resources to understand the structure of molecules based on their measurements and derived properties.
+
+  + Microscope beside computer by Tima Miroshnichenko from Pexels.
+ Microscope beside computer by Tima Miroshnichenko from Pexels.
+
+
+
+  + Eric Jonas, Assistant professor at UChicago
+ Eric Jonas, Assistant professor at UChicago
+
+
+
Picture this: You have just developed a model that predicts the properties of some molecules and plan to include this model in a section of a research paper. However, just a few days before the paper is to be published on your professional website, you discover an error in the data generation process, which requires you to compute your work again and quickly!
+This scenario was the case with Assistant Professor Eric Jonas, who works in the Department of Computer Science at the University of Chicago (UChicago).
+While this process is normally tedious, he noted how the OSPool helped streamline the steps needed to regenerate results: “The OSPool made it easy to go back and regenerate the data set with about 70 million new molecules in just a matter of days.”
+
+
Although this was a fairly recent incident for Jonas, he is not new to high throughput computing or the OSPool. With usage reaching as far back as his graduate school days, Jonas has utilized resources ranging from cloud computing infrastructures like Amazon Web Services to the National Supercomputing Center for his work with biological signal acquisition, molecular inverse problems, machine learning, and other ways of exploiting scalable computation.
+
+
He soon realized, though, that although these other resources could run large amounts of data in a relatively short time, they required a long, drawn-out sequence of actions to provide results – creating an application, waiting for it to be accepted, and then waiting in line for long periods for a job to run. Faced with this problem in 2021, Jonas found a solution with the OSG Consortium and its OSPool, OSG’s distributed pool of computing resources for running high-throughput jobs.
+
+
In April of 2021, he enlisted the help of HTCondor and the OSPool to run pre-exising computations that allow for the generation of training data and the development of new machine learning techniques to determine molecular structures in mixtures, chemical structures in new plant species, and other related queries.
+
+
Jonas’ decision to transition to the OSPool boiled down to three simple reasons:
+Less red tape involved in getting started.
+Better communication and assistance from staff.
+Greater flexibility with running other people’s software to generate data for his specific research, which, in his words, are a much better fit for his specific research which would otherwise have been too computationally bulky to handle alone.
+
+
In terms of challenges with OSPool utilization, Jonas’ only point of concern is the amount of time it takes for code that has been uploaded to reach the OSPool. “It takes between 8 and 12 hours for that code to get to OSG. The time-consuming containerization process means that any bug in code that prevents it from running isn’t discovered and resolved as quickly, and takes quite a while, sometimes overnight.”
+
+
He and his research team have since continued to utilize OSPool to generate output and share data with other users. They have even become advocates for the resource: “After we build our models, as a next step, we’re like, let’s run our model on the OSPool to allow the community (which constitutes the entirety of OSPool users) also to generate their datasets. I guess my goal, in a way, is to help OSG grow any way I can, whether that involves sharing my output with others or encouraging people to look into it more.”
+
+
Jonas spoke about how he hopes more people would take advantage of OSPool:
+“We’re already working on expanding our use of it at UChicago, but I want even more people to know that OSPool is out there and to know what kind of jobs it’s a good fit for because if it fits the kind of work you’re doing, it’s like having a superpower!”
+
+
+
+
+
+
+
+ HTCondor Team Pictures
+
+
+
2024
+
The CHTC Team at Throughput Computing 2024
+

+
+
+
2023
+
The CHTC Team at Throughput Computing 2023
+

+
+
2022
+
The CHTC Team at HTCondor Week 2022
+

+
+
2021
+
The HTCondor Team at HTCondor Week 2021, May 2020.
+

+
+
2020
+
The HTCondor Team at HTCondor Week 2020, May 2020.
+

+
+
2019
+
The HTCondor Team at HTCondor Week 2019, May 2019.
+

+
+
2018
+
The HTCondor Team at HTCondor Week 2018, May 2018.
+

+
+
2017
+
The HTCondor Team at HTCondor Week 2017, May 2017 (but photo is missing Miron Livny!).
+

+
+
2016
+
The HTCondor Team at HTCondor Week 2016, May 2016.
+

+
+
2015
+
The HTCondor Team at HTCondor Week 2015, May 2015.
+

+
+
2014
+
The HTCondor Team at HTCondor Week 2014, May 2014.
+

+
+
2013
+
The HTCondor Team at HTCondor Week 2013, May 2013.
+

+
+
2012
+
The HTCondor Team at Condor Week 2012, May 2012.
+

+
+
2011
+
The HTCondor Team at Condor Week 2011, May 2011.
+

+
+
2010
+
The HTCondor Team at Condor Week 2010, May 2010.
+

+
+
2009
+
The HTCondor Team at Condor Week 2009, May 2009.
+

+
+
2008
+
The HTCondor Team at Condor Week 2008, May 2008.
+

+
+
2007
+
The HTCondor Team at Condor Week 2007, May 2007.
+

+
+
2006
+
The HTCondor Team at Condor Week 2006, April 2006.
+

+
+
2003
+

+

+
+
2002
+
Taken in October.
+

+
+
+
1999
+
+
+
+  +
+
+
+
1998
+
+

+
+
+
+
+
+
+ Advancing computational throughput of NSF funded projects with the PATh Facility
+
+
Since 2022, the Partnership to Advance Throughput Computing (PATh) Facility has provided dedicated high throughput
+computing (HTC)
+capacity to researchers nationwide. Following a year of expansion, here’s a look into the researchers’ work and how it has been enabled by
+the PATh Facility.
+
+
Searching for more computing capacity, Dr. Himadri Chakraborty of
+Northwest Missouri State University first heard of the PATh Facility, a
+purpose-built,
+national-scale distributed high throughput computing (HTC) resource, from his NSF program director. After approaching PATh Research
+Facilitators to
+acquire an account and computing “credits,” Chakraborty’s team was able to advance their work in physics using computing resources from the
+PATh Facility.
+Christina Koch, Lead Research Facilitator at CHTC guided Chakraborty’s team in
+transitioning workflows to run within HTCondor.
+
+
“As our ambition grew, we were looking out for a larger system, PATh came as a blessing,” Chakraborty reflected. “The ultimate satisfaction is to get
+some new understanding and learning of the science we are working on. We hope that this will be one of our first major achievements using
+the PATh Facility.”
+
+
+  + Himadri Chakraborty.
+ Himadri Chakraborty.
+
+
+
The PATh Facility, created through the Partnership to Advance Throughput Computing (PATh; NSF grant #2030508), enables researchers of an NSF-funded project
+to use its dedicated distributed high-throughput computing (HTC) capacity. PATh is an ongoing collaboration between the Center for High Throughput Computing (CHTC)
+and the OSG Consortium to support HTC-enabled research by improving national cyberinfrastructure. The PATh Facility acquires capacity
+from six institutions — the University of Nebraska-Lincoln, Syracuse University, Florida International University,
+the San Diego Supercomputing Center, the University of Wisconsin-Madison and the Texas Advanced Computing Center
+— integrated into a single pool of capacity available to users. After becoming fully operational in mid-2022, it saw “the most growth in terms of new projects
+joining and getting started in 2023,” Koch said.
+
+
The PATh Facility guarantees users access through credits. Credits operate as a stand-in unit to ensure no
+one user is monopolizing the Facility’s capacity. Users receive 1,000 start-up credits to test if the PATh Facility is a good fit for them, available as Central
+Processing Unit (CPU) or Graphic Processing Unit (GPU) charges. After this initial testing period, they can apply for supplemental credits by contacting PATh
+Facilitators and their NSF officer. If users run through all their credits, they are still able to keep running and facilitators will work with them to request
+additional credits.
+
+
In comparison to the PATh Facility, the Open Science Pool (OSPool) — another distributed HTC resource created
+by the OSG Consortium — acquires available capacity from idle resources across 70 institutions
+nationwide. Projects may be better suited for the PATh Facility than the OSPool if they need additional cores, memory, data or dedicated time. “Since the PATh
+Facility is hardware owned and operated by PATh, we can make more guarantees about how long individual computations can run, that people will be able to get certain
+resources and run computations of a certain size,” Koch explained.
+
+
Following the PATh Facility’s growth, some OSPool users have begun to use the Facility’s dedicated capacity in tandem. One example is North Carolina State University
+Ph.D. candidate Matthew Dorsey, who relied on capacity from the OSPool for two years before expanding his research to the newer PATh Facility.
+In doing so, he was able to run larger jobs without worrying about running out of capacity. “The transition to the PATh Facility was extremely easy,” Dorsey said.
+“I was pleased with how seamless it was.”
+
+
Dorsey became interested in the OSPool after attending OSG School in the summer of 2022. There, he learned the basics of
+HTCondor and got to know Koch and other facilitators. Dorsey’s research specializes in statistical physics. He uses computational models
+to study magnetic materials and how magnetic fields can be used to alter properties made from different kinds of magnetic nanoparticles. His work benefits from the
+consistent access to computing for runs that accumulate over a long period of time.
+
+
+  + Matthew Dorsey.
+ Matthew Dorsey.
+
+
+
Dorsey acknowledges that each system has its advantages and disadvantages. For Dorsey, the PATh Facility is better equipped for more complex jobs due to capacity and
+allocated time, while the OSPool is better for testing out comparatively smaller runs. “It was really easy to translate what I was doing on the OSPool to the PATh
+Facility and quickly scale it up,” Dorsey said.
+
+
A testament to the strength of the PATh Facility, the National Radio Astronomy Observatory (NRAO) used its capacity with the OSPool to
+develop one of the oldest radio images of a well-studied area in space. Working alongside PATh and CHTC staff, the capacity
+from the PATh Facility was instrumental in planning when certain jobs would run, without the risk of reduced capacity from the OSPool.
+
+
The PATh Facility makes it possible to support projects with larger computational requirements. Dr. Vladan Stevanovic
+of the Colorado School of Mines is studying computational material science and relies heavily on the PATh Facility to plan and run data workflows.
+Stevanovic became familiar with the PATh Facility after receiving a Dear Colleague Letter from the NSF.
+
+
His work requires more cores than what the OSPool alone could offer, and he was drawn to the PATh Facility due to its specialization in HTC and ability to guarantee
+availability. Stevanovic and his team hope to develop computational tools to reliably predict the metastable states of solid matter. He describes this work as very
+computational, and has worked with HTC workflows for over 12 years. “PATh is amazing for my research because its primary purpose is HTC, which I rely on,” he said.
+“I’m grateful because my project critically depends on PATh,” he said.
+
+
Stevanovic also appreciates how easy it was to start using the PATh Facility. Start-up credits are typically granted while or directly after meeting with the Facilitation
+team, and Koch’s team continues to support researchers as they ask the NSF for more credits. “The onboarding process was great, and the support from Christina was amazing.
+We were able to get running and get up to speed quickly.”
+
+
Chakraborty’s team faced some initial challenges in switching workflows from in-house to distributed, but coming to the PATh Facility nonetheless expanded the capacity
+available to his team. He recounted that his previous in-house system provided about 28 CPUs per node, while the PATh Facility offers up to 150 CPUs. Overall, Chakraborty
+is optimistic that the new capacity will improve his findings for the future. “We are happy we took that plunge because we went through some difficult times and got help
+from the PATh folks,” he said. “We’ve made some pretty good progress in making our codes run on PATh. It was mostly to be able to use a larger pool of computer power.”
+
+
His work focuses on the simulation of electronic and phonontonic coupled ultrafast relaxation of photoexcited large molecules. The PATh Facility’s new capacity allowed
+his team to make new advances “of a polymer functional system that has a lot of applications,” he said. “It’s not the end, it’s still preliminary and intermediate, and
+they are so exciting. We are looking forward to the final results and finding out new physics.”
+
+
Interested PIs can submit an interest form on the PATh website, to then meet a research computing facilitator for a consultation.
+If the researcher is a good fit, PATh Facilitators help the researcher log in and begin using their start-up credits. If they wish to continue, the researcher begins
+drafting a proposal letter to the NSF, requesting credits. Koch notes that the credit proposal is simpler than a typical project proposal, and the Facilitation team provides
+a multitude of resources such as credit calculators and proposal templates. For users who encounter issues, the facilitation team is available through support email address,
+and weekly support hours, as well as maintaining documentation on the website.
+
+
+
+
+
+
Leadership
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+
+ Brian Bockelman
+
+
+
FoCaS co-lead
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ University of Wisconsin–Madison
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+
+ Todd Tannenbaum
+
+
+
HTCondor Software Lead
+
+
+ University of Wisconsin–Madison
+
+
+
+
+
+
+
+
+
+
Staff
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Aaron Moate
+
+
+
Lead Systems Administrator
+
+
+ University of Wisconsin–Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Aaryan Patel
+
+
+
Research Computing Facilitation Assistant
+
+
+ Morgridge Insititute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Abhinandan Saha
+
+
+
Systems Administration Intern
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Amber Lim
+
+
+
Research Computing Facilitator
+
+
+ University of Wisconsin–Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Andrew Owen
+
+
+
Research Computing Facilitator
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Brian Aydemir
+
+
+
Systems Integration Developer
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Brian Lin
+
+
+
OSG Software Area Coordinator
+
+
+ University of Wisconsin–Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Cannon Lock
+
+
+
Web Developer
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+
+
Lead Research Computing Facilitator
+
+
+ University of Wisconsin - Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Cole Bollig
+
+
+
HTCondor Core Developer
+
+
+ University of Wisconsin - Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ David Baik
+
+
+
System Administrator
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Emile Turatsinze
+
+
+
Systems Administrator
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Emma Turetsky
+
+
+
Research Software Engineer
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Greg Thain
+
+
+
HTCondor Core Developer
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Ian Ross
+
+
+
Systems Integration Developer
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Irene Landrum
+
+
+
Project Manager
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Jaime Frey
+
+
+
HTCondor Core Developer
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Janet Stathas
+
+
+
Project Manager
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Jason Patton
+
+
+
Software Integration Developer
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+
+
System Administrator
+
+
+ Morgridge Institute
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Joe Bartkowiak
+
+
+
Systems Administrator
+
+
+ University of Wisconsin Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ John TJ Knoeller
+
+
+
HTCondor Core Developer
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Justin Hiemstra
+
+
+
Research Software Engineer
+
+
+ Morgridge Institute For Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Kent Cramer III
+
+
+
Network Infrastructure Support Specialist
+
+
+ Morgridge Institute For Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Matt Westphall
+
+
+
Research Cyberinfrastructure Specialist
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Mátyás Selmeci
+
+
+
Software Integration Developer
+
+
+ University of Wisconsin–Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Rachel Lombardi
+
+
+
Research Computing Facilitator
+
+
+ University of Wisconsin–Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Tae Kidd
+
+
+
Project Manager
+
+
+ Morgridge Institute For Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Theng Vang
+
+
+
System Administrator
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+
+
OSG Deputy Director/XO
+
+
+ University of Wisconsin–Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Tim Theisen
+
+
+
Release Manager
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Todd L Miller
+
+
+
HTCondor Core Developer
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ William Swanson
+
+
+
Research Cyberinfrastructure Specialist
+
+
+ University of Wisconsin–Madison
+
+
+
+
+
+
+
+
Students
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Ben Staehle
+
+
+
Fellow
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Bocheng Zou
+
+
+
System Administrator Intern
+
+
+ University of Wisconsin–Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Cristina Encarnacion
+
+
+
Student Science Writer
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Frank Zhang
+
+
+
System Administrator Intern
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Jordan Sklar
+
+
+
Student Science Writer
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Kristina Zhao
+
+
+
Fellow
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Marissa (Yujia) Zhang
+
+
+
System Administrator Intern
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Neha Talluri
+
+
+
Fellow
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Patrick Brophy
+
+
+
Fellow
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Pratham Patel
+
+
+
Fellow
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Ryan Boone
+
+
+
Fellow
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Thinh Nguyen
+
+
+
Fellow
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Wil Cram
+
+
+
Fellow
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
Previous Staff
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Alperen Bakirci
+
+
+
Student Web Developer
+
+
+ Morgridge Institute For Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Bryna Goeking
+
+
+
Student Writer
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Cameron Abplanalp
+
+
+
Research Computing Facilitation Assistant
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Emily Yao
+
+
+
System Administrator Intern
+
+
+ University on Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Haoming Meng
+
+
+
Research Software Engineer
+
+
+ Morgridge Institute For Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Joe Reuss
+
+
+
Software Engineer
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ John Parsons
+
+
+
System Administrator Intern
+
+
+ University of Wisconsin Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Lili Bicoy
+
+
+
Student Science Writer
+
+
+ Morgridge Institute For Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Max Hartke
+
+
+
Student Programming Intern
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Michael Collins
+
+
+
Systems Administrator
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Mihir Manna
+
+
+
System Administrator Intern
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Molly McCarthy
+
+
+
Student Web Developer
+
+
+ Morgridge Institute for Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Rishideep Rallabandi
+
+
+
Student Programming Intern
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Ryan Jacob
+
+
+
System Administrator Intern
+
+
+ University of Wisconsin-Madison
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Shirley Obih
+
+
+
Communications Specialist
+
+
+ Morgridge Institute For Research
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+ Yuxiao Qu
+
+
+
Research Software Engineer
+
+
+ Morgridge Institute For Research
+
+
+
+
+
+
+
+
+
+
+
+
+ Harnessing HTC-enabled precision mental health to capture the complexity of smoking cessation
+
+
Collaborating with CHTC research computing facilitation staff, UW-Madison researcher Gaylen Fronk is using HTC to improve cigarette cessation treatments by accounting for the complex differences among patients.
+
+
+  + Gaylen Fronk. Image credit: UW ARC.
+ Gaylen Fronk. Image credit: UW ARC.
+
+
+
Working at the crossroads of mental health and computing is Gaylen Fronk, a graduate student at the University of Wisconsin-Madison’s Addiction Research Center. By examining treatments for substance use disorders with machine learning models that are enabled by High Throughput Computing (HTC), Fronk captures the array of differences among individuals while still ensuring that her models are applicable to new patients. Her work is embedded within the larger context of precision mental health, an emerging field that relies on computational tools to evaluate complex, individual-level data in determining the fastest and most effective treatment plan for a given patient.
+
+
Fronk’s pursuit of precision mental health has recently led her to the world of computing that involves high-throughput workloads. Currently, she’s using HTC to predict treatment responses for people who are quitting cigarette smoking.
+
+
“I feel like [HTC] has been critical for my entire project,” Fronk reasons. “It removes so many constraints from how I have to think about my research. It keeps so many possibilities open because, within reason, I just don’t have to worry about computational time –– it allows me to explore new questions and test out ideas. It allows me to think bigger and add complexity rather than only having to constrain.”
+
+
Embarking on this project in August of 2019, Fronk began by reaching out to the research computing facilitators at UW-Madison’s Center for High Throughput Computing (CHTC). Dedicated to bringing the power of HTC to all fields of research, CHTC staff provided Fronk with the advice and resources she needed to get up and running. Soon, she was able to access hundreds of concurrent cores on CHTC’s HTC system through the HTCondor Software Suite (HTCSS), which was developed at UW-Madison and is used internationally for automating and managing batch HTC workloads. This computing capacity has been undeniably impactful on Fronk’s research, yet when reflecting on the beginnings of her project today, Fronk considers the collaborative relationships she’s developed along the way to be particularly powerful.
+
+
“I am never going to be a computer scientist,” explains Fronk. “I’m doing my best and I’m learning, but that’s not what my focus is and that’s never going to be my area of expertise. I think it’s really wonderful to be able to lean on people for whom that is their area of expertise, and have those collaborative relationships.” This type of collaboration among computing experts and researchers will be vital as computational advances continue to spread throughout the social sciences. Computing staff like CHTC’s research computing facilitators help researchers to transform, expand, and accelerate their work; and specialized researchers like Fronk provide their domain expertise to ensure these computational methods are incorporated in ways that preserve the conceptual and theoretical basis of their discipline.
+
+
+  + Christina Koch.
+ Christina Koch.
+
+
+
CHTC research computing facilitator Christina Koch has worked closely with Fronk since the beginning of her project, and elaborates on the benefits arising from this synergistic relationship: “Instead of every research group on campus needing to have their own in-house large-scale computing expert, they can meet with our facilitation team and we provide them with the information they need to expand their research computing vision and apply it to their work. But we also learn a lot ourselves from the wide variety of researchers we consult with. Since our experience isn’t siloed to a particular research domain, we take lessons learned from one group and share them with another group, where normally those groups would never have thought to connect with each other.”
+
+
For fellow social scientists who are considering reaching out to people like Christina and incorporating HTC into their work, Fronk urges them to do just that: “There’s a lot you can teach yourself, but you also don’t have to be on your own. Reach out to the people who know more than you. For me, people like Christina and others on the CHTC team have been invaluable.”
+
+
Fronk’s collaborations with Christina all have revolved around the ongoing project that she first began in August of 2019 –– predicting which cigarette cessation treatments will be most effective for a given individual. Data from a Center for Tobacco Research and Intervention (CTRI) 6-month clinical trial serve as a rich and comprehensive foundation to begin building machine learning models from. With the CTRI data in hand, Fronk not only has access to the treatment type and whether it was successful at the end of the trial, but also to approximately 400 characteristics that capture the fine-tuned individual differences among patients. These include demographic information, physical and mental health records, smoking histories, and social pressures, such as the smoking habits of a patient’s friends, roommates, or spouse.
+
+
All these individual-level differences paint valuable complexity onto the picture, and Fronk is able to embrace and dive into that complexity with the help of HTC. Each job she sends over to CHTC’s cores contains a unique model configuration run against a single cross-validation iteration, meaning that part of the CTRI data is used for model fitting while the unused, ‘new’ data is used for model evaluation. For instance, Fronk might start with as many as 200 unique configurations for a given model. If each of these model configurations is fit and evaluated using a cross-validation technique that has 100 unique splits of data, Fronk would then submit the resulting 20,000 jobs to CHTC.
+
+
Before submitting, Fronk alters her code so that each job runs just a single configuration, single iteration context; effectively breaking the comprehensive CTRI data down into small, manageable pieces. Ultimately, when delegated to hundreds of CHTC cores in concurrent use, Fronk’s largest submissions finish in mere hours, as opposed to days on a local computer.
+
+
Thousands of small jobs are handled easily by HTCSS and CHTC’s distributed resources, after which Fronk can aggregate this multitude of output files on her own computer to average the performance of the model configuration across the array of cross-validation iterations. This aggregated output represents how accurately the model predicts whether a certain cigarette cessation treatment will work for a specific individual. After receiving the output, Fronk evaluates it, learns from it, and repeats –– but this time with new insight.
+
+
After her experience with HTC, Fronk now sees computing as an integral part of her work. In fact, the ideal of precision mental health as a compelling alternative to traditional treatment methods has actually been around for a while –– though scalable computing methods that enable it are just beginning to enter the toolboxes of mental health researchers everywhere. “I feel like high-throughput computing really fills a lot of the holes that are needed to move precision mental health forward,” Fronk expresses. “It makes me really excited to be working at that intersection.”
+
+
And at that intersection, Fronk isn’t alone. As computational resources are becoming more accessible, increasingly more researchers are investigating the frontiers of precision mental health and its potential to improve treatment success. But before this approach moves from the research space and into a clinical setting, careful thought is needed to assess how these experimental models will fare in the real world.
+
+
Approaches that require intensive and expensive data, like neuroimaging or genetic analysis for instance, may not be feasible –– especially for clinics located in low-income communities. Elaborating on this idea, Fronk explains, “It’s really exciting to think that neuroimaging or genetic data might hold a lot of predictive potential –– yet if a person can’t get genotyped or imaged, then they’re not going to be able to be split into treatments. And those problems get compounded in lower income areas, or for people who have been historically excluded and underrepresented both in terms of existing research and access to healthcare.”
+
+
It will take time, research, and ethical forethought before precision mental health approaches can reach local clinics, but when that time comes –– the impact will ripple through the lives of people seeking treatment everywhere. “I think precision mental health can really help people on a much shorter timeline than traditional treatment approaches, and that feels very meaningful to me,” says Fronk. In terms of her focus on cigarette smoking cessation, timing is everything. Cigarette smoking –– as well as other substance use disorders like it –– have extremely high costs of failed treatments at both the personal and societal level. If someone is given the right treatment from the start when they’re most motivated to quit, it mitigates not only their own health and financial risks, but also those of society’s.
+
+
Ultimately, these impacts stem from the collaborative relationships seen today between researchers like Fronk and computing facilitators like Christina at CHTC. There’s still much to be done before precision mental health approaches can be brought to bear in the clinical world, but high-throughput computing is powering the research to move that direction in a way that never was possible before. Complexity –– which used to limit Fronk’s research –– now seems to be absolutely central to it.
+
+
…
+
+
A research article about Fronk’s project is forthcoming. In the meantime, watch her presentation from HTCondor Week 2021 or check out the UW-Madison Addiction Research Center to learn more.
+
+
+
+
+
+ CHTC Partners Using CHTC Technologies and Services
+
+
+
+
+
+
+
+
+
+
+
+
+
+
IceCube and Francis Halzen
+
+ Francis Halzen, principal investigator of IceCube and the Hilldale and Gregory Breit Distinguished Professor of Physics.
IceCube has transformed a cubic kilometer of natural Antarctic ice into a neutrino detector. We have discovered a flux of high-energy neutrinos of cosmic origin, with an energy flux that is comparable to that of high-energy photons. We have also identified its first source: on September 22, 2017, following an alert initiated by a 290-TeV neutrino, observations by other astronomical telescopes pinpointed a flaring active galaxy, powered by a supermassive black hole. We study the neutrinos themselves, some with energies exceeding by one million those produced by accelerators. The IceCube Neutrino Observatory is managed and operated by the Wisconsin IceCube Astroparticle Physics Center (WIPAC) in the Office of the Vice Chancellor of Graduate Education and Research and funded by a cooperative agreement with the National Science Foundation. We have used CHTC and the Open Science Pool for over a decade to perform all large-scale data analysis tasks and generate Monte Carlo simulations of the instrument's performance. Without CHTC and OSP resources we would simply be unable to make any of IceCube's groundbreaking discoveries. Francis Halzen is the Principal Investigator of IceCube. See the IceCube web site for project details.
+
+
+
+
+
+
+

+
+
+
+
+
+
+
David O’Connor
+
+ David H. O’Connor, Ph.D., UW Medical Foundation (UWMF) Professor, Department of Pathology and Laboratory Medicine at the AIDS Vaccine Research Laboratory.
Computational workflows that analyze and process genomics sequencing data have become the standard in Virology and genomics research. The resources provided by CHTC allow us to scale up the amount of sequence analysis performed while decreasing sequence processing time. An example of how we use CHTC is generating consensus sequences for COVID-19 samples. Part of this is a step that separates and sorts a multisample sequencing run into individual samples, maps the reads of these individual samples to a reference sequence, and then forms a consensus sequence for each sample. Simultaneously, different metadata and other accessory files are generated on a per-sample basis, and data is copied to and from local machines. This workflow is cumbersome when there are, for example, 4 sequencing runs with each run containing 96 samples. CHTC allows us to cut the processing time from 16 hours to 40 minutes due to the distribution of jobs to different CPUs. Overall, being able to use CHTC resources gives us a major advantage in producing results faster for large-scale and/or time-sensitive projects.
+
+
+
+
+
+
+

+
+
+
+
+
+
+
Small Molecule Screening Facility and Spencer Ericksen
+
+ Spencer Ericksen, Scientist II at the Small Molecule Screening Facility, part of the Drug Development Core – a shared resource in the UW Carbone Cancer Center.
I have been working on computational methods for predicting biomolecular recognition processes. The motivation is to develop reliable models for predicting binding interactions between drug-like small molecules and therapeutic target proteins. Our team at SMSF works with campus investigators on early-stage academic drug discovery projects. Computational models for virtual screening could prioritize candidate molecules for faster, cheaper focused screens on just tens of compounds. To perform a virtual screen, the models evaluate millions to billions of molecules, a computationally daunting task. But CHTC facilitators have been with us through every obstacle, helping us to effectively scale through parallelization over HTC nodes, matching appropriate resources to specific modeling tasks, compiling software, and using Docker containers. Moreover, CHTC provides access to vast and diverse compute resources.
+
+
+
+
+
+
+

+
+
+
+
+
+
+
Natalia de Leon
+
+ Natalia de Leon, Professor of Agronomy, Department of Agronomy.
The goal of her research is to identify efficient mechanisms to better understand the genetic constitution of economically relevant traits and to improve plant breeding efficiency. Her research integrates genomic, phenomic, and environmental information to accelerate translational research for enhanced sustainable crop productivity.
+
+
+
+
+
+
+

+
+
+
+
+
+
+
xDD project and Shanan Peters
+
+ Shanan Peters, project lead for xDD, Dean L. Morgridge Professor of Geology, Department of Geoscience.
Shanan’s primary research thrust involves quantifying the spatial and temporal distribution of rocks in the Earth’s crust in order to constrain the long-term evolution of life and Earth’s surface environment. Compiling data from scientific publications is a key component of this work and Peters and his collaborators are developing machine reading systems deployed over the xDD digital library ad cyberinfrastructure hosted in the CHTC for this purpose.
+
+
+
+
+
+
+

+
+
+
+
+
+
+
Susan Hagness
+
+ In our research we're working on a novel computational tool for THz-frequency characterization of materials with high carrier densities, such as highly-doped semiconductors and metals. The numerical technique tracks carrier-field dynamics by combining the ensemble Monte Carlo simulator of carrier dynamics with the finite-difference time-domain technique for Maxwell's equations and the molecular dynamics technique for close-range Coulomb interactions. This technique is computationally intensive and each test runs long enough (12-20 hours) that our group's cluster isn't enough. This is why we think CHTC can help, to let us run more jobs than we're able to run now.
+
+
+
+
+
+
+

+
+
+
+
+
+
+
Joao Dorea
+
+ Joao Dorea, Assistant Professor in the Department of Animal and Dairy Sciences/Department of Biological Systems Engineering.
The Digital Livestock Lab develops research focused on high-throughput phenotyping strategies to optimize farm management decisions. Our research group is interested in the large-scale development and implementation of computer vision systems, wearable sensors, and infrared spectroscopy (NIR and MIR) to monitor animals in livestock systems. We have a large computer vision system implemented in two UW research farms that generate large datasets. With the help of CHTC, we can train deep learning algorithms with millions of parameters using large image datasets and evaluate their performance in farm settings in a timely manner. We use these algorithms to monitor animal behavior, growth development, social interaction, and to build predictive models for early detection of health issues and productive performance. Without access to the GPU cluster and the facilitation made by CHTC, we would not be able to quickly implement AI technologies in livestock systems.
+
+
+
+
+
+
+

+
+
+
+
+
+
+
Paul Wilson
+
+ Paul Wilson, head of The Computational Nuclear Engineering Research Group (CNERG), the Grainger Professor for Nuclear Engineering, and the current chair of the Department of Engineering Physics.
CNERG’s mission is to foster the development of new generations of nuclear engineers and scientists through the development and deployment of open and reliable software tools for the analysis of complex nuclear energy systems. Our inspiration and motivation come from performing those analyses on large, complex systems. Such simulations require ever-increasing computational resources and CHTC has been our primary home for both HPC and HTC computation for nearly a decade. In addition to producing our results faster and without the burden of administering our computer hardware, we rely on CHTC resources to demonstrate performance improvements that arise from our methods development. The role-defining team of research computing facilitators has ensured a smooth onboarding of each of them and helped them find the resources they need to be most effective.
+
+
+
+
+
+
+

+
+
+
+
+
+
+
Barry Van Veen
+
+ The bio-signal processing laboratory develops statistical signal processing methods for biomedical problems. We use CHTC for casual network modeling of brain electrical activity. We develop methods for identifying network models from noninvasive measures of electric/ magnetic fields at the scalp, or invasive measures of the electric fields at or in the cortex, such as electrocorticography. Model identification involves high throughput computing applied to large datasets consisting of hundreds of spatial channels each containing thousands of time samples.
+
+
+
+
+
+

+
+
+
+
+
+
+
Biomagnetic Resonance Data Bank
+
+ The Biomagnetic Resonance Data Bank (BMRB) is headquarted within UW-Madison's National Magnetic Resonance Facility at Madison (NMRFAM) and uses the CHTC for research in connection with the Biological Magnetic Resonance Data Bank (BMRB).
+
+
+
+
+
+
+

+
+
+
+
+
+
+
CMS LHC Compact Muon Solenoid
+
+ The UW team participating in the Compact Muon Solenoid (CMS) experiment analyzes petabytes of data from proton-proton collisions in the Large Hadron Collider (LHC). We use the unprecedented energies of the LHC to study Higgs Boson signatures, Electroweak Physics, and the possibility of exotic particles beyond the Standard Model of Particle Physics. Important calculations are also performed to better tune the experiment's trigger system, which is responsible for making nanosecond-scale decisions about which collisions in the LHC should be recorded for further analysis.
+
+
+
+
+
+

+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+
Phil Townsend
+
+ Professor Phil Townsend of Forestry and Wildlife Ecology says Our research (NASA & USDA Forest Service funded) strives to understand the outbreak dynamic of major forest insect pests in North America through simulation modeling. As part of this effort, we map forest species and their abundance using multi-temporal Landsat satellite data. My colleagues have written an automatic variable selection routine in MATLAB to preselect the most important image variables to model and map forest species abundance. However, depending on the number of records and the initial variables, this process can take weeks to run. Hence, we seek resources to speed up this process.
+
+
+
+
+
+

+
+
+
+
+
+
+
+
+
+ Research Facilitation
+
+
+
RC facilitators serve as proactive and personalized guides, helping researchers
+identify and implement computational approaches that result in the greatest impact to their projects. Rather
+than possessing a significant depth of expertise in computational technologies, RC facilitators build and
+leverage their team of expert technical staff and translate the details of computational options for individual
+researchers. Through this two-way relationship-building approach, dedicated RC facilitators have enabled
+previously unimagined and significant scholarship outcomes of scale and scope across a variety of research
+domains, especially within the space of campus-supported research computing centers.
+
+
Impact
+
+
Since the hiring of the first Research Computing Facilitator in 2013 usage of computing services by previously underserved researchers
+increased significantlyy (see figure below). Importantly, more than 95% of usage from the life sciences and
+social sciences has been on an HTC-optimized compute configuration rather than a traditional HPC
+cluster, emphasizing the applicability of multiple compute configurations to meet needs across domains.
+
+

+
+
Goals
+
+
The following outlines the primary goals (the needs) of successful RC facilitation and identifies the related
+major activities for achieving those goals.
+
+
+ - Proactive Engagement
+ - Personalized Guidance
+ - Teaching Researchers to Fish
+ - Building Relationships
+ - Advocating for Research Needs
+ - Developing Connections among Staff
+
+
+
Skills and Backgrounds
+
+
Three key areas of experience and interest are relevant for successful RC facilitators: individual interests
+and motivation, communication and interpersonal skills, and technical knowledge.
+
+
Interests and Motivation
+
+
+ - A desire to enable and support the scholarly work of others
+ - Interest in a wide set of research domains beyond their own area of expertise
+ - The ability and the desire to work in a team environment
+ - A desire to further develop the skills and interests relevant to effective facilitation
+
+
+
Communication and Interpersonal Skills
+
+
+ - Excellent written and verbal communication, including active and empathetic listening skills and an
+ability to translate complex and domain-specific information for nonspecialists
+ - Demonstrated effectiveness and comfort in teaching and public speaking
+ - Success and demonstrated interest in interpersonal networking and liaising
+ - The desire to work in a team environment, where staff frequently depend on one another
+ - Leadership skills that inspire action and coordinate the activities of shared contributions
+
+
+
Technical Abilities
+
+
+ - Prior experience conducting research projects or other significant scholarly work with some
+integration of relevant computational systems and tools
+ - A demonstrated ability to understand multiple aspects of a problem and identify appropriate solutions
+ - The ability to provide solution-agnostic support by focusing on research requirements and desired
+outcomes
+ - A desire for continuous learning of relevant technology topics
+
+
+
+
+
More information on Research Facilitators can be in the paper
+“Research Computing Facilitators: The Missing Human Link in Needs-Based Research Cyberinfrastructure”
+co-written by CHTC’s own Lauren Micheal.
+
+
+
+
+
+
+
+
Overview
+
+ -
+ Douglas Thain, Todd Tannenbaum, and Miron Livny,
+ "Distributed Computing in Practice: The Condor Experience"
+ Concurrency and Computation: Practice and Experience,
+ Vol. 17, No. 2-4, pages 323-356, February-April, 2005.
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Douglas Thain and Miron Livny,
+ "Building Reliable Clients and Servers",
+ in Ian Foster and Carl Kesselman, editors,
+ The Grid: Blueprint for a New Computing Infrastructure,
+ Morgan Kaufmann, 2003, 2nd edition. ISBN: 1-55860-933-4.
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Douglas Thain, Todd Tannenbaum, and Miron Livny,
+ "Condor and the Grid",
+ in Fran Berman, Anthony J.G. Hey, Geoffrey Fox, editors,
+ Grid Computing: Making The Global Infrastructure a Reality,
+ John Wiley, 2003.
+ ISBN: 0-470-85319-0
+ [PDF]
+
+ [BibTeX Source for Citation]
+
+ -
+ Todd Tannenbaum, Derek Wright, Karen Miller, and Miron Livny,
+ "Condor - A Distributed Job Scheduler",
+ in Thomas Sterling, editor,
+ Beowulf Cluster Computing with Linux,
+ The MIT Press, 2002.
+ ISBN: 0-262-69274-0
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+ [MIT Press' Web Page]
+
+ The MIT Press is pleased to present material from a preliminary draft of
+ Beowulf Cluster Computing with Linux.
+ This material is Copyright 2002 Massachusetts Institute of Technology, and
+ may not be used or distributed for any commercial purpose without the
+ express written consent of The MIT Press. Because
+ this material was a draft chapter, neither
+ The MIT Press nor the authors can be held liable for changes or
+ alternations in the final edition.
+
+ -
+ Jim Basney and Miron Livny,
+ "Deploying a High Throughput Computing Cluster",
+ High Performance Cluster Computing, Rajkumar Buyya, Editor,
+ Vol. 1, Chapter 5, Prentice Hall PTR, May 1999.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Miron Livny, Jim Basney, Rajesh Raman, and Todd Tannenbaum,
+ "Mechanisms for High Throughput Computing",
+ SPEEDUP Journal, Vol. 11, No. 1, June 1997.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Jim Basney, Miron Livny, and Todd Tannenbaum,
+ "High Throughput Computing with Condor",
+ HPCU news, Volume 1(2), June 1997.
+
+
+ -
+ D. H. J Epema, Miron Livny, R. van Dantzig, X. Evers, and Jim Pruyne,
+ "A Worldwide Flock of Condors : Load Sharing among Workstation Clusters"
+ Journal on Future Generations of Computer Systems, Volume 12, 1996
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Scott Fields,
+ "Hunting for Wasted Computing Power",
+ 1993 Research Sampler, University of Wisconsin-Madison.
+ [HTML]
+
+ -
+ Michael Litzkow, Miron Livny, and Matt Mutka,
+ "Condor - A Hunter of Idle Workstations",
+ Proceedings of the 8th International Conference of Distributed Computing Systems,
+ pages 104-111, June, 1988.
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Michael Litzkow,
+ "Remote Unix - Turning Idle Workstations into Cycle Servers",
+ Proceedings of Usenix Summer Conference, pages 381-384, 1987.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+
+
+
Matchmaking and ClassAds
+
+ -
+ Nicholas Coleman, "Distributed Policy Specification and Interpretation with
+ Classified Advertisements", Practical Aspects of Declarative Languages, Lecture
+ Notes in Computer Science Volume 7149, 2012, pp 198-211, January 2012.
+ [PDF]
+
+
+ -
+ Rajesh Raman, Miron Livny, and Marvin Solomon,
+ "Policy Driven Heterogeneous Resource Co-Allocation with Gangmatching",
+ Proceedings of the Twelfth IEEE International Symposium on
+ High-Performance Distributed Computing, June, 2003, Seattle, WA
+ [Postscript]
+ [PDF]
+
+ -
+ Nicholas Coleman, Rajesh Raman, Miron Livny and Marvin Solomon,
+ "Distributed Policy Management and Comprehension with Classified
+ Advertisements",
+ University of Wisconsin-Madison Computer Sciences Technical Report #1481,
+ April 2003.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Nicholas Coleman, "An Implementation of Matchmaking Analysis in Condor",
+ Masters' Project report,University of Wisconsin, Madison, May 2001.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Rajesh Raman, Miron Livny, and Marvin Solomon,
+ "Resource Management through Multilateral Matchmaking",
+ Proceedings of the Ninth IEEE Symposium on High Performance Distributed Computing (HPDC9),
+ Pittsburgh, Pennsylvania, August 2000, pp 290-291.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Rajesh Raman, Miron Livny, and Marvin Solomon,
+ "Matchmaking: Distributed Resource Management for High Throughput Computing",
+ Proceedings of the Seventh IEEE International Symposium on High Performance Distributed Computing, July 28-31, 1998, Chicago, IL.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+
+
+
Workflow and DAGMan
+
+ - Peter Couvares, Tevik Kosar, Alain Roy, Jeff Weber and Kent
+ Wenger, "Workflow in Condor", in In Workflows for e-Science, Editors: I.Taylor, E.Deelman, D.Gannon,
+ M.Shields, Springer Press, January 2007 (ISBN: 1-84628-519-4)
+ [PDF]
+
+
+
+
+
Resource Management
+
+ -
+ Zhe Zhang, Brian Bockelman, Dale Carder, and Todd Tannenbaum,
+ "Lark: Bringing Network Awareness to High Throughput Computing",
+ Proceedings of the 15th IEEE/ACM International Symposium on Cluster, Cloud and Grid Computing (CCGrid 2015), Shenzhen, Guangdong, China, May 2015.
+ [PDF]
+
+ -
+ Jim Basney and Miron Livny,
+ "Managing Network Resources in Condor",
+ Proceedings of the Ninth IEEE Symposium on High Performance Distributed Computing (HPDC9),
+ Pittsburgh, Pennsylvania, August 2000, pp 298-299.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Jim Basney and Miron Livny,
+ "Improving Goodput by Co-scheduling CPU and Network Capacity",
+ International Journal of High Performance Computing Applications,
+ Volume 13(3), Fall 1999.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Miron Livny and Rajesh Raman,
+ "High Throughput Resource Management",
+ chapter 13 in The Grid: Blueprint for a New Computing Infrastructure,
+ Morgan Kaufmann, San Francisco, California, 1999.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ Morgan Kaufmann is pleased to present material from a preliminary draft of
+ High Performance Distributed Computing: Building a Computational Grid;
+ the material is Copyright 1997 Morgan Kaufmann Publishers. This
+ material may not be used or distributed for any commercial purpose without the
+ express written consent of Morgan Kaufmann Publishers. Please note that
+ this material is a draft of forthcoming publication, and as such neither
+ Morgan Kaufmann nor the author can be held liable for changes or
+ alternations in the final edition.
+
+ -
+ Matt Mutka and Miron Livny,
+ "The Available Capacity of a Privately Owned Workstation Environment",
+ Performance Evaluation, vol. 12, no. 4 pp. 269-284, July, 1991.
+ [BibTeX Source for Citation]
+
+ -
+ Matt Mutka and Miron Livny,
+ "Profiling Workstations' Available Capacity for Remote Execution",
+ Performance '87,12th IFIP WG 7.3, pp. 529-544, December 1987.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+
+
+
Checkpointing
+
+ -
+ Joe Meehean and Miron Livny,
+ "A Service Migration Case Study: Migrating the Condor Schedd",
+ Midwest Instruction and Computing Symposium, April 2005.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Jim Basney, Miron Livny, and Paolo Mazzanti,
+ "Utilizing Widely Distributed Computational Resources Efficiently with Execution Domains",
+ Computer Physics Communications, 2001.
+ (This is an extended version of the CHEP 2000 paper below.)
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Jim Basney, Miron Livny, and Paolo Mazzanti,
+ "Harnessing the Capacity of Computational Grids for High Energy Physics",
+ Proceedings of the International Conference on Computing in High Energy and
+ Nuclear Physics (CHEP 2000),
+ February 2000, Padova, Italy.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Michael Litzkow, Todd Tannenbaum, Jim Basney, and Miron Livny,
+ "Checkpoint and Migration of UNIX Processes in the Condor Distributed Processing System",
+ University of Wisconsin-Madison Computer Sciences Technical Report #1346,
+ April 1997.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Jim Pruyne and Miron Livny,
+ "Managing Checkpoints for Parallel Programs",
+ Workshop on Job Scheduling Strategies for Parallel Processing IPPS '96.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Todd Tannenbaum and Michael Litzkow,
+ "Checkpointing and Migration of UNIX Processes in the Condor Distributed Processing System",
+ Dr Dobbs Journal, Feb 1995.
+ [HTML]
+ [Postscript]
+ [BibTeX Source for Citation]
+
+ -
+ Michael Litzkow and Marvin Solomon,
+ "Supporting Checkpointing and Process Migration Outside the UNIX Kernel",
+ Usenix Conference Proceedings,
+ San Francisco, CA, January 1992, pages 283-290.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+
+
+
Data Intensive Computing
+
+
+ -
+ Parag Mhashilkar, Zachary Miller, Rajkumar Kettimuthu, Gabriele Garzoglio, Burt
+ Holzman, Cathrin Weiss, Xi Duan, and Lukasz Lacinski, "End-To-End Solution for
+ Integrated Workload and Data Management using GlideinWMS and Globus Online",
+ Journal of Physics: Conference Series, Volume 396, Issue 3, Year 2012
+ [PDF]
+
+
+ -
+ Ian T. Foster, Josh Boverhof, Ann Chervenak, Lisa Childers, Annette DeSchoen,
+ Gabriele Garzoglio, Dan Gunter, Burt Holzman, Gopi Kandaswamy, Raj Kettimuthu,
+ Jack Kordas, Miron Livny, Stuart Martin, Parag Mhashilkar, Zachary Miller,
+ Taghrid Samak, Mei-Hui Su, Steven Tuecke, Vanamala Venkataswamy, Craig Ward,
+ Cathrin Weiss,
+ "Reliable high-performance data transfer via Globus Online",
+ in Proc. SciDAC 2011, Denver, CO, July 10-14.
+ [PDF]
+
+
+ -
+ Ann Chervenak, Ewa Deelman, Miron Livny, Mei-Hui Su, Rob Schuler, Shishir Bharathi, Gaurang Mehta, Karan Vahi,
+ "Data Placement for Scientific Applications in Distributed Environments",
+ In Proceedings of the 8th IEEE/ACM International Conference on Grid
+ Computing (Grid 2007), Austin, TX, September 2007.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+
+ -
+ George Kola, Tevfik Kosar, Jaime Frey, Miron Livny, Robert J. Brunner and Michael Remijan,
+ "DISC: A System for Distributed Data Intensive Scientific Computing",
+ In Proceedings of the First Workshop on Real, Large Distributed Systems (WORLDS'04), San Francisco, CA, December 2004, in conjunction with OSDI'04
+ [PostScript]
+ [PDF]
+
+
+
+ -
+ George Kola, Tevfik Kosar and Miron Livny,
+ "Profiling Grid Data Transfer Protocols and Servers",
+ In Euro-Par 2004, Pisa, Italy, September 2004.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ George Kola, Tevfik Kosar and Miron Livny,
+ "A Fully Automated Fault-tolerant System for Distributed Video Processing and Off-site Replication",
+ In
+ The 14th ACM International Workshop on Network and Operating Systems Support for Digital Audio and Video (NOSSDAV 2004), Kinsale, Ireland, June 2004.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ George Kola, Tevfik Kosar and Miron Livny,
+ "Run-time Adaptation of Grid Data-placement Jobs",
+ In Parallel and Distributed Computing Practices, 2004.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Tevfik Kosar and Miron Livny, "Stork: Making Data Placement a First Class Citizen in the Grid",
+ In Proceedings of 24th IEEE Int. Conference on Distributed Computing Systems (ICDCS2004), Tokyo, Japan, March 2004.
+ [PDF]
+
+
+ -
+ Tevfik Kosar, George Kola and Miron Livny, "A Framework for Self-optimising, Fault-tolerant, High Performance Bulk Data Transfers in a Heterogeneous Grid Environment",
+ Proceedings of 2nd Int. Symposium on Parallel and Distributed Computing (ISPDC2003), Ljubljana, Slovenia, October 2003.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ George Kola, Tevfik Kosar and Miron Livny,
+ "Run-time Adaptation of Grid Data-placement Jobs",
+ Proceedings of Int. Workshop on Adaptive Grid Middleware (AGridM2003), New Orleans, LA, September 2003.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Tevfik Kosar, George Kola and Miron Livny,
+ "Building Data Pipelines for High Performance Bulk Data Transfers in a Heterogeneous Grid Environment",
+ Technical Report CS-TR-2003-1487, University of Wisconsin-Madison Computer Sciences, August 2003.
+ [PDF]
+
+
+
+
Grid Computing
+
+ -
+ C. Acosta-Silva, A. Delgado Peris, J. Flix, J. Frey, J.M. Hernández, A. Pérez-Calero Yzquierdo, and T. Tannenbaum
+ "Exploitation of network-segregated CPU resources in CMS",
+ Proceedings of the 25th International Conference on Computing in High Energy and Nuclear Physics (CHEP 2021), May 2021.
+ [PDF]
+
+ -
+ Brian Bockelman, Miron Livny, Brian Lin, Francesco Prelz
+ "Principles, technologies, and time: The translational journey of the HTCondor-CE",
+ Journal of Computational Science, 2020
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ B Bockelman, T Cartwright, J Frey, E M Fajardo, B Lin, M Selmeci, T Tannenbaum and M Zvada
+ "Commissioning the HTCondor-CE for the Open Science Grid",
+ Journal of Physics: Conference Series, Vol. 664, 2015
+ [PDF]
+ [BibTeX /Source for Citation]
+
+
+ -
+ I Sfiligoi, D C Bradley, Z Miller, B Holzman, F Würthwein, J M Dost, K Bloom,
+ and C Grandi, "glideinWMS experience with glexec",
+ Journal of Physics: Conference Series, Volume 396, Issue 3, Year 2012
+ [PDF]
+
+
+ -
+ W Andrews, B Bockelman, D Bradley, J Dost, D Evans, I Fisk, J Frey, B Holzman, M Livny, T Martin, A McCrea, A Melo, S Metson, H Pi, I Sfiligoi, P Sheldon, T Tannenbaum, A Tiradani, F Würthwein and D Weitzel,
+ "Early experience on using glideinWMS in the cloud",
+ Journal of Physics: Conference Series, Vol. 331, No. 6, 2011
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Igor Sfiligoi, Greg Quinn, Chris Green, Greg Thain,
+ "Pilot Job Accounting and Auditing in Open Science Grid",
+ The 9th IEEE/ACM International Conference on Grid Computing,
+ Tsukuba, Japan, 2008
+ [PDF]
+
+
+ -
+ Alexandru Iosup, Dick H.J. Epema, Todd Tannenbaum, Matthew Farrellee, Miron Livny,
+ "Inter-Operating Grids through Delegated MatchMaking",
+ in proceedings of the International Conference for High Performance
+ Computing, Networking, Storage and Analysis (SC07),
+ Reno, Nevada, November 2007.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Sander Klous, Jamie Frey, Se-Chang Son, Douglas Thain, Alain Roy,
+ Miron Livny, and Jo van den Brand, "Transparent Access to Grid
+ Resources for User Software", in Concurrency and Computation:
+ Practice and Experience, Volume 18, Issue 7, pages 787-801, 2006.
+
+
+ -
+ Sechang Son, Matthew Farrellee, and Miron Livny,
+ "A Generic Proxy Mechanism for Secure Middlebox Traversal",
+ CLUSTER 2005,
+ Boston, MA, September 26-30, 2005.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Bruce Beckles, Sechang Son, and John Kewley,
+ "Current methods for negotiating firewalls for the Condor system",
+ Proceedings of the 4th UK e-Science All Hands Meeting 2005,
+ Nottingham, UK, September 19-22, 2005.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Sechang Son, Bill Allcock and Miron Livny,
+ "CODO: Firewall Traversal by Cooperative On-Demand Opening",
+ Proceedings of the 14th IEEE Symposium on High Performance Distributed Computing (HPDC14),
+ Research Triangle Park, NC, July 24-27, 2005.
+ [PDF]
+ [MS Word]
+ [BibTeX Source for Citation]
+
+
+ -
+ Clovis Chapman, Paul Wilson, Todd Tannenbaum, Matthew Farrellee, Miron Livny, John Brodholt, and Wolfgang Emmerich,
+ "Condor services for the global grid: Interoperability between Condor and OGSA",
+ Proceedings of the 2004 UK e-Science All Hands Meeting, ISBN 1-904425-21-6, pages 870-877, Nottingham, UK, August 2004.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Clovis Chapman, Charaka Goonatilake, Wolfgang Emmerich, Matthew Farrellee, Todd Tannenbaum, Miron Livny, Mark Calleja, and Martin Dove,
+ "Condor BirdBath: Web Service interfaces to Condor",
+ Proceedings of the 2005 UK e-Science All Hands Meeting, ISBN 1-904425-53-4, pages 737-744, Nottingham, UK, September 2005.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Sriya Santhanam, Pradheep Elango, Andrea Arpaci-Dusseau, and Miron Livny,
+ "Deploying Virtual Machines as Sandboxes for the Grid",
+ WORLDS 2005, San Francisco, CA, December 2004
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+
+ -
+ George Kola, Tevfik Kosar and Miron Livny,
+ "Phoenix: Making Data-intensive Grid Applications Fault-tolerant",
+ In Grid 2004, Pittsburgh, PA, November 2004
+ [PostScript]
+ [PDF]
+
+
+
+ -
+ Andrew Baranovski, Gabriele Garzoglio, Igor Terekhov, Alain Roy and Todd Tannenbaum,
+ "Management of Grid Jobs and Data within SAMGrid",
+ Proceedings of the 2004 IEEE International Conference on Cluster Computing,
+ pages 353-360,
+ San Diego, CA, September 2004.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ George Kola, Tevfik Kosar and Miron Livny,
+ "A Client-centric Grid Knowledgebase",
+ Proceedings of the 2004 IEEE International Conference on Cluster Computing,
+ pages 431-438,
+ San Diego, CA, September 2004.
+ [PostScript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ George Kola, Tevfik Kosar and Miron Livny,
+ "Profiling Grid Data Transfer Protocols and Servers",
+ In Euro-Par 2004, Pisa, Italy, September 2004.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ John Bent, Douglas Thain, Andrea Arpaci-Dusseau, Remzi Arpaci-Dusseau, and Miron Livny,
+ "Explicit Control in a Batch Aware Distributed File System",
+ Proceedings of the First USENIX/ACM Conference on Networked Systems Design and Implementation,
+ San Francisco, CA, March 2004.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Sechang Son and Miron Livny, "Recovering Internet Symmetry in Distributed Computing",
+ Proceedings of the 3rd International Symposium on Cluster Computing and the Grid, Tokyo, Japan, May 2003.
+ [PDF]
+ [MS Word]
+ [BibTeX Source for Citation]
+
+
+ -
+ Douglas Thain, John Bent, Andrea Arpaci-Dusseau, Remzi Arpaci-Dusseau and Miron Livny,
+ "Pipeline and Batch Sharing in Grid Workloads",
+ in Proceedings of the Twelfth IEEE Symposium on High Performance Distributed Computing,
+ Seattle, WA, 2003.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Douglas Thain and Miron Livny,
+ "The Ethernet Approach to Grid Computing",
+ in Proceedings of the Twelfth IEEE Symposium on High Performance Distributed Computing,
+ Seattle, WA, 2003.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ John Bent, Venkateshwaran Venkataramani, Nick LeRoy,
+ Alain Roy, Joseph Stanley, Andrea Arpaci-Dusseau,
+ Remzi Arpaci-Dusseau, and Miron Livny,
+ "NeST - A Grid Enabled Storage Appliance",
+ in Jan Weglarz and Jarek Nabrzyski and Jennifer Schopf and
+ Macief Stroinkski, editors,
+ Grid Resource Management,
+ Kluwer Academic Publishers, 2003.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Douglas Thain, Todd Tannenbaum, and Miron Livny,
+ "Condor and the Grid",
+ in Fran Berman, Anthony J.G. Hey, Geoffrey Fox, editors,
+ Grid Computing: Making The Global Infrastructure a Reality,
+ John Wiley, 2003.
+ ISBN: 0-470-85319-0
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Francesco Giacomini,
+ Francesco Prelz,
+ Massimo Sgaravatto,
+ Igor Terekhov,
+ Gabriele Garzoglio,
+ and Todd Tannenbaum,
+ "Planning on the Grid: A Status Report [DRAFT]",
+ Technical Report PPDG-20,
+ Particle Physics Data Grid collaboration (http://www.ppdg.net),
+ October 2002.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ John Bent, Venkateshwaran Venkataramani,
+ Nick LeRoy,
+ Alain Roy,
+ Joseph Stanley,
+ Andrea Arpaci-Dusseau,
+ Remzi H. Arpaci-Dusseau,
+ and Miron Livny,
+ "Flexibility, Manageability, and Performance in a Grid Storage Appliance",
+ Proceedings of the Eleventh IEEE Symposium on High Performance Distributed Computing,
+ Edinburgh, Scotland, July 2002.
+ [Abstract]
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Douglas Thain and Miron Livny,
+ "Error Scope on a Computational Grid: Theory and Practice",
+ Proceedings of the Eleventh IEEE Symposium on High Performance Distributed Computing (HPDC11),
+ Edinburgh, Scotland, July 2002.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+ (This paper also describes aspects of Condor's Java Universe)
+
+
+ -
+ Douglas Thain, John Bent, Andrea Arpaci-Dusseau, Remzi Arpaci-Dusseau, and Miron Livny,
+ "Gathering at the Well: Creating Communities for Grid I/O",
+ in Proceedings of Supercomputing 2001,
+ Denver, Colorado, November 2001.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ James Frey, Todd Tannenbaum, Ian Foster, Miron Livny, and Steven Tuecke,
+ "Condor-G: A Computation Management Agent for Multi-Institutional Grids",
+ Journal of Cluster Computing
+ volume 5, pages 237-246, 2002.
+ [BibTeX Source for Citation]
+
+
+ -
+ James Frey, Todd Tannenbaum, Ian Foster, Miron Livny, and Steven Tuecke,
+ "Condor-G: A Computation Management Agent for Multi-Institutional Grids",
+ Proceedings of the Tenth IEEE Symposium on High Performance Distributed Computing (HPDC10)
+ San Francisco, California, August 7-9, 2001.
+ [Postscript]
+ [PDF]
+ [MS Word]
+ [BibTeX Source for Citation]
+
+
+ -
+ Douglas Thain, Jim Basney, Se-Chang Son, and Miron Livny,
+ "The Kangaroo Approach to Data Movement on the Grid",
+ in Proceedings of the Tenth IEEE Symposium on High Performance Distributed Computing (HPDC10),
+ San Francisco, California, August 7-9, 2001.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ John Bent, "Building Storage Appliances for the Grid and Beyond",
+ Masters' Project report, University of Wisconsin, Madison, May 2001.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+
+
+
Master-Worker Computing (MW, PVM, MPI, CARMI)
+
+
+ - Elisa Heymann, Miquel A. Senar, Emilio Luque, and Miron Livny,
+ "Adaptive Scheduling for Master-Worker Applications on the Computational Grid".
+ in Proceedings of the First IEEE/ACM International Workshop on Grid Computing (GRID 2000), Bangalore, India, December 17, 2000.
+ [Postscript]
+ [PDF]
+ [MS Word]
+ [BibTeX Source for Citation]
+
+
+ - Elisa Heymann, Miquel A. Senar, Emilio Luque, and Miron Livny,
+ "Evaluation of an Adaptive Scheduling Strategy for Master-Worker Applications on Clusters of Workstations".
+ in Proceedings of the 7th International Conference on High Performance Computing (HiPC 2000), Bangalore, India, December 17, 2000.
+ [Postscript]
+ [PDF]
+ [MS Word]
+ [BibTeX Source for Citation]
+
+
+ - Jeff Linderoth, Sanjeev Kulkarni, Jean-Pierre Goux, and Michael Yoder,
+ "An Enabling Framework for Master-Worker Applications on the Computational Grid",
+ Proceedings of the Ninth IEEE Symposium on High Performance Distributed Computing (HPDC9),
+ Pittsburgh, Pennsylvania, August 2000, pp 43-50.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Jeff Linderoth, Jean-Pierre Goux, and Michael Yoder,
+ "Metacomputing and the Master-Worker Paradigm",
+ Preprint ANL/MCS-P792-0200,
+ Mathematics and Computer Science Division, Argonne National Laboratory, February 2000.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Jim Pruyne and Miron Livny,
+ "Providing Resource Management Services to Parallel Applications",
+ Proceedings of the Second Workshop on Environments and Tools for Parallel Scientific Computing, May, 1994.
+ [Postscript]
+ [BibTeX Source for Citation]
+
+
+
+
+
Java
+
+ -
+ Douglas Thain and Miron Livny,
+ "Error Scope on a Computational Grid: Theory and Practice",
+ Proceedings of the Eleventh IEEE Symposium on High Performance Distributed Computing (HPDC11),
+ Edinburgh, Scotland, July 2002.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+ (This paper describes aspects of error handling in Condor's Java Universe)
+
+
+ -
+ Al Globus, Eric Langhirt, Miron Livny, Ravishankar Ramamurthy, Marvin Solomon, and Steve Traugott,
+ "JavaGenes and Condor: cycle-scavenging genetic algorithms",
+ Proceedings of the ACM Conference on JavaGrande,
+ San Francisco, California, 2000.
+ [PDF]
+ [BibTeX Source for Citation]
+ (This paper describes checkpointing Java applications for opportunistic computing.)
+
+
+
+
+
Remote Execution and Interposition Agents
+
+
+ - Douglas Thain and Miron Livny,
+ "Parrot: Transparent User-Level Middleware for Data-Intensive
+ Computing",
+ Scalable Computing: Practice and Experience,
+ Volume 6, Number 3, Pages 9-18, 2005.
+ [PDF]
+
+
+
+
+ -
+ Douglas Thain and Miron Livny,
+ "Parrot: Transparent User-Level Middleware for Data-Intensive Computing",
+ Workshop on Adaptive Grid Middleware,
+ New Orleans, Louisiana,
+ September 2003.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Douglas Thain and Miron Livny,
+ "Error Management in the Pluggable File System",
+ Technical Report 1448,
+ Computer Sciences Department, University of Wisconsin, October 2002.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Douglas Thain and Miron Livny,
+ "Multiple Bypass: Interposition Agents for Distributed Computing",
+ The Journal of Cluster Computing,
+ Volume 4, 2001, pp 39-47.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Douglas Thain and Miron Livny,
+ "Bypass: A Tool for Building Split Execution Systems",
+ Proceedings of the Ninth IEEE Symposium on High Performance Distributed Computing (HPDC9),
+ Pittsburgh, Pennsylvania, August 2000, pp 79-86.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Victor C. Zandy, Barton P. Miller, and Miron Livny,
+ "Process Hijacking",
+ The Eighth IEEE International Symposium on High Performance Distributed Computing (HPDC8),
+ Redondo Beach, California, August 1999, pp. 177-184.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Miron Livny and Michael Litzkow,
+ "Making Workstations a Friendly Environment for Batch Jobs",
+ Third IEEE Workshop on Workstation Operating Systems,
+ April 1992, Key Biscayne, Florida.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+
+
+
Security
+
+
+ -
+ Zach Miller, Dan Bradley, Todd Tannenbaum, Igor Sfiligoi,
+ "Flexible Session Management in a Distributed Environment",
+ Journal of Physics: Conference Series Volume 219, Issue 4, Year 2010.,
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Gabriele Garzoglio, Ian Alderman, Mine Altunay, Rachana Ananthakrishnan, Joe
+ Bester, Keith Chadwick, Vincenzo Ciaschini, Yuri Demchenko, Andrea Ferraro,
+ Alberto Forti, David L. Groep, Ted Hesselroth, John Hover, Oscar Koeroo, Chad
+ La Joie, Tanya Levshina, Zach Miller, Jay Packard, Håkon Sagehaug, Valery
+ Sergeev, Igor Sfiligoi, Neha Sharma, Frank Siebenlist, Valerio Venturi, John
+ Weigand,
+ "Definition and Implementation of a SAML-XACML Profile for Authorization
+ Interoperability Across Grid Middleware in OSG and EGEE",
+ Journal of Grid Computing, Volume 7, Issue 3, Year 2009.
+ [PDF]
+
+
+ -
+ Hao Wang, Somesh Jha, Miron Livny, and Patrick D. McDaniel,
+ "Security Policy Reconciliation in Distributed Computing Environments",
+ IEEE Fifth International Workshop on Policies for Distributed
+ Systems and Networks (POLICY 2004),
+ June 2004, Yorktown Heights, New York.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+
+
+
Scalability and Performance
+
+
+ -
+ E M Fajardo, J M Dost, B Holzman, T Tannenbaum, J Letts, A Tiradani, B Bockelman, J Frey and D Mason,
+ "How much higher can HTCondor fly?",
+ Journal of Physics: Conference Series, Vol. 664, 2015
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Dan Bradley, Timothy St Clair, Matthew Farrellee, Ziliang Guo, Miron Livny, Igor Sfiligoi,
+ and Todd Tannenbaum,
+ "An update on the scalability limits of the Condor batch system",
+ Journal of Physics: Conference Series, Vol. 331, No. 6, 2011.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ D Bradley, I Sfiligoi, S Padhi, J Frey and T Tannenbaum,
+ "Scalability and interoperability within glideinWMS",
+ Journal of Physics: Conference Series, Vol. 219, No. 6, 2010
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ D Bradley, S Dasu, M Livny, A Mohapatra, T Tannenbaum and G Thain,
+ "Condor enhancements for a rapid-response adaptive computing environment for LHC",
+ Journal of Physics: Conference Series Vol. 219, No. 6, 2010.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+
+
+
Experience
+
+ -
+ Michael Litzkow and Miron Livny,
+ "Experience With The Condor Distributed Batch System",
+ IEEE Workshop on Experimental Distributed Systems, Oct 1990, Huntsville, Al.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+
+
+
Scientific Applications
+
+
+ -
+ Douglas Thain, John Bent, Andrea Arpaci-Dusseau, Remzi Arpaci-Dusseau and Miron Livny,
+ "Pipeline and Batch Sharing in Grid Workloads",
+ in Proceedings of the Twelfth IEEE Symposium on High Performance Distributed Computing,
+ Seattle, WA, 2003.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Jim Basney, Rajesh Raman, and Miron Livny,
+ "High Throughput Monte Carlo",
+ Proceedings of the Ninth SIAM Conference on Parallel Processing for Scientific Computing,
+ March 22-24, 1999, San Antonio, Texas.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+ -
+ Chungmin Chen, Kenneth Salem, and Miron Livny,
+ "The DBC: Processing Scientific Data Over the Internet",
+ 16th International Conference on Distributed Computing Systems,
+ May 1996.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+
+
+
Scheduling
+
+
+
+ -
+ Mark Silberstein, Dan Geiger, Assaf Schuster, and Miron Livny,
+ "Scheduling Mixed Workloads in Multi-grids: The Grid Execution Hierarchy",
+ Proceedings of the 15th IEEE Symposium on High Performance Distributed Computing (HPDC),
+ city, state, month 2006.
+ [PDF]
+ [BibTeX Source for Citation]
+
+ - Derek Wright, "Cheap cycles from the desktop to the dedicated
+ cluster: combining opportunistic and dedicated scheduling with
+ Condor", Conference on Linux Clusters: The HPC Revolution,
+ June, 2001, Champaign - Urbana, IL.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+ [Power Point slides of talk presentation]
+
+
+ -
+ P. E. Krueger and Miron Livny,
+ "A Comparison of Preemptive and Non-Preemptive Load Distributing",
+ Proc. of the 8th International Conference on Distributed Computing Systems,
+ pp. 123-130, June 1988.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ -
+ Matt Mutka and Miron Livny,
+ "Scheduling Remote Processing Capacity In A Workstation-Processing Bank Computing System",
+ Proceedings of the 7th International Conference of Distributed Computing Systems,
+ pp. 2-9, September, 1987.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+ - Alain Roy and Miron Livny, "Condor and Preemptive Resume
+ Scheduling", Published in Grid Resource Management: State of the Art and
+ Future Trends, Fall 2003, pages 135-144, Fall 2003, Edited by Jarek
+ Nabrzyski, Jennifer M. Schopf and Jan Weglarz, published by Kluwer
+ Academic Publishers.
+ [PDF]
+
+
+
+
+
+
NMI Build & Test Laboratory
+
+
+ - Andrew Pavlo, Peter Couvares, Rebekah Gietzel, Anatoly Karp, Ian D. Alderman, Miron Livny, and Charles Bacon,
+ "The NMI Build & Test Laboratory: Continuous Integration Framework for Distributed Computing Software",
+ Proceedings of LISA '06: Twentieth Systems Administration Conference,
+ Washington, DC, December 2006, pp. 263 - 273.
+ [Postscript]
+ [PDF]
+ [BibTeX Source for Citation]
+ [PDF of presentation slides]
+
+ - A. Iosup, D.H.J. Epema, P. Couvares, A. Karp, and M. Livny,
+ "Build-and-Test Workloads for Grid Middleware: Problem, Analysis, and Applications",
+ Seventh IEEE International Symposium on Cluster Computing and the Grid (CCGRID),
+ IEEE Computer Society, Pages 205-213, May 2007.
+ [PDF]
+ [BibTeX Source for Citation]
+ [PDF of presentation slides]
+
+
+
+
+
Background Work
+
+
+ -
+ Miron Livny and Myron Melman,
+ "Load Balancing in Homogeneous Broadcast Distributed Systems",
+ Proceedings of Computer Network Performance Symposium,
+ April 13-14, 1982,
+ College Park, Maryland.
+ [PDF]
+ [BibTeX Source for Citation]
+
+
+
+
+
Miscellaneous
+
+ - Douglas Thain, Todd Tannenbaum, and Miron Livny, "How to Measure a
+ Large Open Source Distributed System", in Concurrency and
+ Computation: Practice and Experience, to appear in 2006.
+
+ - Zach Miller, Todd Tannenbaum, and Ben Liblit, "Enforcing Murphy's
+ Law for Advance Identification of Run-time Failures", Proceedings of
+ USENIX 2012.
+ [PDF]
+
+
+
+
+
PhD Dissertations from HTCondor team members at UW-Madison
+
+
+
+
+
+
+
+ Plant physiologists used high throughput computing to remedy research “bottleneck”
+
+
HTC resources increased the efficiency of the Spalding group’s data analyses, which enabled an increase in the scope of their research.
+
+
Enhancing his research with high throughput computing was a pivotal moment for University of Wisconsin–Madison molecular plant physiologist Edgar Spalding when his
+research group adopted it in 2006. Over the past five years, the research group has used more than 200,000 computing hours, including to facilitate “the development of the measurement algorithm and the automatic processing of tens-of-thousands of images” of maize seedling root growth, Spalding says.
+
+
+ 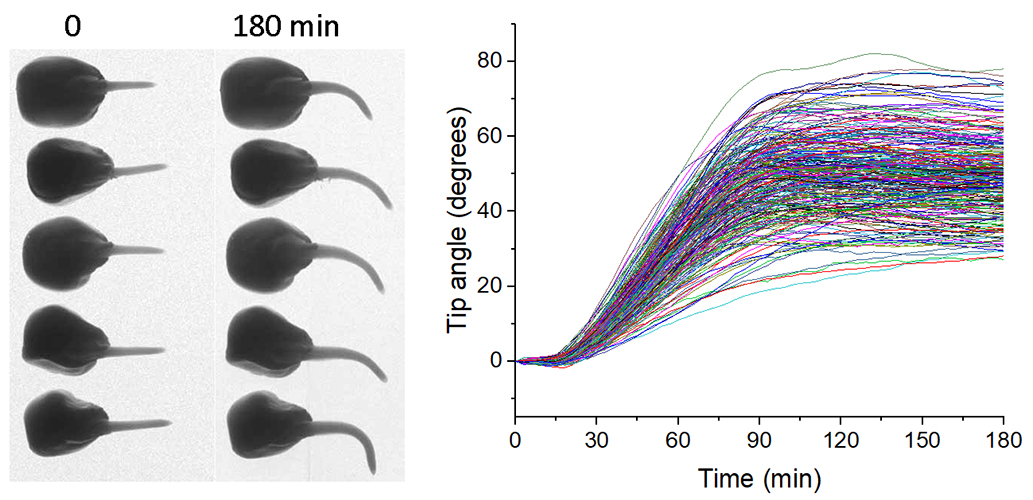 + The graph shows the average gravitropic response of each of the maize types. Statistical genetics techniques mapped variation in these curves (the phenotype) to genes that control the process. HTCondor scheduling software and computing resources at CHTC were used to develop the measurement algorithm and to process the tens of thousands of images automatically.
+ The graph shows the average gravitropic response of each of the maize types. Statistical genetics techniques mapped variation in these curves (the phenotype) to genes that control the process. HTCondor scheduling software and computing resources at CHTC were used to develop the measurement algorithm and to process the tens of thousands of images automatically.
+
+
+
Spalding’s research group was studying Arabidopsis plant populations with genetically diverse members and tracking their response to light or gravity due to a mutation — one seedling at a time. Since Arabidopsis seedlings are only a few millimeters tall, Spalding says his research group found that obtaining high-resolution digital images was the best approach to measure the direction of their growth. A computer collected images every few minutes as the seedlings grew. “If we could characterize this whole genetically diverse population, we could use the powerful techniques of statistical genetics to track down the genes affecting the process. That meant we now had thousands and thousands of images to measure,” Spalding explains.
+
+
The thousands of digital images to measure created a bottleneck in Spalding’s research. That was before he led an effort with the Center for High Throughput Computing (CHTC) Director Miron Livny, other plant biologists, and computer scientists to develop a proposal for a competitive National Science Foundation (NSF) grant that would produce cyberinfrastructure to support plant biology research. Though the application wasn’t successful, the connections Spalding made from that meeting were meaningful nonetheless.
+
+
Speaking with Livny at the meeting — from whom he learned about the capabilities of the HTC approach that was pioneered on our campus — helped Spalding realize the inefficiencies of his group in analyzing thousands of seedlings. “[O]ur research up until that point had been focused on one seedling at a time. Faced with large numbers of seedlings to do a broader scale of investigation meant that we had to find computing methodologies that matched our new data type, which was tens of thousands of images instead of a couple of dozen. That drove our need for a different way of computing,” Spalding describes.
+
+
When asked about which accomplishment using HTC was most impactful, Spalding said “The way we measure yield-related features from maize ears and several thousand kernels has had a large impact.” Others from around the world began asking for their help with making similar measurements. “In many cases, we can use our workflow [algorithms] running on CHTC to process their images of maize ears and kernels and return data that helps them answer their scientific or crop breeding questions,” Spalding says.
+
+
Since the goals of the experiments determine the type of data the researchers collect, they did not need to adjust the type of data they collected. Rather, adopting the HTC approach changed the way they created tools to analyze the data. Today, Spalding says his research group continues to use HTC in three ways: “from tool development to extracting the features from the images with the tool that you developed to applying it in the challenge of statistically matching it to elements of the results to elements of the genome.” As his team became more experienced in writing new algorithms to make measurements, they realized that HTC was useful in developing new methodologies; it was more than just more automation and increased computing capacity.
+
+
In other words, HTC is useful as both a development resource and a production resource. Making measurements on seedlings and then matching processes to the genome elements that control those processes involved an ever-growing amount of computing capacity. “We realized that statistical modeling of the measurements from the biology to the genetic information in the population also benefited from high throughput computing.” HTC in all these cases, Spalding elaborates, “was beneficial and changed the way we work. It changed the nature of the questions we asked.” In addition to these uses of HTC, the research group’s uses of machine learning (ML) also continue to become a bigger part of the tool development stage and in driving the methods to train a model to recognize a feature in a seedling.
+
+
Spalding has also shared his HTC experience with the attendees of the annual OSG School. Spalding emphasizes that students “should not hold back on doing something because they think computing will be a bottleneck. There are ways to bring the computing they need to their problem and they should not shy away from a question just because they think it might be difficult to compute. There are people like the CHTC staff that can remove that bottleneck if the person’s willing to learn about it.”
+
+
“Engaged and motivated collaborators like Spalding and his group is what guides CHTC in advancing the state of the art of HTC and drives our commitment to bring these advances to researchers on the UW-Madison campus and around the world,” says Livny.
+
+
+
+
+
+
+
+
+ Adding News Articles
+
+
+
Table of Contents
+
+
+
Using Markdown
+
+
You will be using Markdown to write all news articles. Markdown is a popular markup
+language that is converted to HTML before being displayed on the website.
+
+
A good cheatsheet can be found here which contains
+the markdown syntax and examples of how it looks when converted to html.
+
+
Adding Article To the Website
+
+
After you have written your article in the text editor of your choice and are ready to have it on the website you will first need to create a preview branch.
+
+
All of our websites have a preview location where you can view changes before adding them to the main website. I will use PATh for the example below, but this is the same process for CHTC, HTCondor and OSG as well.
+
+
+
+
+ - Go to the Github repo and Create a preview branch
+
+ - Branch name must start with ‘preview-‘ followed by a descriptive term.
+
+ - Example: You write an article about HTC and Genes, you name the branch ‘preview-gene-article’.
+
+
+ 
+
+ - Add your News article
+
+ - Check that are in your new branch
+
+ - The previous step will put you in your new preview branch. You can check by looking at the branch name displayed.
+
+
+ - Go into the news article directory
+ - Add new file with title ‘YYYY-MM-DD-title.md’
+
+ - Example: For the HTC and Genes article -> ‘2021-12-25-htc-and-genes.md’ if you are going to publish the article on Christmas 2021.
+
+
+ - Copy and Paste in the template
+
---
+ title: # Article Title
+ date: 9999-12-31 # Article Date - In format YYYY-MM-DD - Article will not show on website until Article Data >= Current Date
+ excerpt: # Article Excerpt - An abstract of the article
+ image_src: # Path to the image to be displayed in article cards
+ image_alt: # A description of this image
+ author: # Article Author
+ published: false # If this article should be on the website, change to true when ready to publish
+ ---
+
+ Content
+
+
+ - Fill in all the front matter and replace ‘Content’ with your article.
+

+
+
+ - Review your Preview
+
+ - Look for your article preview at
+ - Example for PATh: https://path-cc.io/web-preview/preview-helloworld
+
+ - Project Preview URL Prefix: https://path-cc.io/web-preview/
+ - Branch Name: preview-helloworld
+

+
+
+
+
+ - Create a Pull Request
+
+ - When the article is ready to go live you must create a pull request from your branch into ‘master’.
+ - Comment the preview URL in the Pull Request for easy review.
+

+
+
+
+
+
Using Images
+
+
Adding Images
+
+
Images can be added using either MD or HTML.
+
+
+ - Markdown
+
+
+ - HTML
+
+ - Images added with html will use the ‘img’ tag.
+ - The syntax for this is:
+
<img class="optional-class" src="/path/to/image" alt="Description of image for the visually impaired"
+
+
+ - Using HTML gives you more options such as having figure captions
+
<figure>
+ <img src="/path/to/image" alt="Description">
+ <figcaption>The image caption</figcaption>
+ </figure>
+
+
+
+
+
+
+
Reducing Image Size
+
+
High definition images can take up space that slows down the website when it loads, because of this it is important to reduce this footprint before adding them to the website.
+
+
To keep the image size reasonable follow the rules below.
+
+
+ - <= 1000 pixels wide
+
+ - This is the maximum website width, so any images wider will have unused resolution.
+
+
+ - Convert to jpg and reduce to reasonable size
+
+ - This is up to you and changes picture to picture. Some pictures look fine when compressed, some don’t.
+ - Reasonable target is ~200kb
+
+
+
+
+
Example
+
+

+
+
We will reduce this 2MB image to demonstrate.
+
+
+ -
+
Pull up the image in photoshop. If you don’t have photoshop contact IT as you do have free access.
+
+ -
+
Go to Export as…
+
+
+ -
+
Update the Values to reduce size.
+
+
+ -
+
Use your new compressed image in the article.
+
+
+
+
+
Positioning Images
+
+
To position images on the page you must use classes.
+For markdown this means including [: .<class> } above the image syntax, and
+for html this means adding class="<class>" inside the html tag.
+
+
Potential classes that can be used to position the image come from Bootstrap Utilities.
+
+
The ones you will find the most helpful are:
+
+ - Floats
+ - Image Specific
+
+
+
+
+
+
+
+
+
+ Schedule Calendar Guide
+
+
+
+
+
+
+
Moving From Previous AFS File
+
+
This section shows how one might transfer from entering their times in afs to entering their times into a Google calendar.
+
+
Save a copy of your schedule file, the google calendar will overwrite it
+
+
If you did not do this and now need it back, Cannon has them all saved as of 2022-07-15.
+
+
Create an ics file from your current file in afs
+
+
Login to moria
+
+
cd /p/condor/public/html/htcondor/developers/schedules
+
+source generate_personal_ics.sh <your_file_name>
+
+
+
If an error is reported in your schedule file you can fix it or ignore it and that event just won’t be added.
+
+
Grab the ics file from this build and import it into your calendar you create below.
+
+
This can be done on the import page.
+
+
Don’t forget to also create your new schedule file which holds your schedule meta-data.
+
+
Creating The Schedule File
+
+
This section will describe how to create your yaml file, you can find a verbose template file on afs or use the one below.
+
+
<filename>.yml
+
Name: "name"
+ShortName: "shortname"
+corehours: "corehours"
+DailyEmail: "Yes" # ( or omit line entirely )
+Email: "email"
+Office: "location"
+Phone: "phone" # Office and/or Cell - This is read as a string so format how you want
+calendarurl: "calendarurl"
+default:
+ starttime: "starttime"
+ endtime: "endtime"
+ status: "status"
+
+
File Details
+
+
+ - Name: First Last
+ - ShortName: (Optional, defaults to First Name if not specified) Should be unique and obviously you. If your name is “George Washington” and George Foreman also works in your group, “GeorgeW” would be a good choice.
+ - corehours: A description of your corehours that is displayed. Format is not important. Example is “9:00 AM to 5:00 PM”
+ - DailyEmail: If ‘Yes’ then you will receive a daily email with who is out, otherwise should be omitted entirely.
+ - Email: Your preferred email address. Defaults to filename@cs.wisc.edu so you will likely want to change this
+ - Office: Your office location. Example => “4261 CS”
+ - Phone: Your phone number(s). Example => “+1 608 265 5736 (office)
+1 608 576 0351 (cell)”
+ - DailyEmail: Do you want a daily email with information about who is gone?
+ - calendarurl: The url to your outage calendar. Details on obtaining this found below.
+ - starttime: Your typical start time, use military format. Example => “09:00”
+ - endtime: Your typical end time, use military format. Example => “17:00”
+ - status: Your status during these hours. If you are unsure use “Office”.
+ - default[Monday, Tuesday, Wednesday, Thursday, Friday]: This overwrites the default for that day. Use the same format as default.
+
+
+
Important
+
+
All of these data strings have to be encased in double quotations to be valid yaml. This encasement can be seen in the template file.
+
+
Creating Your ICAL URL
+
+
To power your outage calendar you need to create a google calendar which is solely used to populate your outages.
+
+
+ - Go to https://calendar.google.com/ and sign in with your preferred account. You can use @morgridge.org and @wisc.edu.
+ - Create a new calendar
+
+ - Name and Description do not matter
+ 
+
+ - Go into Calendar settings and retrieve the Secret Address
+
+ - Go to calendar Settings
+

+ - Get the secret calendar url ( Will warn not to give this out )
+
+
+
+ - Post this address into your yaml file as the calendarurl
+
+
+
Populating Your Days Off
+
+
Event Title
+
+
The event title should be one of the statuses bolded below. These statuses are
+used to key the type of outage so anything but a approved status should be in the event title.
+
+
+ - Travel: Working, but not at the office. Perhaps a conference
+ - Vacation: Taking vacation (“vacation” and “personal holiday” on the leave report)
+ - Sick: Taking sick leave (“sick leave” on the leave report)
+ - Holiday: Taking floating holiday (“legal holiday” on the leave report)
+ - Furlough: State- or UW-mandated furlough (as required). Includes both fixed (“mandatory”) and floating time.
+ - Off: Days not worked on a part-time employment
+ - WFH: Work From Home
+
+
+
Event Description
+
+
Any description of the outage you would like to add can be added in the event
+description.
+
+
Marking Event Time
+
+
Marking Full day/days Out
+
+
To mark full day outages you create an event with the “All day” attribute ticked ( This is used in the demo above ). Populate the title and description as expected.
+
+
Do not use the recurring event feature for multiple outage days.
+
+
Marking Partial Outages
+
+
To mark partial time you must do two different things.
+
+
+ - Append the amount of hours this outage is taking with a colon separating the title.
+
+ - For Example, if you have a four hour doctor appt. you would mark SICK:4
+ - For Example, if you leave for vacation half a day early you would mark VACATION:4
+
+
+ - Mark the time you are *in* the office on Google
+
+ - This is non-intuitive but when you are marking time you mark the time you are in.
+ - For Example, if I am normally in 9-5 and am leaving 4 hours early I will mark my event to go from 9:00 AM to 1:00 PM.
+
+
+
+
+
Example
+
+
If you mark your title in Google as “Sick” and the description as “Wisdom Tooth Surgery and Recovery” the schedule output will be as so.
+
+

+
+

+
+
+
+
+
+
+
+
+ CHTC Staff Page
+
+
+
Docs:
+
+
+
Schedule Calendar
+
+
+
+
+
+
+
+
+ Tackling Strongly Correlated Quantum Systems on OSPool
+
+
Duke University Associate Professor of Physics Shailesh Chandrasekharan and his graduate student Venkitesh Ayyar are
+using the OSpool to tackle notoriously difficult problems in quantum systems.
+
+
+
+
+  + Shailesh Chandrasekharan, courtesy photo
+
+ Shailesh Chandrasekharan, courtesy photo
+
+
+
+
+  +
+
+ Venkitesh Ayyar, courtesy photo
+
+
+
+
+
These quantum systems are the physical systems of our universe, being investigated at the fundamental level where
+elemental units carrying energy behave according to the laws of quantum mechanics.
+In many cases, these units might be referred to as particles or more generally as “quantum degrees of freedom.”
+The most exciting physics arises when these units are strongly correlated: the behavior of each one depends on the
+system as a whole; they cannot be taken and studied independently.
+Such systems arise naturally in many areas of fundamental physics, ranging from condensed matter (many materials
+fabricated in laboratories contain electrons that are strongly correlated and show exotic properties) to nuclear and
+particle physics.
+
+
The proton, one of the particles inside an atom’s nucleus, is itself a strongly correlated bound state involving many
+quarks and gluons.
+Understanding its properties is an important research area in nuclear physics.
+The origin of mass and energy in the universe could be the result of strong correlations between fundamental quantum
+degrees.
+
+
“Often we can write down the microscopic theory that describes a physical system.
+For example, we believe we know how quarks and gluons interact with each other to produce a proton.
+But then to go from there to calculate, for instance, the spin of the proton or its structure is non-trivial,” said
+Chandrasekharan.
+“Similarly, in a given material we have a good grasp of how electrons hop from one atom to another.
+However, from that theory to compute the conductivity of a strongly correlated material is very difficult.
+The final answer—that helps us understand things better—requires a lot of computation.
+Typically the computational cost grows exponentially with the number of interacting quantum degrees of freedom.”
+
+
According to Chandrasekharan, the main challenge is to take this exponentially hard problem and convert it to something
+that scales as a polynomial and can be computed on a classical computer.
+“This step is often impossible for many strongly correlated quantum systems, due to the so-called
+sign problem which arises due to quantum mechanics,” added
+Chandrasekharan.
+“Once the difficult sign problem is solved, we can use Monte Carlo calculations to obtain answers.
+Computing clusters like the OSG can be used at that stage.”
+
+
Chandrasekharan has proposed an idea, called the fermion bag approach,
+that has solved numerous sign problems that seemed unsolvable in systems containing fermions (electrons and quarks are
+examples of fermions).
+In order to understand a new mechanism for the origin of mass in the universe, Ayyar is specifically using the OSG to
+study an interacting theory of fermions using the fermion bag approach.
+
+
+  + Illustration of a fermion bag configuration. Image credit: Shailesh Chandrasekharan
+
+ Illustration of a fermion bag configuration. Image credit: Shailesh Chandrasekharan
+
+
+
“We compute correlation functions on lattices and look at their behavior as the lattice size increases,” Ayyar explained.
+In the presence of a mass, the correlation functions decay exponentially.
+“Ideally, we would want to perform computations on very large lattices (>100x100x100).
+Each calculation involves computing the inverse of large matrices millions of times.
+The matrix size scales with the lattice size and so the time taken increases very quickly (from days to weeks to months).
+This is what limits the size of the lattice used in our computation and the precision of the quantities calculated.
+”In a recent publication, Ayyar and Chandrasekharan performed computations on lattices of sizes up to 28x28x28, and
+more recently they have been able to push these to lattices of size 40x40x40.
+
+
Since their computation is parallelizable, they can run several calculations at the same time.
+Ayyar says this makes the OSG perfect for their work.
+“Instead of running a job for 100 days sequentially,” he noted, “we can run 100 jobs simultaneously for one day to get
+the same information.
+This not only helps us speed up our calculation several times, but we also get very high precision.”
+
+
Ayyar uses simple scripts to submit a large number of jobs and monitor their progress.
+One challenge he faced was the check-pointing of jobs.
+“Some of our jobs run long, say two to six days, and we found these getting terminated before completion due to the
+queuing system,” Ayyar said.
+To solve this, he developed what he calls ‘manual check-pointing’ to execute jobs in pieces.
+“This extends the completed processes and submits them so that long-running processes can be completed.
+Being able to control the memory and disk-space requirements on the target nodes has proved to be extremely useful.”
+
+
Ayyar also noted that many individual research groups cannot afford the luxury of having thousands of computing nodes.
+“This kind of resource sharing on the OSG has helped computational scientists like us attempt calculations that could
+not be done before,” he added.
+“For example, we are now attempting computations on lattices of size 60x60x60.
+One sweep should only take a few hours on each core.”
+
+
Chandrasekharan points out that past technology breakthroughs like the kind that revolutionized processor chip
+manufacturing have largely been based on basic quantum mechanics learned in the 1940s and 1950s.
+“We still need to understand the complexity that can be produced when many quantum degrees of freedom interact with each
+other strongly,” said Chandrasekharan.
+“The physics we learn could be quite rich.”
+
+
He says this next phase of research is already happening in nanoelectronics.
+“If the computational quantum many-body challenge that we face today is solved, it may help revolutionize the next
+generation of technology and give us a better understanding of the physics.”
+
+
Ayyar and Chandrasekharan recently submitted a paper based on their work using the OSG.
+Titled Massive fermions without fermion bilinear condensates,
+it has been published in the journal Physical Review D of the American Physical Society.
+
+
– Greg Moore
+
+
+
+
+
+
+
+ Technologies
+
+
The CHTC offers a suite of open-source software tools that manage HTC
+workloads and enable organizations to form distributed HTC pools. The
+HTCondor Software Suite (HTCSS)
+is the product of over three decades of
+research and development at the Computer Sciences Department of the
+University of Wisconsin-Madison. It has been adopted by academic and
+commercial entities around the world in support of their HTC workloads.
+
+
+
+
+
+
+
+
+ The Pelican Project: Building a universal plug for scientific data-sharing
+
+
From its founding, the Morgridge Institute for Research has driven the idea that open sharing of research computing resources will be a great enabler of scientific discovery, powering everything from black hole astronomy to stem cell biology.
+
+
Increasingly, the principle of sharing is being applied not only to computing resources, but to the wealth of data those projects are producing. Resources such as high-throughput computing and the OSG Consortium have been incorporating more tools for scientists to share their raw data for further exploration.
+
+
This principle is now getting traction on a national policy scale. The White House Office of Science and Technology Policy (OSTP) established new requirements in 2022 that any research supported by federal funds must be made available to the public without embargoes or paywalls.
+
+
This mandate applies not only to published findings, but to the core data those findings are based upon. Within the scientific community, the approach is referred to as the “FAIR” principles, which means that scientific data should be “findable, accessible, interoperable and reusable.”
+
+
Obviously, applying this new standard to data is as much a technical challenge as it is a cultural one. A new project at the Morgridge, led by research computing investigators Brian Bockelman and Miron Livny, is working toward creating a software platform that can facilitate the sharing of diverse research datasets.
+
+
Nicknamed “Pelican,” the project is supported through a $7 million grant from the National Science Foundation (NSF). The award (OAC-2331489) will strive to make data produced by researchers, from single-investigator labs to international collaborations, more accessible for computing and remote clients for viewing. Pelican supports and extends the work Bockelman and Livny have been doing as part of the OSG Consortium for over a decade.
+
+
Bockelman says that public research data-sharing has been a growing movement the past decade, but the COVID-19 pandemic served as a potent catalyst. The pandemic made the benefits of sharing abundantly clear, including the development of a vaccine at an unprecedented pace — 6 months compared to a typical multi-year process.
+
+
“Our philosophy is that not only should your research paper be public and readable, but your data should be as well,” Bockelman says. “If scientists just say, ‘here are the results in a pretty graph,’ and don’t share the underlying dataset, we lose a lot of value when others can’t access the data, can’t interpret it, or use it for their own research.”
+
+
Bockelman says there are some other core benefits that may come from the open science push. By making data more readily accessible, it should improve the reproducibility of experiments and potentially reduce scientific fraud. It can also narrow the gap between the “haves” and “have-nots” in the research world by providing data access regardless of institutional resources.
+
+
Bockelman likens the Pelican project to developing a “universal adapter plug” that can accommodate all different types of data. Just like homes have standard outlets that work for all different household appliances, that same approach should help individual scientists plug into a sharable data platform regardless of the nature of their data.
+
+
One of the first proving grounds for Pelican will be its participation within the National Discovery Cloud for Climate, an effort to bring together compute, data, and network resources to democratize access and advance the climate-related science and engineering. Bockelman says the Pelican project will help optimize this data sharing effort with the climate science community and provide a proof of concept for other research areas.
+
+
But ultimately, the best benefit may be enhancing public trust in high-impact science.
+
+
“Even for people who may not go digging into the data, they want to know that science has been done responsibly, especially for fields where it directly affects their lives,” Bockelman says. “Climate is a great example of where the science can really drive regulations that affect people. Getting data out as open and following the FAIR principles … is part of that relationship between the scientific community and the society at large.”
+
+
Bockelman says making data accessible is more than just downloading from a webserver. Pelican works to establish approaches that help people utilize the data effectively from anywhere in the nation’s computing infrastructure — essential so anyone from a tribal college to the largest university can understand and interpret the climate data.
+
+
The original memo was written in 2022 by then OSTP Director Alondra Nelson, and today the “Nelson memo” is viewed as a watershed document in federal research policy.
+
+
“When research is widely available to other researchers and the public, it can save lives, provide policy makers with the tools to make critical decisions, and drive more equitable outcomes across every sector of society,” Nelson wrote. “The American people fund tens of billions of dollars of cutting-edge research annually. There should be no delay or barrier between the American public and the returns on their investments in research.”
+
+
+
+
+
+
+
+ Tribal College and CHTC pursue opportunities to expand computing education and infrastructure
+
+
Salish Kootenai College and CHTC take steps toward bringing underrepresented communities to cyberinfrastructure.
+
+
Access to cyberinfrastructure (CI) is the bedrock foundation essential for students and researchers determined to contribute to science.
+That’s why Lee Slater,
+the Cyberinfrastructure Facilitator at Salish Kootenai College (SKC), a tribal community college in northwest Montana, first brought
+up the “missing millions.” The term was coined after the National Science Foundation (NSF) reported
+that users and providers of the CI as a whole do not accurately represent society. Underrepresented racial and gender demographics were largely missing from
+the field. “[The missing millions] just don’t have access to high performance computing platforms and so they’re not contributing greatly to the scientific
+body of knowledge that other privileged students have access to,” Slater explained. “It’s a real serious deficit for these students. One of the goals we’re
+trying to get accomplished is to bring these educational and research platforms to students and faculty to really enrich the experience they have as students.”
+
+
SKC inhabits an indigenous reserve known as the Flathead Reservation, which includes territory in four western states. Established in 1855, the reservation
+is home to the Confederated Salish and Kootenai Tribes. SKC — with just over 600 students — makes up a
+small, but vital portion of the much larger reservation. The college consists largely of tribal descendents or members, making up almost 80 percent of the
+school population.
+
+
+  + TCU Salish Kootenai College in Montana.
+
+ TCU Salish Kootenai College in Montana.
+
+
+
+
The Center for High Throughput Computing (CHTC) Director Miron Livny traveled
+to Montana this past October to meet with Salish Kootenai College faculty and staff. The four-day trip was coordinated by International Networking
+Coordinator Dale Smith from the University of Oregon, who also works for the American Indian Higher Education Consortium.
+The visit was meant for Livny to experience one of the nation’s tribal colleges and universities (TCUs) and to further the discourse between CHTC and SKC.
+“The main goal was for him to see our infrastructure, meet the faculty and see research opportunities,” Slater recalled.
+
+
SKC’s biggest and most immediate computing goal is to provide the access and training to utilize a web platform for JupyterHub that would be available
+for faculty and student use. The Jupyter Notebook connects with an OSPool Access Point, where students can place their workloads and data and which
+automates the execution of jobs and data movement across associated resources. Slater believes this would be beneficial, as many SKC faculty members do
+computing and data analysis within their specialties. “The fact that we could have a web platform with JupyterHub that students could access and faculty
+could access would really be a great facilitation,” Slater explained.
+
+
Slater would also like to collaborate with other TCUs, train faculty in computing software and overall increase their cyberinfrastructure capabilities.
+SKC Chief Information Officer (CIO) Al Anderson would
+like to leverage storage capacity for a faculty researcher who is examining the novel behavior of elk on the National Bison Range. This work requires taking a
+vast amount of photographs that then must be processed and stored. “We found that we have this storage issue — right now they’re using portable hard drives
+and it’s just a mess,” Anderson said.
+
+
Engagements like this are an early, but important step in bringing underserved communities to cyberinfrastructure and thus to science and research.
+The NSF “Missing Millions” report focused on the need for democratizing access to
+computing and showed a deficiency of engagement with institutions created for marginalized groups. Institutions like historically black colleges and universities (HBCUs)
+and TCUs tend to lack cyberinfrastructure capabilities that can be hard to implement without engagement from outside institutions.
+SKC’s engagement with CHTC is an example of steps both are taking in addressing this deficiency.
+
+
Longer term goals for the college are largely educational-focused. “We’re a small school, traditionally we have more educational needs than really heavy
+research needs,” Slater said. Anderson agreed stating, “I think a lot of our focus is the educational side of computing and how to get people hooked into
+those things.”
+
+
Anderson and Slater are also focused on relationship-building with faculty and discovering what they need to educate their students.
+They believe hearing from the SKC community should be first and foremost. “We’re still in that formative stage of asking, what do we need to support?”
+Anderson explained, “Through these conversations we’re slowly discovering.”
+
+
+
+
+
+
+
+ PATh Extends Access to Diverse Set of High Throughput Computing Research Programs
+
+
Finding the right road to research results is easier when there is a clear PATh to follow. The Partnership to Advance Throughput Computing (PATh)—a partnership between the OSG Consortium and the University of Wisconsin-Madison’s Center for High Throughput Computing (CHTC) supported by the National Science Foundation (NSF)—has cleared the way for science and engineering researchers for years with its commitment to advancing distributed high throughput computing (dHTC) technologies and methods.
+
+
HTC involves running a large number of independent computational tasks over long periods of time—from hours and days to week or months. dHTC tools leverage automation and build on distributed computing principles to save researchers with large ensembles incredible amounts of time by harnessing the computing capacity of thousands of computers in a network—a feat that with conventional computing could take years to complete.
+
+
Recently PATh launched the PATh Facility, a dHTC service meant to handle HTC workloads in support and advancement of NSF-funded open science. It was announced earlier this year via a Dear Colleague Letter issued by the NSF and identified a diverse set of eligible research programs that range across 14 domain science areas including geoinformatics, computational methods in chemistry, cyberinfrastructure, bioinformatics, astronomy, arctic research and more. Through this 2022-2023 fiscal year pilot project, the NSF awards credits for access to the PATh Facility, and researchers can request computing credits associated with their NSF awards. There are two ways to request credit: 1) within new proposals or 2) with existing awards via an email request for additional credits to participating program officers.
+
+
“It is a remarkable program because it spans almost the entirety of the NSF’s directorates and offices,” said San Diego Supercomputer Center (SDSC) Director Frank Würthwein, who also serves as executive director of the OSG Consortium.
+
+
Access to the PATh Facility offers researchers approximately 35,000 modern cores and up to 44 A100 GPUs. Recently SDSC, located at UC San Diego, added PATh Facility hardware on its Expanse supercomputer for use by researchers with PATh credits. According to SDSC Deputy Director Shawn Strande: “Within the first two weeks of operations, we saw researchers from 10 different institutions, including one minority serving institution, across nearly every field of science. The beauty of the PATh model of system integration is that researchers have access as soon as the resource is available via OSG. PATh democratizes access by lowering barriers to doing research on advanced computing resources.”
+
+
While the PATh credit ecosystem is still growing, any PATh Facility capacity not used for credit will be available to the Open Science Pool (OSPool) to benefit all open science under a Fair-Share allocation policy. “For researchers familiar with the OSPool, running HTC workloads on the PATh Facility should feel like second-nature” said Christina Koch, PATh’s research computing facilitator.
+
+
“Like the OSPool, the PATh Facility is nationally spanning, geographically distributed and ideal for HTC workloads. But while resources on the OSPool belong to a diverse range of campuses and organizations that have generously donated their resources to open science, the allocation of capacity in the PATh Facility is managed by the PATh project itself,” said Koch.
+
+
PATh will eventually reach over six national sites: SDSC at UC San Diego, CHTC at the University of Wisconsin-Madison, the Holland Computing Center at the University of Nebraska-Lincoln, Syracuse University’s Research Computing group, the Texas Advanced Computing Center at the University of Texas at Austin and Florida International University’s AMPATH network in Miami.
+
+
PIs may contact credit-accounts@path-cc.io with questions about PATh resources, using HTC, or estimating credit needs. More details also are available on the PATh credit accounts web page.
+
+
+  +
+ A diverse set of PATh national and international users benefit from the resource, and the recent launch of the PATh Facility further supports HTC workloads in an effort to advance NSF-funded open science. The colors on the chart correspond to the total number of core hours – nearly 884,000 – utilized by researchers at participating universities on PATh Facility hardware located at SDSC. Credit: Ben Tolo, SDSC
+
+
+
+ A diverse set of PATh national and international users benefit from the resource, and the recent launch of the PATh Facility further supports HTC workloads in an effort to advance NSF-funded open science. The colors on the chart correspond to the total number of core hours – nearly 884,000 – utilized by researchers at participating universities on PATh Facility hardware located at SDSC. Credit: Ben Tolo, SDSC
+
+
+
+
+
+
+
+
+
+ Protecting ecosystems with HTC
+
+
Researchers at the USGS are using HTC to pinpoint potential invasive species for the United States.
+
+
+  + From left to right: Mississippi River Delta, Colorado Rocky Mountains, Kansas’s Milford Lake. Images by USGS on Unsplash.
+ From left to right: Mississippi River Delta, Colorado Rocky Mountains, Kansas’s Milford Lake. Images by USGS on Unsplash.
+
+
+
Benjamin Franklin famously advised that an ounce of prevention is worth a pound of cure, and researcher Richard Erickson has taken this advice to heart in his mission to protect our lakes and wildlife from invasive species. As a research ecologist at the United States Geological Survey’s (USGS) Upper Midwest Environmental Sciences Center, Erickson uses computation to identify invasive species before they pose a threat to U.S. ecosystems.
+
+
Instrumental to his preventative mission is the HTCondor Software Suite (HTCSS) and consulting from UW-Madison’s Center for High Throughput Computing (CHTC), which have been integral to the USGS’s in-house computing infrastructure. Equipped with the management capabilities of HTCSS and guidance from CHTC, Erickson recently completed a high-throughput horizon scan of over 8000 different species in less than two days.
+
+
Explaining how his team was able to accomplish such a feat in merely one weekend, Erickson reasons: ”High throughput computing software allows [big problems] to be broken into small jobs. Rather than having to worry about everything, I just have to worry about a small thing, and then high throughput computing does the small thing many times over, to solve big problems through small steps.”
+
+
Erickson’s big problem first began to take shape in 2020 when the U.S. Fish and Wildlife Service (FWS) provided the USGS with a list of over 8000 species currently being bought and sold in the United States, from Egyptian Geese, to Algerian hedgehogs, to Siberian weasels. If these animals proliferate in U.S. environments, they could potentially threaten native species, the ecosystem as a whole, and the societal and economic value associated with it. Erickson’s job? To determine which species are a threat, and to what areas –– a tall order when faced with 8000 unique species and roughly 900 different ecological regions across the United States.
+
+
With HTC, Erickson could approach this task by breaking it down into small, manageable steps. Each species was independent of one another, meaning that the colossal collection of 8000 plants and animals could be organized into 8000 different jobs for HTCSS to run in parallel. Each job contained calculations comparing the US and non-US environments across sixteen different climate metrics. Individually, the jobs took anywhere from under thirty minutes to over two hours to run.
+
+
To analyze this type of data, the team created their own R package, climatchR. The package was released to the public in early September, and the team plans to make their HTCondor code publicly available after it undergoes USGS review.
+
+
But the HTC optimization didn’t end there. Because the project also required several complex GIS software dependencies, the group used Docker to build a container that could hold the various R and GIS dependencies in the context of a preferred operating system. Such containers make the software portable and consistent between diverse users and their computers, and can also be easily distributed by HTCSS to provide a consistent and custom environment for each computed job running across a cluster.
+
+
By the end of their computing run, the 8000 jobs had used roughly a year of computing in less than two days. The output included a climate score between zero and ten for each of the 8000 species, corresponding to how similar a species’ original climate is to the climates of the United States.
+
+
Currently, different panels of experts are reviewing species with climate scores above 6 to determine which of them could jeopardize US ecosystems. This expert insight will inform FWS’s regulation and management of the species traded in the United States, ultimately preventing the arrival of those that are likely to be invasive.
+
+
Invasive species disrupt ecological interactions, contributing to the population decline and extinction of native species. But beyond their environmental consequences, these non-native species impact property values, tourism activities, and agricultural yields. Hopefully, the results of Erickson’s high-throughput horizon screen will prevent these costs before they’re endured –– all by using HTC to solve big problems, through small steps.
+
+
…
+
+
Erickson co-authored an open-access tutorial to help other environmental scientists and biologists who are getting started with HTCondor.
+Erickson’s team hopes to make the results from this project publicly available in 2022.
+
+
+
+
+
+
+
+
+ !!!This is the content of the dropdown!!!
+
+
+
+
+
+
+
+
+ !!!This is the content of the dropdown!!!
+
+
+
+
+
+
+
+
+ How to Request a CHTC Account
+
+
+
The following sections detail the processes for requesting a new CHTC account,
+or for continuing to use an existing CHTC account. Use of CHTC services are free
+to use in support of University of Wisconsin - Madison’s research and teaching mission.
+
+
Current Member of UW - Madison
+
+
If you are a current student, staff, or faculty at UW - Madison, you can request an account
+by completing the Account Request Form. A staff member from CHTC will follow
+up with next steps.
+
+
All accounts require an active NetID and a faculty sponsor (typically the PI that is leading
+your research project.)
+
+
Graduating from UW - Madison
+
+
We understand that some users may need to continue carrying out their computational
+analyses after graduation and the subsequent expiration of their NetID.
+
+
Once you are no longer enrolled in or employed by the University, you can continue
+to use your CHTC account as an “External Collaborator”. Follow the instructions in
+the section below to have your faculty advisor sponsor your continued access to
+CHTC.
+
+
We highly recommend reaching out to CHTC staff before your NetID expires, if possible.
+
+
+ - Our policy is that CHTC accounts are deactivated and user data is erased after a user
+is no longer actively using their account (~1 year of inactivity). It is your responsibility
+to maintain your data and important files in a location that is not CHTC’s file systems.
+
+
+
External Collaborator
+
+
If you are not a current member of UW - Madison, you can gain access to CHTC provided
+that you are sponsored by a faculty member of UW - Madison. To begin the account
+request process, have your Faculty Sponsor email CHTC (chtc@cs.wisc.edu) and provide:
+
+
+ - Your name,
+ - The reason you need (continued) access to CHTC resources,
+ - The amount of time they would like to sponsor your account,
+ - Your city/country of residence, and
+ - Your institution.
+
+
+
CHTC staff will then follow up with next steps to create or extend your account.
+
+
+ - Your faculty sponsor can sponsor your account for up to one year at a time. If
+you need continued access past one year, your faculty sponsor must contact us and
+re-confirm that you should have continued access.
+ - Our policy is that CHTC accounts are deactivated and user data is erased after a user
+is no longer actively using their account (~1 year of inactivity). It is your responsibility
+to maintain your data and important files in a location that is not CHTC’s file systems.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Building an Apptainer Container
+
+
+
This guide describes the general process for creating an Apptainer container.
+Specifically, we discuss the components of the “definition file” and how that file is used to construct or “build” the container itself.
+
+
For instructions on using and building Apptainer containers
+
+
+
+
+
+
Table of Contents
+
+
+
+
+
The Apptainer Definition File
+
+
The instructions for how Apptainer should build a container are located in the definition file (typically suffixed with .def).
+The following table summarizes the sections that are routinely used when constructing a container:
+
+
+
+
+ | Section |
+ Description |
+
+
+
+
+ | “Header” |
+ Choose an existing container to start from. |
+
+
+ %files |
+ Add existing files (e.g., pre-downloaded source code) to use in the container. |
+
+
+ %post |
+ Installation commands for adding libraries/programs to the container. |
+
+
+ %environment |
+ Automatically set environment variables when the container is started to help find installed programs. |
+
+
+ %labels |
+ Add information or metadata to help identify the container and its contents. |
+
+
+ %help |
+ Add text to help others use the container and its contents. |
+
+
+
+
+
With the exception of the “Header”, sections in the definition file begin with a line starting with %name_of_section and all subsequent lines belong to that section until the end of the file or the next %section line is encountered.
+Typically the contents of a section are indented to help visually distinguish the different parts.
+
+
Additional sections can be specified, though not all may be functional when using the container on CHTC systems.
+For additional information on Apptainer definition files, see the Apptainer documentation.
+The manual page provides a full reference on the different sections of the definition file.
+
+
+ Note that the %runscript section is ignored when the container is executed on the High Throughput system.
+
+
+
+
+
This must be the first section of the definition file.
+The header specifies the container image that Apptainer should start with.
+Apptainer will load this container image before attempting to execute the build commands.
+
+
Most users will use
+
+
Bootstrap: docker
+From: user/repo:tag
+
+
+
where user/repo:tag is any valid address to a Docker-based container registry.
+For example,
+
+
Bootstrap: docker
+From: rocker/tidyverse:4.1.3
+
+
+
would use the Rocker tidyverse v4.1.3 container hosted on DockerHub as the base for the current build.
+
+
Alternatively,
+
+
Bootstrap: docker
+From: nvcr.io/nvidia/tensorflow:24.02-tf2-py3
+
+
+
would use the NVIDIA TensorFlow 2 v24.02 container hosted on the NVIDIA Container Registry (nvcr).
+
+
+ If you just want to convert an existing Docker container into an Apptainer container, you do not need to use a definition file.
+Instead, you can directly run the apptainer build command using the Docker address, as described below.
+
+
+
Files section
+
+
The %files section is used to copy files from the machine that is running Apptainer (the “host”) into the container that Apptainer is building.
+This section is typically used when you have the source code saved on the host and want to extract/compile/install it inside of the container image.
+
+
+ While the container is being built on the host system, by default it does not have direct access to files located on the host system.
+The %files section serves as the bridge between the host system and the container being built.
+
+
+
The syntax for use is
+
+
%files
+ file_on_host file_in_container
+
+
+
where file_on_host is in the same directory as the .def definition file, and where file_in_container will be copied to the container’s root (/) by default.
+You can instead provide absolute paths to the files on the host or in the container, or both.
+For example:
+
+
%files
+ /home/username/my-source-code.tar.gz /opt/my-program-build/my-source-code.tar.gz
+
+
+
+ If the directories in the path in the container do not already exist, they will be created.
+
+
+
Post section
+
+
The %post section contains any and all commands to be executed when building the container.
+Typically this involves first installing packages using the operating system’s package manager and then compiling/installing your custom programs.
+Environment variables can be set as well, but they will only be active during the build (use the %environment section if you need them active during run time).
+
+
For example, if using an ubuntu based container, then you should be able to use the apt package manager to install your program’s dependencies.
+
+
%post
+ apt update -y
+ apt install -y gcc make wget
+
+
+
Note that we have used the -y option for apt to pre-emptively agree to update apt and to install the gcc, make, and wget packages.
+Otherwise, the apt command will prompt you to confirm the executions via the command line.
+But since the Apptainer build process is executed non-interactively, you will be unable to enter a response via the command line, and the commands will eventually time out and the build fail.
+
+
Once you install the dependencies you need using the operating system’s package manager, you can use those packages to obtain and install your desired program.
+For example, the following commands will install the GNU Units command units.
+
+
mkdir -p /opt/units-source
+ cd /opt/units-source
+ wget https://ftp.gnu.org/gnu/units/units-2.23.tar.gz
+ tar -xzf units-2.23.tar.gz
+ cd units-2.23
+ ./configure
+ make
+ make install
+
+
+
If using the default installation procedure, your program should be installed in and detectable by the operating system.
+If not, you may need to manually environment variables to recognize your program.
+
+
Environment section
+
+
The %environment section can be used to automatically set environment variables when the container is actually started.
+
+
For example, if you installed your program in a custom location /opt/my-program and the binaries are in the bin/ folder, you could use this section to add that location to your PATH environment variable:
+
+
%environment
+ export PATH="/opt/my-program/bin:$PATH"
+
+
+
+ Effectively, this section can be used like a .bashrc or .bash_profile file.
+
+
+
Labels section
+
+
The %labels section can be used to provide custom metadata about the container, which can make it easier for yourself and others to identify the nature and provenance of a container.
+
+
The syntax for this section is
+
+
%labels
+ LabelNameA LabelValueA
+ LableNameB LabelValueB
+
+
+
where LabelName is the name of the label, and LabelValue is the corresponding value.
+For example,
+
+
%labels
+ Author Bucky Badger
+ ContactEmail bbadger@wisc.edu
+ Name Bucky's First Container
+
+
+
will generate the metadata in the container showing the Author as Bucky Badger, the ContactEmail as bbadger@wisc.edu, and the container Name as Bucky's First Container.
+
+
For an existing container, you can inspect the metadata with the command apptainer inspect my_container.sif.
+
+
+ For a container with the %labels in the above example, you should see the following output:
+
+ $ apptainer inspect my_container.sif
+
+Author: Bucky Badger
+ContactEmail: bbadger@wisc.edu
+Name: Bucky's First Container
+
+
+ along with some automatically generated labels.
+
+
+
Help section
+
+
The %help section can be used to provide custom help text about how to use the container.
+This can make it easier for yourself and others to interact and use the container.
+
+
For example,
+
+
%help
+ This container is based on Ubuntu 22.04 and has the GNU Units command installed.
+ You can use the command `units` inside this container to convert from one unit of measurement to another.
+ For example,
+ $ units '1 GB' 'MB'
+ returns
+ * 1000
+ / 0.001
+
+
+
For an existing container, you can inspect the help text with the command apptainer run-help my-container.sif.
+
+
The Apptainer Container Image
+
+
The actual container image, which can be executed by Apptainer as a stand-alone operating system, is stored in a .sif file.*
+The instructions for constructing the .sif file are provided by the .def definition file, as described above.
+Basically, the .sif file is a compression of all of the files in the stand-alone operating system that comprises a “container”.
+Apptainer can use this one file to reconstitute the container at runtime.
+
+
* sif stands for “Singularity Image File”; Apptainer is formerly an open-source fork of the original program called Singularity.
+
+
Building the container
+
+
To create the .sif file from the .def file, you need to run the command
+
+
apptainer build my-container.sif my-container.def
+
+
+
Here the syntax is to provide the name of the .sif file that you want to create and then provide the name of the existing .def definition file.
+
+
+ Don’t run the apptainer build command on the login server!
+Building the container image can be an intensive process and can consume the resources of the login server.
+
+
+ - On the High Throughput system, first start an interactive build job as described in our Use Apptainer Containers guide.
+ - On the High Performance system, first launch an interactive Slurm session as described here.
+
+
+
+
Converting a Docker image to an Apptainer container image
+
+
You can directly convert an existing Docker container into an Apptainer container image without having to provide a definition file.
+To do so, use the command
+
+
apptainer build my-container.sif docker://user/repo:tag
+
+
+
where user/repo:tag is any valid address to a Docker-based container registry. (For example, rocker/tidyverse:4.1.3 from DockerHub or nvcr.io/nvidia/tensorflow:24.02-tf2-py3 from NVIDIA Container Registry.)
+
+
Testing the container interactively
+
+
After building your container, we strongly recommend that you start it up and check that your program has been installed correctly.
+Assuming that you are in an interactive session (i.e., not on the login server), then you can run
+
+
apptainer shell my-container.sif
+
+
+
This command should log you into a terminal that is backed by the container’s operating system.
+
+
+ On the High Throughput system, you can instead submit an interactive job that uses the .sif file as the container_image.
+In this case, you do not need to run any apptainer commands, as HTCondor has automatically done so before you load into the interactive session.
+
+
+
Then you can check that the files are in the right place, or that your program can be found.
+An easy way to check if your program is at least in recognized by the container is to try to print the help text for the program.
+
+
For example,
+
+
[username@hostname ~]$ apptainer shell units.sif
+Apptainer> units --help
+
+Usage: units [options] ['from-unit' 'to-unit']
+
+<additional output truncated>
+
+
+
+ By default, only your current directory will be mounted into the container, meaning the only files you can see from the host system are those in the directory where you ran the command.
+
+ Furthermore, the interactive container session may inherit environment variables from your terminal session on the host system, which may conflict with the container environment.
+In this case, use the -e option to use a “clean” environment for the interactive session: apptainer shell -e my-container.sif.
+
+
+
Special Considerations for Building Your Container
+
+
+
+
+ -
+
Non-interactive
+
+ Because the container build is a non-interactive process, all commands within the .def file must be able to execute without user intervention.
+
+ -
+
Be prepared to troubleshoot
+
+ A consequence of the non-interactive build is that when something goes wrong, the build process will fail without creating a .sif file.
+That in turn means that when the build is restarted, it does so from completely from scratch.
+
+ It is rare to correctly write your .def file such that the container builds successfully on your first try!
+Do not be discouraged - examine the build messages to determine what went wrong and use the information to correct your .def file, then try again.
+
+ -
+
Multi-stage build
+
+ It is possible to have a multi-stage build.
+In this scenario, you have two .def files.
+You use the first one to construct an intermediary .sif file, which you can then use as the base for the second .def file.
+In the second .def file, you can specify
+
+ Bootstrap: localimage
+From: path/to/first.sif
+
+
+ -
+
.sif files can be large
+
+ If you are installing a lot of programs, the final .sif image can be large, on the order of 10s of gigabytes.
+Keep that in mind when requesting disk space.
+On the High Throughput system, we encourage you to place your container image on the /staging system.
+
+ -
+
Files cannot be created or modified after the container has been built
+
+ While you can read and execute any file within the container, you will not be able to create or modify files in the container once it has been built.
+The exception is if the location is “mounted” into the container, which means that there is a corresponding location on the host system where the files will be stored.
+Even then, you will only be allowed to create/modify files in that location if you would be able to normally without a container.
+
+ This behavior is intentional as otherwise it would be possible for users to modify files on the host machine’s operating system, which would be a signicant security, operations, and privacy risk.
+
+ -
+
Manually set a HOME directory
+
+ Some programs create .cache directories and may attempt to do so in the user’s “HOME” directory.
+When executing in a container, however, the user typically does NOT have a “HOME” directory.
+In this case, some programs default to creating the directory in the root / directory.
+This will not work for reasons in the previous item.
+
+ One workaround may be to manually set the HOME environment variable after the container has started.
+On CHTC systems, the following should address this issue:
+
+ export HOME=$(pwd)
+
+
+ If this does not address the issue, examine the error messages and consult the program documentation for how configure the program to use an alternate location for cache or temporary directories.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Using Apptainer Containers on HPC
+
+
+
Introduction
+
+
Similar to Docker containers, Apptainer environments allow users to prepare portable software and computing environments that can be sent to many jobs.
+This means your jobs will run in a more consistent environment that is easily reproducible by others.
+
+
+
+
Table of Contents
+
+
+
+
+
Create a Definition File
+
+
The definition (.def) file contains the instructions for what software to install while building the container.
+CHTC provides example definition files in the software folder of our Recipes GitHub repository. We strongly recommend that you use one of the existing examples as the starting point for creating your own container.
+
+
To create your own container using Apptainer, you will need to create a definition (.def) file.
+We encourage you to read our Building an Apptainer Container guide to learn more about the components of the Apptainer definition file.
+
+
Regarding MPI
+
+
We are still in the process of developing guidance for deploying MPI-based software in containers on the High Performance system.
+The instructions in this guide should work for single-node jobs.
+Multi-node jobs require the MPI installed in the container to integrate with Slurm and/or the cluster installation of MPI, and we are still exploring how to do so.
+
+
Start an Interactive Session
+
+
Building a container can be a computationally intense process.
+As such, we require that you only build containers while in an interactive session.
+On the High Performance system, you can use the following command to start the interactive session:
+
+
srun --mpi=pmix -n4 -N1 -t 240 -p int --pty bash
+
+
+
Build Your Container
+
+
We recommend working from your /scratch directory when first building your container.
+
+
Once the interactive session starts, set the Apptainer temporary directory:
+
+
export APPTAINER_TMPDIR=/scratch/$USER/apptainer/tmp
+mkdir -p $APPTAINER_TMPDIR
+
+
+
To build a container, Apptainer uses the instructions in the .def file to create a .sif file. The .sif file is the compressed collection of all the files that comprise the container.
+
+
To build your container, run this command:
+
+
apptainer build my-container.sif image.def
+
+
+
Feel free to rename the .sif file as you desire; for the purposes of this guide we are using my-container.sif.
+
+
As the command runs, a variety of information will be printed to the terminal regarding the container build process.
+Unless something goes wrong, this information can be safely ignored.
+Once the command has finished running, you should see INFO: Build complete: my-container.sif.
+Using the ls command, you should now see the container file my-container.sif.
+
+
If the build command fails, examine the output for error messages that may explain why the build was unsuccessful.
+Typically there is an issue with a package installation, such as a typo or a missing but required dependency.
+Sometimes there will be an error during an earlier package installation that doesn’t immediately cause the container build to fail.
+But, when you test the container, you may notice an issue with the package.
+
+
If you are having trouble finding the error message, edit the definition file and remove (or comment out) the installation commands that come after the package in question.
+Then rebuild the image, and now the relevant error messages should be near the end of the build output.
+
+
For more information on building Apptainer containers, see our Building an Apptainer Container guide.
+
+
Test Your Container
+
+
Once your container builds successfully, it is important to test it to make sure you have all software, packages, and libraries installed correctly.
+
+
To test your container, use the command
+
+
apptainer shell -e my-container.sif
+
+
+
You should see your command prompt change to Apptainer>.
+
+
The shell command logs you into a terminal “inside” the container, with access to the libraries, packages, and programs that were installed in the container following the instructions in your image.def file.
+(The -e option is used to prevent this terminal from trying to use the host system’s programs.)
+
+
While “inside” the container, try to run your program(s) that you installed in the container.
+Typically it is easiest to try to print your program’s “help” text, e.g., my-program --help.
+If using a programming language such as python3 or R, try to start an interactive code session and load the packages that you installed.
+
+
If you installed your program in a custom location, consider using ls to verify the files are in the right location.
+You may need to manually set the PATH environment variable to point to the location of your program’s executable binaries.
+For example,
+
+
export PATH=/opt/my-program/bin:$PATH
+
+
+
Consult the “Special Considerations” section of our Building an Apptainer Container guide for additional information on setting up and testing your container.
+
+
When you are finished running commands inside the container, run the command exit to exit the container.
+Your prompt should change back to something like [username@spark-a006 directory]$.
+If you are satisfied with the container that you built, run the exit command again to exit the interactive Slurm session.
+
+
Use an Apptainer Container in HPC Jobs
+
+
Now that you have the container image saved in the form of the .sif file, you can use it as the environment for running your HPC jobs.
+
+
For execution on a single node, we recommend adding the following commands to your sbatch script:
+
+
export TMPDIR=/scratch/$USER/apptainer_tmp/${SLURM_JOB_ID}
+mkdir -p $TMPDIR
+
+srun apptainer exec -e \
+ --bind /home/$USER \
+ --bind /scratch/$USER \
+ --bind $TMPDIR \
+ my-container.sif my-job-script.sh
+
+rm -rf $TMPDIR
+
+
+
All of the commands that you want to execute using the container should go into my-job-script.sh.
+
+
Then submit your job to Slurm as usual, as described in our Submitting and Managing Jobs Using SLURM guide.
+
+
On multiple nodes
+
+
We are still in the early stages of deploying containers on the High Performance system.
+A complicating factor is the construction of the .def file to deploy MPI on the system to allow for execution across multiple nodes.
+If you are interested in mutli-node execution using containers, contact a facilitator for more information.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Advanced Apptainer Example - SUMO
+
+
+
Sometimes the program you want to use does not have a pre-existing container that you can build on top of.
+Then you will need to install the program and its dependencies inside of the container.
+In this example, we will show how to install the program SUMO in a container, as an illustration of how to build a container more-or-less from scratch.
+
+
+
+
Table of Contents
+
+
+
+
+
1. Choose the Base Container Image
+
+
First, you will need to choose a base image for the container.
+Consult the documentation for the program you want to install to make sure you select a compatible operating system.
+
+
For this example, we will use the most recent LTS version of Ubuntu from Docker.
+The beginning of the image.def file should look like this:
+
+
Bootstrap: docker
+From: ubuntu:22.04
+
+
+
2. Add the Installation Commands
+
+
All of the installation commands that you want Apptainer to execute during the container build step are provided in the %post section of the definition file.
+
+
Setting up non-interactive installation
+
+
First, you may need to instruct programs that you are executing commands in a non-interactive environment.
+There can be issues with installing packages in a container that would not normally occur when installing manually in the terminal.
+
+
On the HTC system in particular, the /tmp directory inside of the container needs to be given global read/write permissions.
+This can be done by adding the following line at the start of the %post section:
+
+
chmod 777 /tmp
+
+
+
Similarly, some packages require that the user answer interactive prompts for selecting various options.
+Since the Apptainer build is non-interactive, this can cause the package installation to hang.
+While this isn’t an issue in the present example, the issue can be avoided by adding the following line near the start of the %post section:
+
+
DEBIAN_FRONTEND=noninteractive
+
+
+
Note that this particular command only applies to Debian-based container images, such as Ubuntu.
+
+
The image.def so far looks like this:
+
+
Bootstrap: docker
+From: ubuntu:22.04
+
+%post
+ chmod 777 /tmp
+ DEBIAN_FRONTEND=noninteractive
+
+
+
Install dependencies
+
+
First, you should install the dependencies that your program requires.
+
+
Following the program’s instructions, we can do install the dependencies with the following set of commands.
+
+
apt-get update -y
+apt-get install -y \
+ git \
+ cmake \
+ python3 \
+ g++ \
+ libxerces-c-dev \
+ libfox-1.6-dev \
+ libgdal-dev \
+ libproj-dev \
+ libgl2ps-dev \
+ python3-dev \
+ swig \
+ default-jdk \
+ maven \
+ libeigen3-dev
+
+
+
Note that we are using the built-in package manager (apt) of Ubuntu, since that is the base operating system we chose to build on top of.
+If you choose a different operating system, you may need to use a different package manager.
+
+
In this case, the first command is apt-get update which will update the list of available packages.
+This is necessary to get the latest versions of the packages in the following apt-get install command.
+
+
The apt-get install command will install the dependencies required by the SUMO program.
+
+
+ Note that these installation commands do not use sudo, as Apptainer already has permissions to install programs in the container.
+
+
+
The image.def file now looks like this:
+
+
Bootstrap: docker
+From: ubuntu:22.04
+
+%post
+ chmod 777 /tmp
+ DEBIAN_FRONTEND=noninteractive
+
+ apt-get update -y
+ apt-get install -y \
+ git \
+ cmake \
+ python3 \
+ g++ \
+ libxerces-c-dev \
+ libfox-1.6-dev \
+ libgdal-dev \
+ libproj-dev \
+ libgl2ps-dev \
+ python3-dev \
+ swig \
+ default-jdk \
+ maven \
+ libeigen3-dev
+
+
+
Compile the program
+
+
After installing the dependencies, you can provide the commands for actually compiling your program.
+
+
We now add the commands for compiling the SUMO program itself:
+
+
git clone --recursive https://github.com/eclipse/sumo
+ export SUMO_HOME=/sumo
+ mkdir sumo/build/cmake-build && cd sumo/build/cmake-build
+ cmake ../..
+ make
+
+
+
The %post section is now complete and will install SUMO and its dependencies in the container at build time.
+
+
The image.def file now looks like this:
+
+
Bootstrap: docker
+From: ubuntu:22.04
+
+%post
+ chmod 777 /tmp
+ DEBIAN_FRONTEND=noninteractive
+
+ apt-get update -y
+ apt-get install -y \
+ git \
+ cmake \
+ python3 \
+ g++ \
+ libxerces-c-dev \
+ libfox-1.6-dev \
+ libgdal-dev \
+ libproj-dev \
+ libgl2ps-dev \
+ python3-dev \
+ swig \
+ default-jdk \
+ maven \
+ libeigen3-dev
+
+ git clone --recursive https://github.com/eclipse/sumo
+ export SUMO_HOME=/sumo
+ mkdir sumo/build/cmake-build && cd sumo/build/cmake-build
+ cmake ../..
+ make
+
+
+
3. Add Environment Variables
+
+
While the %post section now contains all of the instructions for installing and compiling your desired program,
+you likely need to add commands for setting up the environment so that the shell recognizes your program.
+This is typically the case if your program compiled successfully but you still get a “command not found” error when you try to execute it.
+
+
To set environment variables automatically when your container runs, you need to add them to the %environment section before you build the container.
+
+
For example, in the %post section there is the command export SUMO_HOME=/sumo, which sets the environment variable SUMO_HOME to the location of the sumo directory.
+This environment variable, however, is only active during the installation phase of the container build, and will not be set when the container is actually run.
+Thus, we need to set SUMO_HOME and update PATH with the location of the SUMO bin folder by using the %environment section.
+
+
We therefore add the following lines to the image.def file:
+
+
%environment
+ export SUMO_HOME=/sumo
+ export PATH=/sumo/bin:$PATH
+
+
+
These environment variables will be set when the container starts, so that the sumo command is automatically found when we try to execute it.
+
+
Summary
+
+
The full image.def file for this advanced example is now:
+
+
Bootstrap: docker
+From: ubuntu:22.04
+
+%post
+ chmod 777 /tmp
+ DEBIAN_FRONTEND=noninteractive
+
+ apt-get update -y
+ apt-get install -y \
+ git \
+ cmake \
+ python3 \
+ g++ \
+ libxerces-c-dev \
+ libfox-1.6-dev \
+ libgdal-dev \
+ libproj-dev \
+ libgl2ps-dev \
+ python3-dev \
+ swig \
+ default-jdk \
+ maven \
+ libeigen3-dev
+
+ git clone --recursive https://github.com/eclipse/sumo
+ export SUMO_HOME=/sumo
+ mkdir sumo/build/cmake-build && cd sumo/build/cmake-build
+ cmake ../..
+ make
+
+%environment
+ export SUMO_HOME=/sumo
+ export PATH=/sumo/bin:$PATH
+
+
+
We can now build the container using this definition file.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Use Apptainer Containers
+
+
+
Introduction
+
+
HTCondor supports the use of Apptainer (formerly known as Singularity) environments for jobs on the High Throughput Computing system.
+
+
Similar to Docker containers, Apptainer environments allow users to prepare portable software and computing environments that can be sent to many jobs.
+This means your jobs will run in a more consistent environment that is easily reproducible by others.
+
+
Container jobs are able to take advantage of more of CHTC’s High Throughput resources because the operating system where the job is running does not need to match the operating system where the container was built.
+
+
+
+
Table of Contents
+
+
+
+
+
Quickstart
+
+
Use an existing container
+
+
If you or a group member have already created the Apptainer .sif file, or are using a container from reputable sources such as the OSG, follow these steps to use it in an HTCondor job.
+
+
1. Add the container .sif file to your submit file
+
+
If the .sif file is in a /home directory:
+
+
container_image = path/to/my-container.sif
+
+
+
If the .sif file is in a /staging directory:
+
+
container_image = file:///staging/path/to/my-container.sif
+
+
+
If the .sif file is in a /staging directory AND you are using +WantFlocking or +WantGliding:
+
+
container_image = osdf:///chtc/staging/path/to/my-container.sif
+
+
+
Jump to more information
+
+
2. Test your container job
+
+
As always with the High Throughput system, submit a single test job and confirm that your job behaves as expected.
+If there are issues with the job, you may need to modify your executable, or even (re)build your own container.
+
+
Build your own container
+
+
If you need to create your own container for the software you want to use, follow these steps.
+For more information on any particular step, jump to the corresponding section later in this guide.
+
+
+
+
1. Create a definition file
+
+
The definition (.def) file contains the instructions for what software to install while building the container.
+CHTC provides example definition files in the software folder of our Recipes GitHub repository. Choose from one of the existing examples, or create your own using the instructions later in this guide.
+
+
Jump to more information
+
+
2. Start an interactive build job
+
+
Start an interactive build job (an example submit file build.sub is provided below).
+Be sure to include your .def file in the transfer_input_files line, or else create the file once the interactive job starts using a command line editor.
+
+
Then submit the interactive build job with
+
+
condor_submit -i build.sub
+
+
+
Jump to more information
+
+
3. Build your container
+
+
While in an interactive build job, run the command
+
+
apptainer build my-container.sif image.def
+
+
+
If the container build finishes successfully, then the container image (.sif) file is created.
+This file is used for actually executing the container.
+
+
Jump to more information
+
+
4. Test your container
+
+
While still in the interactive build job, run the command
+
+
apptainer shell -e my-container.sif
+
+
+
This command will start the container and log you into it, allowing you to test your software commands.
+
+
Once you are done testing the container, enter
+
+
exit
+
+
+
once to exit the container.
+
+
Jump to more information
+
+
5. Move the container .sif file to staging
+
+
Once you are satisfied that your container is built correctly, copy your .sif file to your staging directory.
+
+
mv my-container.sif /staging/$USER
+
+
+
Once the file has transferred, exit the interactive job with
+
+
exit
+
+
+
Jump to more information
+
+
Once you’ve built the container, use the instructions above to use the container in your HTCondor job.
+
+
Create a Definition File
+
+
To create your own container using Apptainer, you will need to create a definition (.def) file.
+For the purposes of this guide, we will call the definition file image.def.
+
+
CHTC provides example definition files in the software folder of our Recipes GitHub repository. We strongly recommend that you use one of the existing examples as the starting point for creating your own container.
+
+
If the software you want to use is not in the CHTC Recipes repository, you can create your own container. Here is general process for creating your own definition file for building your custom container:
+
+
+ -
+
Consult your software’s documentation
+
+ Determine the requirements for installing the software you want to use.
+In particular you are looking for (a) the operating systems it is compatible with and (b) the prerequisite libraries or packages.
+
+ -
+
Choose a base container
+
+ The base container should at minimum use an operating system compatible with your software.
+Ideally the container you choose also has many of the prerequisite libraries/programs already installed.
+
+ -
+
Create your own definition file
+
+ The definition file contains the installation commands needed to set up your software.
+We encourage you to read our Building an Apptainer Container guide to learn more about the components of the Apptainer definition file.
+An advanced example of a definition file is provided in our Advanced Apptainer Example - SUMO guide.
+
+
+
+
A simple definition file
+
+
As a simple example, here is the .def file that uses an existing container with python installed inside (python:3.11, from DockerHub),
+and furthermore installs the desired packages cowsay and tqdm:
+
+
Bootstrap: docker
+From: python:3.11
+
+%post
+ python3 -m pip install cowsay tqdm
+
+
+
Remember that the .def file contains the instructions for creating your container and is not itself the container.
+To use the software defined within the .def file, you will need to first “build” the container and create the .sif file, as described in the following sections.
+
+
Jump back to Quickstart
+
+
Start an Interactive Build Job
+
+
Building a container can be a computationally intense process.
+As such, we require that you only build containers while in an interactive build job.
+On the High Throughput system, you can use the following submit file build.sub:
+
+
+
# build.sub
+# For building an Apptainer container
+
+universe = vanilla
+log = build.log
+
+# In the latest version of HTCondor on CHTC, interactive jobs require an executable.
+# If you do not have an existing executable, use a generic linux command like hostname as shown below.
+executable = /usr/bin/hostname
+
+# If you have additional files in your /home directory that are required for your container, add them to the transfer_input_files line as a comma-separated list.
+transfer_input_files = image.def
+
+requirements = (HasCHTCStaging == true)
+
++IsBuildJob = true
+request_cpus = 4
+request_memory = 16GB
+request_disk = 16GB
+
+queue
+
+
+
Note that this submit file assumes you have a definition file named image.def in the same directory as the submit file.
+
+
Once you’ve created the submit file, you can submit an interactive job with the command
+
+
condor_submit -i build.sub
+
+
+
+ Apptainer .sif files can be fairly large, especially if you have a complex software stack.
+If your interactive job abruptly fails during the build step, you may need to increase the value of request_disk in your submit file.
+In this case, the .log file should have a message about the reason the interactive job was interrupted.
+
+
+
Jump back to Quickstart
+
+
Build Your Container
+
+
Once the interactive build job starts, confirm that your image.def was transferred to the current directory.
+
+
To build a container, Apptainer uses the instructions in the .def file to create a .sif file. The .sif file is the compressed collection of all the files that comprise the container.
+
+
To build your container, run this command:
+
+
apptainer build my-container.sif image.def
+
+
+
Feel free to rename the .sif file as you desire; for the purposes of this guide we are using my-container.sif.
+
+
As the command runs, a variety of information will be printed to the terminal regarding the container build process.
+Unless something goes wrong, this information can be safely ignored.
+Once the command has finished running, you should see INFO: Build complete: my-container.sif.
+Using the ls command, you should now see the container file my-container.sif.
+
+
If the build command fails, examine the output for error messages that may explain why the build was unsuccessful.
+Typically there is an issue with a package installation, such as a typo or a missing but required dependency.
+Sometimes there will be an error during an earlier package installation that doesn’t immediately cause the container build to fail.
+But, when you test the container, you may notice an issue with the package.
+
+
If you are having trouble finding the error message, edit the definition file and remove (or comment out) the installation commands that come after the package in question.
+Then rebuild the image, and now the relevant error messages should be near the end of the build output.
+
+
Once the image is built, it is important to test it to make sure you have all software, packages, and libraries installed correctly.
+
+
For more information on building Apptainer containers, see our Building an Apptainer Container guide.
+
+
Jump back to Quickstart
+
+
Test Your Container
+
+
Once your container builds successfully, we highly encourage you to immediately test the container while still in the interactive build session.
+
+
To test your container, use the command
+
+
apptainer shell -e my-container.sif
+
+
+
You should see your command prompt change to Apptainer>.
+
+
The shell command logs you into a terminal “inside” the container, with access to the libraries, packages, and programs that were installed in the container following the instructions in your image.def file.
+(The -e option is used to prevent this terminal from trying to use the host system’s programs.)
+
+
While “inside” the container, try to run your program(s) that you installed in the container.
+Typically it is easiest to try to print your program’s “help” text, e.g., my-program --help.
+If using a programming language such as python3 or R, try to start an interactive code session and load the packages that you installed.
+
+
If you installed your program in a custom location, consider using ls to verify the files are in the right location.
+You may need to manually set the PATH environment variable to point to the location of your program’s executable binaries.
+For example,
+
+
export PATH=/opt/my-program/bin:$PATH
+
+
+
Consult the “Special Considerations” section of our Building an Apptainer Container guide for additional information on setting up and testing your container.
+
+
When you are finished running commands inside the container, run the command exit to exit the container.
+Your prompt should change back to something like [username@build4000 ~]$.
+
+
Jump back to Quickstart
+
+
Move the Container .sif File to Staging
+
+
Since Apptainer .sif files are routinely more than 1GB in size, we recommend that you transfer my-container.sif to your /staging directory.
+It is usually easiest to move the container file directly to staging while still in the interactive build job:
+
+
mv my-container.sif /staging/$USER
+
+
+
If you do not have a /staging directory, you can skip this step and the .sif file will be automatically transferred back to the login server when you exit the interactive job.
+We encourage you to request a /staging directory, especially if you plan on running many jobs using this container.
+See our Managing Large Data in Jobs guide for more information on using staging.
+
+
Jump back to Quickstart
+
+
Use an Apptainer Container in HTC Jobs
+
+
Now that you have the container image saved in the form of the .sif file, you can use it as the environment for running your HTCondor jobs.
+In your submit file, specify the image file using the container_image command.
+HTCondor will automatically transfer the .sif file and automatically execute your executable file inside of the container; you do not need to include any apptainer commands in your executable file.
+
+
If the .sif file is located on the login server, you can use
+
+
container_image = my-container.sif
+
+
+
although we generally don’t recommend this, since .sif files are large and should instead be located in staging.
+
+
Therefore, we recommend using
+
+
container_image = file:///staging/path/to/my-container.sif
+
+
+
The full submit file otherwise looks like normal, for example:
+
+
# apptainer.sub
+
+# Provide HTCondor with the name of your .sif file and universe information
+container_image = file:///staging/path/to/my-container.sif
+
+executable = myExecutable.sh
+
+# Include other files that need to be transferred here.
+# transfer_input_files = other_job_files
+
+log = job.log
+error = job.err
+output = job.out
+
+requirements = (HasCHTCStaging == true)
+
+# Make sure you request enough disk for the container image in addition to your other input files
+request_cpus = 1
+request_memory = 4GB
+request_disk = 10GB
+
+queue
+
+
+
Then use condor_submit with the name of your submit file:
+
+
condor_submit apptainer.sub
+
+
+
If you are using +WantFlocking or +WantGliding as described in our Scale Beyond Local HTC Capacity guide, then you should instead use
+
+
container_image = osdf:///chtc/staging/path/to/my-container.sif
+
+
+
to enable transferring of the .sif file via the OSDF to compute capacity beyond CHTC.
+
+
Jump back to Quickstart
+
+
+ From the user’s perspective, a container job is practically identical to a regular job.
+The main difference is that instead of running on the execute point’s default operation system, the job is run inside the container.
+
+ When you submit a job to HTCondor using a submit file with container_image set, HTCondor automatically handles the process of obtaining and running the container.
+The process looks roughly like
+
+
+ - Claim machine that satisifies submit file requirements
+ - Pull (or transfer) the container image
+ - Transfer input files, executable to working directory
+ - Run the executable script inside the container, as the submit user, with key directories mounted inside (such as the working directory, /staging directories, etc.)
+ - Transfer output files back to the submit server
+
+
+ For testing purposes, you can replicate the behavior of a container job with the following command.
+First, start an interactive job.
+Then run this command but change my-container.sif and myExecutable.sh to the names of the .sif and .sh files that you are using:
+
+ apptainer exec \
+ --scratch /tmp \
+ --scratch /var/tmp \
+ --workdir $(pwd) \
+ --pwd $(pwd) \
+ --bind $(pwd) \
+ --no-home \
+ --containall \
+ my-container.sif \
+ /bin/bash myExecutable.sh 1> job.out 2> job.err
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Transitioning to a New HPC Cluster
+
+
+
The Center for High Throughput Computing’s High Performance Cluster is being
+replaced by a new High Performance System! All components of the system (execute
+nodes, network, shared file system) are new and we expect an improved experience for our HPC users.
+
+
ALL USERS OF THE EXISTING HPC SYSTEM WILL NEED TO MIGRATE TO THIS NEW CLUSTER.
+Importantly, access to the existing cluster will be phased out in early 2022.
+CHTC staff are here to assist with your transition.
+
+
Highlights
+
+
+ - The existing HPC cluster is being replaced by a new cluster. After February 2023
+ALL users will lose access to the existing cluster, and all user files will be
+deleted.
+ - Custom software will need to be reinstalled and jobs will need to be tested on
+the new cluster.
+ - The
univ2 partition is being renamed, and partition policies have changed.
+ - Users should avoid using
mpirun and instead should use srun to execute their
+MPI code.
+
+ Note: At this time, interactive jobs on the “Spark” HPC Cluster can not be
+used to run MPI code.
+
+
+ - File system conventions have changed - jobs will now use
/scratch/$USER to run,
+and /home/$USER will be mainly used for software installations and reference
+files.
+
+
+
Important Dates
+
+
+ - Mid January 2023: New cluster available for general use
+ - February 28, 2023: Jobs will no longer run on the old cluster
+ - March 15, 2023: Access to
hpclogin1.chtc.wisc.edu login node and old file
+system removed, Data for all users will be deleted on the old HPC system.
+
+
+
What You Need to Do
+
+
Move Data
+
+
Back up files from the old cluster to another system (e.g. your laptop), copy
+files you are actively working with to the new cluster, and delete all data off
+the old HPC system. All files in /home and /software will be deleted off the
+existing system starting March 15, 2023.
+
+
Log In and Quota Check
+
+
Confirm you can access the new cluster by logging into the new login node.
+
+
Prepare and Submit Test Jobs
+
+
After logging in, prepare and submit a few test jobs to confirm that your work
+will run, paying attention to these important changes:
+
+
+ - Appropriate usage of
/home and /scratch:
+
+ - Jobs should be run out of
/scratch/$USER. Your scratch directory has a quota of 100GB disk space and 250,000 items
+ - Only use your
/home directory for software installations and general job files and templates.
+Your /home directory has a quota of 20GB disk space and 250,000 items.
+ - The
/software directory is being phased out.
+
+
+ -
+
Build software with new modules: users will need to reinstall and/or rebuild
+their software on the new HPC cluster. Users may encounter different versions of
+common tools on the new cluster, so it is important to try installing your
+software early to ensure compatibility. If a software or library is not available
+that is necessary for your installation is not installed, contact CHTC staff (see
+our get help page).
+
+ -
+
Change MPI execution: Our systems administrators now recommend using srun
+with the --mpi=pmix flag instead of mpirun or mpiexec to execute MPI type code. It
+should look like this:
+ srun –mpi=pmix mpi_program
+
+ - Change #SBATCH options: The new cluster has different partition names and
+different sized nodes. The main general use partition is now called
shared
+instead of univ2 or univ3. We also recommend the following changes because
+most of our nodes now have 128 cores, so requesting multiple nodes is not
+advantageous if your jobs are smaller than 128 cores. We also now recommend requesting
+memory per core instead of memory per node, for similar reasons, using the --mem-per-cpu
+flag with units of MB. Here are our recommendations for different sized jobs:
+
+
+
+
+ | Job size |
+ Recommended #SBATCH flags |
+
+
+ | 32-128 cores |
+ Example for 32 cores:
+#SBATCH --nodes=1
+#SBATCH --ntasks-per-node=32 # recommend multiples of 16
+#SBATCH --mem-per-cpu=4000 |
+
+
+ | 96 - 256 cores |
+ Split over a few nodes, for example for 160 cores:
+#SBATCH --nodes=2
+#SBATCH --ntasks-per-node=80 # designate cores per node
+#SBATCH --mem-per-cpu=4000
+ OR:
+#SBATCH --nodes=2
+#SBATCH --ntasks=160 # designate overall cores
+#SBATCH --mem-per-cpu=4000 |
+
+
+ | 128 or 256 cores (whole nodes) |
+ Example for 256 cores:
+#SBATCH --nodes=2
+#SBATCH --ntasks-per-node=128
+#SBATCH --mem-per-cpu=4000 |
+
+
+
+
Optimizing Jobs (Optional)
+
+
The new cluster nodes have very fast local disk space on each node. If your code
+is able to use local space for certain parts of its calculations or is able to
+sync data between local spaces, it may be advantageous to use this disk to speed
+up your jobs. It is located at the following path on each node:
+
+
/local/$USER
+
+
+
New Cluster Specifications
+
+
Execute Nodes
+
+
We have 40 general use execute nodes, representing 5,120 cores of capacity.
+Server specs (Dell Poweredge R6525):
+
+ - 128 cores using the AMD Epyc 7763 processor
+ - 512GB of memory
+ - 1.5TB of local (not shared) fast NVME disk
+
+
+
Operating System: CentOS Stream 8
+
+
Scheduler: SLURM 22.05.6
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+While our High Throughput system has little in the way of pre-installed software,
+we've created resources to help users set up the software they want to use for running their jobs.
+
+
+{% capture content %}
+
+Quickstart
+CHTC Recipes Repository
+Containers
+
+{% endcapture %}
+{% include /components/directory.html title="Table of Contents" %}
+
+Quickstart
+
+
+Click the button that corresponds to the language/program/software that you want to use.
+More information is provided in the Recipes repository and Containers sections.
+
+
+
+
+
+ -
+
+
+ -
+
+
+ -
+
+
+
+
+
Home tab content
+
Profile tab content
+
Contact tab content
+
CHTC Recipes Repository
+
+
+CHTC provides examples for software and workflows for use on our systems in our "Recipes" repository on Github:
+https://github.com/CHTC/recipes.
+
+
+Containers
+
+
+Many of the recipes in our Recipes repository involve building your own container.
+In this section, we provide a brief introduction into how to use containers for setting up your own software to run on the High Throughput system.
+
+
+
\ No newline at end of file
diff --git a/preview-fall2024-info/uw-research-computing/archived/tensorflow-singularity-wait.html b/preview-fall2024-info/uw-research-computing/archived/tensorflow-singularity-wait.html
new file mode 100644
index 000000000..f36553d2a
--- /dev/null
+++ b/preview-fall2024-info/uw-research-computing/archived/tensorflow-singularity-wait.html
@@ -0,0 +1,483 @@
+
+
+
+
+
+
+Running Tensorflow Jobs
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Running Tensorflow Jobs
+
+
+
This guide describes how to use a pre-built Tensorflow environment
+(implemented as a Singularity container)
+to run Tensorflow jobs in CHTC and on the OS Pool.
+
+
Overview
+
+
Typically, software in CHTC jobs is installed or compiled locally by
+individual users and then brought along to each job, either using the
+default file transfer or our SQUID web server. However, another option
+is to use a container system, where the software is installed in a
+container image. CHTC (and the OS Pool) have capabilities to access and
+start containers and run jobs inside them. One container option
+available in CHTC is Docker; another is
+Singularity.
+
+
In CHTC, our Singularity support consists of running jobs inside a
+pre-made Singularity container with an installation of Tensorflow. This
+Singularity set up is very flexible: it is accessible both in CHTC and
+on the OS Pool, and can be used to run Tensorflow either with
+CPUs or GPUs. This guide starts with a basic CPU example, but then goes
+on to describe how to use the Singularity Tensorflow container for GPUs,
+and also how to run on the OS Pool.
+
+
+ - Basic Tensorflow Job Template
+ - Using Tensorflow on GPUs
+ - Using Tensorflow on the OS Pool
+
+
+
+
+
1. Basic Tensorflow Job Template
+
+
The submit file for jobs that use the Tensorflow singularity container
+will look similar to other CHTC jobs, except for the additional
+Singularity options seen below.
+
+
Submit File
+
+
# Typical submit file options
+universe = vanilla
+log = $(Cluster).$(Process).log
+error = $(Cluster).$(Process).err
+output = $(Cluster).$(Process).out
+
+# Fill in with your own script, arguments and input files
+# Note that you don't need to transfer any software
+executable = run_tensorflow.sh
+arguments =
+transfer_input_files =
+
+# Singularity settings
++SingularityImage = "/cvmfs/singularity.opensciencegrid.org/opensciencegrid/tensorflow:latest"
+Requirements = HAS_SINGULARITY == True
+
+# Resource requirements
+request_cpus = 1
+request_memory = 2GB
+request_disk = 4GB
+
+# Number of jobs
+queue 1
+
+
+
Sample Executable (Wrapper Script)
+
+
Your job will be running inside a container that has Tensorflow
+installed, so there should be no need to set any environment variables.
+
+
#!/bin/bash
+
+# your own code here
+python test.py
+
+
+
+
+
2. CPUs vs GPUs
+
+
The submit file above use a CPU-enabled version of Tensorflow. In order
+to take advantage of GPUs, make the following changes to the submit file
+above:
+
+
+ -
+
Request GPUs in addition to CPUs:
+
+ request_gpus = 1
+
+
+ -
+
Change the Singularity image to tensorflow with GPUs:
+
+ +SingularityImage = "/cvmfs/singularity.opensciencegrid.org/opensciencegrid/tensorflow-gpu:latest"
+
+
+ -
+
Add a GPU card requirement to the requirements line:
+
+ requirements = HAS_SINGULARITY == True && CUDACapability >= 3
+
+
+
+
+
For more information about GPUs and how GPU jobs work in CHTC, see our
+GPU Jobs guide.
+
+
+ Limited GPU availablity in CHTC
+This Singularity/Tensorflow functionality is not yet available on
+CHTC's newer GPUs with a sufficiently high CUDA Capability.
+Therefore, for now, the best way to use this Singularity/Tensorflow
+environment with GPUs is by running jobs on the OS Pool (see
+below). We are working on having Singularity support on all CHTC GPUs
+soon.
+
+
+
+
+
3. Running on OS Pool
+
+
This Tensorflow environment can also be run on the OS Pool either as the CPU or GPU version.
+
+
For more details on accessing the OS Pool, see our guide for running
+outside CHTC, sections 3 and 4.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Check Disk Quota and Usage
+
+
+
The following commands will allow you to monitor the amount of disk
+space you are using in your home directory on our (or another) submit node and to determine the
+amount of disk space you have been allotted (your quota).
+
+
If you also have a /staging directory on the HTC system, see our
+staging guide for
+details on how to check your quota and usage.
+
+The default quota allotment on CHTC submit nodes is 20 GB with a hard
+limit of 30 GB (at which point you cannot write more files).
+
+Note: The CHTC submit nodes are not backed up, so you will want to
+copy completed jobs to a secure location as soon as a batch completes,
+and then delete them on the submit node in order to make room for future
+jobs. If you need more disk space to run a single batch or concurrent
+batches of jobs, please contact us (Get Help!). We have multiple ways of dealing with large disk space
+requirements to make things easier for you.
+
+
If you wish to change your quotas, please see Request a Quota Change.
+
+
1. Checking Your User Quota and Usage
+
+
From any directory location within your home directory, type
+quota -vs. See the example below:
+
+
[alice@submit]$ quota -vs
+Disk quotas for user alice (uid 20384):
+ Filesystem space quota limit grace files quota limit grace
+ /dev/sdb1 12690M 20480M 30720M 161k 0 0
+
+
+
The output will list your total data usage under blocks, your soft
+quota, and your hard limit at which point your jobs will no longer
+be allowed to save data. Each of the values given are in 1-kilobyte
+blocks, so you can divide each number by 1024 to get megabytes (MB), and
+again for gigabytes (GB). (It also lists information for ` files`, but
+we don't typically allocate disk space by file count.)
+
+
2. Checking the Size of Directories and Contents
+
+
Move to the directory you'd like to check and type du . After several
+moments (longer if you're directory contents are large), the command
+will add up the sizes of directory contents and output the total size of
+each contained directory in units of kilobytes with the total size of
+that directory listed last. See the example below:
+
+
[alice@submit]$ du ./
+4096 ./dir/subdir/file.txt
+4096 ./dir/subdir
+7140 ./dir
+74688 .
+
+
+
As for quota usage above, you can divide each value by 1024 to get
+megabytes, and again for gigabytes.
+
+
Using du with the -h or --human-readable flags will display the
+same values with only two significant digits and a K, M, or G to denote
+the byte units. The -s or --summarize flags will total up the size
+of the current directory without listing the size of directory contents
+. You can also specify which directory you'd like to query, without
+moving to it, by adding the relative filepath after the flags. See the
+below example from the home directory which contains the directory
+dir:
+
+
[alice@submit]$ du -sh dir
+7.1K dir
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Checkpointing Jobs
+
+
+
What is Checkpointing?
+
+
Checkpointing is a technique that provides fault tolerance for a user’s analysis. It consists of saving snapshots of a job’s progress so the job can be restarted without losing its progress and having to restart from the beginning. We highly encourage checkpointing as a solution for jobs that will exceed the 72 hour maximum default runtime on the HTC system.
+
+
This section is about jobs capable of periodically saving checkpoint information, and how to make HTCondor store that information safely, in case it’s needed to continue the job on another machine or at a later time.
+
+
There are two types of checkpointing: exit driven and eviction driven. In a vast majority of cases, exit driven checkpointing is preferred over eviction driven checkpointing. Therefore, this guide will focus on how to utilize exit driven checkpointing for your analysis.
+
+
Note that not all software, programs, or code are capable of creating checkpoint files and knowing how to resume from them. Consult the manual for your software or program to determine if it supports checkpointing features. Some manuals will refer this ability as “checkpoint” features, as the ability to “resume” mid-analysis if a job is interrupted, or as “checkpoint/restart” capabilities. Contact a Research Computing Facilitator if you would like help determining if your software, program, or code is able to checkpoint.
+
+
Why Checkpoint?
+
+
Checkpointing allows a job to automatically resume from approximately where it left off instead of having to start over if interrupted. This behavior is advantageous for jobs limited by a maximum runtime policy (72 hours on the HTC system). It is also advantageous for jobs submitted to backfill resources with no runtime guarantee (e.g. for +WantFlocking or +WantGliding jobs) where the compute resources may also be more prone to hardware or networking failures.
+
+
For example, checkpointing jobs that are limited by a runtime policy can enable HTCondor to exit a job and automatically requeue it to avoid hitting the maximum runtime limit. By using checkpointing, jobs circumvent hitting the maximum runtime limit and can run for extended periods of time until the completion of the analysis. This behavior avoids costly setbacks that may be caused by losing results mid-way through an analysis due to hitting a runtime limit.
+
+
Process of Exit Driven Checkpointing
+
+
Using exit driven checkpointing, a job is specified to time out after a user-specified amount of time with an exit code value of 85 (more on this below). Upon hitting this time limit, HTCondor transfers any checkpoint files listed in the submit file attribute transfer_checkpoint_files to a directory called /spool. This directory acts as a storage location for these files in case the job is interrupted. HTCondor then knows that jobs with exit code 85 should be automatically requeued, and will transfer the checkpoint files in /spool to your job’s working directory prior to restarting your executable.
+
+
The process of exit driven checkpointing relies heavily on the use of exit codes to determine the next appropriate steps for HTCondor to take with a job. In general, exit codes are used to report system responses, such as when an analysis is running, encountered an error, or successfully completes. HTCondor recognizes exit code 85 as checkpointing jobs and therefore will know to handle these jobs differently than non-checkpoiting jobs.
+
+
Requirements for Exit Driven Checkpointing
+
+
Requirements for your code or software:
+
+
+ - Checkpoint: The software, program, or code you are using must be able to generate checkpoint files (i.e. snapshots of the progress made thus far) and know how to resume from them.
+ - Resume: This means your code must be able to recognize checkpoint files and know to resume from them instead of the original input data when the code is restarted.
+ - Exit: Jobs should exit with an exit code value of
85 after successfully creating checkpoint files. Additionally, jobs need to be able to exit with a non-85 value if they encounter an error or write the writing the final outputs.
+
+
+
In some cases, these requirements can be achieved by using a wrapper script. This means that your executable may be a script, rather than the code that is writing the checkpoint. An example wrapper script that enables some of these behaviors is below.
+
+
Contact a Research Computing Facilitator for help determining if your job is capable of using checkpointing.
+
+
Changes to the Submit File
+
+
Several modifications to the submit file are needed to enable HTCondor’s checkpointing feature.
+
+
+ - The line
checkpoint_exit_code = 85 must be added. HTCondor recognizes code 85 as a checkpoint job. This means HTCondor knows to end a job with this code but to then to requeue it repeatedly until the analysis completes.
+ - The value of
when_to_transfer_output should be set to ON_EXIT.
+ - The name of the checkpoint files or directories to be transferred to
/spool should be specified using transfer_checkpoint_files.
+
+
+
Optional
+In some cases, it is necessary to write a wrapper script to tell a job when to timeout and exit. In cases such as this, the executable will need to be changed to the name of that wrapper script. An example of a wrapper script that enables a job to checkout and exit with the proper exit codes can be found below.
+
+
An example submit file for an exit driven checkpointing job looks like:
+
+
# exit-driven-example.submit
+
+executable = exit-driven.sh
+arguments = argument1 argument2
+
+checkpoint_exit_code = 85
+transfer_checkpoint_files = my_output.txt, temp_dir, temp_file.txt
++is_resumable = true
+
+should_transfer_files = yes
+when_to_transfer_output = ON_EXIT
+
+output = example.out
+error = example.err
+log = example.log
+
+cpu = 1
+request_disk = 2 GB
+request_memory = 2 GB
+
+queue 1
+
+
+
Example Wrapper Script for Checkpointing Job
+
+
As previously described, it may be necessary to use a wrapper script to tell your job when and how to exit as it checkpoints. An example of a wrapper script that tells a job to exit every 4 hours looks like:
+
+
#!/bin/bash
+
+timeout 4h do_science arg1 arg2
+
+timeout_exit_status=$?
+
+if [ $timeout_exit_status -eq 124 ]; then
+ exit 85
+fi
+
+exit $timeout_exit_status
+
+
+
Let’s take a moment to understand what each section of this wrapper script is doing:
+
+
#!/bin/bash
+
+timeout 4h do_science argument1 argument2
+# The `timeout` command will stop the job after 4 hours (4h).
+# This number can be increased or decreased depending on how frequent your code/software/program
+# is creating checkpoint files and how long it takes to create/resume from these files.
+# Replace `do_science argument1 argument2` with the execution command and arguments for your job.
+
+timeout_exit_status=$?
+# Uses the bash notation of `$?` to call the exit value of the last executed command
+# and to save it in a variable called `timeout_exit_status`.
+
+
+
+if [ $timeout_exit_status -eq 124 ]; then
+ exit 85
+fi
+
+exit $timeout_exit_status
+
+# Programs typically have an exit code of `124` while they are actively running.
+# The portion above replaces exit code `124` with code `85`. HTCondor recognizes
+# code `85` and knows to end a job with this code once the time specified by `timeout`
+# has been reached. Upon exiting, HTCondor saves the files from jobs with exit code `85`
+# in the temporary directory within `/spool`. Once the files have been transferred,
+# HTCondor automatically requeues that job and fetches the files found in `/spool`.
+# If an exit code of `124` is not observed (for example if the program is done running
+# or has encountered an error), HTCondor will end the job and will not automaticlally requeue it.
+
+
+
+
The ideal timeout frequency for a job is every 1-5 hours with a maximum of 10 hours. For jobs that checkpoint and timeout in under an hour, it is possible that a job may spend more time with checkpointing procedures than moving forward with the analysis. After 10 hours, jobs that checkpoint and timeout are less able to take advantage of submitting jobs outside of CHTC to run on other campus resources or on the OSPool.
+
+
Checking the Progress of Checkpointing Jobs
+
+
Always test a single checkpointing job before scaling up to identify odd or unintentional behaviors in your analysis.
+
+
To determine if your job is successfully creating and saving checkpoint files, you can investigate checkpoint files once they have been transferred to /spool.
+
+
You can explore the checkpointed files in /spool by navigating to /var/lib/condor/spool. The directories in this folder are the last four digits of a job’s cluster ID with leading zeros removed. Sub folders are labeled with the process ID for each job. For example, to investigate the checkpoint files for 17870068.220, the files in /spool would be found in folder 68 in a subdirectory called 220.
+
+
It is also possible to intentionally evict a running job and have it rematch to an execute server to test if your code is successfully resuming from checkpoint files or not. To test this, use condor_vacate_job <JobID>. This command will evict your job intentionally and have it return to “Idle” state in the queue. This job will begin running once it rematches to an execute server, allowing you to test if your job is correctly resuming from checkpoint files or incorrectly starting over with the analysis.
+
+
+
+
More information on checkpointing HTCondor jobs can be found in HTCondor’s manual: https://htcondor.readthedocs.io/en/latest/users-manual/self-checkpointing-applications.html This documentation contains additional features available to checkpointing jobs, as well as additional examples such as a python checkpointing job.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ CHTC Services for Research Computing
+
+
+
This page outlines CHTC services offered to UW - Madison affiliates and general guidelines for their use. Users with existing accounts should also refer to our User Expectations pages for more specific limits and guidelines for using CHTC services. To apply for a CHTC account, fill out this form: Getting Started
+
+
Overview
+
+
CHTC’s Research Computing activities aim to empower the research and teaching mission of the University of Wisconsin - Madison by providing access to scalable computing and data capacity.
+
+
Access to standard CHTC services is free of charge.
+
+
Who can use CHTC services?
+
+
Access to CHTC services is available to:
+
+
+ - Current UW Madison affiliates (faculty, students, staff, post-docs)
+ - Current UW System affiliates
+ - Collaborators of UW Madison affiliates, where collaborator access benefits the work of the UW - Madison affiliate, e.g. recently graduated students, collaborators on multi-institution grants
+
+
+
Computing
+
+
CHTC operates two large-scale computing systems.
+
+
High Throughput Computing
+
+
+ - Roughly 15k CPU cores, 100+ GPUs
+ - Additional capacity available via campus HTC systems and the national OSPool.
+ - Single user: 10s - 1000s of tasks (jobs) running at once
+ - Scheduled using HTCondor
+
+
+
+
+ - About 8k CPU cores
+ - Infiniband networking for multi-node capability
+ - Single user: up to 10 jobs running or 720 cores in use
+ - Jobs are scheduled using SLURM job scheduling software
+
+
+
CHTC computing capacity is allocated via a fair-share scheduling algorithm. For groups that require additional or dedicated computing capacity, there is the option to purchase hardware. See our description of buy-in options here: CHTC Buy-In Overview
+
+
Data
+
+
CHTC provides space for data, software and other files that are being used for active computational work. Our file systems have no backup or other redundancy and should not be used as a storage solution. Researchers are assigned a default space quota when their account is created that can be increased upon request. For needs greater than 2TB or for longer-term projects, contact us to sign a data use memorandum of understanding.
+
+
Software
+
+
CHTC systems support software that runs on Linux. Whether or not licensed software
+can be run on CHTC depends significantly on the type of license.
+
+
Both CHTC computing systems support containers.
+
+
Citing CHTC
+
+
In order to track our scientific impact we ask that users cite a DOI in all publications that have benefited from our services. See Citing CHTC for more details.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Citing CHTC Resources
+
+
+
In a Publication
+
+
In order to track our scientific impact we ask that users cite the following DOI in all publications
+that have benefited from our services.
+
+
+ Center for High Throughput Computing. (2006). Center for High Throughput Computing. doi:10.21231/GNT1-HW21
+
+
+
For your convenience, a BibTex file containing the above reference is been provided.
+
+
(Last updated Feb, 2023)
+
+
Appropriate acknowledgement of OSG resources is described here
+
+
+
+
For a Grant Proposal
+
+
(Feel free to modify the below text, use only certain paragraphs, or contact us for more input or customizable letters of support.)
+
+
The University of Wisconsin-Madison (UW-Madison) campus is an excellent match for meeting the computational needs of this project. Existing UW-Madison technology infrastructure supported by the CHTC can be readily leveraged, including CPU capacity, network connectivity, storage availability, and middleware connectivity. The UW-Madison has invested in the CHTC as the primary provider of shared, computing resources to campus researchers. All standard CHTC services are provided free-of-charge to UW-Madison researchers, their projects, and collaborators. But perhaps most important, the UW-Madison has significant staff experience and core competency in deploying, managing, and using computational technology.
+
+
The CHTC is home to over 20 full-time staff with a proven track record of making compute middleware work for scientists. Far beyond just being familiar with the deployment and use of such software, UW staff has been intimately involved in its design and implementation. Dedicated Research Computing Facilitators are available to provide training to all CHTC users and are available to consult on computational practices for achieving the best scientific throughput. As always, CHTC will be happy to provide consulting to ensure optimal use of its facilities, and development of robust, reproducible methods for scalable computing.
+
+
The UW-Madison maintains multiple compute clusters (including the largest of these operated by CHTC) across campus that are managed using either HTCondor or SLURM with support from CHTC. These clusters are connected by HTCondor technology to share resources with each other and with other institutions around the world via OSG services. Local computing capacity directly enabled by CHTC includes:
+
+
+ -
+
High-Throughput Computing (HTC) resources totaling about 30,000 CPU cores in support of research. Temporary file space for large individual files can support up to hundreds of terabytes of total working data. For single computing runs needing significant memory on a single server, the CHTC maintains several multi-core servers with terabytes of memory.
+
+
+ - When on-campus resources are fully utilized, CHTC leverages OSG services to provision additional opportunistic resources from multiple external sites.
+
+
+ -
+
A High-Performance Computing (HPC) cluster consisting of roughly 7,000 tightly coupled cores. Compute nodes have 64 or 128 cores each, and 512 GB RAM, and are networked with 200 Gbps Infiniband, with access to a shared file system and resources managed via Slurm.
+
+ -
+
An origin server where users can make research data available through the Open Science Data Federation and an on-campus cache server which allows external jobs to cache files locally.
+
+
+
+
In the last year, CHTC made possible the use of more than 40,000 core years of computing work for campus researchers, supporting over 300 projects across a wide range of research domains. Temporary storage space for large files can support up to hundreds of terabytes of total working data. Should these resources not be sufficient for the project, the CHTC can also engage computing resources from across the campus grid and the OS Pool, an NSF-supported and expanding alliance of more than 100 universities, national laboratories, scientific collaborations, and software developers.
+
+
The UW–Madison network currently comprises a 200Gbps backbone and WAN connectivity with 160Gbps to the Discovery building. The equipment is located on the “Research Backbone Network”, which allows for friction-free (e.g., no middlebox devices such as firewalls on the data path) to the nation’s research and education networks. Redundancy is built into the network and its supporting infrastructure. An equitable funding model assures that network resources are kept current. The UW has been fundamental to the establishment of the Broadband Optical Research Education And Science network (BOREAS). This Regional Optical Network (RON) connects to the CIC OmniPoP in Chicago, providing a high-speed gateway to various research networks, including Internet2, ESNet, CERN, and other global research networks. BOREAS, along with our participation in the Northern Tier Network Consortium, provides various options to connect at very high speeds to research partners with shared or dedicated bandwidth
+
+
(Last updated May 3, 2024)
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Create a Portable Python Installation with Miniconda
+
+
+
Quickstart: Conda
+
+
Option A (recommended)
+
+
Build a container with Conda packages installed inside:
+
+
+ - How to build your own container
+ - Example container recipes for Conda
+ - Use your container in your HTC jobs
+
+
+
Option B
+
+
Create your own portable copy of your Conda packages:
+
+
+ - Follow the instructions in our guide
+
+
+
+ This approach may be sensitive to the operating system of the execution point.
+We recommend building a container instead, but are keeping these instructions as a backup.
+
+
+
+
+
+
+
The above instructions are intended for if you have package(s) that need to be installed using conda install.
+Miniconda can be used to install Python and R and corresponding packages.
+But if you only need to install Python or R, and do not otherwise need to use a conda install command to set up the packages,
+you should see the instructions specifically for setting up Python or R because there is less chance of obscure errors when building your container.
+
+
When building or using a Miniconda container, you do not need to create or activate a conda environment.
+For the build process, you skip directly to the conda install commands you want to run.
+Similarly, when executing a script in a Miniconda container, the packages are loaded when the container starts.
+
+
Executable
+
+
If you are planning to execute a python .py script using your Miniconda container, you can follow the instructions in the Python guide.
+
+
If you are planning to execute a .R script using your Miniconda container, you can follow the instructions in the R guide.
+
+
Otherwise, you can use a bash .sh script as the submit file executable:
+
+
#!/bin/bash
+
+<your commands go here>
+
+
+
where the contents of the file are the commands that you want to execute using your conda environment.
+You do not and should not try to activate the conda environment in the executable if you are using a container.
+
+
Specifying Exact Dependency Versions
+
+
An important part of improving reproducibility and consistency between runs
+is to ensure that you use the correct/expected versions of your dependencies.
+
+
When you run a command like conda install numpy, conda tries to install
+the most recent version of numpy. For example, numpy version 1.18.2
+was released on March 17, 2020. To install exactly this version of numpy, you
+would run conda install numpy=1.18.2
+(the same works for pip, if you replace = with ==). We
+recommend installing with an explicit version to make sure you have exactly
+the version of a package that you want. This is often called
+“pinning” or “locking” the version of the package.
+
+
If you want a record of what is installed in your environment, or want to
+reproduce your environment on another computer, conda can create a file, usually
+called environment.yml, that describes the exact versions of all of the
+packages you have installed in an environment.
+This file can be re-used by a different conda command to recreate that
+exact environment on another computer.
+
+
To create an environment.yml file from your currently-activated environment, run
+
+
[alice@submit]$ conda env export > environment.yml
+
+
+
This environment.yml will pin the exact version of every dependency in your
+environment. This can sometimes be problematic if you are moving between
+platforms because a package version may not be available on some other platform,
+causing an “unsatisfiable dependency” or “inconsistent environment” error.
+A much less strict pinning is
+
+
[alice@submit]$ conda env export --from-history > environment.yml
+
+
+
which only lists packages that you installed manually, and does not pin their
+versions unless you yourself pinned them during installation.
+If you need an intermediate solution, it is also possible to manually edit
+environment.yml files; see the
+conda environment documentation
+for more details about the format and what is possible.
+In general, exact environment specifications are simply not guaranteed to be
+transferable between platforms (e.g., between Windows and Linux).
+We strongly recommend using the strictest possible pinning available to you.
+
+
To create an environment from an environment.yml file, run
+
+
[alice@submit]$ conda env create -f environment.yml
+
+
+
By default, the name of the environment will be whatever the name of the source
+environment was; you can change the name by adding a -n <name> option to the
+conda env create command.
+
+
If you use a source control system like git, we recommend checking your
+environment.yml file into source control and making sure to recreate it
+when you make changes to your environment.
+Putting your environment under source control gives you a way to track how it
+changes along with your own code.
+
+
If you are developing software on your local computer for eventual use on
+the CHTC pool, your workflow might look like this:
+
+ - Set up a conda environment for local development and install packages as desired
+(e.g.,
conda create -n science; conda activate science; conda install numpy).
+ - Once you are ready to run on the CHTC pool, create an
environment.yml file
+from your local environment (e.g., conda env export > environment.yml).
+ - Move your
environment.yml file from your local computer to the submit machine
+and create an environment from it (e.g., conda env create -f environment.yml),
+then pack it for use in your jobs, as per
+Create Software Package.
+
+
+
More information on conda environments can be found in
+their documentation.
+
+
+
+
Option B: Create your own portable copy
+
+
1. Create a Miniconda installation
+
+
On the submit server,
+download the latest Linux miniconda installer and run it.
+
+
[alice@submit]$ wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
+[alice@submit]$ sh Miniconda3-latest-Linux-x86_64.sh
+
+
+
Accept the license agreement and default options. At the end, you can choose whether or
+not to “initialize Miniconda3 by running conda init?”
+We recommend that you enter “yes”.
+Once you’ve completed the installer, you’ll be prompted to restart your terminal.
+Log out and log back in, and conda will be ready to use to set up your software.
+
+
+ If you choose “no” you’ll want to save the eval command shown by the installer so that you can reactivate the
+Miniconda installation when needed in the future.
+
+
+
2. Create a conda “environment” with your software
+
+
+ (If you are using an environment.yml file as described
+later, you should instead create
+the environment from your environment.yml file. If you don’t have an
+environment.yml file to work with, follow the install instructions in this
+section. We recommend switching to the environment.yml method of creating
+environments once you understand the “manual” method presented here.)
+
+
+
Make sure that you’ve activated the base Miniconda environment if you haven’t
+already. Your prompt should look like this:
+
+
(base)[alice@submit]$
+
+
+
To create an environment, use the conda create command and then activate the
+environment:
+
+
(base)[alice@submit]$ conda create -n env-name
+(base)[alice@submit]$ conda activate env-name
+
+
+
Then, run the conda install command to install the different packages and
+software you want to include in the installation. How this should look is often
+listed in the installation examples for software
+(e.g. Qiime2,
+Pytorch).
+
+
(env-name)[alice@submit]$ conda install pkg1 pkg2
+
+
+
Some Conda packages are only available via specific Conda channels
+which serve as repositories for hosting and managing packages. If Conda is
+unable to locate the requested packages using the example above, you may
+need to have Conda search other channels. More detail are available at
+https://docs.conda.io/projects/conda/en/latest/user-guide/concepts/channels.html.
+
+
Packages may also be installed via pip, but you should only do this
+when there is no conda package available.
+
+
Once everything is installed, deactivate the environment to go back to the
+Miniconda “base” environment.
+
+
(env-name)[alice@submit]$ conda deactivate
+
+
+
For example, if you wanted to create an installation with pandas and
+matplotlib and call the environment py-data-sci, you would use this sequence
+of commands:
+
+
(base)[alice@submit]$ conda create -n py-data-sci
+(base)[alice@submit]$ conda activate py-data-sci
+(py-data-sci)[alice@submit]$ conda install pandas matplotlib
+(py-data-sci)[alice@submit]$ conda deactivate
+(base)[alice@submit]$
+
+
+
+ More about Miniconda
+
+ See the official conda documentation for
+more information on creating and managing environments with conda.
+
+
+
3. Create Software Package
+
+
Make sure that your job’s Miniconda environment is created, but deactivated, so
+that you’re in the “base” Miniconda environment:
+
+
(base)[alice@submit]$
+
+
+
Then, run this command to install the conda pack tool:
+
+
(base)[alice@submit]$ conda install -c conda-forge conda-pack
+
+
+
Enter y when it asks you to install.
+
+
Finally, use conda pack to create a zipped tar.gz file of your environment
+(substitute the name of your conda environment where you see env-name),
+set the proper permissions for this file using chmod, and check the size of
+the final tarball:
+
+
(base)[alice@submit]$ conda pack -n env-name --dest-prefix='$ENVDIR'
+(base)[alice@submit]$ chmod 644 env-name.tar.gz
+(base)[alice@submit]$ ls -sh env-name.tar.gz
+
+
+
When this step finishes, you should see a file in your current directory named
+env-name.tar.gz
+
+
4. Check Size of Conda Environment Tar Archive
+
+
The tar archive, env-name.tar.gz, created in the previous step will be used as input for
+subsequent job submission. As with all job input files, you should check the size of this
+Conda environment file. If >100MB in size, you should NOT transfer the tar ball using
+transfer_input_files. Instead, you should plan to use either CHTC’s web proxy, SQUID or
+large data filesystem Staging. Please contact a research computing facilitators at
+chtc@cs.wisc.edu to determine the best option for your jobs.
+
+
More information is available at File Availability with Squid Web Proxy
+and Managing Large Data in HTC Jobs.
+
+
5. Create a Job Executable
+
+
The job will need to go through a few steps to use this “packed” conda environment;
+first, setting the PATH, then unzipping the environment, then activating it,
+and finally running whatever program you like. The script below is an example
+of what is needed (customize as indicated to match your choices above).
+
+
#!/bin/bash
+
+# have job exit if any command returns with non-zero exit status (aka failure)
+set -e
+
+# replace env-name on the right hand side of this line with the name of your conda environment
+ENVNAME=env-name
+# if you need the environment directory to be named something other than the environment name, change this line
+export ENVDIR=$ENVNAME
+
+# these lines handle setting up the environment; you shouldn't have to modify them
+export PATH
+mkdir $ENVDIR
+tar -xzf $ENVNAME.tar.gz -C $ENVDIR
+. $ENVDIR/bin/activate
+
+# modify this line to run your desired Python script and any other work you need to do
+python3 hello.py
+
+
+
6. Submit Jobs
+
+
In your submit file, make sure to have the following:
+
+
+ - Your executable should be the the bash script you created in step 5.
+ - Remember to transfer your Python script and the environment
tar.gz file via
+ transfer_input_files.
+Since the tar.gz file will almost certainly be larger than 100MB,
+please email us about different tools for
+delivering the installation to your jobs,
+likely our SQUID web proxy.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Learn About Your Jobs Using condor_q
+
+
+
The condor_q command can be used for much more than just
+checking on whether your jobs are running or not! Read on to learn how
+you can use condor_q to answer many common questions about running
+jobs.
+
+
Summary
+
+
+ condor_q: Show my jobs that have been submitted on this server.
+Useful options:
+
+ -nobatch: Starting in version HTCondor 8.6.0 installed in July
+2016, data is displayed in a compact mode (one line per
+cluster). With this option output will be displayed in the old
+format (one line per process)-all: Show all the jobs submitted on the submit server.-hold: Show only jobs in the "on hold" state and the reason
+for that. Held jobs are those that got an error so they could
+not finish. An action from the user is expected to solve the
+problem.-better-analyze JobId: -better-analyze : Analyse a specific
+job and show the reason why it is in its current state.-run: Show your running jobs and related info, like how much
+time they have been running, in which machine, etc.-dag: Organize condor_q output by DAG.-long JobId: Show all information related to that job.-af Attr1 Attr2 ...: List specific attributes of jobs, using
+autoformat.
+
+
+
Examples and Further Explanation
+
+
+
+
1. Default condor_q output
+
+
As of July 19, 2016, the default condor_q output will show a single
+user's jobs, grouped in "batches", as shown below:
+
+
[alice@submit]$ condor_q
+OWNER BATCH_NAME SUBMITTED DONE RUN IDLE HOLD TOTAL JOB_IDS
+alice CMD: sb 6/22 13:05 _ 32 _ _ _ 14297940.23-99
+alice DAG: 14306351 6/22 13:47 27 113 65 _ 205 14306411.0 ...
+alice CMD: job.sh 6/22 13:56 _ _ 12 _ _ 14308195.6-58
+alice DAG: 14361197 6/22 16:04 995 1 _ _ 1000 14367836.0
+
+
+
HTCondor will automatically group jobs into "batches" for this
+display. However, it's also possible for you to specify groups of jobs
+as a "batch" yourself. You can either:
+
+
+ -
+
Add the following line to your submit file:
+
+ batch_name = "CoolJobs"
+
+
+ -
+
Use the -batch-name option with condor_submit:
+
+ [alice@submit]$ condor_submit submit_file.sub -batch-name CoolJobs
+
+
+
+
+
Either option will create a batch of jobs with the label "CoolJobs".
+
+
+
+
2. View all jobs.
+
+
To display more detailed condor_q output (where each job is listed on a
+separate line), you can use the batch name or any existing grouping
+constraint (ClusterId or other "-constraint" options - see
+below for more on constraints) and the -nobatch flag.
+
+
Looking at a batch of jobs with the same ClusterId would look like
+this:
+
+
[alice@submit]$ condor_q -nobatch 195
+
+ ID OWNER SUBMITTED RUN_TIME ST PRI SIZE CMD
+195.10 alice 6/22 13:00 0+00:00:00 H 0 0.0 job.sh
+195.14 alice 6/22 13:00 0+00:01:44 R 0 0.0 job.sh
+195.16 alice 6/22 13:00 0+00:00:26 R 0 0.0 job.sh
+195.39 alice 6/22 13:00 0+00:00:05 R 0 0.0 job.sh
+195.40 alice 6/22 13:00 0+00:00:00 I 0 0.0 job.sh
+195.41 alice 6/22 13:00 0+00:00:00 I 0 0.0 job.sh
+195.53 alice 6/22 13:00 0+00:00:00 I 0 0.0 job.sh
+195.57 alice 6/22 13:00 0+00:00:00 I 0 0.0 job.sh
+195.58 alice 6/22 13:00 0+00:00:00 I 0 0.0 job.sh
+
+9 jobs; 0 completed, 0 removed, 5 idle, 3 running, 1 held, 0 suspended
+
+
+
This was the default view for condor_q from January 2016 until July
+2016.
+
+
+
+
3. View jobs from all users.
+
+
By default, condor_q will just show you information about your
+jobs. To get information about all jobs in the queue, type:
+
+
[alice@submit]$ condor_q -all
+
+
+
This will show a list of all job batches in the queue. To see a list of
+all jobs (individually, not in batches) for all users, combine the
+-all and -nobatch options with condor_q. This was the default view
+for condor_q before January 2016.
+
+
+
+
4. Determine why jobs are on hold.
+
+
If your jobs have gone on hold, you can see the hold reason by running:
+
+
[alice@submit]$ condor_q -hold
+
+
+
or
+
+
[alice@submit]$ condor_q -hold JobId
+
+
+
The first will show you the hold reasons for all of your jobs that
+are on hold; the second will show you the hold reason for a specific
+job. The hold reason is sometimes cut-off; try the following to see the
+entire hold reason:
+
+
[alice@submit]$ condor_q -hold -af HoldReason
+
+
+
If you aren't sure what your hold reason means email
+chtc@cs.wisc.edu.
+
+
+
+
5. Find out why jobs are idle
+
+
condor_q has an option to describe why a job hasn't matched and
+started running. Find the JobId of a job that hasn't started running
+yet and use the following command:
+
+
$ condor_q -better-analyze JobId
+
+
+
After a minute or so, this command should print out some information
+about why your job isn't matching and starting. This information is not
+always easy to understand, so please email us with the output of this
+command if you have questions about what it means.
+
+
+
+
6. Find out where jobs are running.
+
+
To see which computers your jobs are running on, use:
+
+
[alice@submit]$ condor_q -nobatch -run
+428.0 alice 6/22 17:27 0+00:07:17 slot1_12@e313.chtc.wisc.edu
+428.1 alice 6/22 17:27 0+00:07:11 slot1_8@e376.chtc.wisc.edu
+428.2 alice 6/22 17:27 0+00:07:16 slot1_15@e451.chtc.wisc.edu
+428.3 alice 6/22 17:27 0+00:07:16 slot1_17@e277.chtc.wisc.edu
+428.5 alice 6/22 17:27 0+00:07:16 slot1_9@e351.chtc.wisc.edu
+428.7 alice 6/22 17:27 0+00:07:16 slot1_1@e373.chtc.wisc.edu
+428.8 alice 6/22 17:27 0+00:07:16 slot1_5@e264.chtc.wisc.edu
+
+
+
+
+
7. View jobs by DAG.
+
+
If you have submitted multiple DAGs to the queue, it can be hard to tell
+which jobs belong to which DAG. The -dag option to condor_q will
+sort your queue output by DAG:
+
+
[alice@submit]$ condor_q -nobatch -dag
+ ID OWNER/NODENAME SUBMITTED RUN_TIME ST PRI SIZE CMD
+460.0 alice 11/18 16:51 0+00:00:17 R 0 0.3 condor_dagman -p 0
+462.0 |-0 11/18 16:51 0+00:00:00 I 0 0.0 print.sh
+463.0 |-1 11/18 16:51 0+00:00:00 I 0 0.0 print.sh
+464.0 |-2 11/18 16:51 0+00:00:00 I 0 0.0 print.sh
+461.0 alice 11/18 16:51 0+00:00:09 R 0 0.3 condor_dagman -p 0
+465.0 |-0 11/18 16:51 0+00:00:00 I 0 0.0 print.sh
+466.0 |-1 11/18 16:51 0+00:00:00 I 0 0.0 print.sh
+467.0 |-2 11/18 16:51 0+00:00:00 I 0 0.0 print.sh
+
+8 jobs; 0 completed, 0 removed, 6 idle, 2 running, 0 held, 0 suspended
+
+
+
+
+
8. View all details about a job.
+
+
Each job you submit has a series of attributes that are tracked by
+HTCondor. You can see the full set of attributes for a single job by
+using the "long" option for condor_q like so:
+
+
[alice@submit]$ condor_q -l JobId
+...
+Iwd = "/home/alice/analysis/39909"
+JobPrio = 0
+RequestCpus = 1
+JobStatus = 1
+ClusterId = 19997268
+JobUniverse = 5
+RequestDisk = 10485760
+RequestMemory = 4096
+DAGManJobId = 19448402
+...
+
+
+
Attributes that are often useful for checking on jobs are:
+
+
+ Iwd: the job's submission directory on the submit nodeUserLog: the log file for a jobRequestMemory, RequestDisk: how much memory and disk you've
+requested per jobMemoryUsage: how much memory the job has used so farJobStatus: numerical code indicating whether a job is idle,
+running, or heldHoldReason: why a job is on holdDAGManJobId: for jobs managed by a DAG, this is the JobId of the
+parent DAG
+
+
+
+
+
+
If you would like to see specific attributes (see above) for a job or
+group of jobs, you can use the "auto-format" (-af) option to
+condor_q which will print out only the attributes you name for a
+single job or group of jobs.
+
+
For example, if I would like to see the amount of memory and disk I've
+requested for all of my jobs, and how much memory is currently behing
+used, I can run:
+
+
[alice@submit]$ condor_q -af RequestMemory RequestDisk MemoryUsage
+1 325 undefined
+1 325 undefined
+2000 1000 245
+2000 1000 220
+2000 1000 245
+
+
+
+
+
10. Constraining the output of condor_q.
+
+
If you would like to find jobs that meet certain conditions, you can use
+condor_q's "constraint" option. For example, suppose you want to
+find all of the jobs associated with the DAGMan Job ID "234567". You
+can search using:
+
+
[alice@submit]$ condor_q -constraint "DAGManJobId == 234567"
+
+
+
To use a name (for example, a batch name) as a constraint, you'll need
+to use multiple sets of quotation marks:
+
+
[alice@submit]$ condor_q -constraint 'JobBatchName == "MyJobs"'
+
+
+
One common use of constraints is to find all jobs that are running,
+held, or idle. To do this, use a constraint with the JobStatus
+attribute and the appropriate status number - the status codes can be
+found in Appendix
+A
+of the HTCondor Manual.
+
+
Remember condor_q -hold from before? In the background, the
+-hold option is constraining the list of jobs to jobs that are on hold
+(using the JobStatus attribute) and then printing out the HoldReason
+attribute. Try running:
+
+
[alice@submit]$ condor_q -constraint "JobStatus == 5" -af ClusterId ProcId HoldReason
+
+
+
You should see something very similar to running condor_q -hold!
+
+
+
+
11. Remove a held job from the queue
+
+
To remove a job held in the queue, run:
+
+
[alice@submit]$ condor_rm <JobID>
+
+
+
This will remove the job in the queue. Once you have made changes to allow the job to run successfully, the job can be resubmitted using condor_submit.
+
+
+
+
This page takes some of its content and formatting from this HTCondor
+reference
+page.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Automate CHTC Log In
+
+
+
This guide describes
+
+ - how to authenticate with Duo when logging into CHTC’s HTC and HPC systems
+ - how to set your login (SSH) configuration to “reuse” a two-factor authenticated
+connection over a certain period of time.
+ - terminals and applications that are known to support persistent connections
+
+
+
Authentication with Duo
+
+
As of December 2022, accessing CHTC resources
+now requires two-factor authentication. The first “factor” uses your NetID password
+(or SSH keys) and the second “factor” is authentication using Duo, via either a
+Duo fob or the Duo app.
+
+
See the following video for an demonstration of two-factor authentication with Duo
+when logging into CHTC:
+
+
+
+
Re-Using SSH Connections
+
+
To reduce the number of times it is necessary to enter your credentials, it’s possible
+to customize your SSH configuration in a way that allows you to “reuse” a connection
+for logging in again or moving files. This configuration is optional, and
+most useful if you will connect to
+the same server multiple times in a short window, for example, when uploading or
+downloading files.
+
+
WARNING: This guide describes how to configure your local machine to not require
+reentering your NetID password or Duo authentication each time you login.
+This should ONLY be used on secure devices that you manage - it should
+not be used on any shared laptop, desktop, or research group resource. Users
+found violating this policy risk having their CHTC account permanently deactivated.
+
+
The instructions below are meant for users who can use a terminal (Mac, Linux, newer Windows operating systems):
+
+
+ -
+
Open a terminal window.
+
+ - Create (or edit) your personal SSH configuration file at
~/.ssh/config to use
+what’s called “ControlMaster”
+This is the text that should be added to a file called config in the .ssh directory in your computer’s home directory:
+ Host *.chtc.wisc.edu
+ # Turn ControlMaster on
+ ControlMaster auto
+ # ControlMaster connection will persist
+ # for 2 hours of idleness, after which
+ # it will disconnect
+ ControlPersist 2h
+ # Where to store files that represent
+ # the ControlMaster persistent connections
+ ControlPath ~/.ssh/connections/%r@%h:%p
+
+ If you’re not able to find or create the config file, executing the code below from a terminal on your computer
+ will add the right information to the config file
+ # Let's create (or add to) our SSH client configuration file.
+ echo "
+ Host *.chtc.wisc.edu
+ # Turn ControlMaster on
+ ControlMaster auto
+ # ControlMaster connection will persist
+ # for 2 hours of idleness, after which
+ # it will disconnect
+ ControlPersist 2h
+ # Where to store files that represent
+ # the ControlMaster persistent connections
+ ControlPath ~/.ssh/connections/%r@%h:%p" >> ~/.ssh/config
+
+
+ - You also create a directory that will be used to track connections. In
+the same .ssh directory, make a folder called
connections by typing:
+ $ mkdir -p ~/.ssh/connections
+
+
+ Once you login to a CHTC server, this is where the system will store information
+ about your previous connection information so that you do not have to reenter your
+ password or Duo authenticate.
+
+ - Now, log into your CHTC submit server or login node as normal. The first time you log in, you will need to use
+two-factor authentication, but subsequent logins to that machine will not require
+authentication as long as they occur within the time value used in
+the
ControlPersist configuration option (so in this example, 2 hours).
+
+
+
For Windows users who use PuTTY to log in, you need to go to
+the Connection -> SSH section in the “Category” menu on the left side,
+and then check the “Share SSH Connection if possible” box. If you don’t
+see this option, try downloading a newer version of PuTTY.
+
+
Ending “Stuck” Connections
+
+
Sometimes a connection goes stale and you can’t reconnect using it, even if
+it is within the timeout window. In this case, you can avoid using the existing
+connection by removing the relevant file in ~/.ssh/connections; This will probably
+look something like:
+
+
$ ls ~/.ssh/connections/
+alice@submit.chtc.wisc.edu:22
+$ rm ~/.ssh/connections/alice@submit.chtc.wisc.edu:22
+
+
+
Connection settings
+
+
Note that all port forwarding, including X display forwarding, must be setup by
+the initial connection and cannot be changed. If you forget to use -Y on the initial
+connection, you will not be able to open X programs on subsequent connections.
+
+
+
+
There are a variety of tools that people use for transferring and editing files
+like WinSCP and MobaXTerm. Some of these tools are able to use ssh configuration
+or have options that do not require Duo 2FA every time a file is uploaded or
+downloaded or edited, but some do not.
+
+
Known to support persistent connections
+
+
+ -
+
Linux, Mac, and Windows Subsystem for Linux (WSL) terminals
+
+ -
+
WinSCP
+
+ May need to adjust preferences. Within WinSCP:
+
+
+ - Go to Options, then Preferences, and click on Background under the Transfer section.
+ - Set ‘Maximal number of transfers at the same time:’ to 1.
+ - Make sure ‘Use multiple connections for single transfer’ checkbox is checked.
+ - Click ‘OK’ to save.
+
+
+ -
+
Cyberduck (taken from these docs
+
+ Cyberduck does not use SSH configurations, therefore the following setting
+ can be used to enable connection persistence. Within Cyberduck:
+
+
+ - Select Preferences, then the Transfers button, and then the General section.
+ - Under “Transfers”, use the “Transfer Files” drop-down to select “Use browser
+connection”.
+
+
+
+
+
Known to NOT support ControlMaster or similar persistent connections
+
+
+
+
+
+
For those on spotty wireless or those who move a lot with their connection
+(and on *nix) then the open source shell Mosh (https://mosh.org/) has capabilities
+to keep sessions open as you change connections. Note that Mosh doesn’t support the
+following SSH features:
+
+ - ControlMaster (connection multiplexing)
+ - X11 forwarding
+ - Port forwarding
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Log In to CHTC Resources
+
+
+
This guide assumes
+that you have already gotten a CHTC account for either our high
+throughput or high performance compute systems. If you haven't, see our
+getting started page.
+
+
+
+
Table of Contents
+
+
+
+
+
+
+
1. Accessing the Submit Servers
+
+
You will need the following information to log into our CHTC submit
+servers or head nodes:
+
+
Username and Password
+
+
+ - UW - Madison NetId and password
+
+
+
Hostname
+
+
+
+
+ | HTC System |
+
+
+
+
+ ap2001.chtc.wisc.edu (formerly submit1.chtc.wisc.edu) - typically for accounts created before June 2019, between March 2021 - June 2022, or after Feb 1, 2023 |
+
+
+ ap2002.chtc.wisc.edu (formerly submit2.chtc.wisc.edu) - typically for accounts created between June 2019 - February 2021 or between July 1, 2022 - Jan 31, 2023 |
+
+
+
+
+
+
+
+ | HPC Cluster |
+
+
+
+
+ spark-login.chtc.wisc.edu |
+
+
+ hpclogin3.chtc.wisc.edu - access the old HPC cluster, slated to be decommissioned |
+
+
+
+
+
As of December 2022, we also require two-factor authentication with Duo to
+access CHTC resources.
+
+
+ Are you off-campus?
+All of our CHTC submit servers and head nodes are firewalled to block
+log-ins from off-campus. If you are off-campus and want to log in, you
+can either:
+
+
+ - Activate the campus Virtual Private Network (VPN) (more details on how to set this up
+DoIT’s VPN webpage). This will allow you join the campus network when working off-campus.
+ - Log into another computer that is on campus (typically by SSH-ing into that computer) and then SSH to our submit server.
+
+
+ In either case, it will appear like you are on-campus, and you should
+then be able to log into CHTC as usual.
+
+
+
+
+
2. Logging In
+
+
Using the information described above, you can log in to servers two
+different ways -- from the command line or using an SSH program:
+
+
+
+
A. On the command line
+
+
On Mac, Linux, and modern Windows (10+) systems, you can use the "Terminal" application to
+log in. Open a terminal window and use the following command to connect
+to the appropriate server:
+
+
$ ssh username@hostname
+
+
+
You will be prompted for your password, and then for Duo
+authentication.
+
+
+
+
B. Using an SSH program (Windows)
+
+
There are multiple programs to connect to remote servers for Windows. We
+recommend "PuTTy", which can be downloaded
+here.
+To log in, click on the PuTTy executable (putty.exe). You should see a
+screen like this:
+
+

+
+
Fill in the hostname as described in part 1. You should use Port 22 and
+connect using "ssh" -- these are usually the defaults. After you
+click "connect" you will be prompted to fill in your username and
+password, and then to authenticate with Duo.
+
+
Note that once you have submitted jobs to the queue, you can leave your
+logged in session (by typing exit). Your jobs will run and return
+output without you needing to be connected.
+
+
+
+
C. Re-Using SSH Connections
+
+
To reduce the number of times it is necessary to enter your credentials, it’s
+possible to customize your SSH configuration in a way that allows you to “reuse”
+a connection for logging in again or moving files. More details are shown
+in this guide: Automating CHTC Log In
+
+
+
+
3. Learning About the Command Line
+
+
Why learn about the command line? If you haven't used the command
+line before, it might seem like a big challenge to get started, and
+easier to use other tools, especially if you have a Windows computer.
+However, we strongly recommend learning more about the command line for
+multiple reasons:
+
+
+ - You can do most of what you need to do in CHTC by learning a few
+basic commands.
+ - With a little practice, typing on the command line is significantly
+faster and much more powerful than using a point-and-click graphic
+interface.
+ - Command line skills are useful for more than just large-scale
+computing.
+
+
+
For a good overview of command line tools, see the Software Carpentry
+Unix Shell lesson. In
+particular, we recommend the sections on:
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Workflows with HTCondor's DAGMan
+
+
+
Overview
+
+
If your work requires jobs that run in a particular sequence, you may benefit
+from a workflow tool that submits and monitors jobs for you in the correct
+order. A simple workflow manager that integrates with HTCondor is DAGMan,
+or “DAG Manager” where DAG stands for the typical picture of a workflow, a
+directed acyclic graph.
+
+
Learning Resources
+
+
This talk (originally presented at HTCondor Week 2020) gives a good overview of
+when to use DAGMan and its most useful features:
+
+
+ +
+
+
+
For full details on various DAGMan features, see the HTCondor manual page:
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Using Dask at CHTC
+
+
+
Dask
+is a Python library for parallel computing.
+Though it is not the
+traditional HTCondor workflow, it is possible to use
+Dask on the CHTC pool through a special adapter package provided by CHTC.
+This guide describes the situations in which you should consider using
+Dask instead of the traditional workflow, and will point you toward the
+documentation for the adapter package (which will guide you through
+actually using it).
+
+
+ This is a new how-to guide on the CHTC website. Recommendations and
+feedback are welcome via email (chtc@cs.wisc.edu) or by creating an
+issue on the CHTC website GitHub repository: Create an issue
+
+
+
What is Dask?
+
+
Dask
+is a Python library that can “scale up” Python code in two ways:
+
+ - “Low-level” parallelism, through transparently-parallel calculations on familiar interfaces like
numpy arrays.
+ - “High-level” parallelism, through an explicit run-functions-in-parallel interface.
+
+
+
Both kinds of parallelism can be useful, depending on your work.
+For example, Dask could be used to perform data analysis on a single multi-TB
+dataframe stored in distributed memory, as if it was all stored locally.
+It could also be used to run thousands of independent simulations across
+a cluster, aggregating their results locally as they finish.
+Dask can also smoothly handle cases between these extremes (perhaps each of your
+independent simulations also needs a large amount of memory?).
+
+
Dask also “scales down”: it runs the same way on your laptop as it does on
+a cluster thereby providing a smooth transition between running on
+local resources and running on something like the CHTC pool.
+
+
When should I use Dask at CHTC?
+
+
Several use cased are described below for considering the use of Dask for parallelism
+in CHTC instead of the traditional HTCondor workflow
+of creating jobs and DAGs:
+
+
+ - You are already using Dask for parallelism and want to smoothly scale
+up your computing resources. Note that many foundational libraries in the
+scientific Python ecosystem, like xarray,
+now use Dask internally.
+ - You are already using something like
+multiprocessing or
+joblib
+for high-level parallelism.
+Dask’s high-level parallelism interface is fairly similar to these libraries,
+and switching from them to Dask should not involve too much work.
+ - You can make your overall workflow more efficient by adjusting it based
+on intermediate results.
+For example,
+adaptive hyperparameter optimization
+can be significantly more efficient than something like a random grid search,
+but requires a “controller” to guide the process at every step.
+ - You want to operate on single arrays or dataframes that are larger
+than can be stored in the memory of a single average CHTC worker
+(more than a few GB). Dask can store this kind of data in “chunks” on workers
+and seamlessly perform calculations on the chunks in parallel.
+ - You want your workflow to “scale down” to local resources. Being able to run
+your workflow locally may make developing and testing it easier.
+ - You want a more interactive way of using the CHTC pool.
+The adapter package provides tools for running Jupyter Notebooks on the
+CHTC pool, connected to your Dask cluster.
+This can be useful for debugging or inspecting the progress of your workflows.
+
+
+
You may also be interested in Dask’s own
+“Why Dask?” page.
+
+
If you are unsure whether you should use Dask or the traditional workflow,
+please get in touch with a research computing facilitator by emailing
+chtc@cs.wisc.edu to set up a consultation.
+
+
How do I use Dask at CHTC?
+
+
Dask integration with the CHTC pool is provided by the
+Dask-CHTC package.
+See that package’s documentation
+for details on how to get started.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Build a Docker Container Image
+
+
+
Linux containers are a way to build a self-contained environment that
+includes software, libraries, and other tools. CHTC currently supports
+running jobs inside Docker
+containers. This guide describes how to build a Docker image
+that you can use for running jobs in CHTC. For information on using
+this image for jobs, see our Docker Jobs guide.
+
+
Overview
+
+
Note that all the steps below should be run on your own computer, not
+in CHTC.
+
+
Docker images can be created using a special file format
+called a “Dockerfile”. This file has commands that allow you to:
+
+
+ - use a pre-existing Docker image as a base
+ - add files to the image
+ - run installation commands
+ - set environment variables
+
+
+
You can then “build” an image from this
+file, test it locally, and push it to DockerHub, where
+HTCondor can then use the image to build containers to run jobs in.
+Different versions of the image can be labeled with different version
+“tags”.
+
+
This guide has:
+
+
+ - Step by Step Instructions
+ - Examples
+
+
+
A. Step by Step Instructions
+
+
1. Set Up Docker on Your Computer
+
+
If you haven’t already, create a DockerHub account and install
+Docker on your computer. You’ll want to look for the Docker Community
+Edition
+for your operating system. It sometimes takes some time for Docker to
+start, especially the first time. Once Docker starts, it won’t open a
+window; you’ll just see a little whale and container icon in one of your
+computers toolbars. In order to actually use Docker, you’ll need to
+open a command line program (like Terminal, or Command Prompt) and run
+commands there.
+
+
2. Explore Docker Containers (optional)
+
+
If you have never used Docker before, we recommend exploring a pre-existing container
+and testing out installation steps interactively before creating a Dockerfile. See the
+first half of this guide: Exploring and Testing a Docker Container
+
+
3. Create a Dockerfile
+
+
A Dockerfile is a plain text file with keywords that add elements to a
+Docker image. There are many keywords that can be used in a Dockerfile (documented on
+the Docker website here: Dockerfile
+keywords), but we will use a
+subset of these keywords following this basic outline:
+
+
+ - Starting point: Which Docker image do you want to start with?
+ - Additions: What needs to be added? Folders? Data? Other software?
+ - Environment: What variables (if any) are set as part of the software installation?
+
+
+
Create the file
+
+
Create a blank text file named Dockerfile. If you are planning on making
+multiple images for different parts of your workflow,
+you should create a separate folder for each
+new image with the a Dockerfile inside each of them.
+
+
Choose a base image with FROM
+
+
Usually you don’t want to start building your image from scratch.
+Instead you’ll want to choose a “base” image to add things to.
+
+
You can find a base image by searching DockerHub. If you’re
+using a scripting language like Python, R or perl, you could start with
+the “official” image from these languages. If you’re not sure what to
+start with, using a basic Linux image (Debian, Ubuntu and CentOS are common
+examples) is often a good place to start.
+
+
Images often have tagged versions. Besides choosing the image
+you want, make sure to choose a version by clicking on the “Tags” tab of
+the image.
+
+
Once you’ve decided on a base image and version, add it as the first
+line of your Dockerfile, like this:
+
+
FROM repository/image:tag
+
+
+
Some images are maintained by DockerHub itself
+(these are the “official” images mentioned above),
+and do not have a repository.
+For example, to start with Centos 7,
+you could use
+
+
FROM centos:7
+
+
+
while starting from one of
+HTCondor’s HTC Jupyter notebook images
+might look like
+
+
FROM htcondor/htc-minimal-notebook:2019-12-02
+
+
+
When possible, you should use a specific tag
+(not the automatic latest tag)
+in FROM statements.
+
+
Here are some base images you might find useful to build off of:
+
+
+
+
Install packaged software with RUN
+
+
The next step is the most challenging. We need to add commands to the
+Dockerfile to install the desired software. There are a few standard ways to
+do this:
+
+
+ - Use a Linux package manager. This is usually
apt-get for Debian-based
+containers (e.g, Ubuntu) or yum for RedHat Linux containers (e.g., CentOS).
+ - Use a software-specific package manager (like
pip or conda for Python).
+ - Use installation instructions (usually a progression of
configure,
+make, make install).
+
+
+
Each of these options will be prefixed by the RUN keyword. You can
+join together linked commands with the && symbol; to break lines, put
+a backslash \ at the end of the line. RUN can execute any command inside the
+image during construction, but keep in mind that the only thing kept in the final
+image is changes to the filesystem (new and modified files, directories, etc.).
+
+
For example, suppose that your job’s executable ends up running Python and
+needs access to the packages numpy and scipy, as well as the Unix tool wget.
+Below is an example of a Dockerfile that uses RUN to install these packages
+using the system package manager and Python’s built-in package manager.
+
+
# Build the image based on the official Python version 3.8 image
+FROM python:3.8
+
+# Our base image happens to be Debian-based, so it uses apt-get as its system package manager
+# Use apt-get to install wget
+RUN apt-get update \
+ && apt-get install wget
+
+# Use RUN to install Python packages (numpy and scipy) via pip, Python's package manager
+RUN pip3 install numpy scipy
+
+
+
If you need to copy specific files (like source code) from your computer into the
+image, place the files in the same folder as the
+Dockerfile and use the COPY keyword. You could also download files
+within the image by using the RUN keyword and commands like wget
+or git clone.
+
+
For example, suppose that you need to use
+JAGS
+and the
+rjags package for R.
+If you have the
+JAGS source code
+downloaded next to the Dockerfile, you could compile and
+install it inside the image like so:
+
+
FROM rocker/r-ver:3.4.0
+
+# COPY the JAGS source code into the image under /tmp
+COPY JAGS-4.3.0.tar.gz /tmp
+
+# RUN a series of commands to unpack the JAGS source, compile it, and install it
+RUN cd /tmp \
+ && tar -xzf JAGS-4.3.0.tar.gz \
+ && cd JAGS-4.3.0 \
+ && ./configure \
+ && make \
+ && make install
+
+# install the R package rjags
+RUN install2.r --error rjags
+
+
+
Set up the environment with ENV
+
+
Your software might rely on certain environment variables being set correctly.
+
+
One common situation is that if you’re installing a program to a custom location
+(like a home directory), you may need to add that directory to the image’s system
+PATH. For example, if you installed some scripts to /home/software/bin, you
+could use
+
+
ENV PATH="/home/software/bin:${PATH}"
+
+
+
to add them to your PATH.
+
+
You can set multiple environment variables at once:
+
+
ENV DEBIAN_FRONTEND=noninteractive \
+ LC_ALL=en_US.UTF-8 \
+ LANG=en_US.UTF-8 \
+ LANGUAGE=en_US.UTF-8
+
+
+
4. Build, Name, and Tag the Image
+
+
So far we haven’t actually created the image – we’ve just been
+listing instructions for how to build the image in the Dockerfile.
+Now we are ready to build the image!
+
+
First, decide on a name for the image, as well as a tag. Tags are
+important for tracking which version of the image you’ve created (and
+are using). A simple tag scheme would be to use numbers (e.g. v0, v1,
+etc.), but you can use any system that makes sense to you.
+
+
Because HTCondor caches Docker images by tag, we strongly recommend that you
+never use the latest tag, and always build images with a new, unique tag that
+you then explicitly specify in new jobs.
+
+
To build and tag your image, open a Terminal (Mac/Linux) or Command
+Prompt (Windows) and navigate to the folder that contains your
+Dockerfile:
+
+
$ cd directory
+
+
+
(Replace directory with the path to the appropriate folder.)
+
+
Then make sure Docker is running (there should be an icon on
+your status bar, and running docker info shouldn’t indicate any errors) and run:
+
+
$ docker build -t username/imagename:tag .
+
+
+
Replace username with your Docker Hub username and replace
+imagename and tag with the values of your choice. Note the . at the end
+of the command (to indicate “the current directory”).
+
+
If you get errors, try to determine what you may need to add or change
+to your Dockerfile and then run the build command again. Debugging a Docker
+build is largely the same as debugging any software installation process.
+
+
5. Test Locally
+
+
This page describes how to interact with your new Docker image on your
+own computer, before trying to run a job with it in CHTC:
+
+
+
+
6. Push to DockerHub
+
+
Once your image has been successfully built and tested, you
+can push it to DockerHub so that it will be available to run jobs in
+CHTC. To do this, run the following command:
+
+
$ docker push username/imagename:tag
+
+
+
(Where you once again replace username/imagename:tag with what you used in
+previous steps.)
+
+
The first time you push an image to DockerHub, you may need to run this
+command beforehand:
+
+
$ docker login
+
+
+
It should ask for your DockerHub username and password.
+
+
+ Reproducibility
+
+ If you have a free account on Docker Hub, any container image that you
+have pushed there will be scheduled for removal if it is not used (pulled) at least once
+every 6 months (See the Docker Terms of Service).
+
+ For this reason, and just because it’s a good idea in general, we recommend
+creating a file archive of your container image and placing it in whatever space
+you use for long-term, backed-up storage of research data and code.
+
+ To create a file archive of a container image, use this command,
+changing the name of the archive file and container to reflect the
+names you want to use:
+ docker save --output archive-name.tar username/imagename:tag
+
+
+ It’s also a good idea to archive a copy of the Dockerfile used to generate a
+container image along with the file archive of the container image itself.
+
+
+
7. Running Jobs
+
+
Once your Docker image is on Docker Hub, you can use it to run
+jobs on CHTC’s HTC system. See this guide for more details:
+
+
+
+
B. Examples
+
+
This section holds various example Dockerfile that cover more advanced use cases.
+
+
Installing a Custom Python Package from GitHub
+
+
Suppose you have a custom Python package hosted on GitHub, but not available
+on PyPI.
+Since pip can install packages directly from git repositories, you could
+install your package like this:
+
+
FROM python:3.8
+
+RUN pip3 install git+https://github.com/<RepositoryOwner>/<RepositoryName>
+
+
where you would replace <RepositoryOwner> and <RepositoryName> with your
+desired targets.
+
+
QIIME
+
+
This Dockerfile installs QIIME2 based on
+these instructions.
+It assumes that the Linux 64-bit miniconda
+installer has been downloaded into the
+directory with the Dockerfile.
+
+
FROM python:3.6-stretch
+
+COPY Miniconda3-latest-Linux-x86_64.sh /tmp
+
+RUN mkdir /home/qiimeuser
+ENV HOME=/home/qiimeuser
+
+RUN cd /tmp \
+ && ./Miniconda3-latest-Linux-x86_64.sh -b -p /home/qiimeuser/minconda3 \
+ && export PATH=/home/qiimeuser/minconda3/bin:$PATH \
+ && conda update conda \
+ && conda create -n qiime2-2017.10 --file https://data.qiime2.org/distro/core/qiime2-2017.10-conda-linux-64.txt
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Running HTC Jobs Using Docker Containers
+
+
+
Linux containers are a way to build a self-contained environment that
+includes software, libraries, and other tools. This guide shows how to
+submit jobs that use Docker containers.
+
+
Overview
+
+
Typically, software in CHTC jobs is installed or compiled locally by
+individual users and then brought along to each job, either using the
+default file transfer or our SQUID web server. However, another option
+is to use a container system, where the software is installed in a
+container image. Using a container to handle software can be
+advantageous if the software installation 1) has many dependencies, 2)
+requires installation to a specific location, or 3) “hard-codes” paths
+into the installation.
+
+
CHTC has capabilities to access and start containers and
+run jobs inside them. This guide shows how to do this for
+Docker containers.
+
+
1. Use a Docker Container in a Job
+
+
Jobs that run inside a Docker container will be almost exactly the same
+as “vanilla” HTCondor jobs. The main change is indicating which Docker
+container to use and an optional “container universe” option:
+
+
# HTC Submit File
+
+# Provide HTCondor with the name of the Docker container
+container_image = docker://user/repo:tag
+universe = container
+
+executable = myExecutable.sh
+transfer_input_files = other_job_files
+
+log = job.log
+error = job.err
+output = job.out
+
+request_cpus = 1
+request_memory = 4GB
+request_disk = 2GB
+
+queue
+
+
+
In the above, change the address of the Docker container image as
+needed based on the container you are using. More information on finding
+and making container is below.
+
+
Integration with HTCondor
+
+
When your job starts, HTCondor will pull the indicated image from
+DockerHub, and use it to run your job. You do not need to run any
+Docker commands yourself.
+
+
Other pieces of the job (your executable and input files) should be just
+like a non-Docker job submission.
+
+
The only additional change may be that your
+executable no longer needs to install or unpack your software, since it
+will already be present in the Docker container.
+
+
2. Choose or Create a Docker Container Image
+
+
To run a Docker job, you will first need access to a Docker container
+image that has been built and placed onto the
+DockerHub website. There are two primary ways
+to do this.
+
+
A. Pre-existing Images
+
+
The easiest way to get a Docker container image for running a job is to
+use a public or pre-existing image on DockerHub. You can find images by
+getting an account on DockerHub and searching for the software you want
+to use.
+
+
Sample images:
+
+
+
+
An image supported by a group will be continuously updated and the
+versions will be indicated by “tags”. We recommend choosing a specific
+tag (or tags) of the container to use in CHTC.
+
+
B. Build Your Own Image
+
+
You can also build your own Docker container image and upload it to
+DockerHub. See this link to our guide on building containers or the Docker
+documentation for more
+information.
+
+
Similarly, we recommend using container tags. Importantly, whenever you make a significant change
+to your container, you will want to use a new tag name to ensure that your jobs are getting an
+updated version of the container, and not an ‘old’ version that has been cached by DockerHub
+or CHTC.
+
+
3. Testing
+
+
If you want to test your jobs, you have two options:
+
+
+ - We have a guide on exploring and testing Docker containers on your own computer here:
+
+
+ - You can test a container interactively in CHTC by using a normal Docker job submit file and using the
+interactive flag with
condor_submit:
+ [alice@submit]$ condor_submit -i docker.sub
+
+ This should start a session inside the indicated Docker container and connect you to it using ssh. Type exit to end the interactive job. Note: Files generated during your interactive job with Docker will not be transfered back to the submit node. If you have a directory on staging, you can transfer the files there instead; if you have questions about this, please contact a facilitator.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Explore and Test Docker Containers
+
+
+
Linux containers are a way to build a self-contained environment that
+includes software, libraries, and other tools. This guide shows how to
+explore and test a Docker container on your own computer.
+
+
A. Overview
+
+
Note that all the steps below should be run on your own computer, not
+in CHTC.
+
+
This guide has two sections:
+
+
+
+
If you’ve never used Docker before, and/or are getting ready to build your own
+container image, we recommend starting with the first part of the
+guide.
+
+
If you’ve explored Docker already or built your own image and you want to test if it
+will work successfully in CHTC’s HTC system,
+you can follow the directions in the second section.
+
+
A. Set Up Docker on Your Computer
+
+
If you haven’t already, create a DockerHub account and install
+Docker on your computer. You’ll want to look for the Docker Community
+Edition
+for your operating system. It sometimes takes some time for Docker to
+start, especially the first time. Once Docker starts, it won’t open a
+window; you’ll just see a little whale and container icon in one of your
+computers toolbars. In order to actually use Docker, you’ll need to
+open a command line program (like Terminal, or Command Prompt) and run
+commands there.
+
+
B. Explore Docker Containers
+
+
1. Get a Docker Container Image
+
+
We need to have a local copy of the Docker container image in order to
+test it. You can see what container images you already have on your
+computer by running:
+
+
$ docker image ls
+
+
+
If you just installed Docker on your computer
+and are using it for the first time, this list is probably empty.
+If you want to use a pre-made container from Docker Hub,
+you will need to “pull” it down to your computer.
+If you created a container on your computer, it should already
+be in the list of container images.
+
+
If using a container from Docker Hub, find the container and its name, which
+will be of the format: username/imagename:tag. Then pull a copy of the container
+image to your computer by running the following from either a Terminal
+(Mac/Linux) or Command Prompt (Windows):
+
+
$ docker pull username/image:tag
+
+
+
If you run docker image ls again, you should see the container you downloaded
+listed.
+
+
2. Explore the Container Interactively
+
+
To actually explore a container, run this command:
+
+
$ docker run -it --rm=true username/image:tag /bin/bash
+
+
+
This will start a running copy of the container and start a command line shell
+inside. You should see your command line prompt change to something like:
+
+
root@2191c1169757:/#
+
+
+
+ What Do All the Options Mean?
+
+
+ -it: interactive flag--rm=true: after we exit, this will clean up the runnining container so Docker uses less disk space.username/image:tag: which container to start/bin/bash: tells Docker that when the container starts, we want a command line (bash) inside to run commands
+
+
+
If you explore the container using cd and ls, you’ll see that this is a whole,
+self-contained file system, separate from your computer. Try running commands with their
+ --help or --version options to see what’s installed. If you’re planning to create
+ your own container, try following a few of the installation instructions for the software
+ you want to use and see what happens.
+
+
3. Exit the Container
+
+
Once you’re done exploring, type exit to leave the container.
+
+
root@2191c1169757:/# exit
+
+
+
Note that any changes or
+commands you ran in the container won’t be saved! Once you exit the
+running container is shut down and removed (although the container image will still be
+on your computer, which you can see if you type docker image ls again).
+
+
C. Simulate a CHTC Docker Job
+
+
The directions above were about simply exploring a container. If you want to
+simulate what happens in a CHTC job more specifically, we’ll want to do a few things:
+
+
+ - create a test working directory, with needed files
+ - have a list of commands to run or a script you want to use as the executable.
+ - use some extra options when running the container.
+
+
+
1. Create Working Directory
+
+
For testing, we need a folder on your computer to stand in for the
+working directory that HTCondor creates for running your job. Create a folder
+for this purpose on your Desktop. The folder’s name shouldn’t include
+any spaces. Inside this folder, put all of the files that are normally
+inside the working directory for a single job – data, scripts, etc. If
+you’re using your own executable script, this should be in the folder.
+
+
Open a Windows Command Prompt or Mac/Linux Terminal to access that
+folder, replacing “folder” with the name of the folder you created.
+
+
+ - Mac/Linux:
+
$ cd ~/Desktop/folder
+
+
+
+
+
+
2. Plan What to Run
+
+
Once the container starts, you have a few options for testing your job:
+
+
+ - Run Commands Directly
+
+ - When you start the container, you’ll be able to run each command you
+ want to use, step-by-step. If you have multiple commands, these will eventually
+ need to be put into a shell script as your executable.
+ - Example: Running multiple steps of a bioinformatics pipeline
+
+
+ - Run an Executable
+
+ - If you’ve already written a script with all your commands or code, you can
+ test this in the container.
+ - Examples: Running a shell script with multiple steps, running a machine learning Python script
+
+
+ - Run a Single Command
+
+ - If you only want to run one command, using a program installed in the Docker
+ container, you can run this in the container.
+ - Example: Running GROMACS from a container
+
+
+
+
+
3. Start the Docker Container
+
+
We’ll use a similar docker run command to start the Docker container,
+with a few extra options to better emulate how containers are run in
+the HTC system with HTCondor.
+
+
This command can be run verbatim except for the
+username, imagename and tag; these should be whatever you used to
+pull or tag the container image.
+
+
+
+
+
For Windows users, a window may pop up, asking for permission to share
+your main drive with Docker. This is necessary for the files to be
+placed inside the container. As in the previous section, the docker run command
+will start a running copy of the container and start a command line shell
+inside.
+
+
+ What Do All the Options Mean? Part 2
+
+ The options that we have added for this example are used in CHTC to make jobs run
+successfully and securely.
+
+
+ --user $(id -u):$(id -g): runs the container with more restrictive permissions-v $(pwd):/scratch: Put the current working directory (pwd) into the container but call it /scratch.
+In CHTC, this working directory will be the job’s usual working directory.-w /scratch: when the container starts, make /scratch the working directory
+
+
+
4. Test the job
+
+
Your command line prompt should have changed to look like this:
+
+
I have no name!@5a93cb:/scratch$
+
+
+
We can now see if the job would complete successfully!
+
+
If you have a single command or list of commands to run, start running them one by one.
+If you have an executable script, you can run it like so:
+
+
I have no name!@5a93cb:/scratch$ ./exec.sh
+
+
+
If your “executable” is software already in the container, run the
+appropriate command to use it.
+
+
+ Permission Errors
+
+ The following commands may not be necessary, but if you see messages
+about “Permission denied” or a bash error about bad formatting, you
+may want to try one (or both) of the following (replacing exec.sh
+with the name of your own executable.)
+
+ You may need to add executable permissions to the script for it to run
+correctly:
+
+ I have no name!@5a93cb:/scratch$ chmod +x exec.sh
+
+
+ Windows users who are using a bash script may also need to run the
+following two commands:
+
+ I have no name!@5a93cb:/scratch$ cat exec.sh | tr -d \\r > temp.sh
+I have no name!@5a93cb:/scratch$ mv temp.sh exec.sh
+
+
+
+
When your test is done, type exit to leave the container:
+
+
If the program didn’t work, try searching for the cause of the error
+messages, or email CHTC’s Research Computing Facilitators.
+
+
If your local test did run successfully, you are now ready to set up
+your Docker job to run on CHTC.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Windows / Linux Incompatibility
+
+
+
If your job is running a bash or shell script (includes the header
+#!/bin/bash), and it goes on hold, you might be experiencing a
+Windows/Linux incompability error. Files written in Windows (based on
+the DOS operating system) and files written in Mac/Linux (based on the
+UNIX operating system) use different invisible characters to mean "end
+of a line" in a file. Normally this isn't a problem, except when
+writing bash scripts; bash will not be able to run scripts if they have
+the Windows/DOS line endings.
+
+
To find why the job went on hold, look for the hold reason, either by
+running
+
+
[alice@submit]$ condor_q -af HoldReason
+
+
+
or by looking in the log file.
+
+
If a Windows/Linux incompatibility is the problem, the hold reason will
+look something like this:
+
+
Error from slot1_11@e189.chtc.wisc.edu: Failed to execute
+'/var/lib/condor/execute/slot1/dir_4086540/condor_exec.exe' with
+arguments 2: (errno=2: 'No such file or directory')
+
+
+
To check if this is the problem, you can open the script in the vi text
+editor, using its "binary" mode:
+
+
[alice@submit]$ vi -b hello-chtc.sh
+
+
+
(Replace hello-chtc.sh with the name of your script.) If you see ^M
+characters at the end of each line, those are the DOS line endings and
+that's the problem.
+(Type :q to quit vi)
+
+
Luckily, there is an easy fix! To convert the script to unix line
+endings so that it will run correctly, you can run:
+
+
[alice@submit]$ dos2unix hello-chtc.sh
+
+
+
on the submit node and it will change the format for you. If you release
+your held jobs (using condor_release) or re-submit the jobs, you
+should no longer get the same error.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Managing Large Data in HTC Jobs
+
+
+
Which Option is the Best for Your Files?
+
+
+
+ | Input Sizes |
+ Output Sizes |
+ Link to Guide |
+ File Location |
+ How to Transfer |
+ Availability, Security |
+
+
+
+
+ | 0 - 100 MB per file, up to 500 MB per job |
+ 0 - 5 GB per job |
+ Small Input/Output File Transfer via HTCondor |
+ /home |
+ submit file; filename in transfer_input_files |
+ CHTC, UW Grid, and OSG; works for your jobs |
+
+
+
+
+ | 100 MB - 1 GB per repeatedly-used file |
+ Not available for output |
+ Large Input File Availability Via Squid |
+ /squid |
+ submit file; http link in transfer_input_files |
+ CHTC, UW Grid, and OSG; files are made *publicly-readable* via an HTTP address |
+
+
+
+
+ | 100 MB - TBs per job-specific file; repeatedly-used files > 1GB |
+ 4 GB - TBs per job |
+ Large Input and Output File Availability Via Staging |
+ /staging |
+ job executable; copy or move within the job |
+ a portion of CHTC; accessible only to your jobs |
+
+
+
+
+
+
+
+
When submitting jobs to CHTC’s High Throughput Computing (HTC) system,
+there is a distinct location for staging data that is too large to be
+handled at scale via the default HTCondor file transfer mechanism. This
+location should be used for jobs that require input files larger than 100MB
+and/or that generate output files larger than 3-4GB.
+
+
To best understand the below information, users should already be
+familiar with:
+
+
+ - Using the command-line to: navigate directories,
+create/edit/copy/move/delete files and directories, and run intended
+programs (aka “executables”).
+ - CHTC’s Intro to Running HTCondor Jobs
+ - CHTC’s guide for Typical File Transfer
+
+
+
+
+
Table of Contents
+
+
+
+
+
1. Policies and Intended Use
+
+
+ USERS VIOLATING ANY OF THE POLICIES IN THIS GUIDE WILL
+HAVE THEIR DATA STAGING ACCESS AND/OR CHTC ACCOUNT REVOKED UNTIL CORRECTIVE
+MEASURES ARE TAKEN. CHTC STAFF RESERVE THE RIGHT TO REMOVE ANY
+PROBLEMATIC USER DATA AT ANY TIME IN ORDER TO PRESERVE PERFORMANCE.
+
+
+
A. Intended Use
+
+
Our large data staging location is only for input and output files that
+are individually too large to be managed by our other data movement
+methods, HTCondor file transfer or SQUID. This includes individual input files
+greater than 100MB and individual output files greater than 3-4GB.
+
+
Users are expected to abide by this intended use expectation and follow the
+instructions for using /staging written in this guide (e.g. files placed
+in /staging should NEVER be listed in the submit file, but rather accessed
+via the job’s executable (aka .sh) script).
+
+
B. Access to Large Data Staging
+
+
Any one with a CHTC account whose data meets the intended use above can request
+space in our large data staging area. A Research Computing Facilitator will
+review the request and follow up. If appropriate, access will be granted via
+a directory in the system and a quota. Quotas are based on individual user needs;
+if a larger quota is needed, see our Request a Quota Change guide.
+
+
We can also create group or shared spaces by request.
+
+
C. User Data Management Responsibilities
+
+
As with all CHTC file spaces:
+
+
+ - Keep copies: Our large data staging area is not backed up and has the
+possibility of data loss; keep copies of ANY and ALL data in
/staging in another, non-CHTC
+location.
+ - Remove data: We expect that users remove data from
/staging AS
+SOON AS IT IS NO LONGER NEEDED FOR ACTIVELY-RUNNING JOBS.
+ - Monitor usage and quota: Each
/staging folder has both a size and “items” quota. Quota changes
+can be requested as described in our Request a Quota Change guide.
+
+
+
CHTC staff reserve the right to remove data from our large data staging
+location (or any CHTC file system) at any time.
+
+
D. Data Access Within Jobs
+
+
Staged large data will
+be available only within the the CHTC pool, on a subset of our total
+capacity.
+
+
Staged data are owned by the user, and only the user’s own
+jobs can access these files (unless the user specifically modifies unix
+file permissions to make certain files available for other users).
+
+
2. Staging Large Data
+
+
In order to stage large data for use on CHTC’s HTC system:
+
+
+ - Get a directory: Large data staging is available by request.
+ - Reduce file counts: Combine and compress files that are used together.
+ - Use the transfer server: Upload your data via our dedicated file transfer server.
+ - Remove files after jobs complete: our data staging space is quota controlled and not backed up.
+
+
+
A. Get a Directory
+
+
Space in our large data staging area is granted by request. If you think you need
+a directory, email CHTC’s Research Computing Facilitators (chtc@cs.wisc.edu).
+
+
The created directory will exist at this path: /staging/username
+
+
B. Reduce File Counts
+
+
Data placed in our large data /staging location
+should be stored in as few files as possible (ideally,
+one file per job), and will be used by a job only after being copied
+from /staging into the job working directory (see below).
+Similarly, large output should first be written to the
+job working directory then compressed in to a single file before being
+copied to /staging at the end of the job.
+
+
To prepare job-specific data that is large enough to pre-staging
+and exists as multiple files or directories (or a directory of multiple
+files), first create a compressed tar package before placing the file in
+/staging (either before submitting jobs, or within jobs before
+moving output to /staging). For example:
+
+
$ tar -czvf job_package.tar.gz file_or_dir
+
+
+
C. Use the Transfer Server
+
+
Movement of data into/out of /staging before and after jobs should
+only be performed via CHTC’s transfer server, as below, and not via a
+CHTC submit server. After obtaining a user directory within
+/staging and an account on the transfer server, copy relevant
+files directly into this user directory from your own computer:
+
+
+
+
D. Remove Files After Jobs Complete
+
+
As with all CHTC file spaces, data should be removed from /staging AS
+SOON AS IT IS NO LONGER NEEDED FOR ACTIVELY-RUNNING JOBS. Even if it
+will be used it the future, it should be deleted from and copied
+back at a later date. Files can be taken off of /staging using similar
+mechanisms as uploaded files (as above).
+
+
3. Using Staged Files in a Job
+
+
As shown above, the staging directory for large data is /staging/username.
+All interaction with files in this location should occur within your job’s
+main executable.
+
+
+
+
To use large data placed in the /staging location, add commands to your
+job executable that copy input
+from /staging into the working directory of the job. Program should then use
+files from the working directory, being careful to remove the coiped
+files from the working
+directory before the completion of the job (so that they’re not copied
+back to the submit server as perceived output).
+
+
Example, if executable is a shell script:
+
+
#!/bin/bash
+#
+# First, copy the compressed tar file from /staging into the working directory,
+# and un-tar it to reveal your large input file(s) or directories:
+cp /staging/username/large_input.tar.gz ./
+tar -xzvf large_input.tar.gz
+#
+# Command for myprogram, which will use files from the working directory
+./myprogram large_input.txt myoutput.txt
+#
+# Before the script exits, make sure to remove the file(s) from the working directory
+rm large_input.tar.gz large_input.txt
+#
+# END
+
+
+
B. Moving Large Output Files
+
+
If jobs produce large (more than 3-4GB) output files, have
+your executable write the output file(s) to a location within
+the working directory, and then make sure to move this large file to
+the /staging folder, so that it’s not transferred back to the home directory, as
+all other “new” files in the working directory will be.
+
+
Example, if executable is a shell script:
+
+
#!/bin/bash
+#
+# Command to save output to the working directory:
+./myprogram myinput.txt output_dir/
+#
+# Tar and mv output to staging, then delete from the job working directory:
+tar -czvf large_output.tar.gz output_dir/ other_large_files.txt
+mv large_output.tar.gz /staging/username/
+rm other_large_files.txt
+#
+# END
+
+
+
C. Handling Standard Output (if needed)
+
+
In some instances, your software may produce very large standard output
+(what would typically be output to the command screen, if you ran the
+command for yourself, instead of having HTCondor do it). Because such
+standard output from your software will usually be captured by HTCondor
+in the submit file “output” file, this “output” file WILL still be
+transferred by HTCondor back to your home directory on the submit
+server, which may be very bad for you and others, if that captured
+standard output is very large.
+
+
In these cases, it is useful to redirect the standard output of commands
+in your executable to a file in the working directory, and then move it
+into /staging at the end of the job.
+
+
Example, if “myprogram” produces very large standard output, and is
+run from a script (bash) executable:
+
+
#!/bin/bash
+#
+# script to run myprogram,
+#
+# redirecting large standard output to a file in the working directory:
+./myprogram myinput.txt myoutput.txt > large_std.out
+#
+# tar and move large files to staging so they're not copied to the submit server:
+tar -czvf large_stdout.tar.gz large_std.out
+cp large_stdout.tar.gz /staging/username/subdirectory
+rm large_std.out large_stdout.tar.gz
+# END
+
+
+
4. Submit Jobs Using Staged Data
+
+
In order to properly submit jobs using staged large data, always do the following:
+
+
+ - Submit from
/home: ONLY submit jobs from within your home directory
+ (/home/username), and NEVER from within /staging.
+
+
+
In your submit file:
+
+
+ - No large data in the submit file: Do NOT list any
/staging files in any of the submit file
+ lines, including: executable, log, output, error, transfer_input_files. Rather, your
+ job’s ENTIRE interaction with files in /staging needs to occur
+ WITHIN each job’s executable, when it runs within the job (as shown above)
+ - Request sufficient disk space: Using
request_disk, request an amount of disk
+space that reflects the total of a) input data that each job will copy into
+ the job working directory from /staging, and b) any output that
+ will be created in the job working directory.
+ - Require access to
/staging: Include the CHTC specific attribute that requires
+servers with access to /staging
+
+
+
See the below submit file, as an example, which would be submitted from
+within the user’s /home directory:
+
+
### Example submit file for a single job that stages large data
+# Files for the below lines MUST all be somewhere within /home/username,
+# and not within /staging/username
+
+executable = run_myprogram.sh
+log = myprogram.log
+output = $(Cluster).out
+error = $(Cluster).err
+
+## Do NOT list the large data files here
+transfer_input_files = myprogram
+
+# IMPORTANT! Require execute servers that can access /staging
+Requirements = (Target.HasCHTCStaging == true)
+
+# Make sure to still include lines like "request_memory", "request_disk", "request_cpus", etc.
+
+queue
+
+
+
+ Note: in no way should files on /staging be specified in the submit file,
+directly or indirectly! For example, do not use the initialdir option (
+Submitting Multiple Jobs in Individual Directories)
+to specify a directory on /staging.
+
+
+
5. Checking your Quota, Data Use, and File Counts
+
+
You can use the command get_quotas to see what disk
+and items quotas are currently set for a given directory path.
+This command will also let you see how much disk is in use and how many
+items are present in a directory:
+
+
[username@transfer ~]$ get_quotas /staging/username
+
+
+
Alternatively, the ncdu command can also be used to see how many
+files and directories are contained in a given path:
+
+
[username@transfer ~]$ ncdu /staging/username
+
+
+
When ncdu has finished running, the output will give you a total file
+count and allow you to navigate between subdirectories for even more
+details. Type q when you're ready to exit the output viewer. More
+info here: https://lintut.com/ncdu-check-disk-usage/
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Managing Large Data in HTC Jobs with S3 Buckets
+
+
+
Which Option is the Best for Your Files?
+
+
+
+ | Input Sizes |
+ Output Sizes |
+ Link to Guide |
+ File Location |
+ How to Transfer |
+ Availability, Security |
+
+
+
+
+ | 0 - 100 MB per file, up to 500 MB per job |
+ 0 - 5 GB per job |
+ Small Input/Output File Transfer via HTCondor |
+ /home |
+ submit file; filename in transfer_input_files |
+ CHTC, UW Grid, and OSG; works for your jobs |
+
+
+
+
+ | 100 MB - 1 GB per repeatedly-used file |
+ Not available for output |
+ Large Input File Availability Via Squid |
+ /squid |
+ submit file; http link in transfer_input_files |
+ CHTC, UW Grid, and OSG; files are made *publicly-readable* via an HTTP address |
+
+
+
+
+ | 100 MB - TBs per job-specific file; repeatedly-used files > 1GB |
+ 4 GB - TBs per job |
+ Large Input and Output File Availability Via Staging |
+ /staging |
+ job executable; copy or move within the job |
+ a portion of CHTC; accessible only to your jobs |
+
+
+
+
+
+
+
+
When submitting jobs to CHTC’s High Throughput Computing (HTC) system,
+there is a distinct location for staging data that is too large to be
+handled at scale via the default HTCondor file transfer mechanism
+but needs to be accessed outside of CHTC
+(for example, data for jobs that run on the OS Pool).
+
+
To best understand the below information, users should already be
+familiar with:
+
+
+ - Using the command-line to: navigate directories,
+create/edit/copy/move/delete files and directories, and run intended
+programs (aka “executables”).
+ - CHTC’s Intro to Running HTCondor Jobs
+ - CHTC’s guide for Typical File Transfer
+
+
+
Contents
+
+
+ - Policies and Intended Use
+ - Staging Large Data in S3 Buckets
+ - Using Staged Files in a Job
+
+
+ - Checking your Data Use and File Counts
+
+
+
1. Policies and Intended Use
+
+
+ USERS VIOLATING ANY OF THE POLICIES IN THIS GUIDE WILL
+HAVE THEIR DATA STAGING ACCESS AND/OR CHTC ACCOUNT REVOKED UNTIL CORRECTIVE
+MEASURES ARE TAKEN. CHTC STAFF RESERVE THE RIGHT TO REMOVE ANY
+PROBLEMATIC USER DATA AT ANY TIME IN ORDER TO PRESERVE PERFORMANCE.
+
+
+
A. Intended Use
+
+
Our S3 data storage is only for input and output files that
+are individually too large to be managed by our other data movement
+methods, HTCondor file transfer or SQUID, and when these files are
+expected to be accessed outside of CHTC. This includes individual input files
+greater than 100MB and individual output files greater than 3-4GB.
+
+
Files in our S3 data storage are organized in storage units called
+“buckets.” You can think of an S3 bucket like a folder containing a
+set of data. Each bucket has a unique name of your choosing and can
+contain folders, executable files, data files, and most other types of
+files. S3 buckets are protected with a key that is unique to you
+(similar to a password) and, when provided with the key, buckets
+can be accessed from any machine with an internet connection. CHTC
+automatically creates and manages keys for users, so you do not have
+to provide your key when manging files in your S3 buckets on CHTC
+transfer servers or when submitting jobs on CHTC submit servers that
+transfer data from S3 buckets.
+
+
Users are expected to abide by this intended use expectation and follow the
+instructions for using S3 buckets written in this guide (e.g. files placed
+in S3 buckets should ALWAYS be listed in the submit file).
+
+
B. Getting Access to Create S3 Buckets
+
+
Any one with a CHTC account whose data meets the intended use above
+can request access to create S3 buckets inside CHTC’s S3 data
+storage. A Research Computing Facilitator will review the request and
+follow up. If appropriate, S3 bucket creation will be enabled for and
+a quota will be set on your account. Quotas are based on individual
+user needs; if a larger quota is needed, email chtc@cs.wisc.edu with
+your request.
+
+
C. User Data Management Responsibilities
+
+
As with all CHTC file spaces:
+
+
+ - Keep copies: Our S3 buckets are not backed up and have the
+possibility of data loss; keep copies of ANY and ALL data in S3
+buckets in another, non-CHTC location.
+ - Remove data: We expect that users remove data from S3 buckets AS
+SOON AS IT IS NO LONGER NEEDED FOR ACTIVELY-RUNNING JOBS.
+ - Monitor usage and quota: Your account has both a size and
+number of files quota that applies across all buckets owned by your
+account. Quota changes can be requested by emailing chtc@cs.wisc.edu.
+
+
+
CHTC staff reserve the right to remove S3 buckets or revoke bucket
+creation permission at any time.
+
+
D. Data Access Within Jobs
+
+
Data in a CHTC S3 bucket can be accessed from jobs running almost
+anywhere (including most of OS Pool). HTCondor automatically matches and
+runs jobs that use S3 buckets only on machines that support S3 data
+transfers.
+
+
Data in CHTC S3 buckets are owned by the user (or a set of users), and
+only the user’s (or users’) own jobs can access these files.
+
+
2. Staging Large Data in S3 Buckets
+
+
In order to stage data in an S3 bucket for use on CHTC’s HTC system:
+
+
+ - Get S3 bucket creation access: Bucket creation access is granted by request.
+ - Create an S3 bucket: Create a bucket that will contain the data for your project.
+ - Reduce file counts: Combine and compress files that are used together.
+ - Use the transfer server: Upload your data to your bucket via our dedicated file transfer server.
+ - Remove files after jobs complete: Data in S3 buckets are quota controlled and not backed up.
+
+
+
A. Get S3 Bucket Creation Access
+
+
CHTC S3 bucket creation access is granted by request. If you think you need
+to create S3 buckets, email CHTC’s Research Computing Facilitators (chtc@cs.wisc.edu).
+
+
B. Create an S3 Bucket
+
+
Buckets can be created on a CHTC submit server or the CHTC transfer server
+using the mc command:
+
+
[alice@transfer]$ mc mb chtc/my-bucket-name
+
+
+
Each bucket in CHTC must have a unique name, so be descriptive! We
+recommend creating a bucket per dataset or per batch of jobs.
+
+
C. Reduce File Counts
+
+
Data placed in S3 buckets should be stored in as few files as possible
+(ideally, one file per job). Similarly, large output should first be
+written to the job working directory then compressed in to a single
+file before being transferred back to an S3 bucket at the end of the job.
+
+
To prepare job-specific data that is large enough
+and exists as multiple files or directories (or a directory of multiple
+files), first create a compressed tar package before placing the file in
+an S3 bucket (either before submitting jobs, or within jobs before
+transferring output to). For example:
+
+
$ tar -czvf job_package.tar.gz file_or_dir
+
+
+
D. Use the Transfer Server
+
+
Movement of large data into/out of S3 buckets before and after jobs
+should be performed via CHTC’s transfer server, as below, and
+not via a CHTC submit server. After obtaining an account on the
+transfer server and creating an S3 bucket, copy relevant files directly into your
+home directory from your own computer:
+
+
+
+
Then in an SSH session on the transfer server, copy files in to your
+S3 bucket:
+
+
[alice@transfer]$ mc cp large-input.file chtc/my-bucket
+
+
+
E. Remove Files After Jobs Complete
+
+
As with all CHTC file spaces, data should be removed from S3 buckets AS
+SOON AS IT IS NO LONGER NEEDED FOR ACTIVELY-RUNNING JOBS. Even if it
+will be used again in the future, it should be deleted from and copied
+back at a later date. Files can be taken out of S3 buckets using similar
+mechanisms as uploaded files. In an SSH session on the transfer
+server, copy files from your bucket to your home directory:
+
+
[alice@transfer]$ mc cp chtc/my-bucket/large-output.file .
+
+
+
Then copy files from the transfer server to your own computer:
+
+
+
+
To remove a file inside your S3 bucket, in an SSH session on the
+transfer server:
+
+
[alice@transfer]$ mc rm chtc/my-bucket/large-input.file
+[alice@transfer]$ mc rm chtc/my-bucket/large-output.file
+
+
+
To remove an entire bucket (only do this if you are certain the
+bucket is no longer needed):
+
+
[alice@transfer]$ mc rb chtc/my-bucket
+
+
+
3. Using Staged Files in a Job
+
+
+
+
To use data placed in a CHTC S3 bucket, add files to your submit
+file’s transfer_input_files that point to the filename
+(e.g. large-input.file) inside your bucket (e.g. my-bucket) on
+CHTC’s S3 storage (s3dev.chtc.wisc.edu):
+
+
...
+executable = my_script.sh
+transfer_input_files = s3://s3dev.chtc.wisc.edu/my-bucket/large-input.file
+arguments = large-input.file
+...
+
+
+
B. Moving Large Output Files
+
+
To have your job automatically copy data back to your CHTC S3 bucket,
+add file mappings to a transfer_output_remaps command inside your
+submit file:
+
+
transfer_output_remaps = "large-output.file = s3://s3dev.chtc.wisc.edu/my-bucket/large-output.file"
+
+
+
4. Checking Your Data Use and File Counts
+
+
To check what files are in your bucket and the size of the files:
+
[alice@submit]$ mc ls chtc/my-bucket
+
+
+
To check your bucket’s total data usage:
+
[alice@submit]$ mc du chtc/my-bucket
+
+
+
To check your bucket’s file count:
+
[alice@submit]$ mc find chtc/my-bucket | wc -l
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Transfer Large Input Files Via Squid
+
+
+
Which Option is the Best for Your Files?
+
+
+
+ | Input Sizes |
+ Output Sizes |
+ Link to Guide |
+ File Location |
+ How to Transfer |
+ Availability, Security |
+
+
+
+
+ | 0 - 100 MB per file, up to 500 MB per job |
+ 0 - 5 GB per job |
+ Small Input/Output File Transfer via HTCondor |
+ /home |
+ submit file; filename in transfer_input_files |
+ CHTC, UW Grid, and OSG; works for your jobs |
+
+
+
+
+ | 100 MB - 1 GB per repeatedly-used file |
+ Not available for output |
+ Large Input File Availability Via Squid |
+ /squid |
+ submit file; http link in transfer_input_files |
+ CHTC, UW Grid, and OSG; files are made *publicly-readable* via an HTTP address |
+
+
+
+
+ | 100 MB - TBs per job-specific file; repeatedly-used files > 1GB |
+ 4 GB - TBs per job |
+ Large Input and Output File Availability Via Staging |
+ /staging |
+ job executable; copy or move within the job |
+ a portion of CHTC; accessible only to your jobs |
+
+
+
+
+
+
+
+
+
+
SQUID Web Proxy
+
+
CHTC maintains a SQUID web proxy from which pre-staged input files and
+executables can be downloaded into jobs using CHTC's proxy HTTP
+address.
+
+
+
+
Table of Contents
+
+
+
+
+
+
+
1. Applicability
+
+
+ -
+
Intended Use:
+ The SQUID web proxy is best for cases where many jobs will use the
+ same large file (or few files), including large software. It is not
+ good for cases when each of many jobs needs a different large
+ input file, in which case our large data staging
+ location should be used. Remember that
+ you're always better off by pre-splitting a large input file into
+ smaller job-specific files if each job only needs some of the large
+ files's data. If each job needs a large set of many files, you
+ should create a .tar.gz file containing all the files, and this
+ file will still need to be less than 1 GB.
+
+ -
+
Access to SQUID:
+ is granted upon request to chtc@cs.wisc.edu. A user on CHTC submit
+ servers may will be granted a user directory within /squid, which
+ users should transfer data into via the CHTC transfer server
+ (transfer.chtc.wisc.edu). As for all CHTC file space, users should
+ minimize the amount of data on the SQUID web proxy, and should clean
+ files from the /squid location regularly. CHTC staff reserve the
+ right to remove any file from /squid when needed to preserve
+ availability and performance for all users.
+
+ -
+
Advantages:
+ Files placed on the SQUID web proxy can be downloaded by jobs
+ running anywhere, because the files are world-readable.
+
+ - Limitations and Policies:
+
+ - SQUID cannot be used for job output, as there is no way to
+change files in SQUID from within a job.
+ - SQUID is also only capable of delivering individual files up to
+1 GB in size.
+ - A change you make to a file within your
/squid directory may
+not take effect immediately on the SQUID web proxy if you use
+the same filename. Therefore, it is important to use a new
+filename when replacing a file in your /squid directory.
+ - Jobs should still ALWAYS and ONLY be submitted from within the
+user's
/home location.
+ - Only the "http" address should be listed in the
+"
transfer_input_files" line of the submit file. File
+locations starting with "/squid" should NEVER be listed in
+the submit file.
+ - Users should only have data in /squid that is being use for
+currently-queued jobs; CHTC provides no back ups of any data in
+CHTC systems, and our staff reserve the right to remove any data
+causing issues. It is the responsibility of users to keep copies
+of all essential data in preparation for potential data loss or
+file system corruption.
+
+
+
+ - Data Security:
+ Files placed in SQUID can only be edited by the owner of the user
+ directory within /squid, but will end up being world-readable on
+ the SQUID web proxy in order to be readily downloadable by jobs
+ (with the proper HTTP address); thus, large files that should be
+ "private" should not be placed in your user directory in /squid,
+ and should instead use CHTC's large data staging
+ space for large-file staging.
+
+
+
+
+
+
+
+ -
+
Request a directory in SQUID. Write to chtc@cs.wisc.edu describing the data you'd like to place in SQUID, and indicating your username and submit server hostname (i.e. submit-5.chtc.wisc.edu).
+
+ -
+
Place files within your /squid/username directory via a CHTC
+transfer server (if from your laptop/desktop) or on the submit
+server.
+
+ From your laptop/desktop:
+ [username@computer]$ scp large_file.tar.gz username@transfer.chtc.wisc.edu:/squid/username/
+
+
+ If the file already exists within your /home directory on a submit
+server:
+
+ [username@submit]$ cp large_file.tar.gz /squid/username/
+
+
+ Check the file from the submit server:
+
+ [username@submit]$ ls /squid/username/
+
+
+ -
+
Have HTCondor download the file to the working job using the
+http://proxy.chtc.wisc.edu/SQUID address in the
+transfer_input_files line of your submit file:
+
+ transfer_input_files = other_file1,other_file2,http://proxy.chtc.wisc.edu/SQUID/username/large_file.txt
+
+
+ Important:Make sure to replace "username" with your username
+in the above address. All other files should be staged before job
+submission.
+
+If your large file is a .tar.gz file that untars to include other
+files, remember to remove such files before the end of the job;
+otherwise, HTCondor will think that such files are new output that
+needs to be transferred back to the submit server. (HTCondor will
+not automatically transfer back directories.)
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Small Input and Output File Availability Via HTCondor
+
+
+
Which Option is the Best for Your Files?
+
+
+
+ | Input Sizes |
+ Output Sizes |
+ Link to Guide |
+ File Location |
+ How to Transfer |
+ Availability, Security |
+
+
+
+
+ | 0 - 100 MB per file, up to 500 MB per job |
+ 0 - 5 GB per job |
+ Small Input/Output File Transfer via HTCondor |
+ /home |
+ submit file; filename in transfer_input_files |
+ CHTC, UW Grid, and OSG; works for your jobs |
+
+
+
+
+ | 100 MB - 1 GB per repeatedly-used file |
+ Not available for output |
+ Large Input File Availability Via Squid |
+ /squid |
+ submit file; http link in transfer_input_files |
+ CHTC, UW Grid, and OSG; files are made *publicly-readable* via an HTTP address |
+
+
+
+
+ | 100 MB - TBs per job-specific file; repeatedly-used files > 1GB |
+ 4 GB - TBs per job |
+ Large Input and Output File Availability Via Staging |
+ /staging |
+ job executable; copy or move within the job |
+ a portion of CHTC; accessible only to your jobs |
+
+
+
+
+
+
+
+
+
+
+
Table of Contents
+
+
+
+
+
+
+
HTCondor File Transfer
+
+
Due to the distributed configuration of the CHTC HTC pool, more often than not,
+your jobs will need to bring along a copy (i.e. transfer a copy) of
+data, code, packages, software, etc. from the submit server where the job
+is submitted to the execute node where the job will run. This requirement
+applies to any and all files that are needed to successfully execute and
+complete your job.
+
+
Any output that gets generated by your jobs is specifically written to
+the execute node on which the job ran. In order to get access to
+your output files, a copy of the output must be transferred back
+to an user accessible location like the submit server.
+
+
The mechanism that you use for file transfers will depend on the size
+of the individual input and output files of your jobs. This guide
+specifically describes input and output file transfer for input files
+<100MB in size (and <500MB of total input file transfer) and output
+files <4GB in size using the standard solution built into HTCondor
+job scheduling. More information about file transfer on a system
+without a shared filesystem is available in the
+HTCondor manual.
+
+
+
+
Applicability
+
+
+ -
+
Intended use:
+Good for delivering any type of data to jobs, but with file-size
+limitations (see below). Remember that you can/should split up a large
+input file into many smaller files for cases where each job only needs a
+portion of the data. By default, the submit file executable,
+output, error, and log files are ALWAYS transferred.
+
+ -
+
Advantages:
+HTCondor file transfer is robust and is available on ANY of CHTC's
+accessible HTC resources including the UW Grid of campus pools, and the
+OS Pool.
+
+ -
+
Data Security:
+Files transferred with HTCondor transfer are owned by the job and
+protected by user permissions in the CHTC pool. When signaling your jobs
+to run on the UW Grid (Flocking) or the OS Pool (Glidein),
+your files will exist on someone else's computer only for the duration
+of each job. Please feel free to email us if you have data security
+concerns regarding HTCondor file transfer, as encryption options are
+available.
+
+
+
+
+
+
+
+
To have HTCondor transfer small (<100MB) input files needed by
+your job, include the following attributes in your CHTC HTCondor submit files:
+
+
# my job submit file
+
+should_transfer_files = YES
+when_to_transfer_output = ON_EXIT
+transfer_input_files = file1, ../file2, /home/username/file3, dir1, dir2/
+
+... other submit file details ...
+
+
+
+
By default, the submit file executable, output, and
+error files are ALWAYS transferred.
+
+
Important Considerations
+
+
+ -
+
DO NOT use transfer_input_files for files within /staging;
+for files in /squid only http links (e.g. http://proxy.chtc.wisc.edu/SQUID/username/file) should be
+used instead of direct file paths. These policies are in place to prevent severe performance issues for your
+jobs and those of other users. Jobs should should never be submitted
+from within /squid or /staging.
+
+ -
+
HTCondor's file transfer can cause issues for submit server performance
+when too many jobs are transferring too much data at the same time.
+Therefore, HTCondor file transfer is only good for input files up to
+~20 MB per file IF the number of concurrently-queued jobs will be 100
+or greater. Even when individual files are small, there are issues when
+the total amount of input data per-job approaches 500 MB. For cases
+beyond these limitations, one of our other CHTC file delivery methods
+should be used. Remember that creating a tar.gz file of directories
+and files can give your input and output data a useful amount of
+compression.
+
+ -
+
Comma-separated files and directories to-be-transferred should be
+listed with a path relative to the submit directory, or can be
+listed with the absolute path(s), as shown above for file3. The
+submit file executable is automatically transferred and does not
+need to be listed in transfer_input_files.
+
+ -
+
All files that are transferred to a job will appear within the top
+of the working directory of the job, regardless of how they are
+arranged within directories on the submit server.
+
+ -
+
A whole directory and it's contents will be transferred when listed
+without the trailing forward slash ("/") after the directory name. When a directory is
+listed with the trailing forward slash ("/") after the directory name, only the directory
+contents will be transferred. Care should be taken when transferring whole directories
+so that only the files needed by your jobs will be transferred.
+Generally, we recommend creating a tar.gz file of directories
+and files to be used a job inputs - this will help streamline the process of input
+file transfer and help speed up transfer times by reducing the overall size of
+files that will be transferred.
+
+ -
+
Jobs will be placed on hold by HTCondor if any of the files or
+directories do not exist or if you have a typo.
+
+ -
+
Learn more about HTCondor input files transfer.
+
+
+
+
+
+
Transferring Output Files
+
+
All of your HTCondor submit files should have the following attributes:
+
+
# my job submit file
+
+should_transfer_files = YES
+when_to_transfer_output = ON_EXIT
+
+
+
+
when_to_transfer_output = ON_EXIT will instruct HTCondor to automatically transfer
+ALL new or modified files in the top level directory of the job (where it ran on the execute
+server), back to the job’s initial directory on the submit server. Please note: this behavior
+only applies to files in the job’s top-level working directory, meaning HTCondor will ignore
+any files created in subdirectories of the job’s main working directory. Several options exist for modifying
+this default output file transfer behavior - see below for some examples.
+
+
Only individual output files <4GB should be transferred back to your home directory
+using HTCondor’s default behavior described here. Large output files >4GB should instead
+use CHTC’s large data filesystem called staging, more information is available at
+Managing Large Data in HTC Jobs. To help reduce output file
+sizes, and help speed up file transfer times, we recommend creating a tar.gz file of all
+desired output before job completion (and to also delete the “un-tar'd”
+files so they are not also transferred back); see our example below.
+
+
+
+
Group Multiple Output Files For Convenience
+
+
If your jobs will generate multiple output files, we recommend combining all output into a compressed
+tar archive for convenience, particularly when transferring your results to your local computer from
+the submit server. To create a compressed tar archive, include commands in your your bash executable script
+to create a new subdirectory, move all of the output to this new subdirectory, and create a tar archive.
+For example:
+
+
#! /bin/bash
+
+# various commands needed to run your job
+
+# create output tar archive
+mkidr my_output
+mv my_job_output.csv my_job_output.svg my_output/
+tar -czf my_job.output.tar.gz my_ouput/
+
+
+
The example above will create a file called my_job.output.tar.gz that contains all the output that
+was moved to my_output. Be sure to create my_job.output.tar.gz in the top-level directory of where
+your job executes and HTCondor will automatically transfer this tar archive back to your /home
+directory.
+
+
+
+
Select Specific Output Files to Transfer to /home
+
+
As described above, HTCondor will transfer ALL new or modified files in the top level
+directory of the job (where it ran on the execute server), back to the job’s initial directory
+on the submit server. If your jobs will produce multiple output
+files but you only need to retain a subset of these output files, we recommend deleting the unrequired
+output files or moving them to a subdirectory as a step in the bash
+executable script of your job - only the output files that remain in the top-level
+directory will be transferred back to your /home directory. This will help keep ample
+space free and available on your /home directory on the submit server and help prevent
+you from exceeding the disk quota.
+
+
For jobs that use large input files from /staging, you must include steps in your bash script
+to either remove these files or move them to a subdirectory before the job terminates. Else,
+these large files will be transferred back to your /home directory. For more details, please
+see Managing Large Data in HTC Jobs.
+
+
In cases where a bash script is not used as the excutable of your job and you wish to have only specific
+output files transferred back, please contact us.
+
+
+
+
Get Additional Options For Managing Job Output
+
+
Several options exist for managing output file transfers back to your /home directory and we
+encourage you to get in touch with us at chtc@cs.wisc.edu to
+help identify the best solution for your needs.
+
+
Request a Quota Change
+
+
If you find that you are need of more space in you /home directory to handle the number
+of jobs that you want to run, please see our Request a Quota Change guide.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Account Request Form
+
+
+
Please fill out the below form in order to provide us with
+information about the computing work you would like
+to do (to the best of your knowledge). If you are unsure of an answer to a
+below question, leave the question blank or indicate that you don't know.
+
+
After filling out this form, CHTC Facilitation staff will follow up to
+create your account and offer times for an initial consultation. The consultation
+is only mandatory for new groups to CHTC, but we strongly encourage anyone who
+is getting started to take advantage of this valuable opportunity to discuss your
+work one-on-one with a CHTC Research Computing Facilitator.
+
+
For more information, see How to Request a CHTC Account.
+
+
Account Request Form
+
+
The following link leads to a Qualtrics form that we use for the account request process.
+
+
+
+
If you do not receive an automated email from chtc@cs.wisc.edu within a few hours of completing the form,
+ OR if you do not receive a response from a human within two business days (M-F), please email chtc@cs.wisc.edu.
+
+
+
+
+
+
+
+ Get Help
+
+
+
There are multiple ways to get help from CHTC staff. See below:
+
+
Research Computing Facilitators
+
+
+
+
+
+
+  + Andrew Owen
+ Andrew Owen
Research Computing Facilitator
+
+
+
+
+
+  + Christina Koch
+ Christina Koch
Research Facilitator Manager
+
+
+
+
+
+  + Rachel Lombardi
+ Rachel Lombardi
Research Computing Facilitator
+
+
+
+
+
+
To help researchers effectively utilize computing resources, our Research
+Computing Facilitators (RCFs) not only assist your in
+implementing your computational work on CHTC compute resources
+resources, but can also point you to other on- and off-campus services
+related to research computing and data needs. For example, RCFs can:
+
+
+ - Assist with planning your computational approach for a research
+problem
+ - Teach you to submit jobs to CHTC compute systems
+ - Help you with troubleshooting on CHTC compute systems
+ - Connect you with other researchers using similar software or methods
+ - Point to learning materials for programming and software development
+ - Help you identify applicable non-CHTC data storage options provided by DoIT
+ - Find the person who knows the answer to your question, even if the RCF doesn’t
+ - … and other helpful activities to facilitate your use of cyberinfrastructure
+
+
+
Get An Account
+
+
If you don’t have an account yet, please fill out our Request
+Form, and we’ll follow up quickly to set up a meeting time
+and create accounts. If you don’t have an account but just have general
+questions, feel free to send an email to chtc@cs.wisc.edu (see below).
+
+
Request a Quota Change
+
+
If you’d like to request a change in your quotas for one of our data
+storage locations, please see our Request a Quota Change guide.
+
+
Help Via Email
+
+
We provide support via email at the address
+chtc@cs.wisc.edu, and it’s never a bad idea
+to start by sending questions or issues via email. You can typically
+expect a first response within a few business hours.
+
+
When emailing us for assistance in troubleshooting an issue, please provide which system you are using,
+an explanation of what you expected to happen versus what actually happened, and
+include relevant files (or provide the locations of them on the system), such as:
+
+
+ - The job submit file (.sub)
+ - The job executable (.sh) or list of commands used in an interactive job
+ - Standard error and standard output files (usually .out or .err)
+ - If on the HTC system, the HTCondor log file (.log)
+
+
+
We will use this information to give you more effective responses and solutions.
+
+
Office Hours
+
+
+
+
+
+
For users who already have accounts, we have drop-in office hours, online, during the following times:
+
+
+ - Tuesday morning: 10:30 am - 12:00 pm. CANCELED OVER THE SUMMER (May 28 through August 27)
+ - Thursday afternoon: 3:00 - 4:30 pm.
+
+
+
To drop in, find the videoconference link in either your email or in the
+login message when you log into a CHTC server.
+
+
As always, if the times above don’t work for you, please email us
+at our usual support address to schedule a separate meeting.
+
+
Click to sign-in for office hours
+
+
+
+
Make an Appointment
+
+
We are happy to arrange meetings outside of designated Office Hours, per
+your preference. Simply email us at the address above, and we will set
+up a time to meet!
+
+
+
+
+
+
+
+ Getting Started With CHTC
+
+
+
+
Anyone on the UW-Madison campus, and even off-campus collaborators,
+may use the CHTC. In order to get started, we need some information
+to understand how best to help you.
+Please fill out the form here,
+being as thorough as you can.
+
+
+
+
+
+
+
+
+ Getting a Submit Node
+
+
+
In order to submit jobs to our campus-wide collection of resources, you
+will need access to a submit node. There are several options for getting
+access to a submit node:
+
+
+ - Use ours. We operate a submit node that
+is shared by many researchers. This is a great way to get started
+quickly, and it is sufficient if you do not need to run tens of
+thousands of jobs with heavy data transfer requirements.
+ - Use your department's. Perhaps your department already has its
+own submit node, in which case you can contact your local
+administrator for an account. You will still need to provide all the
+info requested on the getting started form, so
+we can set up things on our end. The benefits of using a
+departmental or group submit node are: access to data on local file
+systems; limited impact from other, potentially new users; and,
+greater scalability in the number of simultaneous jobs you can run,
+as well as the amount of data you can transfer.
+ -
+
Set up a new submit node on a server. If you do not already have
+one and need access to data on local file systems, or if you believe
+that you will have a significant job and/or data volume, getting
+your own submit node is probably the best way to go. Here's an
+example system configuration that we've found works well for a
+variety of submit work loads. You can expect to spend around
+$4,000 - $5,000 for such a system.
+
+ Typical submit node configuration
+
+
+ - A 1U rack-mount enclosure, like a Dell PowerEdge 410.
+ - Two processors with 12 cores total, for example Intel Xeon
+E5645, 2.4GHz 6-core processors
+ - 24GB of 1.3 GHz RAM
+ - Two drives for the operating system. 500GB each is enough. You
+can use mirroring or a RAID configuration like RAID-6 for
+reliability.
+ - Two or more 2-3TB drives for data, depending on your needs.
+
+
+ - Use your desktop. Depending on your department's level of
+system adminstration support, you may be able to have HTCondor
+installed on your desktop and configured to submit into our campus
+resources. Another option that is under development is
+Bosco, a
+user-installable software package that lets you submit jobs into
+resources managed by HTCondor, PBS or SGE.
+
+
+
Still not sure what option is right for you? No worries. This is one of
+the topics we discuss in our initial consultation. To schedule an
+initial consultation, fill out our getting started
+form.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Access a Private GitHub Repository Remotely
+
+
+
This guide describes how to remotely access a private GitHub repository from the HTC and HPC clusters, specifically
+
+ -
+
how to generate an SSH key pair
+
+ -
+
how to add a public SSH key to your GitHub account
+
+ -
+
how to remotely access your private GitHub repository
+
+
+
+
You will need to have access to a CHTC cluster. You will also need to have a GitHub account with access to the private repository of interest.
+
+
A. Generate the SSH Key Pair
+
+
We will be following the instructions provided by GitHub to generate the SSH key pair (Generating a new SSH key…).
+
+
+ -
+
Log in to the submit node as usual (Connecting to CHTC).
+
+ - Generate the SSH key by running the following command, where the example email is replaced with the email that you use for your GitHub account.
+
ssh-keygen -t ed25519 -C "your_email@example.com"
+
+
+ A message will appear stating that the key pair is being generated.
+
+ - A second message will appear prompting you to enter the location where the SSH keys should be stored:
+
Enter a file in which to save the key (/home/your_NetID/.ssh/ed25519):
+
+
+ Simply hit the enter key to accept the specified file path.
+
+ Note: If a SSH key already exists at the displayed path it will be overwritten by this action.
+This can be avoided by typing in an alternate path before pressing the enter key.
+
+
+ - You will be prompted to create a passphrase. Type your desired passphrase and then hit enter. Repeat a second time when asked to confirm your passphrase.
+
+ Warning: If you leave the passphrase empty (hit enter without typing anything), a passphrase will not be created nor required for using the SSH connection. In principle, this means anyone with access to the private key can access and modify your GitHub account remotely.
+
+
+ - A message will appear confirming the creation of the SSH key pair, as well as the paths and names of the private and public keys that were generated. Make note of these paths for use in the following steps.
+
+
+
B. Add the SSH Key to Your GitHub Account
+
+
Now we will be adding the SSH public key to your GitHub account, following the instructions provided by GitHub (Adding a new SSH key to your GitHub account).
+
+
+ -
+
Copy the contents of the public SSH key file (id_ed25519.pub) created in Part A. There are several ways of doing this.
+
+
+ If you provided an alternate file name in Step 3. of Part A., then the public SSH key will be the name of that file plus the .pub extension.
+
+
+
+
+ - Next, log in to github.com using the same email that you used in Step 2. of Part A.
+ - Go to your account settings by clicking on your profile icon in the top right corner of the webpage, then click on Settings within the drop-down menu. If your browser window is small, the Settings button can be found by clicking the menu button at the top left of the webpage.
+ - Go to the SSH and GPG keys section. Under the SSH keys section, click New SSH key.
+ - Paste the contents of the SSH public key from Step 1. into the Key textbox.
+ - Name the SSH key using the Title textbox. We recommend “CHTC” plus the name of the login node. For example: “CHTC ap2001”.
+ - Click Add SSH key. The SSH key will now appear in the SSH keys section in your GitHub account settings.
+
+
+
C. Accessing Your Private GitHub Repository from the Cluster
+
Once the SSH key has been added to your GitHub account, you can access your private repository using the repository’s SSH address.
+
+
+ - In your web browser and while logged in to your GitHub account, go to webpage for the private repository.
+ - Click the <>Code button, then select the Local tab and then the SSH tab.
+ - Copy the SSH address that is shown.
+ -
+
On the CHTC submit node, you can now access the repository using git commands by using the SSH address in place of the HTTPS address. For example,
+
+ git clone git@github.com:username/user-private-repository.git
+
+
+ - If prompted for a passphrase when running commands with the SSH address, provide the passphrase you created in Step 4. of Part A.
+
+
+
From an interactive job
+
+
Because the interactive job takes place on a different node than the submit node, it will not know about the SSH key that you set up above. Use the following instructions to transfer and use the private identity key in the interactive job (see Compiling or Testing Code with an Interactive Job for more information on interactive jobs).
+
+
+ -
+
When creating the submit file for your interactive job, include the path to the private SSH key identity file as a value for the transfer_input_files keyword. This will ensure that the identity file is copied to the interactive job directory. For example,
+
+ transfer_input_files = /home/your_NetID/.ssh/id_ed25519, /path/to/include/other/files
+
+
+
+ Note: Make sure that you are transferring the private SSH key file, not the public. The public SSH key should have the .pub extension, while the private SSH key does not.
+
+
+ - Once your submit file is set up, start the interactive job using
condor_submit -i and then the name of your submit file. When the interactive job has started, you will see that the private SSH key file is included in the initial directory. The SSH program, however, still needs to be told to use it.
+ -
+
Initialize an SSH agent using the command
+
+ eval "$(ssh-agent -s)"
+
+
+ -
+
Add the private SSH to the SSH agent by using the ssh-add command followed by the name of the private SSH key file that you transferred. You will be prompted to enter the passphrase that you created when you created the SSH key pair. For example,
+
+ ssh-add id_ed25519
+
+
+ You will now be able to access the repository during the interactive job.
+
+
+
+
Additional Notes
+
+
+ - If you forget the passphrase you created in Step 4. of Part A., you will need to repeat this guide to create a new SSH key pair to replace the previous one.
+ -
+
When using the SSH address to your repository with non-git commands, you may need to replace the colon (:) in the address with a forward slash (/). For example,
+
+ Original SSH address
+ git@github.com:username/user-private-repository.git
+
+
+ Modified SSH address
+ git@github.com/username/user-private-repository.git
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Use Globus to Transfer Files to and from CHTC
+
+
+
Globus is a data management service that lets you
+move files files between endpoints, computers that are connected to the
+Globus file transfer network.
+Globus is primarily useful when you need to move large amounts of data to or
+from somewhere that is already providing a Globus endpoint.
+For example, a collaborator might provide shared data through Globus,
+or might expect you to send data to them through Globus.
+
+
This guide will show you how to execute such transfers to and from CHTC using the CHTC
+Globus endpoint, which may be simpler than trying to move the files to your
+own computer first.
+
+
Prerequisites
+
+
All file transfer via Globus at CHTC requires:
+
+
+ - access to a directory in the
/staging or /projects folders
+ - login access to the
transfer.chtc.wisc.edu server.
+
+
+
Contact us at chtc@cs.wisc.edu if you need either of the above.
+
+
You will also need to be able to
+log in to the Globus web interface;
+you can use your UW-Madison NetID (if you have one, or similar) by selecting
+University of Wisconsin-Madison from the drop down and pressing “Continue”.
+
+
Using the CHTC Globus Endpoints
+
+
You can use the Globus web interface to transfer files to and from CHTC.
+In the web interface, you can select two endpoints and then initiate a transfer
+between them.
+
+
The first step is to find the CHTC Globus endpoints. They can be found in the Globus web interface
+by searching endpoints for “CHTC Staging” or “CHTC Projects”.
+
+

+
+
Or can be found at these links:
+
+
+
+
If you need the actual endpoint UUID, it is listed on the above pages near the bottom
+of the “Overview”.
+
+
To use an endpoint, you must first activate it.
+Activations are usually time-limited, and transfers can only proceed while
+both the source and destination endpoints are activated.
+Activating an endpoint generally requires logging in.
+You should log in using your UW - Madison NetID.
+You can see how long your activation will last on the endpoint information page
+in the Globus web interface.
+
+
To begin a file transfer, go to the
+File Manager.
+In the top-right corner of the page, make sure you are in the “two panel” view.
+Select the two endpoints you want to transfer between
+(they are called “Collections” on this page).
+You should see a directory listing appear in the middle of each of the panes;
+select a directory or file and click “Start” at the bottom of the page to
+move that directory or file to the other endpoint.
+The item will be moved to the currently-selected directory on the other endpoint.
+
+
Globus transfers are asynchronous, and you do not need to leave the web
+interface open while they run.
+You will receive emails updates on the progress of the transfer, and you can
+view the status of in-progress and historical transfers
+on the Activity page.
+
+
You may find some of the “transfer settings”, available by clicking the
+“Transfer & Sync Options” dropdown, useful.
+In particular, sync will help reduce the amount of time it takes to transfer
+when some data has already been transferred.
+
+
Running a Personal Globus Endpoint
+
+
The CHTC Globus endpoint is a “Globus Connect Server”, designed for shared use
+on a dedicated machine.
+It is also possible to run
+Globus Connect Personal,
+a lighter-weight package that adds a Globus endpoint to your own computer,
+like a laptop or lab computer.
+Installers are available at that link for Mac, Linux, and Windows.
+
+
We only recommend using Globus Connect Personal if you are also working with
+some other Globus endpoint (not just CHTC and your computer).
+If you are just moving files between CHTC and your own computer, traditional
+file transfer tools like rsync will likely be more efficient.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Use GPUs
+
+
+
Overview
+
+
GPUs (Graphical Processing Units) are a special kind of computer
+processor that are optimized for running very large numbers of simple
+calculations in parallel, which often can be applied to problems related
+to image processing or machine learning. Well-crafted GPU programs for
+suitable applications can outperform implementations running on CPUs by
+a factor of ten or more, but only when the program is written and
+designed explicitly to run on GPUs using special libraries like CUDA.
+For researchers who have problems that are well-suited to GPU
+processing, it is possible to run jobs that use GPUs in CHTC. Read on to
+determine:
+
+
+
+
A. Available CHTC GPUs
+
+
1. GPU Lab
+
+
CHTC has a set of GPUs that are available for use by any CHTC user with an
+account on our high throughput computing (HTC) system
+via the CHTC GPU Lab, which includes templates and a campus GPU community.
+
+
Our expectation is that most, if not all, of CHTC users running GPU jobs should utilize
+the capacity of the GPU Lab to run their work.
+
+
+
+ | Number of Servers |
+ Names |
+ GPUs / Server |
+ GPU Type (DeviceName) |
+ Hardware Generation Capability |
+ GPU Memory GlobalMemoryMB |
+
+
+
+ | 2 |
+ gpu2000, gpu2001 |
+ 2 |
+ Tesla P100-PCIE-16GB |
+ 6.0 |
+ 16GB |
+
+
+ | 4 |
+ gpulab2000 - gpulab2003 |
+ 8 |
+ GeForce RTX 2080 Ti |
+ 7.5 |
+ 10GB |
+
+
+ | 2 |
+ gpulab2004, gpulab2005 |
+ 4 |
+ A100-SXM4-40GB |
+ 8.0 |
+ 40GB |
+
+
+ | 10 |
+ gpu2002 - gpu2011 |
+ 4 |
+ A100-SXM4-80GB |
+ 8.0 |
+ 80GB |
+
+
+ | 3 |
+ gpu4000 - gpu4002 |
+ 10 |
+ L40 |
+ 8.9 |
+ 45GB |
+
+
+
+
Special GPU Lab Policies
+
+
Jobs running on GPU Lab servers have time limits and job number limits
+(differing from CHTC defaults across the rest of the HTC System).
+
+
+
+
+ | Job type |
+ Maximum runtime |
+ Per-user limitation |
+
+
+
+
+ | Short |
+ 12 hrs |
+ 2/3 of CHTC GPU Lab GPUs |
+
+
+ | Medium |
+ 24 hrs |
+ 1/3 of CHTC GPU Lab GPUs |
+
+
+ | Long |
+ 7 days |
+ up to 4 GPUs in use |
+
+
+
+
+
There are a certain number of slots in the GPU Lab reserved for interactive use. Interactive
+jobs that use GPU Lab servers are restricted to using a single GPU and a 4 hour runtime.
+
+
2. Other Capacity
+
+
There is additional dedicated and backfill GPU capacity available in CHTC and beyond;
+see GPU capacity beyond the GPU Lab for details.
+
+
B. Submit Jobs Using GPUs in CHTC
+
+
+
+
The following options are needed in your HTCondor submit file in order
+to access the GPUs in the CHTC GPU Lab and beyond:
+
+
+ - Request GPUs (required): All jobs that use GPUs must request GPUs in their submit file (along
+with the usual requests for CPUs, memory, and disk).
+
request_gpus = 1
+
+
+ - Request the CHTC GPU Lab: To use CHTC’s shared use GPUs, you need to opt-in to the GPU Lab. To
+do so, add the
+following line to your submit file:
+
+WantGPULab = true
+
+
+ - Indicate Job Type: We have categorized three “types”
+of GPU jobs, characterized in the table above. Indicate which job type you would
+like to submit by using the submit file option below.
+
+GPUJobLength = "short"
+# Can also request "medium" or "long"
+
+ If you do not specify a job type, the medium job type will be used as the default. If
+ your jobs will run in less than 12 hours, it is advantageous to indicate that they are
+ “short” jobs because you will be able to have more jobs running at once.
+
+ - Request Specific GPUs or CUDA Functionality Using
require_gpus (optional): If your software or code requires a certain
+type of GPU, or has some other special requirement, there is a special submit file line
+to request these capabilities, require_gpus. For example, if you want a certain
+class of GPU, represented by
+the attribute Capability, your require_gpus statement would look like this:
+ require_gpus = (Capability > 7.5)
+
+
+ You can see a table of the different attributes that HTCondor tracks
+ about the GPU nodes, and how to explore their values, in the section
+ on Using condor_status to explore GPUs.
+
+ It may be tempting to add requirements for specific GPU servers or
+ types of GPU cards. However, when possible, it is best to write your
+ code so that it can run across GPU types and without needing the
+ latest version of CUDA.
+
+ - Specify Multiple GPU Requirements (optional): Multiple requirements can be specified by using && statements:
+
require_gpus = (Capability >= 7.5) && (GlobalMemoryMb >= 11000)
+
+ Ensure all specified requirements match the attributes of the GPU/Server of interest. HTCondor matches jobs to GPUs that match all specified requirements. Otherwise, the jobs will sit idle indefinitely.
+
+
+ We are testing a new set of submit commands for specifying the requirements of the GPU:
+
+ gpus_minimum_capability = <version>
+gpus_maximum_capability = <version>
+gpus_minimum_memory = <quantity in MB>
+
+
+ More information on these commands can be found in the HTCondor manual.
+
+
+ -
+
Indicate Software or Data Requirements Using requirements: If your data is large enough to
+ use our /staging data system (see more information here),
+ or you are using modules or other software in our shared /software system, include
+ the needed requirements.
+
+ - Indicate Shorter/Resumable Jobs: if your jobs are shorter than 4-6 hours, or have
+ the ability to checkpoint at least that frequently, we highly recommend taking
+ advantage of the additional GPU servers in CHTC that can run these kind of jobs
+ as backfill! Simply add the following option to your submit file:
+
+is_resumable = true
+
+
+ For more information about the servers that you can run on with this option,
+ and what it means to run your jobs as “backfill” see
+ the section below on Accessing Research Group GPUs.
+
+
+
+
2. Sample Submit File
+
+
A sample submit file is shown below. There are also example submit files and
+job scripts in this GPU Job Templates repository
+in CHTC’s Github organization.
+
+
# gpu-lab.sub
+# sample submit file for GPU Lab jobs
+
+universe = vanilla
+log = job_$(Cluster)_$(Process).log
+error = job_$(Cluster)_$(Process).err
+output = job_$(Cluster)_$(Process).out
+
+# Fill in with whatever executable you're using
+executable = run_gpu_job.sh
+#arguments =
+
+should_transfer_files = YES
+when_to_transfer_output = ON_EXIT
+# Uncomment and add input files that are in /home
+# transfer_input_files =
+
+# Uncomment and add custom requirements
+# requirements =
+
++WantGPULab = true
++GPUJobLength = "short"
+
+request_gpus = 1
+request_cpus = 1
+request_memory = 1GB
+request_disk = 1GB
+
+queue 1
+
+
+
+
3. Notes
+
+
It is important to still request at least one CPU per job to do the
+processing that is not well-suited to the GPU.
+
+
Note that HTCondor will make sure your job has access to the GPU; it will
+set the environment variable CUDA_VISIBLE_DEVICES to indicate which GPU(s)
+your code should run on. The environment variable will be read by CUDA to select the appropriate
+GPU(s). Your code should not modify this environment variable or manually
+select which GPU to run on, as this could result in two jobs sharing a GPU.
+
+
It is possible to request multiple GPUs. Before doing so, make sure you’re
+using code that can utilize multiple GPUs and then submit a test job to confirm
+success before submitting a bigger job. Also keep track of how long jobs
+are running versus waiting; the time you save by using multiple GPUs may be
+not worth the extra time that the job will likely wait in the queue.
+
+
C. GPU Capacity Beyond the CHTC GPU Lab
+
+
The following resources are additional CHTC-accessible servers with GPUs. They do not have the
+special time limit policies or job limits of the GPU Lab. However, some of them are
+owned or prioritized by specific groups. The implications of this
+on job runtimes is noted in each section.
+
+
Note that all GPU jobs need to include the request_gpus option in their submit file,
+even if they are not using the GPU Lab.
+
+
1. Access Research Group GPUs
+
+
Certain GPU servers in CHTC are prioritized for the
+research groups that own them, but are available to run other jobs when
+not being used by their owners. When running on these servers, jobs
+forfeit our otherwise guaranteed runtime of 72 hours, and have the potential to be interrupted. However, for
+shorter jobs or jobs that have implemented self-checkpointing, this is not a drawback and allowing jobs to run on these
+additional servers opens up more capacity.
+
+
Therefore, these servers are a good fit for GPU jobs that run in a few hours
+or less, or have implemented self-checkpointing (the capability to save progress
+to a file and restart from that progress). Use the is_resumable option shown
+above in the list of submit file options.
+
+
2. Use the gzk Servers
+
+
These are servers that are similar to the GPU Lab severs with two important differences
+for running GPU jobs:
+
+ - they do not have access to CHTC’s large data
/staging file system
+ - they do not have Docker capability
+
+
+
You do not need to do anything specific to allow jobs to run on these servers.
+
+
3. Using GPUs in CHTC’s OSG Pool and the UW Grid
+
+
CHTC, as a member of the OSG Consortium can access GPUs that
+are available on the OS Pool. CHTC is
+also a member of a campus computing network called the UW Grid, where groups on campus
+share computing capacity, including access to idle GPUs.
+
+
See this guide to know
+whether your jobs are good candidates for the UW Grid or OS Pool and then get in touch
+with CHTC’s Research Computing Facilitators to discuss details.
+
+
D. Using condor_status to explore CHTC GPUs
+
+
You can find out information about GPUs in CHTC through the
+condor_status command. All of our servers with GPUs have a TotalGPUs
+attribute that is greater than zero; thus we can query the pool to find
+GPU-enabled servers by running:
+
+
[alice@submit]$ condor_status -compact -constraint 'TotalGpus > 0'
+
+
+
To print out specific information about a GPU server and its GPUs, you
+can use the “auto-format” option for condor_status and the names of
+specific server attributes. In general, when querying attributes using
+condor_status, a “GPUs_” prefix needs to be added to the attribute name.
+For example, the tables at the top of the guide can be mostly
+recreated using the attributes Machine, TotalGpus,
+GPUs_DeviceName and GPUs_Capability:
+
+
[alice@submit]$ condor_status -constraint 'Gpus > 0' \
+ -af Machine TotalGpus GPUs_DeviceName GPUs_Capability
+
+
+
In addition, HTCondor tracks other GPU-related attributes for each
+server, including:
+
+
+
+ | Attribute |
+ Explanation |
+
+
+ Gpus |
+ Number of GPUs in an individual job slot on a server (one server can be divided into slots to run multiple jobs). |
+
+
+ TotalGPUs |
+ The total number of GPUs on a server. |
+
+
+ (GPUs_)DeviceName |
+ The type of GPU card. |
+
+
+ (GPUs_)Capability |
+ Represents various capabilities of the GPU. Can be used as a proxy for the GPU card type when
+ requiring a specific type of GPU. Wikipedia
+ has a table showing the compute capability for specific GPU architectures and cards.
+ More details on what the capability numbers mean can be found on the
+
+ NVIDIA website. |
+
+
+ (GPUs_)DriverVersion |
+ Not the version of CUDA on the server or the NVIDIA driver version, but the maximum CUDA runtime version supported by the NVIDIA driver on the server. |
+
+
+ (GPUs_)GlobalMemoryMb |
+ Amount of memory available on the GPU card. |
+
+
+
+
E. Prepare Software Using GPUs
+
+
Before using GPUs in CHTC you should ensure that the use of GPUs will
+actually help your program run faster. This means that the code or
+software you are using has the special programming required to use GPUs
+and that your particular task will use this capability.
+
+
If this is the case, there are several ways to run GPU-enabled software
+in CHTC:
+
+
+ Machine Learning
+ For those using machine learning code specifically, we have a guide
+with more specific recommendations here: Run Machine Learning Jobs on
+HTC
+
+
+
1. Compiled Code
+
+
You can use our conventional methods of creating a portable installation
+of a software package (as in our R/Python guides) to run on GPUs. Most
+of our build servers or GPU servers have copies of the CUDA Runtime that
+can be used to compile code. To access these servers, submit an
+interactive job, following the instructions in our Build Job
+Guide or by submitting a GPU job submit file with the
+interactive flag for condor_submit. Once on a build or GPU server, see
+what CUDA versions are available by looking at the path
+/user/local/cuda-*.
+
+
Note that we strongly recommend software installation strategies that
+incorporate the CUDA runtime into the final installed code, so that jobs
+are able to run on servers even if a different version of the CUDA
+runtime is installed (or there’s no runtime at all!). For compiled code,
+look for flags that enable static linking or use one of the solutions
+listed below.
+
+
2. Docker
+
+
CHTC’s GPU servers have “nvidia-docker” installed, a specific version of
+Docker that integrates Docker containers with GPUs. If you can find or
+create a Docker image with your software that is based on the
+nvidia-docker container, you can use this to run your jobs in CHTC. See
+our Docker guide for how to use Docker in CHTC.
+
+
Currently we recommend using
+“nvidia/cuda” containers with a tag beginning with “12.1.1-devel”
+for best integration with our system.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Welcome to the CHTC GPU Lab
+
+
+
+
+
+
+
+
+
The CHTC GPU Lab was created by a UW2020-funded project to expand shared GPU
+computing infrastructure at UW-Madison. It includes:
+
+
+
+
+
+
+
+
+

+
+
Hardware
+
A pool of shared GPU servers managed by CHTC.
+
+
+
+
+
+
+
+

+
+
Expertise
+
A community of technical experts.
+
+
+
+
+
+
+
+

+
+
Documentation
+
A library of sharable software and documentation.
+
+
+
+
+
+
+
+
+
+
Get Involved
+
+
If you want to use GPU resources in CHTC for your research:
+
+
+
+
The CHTC GPU Lab mailing list is used to announce new GPU hardware availability and
+GPU-related events, solicit feedback from GPU users, and share best practices for
+GPU computing in CHTC. Any CHTC user can subscribe to the list by
+emailing chtc-gpu-lab+managers@g-groups.wisc.edu
+and asking to join.
+Their subscription request will be reviewed by the list administrators.
+
+
+ The CHTC GPU Lab is led by Anthony Gitter, Christina Koch, Brian Bockelman, and Miron Livny.
+
+
+
+ The original UW2020 project was led by Anthony Gitter, Lauren Michael, Brian Bockelman, and Miron Livny and
+funded by the Office of the Vice Chancellor for Research and Graduate
+Education and the Wisconsin Alumni Research Foundation.
+
+
+
For more information about the CHTC GPU Lab project contact Anthony Gitter.
+
+
+
+
+
+
+
+ Computing Guides
+
+
+
+Below is a list of guides for some of the most common tasks our users need to
+carry out as they begin and continue to use the resources at the CHTC.
+Some of these are general computing solutions; others are specific to HTCondor
+or to the configuration of CHTC computing resources.
+
+
+Guides will be added to the list as we can provide them. Please contact us
+(email at bottom of page) if you find any of the information to be incorrect.
+
+
+
+
+
User Expectations
+
+Read through these user expectations and policies before using CHTC services.
+
+
+
+
+
+
+
HTC Documentation
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
HPC Documentation
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
External Documentation
+
+
+
+
+
Icon Credits
+
+
+
+
+
+
+

+
+
+
+
+
+
+
+
+ Submit High Memory Jobs
+
+
+
The examples and information in the below guide areuseful ONLY if:
+
+ - you already have an account on a CHTC-administered submit server
+ - your jobs will use more than ~120 GB of memory
+
+
+
To best understand the below information, users should already befamiliar with:
+
+ - Using the command-line to: navigate directories,
+create/edit/copy/move/delete files and directories, and run intended
+programs (aka "executables").
+ - CHTC's Intro to Running HTCondor Jobs
+ - CHTC's guides for handling large data (Guide
+here) and software installation.
+
+
+
Overview
+
+
A high-memory job is one that requires a significantly larger amount of
+memory (also known as RAM) than a typical high throughput job usually
+over 200 GB and up to 1-4 TB. In the following guide, we cover resources
+and recommendations for running high-memory work in CHTC. However,
+please make sure to email us if you believe you will need to run
+"high-memory" work for the first time, or are planning the execution
+of new "high-memory" work that is different from what you've run
+before. We'll happily help you with some personalized tips and
+considerations for getting your work done most efficiently.
+
+
+ - High Memory Resources in CHTC
+ - Getting Started
+ - Running High Memory Jobs
+
+
+
+
+
1. High Memory Resources in CHTC
+
+
Our high memory servers have the following specs:
+
+
+
+
+ | Number of servers |
+ Memory per server |
+ CPUs per server |
+ Local disk space on server |
+ Names |
+
+
+
+
+ | 16 |
+ 512 GB |
+ 40 |
+ 1.2 TB |
+ e2003-e2018 |
+
+
+ | 2 |
+ 2 TB |
+ 80 |
+ 3.5+ TB |
+ mem2001, mem2002 |
+
+
+
+
+
+
+
2. Getting Started
+
+
+
+
A. Identifying High Memory Jobs
+
+
Jobs that request over 200GB of memory in their submit file
+can run on our dedicated high memory machines. However, if your job
+doesn't need quite that much memory, it's good to request less, as
+doing so will allow your job(s) to run on more servers, since CHTC has
+hundreds of servers with up to 100 GB of memory and dozens of servers
+with up to 250 GB of memory.
+
+
+
+
B. Testing
+
+
Before running a full-size high-memory job, make sure to use a small
+subset of data in a test job. Not only will this give you a chance to
+try out the submit file syntax and make sure your job runs, but it can
+help you estimate how much memory and/or disk you will need for a job
+using your full data.
+
+
You can also use interactive jobs to test commands that will end up in
+your "executable" script. To run an interactive job, prepare your
+submit file as usual. Note that for an interactive job, you should use a
+smaller memory request (and possibly lower CPU and disk as well) than
+for the final job (so that the interactive job starts) and plan to
+simply test commands, not run the entire program. To submit interactive
+job, use the -i flag with condor_submit:
+
+
[alice@submit]$ condor_submit -i submit.file
+
+
+
After waiting for the interactive job to start, this should open a bash
+session on an execute machine, which will allow you to test your
+commands interactively. Once your testing is done, make the appropriate
+changes to your executable, adjust your resource requests, and submit
+the job normally.
+
+
+
+
C. Consult with Facilitators
+
+
If you are unsure how to run high-memory jobs on CHTC, or if you're not
+sure if everything in this guide applies to you, get in touch with a
+research computing facilitator by emailing chtc@cs.wisc.edu.
+
+
+
+
3. Running High Memory Job
+
+
+
+
A. Submit File
+
+
The submit file shown in our Hello World example is
+a good starting point for building your high memory job submit file. The
+following are places where it's important to customize:
+
+
+ -
+
request_memory: It is crucial to make this request as accurate
+as you can by testing at a small scale if possible (see
+above). Online documentation/help pages or your colleagues'
+experience is another source of information about required memory.
+
+ - Long running jobs: If your high memory job is likely to run
+longer than our 3-day time limit, please email us for options on how
+to run for longer. In the past, high memory jobs received an extra
+time allowance automatically but this is no longer the case.
+ -
+
request_cpus: Sometimes, programs that use a large amount of
+memory can also take advantage of multiple CPUs. If this is the case
+for your program, you can request multiple CPUs. However, it is
+always easier to start jobs that request fewer number of cores,
+rather than more. We recommend:
+
+
+
+
+ | Requesting ___ of memory? |
+ Request fewer than ___ CPUs |
+
+
+
+
+ | up to 100 GB |
+ 4 |
+
+
+ | 100-500 GB |
+ 8 |
+
+
+ | 500GB-1TB |
+ 16 |
+
+
+ | 1-1.5TB |
+ 20 |
+
+
+ | 1.5-2TB |
+ 20 |
+
+
+ | 2TB or greater |
+ 32 |
+
+
+
+
+ If you think a higher CPU request would significantly improve your
+job's performance, contact a facilitator.
+
+ -
+
request_disk: Request the maximum amount of data your job will
+ever have within the job working directory on the execute node,
+including all output and input (which will take up space before some
+of it is removed from the job working directory at the end of the
+job).
+
+ - Other requirements: if your job uses files from our large data
+space, or Docker for
+software, add the necessary requirements for
+these resources to your submit file.
+
+
+
Altogether, a sample submit file may look something like this:
+
+
### Example submit file for a single staging-dependent job
+
+universe = vanilla
+
+# Files for the below lines will all be somewhere within /home/username,
+# and not within /staging/username
+log = run_myprogram.log
+executable = run_Trinity.sh
+output = $(Cluster).out
+error = $(Cluster).err
+transfer_input_files = trinityrnaseq-2.0.1.tar.gz
+should_transfer_files = YES
+
+# Require execute servers that have large data staging
+Requirements = (Target.HasCHTCStaging == true)
+
+# Memory, disk and CPU requests
+request_memory = 200GB
+request_disk = 100GB
+request_cpus = 4
+
+# Submit 1 job
+queue 1
+### END
+
+
+
+
+
B. Software
+
+
Like any other job, the best option for high memory work is to create a
+portable installation of your software. We have guides for scripting
+languages and using
+Docker, and can otherwise provide individual
+support for program installation during office hours or over
+email.
+
+
+
+
C. "Executable" script
+
+
As described in many of our guides (for
+software or for using large
+data), you will need to write a script
+that will run your software commands for you and that will serve as the
+submit file "executable". Things to note are:
+
+
+ - If using files from our large data staging space, follow the
+recommendations in our guide.
+ - If using multiple cores, make sure that you request the same number
+of "threads" or "processes" in your command as you requested in
+your submit file.
+
+
+
Altogether, a sample script may look something like this (perhaps called
+run_Trinity.sh):
+
+
#!/bin/bash
+# Copy input data from /staging to the present directory of the job
+# and un-tar/un-zip them.
+cp /staging/username/reads.tar.gz ./
+tar -xzvf reads.tar.gz
+rm reads.tar.gz
+
+# Set up the software installation in the job working directory, and
+# add it to the job's PATH
+tar -xzvf trinityrnaseq-2.0.6-installed.tar.gz
+rm trinityrnaseq-2.0.6-installed.tar.gz
+export PATH=$(pwd)/trinityrnaseq-2.0.6:$PATH
+
+# Run software command, referencing input files in the working directory and
+# redirecting "stdout" to a file. Backslashes are line continuation.
+Trinity --seqType fq --left reads_1.fq \
+--right reads_2.fq --CPU 4 --max_memory \
+20G > trinity_stdout.txt
+
+# Trinity will write output to the working directory by default,
+# so when the job finishes, it needs to be moved back to /staging
+tar -czvf trinity_out_dir.tar.gz trinity_out_dir
+cp trinity_out_dir.tar.gz trinity_stdout.txt /staging/username/
+rm reads_*.fq trinity_out_dir.tar.gz trinity_stdout.txt
+
+### END
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ CHTC Tools for Matlab, R, and Python Portability
+
+
+
This page is deprecated. Please see our guide Overview: How to Use
+Software instead.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ HPC System Transition to a New Linux Version (CentOS Stream 9)
+
+
+
Starting in May 2024, CHTC’s high performance computing (HPC) cluster began upgrading
+the Linux distribution and version we use on our servers to CentOS Stream 9 (EL9). This transition is expected to complete in June 2024.
+
+
Note that this page only applies to a transition on the HPC cluster. For information
+on the HTC system transition, see HTC System Transition to a New Linux Version (CentOS Stream 9)
+
+
All updates to the HPC Cluster will be reflected on this page; significant changes may
+also include a notification to the chtc-users mailing list.
+
+
Important Dates
+
+
+ - May 31: Log in to upgraded cluster login node is available. Worker nodes start transitioning from the existing cluster to upgraded cluster partitions.
+ - May 31 - June 17: Users should rebuild their code and test jobs on the upgraded cluster. Users should be running primarily on the upgraded cluster.
+ - June 17: Most nodes will have been upgraded and transitioned.
+ - June 24: The old cluster partitions are closed.
+
+
+
What is Changing
+
+
As part of this transition, there will be a new login node for
+the HPC cluster: spark-login.chtc.wisc.edu.
+
+
If you log into spark-login, you will have access to a new
+module stack, compiled on CentOS Stream 9, and the partitions available will
+have worker nodes that are running CentOS Stream 9.
+
+
The files in your /home and /scratch directories will be unchanged.
+
+
What You Need to Do
+
+
As soon as possible, do the following:
+
+
+ -
+
Log into the new login node spark-login.chtc.wisc.edu.
+
+ -
+
Run a typical job as a test. It is highly likely that certain codes will
+fail on the new worker nodes, as the underlying dependencies of your code, including
+the operating system, and any modules used, have changed.
+
+ -
+
If your jobs no longer run, archive your previous software installation(s) and
+rebuild your software. The Software Changes section below has
+more specific information about how to do this.
+
+ -
+
If you recompiled your code, run a few small test jobs to confirm that the
+code is working correctly.
+
+
+
+
If you are having trouble getting your jobs to run successfully on the new operating system,
+please contact the facilitation team at chtc@cs.wisc.edu or come to office hours
+
+
Software Changes
+
+
Almost all packages and libraries have been upgraded as part of the operating system transition.
+Unless your code is fairly simple, you will likely need to recompile it.
+
+
Remember to always compile your code/programs in a (interactive) Slurm job! How To
+
+
+ Not only does this help avoid stressing the resources of the login server, but the upgraded login server uses a newer CPU architecture than the worker nodes in the cluster.
+Most compilers auto-detect the CPU architecture and adapt the compilation to use that architecture.
+Attempting to use such compiled code on a different/older CPU architecture can lead to “Illegal instruction” errors, among others.
+
+
+
Modules
+
+
Most of the modules on the upgraded cluster have been kept, but with upgraded versions.
+The following table is a comparison of the modules on the old operating system (EL8) versus the new operating system (EL9).
+(Adapted from the output of module avail on the respective servers.)
+
+
You will likely need to recompile your code to use the new module versions.
+Remember to also update any module load commands that specify a particular version of the module,
+otherwise you may encounter “module(s) are unknown” errors.
+
+
Module comparison
+
+
+
+
+ | Module name |
+ Old version (EL8) |
+ New version (EL9) |
+
+
+
+
+ | abaqus |
+ 2018-hotfix-1904 |
+ TBD |
+
+
+ | ansys |
+ 2022r24 |
+ 2024r1 |
+
+
+ | aocc |
+ 3.2.0 |
+ 4.2.0 |
+
+
+ | cmake |
+ 3.27.7 |
+ 3.27.9 |
+
+
+ | comsol |
+ 6.0, 6.1, 6.2 |
+ 6.2 |
+
+
+ | gcc |
+ 11.3.0 |
+ 13.2.0 |
+
+
+ | hdf5 (intel-oneapi-mpi) |
+ 1.12.2 |
+ dropped |
+
+
+ | hdf5 (openmpi) |
+ 1.12.2 |
+ 1.14.3 |
+
+
+ | intel-oneapi-compilers |
+ 2023.2.1 |
+ 2024.1.0 |
+
+
+ | intel-oneapi-mkl |
+ 2023.2.0 |
+ 2024.0.0 |
+
+
+ | intel-oneapi-mpi |
+ 2021.10.0 |
+ 2021.12.1 |
+
+
+ | intel-tbb |
+ 2021.9.0 |
+ deprecated |
+
+
+ | lmstat.comsol |
+ 6.0 |
+ TBD |
+
+
+ | lumerical-fdtd |
+ 2022-r2.4 |
+ 2024-R1.2 |
+
+
+ | matlab |
+ R2021b, R2022b |
+ R2024a |
+
+
+ | mvapich2 |
+ 2.3.7-1 |
+ deprecated |
+
+
+ | mvapich |
+ n/a |
+ 3.0 |
+
+
+ | netcdf-c |
+ 4.8.1 |
+ 4.9.2 |
+
+
+ | netcdf-cxx4 |
+ 4.3.1 |
+ 4.3.1 |
+
+
+ | netcdf-fortran |
+ 4.5.4 |
+ 4.6.1 |
+
+
+ | openmpi (aocc) |
+ 4.1.3 |
+ dropped |
+
+
+ | openmpi (gcc) |
+ 4.1.3 |
+ 5.0.3 |
+
+
+ | patchelf (gcc) |
+ 0.17.2 |
+ 0.17.2 |
+
+
+ | patchelf (intel) |
+ 0.18.0 |
+ dropped |
+
+
+ | patchelf (oneapi) |
+ 0.18.0 |
+ 0.17.2 |
+
+
+ | petsc |
+ 3.18.1 |
+ 3.21.1 |
+
+
+ | pmix |
+ n/a |
+ 5.0.1 |
+
+
+
+
+
+ Different versions of module packages, or packages that are “dropped” or “deprecated” may be manually installed by the user using Spack.
+
+
+
Spack
+
+
Spack is a package manager platform that allows users to install software without admin privileges.
+CHTC also uses Spack to install the software underlying the system-wide modules discussed above.
+
+
+ If you have not used Spack before, you can skip this section and go directly to the Set Up Spack on HPC guide.
+
+
+
Here is the general process for setting up your software on the upgraded EL9 system; detailed instructions are provided after the general process:
+
+
+ -
+
Identify the environments you currently have and which you want to reproduce on the upgraded system.
+
+ -
+
Remove your existing Spack folders.
+
+ -
+
Do a clean installation of Spack.
+
+ -
+
In an interactive job, create your Spack environment(s) and install the packages as you did previously.
+
+ -
+
Update your job submission scripts and/or recompile programs as needed to use the new Spack environment(s).
+
+
+
+
The following instructions assume that you previously installed Spack in your home (~/) directory for individual use.
+
+
1. Identify your environments
+
+
You can see your Spack environments with
+
+
spack env list
+
+
+
Activate an environment that you want to replicate with
+
+
spack env activate environment_name
+
+
+
Then list your package “specs” with the command
+
+
spack find
+
+
+
There is a section “==> Root specs” that lists the package specs you explicity added when you created your environment.
+Save a copy of these specs somewhere safe, so that you can use them to replicate the environment later on.
+You can ignore the “installed packages” section, as that will certainly change on the new system.
+
+
Repeat the above steps for each environment you want to replicate on the upgraded system.
+
+
2. Remove your existing Spack folders
+
+
The easiest way to update Spack for the upgraded system is to remove the current Spack installation and reinstall from scratch.
+
+
+ Before proceeding, you may want to make a backup of each folder using
+
+ tar -czf folder_name.tar.gz ~/folder_name
+
+
+
+
For most users, the following commands should work:
+
+
cd ~/
+rm -rf spack spack_programs spack_modules .spack
+
+
+
The command may take a while to run.
+
+
3. Fresh install of Spack
+
+
Next, follow the instructions in our guide Set Up Spack on HPC to do a fresh installation of Spack.
+The commands in the guide have been updated for setting up Spack on the new operating system.
+
+
4. Recreate your environments
+
+
Follow the instructions in our guide Install Software Using Spack to create your desired environments
+using the “root specs” that you saved earlier.
+
+
NOTE: We’ve made small but important change to this guide: you should always start an interactive Slurm job before creating or modifying a Spack environment.
+The login server uses different hardware than the execute servers, and the mismatch leads to Spack using the wrong settings for installing packages.
+Of course, as before, you should only install packages while in interactive Slurm job.
+
+
Behind the scenes, we’ve made a few changes to the configuration that will hopefully make the package installation much smoother.
+
+
5. Update your workflow
+
+
Finally, remember to update your workflow to use the new Spack environments and the packages installed therein.
+
+
+ -
+
If you explicitly provide paths to packages installed using Spack, be sure to update those paths in your compiler configuration or in your job submission script.
+
+ -
+
If you used Spack to provide dependencies for manually compiling a program, remember to recompile the program.
+
+ -
+
If you changed the name of your environment, be sure to update the name in your job submission script.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Submitting and Managing Jobs Using SLURM
+
+
+
The HPC Cluster uses SLURM to manage jobs on the HPC Cluster. This page describes
+how to submit and manage jobs using SLURM.
+
+
Contents
+
+
+ - Submitting Jobs Using SLURM
+ - Viewing Jobs in the Queue
+ - Viewing Additional Job Information
+ - Removing or Holding Jobs
+
+
+
The following assumes that you have been granted access to the HPC cluster
+and can log into the head node spark-login.chtc.wisc.edu. If this is not
+the case, please see the CHTC account application page or email
+the facilitation team at chtc@cs.wisc.edu.
+
+
1. Submitting Jobs Using SLURM
+
+
A. Submitting a Job
+
+
Jobs can be submitted to the cluster using a submit file, sometimes also
+called a “batch” file. The top half of the file consists of #SBATCH
+options which communicate needs or parameters of the job – these lines
+are not comments, but essential options for the job. The values for
+#SBATCH options should reflect the size of nodes and run time limits
+described here.
+
+
After the #SBATCH options, the submit file should contain the commands
+needed to run your job, including loading any needed software modules.
+
+
An example submit file is given below. It requests 1 nodes of 64 cores
+and 4GB of memory each (so 64 cores and 256 GB of memory total), on the
+shared partition. It also specifies a run time limit of 4.5 hours.
+
+
#!/bin/sh
+#This file is called submit-script.sh
+#SBATCH --partition=shared # default "shared", if not specified
+#SBATCH --time=0-04:30:00 # run time in days-hh:mm:ss
+#SBATCH --nodes=1 # require 1 nodes
+#SBATCH --ntasks-per-node=64 # cpus per node (by default, "ntasks"="cpus")
+#SBATCH --mem=4000 # RAM per node in megabytes
+#SBATCH --error=job.%J.err
+#SBATCH --output=job.%J.out
+# Make sure to change the above two lines to reflect your appropriate
+# file locations for standard error and output
+
+# Now list your executable command (or a string of them).
+# Example for code compiled with a software module:
+module load mpimodule
+srun --mpi=pmix -n 64 /home/username/mpiprogram
+
+
+
Once the submit file is created, it can be submitted using the sbatch command:
+
+
[alice@login]$ sbatch submit-script.sh
+
+
+
B. Optimizing Your Submit File
+
+
The new cluster has different partition names and different sized nodes. We always recommend requesting cores per node (instead of total cores), using a multiple of 32 cores as your request per node. Requesting multiple nodes is not advantageous if your jobs are smaller than 128 cores. We also now recommend requesting memory per core instead of memory per node, for similar reasons, using the --mem-per-cpu flag with units of MB. Here are our recommendations for different sized jobs:
+
+
+
+ | Job size |
+ Recommended #SBATCH flags |
+
+
+ | 32-128 cores |
+ Example for 64 cores:
+#SBATCH --nodes=1
+#SBATCH --ntasks-per-node=64 # recommend multiples of 32
+#SBATCH --mem-per-cpu=4000 |
+
+
+ | 128 - 256 cores |
+ Split over a few nodes, for example for 128 cores:
+#SBATCH --nodes=2
+#SBATCH --ntasks-per-node=64 # designate cores per node
+#SBATCH --mem-per-cpu=4000
+ |
+
+
+ | 128 or 256 cores (whole nodes) |
+ Example for 256 cores:
+#SBATCH --nodes=2
+#SBATCH --ntasks-per-node=128
+#SBATCH --mem-per-cpu=4000 |
+
+
+
+
C. Requesting an Interactive Job ("int" and "pre" partitions)
+
+
If you want to run your job commands yourself, as a test before submitting
+a job as described above, you can request an interactive job on the cluster.
+
+
There is a dedicated partition
+for interactive work called int; you may request up to 16 CPUS and 64GB of memory
+when requesting an interactive session in the "int" partition. By default,
+the session is limited to 60 minutes though you can request up to 4 hours.
+Using another partition (like pre) will
+mean your interactive job is subject to the limits of that partition instead.
+
+
For simple testing or compiling
+
+
The command to request an interactive job is srun --mpi=pmix, and includes the partition
+in which you’d like to run the interactive job.
+
+
[alice@login]$ srun --mpi=pmix -n4 -N1 -p int --pty bash
+
+
+
+ Note: You will not be able to run MPI code in this interactive session.
+
+
+
The above example indicates a request for 4 CPUs (-n4) on a single
+node (-N1) in the "int" partition (-p int). Adding "-t 15" would
+indicate a request for 15 minutes, if desired, rather than the 60-minute
+default. After the interactive shell is created to a compute node with
+the above command, you'll have access to files on the shared file
+system and be able to execute code interactively as if you had directly
+logged in to that node. It is important to exit the interactive shell
+when you're done working by typing exit.
+
+
For running MPI code
+
+
To run an MPI program in an interactive session, you will need to (1) allocate the
+resources using salloc, then (2) use srun to run the MPI code, and finally (3)
+give up the allocated resources.
+
+
+ -
+
Request resources
+
+ [alice@login]$ salloc -n4 -N1 -p int
+
+
+ This command requests 4 CPUs (-n4) on a single node (-N1) in the "int"
+partition (-p int), and assigns the resources to a new terminal session
+on the login node. When the allocation has started, you will see a message
+like this:
+ salloc: Granted job allocation 18701
+ Guest on spark-a005.chtc.wisc.edu
+
+
+ To run code in this allocation, be sure to use srun as described in the next step!
+
+ -
+
Use resources
+
+ At this point, your terminal is still running on the login node. To run
+commands using the resources in the allocation, you will need to use srun.
+ [alice@login]$ srun --mpi=pmix /path/to/mpi/script
+
+
+ This will execute the specified script using the allocated resources.
+When the srun calculation has finished, you will remain in the allocation
+session, allowing you to run srun multiple times in quick succession.
+
+ You can also use the allocated resources interactively with
+ [alice@login]$ srun --mpi=pmix --pty bash
+
+
+ which will start an interactive terminal session in your allocation (this
+is evident by the change in the command prompt from [alice@login] to
+[alice@spark-a###]). Keep in mind that you will not be able to use
+MPI inside the interactive session. You can exit the interactive session
+and return to the allocation by entering exit.
+
+ -
+
Give up resources
+
+ To end your allocation, simply enter exit. You will see a message like
+this:
+ exit
+salloc: Relinquishing job allocation 18701
+salloc: Job allocation 18701 has been revoked.
+
+
+
+
+
+ It can be difficult to remember whether or not you are currently using an
+allocation. A quick way of checking is to see if the SLURM_JOB_ID is set
+by entering echo $SLURM_JOB_ID. If you are in an allocation, this command
+will return the job ID number that corresponds to an entry in your SLURM queue
+(see below).
+
+ A more convenient option is to update your .bashrc file so that the command
+prompt changes when you are in an allocation. This can be done using the
+following commands:
+ echo 'PS1="$SLURM_JOB_ID[\u@\h \W]\$ " ' >> ~/.bashrc
+echo 'export PS1' >> ~/.bashrc
+
+
+ Now when you run salloc, your command prompt will start with the corresponding
+SLURM job ID number. This will also be the case for the interactive srun
+command. For example,
+ [alice@login]$ salloc -n4 -N1 -p int
+salloc: Granted job allocation 18701
+ Guest on spark-a005.chtc.wisc.edu
+
+18701[alice@login]$ echo 'I am running an allocation.'
+I am running an allocation.
+18701[alice@login]$ srun --mpi=pmix --pty bash
+
+18701[alice@spark-a006] echo 'I am using the resources interactively.'
+I am using the resources interactively.
+18701[alice@spark-a006] exit
+exit
+18701[alice@login]$ exit
+exit
+salloc: Relinquishing job allocation 18701
+[alice@login]$
+
+
+
+ - This can be undone by removing the two added lines from the
.bashrc file
+ in your home directory.
+
+
+
+ More advanced users can manipulate their bash prompt further.
+The SLURM_JOB_ID variable is created for the allocation, and
+a SLURM_JOB_UID variable is created for the interactive srun.
+
+
+
+
2. Viewing Jobs in the Queue
+
+
To view your jobs in the SLURM queue, use the following command:
+
+
[alice@login]$ squeue -u username
+
+
+
Issuing squeue alone will show all user jobs in the queue. You can
+view all jobs for a particular partition with squeue -p shared.
+
+
+
+
Accounting information for jobs that are invoked with SLURM are logged. The sacct command displays job accouting data in a variety of forms for your analysis.
+
+
If you are having trouble viewing output from sacct try running this command first
+
+
[alice@login]$ sacct --start=2018-01-01
+
+
+
How To Select Jobs
+
+
+ -
+
To display information about a specific job or list of jobs use -j or --jobs followed by a job number or comma separated list of job numbers.
+
+ [alice@login]$ sacct --jobs job1,job2,job3
+
+
+
+ -
+
To select information about jobs in a certain date range use --start and --end Without it, sacct will only return jobs from the current day.
+
+ [alice@login]$ sacct --start=YYYY-MM-DD
+
+
+ - To select information about jobs in a certian time range use
--starttime and --endtime The default start time is 00:00:00 of the current day, unless used with -j, then the default start time is Unix Epoch 0. The default end time is time of running the command. Valid time formats are
+ HH:MM[:SS] [AM|PM]
+ MMDD[YY] or MM/DD[/YY] or MM.DD[.YY]
+ MM/DD[/YY]-HH:MM[:SS]
+ YYYY-MM-DD[THH:MM[:SS]]
+
+
+ [alice@login]$ sacct --starttime 08/23 --endtime 08/24
+
+
+ -
+
To display another user’s jobs use --user
+
+ [alice@login]$ sacct --user BuckyBadger
+
+
+
+ -
+
To only show statistics relevant to the job allocation itself, not taking steps into consideration use -X. This can be useful when trying to figure out which part of a job errored out.
+
+ [alice@login]$ sacct -X
+
+
+
+
+
+
Displaying Specific Fields
+
+
sacct can display different fields about your jobs. You can use the --helpformat flag to get a full list.
+
+
[alice@login]$ sacct --helpformat
+
+
+
Recommended Fields
+
+
When looking for information about your jobs CHTC recommends using these fields
+
elapsed
+end
+exitcode
+jobid
+ncpus
+nnodes
+nodelist
+ntasks
+partition
+start
+state
+submit
+user
+
+
+
For example run
+
+
sacct --start=2020-01-01 --format=jobid
+
+
+
to see jobIDs of all jobs ran since 1/1/2020.
+
+
4. Removing or Holding Jobs
+
+
You can kill and/or remove your job from the
+queue with the following:
+
+
[alice@login]$ scancel job#
+
+
+
where job# is the number shown for your job in the squeue output.
+
+
If you want to leave a job in the queue, but prevent it from running immediately,
+you can “hold” a submitted job by using:
+
+
[alice@login]$ scontrol hold job#
+
+
+
To release jobs that are held so that they can run, use this command:
+
+
[alice@login]$ scontrol release job#
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ HPC Cluster Overview
+
+
+
+
+
+
Table of Contents
+
+
+
+
+
+
+
The CHTC high-performance computing (HPC) cluster provides dedicated support for large,
+singular computations that use specialized software (i.e. MPI) to achieve internal
+parallelization of work across multiple servers of dozens to hundreds of cores.
+
+
Is high-performance computing right for me? Only computational work that
+fits that above description is appropriate for the HPC Cluster. Computational
+work that can complete on a single node in less than a few days will be
+best supported by our larger high-throughput computing (HTC) system (which also
+includes specialized hardware for extreme memory, GPUs, and other cases). For more
+information, please see Our Approach.
+
+
To get access to the HPC Cluster, please complete our
+New User Consultation Form. After your request is received,
+a Research Computing Facilitator will follow up to discuss the computational needs
+of your research and connect you with computing
+resources (including non-CHTC services) that best fit your needs.
+
+
HPC Cluster User Policies
+
+
See our User Policies and Expectations for details on general CHTC and HPC cluster policies.
+
+
HPC Hardware and Configuration
+
+
The HPC Cluster consists of two login nodes and many compute (aka execute)
+nodes. All users log in at a login node, and all user files on the shared file sytem are accessible on all nodes.
+Additionally, all nodes are tightly networked (200 Gbit/s Infiniband) so
+they can work together as a single "supercomputer", depending on the
+number of CPUs you specify.
+
+
Operating System and Software
+
+
All nodes in the HPC Cluster are running CentOS 8 Stream Linux.
+
+
The SLURM scheduler version is 22.05.6.
+
+
To see more details of other software on the cluster, see the HPC Software page.
+
+
Login Nodes
+
+
The login node for the cluster is: spark-login.chtc.wisc.edu
+
+
For more details on logging in, see the “Connecting to CHTC” guide linked above.
+
+
Execute Nodes and Partitions
+
+
Only execute nodes will be used for performing your computational work.
+The execute nodes are organized into several "partitions", including
+the shared, pre, and int partitions which are available to
+all HPC users as well as research group specific partitions that consist
+of researcher-owned hardware and which all HPC users can access on a
+backfill capacity via the pre partition (more details below).
+
+
+
+
+ | Partition |
+ p-name |
+ # nodes (N) |
+ t-default |
+ t-max |
+ max cores/job |
+ cores/node (n) |
+ RAM/node (GB) |
+
+
+
+
+ | Shared |
+ shared |
+ 45 |
+ 1 day |
+ 7 day |
+ 320 |
+ 64 or 128 |
+ 512 |
+
+
+ | Interactive |
+ int |
+ 2 |
+ 1 hr |
+ 4 hrs |
+ 16 |
+ 64 or 128 |
+ 512 (max 64 per job) |
+
+
+ | Pre-emptable (backfill) |
+ pre |
+ 45 |
+ 4 hrs |
+ 24 hrs |
+ 320 |
+ 64 or 128 |
+ 512 |
+
+
+ | Owners |
+ unique |
+ 19 |
+ 24 hrs |
+ 7 days |
+ unique |
+ 64 or 128 |
+ 512 |
+
+
+
+
+
+ -
+
shared compute nodes each have 64 or 128 cores and 512 GB of RAM.
+Jobs submitted to this partition
+can request and use up to 7 days of running time.
+
+ -
+
int consists of two compute nodes is intended for short and immediate interactive
+testing on a single node (up to 16 CPUs, 64 GB RAM). Jobs submitted to this partition
+can run for up to 4 hours.
+
+ -
+
pre (i.e. pre-emptable) is an under-layed partition encompassing all HPC Cluster
+nodes and is intended for more immediate turn-around of shorter, smaller, and/or
+interactive sessions requiring more than the 4 hour time limit of the int partition.
+Jobs submitted to pre are run as back-fill on any idle nodes, including researcher-owned
+compute nodes, meaning these jobs may be pre-empted by higher priority
+jobs. By default, pre-empted jobs will be re-queued (to run again) if they were submitted with
+an sbatch script.
+
+
+
+
Fair Share Allocation
+
+
To promote fair access to HPC computing resources, all users are limited to 10 concurrently
+running jobs (if you need to queue more, please get in touch). Additionally, users are restricted to a total of 720 cores
+across all running jobs (core limits do not apply on research group partitions of
+more than 720 cores).
+
+
When determining which order to run jobs, the following policies are applies, in order or significance
+to job priority determinations:
+
+
A. User priority decreases as the user accumulates hours of CPU time over the last 21 days, across
+all queues. This “fair-share” policy means that users who have run many/larger jobs in the near-past
+will have a lower priority, and users with little recent activity will see their waiting jobs start sooner.
+(The cluster does not have a strict “first-in-first-out” queue policy.)
+
+
B. Job priority increases with job wait time. After the history-based user priority calculation in (A),
+the next most important factor for each job’s priority is the amount of time that each job has already
+waited in the queue. For all the jobs of a single user, these jobs will most closely follow a “first-in-first-out” policy.
+
+
C. Job priority increases with job size, in cores. This least important factor slightly favors larger jobs, so that
+the scheduler can take advantage when large numbers of newly-available nodes happen to become available (requiring less
+wasted time to deliberately drain nodes for larger jobs). So, among a user’s jobs submitted at roughly the same time,
+a larger job may run first, if the number of nodes necessary for the larger job is already available.
+
+
Data Storage and Management
+
+
Data space in the HPC Cluster filesystem is not backed-up and should be
+treated as temporary by users. Only files necessary for
+actively-running jobs should be kept on the filesystem, and files
+should be removed from the cluster when jobs complete. A primary copy of any
+essential files (e.g. software, submit files, input) should be kept in an
+alternate, non-CHTC storage location.
+
+
Each user will receive two primary data storage locations:
+
+
+ -
+
/home/username with an initial disk quota of 30GB
+and 250,000 items. Your home directory is meant to be used for files
+you use repeatedly, like submit file templates, source code, software
+installations, and reference data files.
+
+ -
+
/scratch/username with an initial disk quota of 100GB and
+250,000 items. Jobs should always be submitted and run out of
+/scratch. It is the space for all working data, including individual
+job inputs, job outputs, and job log/stderr/stdout files.
+
+
+
+
+ What about /software?
+
+ If you are installing software meant to be shared within a group,
+we can create a dedicated folder for you in the /software space
+email us (chtc@cs.wisc.edu) if this is you!
+
+
+
To check how many files and directories you have in
+your /home or /scratch directory see the
+instructions below.
+
+
Changes to quotas for either of these locations are available upon request
+per our Request a Quota Change guide. If you don't
+know how many files your installation creates, because it's more than
+the current items quota, simply indicate that in your request.
+
+
CHTC Staff reserve the right to remove any significant amounts of data
+on the HPC Cluster in our efforts to maintain filesystem performance
+for all users.
+
+
Local scratch space is available on each execute node in /local/$USER.
+This space is NOT automatically cleaned out, so if you use this space,
+be sure to remove the files before the end of your job script or
+interactive session.
+
+
+
+
You can use the command get_quotas to see what disk
+and items quotas are currently set for a given directory path.
+This command will also let you see how much disk is in use and how many
+items are present in a directory:
+
+
[username@hpclogin1 ~]$ get_quotas /home/username /scratch/username
+
+
+
Alternatively, the ncdu command can also be used to see how many
+files and directories are contained in a given path:
+
+
[username@hpclogin1 ~]$ ncdu /home/username
+[username@hpclogin1 ~]$ ncdu /scratch/username
+
+
+
When ncdu has finished running, the output will give you a total file
+count and allow you to navigate between subdirectories for even more
+details. Type q when you're ready to exit the output viewer. More
+info here: https://lintut.com/ncdu-check-disk-usage/
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Use Software on the HPC Cluster
+
+
+
This page describes how to install and run software on CHTC’s HPC Cluster.
+
+
Contents
+
+
+ - General Software Policies
+ - Using Pre-Installed Software in Modules
+ - Installing Software on the Cluster
+ - Using Software in Jobs
+
+
+
1. General Software Policies
+
+
In CHTC, we install a minimal set of software for use
+on our systems. On the HPC Cluster, CHTC staff manage installations of
+the following types of programs:
+
+
+ - Compilation tools and common dependencies (e.g. MPI, different GCC versions)
+ - Software that requires a shared license (e.g. COMSOL)
+
+
+
Information on how to access CHTC-managed installations is in the next
+section of this guide. If you need to use a program not in that group, the instructions
+for creating your own installation follow.
+
+
If you have questions or concerns about installing your own software or
+the available dependencies, contact the facilitation team at chtc@cs.wisc.edu.
+
+
2. Using Pre-Installed Software in Modules
+
+
All software on the cluster that is installed by CHTC staff is available via
+a tool called “modules”.
+
+
A. See Available Software Modules
+
+
There are two ways to search through the software modules on the HPC cluster:
+
+
+ - View all modules
+ This command will show all software modules available:
+
[alice@login]$ module avail
+
+
+ - Search for specific modules
+ If you are searching for a specific software module, you can use the
+
module spider command with part of the software name. For example, to
+ search for Open MPI modules, you would type:
+ [alice@login]$ module spider openmpi
+
+
+
+
+
B. Access Software in Modules
+
+
Once you find a software module that you want to use, you need to “load” it
+into your command line environment to make it active, filling in module_name with
+the name you found through one of the above steps.
+
+
[alice@login]$ module load module_name
+
+
+
+ When to Load Modules
+
+ You can load modules to compile code (see below). If you do this, make sure to load
+the same modules as part of your job script before running the main command.
+
+ You can also load modules to run specific software. If done for interactive
+testing, this should be done in an interactive job; otherwise, the module
+should be loaded in the job submit file.
+
+
+
C. Unload Software in Modules
+
+
If you no longer want to use a specific software installation, you can “unload”
+the software module with the following command:
+
+
[alice@login]$ module unload module_name
+
+
+
If you want to clear your command line environment and start over, run the following:
+
+
[alice@login]$ module purge
+
+
+
3. Installing Software on the Cluster
+
+
A. Overview
+
+
Unless you are using a licensed software program provided via modules, you
+are able to compile and install the software you need on the HPC Cluster.
+
+
Compilation can be done via an interactive job as described in
+our HPC Job Submission Guide.
+Software should be installed to your /home/username
+directory. If using CHTC’s provided compilation tools via modules, make
+sure to load the needed modules before compiling and to load the same
+modules in your job submission.
+
+
For groups that would like to share software installations among group
+members, please contact us about getting a shared “group” directory.
+
+
If you are new to software installation, see the section below for
+a more step-by-step description of the process.
+
+
B. Step by Step Process
+
+
+ - Download Source Code - download the source code for your desired program. We
+ recommend downloading it to your
/home/username directory on the login node.
+ You should only need the source code until the software is properly installed, but if desired, you may keep a zipped copy of
+ the source code in /home.
+ - Read the Docs - try to find the installation instructions, either online or
+ in the downloaded source code. In particular, you’ll want to note if there are
+ any special requirements for dependencies like MPI or the compiler needed.
+ - Load Modules - if you are using software modules to help you build your
+ code, load them now. Keep track of what you use so that you can load them
+ in your job submit file later. We also recommend doing a
module purge before
+ loading your compiling modules to make sure you’re starting from a clean environment.
+ - Install - most scientific software follows the three step installation process
+ of
configure - make - make install.
+
+ configure- this step checks for tools and requirements needed to compile
+ the code. This is the step where you set the final installation location of
+ a program. The option for setting this location is typically called the
+ “prefix”; a common syntax is: $ ./configure --prefix=/home/user.
+ This is where you will want to set the installation location to be your
+ /home directory.make - this step compiles and links the code, turning it from human-readable
+ source code to compiled binary code. This is usually the most time consuming
+ step of the installation process.make install - this step copies compiled files to the final installation location
+ (usually specified in the configure step).
+
+ - Clean Up - the final installation should place all needed files into a
+ subdirectory of your
/home directory. The source code and location where
+ you ran the compilation commands can be removed at this point.
+
+
+
+
+
4. Using Software in Jobs
+
+
The commands to run your software will go in the job’s submit file, as described
+in our HPC job submission guide.
+
+
If you used one of the software modules to compile your code, make sure you
+load it in your job’s submit file before running your main command.
+
+
You can access your software by including the path to its location in your
+/home directory, or by setting the PATH environment variable to include
+the software location and then running the command.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Install Software Using Spack
+
+
+
CHTC uses Spack (https://github.com/spack/spack) for installing and managing software packages on the HPC cluster for all users to use, via the module command. Recently, Spack has developed a feature that allows for users to integrate their local installation of Spack with the system-wide installation. This means that when a user installs software with their local installation of Spack, they can automatically incorporate the system-wide packages to satisfy their software’s dependencies (similar to Conda and Miniconda).
+
+
This guide describes how to install and manage software using Spack, including how to install and use a specific compiler.
+
+
This guide assumes you or your group has already set up your local installation of Spack. If you have not installed Spack, follow the instructions in Setting Up Spack on HPC.
+
+
Contents
+
+
+ - Installing Software Using Spack
+ - Using Software Installed in Spack
+ - Installing and Using a Specific Compiler
+
+
+
1. Installing Software Using Spack
+
+
Once your local installation of Spack has been properly configured, you are now ready to install software using Spack.
+
+
Check the documentation for the program you want to install to see if they have instructions for installation using Spack. Even if your program can’t be installed using Spack, you can still use it to install the dependencies that your program needs.
+
+
+ Note: For a group installation of Spack, you will not be able to modify or remove the packages installed by a different user. We recommend that you consult with the rest of your group for permission before proceeding.
+
+
+
A. Start an Interactive Job
+
+
Before creating a Spack environment or installing packages using Spack, first start an interactive Slurm job:
+
+
srun --mpi=pmix -n4 -N1 -t 240 -p int --pty bash
+
+
+
For more information on interactive Slurm jobs, see our guide Submitting and Managing Jobs Using SLURM.
+
+
+ When creating an environment, Spack automatically detects the hardware of the machine being used at the time and configures the packages as such.
+Since the login server uses newer hardware than the execute servers, creating an environment on the login server (not in an interactive job) is a bad idea.
+
+
+
B. Creating and Using a Spack Environment
+
+
Software installations with Spack should be done inside of a Spack environment, to help manage the shell and the paths to access programs and libraries needed for a particular software installation.
+
+
To create a Spack environment, run the command
+
+
spack env create yourEnvironmentName
+
+
+
where you should replace yourEnvironmentName with your desired name for the environment. You can then activate the environment with
+
+
spack env activate yourEnvironmentName
+
+
+
You will need to activate the environment when you wish to use the software that was installed in that environment.
+
+
+ You can see a list of your available environments using
+
+ spack env list
+
+
+ and you can see which environment you are currently using with
+
+ spack env status
+
+
+ To deactivate the environment, run
+
+ spack env deactivate
+
+
+ or close the terminal session.
+
+
+
C. Finding Program Packages in Spack
+
+
Once inside an active Spack environment, you can run the following command to see what packages are installed in the current environment
+
+
spack find
+
+
+
For a new environment, this will show that there are no programs installed. The output of this command will update after you install program packages in the environment.
+
+
To search for packages to install using Spack, use the command
+
+
spack list nameOfProgram
+
+
+
where you should replace nameOfProgram with the program that you are interested in finding. Spack will search for the package and print out a list of all the packages that match that name. (The first time you run this command may take several minutes while Spack downloads a current list of packages that can be installed.)
+
+
To learn more about an available package, use the exact name of the program and run
+
+
spack info exactNameOfProgram
+
+
+
This will print out information about the program, including a short description of the program, a link to the developer’s website, and the available versions of the program and its dependencies.
+
+
D. Adding Package Specifications to the Environment
+
+
Once you find the packages that you want to install, add their specifications to the environment using
+
+
spack add exactNameOfProgram
+
+
+
Spack will automatically decide which version of the program to use at installation time based on the other packages that you’ve added.
+
+
If you want a specific version of a package, you can specify it by appending @= to the end of the package name, followed by the version number. For example,
+
+
spack add python@=3.10
+
+
+
will tell the environment that you want to install version 3.10 of Python. There are additional ways of defining specifications for package versions, the compiler to be used, and dependencies. The documentation for Spack provides the details on how this is done.
+
+
If you need to install a compiler, or need to use a specific compiler to install the desired packages, see section 3. Installing and Using a Specific Compiler.
+
+
E. Installing Packages in an Environment
+
+
Once you have identified the package(s) you would like to install and have added the specifications to your environment,
+
+
i. Create the local scratch directory
+
+
Using the default configuration from Setting Up Spack on HPC, Spack will try to use the machine’s local disk space for staging and compiling files before transferring the finished results to the final installation directory. Using this space will greatly improve the speed of the installation process. Create the local directory with the command
+
+
mkdir /local/yourNetID/spack_build
+
+
+
where you should replace yourNetID with your NetID. At the end of the session, remember to delete this directory so that other people can use the disk space in their jobs.
+
+
+ If the directory already exists, that means you forgot to remove it after one of your previous Spack installation sessions. Simply remove the directory and make it again.
+
+ rm -rf /local/yourNetID/spack_build
+mkdir /local/yourNetID/spack_build
+
+
+
+
ii. Check the programs/packages to be installed
+
+
If you’ve added the installation specifications to the environment, then you can check the installation plan using the command
+
+
spack spec -lI
+
+
+
(the first letter after the hyphen is a lowercase “L” and the second letter is an uppercase “i”).
+
+
+ This command identifies what dependencies Spack needs in order to install your desired packages along with how it will obtain them. Assuming their are no problems, then it will print a list of the packages and their dependencies, where entries that begin with a green [+] have already been installed somewhere in your local Spack installation, while those that begin with a green [^] are referencing the system installation, and those beginning with a gray - will need to be downloaded and installed.
+
+
+
Most users should see a bunch of packages with a green [^] in the first column.
+If you do not, then there are several possible explanations:
+
+
+
+
If you are satisfied with the results, then you can proceed to install the programs.
+
+
iii. Install the environment packages
+
+
Assuming that you are in an interactive Slurm session and have activated the desired environment containing the package specifications, you can run
+
+
spack install -j 4
+
+
+
to install the packages inside of the Spack environment, where the number that comes after -j needs to match the number that you noted from when you started the interactive session (the one after -n when you ran the srun command for the interactive session). You can also add the -v option to have the installation be verbose, which will cause Spack to print the compile and make outputs in addition to the standard Spack output.
+
+
Depending on the number and complexity of the programs you are installing, and how much can be bootstrapped from the system installation, the installation step can take anywhere from several minutes to several hours.
+
+
+ If something goes wrong or your connection is interrupted, the installation process can be resumed at a later time without having to start over from the beginning. Make sure that you are in an interactive Slurm session and that you have activated the Spack environment, then simply rerun the spack install command again.
+
+
+
iv. Finishing the installation
+
+
After the installation has successfully finished, you should be able to see that the programs have been installed by running
+
+
spack find
+
+
+
which should list the programs under the compiler heading used for installing the programs.
+
+
You may need to deactivate and reactivate the environment in order to properly use the programs that have been installed.
+
+
spack env deactivate
+spack env activate yourEnvironmentName
+
+
+
Once you are satisfied that the programs have been installed properly, you can remove packages that are build-only (not used for running the packages you installed) using the command
+
+
spack gc
+
+
+
Finally, remove the local build directory that Spack used during the installation with
+
+
rm -rf /local/yourNetID/spack_build
+
+
+
and then enter exit to end the interactive session.
+
+
To use the packages that you installed, follow the instructions in the next section, 2. Using Software Installed in Spack. If you want to create custom modules using the installed packages, see our guide Creating Custom Modules Using Spack.
+
+
F. Removing an Environment and Uninstalling Unneeded Packages
+
+
You may find it necessary to remove a Spack environment, or packages installed using Spack. To uninstall a package, simply run
+
+
spack uninstall yourPackageName
+
+
+
where you should replace yourPackageName with the name of the package that you want to remove. This command will only work for packages that you ‘added’ to the Spack environment, as described above.
+
+
To remove an environment, first make sure that you have deactivated the environment with
+
+
spack env deactivate
+
+
+
and then run
+
+
spack env rm yourEnvironmentName
+
+
+
where you should replace yourEnvironmentName with the name of the environment that you want to remove. Note that this will not necessarily remove the packages that were installed in the environment! After the environment has been removed, you can uninstall the packages that are no longer needed using the command
+
+
spack gc
+
+
+
2. Using Software Installed in Spack
+
+
If your account is configured correctly for using Spack, and the software has been installed inside of a Spack environment, then to use the software all you need to do is activate the corresponding environment. Simply use the command
+
+
spack env activate yourEnvironmentName
+
+
+
and Spack will update your shell accordingly. (Remember that you can see the available Spack environments by running the command spack env list). Once the environment has been activated, you should be able to use the packages just as normal. You can confirm you are using a command installed using Spack by running
+
+
which nameOfYourCommand
+
+
+
where you replace nameOfYourCommand with the name of the command. The command will output a path, and you should see something like spack/var/spack/environments/yourEnvironmentName/ in that path.
+
+
For submitting jobs using Slurm, you will need to make sure that you activate the Spack environment near the beginning of your sbatch file before the srun command. For example,
+
+
#!/bin/sh
+# This file is called submit-script.sh
+#SBATCH --partition=shared # default "shared", if not specified
+#SBATCH --time=0-04:30:00 # run time in days-hh:mm:ss
+#SBATCH --nodes=1 # require 1 nodes
+#SBATCH --ntasks-per-node=64 # cpus per node (by default, "ntasks"="cpus")
+#SBATCH --mem-per-cpu=4000 # RAM per node in megabytes
+#SBATCH --error=job.%J.err
+#SBATCH --output=job.%J.out
+
+# v---Remember to activate your Spack environment!!
+spack env activate yourEnvironmentName
+
+srun --mpi=pmix -n 64 /home/username/mpiprogram
+
+
+
When you submit this job to Slurm and it executes the commands in the sbatch file, it will first activate the Spack environment, and then your program will be run using the programs that are installed inside that environment.
+
+
+ Some programs include explicit module load commands in their execution, which may override the paths provided by the Spack environment. If your program appears to use the system versions of the packages instead of the versions installed in your Spack environment, you may need to remove or modify these explicit commands. Consult your program’s documentation for how to do so. You may want to create your own custom modules and modify your program to explicitly load your custom modules. See Creating Custom Modules Using Spack for more information on how to create your own modules using Spack.
+
+
+
3. Installing and Using a Specific Compiler
+
+
By default, Spack will attempt to compile packages it installs using one of the system compilers, most likely with GCC version 11.3.0. Some programs, however, may need to be compiled using a specific compiler, or require that their dependencies be built using a specific compiler. While this is possible using Spack, the process for installing and using a compiler is a bit more complicated than that for installing “regular” packages as was described above.
+
+
In brief, you will first create a separate environment for installing the compiler. Then you will add that compiler to the list of available compilers that Spack can use. Finally, you can install your desired packages as in a new environment, but you will need to specify which compiler to use.
+
+
A. Install the Compiler in its Own Environment
+
+
i. Identify the compiler and version
+
+
The first step is to identify the compiler and version you need for your program. Consult your program’s documentation for the requirements that it has. Then follow the instructions in C. Finding Program Packages in Spack to find the package name and confirm the version is available.
+
+
ii. Create the compiler’s environment
+
+
Next, create and activate an environment for installing the desired compiler. For example,
+
+
spack env create compilerName_compilerVersion
+spack env activate compilerName_compilerVersion
+
+
+
where you should replace compilerName and compilerVersion with the name and version of the desired compiler.
+
+
iii. Add the compiler specification to its environment
+
+
Once you’ve activated the environment, add the exact specification for the compiler to the Spack environment with
+
+
spack add compilerName@=compilerVersion
+
+
+
where you need to replace compilerName and compilerVersion with the name and version of the compiler that you identified above.
+
+
iv. Install the compiler in its environment
+
+
Next, follow the instructions in E. Installing Packages in an Environment to install the desired compiler in this environment. Installing the compiler may take several hours, so consider increasing the number of threads to speed up the installation.
+
+
B. Add the Compiler to Spack
+
+
i. Identify the compiler’s installation path
+
+
After installing the compiler, you need to find its location. First, activate the compiler’s environment with spack env activate compilerName_compilerVersion. Next, use the following command to save the path to the compiler as the shell variable compilerPath:
+
+
compilerPath=$(spack location -i compilerName@=compilerVersion)
+
+
+
where you need to replace compilerName and compilerVersion with the name and version of the compiler that you installed. You can see print out the path using the command echo $compilerPath.
+
+
ii. Give the compiler’s path to Spack
+
+
Now that you know where the compiler is installed, deactivate the environment with spack env deactivate. Then run the following command to tell Spack to add the compiler to its list of available compilers:
+
+
spack compiler add $compilerPath
+
+
+
iii. Confirm compiler has been added to Spack
+
+
The command
+
+
spack compiler list
+
+
+
will print out the list of compilers that Spack can use, and should now show compilerName@compilerVersion in the results.
+
+
C. Install Packages Using the New Compiler
+
+
Once the compiler has been installed and recognized by Spack, you can now create and activate a new environment for installing your desired packages, following the instructions in Installing Software Using Spack.
+
+
To make sure the packages are installed using your desired compiler, you need to include the compiler when you add the package specification to the environment (D. Adding Package Specifications to the Environment). To include the compiler in the specification, you need to add the symbol % followed by the compiler name and version to the end of the spack add command. For example,
+
+
spack add python@=3.10 %gcc@=9.5.0
+
+
+
will use GCC version 9.5.0 to compile Python 3.10 when installing the package. As a general rule, you should use the same compiler for installing all of your packages within an environment, unless your program’s installation instructions say otherwise.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Create Custom Modules Using Spack
+
+
+
CHTC uses Spack (https://github.com/spack/spack) for installing and managing software packages on the HPC cluster for all users to use, via the module command. Recently, Spack has developed a feature that allows for users to integrate their local installation of Spack with the system-wide installation. This means that when a user installs software with their local installation of Spack, they can automatically incorporate the system-wide packages to satisfy their software’s dependencies (similar to Conda and Miniconda).
+
+
This guide describes how to create and use custom personal and shared modules for software packages installed using Spack. For instructions on how to install software using Spack for you and/or your research group, see our guide Installing Software Using Spack.
+
+
Contents
+
+
+ - Location of the Module Files
+ - Using Custom Modules
+ - Creating Custom Modules Using Spack
+ - Working with Multiple Environments
+ - Using Hierarchy Based Modules
+
+
+
1. Location of the Module Files
+
+
In order to load a software package using the module command, there must be a corresponding “module file” containing the information that the module command needs in order to load the software package. Spack will automatically generate the required content of the module files, but Spack will need to know where these module files should be saved. Similarly, the module command will need to know where the module files are stored.
+
+
If you followed the instructions in Setting Up Spack on HPC, then the default location of your module files is /home/yourNetID/spack_modules where yourNetID is your NetID.
+
+
+ If you are using a shared installation of Spack for a group, and if the person who set up the installation followed the instructions in Setting Up Spack on HPC, then the default location of the shared module files is likely /home/groups/yourGroupName/spack_modules. You can confirm this by running the command spack config get modules | grep -A 2 'roots' and examining the listed paths (make sure you do not have a Spack environment activated when you do so). If the paths are /home/$user/spack_modules, then you should follow the instructions in iii. Updating location of module files in Setting Up Spack on HPC before proceeding.
+
+
+
2. Using Custom Modules
+
+
Spack will automatically create module files for the packages that you explicitly install, in the location described above.
+
+
To update the module command with the default location of the new module files, run the command
+
+
module use ~/spack_modules
+
+
+
+ For a group installation of Spack, you’ll need to modify the module use command to specify the path to your group’s directory. The following should work if your group followed our instructions when setting up Spack:
+
+ module use /home/groups/yourGroupName/spack_modules
+
+
+
+
Now if you run module avail you should see the your custom modules listed in the first section, with the system modules listed in the following section. You can then use the module load command as usual to load your custom module for use in the current terminal session.
+
+
Note: Spack will not automatically create module files for the “upstream” dependencies (packages already installed on the system). If your module load test does not work, follow the instructions in the next section to generate these additional module files.
+
+
To have your custom modules found automatically by the module command, add the above module use command to the end of your ~/.bash_profile file.
+
+
3. Creating Custom Modules Using Spack
+
+
You may need to manually create the custom module files, especially after editing any of the modules configuration for Spack. To create the module files, first activate the desired environment with
+
+
spack env activate yourEnvironmentName
+
+
+
(where you should replace yourEnvironmentName with the your environment name) and then enter the following command:
+
+
spack module tcl refresh
+
+
+
Spack will print out a list of all the packages installed in the current environment, and you’ll be asked to confirm if you wish to create module files for all of these packages.
+
+
To remove old module files, or to update the directory structure, add the option --delete-tree, i.e.
+
+
spack module tcl refresh --delete-tree
+
+
+
If you tried to load a module but received the error(s) ‘Executing this command requires loading “{module file}” which failed while processing the following…‘, then you will need to generate the “upstream” module files in order to use your desired module. In this case, the following command should resolve the issue:
+
+
spack module tcl refresh --upstream-modules
+
+
+
+ Note: You should only run this command inside of an activated Spack environment, otherwise you will be prompted to create module files for ALL Spack packages, including those installed system-wide, regardless of whether they are required dependencies!
+
+
+
Lastly, note that Spack will not directly create module files for software installed independently of Spack (for example, using pip install).
+
+
4. Working with Multiple Environments
+
+
If you have more than one Spack environment that you wish to create modules for, we recommend that you modify the above procedure in order to better organize the list of modules.
+
+
For each environment that you wish to create module files for, activate the environment and then edit the configuration so that the module files are saved into a sub-directory named for that environment. For example,
+
+
spack env activate my-first-env
+spack config add modules:default:roots:tcl:/home/\$user/spack_modules/my-first-env
+
+
+
first activates my-first-env and then updates the configuration to save the module files to /home/yourNetID/spack_modules/my-first-env.
+
+
+ For a group installation of Spack, you should instead specify the path to your group’s directory. For example,
+
+ spack env activate my-first-env
+spack config add modules:default:roots:tcl:/home/groups/yourGroupName/spack_modules/my-first-env
+
+
+ You should similarly modify the following commands to account for the different paths.
+
+
+
Repeat the process for your other environments.
+
+
To use the modules for a particular environment, run the module use command but specify the path to the environment’s subdirectory. Continuing with our example,
+
+
module use ~/spack_modules/my-first-env
+
+
+
will update the module command with the location of the modules for using my-first-env.
+
+
If you want to switch environments, we recommend that you “unuse” the first environment and then “use” the second, i.e.
+
+
module unuse ~/spack_modules/my-first-env
+module use ~/spack_modules/my-second-env
+
+
+
While you can have more than one environment in “use” by the module command, this increases the chance of loading modules with conflicting dependencies that could result in unexpected behavior.
+
+
5. Using Hierarchy Based Modules
+
+
There are two “flavors” of the module system: tcl and lmod. We use tcl for managing the system modules, and have recommended using tcl throughout this guide. The main difference between the two “flavors” of modules is that tcl uses a “flat” directory structure (all the module files are located in the same central directory) whereas lmod uses a “hierarchy” directory structure (where the module files are grouped by their compiler or MPI version). The hierarchal structure of lmod can be very useful in organizing duplicate module files that differ only by how they were compiled.
+
+
To use the lmod style module files, you should first edit your modules configuration to enable lmod and disable tcl, then refresh your module files.
+
+
spack config add 'modules:default:enable:lmod'
+spack config remove 'modules:default:enable:tcl'
+spack module refresh lmod --delete-tree
+
+
+
More advanced options regarding the naming and structure of the lmod module files can be configured by editing the modules.yaml (described in iii. Updating location of module files in Setting Up Spack on HPC). See the Spack documentation for more information on how to configure module files: https://spack.readthedocs.io/en/latest/module_file_support.html#.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Set Up Spack on HPC
+
+
+
CHTC uses Spack (https://github.com/spack/spack) for installing and managing software packages on the HPC cluster for all users to use, via the module command. Recently, Spack has developed a feature that allows for users to integrate their local installation of Spack with the system-wide installation. This means that when a user installs software with their local installation of Spack, they can automatically incorporate the system-wide packages to satisfy their software’s dependencies (similar to Conda and Miniconda).
+
+
This guide describes how to set up a local copy of Spack and integrate it with the system installation, either for an individual user or for a group of users. For instructions on how to install packages with Spack, see our other guide, Installing Software Using Spack.
+
+
+ If your group has already set up a shared group installation of Spack, you can skip to the end of this guide: 3. Using a Shared Group Installation.
+
+
+
Contents
+
+
+ - Setting Up Spack for Individual Use
+ - Setting Up Spack for Group Use
+ - Using a Shared Group Installation
+
+
+
1. Setting Up Spack for Individual Use
+
+
A. Downloading Spack (Individual)
+
+
First, log in to the HPC cluster.
+
+
You can then install Spack following its documentation. Download the Spack code from their GitHub repository:
+
+
git clone -c feature.manyFiles=true https://github.com/spack/spack.git
+
+
+
and then activate Spack by sourcing the setup script with the . command
+
+
. spack/share/spack/setup-env.sh
+
+
+
That’s it! You can test that Spack has been installed by entering spack and you should see the help text print out. But before trying to install packages using your Spack installation, you should configure it to recognize the system installation of Spack.
+
+
+ This guide assumes that you ran the git clone command in your home directory, i.e. /home/yourNetID. If you did not, then run the following command to print the full path to your Spack installation.
+
+ echo $SPACK_ROOT
+
+
+ We will refer to this path as the SpackRootPath and you will need to use this path where noted in the instructions below.
+
+
+
B. Using Spack in Future Sessions (Individual)
+
+
While Spack has been installed, for each session that you want to use it you will need to rerun the command
+
+
. /home/yourNetID/spack/share/spack/setup-env.sh
+
+
+
A more convenient option is simply to update your account to run this command whenever you log in. Add the command to the end of the .bash_profile file in your home directory, e.g. nano ~/.bash_profile, with the full path to the file. If you ran the git clone command in your home directory, then the line you add should be
+
+
. /home/yourNetID/spack/share/spack/setup-env.sh
+
+
+
where you need to replace yourNetID with your NetID.
+
+
+ If Spack was not installed to your home directory, use the following command instead, where you need to replace SpackRootPath with the path that you noted above.
+
+ . SpackRootPath/share/spack/setup-env.sh
+
+
+
+
C. Obtain the Provided Configuration Files (Individual)
+
+
To simplify the process of configuring your local installation of Spack, we have provided a folder with the necessary configuration files. All that you need to do is copy it to your home directory using the following command.
+
+
cp -R /software/chtc/el9/spack-user-config ~/.spack
+
+
+
Your local Spack installation will automatically find the configuration files and will now recognize the packages that are installed system-wide. You can confirm this with the command
+
+
spack find
+
+
+
This should show a list of packages, including those you see when you run the module avail command. A total of ~120 packages should be listed.
+
+
You are now ready to use Spack for installing the packages that you need! See the instructions in Installing Software Using Spack.
+
+
2. Setting Up Spack for Group Use
+
+
The following instructions for a group installation of Spack assumes that a shared directory has already been created for your group, and that you have access to this shared folder. We also recommend communicating with your colleagues before proceeding.
+
+
A. Downloading Spack (Group)
+
+
First, log in to the HPC cluster, and navigate to your group’s shared directory in /home with
+
+
cd /home/groups/yourGroupName
+
+
+
where you should replace yourGroupName with your group’s name. Note this path for use throughout this guide, and communicate it to your group members for configuring their access to the installation.
+
+
You can then install Spack following its documentation. Download the Spack code from their GitHub repository:
+
+
git clone -c feature.manyFiles=true https://github.com/spack/spack.git
+
+
+
and then activate Spack by sourcing the setup script with the . command.
+
+
. spack/share/spack/setup-env.sh
+
+
+
That’s it! You can test that Spack has been installed by entering spack and you should see the help text print out. But before trying to install packages using your Spack installation, you should configure it to recognize the system installation of Spack.
+
+
+ This guide assumes that you ran the git clone command in your group’s home directory, i.e. /home/groups/yourGroupName. If you did not, then run the following command to obtain the full path to your Spack installation. We will refer to this path as the SpackRootPath and you will need to use this path where noted in the instructions below.
+
+ echo $SPACK_ROOT
+
+
+
+
B. Using Spack in Future Sessions (Group)
+
+
While Spack has been installed, for each session that you want to use it you will need to rerun the command
+
+
. /home/groups/yourGroupName/spack/share/spack/setup-env.sh
+
+
+
A more convenient option is simply to update your account to run this command whenever you log in. You and your group members should add the command to the end of the .bash_profile file in your respective home directories, e.g. nano ~/.bash_profile, with the full path to the file. For a group installation, the line should look like
+
+
. /home/groups/yourGroupName/spack/share/spack/setup-env.sh
+
+
+
where you need to replace yourGroupName with the name of your group.
+
+
+ If Spack was not installed in your group’s home directory, use the following command instead, where you will need to replace SpackRootPath with the path that you noted above.
+
+ . SpackRootPath/share/spack/setup-env.sh
+
+
+
+
C. Obtain the Provided Configuration Files (Group)
+
+
i. Copy the configuration files
+
+
To simplify the process of configuring your local installation of Spack, we have provided a folder with the necessary configuration files. All that you need to do is copy it to your home directory using the following command.
+
+
cp -R /software/chtc/el9/spack-user-config/ /home/groups/yourGroupName/.spack
+
+
+
where you need to replace yourGroupName with your group’s name.
+
+
ii. Updating location of configuration files
+
+
The group installation of Spack needs to be instructed on where to find these configuration files. You can do this by running the command
+
+
export SPACK_USER_CONFIG_PATH=/home/groups/yourGroupName/.spack
+
+
+
and Spack should now recognize the packages that are installed system-wide. You can confirm this with the command
+
+
spack find
+
+
+
This should show a list of packages similar to what you see when you run the module avail command.
+
+
To ensure that the configuration files are found in future terminal sessions, you and your group members need to edit your respective ~/.bash_profile files to include the above export command. That is, use a command-line text editor to open the file at ~/.bash_profile and add the following line to the end of the file:
+
+
export SPACK_USER_CONFIG_PATH=/home/groups/yourGroupName/.spack
+
+
+
iii. Updating location of module files
+
+
If you or someone in your group is interested in creating custom modules following the instructions in the guide Creating Custom Modules Using Spack, then you should update the location where the module files will be saved. You can update the location with the following commands
+
+
spack config add 'modules:default:roots:lmod:/home/groups/yourGroupName/spack_modules'
+spack config add 'modules:default:roots:tcl:/home/groups/yourGroupName/spack_modules'
+
+
+
where you replace yourGroupName with your group’s name.
+
+
You are now ready to use Spack for installing the packages that you need! See the instructions in Installing Software Using Spack.
+
+
3. Using a Shared Group Installation
+
+
Users who want to use a shared group installation of Spack, but who did not set up the installation, only need to modify their ~/.bash_profile file with instructions regarding the path to the shared group installation and its configuration files.
+
+
+ - Log in to the HPC cluster (Connecting to CHTC).
+
ssh yourNetID@spark-login.chtc.wisc.edu
+
+
+ -
+
Edit the .bash_profile file in your home directory (/home/yourNetID).
+You should be able to simply add the following two lines to the end of the file
+
+ . /home/groups/yourGroupName/spack/share/spack/setup-env.sh
+export SPACK_USER_CONFIG_PATH=/home/groups/yourGroupName/.spack
+
+
+ where yourGroupName should be replaced with the name of your group. Confirm the exact commands with the user who installed Spack for your group.
+
+
+ You should be able to find the requisite paths if necessary. For the first line, the command
+ find /home/groups/yourGroupName -type d -name spack | grep "share/spack"
+
+
+ should give the path you need; simply add “setup-env.sh” to the end of the path. For the second line, the command
+ find /home/groups/yourGroupName -type d -name .spack | sort -n | head -n 1
+
+
+ should give the path you need. If it doesn’t, try again without | sort -n | head -n 1 to see the full list of matches, and choose the appropriate one.
+
+
+ - Source the
.bash_profile with
+ . ~/.bash_profile
+
+
+ or else close the terminal and log in again.
+
+
+
+
Once configured, you can follow the instructions in our guide Installing Software Using Spack to install or use already-installed packages in Spack.
+
+
A. Switching Between Spack Installations
+
+
You can easily switch between different Spack installations by creating scripts containing the commands listed in Step 2. above, and then sourcing the one that you want to use.
+
+
For example, let’s say you want to use a personal installation of Spack for an independent research project, but want to use a group installation of Spack as part of a collaboration. In that case, you would create two scripts, load-my-spack.sh and load-group-spack.sh, and save them to some central location like ~/bin. In each script, you provide the path to the setup-env.sh file and the .spack configuration directory for the respective Spack installations. The example contents of these scripts are provided below, where you should replace yourNetID with your NetID and yourGroupName with the group name of your collaboration.
+
+
load-my-spack.sh
+
+
. /home/yourNetID/spack/share/spack/setup-env.sh
+export SPACK_USER_CONFIG_PATH=/home/yourNetID/.spack
+
+
+
load-group-spack.sh
+
+
. /home/groups/yourGroupName/spack/share/spack/setup-env.sh
+export SPACK_USER_CONFIG_PATH=/home/groups/yourGroupName/.spack
+
+
+
To activate the your personal Spack installation, simply run
+
+
. path/to/load-my-spack.sh
+
+
+
To activate the group Spack installation, run
+
+
. path/to/load-group-spack.sh
+
+
+
For submitting jobs, remember to load the correct Spack installation at the start of the submission script.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ HPC Computing Guides
+
+
+
+Below is a list of guides for some of the most common tasks our users need to
+carry out as they begin and continue to use the HPC resources at the CHTC.
+
+
+
User Expectations
+
+Read through these user expectations and policies before using CHTC services.
+
+
+
+
+
+
+
HPC Documentation
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
External Documentation
+
+
+
+
+
Icon Credits
+
+
+
+
+
+
+
+
+ HTC System Transition to a New Linux Version (CentOS Stream 9)
+
+
+
Starting in March 2024, CHTC’s high throughput computing (HTC) system began upgrading
+the Linux distribution and version we use on our servers to CentOS Stream 9. This transition is expected to complete in May 2024.
+
+
Note that this page only applies to a transition on the HTC system (submitting jobs
+with HTCondor). The high performance computing (HPC) cluster will be upgraded in
+the near future as well and have a separate transition
+page.
+
+
All updates to the HTC system will be reflected on this page; significant changes may
+also include a notification to the chtc-users mailing list.
+
+
Important Dates
+
+
Note: By default, CHTC-managed submit servers automatically add a job
+requirement that requires jobs to run on servers running our primary operating system unless otherwise specified by the user.
+
+
+ - April 2023: HTC system will support CentOS 7, CentOS Stream 8, and CentOS Stream 9. By default,
+all jobs not using a software container will continue to match to servers running CentOS 8, however,
+users should begin using software containers or testing jobs on servers running CentOS Stream 9.
+ - May 2023: Default operating system requirements for jobs will change from CentOS 8 to
+CentOS Stream 9.
+
+
+
What You Need to Do
+
+
If your jobs use containers (Docker, Singularity/Apptainer)
+
+
No action is needed for researchers already using a Docker or Singularity/Apptainer software containers in their jobs. Becuase software containers have a small operating system installed inside of them, these jobs carry everything they need with them and do not rely signifcantly on the host operating system. By default, your jobs will match to any operating system in the HTC pool, including the new CentOS Stream 9 hosts.
+
+
All other jobs (not using containers)
+
+
Researchers not already using a Docker or Apptainer software container will need to either
+
+
+ - (a) test their software/code on a CentOS Stream 9 machine to see their software needs to be reinstalled. See Transition to New Operating System.
+ - or
+ - (b) switch to using a software container (recommended). See the below for additional information.
+
+
+
If you would like to access as much computing capacity as possible, consider using a Docker or Apptainer software container for your jobs so that your jobs can match to a variety of operating systems. See the information below for detailed instructions on creating and using software containers.
+
+
Options For Transitioning Your Jobs
+
+
+ - Use a Container (recommended)
+ - Transition to New Operating System
+
+
+
Option 1: Use a Software Container (Recommended)
+
+
Using a software container to provide a base version of Linux will allow you to
+run on any nodes in the HTC system regardless of the operating system it is running, and not limit you to a subset of nodes.
+
+
CHTC provides helpful information for learning about creating and using Docker and Apptainer software commands:
+
+
Apptainer
+
+
+
Docker
+
+
+
CHTC users are welcome to reach out to the Facilitation team via email or in office hours for help installing their software into a container.
+
+
Option 2: Transition to a New Operating System
+
+
At any time, you can require a specific operating system
+version (or versions) for your jobs. Instructions for requesting a specific operating system(s) are outlined here:
+
+
+
+
This option is more limiting because
+you are restricted to operating systems used by CHTC, and the number of nodes
+running that operating system.
+
+
Researchers that do not wish to use containers for their job should test their jobs on the CentOS Stream 9 machines as soon as possible so that jobs are not significantly disrupted by this transition. We recommend:
+
+
+ - Test your jobs by requesting the new operating system (
chtc_want_el9 = true ) for 1-2 test jobs. (Note that after May 1, this option will not be required as EL9 will be default.)
+ - If needed, recompile your code.
+
+
+
If you are having trouble getting your jobs to run successfully on the new operating system,
+please contact the facilitation team at chtc@cs.wisc.edu or come to office hours
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Known Issues on the HTC
+
+
+
This page documents some common and known issues encountered on the HTC system. While this page can be beneficial in troubleshooting, it does not contain a comprehensive list of errors.
+
+
Visit our Get Help page to find more resources for troubleshooting.
+
+
+
+
Table of Contents
+
+
+
+
+
+
+
[General] When submitting a job, it doesn't run / goes on hold and shows the error "Job credentials are not available".
+
+
Cause:
+
This is a complicated bug that can strike randomly. We’re working on a fix.
+
Solution:
+
To work around this issue, run the following command on the access point before resubmitting the job.
+
echo | condor_store_cred add-oauth -s scitokens -i -
+
+
+
+
+
[Container] When building an Apptainer, "apt" commands in the %post block fail to run.
+
+
Example error message:
+
Couldn't create temporary file /tmp/apt.conf.9vQdLs for passing config to apt-key
+
+
Cause:
+
The container needs global read/write permissions in order to update or install packages using the apt command.
+
Solution:
+
Add chmod 777 /tmp to the front of your %post block. See the example below:
+
Bootstrap: docker
+From: ubuntu:22.04
+
+%post
+ chmod 777 /tmp
+ apt-get update -y
+
+
We also recommend using the -y option to prevent installation from hanging due to interactive prompts.
+
+
+
+
+
+
Cause:
+
The Docker container is likely built on an Apple computer using an ARM processor, which is incompatible with Linux machines.
+
Solution:
+
To resolve this, when building your Docker container, use the command:
+
docker build --platform linux/amd64 .
+
+
+
+
+
[GPU] My GPU job has been in the queue for a long period of time and is not starting.
+
+
Cause:
+
Jobs default to using CentOS9, but most GPU nodes are currently running CentOS8.
+
Solution:
+
To your submit file, add the following line and resubmit:
+
requirements = (OpSysMajorVer > 7)
+
+
+
+
+
+
Can’t find your issue?
+
Visit our Get Help page.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Use Software Available in Modules
+
+
+
This guide describes when and how to use software, using MPI as an example, that is available as pre-installed modules on the HTC system.
+
+
To best understand the below information, users should already have an
+understanding of:
+
+
+ - Using the command line to: navigate within directories,
+create/copy/move/delete files and directories, and run their
+intended programs (aka "executables").
+ - The CHTC's Intro to Running HTCondor Jobs
+
+
+
Overview
+
+
+ - General Software Policies
+ - Using Pre-Installed Software in Modules
+ - Installing Software on the HTC System
+ - Example: Using an MPI module in HTC Jobs
+
+
+
1. General Software Policies
+
+
In CHTC, we install a minimal set of software for use
+on our systems. On the HTC System, CHTC staff manage installations of
+the following types of programs:
+
+
+ - Compilation tools and common dependencies (e.g. MPI, different GCC versions)
+ - Software that requires a shared license (e.g. COMSOL)
+
+
+
Information on how to access CHTC-managed installations is in the next
+section of this guide.
+
+
2. Using Pre-Installed Software in Modules
+
+
All software on the HTC system that is installed by CHTC staff is available via
+a tool called “modules”.
+
+
A. See Available Software Modules
+
+
There are two ways to search through the software modules on the HTC system:
+
+
+ - View all modules
+ This command will show all software modules available:
+
[alice@submit]$ module avail
+
+
+ - Search for specific modules
+ If you are searching for a specific software module, you can use the
+
module spider command with part of the software name. For example, to
+ search for Open MPI modules, you would type:
+ [alice@submit]$ module spider openmpi
+
+
+
+
+
B. Load Software in Modules
+
+
Once you find a software module that you want to use, you need to “load” it
+into your command line environment to make it active, filling in module_name with the name you found through one of the above steps.
+
+
[alice@submit]$ module load module_name
+
+
+
+ When to Load Modules
+
+ You can load modules to compile code (see below). If you do this, make sure to load
+the same modules as part of your job script before running the main command.
+
+ You can also load modules to run specific software. If done for interactive
+testing, this should be done in an interactive job; otherwise, the module
+should be loaded in the job submit file.
+
+
+
C. Unload Software in Modules
+
+
If you no longer want to use a specific software installation, you can “unload”
+the software module with the following command:
+
+
[alice@submit]$ module unload module_name
+
+
+
If you want to clear your command line environment and start over, run the following:
+
+
[alice@submit]$ module purge
+
+
+
3. Installing Software on the HTC System
+
+
A. Overview
+
+
Unless you are using a licensed software program provided via modules, you
+are able to compile and install the software you need on the HTC System.
+
+
Compilation can be done via an interactive job as described in
+our HTC Compiling or Testing Code with an Interactive Job guide.
+If using CHTC’s provided compilation tools via modules, make
+sure to load the needed modules before compiling and to load the same
+modules in your job submission.
+
+
For groups that would like to share software installations among group
+members, please contact us about getting a shared “group” directory.
+
+
If you are new to software installation, see the section below for
+a more step-by-step description of the process.
+
+
B. Step by Step Process
+
The process for installing software is described in more detail in our Compiling or Testing Code with an Interactive Job
+
+ - Start an Interactive Job - it is necessary to build software in an interactive job as noted in Compiling or Testing Code with an Interactive Job
+ - Download Source Code - download the source code for your desired program.
+ You should only need the source code until the software is properly installed, but if desired, you may keep a zipped copy of
+ the source code in your workspace.
+ - Read the Docs - try to find the installation instructions, either online or
+ in the downloaded source code. In particular, you’ll want to note if there are
+ any special requirements for dependencies like MPI or the compiler needed.
+ - Load Modules - if you are using software modules to help you build your
+ code, load them now. Keep track of what you use so that you can load them
+ in your job submit file later. We also recommend doing a
module purge before
+ loading your compiling modules to make sure you’re starting from a clean environment.
+ - Install - most scientific software follows the three step installation process
+ of
configure - make - make install.
+
+ configure- this step checks for tools and requirements needed to compile
+ the code. This is the step where you set the final installation location of
+ a program. The option for setting this location is typically called the
+ “prefix”; a common syntax is: $ ./configure --prefix=/home/user.
+ This is where you will want to set the installation location to be your
+ /home directory.make - this step compiles and links the code, turning it from human-readable
+ source code to compiled binary code. This is usually the most time consuming
+ step of the installation process.make install - this step copies compiled files to the final installation location
+ (usually specified in the configure step).
+
+
+
+
+
4. Example: Using an MPI module in HTC Jobs
+
+
Below is the process of setting up HTC jobs that use the MPI modules to run. This process can be modified for other software available in modules as well.
+
+
Before you begin, review our below discussion of MPI requirements and
+use cases, to make sure that our multi-core MPI capabilities
+are the right solution for your computing problem.
+
+
Once you know that you need to run multi-core jobs that use MPI on our
+HTC system, you will need to do the following:
+
+
+ - Compile your code using our MPI module system
+ - Create a script to that loads the MPI module you used for
+compiling, and then runs your code
+ - Make sure your submit file has certain key requirements
+
+
+
+
+
A. Requirements and Use Cases
+
+
Most jobs on CHTC's HTC system are run on one CPU (sometimes called a
+"processor", or "core") and can be executed without any special
+system libraries. However, in some cases, it may be advantageous to run
+a single program on multiple CPUs (also called multi-core), in order to
+speed up single computations that cannot be broken up and run as
+independent jobs.
+
+
Running on multiple CPUs can be enabled by the parallel programming
+standard MPI. For MPI jobs to compile and run, CHTC has a set of MPI
+tools installed to a shared location that can be accessed via software
+modules.
+
+
+
+
B. View MPI Modules on the HTC System
+
+
MPI tools are accessible on the HTC system through software "modules",
+which are tools to access and activate a software installation. To see
+which MPI packages are supported in the HTC, you can type the following
+command from the submit server:
+
+
[alice@submit]$ module avail
+
+
+
Your software may require newer versions of MPI libraries than those
+available via our modules. If this is the case, send an email to
+chtc@cs.wisc.edu, to find out if we can install
+that library into the module system.
+
+
C. Submitting MPI jobs
+
+
+
+
1. Compile MPI Code
+
+
You can compile your program by submitting an interactive build job to
+one of our compiling servers. Do not compile code on the submit server,
+as doing so may cause performance issues. The interactive job is
+essentially a regular HTCondor job, but without an executable; you
+are the one running the commands instead (in this case, to compile the
+program).
+
+
Instructions for submitting an interactive build/compile job are
+available on our interactive submission guide.
+The only line in the submit file that you need to change is
+transfer_input_files to reflect all the source files on which your
+program depends. Otherwise, go through the steps described in that guide
+until immediately after running condor_submit -i.
+
+
Once your interactive job begins on one of our compiling servers, you
+can confirm which MPI modules are available to you by typing:
+
+
[alice@build]$ module avail
+
+
+
Choose the module you want to use and load it with the following
+command:
+
+
[alice@build]$ module load mpi_module
+
+
+
where mpi_module is replaced with the name of the MPI module you'd
+like to use.
+
+
After loading the module, compile your program. If your program is
+organized in directories, make sure to create a tar.gz file of
+anything you want copied back to the submit server. Once typing exit
+the interactive job will end, and any *files* created during the
+interactive job will be copied back to the submit location for you.
+
+
If your MPI program is especially large (more than 100 MB, compiled), or
+if it can only run from the exact location to which it was installed,
+you may also need to take advantage of CHTC's shared software location
+or our public web proxy called Squid. Email CHTC's Research Computing
+Facilitators at chtc@cs.wisc.edu if this is the case.
+
+
+
+
2. Script For Running MPI Jobs
+
+
To run your newly compiled program within a job, you need to write a
+script that loads an MPI module and then runs the program, like so:
+
+
#!/bin/bash
+
+# The following three commands are **REQUIRED** to enable modules, and then to load the appropriate MP/MPI module
+export PATH
+. /etc/profile.d/modules.sh
+module load mpi_module
+
+# Untar your program installation, if necessary
+tar -xzf my_install.tar.gz
+
+# Command to run your OpenMP/MPI program
+# (This example uses mpirun, other programs
+# may use mpiexec, or other commands)
+mpirun -np 8 ./path/to/myprogram
+
+
+
Replace mpi_module with the name of the module you used to compile
+your code, myprogram with the name of your program, and X with the
+number of CPUs you want the program to use. There may be additional
+options or flags necessary to run your particular program; make sure to
+check the program's documentation about running multi-core processes.
+
+
+
+
3. Submit File Requirements
+
+
There are several important requirements to consider when writing a
+submit file for multicore jobs. They are shown in the sample submit file
+below and include:
+
+
+ -
+
Require access to MPI modules. To ensure that your job will have
+access to CHTC software modules, including MPI modules, you must
+include the following in your submit file.
+
+ requirements = (HasChtcSoftware == true)
+
+
+ -
+
The script you wrote above (shown as run_mpi.sh below) should be
+your submit file "executable", and your compiled program and any
+files should be listed in transfer_input_files.
+
+
+
+
A sample submit file for multi-core jobs is given below:
+
+
# multicore.sub
+# A sample submit file for running a single multicore (8 cores) job
+executable = run_mpi.sh
+# arguments = (if you want to pass any to the shell script)
+
+## Specify the name of HTCondor's log, standard error, and standard out files
+log = mc_$(Cluster).log
+output = mc_$(Cluster).out
+error = mc_$(Cluster).err
+
+# Tell HTCondor how to handle input files
+should_transfer_files = YES
+transfer_input_files = (this should be a comma separate list of input files if needed)
+
+# Requirement for accessing new set of modules
+requirements = ( HasChtcSoftware == true )
+
+## Request resources needed by your job
+request_cpus = 8
+request_memory = 8GB
+request_disk = 2GB
+
+queue
+
+
+
After the submit file is complete, you can submit your jobs using
+condor_submit.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ HTC System Overview
+
+
+
+
+
+
Table of Contents
+
+
+
+
+
+
High-Throughput Computing at CHTC
+
+
The CHTC high-throughput computing (HTC) cluster provides support a variety of computational research tasks. The HTC system offers CPUs/GPUs, high-memory nodes, and other specialized hardware. Workflows that run well on this system include RNA/DNA sequencing, machine learning workflows, weather modeling, monte carlo simulations, etc.
+
+
To get access to the HTC System, please complete our
+New User Consultation Form. After your request is received,
+a Research Computing Facilitator will follow up to discuss the computational needs
+of your research and connect you with computing
+resources (including non-CHTC services) that best fit your needs.
+
+
+
+
HTC System User Policies
+
+
See our User Policies and Expectations for details on general CHTC policies.
+
+
+
+
HTC System Specific Policies
+
+
Below are some of the default limits on CHTC’s HTC system. Note that as a large-scale
+computing center, we want you to be able to run at a large scale - often much larger
+than these defaults. Please contact the facilitation team whenever you encounter one
+of these limits so we can adjust your account settings or discuss alternative ways to
+achieve your computing goals.
+
+
+ - Jobs with long runtimes. There is a default run limit of 72
+hours for each job queued in the HTC System, once it starts running.
+Jobs longer than this will be placed in HTCondor’s “hold” state.
+If your jobs will be longer, contact the CHTC facilitation team, and we’ll help you to determine the
+best solution.
+ - Submitting many jobs from one submit file. HTCondor is designed
+to submit thousands (or more) jobs from one submit file. If you are
+submitting over 10,000 jobs per submit file or want to queue
+more than 50,000 total jobs as a single user,
+please email us as we have strategies to
+submit that many jobs in a way that will ensure you have as many
+jobs running as possible without also compromising queue performance.
+ - Submitting many short jobs from one submit file. While HTCondor
+is designed to submit thousands of jobs at a time, many short jobs
+can overwhelm the submit server, resulting in other jobs taking much
+longer to start than usual. If you plan on submitting over
+1000 jobs per submit file, we ask that you ensure each job has a
+minimum run time of 5 minutes (on average).
+ - The default disk quota is 20 GB in your
/home directory, as a
+starting point. You can track your use of disk space and your quota value,
+using our Quota Guide. If you need more space
+for concurrent work, please send an email to chtc@cs.wisc.edu.
+ - Submitting jobs with "large" files: HTCondor's
+normal file transfer mechanism ("transfer_input_files") is good for
+files up to 100MB in size (or 500MB total, per job). For jobs with larger
+files, please see our guide on File Availability
+Options, and contact us to make arrangements.
+
+
+
+
+
HTC Hardware and Configuration
+
+
The HTC System consists of several submit servers and many compute (aka execute)
+nodes. All users log in at a login node, and submit their workflow as HTCondor jobs that run on execute points.
+
+
+
+
HTC Operating System and Software
+
+
Submit servers in the HTC System are running CentOS 7 Linux.
+
+
Due to the distributed and independent nature of the HTC system’s execute points, there can be a variety of operating systems on the pool of execution point resources (especially for users that opt into running jobs on the globally available OSPool operated by the OSG). However, the default operating system is CentOS 8 Stream Linux unless users request to run on a different operating system using their HTCondor submit file.
+
+
The HTC system is a test bed for the HTCondor Software Suite, and thus is typically running the latest or soon-to-be-released versions of HTCondor.
+
+
To see more details of other software on the cluster, see our HTC Guides page.
+
+
+
+
HTC Submit Servers
+
+
There are multiple submit servers for the HTC system. The two most common submit servers are ap2001.chtc.wisc.edu and ap2002.chtc.wisc.edu (formerly submit1.chtc.wisc.edu and submit2.chtc.wisc.edu, respectively). All users will be notified what submit server they should log into when their account is created.
+
+
+
+
HTC Execute Nodes
+
+
Only execute nodes will be used for performing your computational work.
+
+
By default, when users submit HTCondor jobs, their jobs will only run on execute points owned and managed by CHTC staff. As of January 2024, there are approximately 40,000 CPU slots and 80+ GPU slots available in the CHTC execute pool.
+
+
Some users, particularly those requesting GPUs, may wish to access additional execute points so that they may have more jobs running simultantiously. HTC users can opt in to allowing their jobs to run on additional execute points not owned or managed by CHTC staff. There are two additional execute pools that users can opt into using: the UW Grid and the OSG’s OSPool. There are many advantages to opting into running on these execute pools, such as accessing more GPUs, accessing different computer architectures, and having more jobs running in parallel. However, because these machines are not managed by CHTC and thus are backfilling on hardware owned by other entities, it is recommended that users only opt into using these resources if they have short (<~10 hours), inturruptable jobs. For more information, see the Scaling Beyond Local HTC Capacity guide.
+
+
Fair Share Allocation
+
+
To promote fair access to HTC computing resources, all users are subject to a fair-share policy. This “fair-share” policy means that users who have run many jobs in the near-past will have a lower priority, and users with little recent activity will see their waiting jobs start sooner.
+(The HTC system does not have a strict “first-in-first-out” queue policy.)
+
+
Resource requests will also impact the number of jobs a user has running. Smaller jobs (those requesting smaller amounts of CPUs, memory, and disk) as well as more flexible jobs (those requesting to use a variety of GPUs instead of a specific GPU type) are able to match to more execute points than larger, less flexible jobs. Thus, these jobs will start sooner and more jobs will run in parallel.
+
+
+
Data Storage and Management
+
+
Data space in the HTC system is not backed-up and should be
+treated as temporary by users. Only files necessary for
+actively-running jobs should be kept on the filesystem, and files
+should be removed from the system when jobs complete. A primary copy of any
+essential files (e.g. software, submit files, input) should be kept in an
+alternate, non-CHTC storage location.
+
+
CHTC Staff reserve the right to remove any significant amounts of data
+on the HTC System in our efforts to maintain filesystem performance
+for all users.
+
+
+
+
+
+
+
Check /home Quota and Usage
+
To see what disk and items quotas are currently set for your /home direcotry, use the
+quota -vs command. See the example below:
+
+
[alice@submit]$ quota -vs
+Disk quotas for user alice (uid 20384):
+ Filesystem space quota limit grace files quota limit grace
+ /dev/sdb1 12690M 20480M 30720M 161k 0 0
+
+
+
The output will list your total data usage under blocks on the /dev/sbd1 filesystem that manages user /home data:
+
+ space (MB): the amount of disk space you are currently usingquota (MB): your soft quota. This is the value we recommend you consider to be your “quota”.limit (MB): the hard limit or absolute maximum amount of space you can use. This value is almost always 10GB larger than your soft quota, and is only provided as a helpful spillover space. Once you hit this hard limit value, you and your jobs will no longer be allowed to save data.files: the number of files in your /home directory. /home does not typically restrict the number of files a user can have, which is why there are no values for file quota and limit
+
+
Each of the disk space values are given in megabytes (MB), which can be converted to gigabytes (GB) by dividing by 1024.
+
+
+
+
Check /staging Quota and Usage
+
+
To see your /staging quota and usage, use the get_quotas <NetID> command. For example,
+
[NetID@ap2001 ~]$ get_quotas /staging/NetID
+
+
+
If the output of this command is blank, it means you do not have a /staging directory. Contact CHTC staff to request one at any time.
+
+
+
+
Alternative Commands to Check Quotas
+
Alternatively, the ncdu command can also be used to see how many
+files and directories are contained in a given path:
+
+
[NetID@ap2001 ~]$ ncdu /home/NetID
+[NetID@ap2001 ~]$ ncdu /staging/NetID
+
+
+
When ncdu has finished running, the output will give you a total file
+count and allow you to navigate between subdirectories for even more
+details. Type q when you're ready to exit the output viewer. More
+info here: https://lintut.com/ncdu-check-disk-usage/
+
+
+
Request a Quota Increase
+
Increased quotas on either of these locations are available upon email
+request to chtc@cs.wisc.edu after a user has
+cleared out old data and run relevant test jobs to inform the request. In your request,
+please include both size (in GB) and file/directory counts. If you don't
+know how many files your installation creates, because it's more than
+the current items quota, simply indicate that in your request.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+Introduction to the High Throughput Computing Strategy
+
+Like nearly all large-scale compute systems, users of both CHTC's High Throughput and High Performance systems prepare their computational work and submit them as tasks called "jobs" to run on execution points.
+
+
+High Throughput Computing systems specialize in running many small, independent jobs (< ~20 CPUs/job). On the other hand, High Performance Computing systems speicalize in running a few, very large jobs that run on more than one node (~30+ CPUs/job).
+
+
+It is best to keep this distinction in mind when setting up your jobs. On the HTC system, smaller jobs (i.e., those requesting smaller amounts of CPU, memory, and disk resources per job) are easier to find a slot to run on. This means that users will notice they will have jobs start quicker and will have more running simultanioutsly. It is almost always beneficial to break up your analysis pipeline into smaller pieces to take advantage of getting more jobs up and running, quicker.
+
+
+Unlike the High Performance System, CHTC staff do not limit the number of jobs a user can have running in parallel, thus it is to your advantage to strategize your workflow to take advantage of as many resources as possible.
+
+
+More detailed information regarding CHTC's HTC system can be found in the HTC Overview Guide.
+
+
+
+Step Two
+
+
+Log on to a HTC System Access Point
+
+Once your request for an account has been approved by a Research Computing Facilitator, you will be emailed your login information.
+
+
+For security purposes, every CHTC user is required to be connected to either a University of Wisconsin internet network or campus VPN and to use two-factor authentication when logging in to your CHTC "access point" (also called a "submit server").
+
+
+
+
+Step Three
+
+
+Understand the Basics of Submitting HTCondor Jobs
+
+Computational work is run on the CHTC's execution machines by submitting it as “jobs” to the HTCondor job scheduler. Before submitting your own computational work, it is necessary to understand how HTCondor job submission works. The following guide is a short step-by-step tutorial on how to submit basic HTCondor jobs: Practice: Submit HTC Jobs using HTCondor. It is highly recommended that every user follow this short tutorial as these are the steps you will need to know to complete your own analyses.
+
+
+
+Step Four
+
+
+Learn to Run Many HTCondor Jobs using one Submit File
+
+After following this tutorial, we highly recommend users review the Easily Submit Multiple Jobs guide to learn how you can configure HTCondor to automatically pass files or parameters to different jobs, return output to specific directories, and other easily automated organizational behaviors.
+
+
+
+
+Step Five
+
+
+Access your Data on the HTC System
+
+Upload your data to CHTC
+
+When getting started on the HTC system, it is typically necessary to upload your data files to our system so that they can be used in jobs. For users that do not want to upload data to our system, it is possible to configure your HTCondor jobs to pull/push files using `s3` file transfer, or pull data using standard unix commands (`wget`).
+
+
+To learn how to upload data from different sources, including your laptop, see:
+
+
+Choose a Location to Stage your Data
+
+When uploading data to the HTC system, users need to choose a location to store that data on our system. There are two primary locations: `/home` and `/staging`.
+
+`/home` is more efficient at handling "small" files, while `/staging` is more efficient at handling "large" files. For more information on what is considered "small" and "large" data files and to learn how to use files stored in these locations for jobs, visit our HTC Data guides.
+
+
+
+
+Step Six
+
+
+Install your Software
+
+Our “Software Solutions” guides contain information about how to install and use software on the HTC system.
+
+
+Software Containers
+
+In general, we recommend installing your software into a "container" if your software relies on a specific version of R/Python, can be installed with `conda`, if your software has many dependencies, or if it already has a pre-existing container (which many common software packages do). There are many advantages to using a software container; one example is that software containers contain their own operating system. As a result, jobs with software containers have the most flexibility with where they run on CHTC or the OSPool. The CHTC website provides several guides on building, testing, and using software containers.
+
+
+Use Pre-installed Software in Modules
+
+CHTC's infrastructure team has provided a limited collection of software as modules, which users can load and then use in their jobs. This collection includes tools shared across domains, including COMSOL, ANSYS, ABAQUS, GUROBI, and others. To learn how to load these software into your jobs, our Use Software Available in Modules and Use Licensed Software guides.
+
+
+Access Software Building Tools on CHTC's Software Building Machines
+
+The HTC system contains several machines designed for users to use when building their software. These machines have access to common compilers (e.g., gcc) that are necessary to install many software packages. To learn how to submit an interactive job to log into these machines to build your software, see Compiling or Testing Code with an Interactive Job.
+
+
+
+Step Seven
+
+
+Run Test Jobs
+
+Once you have your data, software, code, and HTCondor submit file prepared, you should submit several test jobs. The table created by HTCondor in the `.log` file will help you determine the amount of resources (CPUs/GPUs, memory, and disk) your job used, which is beneficial for understanding future job resource requests as well as troubleshooting. The `.out` file will contain all text your code printed to the terminal screen while running, while the `.err` file will contain any standard errors that your software printed out while running.
+
+
+Things to look for:
+
+ - Jobs being placed on hold (hold messages can be viewed using `condor_q jobID -hold`)
+ - Jobs producing expected your desired files
+ - Size and number of output files (to make sure output is being directed to the correct location and that your quota is sufficient for all of your output data as you submit more jobs)
+
+
+
+
+ Step Eight
+
+ Submit Your Workflow
+
+Once your jobs succeed and you have confirmed your quota is sufficient to store the files your job creates, you are ready to submit your full workflow. For researchers interested in queuing many jobs or accessing GPUs, we encourage you to consider accessing additional CPUs/GPUs outside of CHTC. Information is provided in the following step.
+
+
+ Step Nine
+
+ Access Additional Compute Capacity
+
+ Researchers with jobs that run for less than ~10 hours, use less than ~20GB of data per job, and do not require CHTC modules, can take advantage of additional CPUs/GPUs to run there jobs. These researchers can typically expect to have more jobs running simultaneously.
+
+
+ To opt into using this additional capacity, your jobs will run on hardware that CHTC does not own. Instead, your jobs will "backfill" on resources owned by research groups, UW-Madison departments and organizations, and a national scale compute system: the OSG's Open Science Pool. This allows researchers to access capacity beyond what CHTC can provide. To learn how to take advantage of additional CPUs/GPUs, visit Scale Beyond Local HTC Capacity.
+
+
+Step Ten
+
+ Move Your Data off CHTC
+
+ Data stored on CHTC systems is not backed up. While CHTC staff try to maintain a stable compute environment, it is possible for unexpected outages to occur that may impact your data on our system. We highly recommend all CHTC users maintain copies of important scripts and input files on another compute system (your laptop, lab server, ResearchDrive, etc.) throughout their analysis. Additionally, as you complete your analysis on CHTC servers, we highly recommend you move your data off our system to a backed up storage location.
+
+
+ CHTC staff periodically delete data of users that have not logged in or submitted jobs in several months to clear up space for new users. Eventually, all users should expect their data to be deleted off CHTC servers and should plan accordingly. Data on CHTC is meant to be used for analyses actively being carried out - CHTC is not a long-term storage solution for your data storage needs.
+
+
diff --git a/preview-fall2024-info/uw-research-computing/htc/guides.html b/preview-fall2024-info/uw-research-computing/htc/guides.html
new file mode 100644
index 000000000..c199f99f4
--- /dev/null
+++ b/preview-fall2024-info/uw-research-computing/htc/guides.html
@@ -0,0 +1,632 @@
+
+
+
+
+
+
+HTC Computing Guides
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ HTC Computing Guides
+
+
+
+Below is a list of guides for some of the most common tasks our users need to
+carry out as they begin and continue to use the HTC resources at the CHTC.
+
+
+
User Expectations
+
+Read through these user expectations and policies before using CHTC services.
+
+
+
+
+
+
+
HTC Documentation
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
Icon Credits
+
+
+
+
+
+
+
+
+ Practice: Submit HTC Jobs using HTCondor
+
+
+
Purpose
+
+
This guide discusses how to run jobs on the CHTC using HTCondor.
+
+
Workflow Overview
+
+
The process of running computational workflows on CHTC resources follows the following outline:
+
+

+
+
Terminology:
+
+
+ - Access point is where you login and stage your data, executables/scripts, and software to use in jobs.
+ - HTCondor is a job scheduling software that will run your jobs out on the execution points.
+ - The Execution Points is the set of resources your job runs on. It is composed of servers, as well as other technologies, that compose the cpus, memory, and disk space that will run the computations of your jobs.
+
+
+
Run Jobs using HTCondor
+
+
We are going to run the traditional ‘hello world’ program with a CHTC twist. In order to demonstrate the distributed resource nature of CHTC’s HTC System, we will produce a ‘Hello CHTC’ message 3 times, where each message is produced within is its own ‘job’. Since you will not run execution commands yourself (HTCondor will do it for you), you need to tell HTCondor how to run the jobs for you in the form of a submit file, which describes the set of jobs.
+
+
+ Note: You must be logged into a CHTC Access Point for the following example to work.
+
+
+
Prepare job executable and submit file on an Access Point
+
+
+ -
+
First, create the executable script you would like HTCondor to run.
+For our example, copy the text below and paste it into a file called hello-world.sh (we recommend using a command line text editor) in your home directory.
+
+ #!/bin/bash
+#
+# hello-world.sh
+# My CHTC job
+#
+# print a 'hello' message to the job's terminal output:
+echo "Hello CHTC from Job $1 running on `whoami`@`hostname`"
+#
+# keep this job running for a few minutes so you'll see it in the queue:
+sleep 180
+
+
+ This script would be run locally on our terminal by typing hello-world.sh <FirstArgument>.
+However, to run it on CHTC, we will use our HTCondor submit file to run the hello-world.sh executable and to automatically pass different arguments to our script.
+
+ -
+
Prepare your HTCondor submit file, which you will use to tell HTCondor what job to run and how to run it.
+Copy the text below, and paste it into file called hello-world.sub.
+This is the file you will submit to HTCondor to describe your jobs (known as the submit file).
+
+ # hello-world.sub
+# My HTCondor submit file
+
+# Specify your executable (single binary or a script that runs several
+# commands) and arguments to be passed to jobs.
+# $(Process) will be a integer number for each job, starting with "0"
+# and increasing for the relevant number of jobs.
+executable = hello-world.sh
+arguments = $(Process)
+
+# Specify the name of the log, standard error, and standard output (or "screen output") files. Wherever you see $(Cluster), HTCondor will insert the
+# queue number assigned to this set of jobs at the time of submission.
+log = hello-world_$(Cluster)_$(Process).log
+error = hello-world_$(Cluster)_$(Process).err
+output = hello-world_$(Cluster)_$(Process).out
+
+# This line *would* be used if there were any other files
+# needed for the executable to use.
+# transfer_input_files = file1,/absolute/pathto/file2,etc
+
+# Tell HTCondor requirements (e.g., operating system) your job needs,
+# what amount of compute resources each job will need on the computer where it runs.
+request_cpus = 1
+request_memory = 1GB
+request_disk = 5GB
+
+# Tell HTCondor to run 3 instances of our job:
+queue 3
+
+
+ By using the “$1” variable in our hello-world.sh executable, we are telling HTCondor to fetch the value of the argument in the first position in the submit file and to insert it in location of “$1” in our executable file.
+
+ Therefore, when HTCondor runs this executable, it will pass the $(Process) value for each job and hello-world.sh will insert that value for “$1” in hello-world.sh.
+
+ More information on special variables like “$1”, “$2”, and “$@” can be found here.
+
+ -
+
Now, submit your job to HTCondor’s queue using condor_submit:
+
+ [alice@ap2002]$ condor_submit hello-world.sub
+
+
+ The condor_submit command actually submits your jobs to HTCondor. If all goes well, you will see output from the condor_submit command that appears as:
+
+ Submitting job(s)...
+3 job(s) submitted to cluster 36062145.
+
+
+ -
+
To check on the status of your jobs in the queue, run the following command:
+
+ [alice@ap2002]$ condor_q
+
+
+ The output of condor_q should look like this:
+
+ -- Schedd: ap2002.chtc.wisc.edu : <128.104.101.92:9618?... @ 04/14/23 15:35:17
+OWNER BATCH_NAME SUBMITTED DONE RUN IDLE TOTAL JOB_IDS
+Alice ID: 3606214 4/14 12:31 2 1 _ 3 36062145.0-2
+
+3 jobs; 2 completed, 0 removed, 0 idle, 1 running, 0 held, 0 suspended
+
+
+ You can run the condor_q command periodically to see the progress of your jobs.
+By default, condor_q shows jobs grouped into batches by batch name (if provided), or executable name.
+To show all of your jobs on individual lines, add the -nobatch option.
+
+ -
+
When your jobs complete after a few minutes, they’ll leave the queue.
+If you do a listing of your /home directory with the command ls -l, you should see something like:
+
+ [alice@submit]$ ls -l
+total 28
+-rw-r--r-- 1 alice alice 0 Apr 14 15:37 hello-world_36062145_0.err
+-rw-r--r-- 1 alice alice 60 Apr 14 15:37 hello-world_36062145_0.out
+-rw-r--r-- 1 alice alice 0 Apr 14 15:37 hello-world_36062145_0.log
+-rw-r--r-- 1 alice alice 0 Apr 14 15:37 hello-world_36062145_1.err
+-rw-r--r-- 1 alice alice 60 Apr 14 15:37 hello-world_36062145_1.out
+-rw-r--r-- 1 alice alice 0 Apr 14 15:37 hello-world_36062145_1.log
+-rw-r--r-- 1 alice alice 0 Apr 14 15:37 hello-world_36062145_2.err
+-rw-r--r-- 1 alice alice 60 Apr 14 15:37 hello-world_36062145_2.out
+-rw-r--r-- 1 alice alice 0 Apr 14 15:37 hello-world_36062145_2.log
+-rw-rw-r-- 1 alice alice 241 Apr 14 15:33 hello-world.sh
+-rw-rw-r-- 1 alice alice 1387 Apr 14 15:33 hello-world.sub
+
+
+ Useful information is provided in the user log, standard error, and standard output files.
+
+ HTCondor creates a transaction log of everything that happens to your jobs.
+Looking at the log file is very useful for debugging problems that may arise.
+Additionally, at the completion of a job, the .log file will print a table describing the amount of compute resources requested in the submit file compared to the amount the job actually used.
+An excerpt from hello-world_36062145_0.log produced due the submission of the 3 jobs will looks like this:
+
+ …
+005 (36062145.000.000) 2023-04-14 12:36:09 Job terminated.
+ (1) Normal termination (return value 0)
+ Usr 0 00:00:00, Sys 0 00:00:00 - Run Remote Usage
+ Usr 0 00:00:00, Sys 0 00:00:00 - Run Local Usage
+ Usr 0 00:00:00, Sys 0 00:00:00 - Total Remote Usage
+ Usr 0 00:00:00, Sys 0 00:00:00 - Total Local Usage
+ 72 - Run Bytes Sent By Job
+ 265 - Run Bytes Received By Job
+ 72 - Total Bytes Sent By Job
+ 265 - Total Bytes Received By Job
+ Partitionable Resources : Usage Request Allocated
+ Cpus : 0 1 1
+ Disk (KB) : 118 1024 1810509281
+ Memory (MB) : 54 1024 1024
+
+ Job terminated of its own accord at 2023-04-14T17:36:09Z with exit-code 0.
+
+
+ And, if you look at one of the output files, you should see something like this:
+Hello CHTC from Job 0 running on alice@e389.chtc.wisc.edu.
+
+
+
+
Congratulations. You’ve run an HTCondor job!
+
+
Important Workflow Elements
+
+
A. Removing Jobs
+
+
To remove a specific job, use condor_rm <JobID, ClusterID, Username>.
+Example:
+
+
[alice@ap2002]$ condor_rm 845638.0
+
+
+
B. Importance of Testing & Resource Optimization
+
+
+ -
+
Examine Job Success Within the log file, you can see information about the completion of each job, including a system error code (as seen in “return value 0”).
+You can use this code, as well as information in your “.err” file and other output files, to determine what issues your job(s) may have had, if any.
+
+ -
+
Improve Efficiency Researchers with input and output files greater than 1GB, should store them in their /staging directory instead of /home to improve file transfer efficiency.
+See our data transfer guides to learn more.
+
+ -
+
Get the Right Resource Requests
+Be sure to always add or modify the following lines in your submit files, as appropriate, and after running a few tests.
+
+
+
+ | Submit file entry |
+ Resources your jobs will run on |
+
+
+ | request_cpus = cpus |
+ Matches each job to a computer "slot" with at least this many CPU cores. |
+
+
+ | request_disk = kilobytes |
+ Matches each job to a slot with at least this much disk space, in units of KB. |
+
+
+ | request_memory = megabytes |
+ Matches each job to a slot with at least this much memory (RAM), in units of MB. |
+
+
+
+ -
+
Determining Memory and Disk Requirements.
+The log file also indicates how much memory and disk each job used, so that you can first test a few jobs before submitting many more with more accurate request values.
+When you request too little, your jobs will be terminated by HTCondor and set to “hold” status to flag that job as requiring your attention.
+To learn more about why a job as gone on hold, use condor_q -hold.
+When you request too much, your jobs may not match to as many available “slots” as they could otherwise, and your overall throughput will suffer.
+
+
+
+
You have the basics, now you are ready to run your OWN jobs!
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
Your Home for
Research Computing
+
+
+
+ Join Hundreds of UW-Madison Researchers using CHTC Resources to Complete:
+
+
+
+
+ RNA/DNA Sequencing Analyses
+
+
+
+ Machine Learning Workflows
+
+
+
+ Economic Simulations & Predictions
+
+
+
+ Weather Modeling Analyses
+
+
+
+ Chemical Reaction Predictions
+
+
+
+ Computer Vision
+
+
+
+ Artificial Intelligence
+
+
+
+ Modeling & Decision Making
+
+
+
+ Forestry and Wildlife Analyses
+
+
+
+
+
+ Research Transformed Annually
+
+
+
+
+
+
+
Who We Are
+
+ We are the University of Wisconsin-Madison's core computational service provider for large scale computing.
+ CHTC services are open to UW-Madison staff, students, faculty, and external collaborators.
+
+ We offer both a High Throughput Computing system and a High Performance Computing cluster.
+ Access to CPUs/GPUs, high-memory servers, data storage capacity, as well as personalized consultations and classroom support,
+ are provided at no-cost.
+
+
+
Departments with Researchers using CHTC Services
+
+
+
+
+
+
+ Impact Stories
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+  +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Compiling or Testing Code with an Interactive Job
+
+
+
To best understand the below information, users should already have an
+understanding of:
+
+
+
+
Overview
+
+
This guide provides a generic overview of steps required to install
+scientific software for use in CHTC. If you are using Python, R, or
+Matlab, see our specific installation and use guides here: Guides for
+Matlab, Python and R.
+
+
It is helpful to understand a little bit about normal “batch” HTCondor jobs
+before submitting interactive jobs. Just like batch jobs, interactive jobs
+can transfer input files (usually copies of source code or the software you
+want to install) and will transfer new/updated files in the main working directory
+back to the submit node when the job completes.
+
+
+ One exception to the file transfers working as usual is when running an interactive
+job that uses a Docker container. If any output files are generated inside an
+interactive Docker job, they will not be copied back to the submit node when you
+exist the interactive job. Contact the facilitation team for workarounds to this behavior.
+
+
+
+
+
1. Building a Software Installation
+
+
You are going to start an interactive job that runs on the HTC build
+servers. You will then install your packages to a folder and zip those
+files to return to the submit server.
+
+
+
+
A. Submit an Interactive Job
+
+
First, download the source code for your software to the submit server.
+Then create the following special submit file on the submit server,
+calling it something like build.sub.
+
+
Note that you’ll want to use +IsBuildJob = true to specifically match to CHTC’s servers designated for compiling code (which include Matlab compilers and other compiling tools you may need). Compiling servers do not include specialized resources like GPUs, extreme amounts of RAM/disk, etc.; to build/test software in these cases, submit an interactive job without +IsBuildJob.
+
+
# Software build file
+
+universe = vanilla
+log = interactive.log
+
+# In the latest version of HTCondor on CHTC, interactive jobs require an executable.
+# If you do not have an existing executable, use a generic linux command like hostname as shown below.
+executable = /usr/bin/hostname
+
+# change the name of the file to be the name of your source code
+transfer_input_files = source_code.tar.gz
+
++IsBuildJob = true
+# requirements = (OpSysMajorVer =?= 8)
+request_cpus = 1
+request_memory = 4GB
+request_disk = 2GB
+
+queue
+
+
+
The only thing you should need to change in the above file is the name
+of the source code tar.gz file - in the "transfer_input_files"
+line.
+
+
Once this submit file is created, you will start the interactive job by
+running the following command:
+
+
[alice@submit]$ condor_submit -i build.sub
+
+
+
The interactive build job should start in about a minute. Once it has
+started, the job has a time limit of four hours - if you need more time
+to compile a particular code, talk to CHTC's Research Computing
+Facilitators.
+
+
B. Install the Software
+
+
Software installation typically goes through a set of standard steps --
+configuration, then compilation (turning the source code into binary
+code that the computer can understand), and finally "installation",
+which means placing the compiled code into a specific location. In most
+install instructions, these steps look something like:
+
+
./configure
+make
+make install
+
+
+
There are two changes we make to this standard process. Because you are
+not an administrator, you will want to create a folder for the
+installation in the build job's working directory and use an option in
+the configuration step that will install the software to this folder.
+
+
In what follows, note that anything in italics is a name that you can
+(and should!) choose to be more descriptive. We use general names as
+an example; see the LAMMPS case study lower down to see what you might
+fill in for your own program.
+
+
+ -
+
In the interactive job, create a new directory to hold your final
+software installation:
+
+ [alice@build]$ mkdir program
+
+
+ -
+
You'll also want to un-tar the source code that you brought along,
+and cd into the source code folder.
+
+ [alice@build]$ tar -xzf source_code.tar.gz
+[alice@build]$ cd source_code/
+
+
+ -
+
Our next step will be to configure the installation. This involves
+changing into the un-tarred source code directory, and running a
+configuration script. It's at this step that we change the final
+installation location of the software from its default, to be the
+directory we created in the previous step. In a typical configure
+script, this option is called the "prefix" and is given by the
+--prefix flag.
+
+ [alice@build]$ ./configure --prefix=$_CONDOR_SCRATCH_DIR/program
+
+
+ Note that there are sometimes different options used. Some program
+use a helper program called cmake as their configuration script.
+Often the installation instructions for a program will indicate what
+to use as a prefix option, or, you can often run the configure
+command with the --help flag, which will have all the options
+which can be added to the configure command.
+
+ -
+
After the configuration step, you'll run the steps to compile and
+install your program. This is usually these two commands:
+
+ [alice@build]$ make
+[alice@build]$ make install
+
+
+ -
+
After this step, you can cd back up to the main working directory.
+
+ [alice@build]$ cd ..
+
+
+ -
+
Right now, if we exit the interactive job, nothing will be
+transferred back because we haven't created any new files in
+the working directory, just the new sub-folder with our software
+installation. In order to transfer back our installation, we will
+need to compress it into a tarball file - not only will HTCondor
+then transfer back the file, it is generally easier to transfer a
+single, compressed tarball file than an uncompressed set of
+directories.
+
+ Run the following command to create your own tarball of your
+packages:
+
+ [alice@build]$ tar -czf program.tar.gz program/
+
+
+
+
+
We now have our packages bundled and ready for CHTC! You can now exit
+the interactive job and the tar.gz file with your software installation
+will return to the submit server with you (this sometimes takes a few
+extra seconds after exiting).
+
+
[alice@build]$ exit
+
+
+
+
+
2. Case Study, Installing LAMMPS
+
+
First download a copy of LAMMPS and copy it to the submit server -- in
+this example, we've used the "stable" version under "Download a
+tarball": LAMMPS download
+page
+
+
Then, make a copy of the submit file above on the submit server,
+changing the name of the file to be transferred to
+lammps-stable.tar.gz. Submit the interactive job as described.
+
+
While waiting for the interactive build job to start, take a look at the
+installation instructions for LAMMPS:
+
+
+
+
You'll see that the install instructions have basically the same steps
+as listed above, with two changes:
+
+
+ -
+
Instead of the "configure" step, LAMMPS is using the "cmake"
+command. This means that we'll need to find the equivalent to the
+--prefix option for cmake. Reading further down in the
+documentation, you can see that there's this option:
+
+ -D CMAKE_INSTALL_PREFIX=path
+
+
+ This is exactly what we need to set the installation prefix.
+
+ -
+
There's extra steps before the configure step -- that's fine,
+we'll just add them to our list of commands to run.
+
+
+
+
With all these pieces together, this is what the commands will look like
+to install LAMMPS in the interactive build job and then bring the
+installed copy back to the submit server.
+
+
Create the folder for the installation:
+
+
[alice@build]$ mkdir lammps
+
+
+
Unzip and cd into a build directory:
+
+
[alice@build]$ tar -xf lammps-stable.tar.gz
+[alice@build]$ cd lammps-stable
+[alice@build]$ mkdir build; cd build
+
+
+
Run the installation commands:
+
+
[alice@build]$ cmake -D CMAKE_INSTALL_PREFIX=$_CONDOR_SCRATCH_DIR/lammps ../cmake
+[alice@build]$ make
+[alice@build]$ make install
+
+
+
Move back into the main job directory and create a tar.gz file of the
+installation folder.
+
+
[alice@build]$ cd ../..
+[alice@build]$ tar -czf lammps.tar.gz lammps
+[alice@build]$ exit
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Running Java Jobs
+
+
+
Quickstart: Java
+
+
To use Java on the HTC system, we recommend that you use the Java Development Kit (JDK).
+
+
+ -
+
Obtain a copy of the pre-compiled JDK for “Linux/x64” from https://jdk.java.net/.
+
+ -
+
Include the JDK .tar.gz file in your submit file with the list of files to be transferred:
+
+ transfer_input_files = openjdk-22_linux-x64_bin.tar.gz, program.jar
+
+
+ -
+
Include instructions for using the JDK in your executable file:
+
+ #!/bin/bash
+
+tar -xzf openjdk-22_linux-x64_bin.tar.gz
+export JAVA_HOME=$PWD/jdk-22
+export PATH=$JAVA_HOME/bin:$PATH
+
+java -jar program.jar
+
+
+
+
+
+
+
+
+
To obtain your copy of the Java Development Kit (JDK), go to https://jdk.java.net/.
+Click the link for the JDK that is “Ready for use”.
+There will be a download link “tar.gz” under the “Builds” section for “Linux/x64”.
+You can then either (a) right-click the download link and copy the link address, sign in to the submit server, and use the wget command with that link,
+or (b) click the link to download to your computer, then manually upload the file from your computer to the submit server.
+
+
The example above uses file names for JDK 22 as of 2024-04.
+Be sure to change the file names for the version that you actually use.
+We recommend that you test and explore your setup using an interactive job.
+
+
Executable
+
+
A bash .sh file is used as the executable file in order to unpack and set up the JDK environment for use by your script.
+Here is the executable from the section with comments:
+
+
#!/bin/bash
+
+# Decompress the JDK
+tar -xzf openjdk-22_linux-x64_bin.tar.gz
+
+# Add the new JDK folder to the bash environment
+export JAVA_HOME=$PWD/jdk-22
+export PATH=$JAVA_HOME/bin:$PATH
+
+# Run your program
+java -jar program.jar
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Running Julia Jobs
+
+
+
Quickstart: Julia
+
+
Option A (recommended)
+
+
Build a container with Julia & packages installed inside:
+
+
+ - How to build your own container
+ - Example container recipes for Julia
+ - Use your container in your HTC jobs
+
+
+
Option B
+
+
Use a portable copy of Julia and create your own portable copy of your Julia packages:
+
+
+ - Follow the instructions in our guide Run Julia Jobs.
+
+
+
+ This approach may be sensitive to the operating system of the execution point.
+We recommend building a container instead, but are keeping these instructions as a backup.
+
+
+
+
+
+
+
No CHTC machine has Julia pre-installed, so you must configure a portable copy of Julia to work on the HTC system.
+Using a container as described above is the easiest way to accomplish this.
+
+
Executable
+
+
When using a container, you can use a .jl script as the submit file executable, provided that the first line (the “shebang”) in the .jl file is
+
+
#!/usr/bin/env julia
+
+
+
with the rest of the file containing the commands you want to run using Julia.
+
+
Alternatively, you can use a bash .sh script as the submit file executable, and in that file you can use the julia command:
+
+
#!/bin/bash
+
+julia my-script.jl
+
+
+
In this case, remember to include your .jl file in the transfer_input_files line of your submit file.
+
+
Arguments
+
+
For more information on passing arguments to a Julia script, see the
+Julia documentation.
+
+
Option B: Create your own portable copy
+
+
Use a portable copy of Julia and create your own portable copy of your Julia packages
+
+
This approach may be sensitive to the operating system of the execution point. We recommend building a container instead, but are keeping these instructions as a backup.
+
+
+ -
+
Download the precompiled Julia software from https://julialang.org/downloads/.
+You will need the 64-bit, tarball compiled for general use on a Linux x86 system. The
+file name will resemble something like julia-#.#.#-linux-x86_64.tar.gz.
+
+
+ - Tip: use
wget to download directly to your /home directory on the
+submit server, OR use transfer_input_files = url in your HTCondor submit files.
+
+
+ -
+
Submit an “interactive build” job to create a Julia project and
+install packages, else skip to the next step.
+
+
+
+ -
+
Submit a job that executes a Julia script using the Julia precompiled binary
+with base Julia and Standard Library.
+
+ #!/bin/bash
+
+ # extract Julia binaries tarball
+ tar -xzf julia-#.#.#-linux-x86_64.tar.gz
+
+ # add Julia binary to PATH
+ export PATH=$_CONDOR_SCRATCH_DIR/julia-#.#.#/bin:$PATH
+
+ # run Julia script
+ julia my-script.jl
+
+
+
+ - For more details on the job submission, see the section
+ below: Submit Julia Jobs
+
+
+
+
+
Install Julia Packages
+
+
If your work requires additional Julia packages, you will need to peform a one-time
+installation of these packages within a Julia project. A copy of the project
+can then be saved for use in subsequent job submissions. For more details,
+please see Julia’s documentation at Julia Pkg.jl.
+
+
Create An Interactive Build Job Submit File
+
+
To install your Julia packages, first create an HTCondor submit for
+submitting an “interactive build” job which is a job that will run
+interactively on one of CHTC’s servers dedicated for building
+(aka compiling) software.
+
+
Using a text editor, create the following file, which can be named build.sub
+
+
# Julia build job submit file
+
+universe = vanilla
+log = julia-build.log
+
+# In the latest version of HTCondor on CHTC, interactive jobs require an executable.
+# If you do not have an existing executable, use a generic linux command like hostname as shown below.
+executable = /usr/bin/hostname
+
+# have job transfer a copy of precompiled Julia software
+# be sure to match the name of the version
+# that you have downloaded to your home directory
+transfer_input_files = julia-#.#.#-linux-x86_64.tar.gz
+
++IsBuildJob = true
+
+request_cpus = 1
+request_memory = 4GB
+request_disk = 2GB
+
+queue
+
+
+
The only thing you should need to change in the above file is the name
+of the Julia tarball file in the "transfer_input_files" line.
+
+
Submit Your Interactive Build Job
+
+
Once this submit file is created, submit the job using the following command:
+
+
[alice@submit]$ condor_submit -i build.sub
+
+
+
It may take a few minutes for the build job to start.
+
+
Install Julia Packages Interactively
+
+
Once the interactive jobs starts you should see the following
+inside the job’s working directory:
+
+
bash-4.2$ ls -F
+julia-#.#.#-linux-x86_64.tar.gz tmp/ var/
+
+
+
Run the following commands
+to extract the Julia software and add Julia to your PATH:
+
+
bash-4.2$ tar -xzf julia-#.#.#-linux-x86_64.tar.gz
+bash-4.2$ export PATH=$_CONDOR_SCRATCH_DIR/julia-#.#.#/bin:$PATH
+
+
+
After these steps, you should be able to run Julia from the command line, e.g.
+
+
julia --version
+
+
+
Now create a project directory to install your packages (we’ve called
+it my-project/ below) and tell Julia its name:
+
+
bash-4.2$ mkdir my-project
+bash-4.2$ export JULIA_DEPOT_PATH=$PWD/my-project
+
+
+
You can choose whatever name to use for this directory -- if you have
+different projects that you use for different jobs, you could
+use a more descriptive name than “my-project”.
+
+
We will now use Julia to install any needed packages to the project directory
+we created in the previous step.
+
+
Open Julia with the --project option set to the project directory:
+
+
bash-4.2$ julia --project=my-project
+
+
+
Once you’ve started up the Julia REPL (interpreter), start the Pkg REPL, used to
+install packages, by typing ]. Then install and test packages by using
+Julia’s add Package syntax.
+
+
_
+ _ _ _(_)_ | Documentation: https://docs.julialang.org
+ (_) | (_) (_) |
+ _ _ _| |_ __ _ | Type "?" for help, "]?" for Pkg help.
+ | | | | | | |/ _` | |
+ | | |_| | | | (_| | | Version 1.0.5 (2019-09-09)
+ _/ |\__'_|_|_|\__'_| | Official https://julialang.org/ release
+|__/ |
+
+julia> ]
+(my-project) pkg> add Package
+(my-project) pkg> test Package
+
+
+
If you have multiple packages to install they can be combined
+into a single command, e.g. (my-project) pkg> add Package1 Package2 Package3.
+
+
If you encounter issues getting packages to install successfully, please
+contact us at chtc@cs.wisc.edu.
+
+
Once you are done, you can exit the Pkg REPL by typing the Delete key and then
+exit()
+
+
(my-project) pkg>
+julia> exit()
+
+
+
Save Installed Packages For Later Jobs
+
+
To use this project, and the associated installed packages, in
+subsequent jobs, we need to have HTCondor return some files to
+the submit server by converting the my-project/ directory
+to a tarball, before exiting the interactive job session:
+
+
bash-4.2$ tar -czf my-project.tar.gz my-project/
+bash-4.2$ exit
+
+
+
After the job exits, you will be returned to your /home directory on the
+submit server (specifically where ever you were located when you submitted
+the interactive build job). A copy of packages.tar.gz will be present. Be
+sure to check the size of the project tarball before proceeding to subsequent job
+submissions. If the file is >100MB please contact us at chtc@cs.wisc.edu so
+that we can get you setup with access to our SQUID web proxy. More details
+are available on our SQUID guide: File Availability with SQUID
+
+
[alice@submit]$ ls
+build.sub julia-#.#.#-linux-x86_64.tar.gz julia-build.log
+my-project.tar.gz
+[alice@submit]$ ls -sh my-project.tar.gz
+
+
+
Submit Julia Jobs
+
+
To submit a job that runs a Julia script, create a bash
+script and HTCondor submit file following the examples in this section.
+These examples assume that you have downloaded a copy of Julia for Linux as a tar.gz
+file and if using packages, you have gone through the steps above to install them
+and create an additional tar.gz file of the installed packages.
+
+
Create Executable Bash Script
+
+
Your job will use a bash script as the HTCondor executable. This script
+will contain all the steps needed to unpack the Julia binaries and
+execute your Julia script (script.jl). Below are two example bash script,
+one which can be used to execute a script with base Julia, and one that
+will use packages installed in Julia project (see Install Julia Packages).
+
+
Example Bash Script For Base Julia Only
+
+
If your Julia script can run without additional packages (other than base Julia and
+the Julia Standard library) use the example script directly below.
+
+
#!/bin/bash
+
+# julia-job.sh
+
+# extract Julia tar.gz file
+tar -xzf julia-#.#.#-linux-x86_64.tar.gz
+
+# add Julia binary to PATH
+export PATH=$_CONDOR_SCRATCH_DIR/julia-#.#.#/bin:$PATH
+
+# run Julia script
+julia script.jl
+
+
+
Example Bash Script For Julia With Installed Packages
+
+
#!/bin/bash
+
+# julia-job.sh
+
+# extract Julia tar.gz file and project tar.gz file
+tar -xzf julia-#.#.#-linux-x86_64.tar.gz
+tar -xzf my-project.tar.gz
+
+# add Julia binary to PATH
+export PATH=$_CONDOR_SCRATCH_DIR/julia-#.#.#/bin:$PATH
+# add Julia packages to DEPOT variable
+export JULIA_DEPOT_PATH=$_CONDOR_SCRATCH_DIR/my-project
+
+# run Julia script
+julia --project=my-project script.jl
+
+
+
Create HTCondor Submit File
+
+
After creating a bash script to run Julia, then create a submit file
+to submit the job to run.
+
+
More details about setting up a submit file, including a submit file template,
+can be found in our hello world example page at Run Your First CHTC Jobs.
+
+
# julia-job.sub
+
+universe = vanilla
+
+log = job_$(Cluster).log
+error = job_$(Cluster)_$(Process).err
+output = job_$(Cluster)_$(Process).out
+
+executable = julia-job.sh
+
+should_transfer_files = YES
+when_to_transfer_output = ON_EXIT
+transfer_input_files = julia-#.#.#-linux-x86_64.tar.gz, script.jl
+
+request_cpus = 1
+request_memory = 2GB
+request_disk = 2GB
+
+queue 1
+
+
+
If your Julia script needs to use packages installed for a project,
+be sure to include my-project.tar.gz as in input file in julia-job.sub.
+For project tar.gz files that are <100MB, you can follow the below example:
+
+
transfer_input_files = julia-#.#.#-linux-x86_64.tar.gz, script.jl, my-project.tar.gz
+
+
+
For project tar.gz files that are larger than 100MB, email a facilitator about
+using SQUID.
+
+
Modify the CPU/memory request lines to match what is needed by the job.
+Test a few jobs for disk space/memory usage in order to make sure your
+requests for a large batch are accurate! Disk space and memory usage can be found in the
+log file after the job completes.
+
+
Submit Your Julia Job
+
+
Once you have created an executable bash script and submit file, you can
+submit the job to run using the following command:
+
+
[alice@submit]$ condor_submit julia-job.sub
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Use Licensed Software
+
+
+
This guide describes when and how to run jobs that use licensed software in
+CHTC’s high throughput compute (HTC) system.
+
+
To best understand the below information, users should already have an
+understanding of:
+
+
+ - Using the command line to: navigate within directories,
+create/copy/move/delete files and directories, and run their
+intended programs (aka "executables").
+ - The CHTC's Intro to Running HTCondor Jobs
+
+
+
Overview
+
+
Once you know that you need to use a licensed software program on our
+HTC system, you will need to do the following:
+
+
+
+
+
+
A. CHTC's Licensed Software Policies on the HTC System
+
+
Our typical practice for software support in CHTC is for users to
+install and manage their own software installations. We have multiple
+guides to help users with common software
+programs and additional support is always
+available through CHTC's research computing
+facilitators.
+
+
However, certain software programs require paid licenses which can make
+it challenging for individual users to install the software and use the
+licenses correctly. As such, we provide support for software
+installation and use on our high throughput system. Installation of
+licensed programs is by request to and at the discretion of CHTC staff.
+
+
We always recommend using a free or open-source software alternative
+whenever possible, as certain software licenses restrict the amount of
+computing that can contribute to your research.
+
+
+
+
B. Viewing Licensed Software on the HTC System
+
+
Software with paid licenses that has been installed on the high
+throughput (HTC) system is accessible through software "modules",
+which are tools to access and activate a software installation. To see
+which software programs are available on the HTC system, run the
+following command on an HTC submit server:
+
+
[alice@submit]$ module avail
+
+
+
+ Note: you should never run a program directly on the submit server.
+Jobs that use licensed software/modules should always be submitted as
+HTCondor jobs as described below.
+
+
+
Note that not all software modules are available to all CHTC users. Some
+programs like ansys have a campus or shared license which makes them
+available to all CHTC users. Other software, like lumerical and
+abaqus, is licensed to a specific group and is only available to
+members of that group.
+
+
+
+
C. Submitting Jobs Using Licensed Software Modules
+
+
The following sections describe how to create a bash script executable
+and HTCondor submit file to run jobs that use software accessible via
+the modules.
+
+
+
+
1. Script For Running Jobs with Modules
+
+
To run a job that uses a licensed software installation on the HTC
+system, you need to write a script that loads the software module and
+then runs the program, like so:
+
+
#!/bin/bash
+
+# Commands to enable modules, and then load an appropriate software module
+export PATH
+. /etc/profile.d/modules.sh
+module load software
+
+# For Lumerical (the license requires a home directory)
+export HOME=$_CONDOR_SCRATCH_DIR
+
+# Command to run your software from the command line
+cmd -options input.file
+
+
+
Replace software with the name of the software module you want to use,
+found via the module avail command described above. Replace
+the final command with the syntax to run your software, with the
+appropriate options.
+
+
For example, to run a Comsol job, the script might look like this:
+
+
#!/bin/bash
+
+export PATH
+. /etc/profile.d/modules.sh
+module load COMSOL/5.4
+
+comsol batch -inputfile test.mph -outputfile test-results.mph
+
+
+
+
+
2. Submit File Requirements
+
+
There are several important requirements to consider when writing a
+submit file for jobs that use our licensed software modules. They are
+shown in the sample submit file below and include:
+
+
+ -
+
Require access to the modules. To ensure that your job will have
+access to CHTC software modules you must include the following in
+your submit file.
+
+ requirements = (HasChtcSoftware == true)
+
+
+ -
+
Add a concurrency limit. For software with limited licenses, we have
+implemented concurrency limits, which control the number of jobs running
+at once in the HTC system. If your software is in the table below, use
+the concurrency limit name in your submit file like this:
+
+ concurrency_limits = LIMIT_NAME:num_of_licenses_used
+
+
+
+
+ | Software |
+ Limit Name |
+ Limit |
+
+
+
+
+ | ABAQUS |
+ ABAQUS |
+ 50 |
+
+
+ | ANSYS |
+ ANSYS_RESEARCH |
+ 20 |
+
+
+ | COMSOL (Physics) |
+ COMSOL_PHYSICS |
+ 2 |
+
+
+ | Lumerical |
+ LUMERICAL |
+ 3 |
+
+
+
+
+ So if you were planning to run a job that used one ANSYS license, you would
+use:
+
+ concurrency_limits = ANSYS_RESEARCH:1
+
+
+ - Request accurate CPUs and memory. Run at least one test job and
+look at the log file produced by HTCondor to determine how much
+memory and disk space your jobs actually use. We recommend
+requesting the smallest number of CPUs where your job will finish in
+1-2 days.
+ - The script you wrote above (shown as
run_job.sh below) should be
+your submit file "executable", and any input files should be
+listed in transfer_input_files.
+
+
+
A sample submit file is given below:
+
+
# software.sub
+# A sample submit file for running a single job using software modules
+
+universe = vanilla
+log = job_$(Cluster).log
+output = job_$(Cluster).out
+error = job_$(Cluster).err
+
+# the executable should be the script you wrote above
+executable = run_job.sh
+# arguments = (if you want to pass any to the shell script)
+should_transfer_files = YES
+when_to_transfer_output = ON_EXIT
+transfer_input_files = (this should be a comma separate list of input files if needed)
+
+# Requirement for accessing new set of software modules
+requirements = ( HasChtcSoftware == true )
+
+# If required, add the concurrency limit for your software and uncomment
+# concurrency_limits = LIMIT_NAME:num_of_licenses_used
+
+request_cpus = 1
+request_memory = 2GB
+request_disk = 2GB
+
+queue
+
+
+
After the submit file is complete, you can submit your jobs using
+condor_submit.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Run Machine Learning Jobs
+
+
+
This guide provides some of our recommendations for success
+in running machine learning (specifically deep learning) jobs in CHTC.
+
+
+ This is a new how-to guide on the CHTC website. Recommendations and
+feedback are welcome via email (chtc@cs.wisc.edu) or by creating an
+issue on the CHTC website Github repository: Create an issue
+
+
+
Overview
+
+
It is important to understand the needs of a machine learning job before submitting
+it and have a plan for managing software. This guide covers:
+
+
+ - Considering job requirements
+ - Recommendations for managing software
+
+
+
1. Job Requirements
+
+
Before digging into the nuts and bolts of software installation in the next section,
+it is important to first consider a few other job requirements that might apply to
+your machine learning job.
+
+
A. Do you need GPUs?
+
+
CHTC has about 4 publicly available GPUs and thousands of CPUs. When possible, using
+CPUs will allow your jobs to start more quickly and to have many running at once. For
+certain calculations, GPUs may provide a different advantage as some machine learning
+algorithms are optimized to run significantly faster on GPUs. Consider whether you
+would benefit from running one or two long-running calculations on a GPU or if your
+work is better suited to running many jobs on CHTC’s available CPUs.
+
+
If you need GPUs for your jobs, you can see a summary of available GPUs in CHTC and
+how to access them here:
+
+
+
+
Note that you may need to use different versions of your software, depending on whether or
+not you are using GPUs, as shown in the software section of this guide.
+
+
B. How big is your data?
+
+
CHTC’s usual data recommendations apply for machine learning jobs. If your job is using
+an input data set larger than a few hundred MB or generating output files larger than
+a few GB, you will likely need to use our large data
+file share. Contact the CHTC Research Computing Facilitators to get access and
+read about the large data location here:
+
+
+
+
C. How long does your job run?
+
+
CHTC’s default job length is 72 hours. If your task is long enough that you will
+encounter this limit, contact the CHTC Research Computing Facilitators (chtc@cs.wisc.edu)
+for potential work arounds.
+
+
D. How many jobs do you want to submit?
+
+
Do you have the ability to break your work into many independent pieces? If so,
+you can take advantage of CHTC’s capability to run many independent jobs at once,
+especially when each job is using a CPU. See our guide for running multiple jobs here:
+
+
+
+
2. Software Options
+
+
Many of the tools used for machine learning, specifically deep learning and
+convolutional neural networks, have enough dependencies that our usual installation
+processes work less reliably. The following options are the best way to handle the complexity
+of these software tools.
+
+
Please be aware of which CUDA library version you are using to run your code.
+
+
A. Using Docker Containers
+
+
CHTC’s HTC system has the ability to run jobs using Docker containers, which package
+up a whole system (and software) environment in a consistent, reproducible, portable
+format. When possible, we recommend using standard, publicly available
+Docker containers to run machine learning jobs in CHTC.
+
+
To see how you can use Docker containers to run jobs in CHTC, see:
+
+
+
You can also test and examine containers on your own computer:
+
+
+
Some machine learning frameworks publish ready-to-go Docker images:
+
+
+
If you can not find a Docker container with exactly the tools you need, you can build your
+own, starting with one of the containers above. For instructions on how to build and
+test your own Docker container, see this guide:
+
+
+
+
B. Using Conda
+
+
The Python package manager conda is a popular tool for installing and
+managing machine learning tools.
+See this guide for information on how
+to use conda to provide dependencies for CHTC jobs.
+
+
Note that when installing TensorFlow using conda, it is important to install
+not the generic tensorflow package, but tensorflow-gpu. This ensures that
+the installation will include the cudatoolkit and cudnn dependencies
+required for TensorFlow ‘s GPU capability.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Running Matlab Jobs
+
+
+
Quickstart: Matlab
+
+
Build a container with Matlab & toolboxes installed inside:
+
+
+ - How to build your own container
+ - Example container recipes for Matlab
+ - Use your container in your HTC jobs
+
+
+
+ Note: Because Matlab is a licensed software, you must add the following line to your submit file:
+
+ concurrency_limits = MATLAB:1
+
+
+ Failure to do so may cause your or other users’ jobs to fail to obtain a license from the license server.
+
+
+
+
+
+
+
CHTC has a site license for Matlab that allows for up to 10,000 jobs to run at any given time across all CHTC users.
+Hence the requirement for adding the line concurrency_limits = MATLAB:1 to your submit files, so that HTCondor can keep track of which jobs are using or will use a license.
+
+
Following the instructions above, you are able to install a variety of Matlab Toolboxes when building the container.
+The Toolboxes available for each supported version of Matlab are described here: https://github.com/mathworks-ref-arch/matlab-dockerfile/blob/main/mpm-input-files/.
+Navigate to the text file for the version of interest, and look at the section named “INSTALL PRODUCTS”.
+The example recipes linked above provide instructions on how to specify the packages you want to install when building the container.
+
+
Executable
+
+
When using the Matlab container, we recommend the following process for executing your Matlab commands in an HTCondor job:
+
+
+ -
+
Put your Matlab commands in a .m script. For this example, we’ll call it my-script.m.
+
+ -
+
Create the file run-matlab.sh with the following contents:
+
+ #!/bin/bash
+
+matlab -batch "my-script"
+
+
+ Note that in the script, the .m extension has been dropped from the file name (uses "my-script" instead of "my-script.m").
+
+ -
+
In your submit file, set the .sh script as the executable and list the .m file to be transferred:
+
+ executable = run-matlab.sh
+transfer_input_files = my-script.m
+
+
+
+
+
Arguments
+
+
You can pass arguments from your submit file to your Matlab code via your executable .sh and the matlab -batch command.
+Arguments in your submit file are accessible inside your executable .sh script with the syntax ${n}, where n is the nth value passed in the arguments line.
+You can use this syntax inside of the matlab -batch command.
+
+
For example, if your Matlab script (my-script.m) is expecting a variable foo, you can add foo=${1} before calling my-script:
+
+
#!/bin/bash
+
+matlab -batch "foo=${1};my-script"
+
+
+
This will use the first argument from the submit file to define the Matlab variable foo.
+By default, such values are read in by Matlab as numeric values (or as a Matlab function/variable that evaluates to a numeric function).
+If you want Matlab to read in the argument as a string, you need to add apostrophes around the value, like this:
+
+
#!/bin/bash
+
+matlab -batch "foo=${1};bar='${2}';my-script"
+
+
+
Here, the value of bar is defined as the second argument from the submit file, and will be identified by Matlab as a string because it’s wrapped in apostrophes ('${2}').
+
+
If you have defined your script to act as a function, you can call the function directly and pass the arguments directly as well.
+For example, if you have constructed your my-script.m as a function, then you can do
+
+
#!/bin/bash
+
+matlab -batch "my-script(${1}, ${2})"
+
+
+
Again, by default Matlab will interpret these value of these variables as numeric values, unless you wrap the argument in apostrophes as described above.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+
+
+ Submitting Multiple Jobs in Individual Directories
+
+
+
This guide demonstrates how to submit multiple jobs, using a specific
+directory structure. It is relevant to:
+
+
+ - Researchers who have used CHTC's "ChtcRun" tools in the past
+ - Anyone who wants to submit multiple jobs, where each job has its own
+directory for input/output files on the submit server.
+
+
+
+
+
The first time you submit jobs, you will need to prepare a portable
+version of your software and a script (what we call the job's
+"executable") that runs your code. We have guides for preparing:
+
+
+
+
Choose the right guide for you and follow the directions for compiling
+your code (Matlab) or building an installation (Python, R). Also follow
+the instructions for writing a shell script that runs your program.
+These are typically steps 1 and 2 of the above guides.
+
+
2. Directory Structure
+
+
Once you've prepared your code and script, create the same directory
+structure that you would normally use with ChtcRun. For a single batch
+of jobs, the directories will look like this:
+
+
project_name/
+ run_code.sh
+ shared/
+ scripts, code_package
+ shared_input
+ job1/
+ input/
+ job_input
+ job2/
+ input/
+ job_input
+ job3/
+ input/
+ job_input
+
+
+
You'll want to put all your code and files required for every job in
+shared/ and individual input files in the individual job directories
+in an input folder. In the submit file below, it matters that the
+individual job directories start with the word "job".
+
+
+ Note: the job directories need to be hosted in your /home directory
+on the submit node. The following instructions will not work for files
+hosted on /staging!
+
+
+
3. Submit File
+
+
+ Note: if you are submitting more than 10,000 jobs at once, you'll
+need to use a different submit file. Please email the CHTC Research
+Computing Facilitators at chtc@cs.wisc.edu if this is the case!
+
+
+
Your submit file, which should go in your main project directory, should
+look like this:
+
+
# Specify the HTCondor Universe (vanilla is the default and is used
+# for almost all jobs), the desired name of the HTCondor log file,
+# and the desired name of the standard error and standard output file.
+universe = vanilla
+log = process.log
+error = process.err
+output = process.out
+#
+# Specify your executable (single binary or a script that runs several
+# commands) and arguments
+executable = run_code.sh
+# arguments = arguments to your script go here
+#
+# Specify that HTCondor should transfer files to and from the
+# computer where each job runs.
+should_transfer_files = YES
+when_to_transfer_output = ON_EXIT
+# Set the submission directory for each job with the $(directory)
+# variable (set below in the queue statement). Then transfer all
+# files in the shared directory, and from the input folder in the
+# submission directory
+initialdir = $(directory)
+transfer_input_files = ../shared/,input/
+#
+# Tell HTCondor what amount of compute resources
+# each job will need on the computer where it runs.
+request_cpus = 1
+request_memory = 1GB
+request_disk = 1GB
+#
+# Create a job for each "job" directory.
+queue directory matching job*
+
+
+
You must change the name of the executable to your own script, and
+in certain cases, add arguments.
+
+
Note that the final line matches the pattern of your directory names
+created in the second step. You can use a different name for the
+directories (like data or seed), but you should use whatever word
+they share in the final queue statement in place of "job".
+
+
Jobs can then be submitted as described in our Introduction to HTC
+Guide, using condor_submit.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Submitting Multiple Jobs Using HTCondor
+
+
+
+
+
+
Table of Contents
+
+
+
+
+
Overview
+
HTCondor has several convenient features for streamlining high-throughput
+job submission. This guide provides several examples
+of how to leverage these features to submit multiple jobs with a
+single submit file.
+
+
Why submit multiple jobs with a single submit file?
+
+
Users should submit multiple jobs using a single submit file, or where applicable, as few
+separate submit files as needed. Using HTCondor multi-job submission features is more
+efficient for users and will help ensure reliable operation of the the login nodes.
+
+
Many options exist for streamlining your submission of multiple jobs,
+and this guide only covers a few examples of what is truly possible with
+HTCondor. If you are interested in a particular approach that isn’t described here,
+please contact CHTC’s research computing facilitators and we will
+work with you to identify options to meet the needs of your work.
+
+
+ Before you continue reading: While HTCondor is designed to submit many jobs at a
+time using a single submit file, the hardware of the submit server can be overwhelmed
+if there are a significant number of jobs submitted at once or rapidly starting and finishing.
+Therefore, plan ahead for the following to scenarios:
+
+ 1) If you plan to submit 10,000+ jobs at a time, please let us
+ know, so we can provide options that will protect the queue’s performance.
+2) If you plan to submit 1000+ jobs, please make sure that each job
+ has a minimum run time of 10 minutes (on average). If your calculations are shorter than
+ 10 minutes, then modify your workflow to run multiple calculations per job.
+
+
+
+
+
1. Submit Multiple Jobs Using queue
+
+
All HTCondor submit files require a queue attribute (which must also be
+the last line of the submit file). By default, queue will submit one job, but
+users can also configure the queue attribute to behave like a for loop
+that will submit multiple jobs, with each job varying as predefined by the user.
+
+
Below are different HTCondor submit file examples for submitting batches of multiple
+jobs and, where applicable, how to indicate the differences between jobs in a batch
+with user-defined variables. Additional examples and use cases are provided further below:
+
+
+ - queue <N> - will submit N number of jobs. Examples
+include performing replications, where the same job must be repeated N number
+of times, looping through files named with numbers, and looping through
+a matrix where each job uses information from a specific row or column.
+ - queue <var> from <list> - will loop through a
+list of file names, parameters, etc. as defined in separate text file (i.e. *
*).
+This `queue` option is very flexible and provides users with many options for
+submitting multiple jobs.
+ - Organizing Jobs Into Individual Directories -
+another option that can be helpful in organizing multi-job submissions.
+
+
+
These queue options are also described in the following video from HTCondor Week 2020:
+
+ 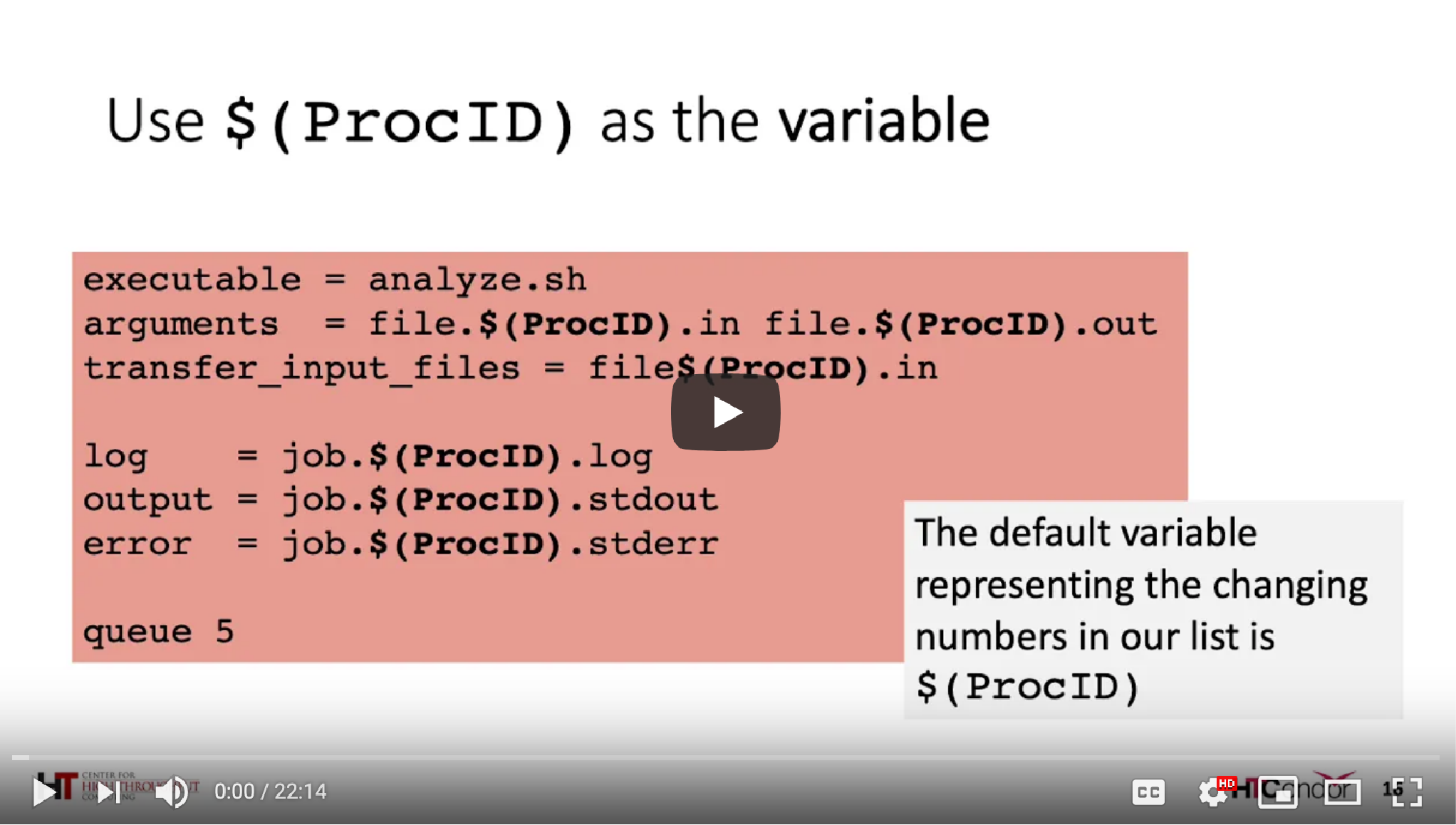
+
+
Submitting Multiple Jobs Using HTCondor Video
+
+
What makes these queue options powerful is the ability to use user-defined
+variables to specify details about your jobs in the HTCondor submit file. The
+examples below will include the use of $(variable_name) to specify details
+like input file names, file locations (aka paths), etc. When selecting a
+variable name, users must avoid bespoke HTCondor submit file variables
+such as Cluster, Process, output, and input, arguments, etc.
+
+
2. Use queue N in your HTCondor submit files
+
+
+
When using queue N, HTCondor will submit a total of N
+jobs, counting from 0 to N - 1 and each job will be assigned
+a unique Process id number spanning this range of values. Because
+the Process variable will be unique for each job, it can be used in
+the submit file to indicate unique filenames and filepaths for each job.
+
+
The most straightforward example of using queue N is to submit
+N number of identical jobs. The example shown below demonstrates
+how to use the Cluster and Process variables to assign unique names
+for the HTCondor error, output, and log files for each job in the batch:
+
+
# 100jobs.sub
+# submit 100 identical jobs
+
+log = job_$(Cluster)_$(Process).log
+error = job_$(Cluster)_$(Process).err
+output = job_$(Cluster)_$(Process).out
+
+... remaining submit details ...
+
+queue 100
+
+
+
For each job, the appropriate number, 0, 1, 2, ... 99 will replace $(Process).
+$(Cluster) will be a unique number assigned to the entire 100 job batch. Each
+time you run condor_submit job.sub, you will be provided
+with the Cluster number which you will also see in the output produced by
+the command condor_q.
+
+
If a uniquely named results file needs to be returned by each job,
+$(Process) and $(Cluster) can also be used as arguments, and anywhere
+else as needed, in the submit file:
+
+
arguments = $(Cluster)_$(Process).results
+
+... remaining submit details ...
+
+queue 100
+
+
+
Be sure to properly format the arguments statement according to the
+executable used by the job.
+
+
What if my jobs are not identical? queue N may still be a great
+option! Additional examples for using this option include:
+
+
+
+
+
[user@login]$ ls *.data
+0.data 1.data 2.data 3.data
+... 97.data 98.data 99.data
+
+
+
In the submit file, use:
+
+
transfer_input_files = $(Process).data
+
+... remaining submit details ...
+
+queue 100
+
+
+
+
2B. Specify a row or column number for each job
+
+
$(Process) can be used to specify a unique row or column of information in a
+matrix to be used by each job in the batch. The matrix needs to then be transferred
+with each job as input. For exmaple:
+
+
transfer_input_files = matrix.csv
+arguments = $(Process)
+
+... remaining submit details ...
+
+queue 100
+
+
+
The above exmaples assumes that your job is set up to use an argument to
+specify the row or column to be used by your software.
+
+
+
2C. Need N to start at 1
+
+
If your input files are numbered 1 - 100 instead of 0 - 99, or your matrix
+row starts with 1 instead of 0, you can perform basic arithmetic in the submit
+file:
+
+
plusone = $(Process) + 1
+NewProcess = $INT(plusone,%d)
+arguments = $(NewProcess)
+
+... remaining submit details ...
+
+queue 100
+
+
+
Then use $(NewProcess) anywhere in the submit file that you would
+have otherwise used $(Process). Note that there is nothing special about the
+names plusone and NewProcess, you can use any names you want as variables.
+
+
+
3. Submit multiple jobs with one or more distinct variables per job
+
+
Think about what’s different between each job that needs to be submitted.
+Will each job use a different input file or combination of software parameters? Do
+some of the jobs need more memory or disk space? Do you want to use a different
+software or script on a common set of input files? Using queue <var> from <list>
+in your submit files can make that possible! <var> can be a single user-defined
+variable or comma-separated list of variables to be used anywhere in the submit file.
+<list> is a plain text file that defines <var> for each individual job to be submitted in the batch.
+
+
Suppose you need to run a program called compare_states that will run on
+on the following set of input files: illinois.data, nebraska.data, and
+wisconsin.data and each input file can analyzed as a separate job.
+
+
To create a submit file that will submit all three jobs, first create a
+text file that lists each .data file (one file per line).
+This step can be performed directly on the login node, for example:
+
+
[user@state-analysis]$ ls *.data > states.txt
+[user@state-analysis]$ cat states.txt
+illinois.data
+nebraska.data
+wisconsin.data
+
+
+
Then, in the submit file, following the pattern queue <var> from <list>,
+replace <var> with a variable name like state and replace <list>
+with the list of .data files saved in states.txt:
+
+
queue state from states.txt
+
+
+
For each line in states.txt, HTCondor will submit a job and the variable
+$(state) can be used anywhere in the submit file to represent the name of the .data file
+to be used by that job. For the first job, $(state) will be illinois.data, for the
+second job $(state) will be nebraska.data, and so on. For example:
+
+
# run_compare_states_per_state.sub
+
+transfer_input_files = $(state)
+arguments = $(state)
+executable = compare_states
+
+... remaining submit details ...
+
+queue state from states.txt
+
+
+
+
3A. Use multiple variables for each job
+
+
Let’s imagine that each state .data file contains data spanning several
+years and that each job needs to analyze a specific year of data. Then
+the states.txt file can be modified to specify this information:
+
+
[user@state-analysis]$ cat states.txt
+illinois.data, 1995
+illinois.data, 2005
+nebraska.data, 1999
+nebraska.data, 2005
+wisconsin.data, 2000
+wisconsin.data, 2015
+
+
+
Then modify the queue to define two <var> named state and year:
+
+
queue state,year from states.txt
+
+
+
Then the variables $(state) and $(year) can be used in the submit file:
+
+
# run_compare_states_by_year.sub
+arguments = $(state) $(year)
+transfer_input_files = $(state)
+executable = compare_states
+
+... remaining submit details ...
+
+queue state,year from states.txt
+
+
+
+
4. Organizing Jobs Into Individual Directories
+
+
+
4A. Submitting Multiple Jobs in Different Directories with queue <variable> from list
+
+
One way to organize jobs is to assign each job to its own directory,
+instead of putting files in the same directory with unique names. To
+continue our "compare_states" example, suppose there's a directory
+for each state you want to analyze, and each of those directories has
+its own input file named input.data:
+
+
[user@state-analysis]$ ls -F
+compare_states illinois/ nebraska/ wisconsin/
+
+[user@state-analysis]$ ls -F illinois/
+input.data
+
+[user@state-analysis]$ ls -F nebraska/
+input.data
+
+[user@state-analysis]$ ls -F wisconsin/
+input.data
+
+
+
The HTCondor submit file attribute initialdir can be used
+to define a specific directory from which each job in the batch will be
+submitted. The default initialdir location is the directory from which the
+command condor_submit myjob.sub is executed.
+
+
Combining queue var from list with initiadir, each line of ** will include
+the path to each state directory and `initialdir` set to this path for
+each job:
+
+
#state-per-dir-job.sub
+initialdir = $(state_dir)
+transfer_input_files = input.data
+executable = compare_states
+
+... remaining submit details ...
+
+queue state_dir from state-dirs.txt
+
+
+
Where state-dirs.txt is a list of each directory with state data:
+
+
[user@state-analysis]$ cat state-dirs.txt
+illinois
+nebraska
+wisconsin
+
+
+
Notice that executable = compare_states has remained unchanged in the above example.
+When using initialdir, only the input and output file path (including the HTCondor log, error, and
+output files) will be changed by initialdir.
+
+
In this example, HTCondor will create a job for each directory in state-dirs.txt and use
+that state's directory as the initialdir from which the job will be submitted.
+Therefore, transfer_input_files = input.data can be used without specifying
+the path to this input.data file. Any output generated by the job will then be returned to the initialdir
+location.
+
+
+
4B. Submitting Multiple Jobs in Different Directories with queue <directory> matching *
+
+
This section demonstrates how to submit multiple jobs, using a specific
+directory structure where folder names have a string of text in common. It is relevant to anyone who wants to submit multiple jobs, where each job has its own directory for input/output files on the submit server.
+
+
Directory Structure
+For a single batch of jobs, the directories will look like this:
+
+
project_name/
+ run_code.sh
+ submit.sub
+ shared/
+ scripts, code_package
+ shared_input
+ job1/
+ input/
+ job_input
+ job2/
+ input/
+ job_input
+ job3/
+ input/
+ job_input
+
+
+
You'll want to put all your code and files required for every job in
+shared/ and individual input files in the individual job directories
+in an input folder. In the submit file below, it matters that the
+individual job directories start with the word "job". Your directories should all have a string of text in common, so that you can use the queue <directory> matching <commonString>* syntax to queue a job for each directory.
+
+
+ Note: the job directories need to be hosted in your /home directory
+on the submit node. The following instructions will not work for files
+hosted on /staging!
+
+
+
Submit File
+Your submit file, which should go in your main project directory, should
+look like this:
+
+
# Specify your executable (single binary or a script that runs several
+# commands) and arguments
+executable = run_code.sh
+# arguments = arguments to your script go here
+#
+# Specify the desired name of the HTCondor log file,
+# and the desired name of the standard error and standard output file.
+log = process.log
+error = process.err
+output = process.out
+#
+# Specify that HTCondor should transfer files to and from the
+# computer where each job runs.
+should_transfer_files = YES
+# Set the submission directory for each job with the $(directory)
+# variable (set below in the queue statement). Then transfer all
+# files in the shared directory, and from the input folder in the
+# submission directory
+initialdir = $(directory)
+transfer_input_files = ../shared/,input/
+#
+# Tell HTCondor what amount of compute resources
+# each job will need on the computer where it runs.
+request_cpus = 1
+request_memory = 1GB
+request_disk = 1GB
+#
+# Create a job for each "job" directory.
+queue directory matching job*
+
+
+
Note that the final line matches the pattern of your directory names that you previously
+created. You can use a different name for the
+directories (like data, sample, or seed), but you should use whatever word
+the directories have in common in the final queue statement in place of "job".
+
+
Jobs can then be submitted using condor_submit.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Policies for Using HTC Submit Servers
+
+
+
If you have not yet requested a CHTC account and met with a Research
+Computing Facilitator, please fill out this form.
+
+
When your account was created, a member of our team emailed you with the
+server address you should use for logging in. See our
+connecting guide for additional information.
+
+
Access to other submit servers is granted for
+specific purposes and will have been indicated to you by CHTC staff.
+
+
Connecting to HTC Submit Servers
+
+
Once your account is active, you can connect to your designated submit
+server with an SSH connection ("Putty" for Windows, putty.org;
+"Terminal" for Linux/Max). Note, however, that our submit servers only
+accept connections from campus networks. Off-campus connections have
+been disabled for security purposes. If you need to connect from off
+campus, you can first SSH to a computer in your department, and then SSH
+to our submit server. You may also be able to use a Virtual Private
+Network (VPN) to join the campus network when working off-campus. DoIT
+provides information on using a
+VPN.
+
+
For more detailed information on connecting to CHTC services, both
+logging in and transferring files, see our Connecting to CHTC
+guide.
+
+
General User Policies
+
+
See our User Policies and Expectations for details on general CHTC policies.
+
+
HTC System Specific Limits
+
+
Below are some of the default limits on CHTC’s HTC system. Note that as a large-scale
+computing center, we want you to be able to run at a large scale - often much larger
+than these defaults. Please contact the facilitation team whenever you encounter one
+of these limits so we can adjust your account settings or discuss alternative ways to
+achieve your computing goals.
+
+
+ - Jobs with long runtimes. There is a default run limit of 72
+hours for each job queued in the HTC System, once it starts running.
+Jobs longer than this will be placed in HTCondor's "hold" state.
+If your jobs will be longer, please email
+us, and we'll help you to determine the
+best solution.
+ - Submitting many jobs from one submit file. HTCondor is designed
+to submit thousands (or more) jobs from one submit file. If you are
+submitting over 10,000 jobs per submit file or want to queue
+more than 50,000 total jobs as a single user,
+please email us as we have strategies to
+submit that many jobs in a way that will ensure you have as many
+jobs running as possible without also compromising queue performance.
+ - Submitting many short jobs from one submit file. While HTCondor
+is designed to submit thousands of jobs at a time, many short jobs
+can overwhelm the submit server, resulting in other jobs taking much
+longer to start than usual. If you plan on submitting over
+1000 jobs per submit file, we ask that you ensure each job has a
+minimum run time of 5 minutes (on average).
+ - The default disk quota is 20 GB in your
/home directory, as a
+starting point. You can track your use of disk space and your quota value,
+using our Quota Guide. If you need more space
+for concurrent work, please see our Request a Quota Change
+guide.
+ - Submitting jobs with "large" files: HTCondor's
+normal file transfer mechanism ("transfer_input_files") is good for
+files up to 100MB in size (or 500MB total, per job). For jobs with larger
+files, please see our guide on File Availability
+Options, and contact us to make arrangements.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Use Custom Linux Versions in CHTC
+
+
+
By default, CHTC-managed submit servers automatically add a job
+requirement that requires jobs to run on servers running our primary operating system unless otherwise specified by the user. There are two options to override this
+default:
+
+
+ - Using a Container (recommended)
+ - Requesting a Specific
+Operating System.
+
+
+
Option 1: Using a Container (Recommended)
+
+
Using a container to provide a base version of Linux will allow you to
+run on any nodes in the HTC system, and not limit you to a subset of nodes.
+
+
After finding a container with the desired version of Linux, just follow our instructions
+for Docker or Singularity/Apptainer jobs.
+
+
Note that the default Linux containers on Docker Hub are often missing commonly installed
+packages. Our collaborators in OSG Services maintain a few curated containers with a
+greater selection of installed tools that
+can be seen here: Base Linux Containers
+
+
Option 2: Requesting a Specific Operating System
+
+
At any time, you can require a specific operating system
+version (or versions) for your jobs. This option is more limiting because
+you are restricted to operating systems used by CHTC, and the number of nodes
+running that operating system.
+
+
Require CentOS Stream 8 (previous default) or CentOS Stream 9
+
+
To request that your jobs run on servers with CentOS 8 only, add the
+following line to your submit file:
+
+
chtc_want_el8 = true
+
+
+
To request that your jobs run on servers with CentOS 9 only, add
+the following line to your submit file:
+
+
chtc_want_el9 = true
+
+
+
+ Note that after May 1, 2024, CentOS9 will be the default and CentOS8 will be phased out
+by the middle of summer 2024. If you think your code relies on CentOS8, make sure to
+see our transition guide or talk to the facilitation
+team about a long-term strategy for running your work.
+
+
+
Use Both CentOS Stream 8 (previous default) and CentOS Stream 9 (current default)
+
+
To request that your jobs run on computers running either version of
+CentOS Linux, add the following requirements line to your submit file:
+
+
requirements = (OpSysMajorVer == 8) || (OpSysMajorVer == 9)
+
+
+ Note: these requirements are not necessary for jobs that use Docker containers;
+these jobs will run on servers with any operating system automatically.
+
+
+
The advantage of this option is that you may be able to access a
+larger number of computers in CHTC. Note that code compiled on a
+newer version of Linux may not run older versions of Linux. Make
+sure to test your jobs specifically on both CentOS Stream 8 and CentOS Stream 9
+before using the option above.
+
+
Does your job already have a requirements statement? If so, you can
+add the requirements above to the pre-existing requirements by using
+the characters &&. For example, if your jobs already require large
+data staging:
+
+
requirements = (Target.HasCHTCStaging == true)
+
+
+
You can add the requirements for using both operating system versions like so:
+
+
requirements = (Target.HasCHTCStaging == true) && ((OpSysMajorVer == 8) || (OpSysMajorVer == 9))
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ HTC Data Storage Locations
+
+
+
[toc]
+
+
Data Storage Locations
+
The HTC system has two primary locations where users can store files: /home and /staging.
+
+
The mechanisms behind /home and /staging that manage data are different and are optimized to handle different file sizes. /home is more efficient at managing small files, while /staging is more efficient at managing larger files. It’s important to place your files in the correct location, as it will improve the speed and efficiency at which your data is handled and will help maintain the stability of the HTC filesystem.
+
+
Understand your file sizes
+
To know whether a file should be placed in /home or in /staging, you will need to know it’s file size (also known as the amount of “disk space” a file uses). There are many commands to print out your file sizes, but here are a few of our favorite:
+
+
Use ls with -lh flags
+
The command ls stands for “list” and, by default, lists the files in your current directory. The flag -l stands for “long” and -h stands for “human-readable”. When the flags are combined and passed to the ls command, it prints out the long metadata associated with the files and converts values such as file sizes into human-readable formats (instead of a computer readable format).
+
+
NetID@submit$ ls -lh
+
+
+
Use du -h
+
Similar to ls -lh, du -h prints out the “disk usage” of directories in a human-readable format.
+
+
NetID@submit$ du -h
+
+
+
Transferring Data to Jobs
+
The HTCondor submit file transfer_input_files = line should always be used to tell HTCondor what files to transfer to each job, regardless of if that file is origionating from your /home or /staging directory. However, the syntax you use to tell HTCondor to fetch files from /home and /staging and transfer to your running job will change:
+
+
+
+
+ | Input Sizes |
+ File Location |
+ Submit File Syntax to Transfer to Jobs |
+
+
+
+
+ | 0-500 MB |
+ /home |
+ transfer_input_files = input.txt |
+
+
+ | 500-10GB |
+ /staging |
+ transfer_input_files = **osdf:///chtc/staging/NetID/input.txt |
+
+
+ | 10GB + |
+ /staging |
+ transfer_input_files = **file:///staging/NetID/input.txt |
+
+
+
+
+
Transfer Data Back from Jobs to /home or /staging
+
+
When a job completes, by default, HTCondor will return newly created or edited files on the top level directory back to your /home directory.
+
+
To transfer files or folders back to /staging, in your HTCondor submit file, use
+transfer_output_remaps = “output1.txt = file:///staging/NetID/output1.txt”, where output1.txt is the name of the output file or folder you would like transfered back to a /staging directory.
+
+
If you have more than one file or folder to transfer back to /staging, use a semicolon (;) to seperate multiple files for HTCondor to transfer back like so:
+transfer_output_remaps = “output1.txt = file:///staging/NetID/output1.txt; output2.txt = file:///staging/NetID/output2.txt”
+
+
Make sure to only include one set of quotation marks that wraps around the information you are feeding to transfer_output_remaps =.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Other Resources
+
+
+
+
+
+
+
+
+
+ Running Python Jobs
+
+
+
Quickstart: Python
+
+
Option A (recommended)
+
+
Build a container with Python & packages installed inside:
+
+
+ - How to build your own container
+ - Example container recipes for Python
+ - Use your container in your HTC jobs
+
+
+
Option B
+
+
Use an existing container with a base installation of Python:
+
+
+ - Choose an existing container. See
+OSG Base Containers
+or
+DockerHub Python Containers.
+ - Use the container in your HTC jobs
+
+
+
+
+
+
+
All CHTC machines have a base installation of Python 3.
+The exact versions and packages installed, however, can vary from machine to machine.
+You should be able to include simple python commands in your calculations, i.e., python3 simple-script.py.
+
+
If you need a specific version of Python 3 or would like to install your own packages, we recommend that you use a container as described above.
+
+
The example recipes provided above for building your own container are intended for python packages that can be installed using python3 -m pip install.
+Additional software can be installed when building your own container.
+
+
For packages that need to be installed with conda install, see the guide on Conda.
+
+
Executable
+
+
When using a container, you can use a python .py script as the submit file executable, provided that the first line (the “shebang”) in the .py file is
+
+
#!/usr/bin/env python3
+
+
+
with the rest of the file containing the commands that you want to run using Python.
+
+
Alternatively, you can use a bash .sh script as the submit file executable, and in that file you can use the python3 command:
+
+
#!/bin/bash
+
+python3 my-script.py
+
+
+
In this case, remember to include your .py file in the transfer_input_files line of your submit file.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Request a Quota Change
+
+
+
To request a change in quota(s) for data storage locations on CHTC systems, please fill out the form below.
+This form applies to the following locations for both individual and shared (group) directories:
+
+
+
+
For other locations, please email us at chtc@cs.wisc.edu.
+Remember, CHTC data locations are not for long-term storage and are NOT backed up.
+Please review our data policies on the Policies and Expectations for Users page.
+
+
How to Check Your Quotas
+
+
The form asks for the current quotas of the folders you wish to change.
+For individual directories, your quotas are printed on login.
+For group directories at HTC /staging, HPC /home, HPC /scratch, you can retrieve your quotas using the command
+
+
get_quotas /path/to/group/directory
+
+
+
+
+
The following link leads to a Qualtrics form that we use for requesting quota changes.
+
+
+
+
If you do not receive an automated email from chtc@cs.wisc.edu within a few hours of completing the form,
+ OR if you do not receive a response from a human within two business days (M-F), please email chtc@cs.wisc.edu.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Running R Jobs
+
+
+
Quickstart: R
+
+
Option A (recommended)
+
+
Build a container with R & packages installed inside:
+
+
+ - How to build your own container
+ - Example container recipes for R
+ - Use your container in your HTC jobs
+
+
+
Option B
+
+
Use an existing container with a base installation of R:
+
+
+ - Choose an existing container. See
+OSG R containers
+or
+Rocker R containers.
+ - Use the container in your HTC jobs
+
+
+
+
+
+
+
No CHTC machine has R pre-installed, so you must configure a portable copy of R to work on the HTC system.
+Using a container as described above is the easiest way to accomplish this.
+
+
Executable
+
+
When using a container, you can use a .R script as the submit file executable, provided that the first line (the “shebang”) in the .R file is
+
+
#!/usr/bin/env Rscript
+
+
+
with the rest of the file containing the commands that you want to run using R.
+
+
Alternatively, you can use a bash .sh script as the submit file executable, and in that file you can use the Rscript command:
+
+
#!/bin/bash
+
+Rscript my-script.R
+
+
+
In this case, remember to include your .R file in the transfer_input_files line of your submit file.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Scale Beyond Local HTC Capacity
+
+
+
This guide provides an introduction to running jobs outside of CHTC: why
+using these resources is beneficial, what resources are available, and
+how to use them.
+
+
Contents
+
+
+ - Why run on additional resources outside CHTC?
+
+
+ - Job Qualifications
+ - Submitting Jobs to Run Beyond CHTC
+
+
+
+
+
1. Why run on additional resources outside CHTC?
+
+
Running on other resources in addition to CHTC has one huge benefit:
+size! The UW Grid and OSG include thousands of computers,
+addition to what's already available in CHTC, including specialized
+hardware resources like GPUs. Most CHTC users who run
+on CHTC, the UW Grid, and the OSG can get more than 100,000 computer
+hours (more than 11 years of computing!) in a single day. Read on to
+learn more about these resources.
+
+
+
+
A. UW Grid
+
+
What we call the "UW Grid" is a collection of all the groups and
+centers on campus that run their own high throughput computing pool that
+uses HTCondor. Some of these groups include departments (Biochemistry,
+Statistics) or large physics projects (IceCube, CMS). Through agreements
+with these groups, jobs submitted in CHTC can opt into running on these
+other campus pools if there is space.
+
+
We call sending jobs to other pools on campus flocking.
+
+
+
+
B. UW-Madison’s OSG Pool
+
+
CHTC maintains an OSG pool for the campus community, which includes
+resources contributed by campuses, national labs, and other institutions
+across and beyond the US.
+
+
When you send jobs to other institutions in our OSG pool, we call that gliding.
+
+
+
+
2. Job Qualifications
+
+
Not all jobs will run well outside of CHTC. Because these jobs are
+running all over the campus or country, on computers that don't belong
+to us, they have two major requirements:
+
+
+ -
+
Moderate Data Sizes: We can support input file sizes of up to
+20 GB per file per job. This covers input files that would normally be
+transferred out of a /home directory or use SQUID, in addition to larger
+files up to 20GB. Outputs per job can be of similar sizes. If your input or
+output files are larger than 1GB, or you have any other questions about
+handling data on resources beyond CHTC, please contact us!
+
+ -
+
Short or interruptable jobs: Your job can complete in under 10 hours
+-- either it finishes in that amount of time, or it
+self-checkpoints at least that frequently. If you would like to implement
+self-checkpointing for a longer code, we are happy to provide resources
+and guidance.
+
+
+
+
+
+
3. Submitting Jobs to Run Beyond CHTC
+
+
If your jobs meet the characteristics above and you would like to use
+either the UW Grid or OS Pool to run jobs, in addition to CHTC, you can add
+the following to your submit file:
+
+
+
+
+ | +WantFlocking = true |
+ Also send jobs to other HTCondor Pools on campus (UW Grid)
Good for jobs that are less than ~8 hours, on average, or checkpointing jobs. |
+
+
+ | +WantGlideIn = true |
+ Also send jobs to the OS Pool.
Good for jobs that are less than ~8 hours, on average, or checkpointing jobs. |
+
+
+
+
+
To guarantee maximum efficiency, please do the following steps
+whenever submitting a new type of job to the UW Grid or OSG:
+
+
+ -
+
Test Your Jobs: You should run a small test (anywhere from
+10-100 jobs) outside CHTC before submitting your full workflow. To
+do this, take a job submission that you know runs successfully on
+CHTC. Then add the following options in the submit file + submit the
+test jobs:
+
+ requirements = (Poolname =!= "CHTC")
+
+
+ (If your submit file already has a requirements = line, you can
+appending the Poolname requirement by using a double ampersand
+(&&) and then the additional requirement.)
+
+ -
+
Troubleshooting: If your jobs don't run successfully on the UW
+Grid or OS Pool, please get in touch with a research computing
+facilitator.
+
+ -
+
Scaling Up: Once you have tested your jobs and they seem to be
+running successfully, you are ready to submit a full batch of jobs
+that uses CHTC and the UW Grid/OS Pool. REMOVE the Poolname
+requirement from the test jobs but leave the +wantFlocking and
++wantGlidein lines.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Sign In for Office Hours
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Using Software in a Container on the HPC Cluster
+
+
+
DEPRECATED
+
+
See instead our “Using Apptainer Containers on HPC” guide: https://chtc.cs.wisc.edu/uw-research-computing/apptainer-hpc.
+
+
Overview
+
+
Software that is packaged in a "container" can
+be run on the HPC cluster. This guide assumes that you are starting with
+an existing Docker container and shows how to use it to run a job on the HPC cluster.
+
+
Note that you may need to install a version of MPI to your container
+when it is initially created. See the notes about this below.
+
+
The two steps to run a container on the HPC cluster:
+
+ - Convert the container to a Singularity image file
+ - Run a job that uses the container
+
+
+
+Notes about MPI and Containers
+==================
+
+
There are two ways to run a Singularity container integrated with MPI: hybrid
+mode and bind mode.
+
+
In hybrid mode, the container has its own copy of MPI that is compatible
+with a version of MPI already installed on the cluster.
+
+
In bind mode, the code in the container has been compiled with MPI that
+exists outside the container and there is no MPI installation in the container itself.
+Again, the version of MPI used needs to be compatible with one already installed
+on the cluster.
+
+
This will be relevant in how the job is executed later on: Using Singularity Container Images
+
+
+
+
+
+
We assume that there is a Docker container (either found
+or created by you) online that you want to use. To use this container
+on the HPC cluster, it needs to be converted to a Singularity-format
+image file. To do this:
+
+
+ - Log in to one of the HPC cluster log in nodes.
+ - Start an interactive job:
+
[alice@login]$ srun -n4 -N1 -p int --pty bash
+
+
+ - Once the interactive job starts, you’ll need to unset a shell environment
+variable that prevents download of the Docker container.
+
[alice@int]$ unset HTTPS_PROXY
+
+
+ - Then, save the Docker container to a Singularity image.
+
[alice@int]$ singularity build /software/alice/name.simg docker://user/image:version
+
+
+ For example, if user "Alice" wanted to use the "Fenics" container
+ provided on DockerHub,
+ and save it to a file named fenics.simg, she would run:
+
+ [alice@int]$ singularity build /software/alice/fenics.simg docker://fenicsproject/stable:latest
+
+
+
+ This command will by default, pull the initial Docker container from
+Docker Hub. If your Docker container is stored elsewhere, or you are
+starting with a Singularity image, contact CHTC staff for specific instructions.
+
+
+ - Once the Singularity command completes, type
exit to leave the interactive job.
+
+
+
+
+
2. Using Singularity Container Images
+
+
To use a Singularity container in a job, the SLURM submit file will remain mostly the
+same; what will change is the job’s primary command at the end of the
+file. This command will run your primary program inside the container
+file you've downloaded. The main MPI command will still be part of the
+singularity command:
+
+
#!/bin/sh
+#SBATCH options
+
+module load MPI/version
+mpirun -np ## singularity exec /path/to/container/file command-to-run
+
+
+
For example, if Alice wanted to run a script she had written
+(poisson.py) inside the downloaded fenics container, using 40 cores, she would use the
+following command at the end of her submit file:
+
+
mpirun -np 40 singularity exec /software/alice/fenics.simg ./poisson.py
+
+
+
The example shown above uses the “hybrid” model for running MPI, which assumes
+that there is a copy of MPI installed in the container that matches what already
+exists on the cluster.
+
+
If your container does not have it’s own copy of MPI installed, you need
+to use the “bind” model for running MPI which requires an additional flag and
+the location of the main MPI directory:
+
+
#!/bin/sh
+#SBATCH options
+
+module load MPI/version
+mpirun -np ## singularity exec --bind /path/to/cluster/MPI/dir/ /path/to/container/file command-to-run
+
+
+
On CHTC’s cluster, the GCC based version of OpenMPI is installed at the path:
+` /software/chtc/easybuild/v2/software/OpenMPI/4.0.5-GCC-9.3.0/`
+So the command(s) to run the “Alice” example above would be:
+
+
MPI_DIR=/software/chtc/easybuild/v2/software/OpenMPI/4.0.5-GCC-9.3.0/
+mpirun -np 40 singularity exec --bind $MPI_DIR /software/alice/fenics.simg ./poisson.py
+
+
+
More details on the difference between using the “hybrid” and “bind” model
+for MPI and Singularity is here: https://sylabs.io/guides/3.8/user-guide/mpi.html
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Use Apptainer (Singularity) Environments
+
+
+
The contents of this page have been updated and split into the following guides:
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Overview: How to Use Software
+
+
+
+
+
+In order to run jobs on the High Throughput Computing (HTC) system, researchers need to set up their software on the system.
+This guide introduces how to build software in a container (our recommended strategy), links to a repository with a selection of software installation “recipes”, and quick links to common software packages and their installation recommendations.
+
+
+
+
Table of Contents
+
+
+
+
+
Quickstart
+
+
Click the link in the table below to jump to the instructions for the language/program/software that you want to use.
+More information is provided in the CHTC Recipes Repository and Containers sections.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
Quickstart: Java
+
+
To use Java on the HTC system, we recommend that you use the Java Development Kit (JDK).
+
+
+ -
+
Obtain a copy of the pre-compiled JDK for “Linux/x64” from https://jdk.java.net/.
+
+ -
+
Include the JDK .tar.gz file in your submit file with the list of files to be transferred:
+
+ transfer_input_files = openjdk-22_linux-x64_bin.tar.gz, program.jar
+
+
+ -
+
Include instructions for using the JDK in your executable file:
+
+ #!/bin/bash
+
+tar -xzf openjdk-22_linux-x64_bin.tar.gz
+export JAVA_HOME=$PWD/jdk-22
+export PATH=$JAVA_HOME/bin:$PATH
+
+java -jar program.jar
+
+
+
+
+
+
More Information
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
Recipes
+
+
CHTC provides specific examples for software and workflows for use on our systems in our “Recipes” repository on Github:
+https://github.com/CHTC/recipes.
+
+
Links to specific recipes are used in the Software section for certain softwares and coding languages.
+
+
+
+
Containers
+
+
Many of the recipes in our Recipes repository involve building your own container.
+In this section, we provide a brief introduction into how to use containers for setting up your own software to run on the High Throughput system.
+
+
What is a Container?
+
+
“A container is a standard unit of software that packages up code and all its dependencies so the application runs quickly and reliably from one computing environment to another.”
+– Docker
+
+
A container is a portable, self-contained operating system and can be easily executed on different computers regardless of their operating systems or programs.
+When building the container you can choose the operating system you want to use, and can install programs as if you were the owner of the computer.
+
+
While there are some caveats, containers are useful for deploying software on shared computing systems like CHTC, where you do not have permission to install programs directly.
+
+
“You can build a container using Apptainer on your laptop, and then run it on many of the largest HPC clusters in the world, local university or company clusters, a single server, in the cloud, or on a workstation down the hall.”
+– Apptainer
+
+
+ What is a Container Image?
+
+ A “container image” is the persistent, on-disk copy of the container.
+When we talk about building or moving or distributing a container, we’re actually talking about the file(s) that constitute the container.
+When a container is “running” or “executed”, the container image is used to create the run time environment for executing the programs installed inside of it.
+
+
+
Container Technologies
+
+
There are two container technologies supported by CHTC: Docker and Apptainer.
+Here we briefly discuss the advantages of each.
+
+
Docker
+
+
https://www.docker.com/
+
+
Docker is a commercial container technology for building and distributing containers.
+Docker provides a platform for distributing containers, called Docker Hub.
+Docker Hub can make it easy to share containers with colleagues without having to worry about the minutiae of moving files around.
+
+
On the HTC system, you can provide the name of your Docker Hub container in your submit file,
+and HTCondor will automatically pull (download) the container and use it to create the software environment for executing your job.
+Unfortunately, however, you are unable to build a Docker container and upload it to Docker Hub from CHTC servers,
+so your container must already exist on Docker Hub in a public repository.
+This requires that you have Docker installed on your computer so that you can build the container and upload it to Docker Hub.
+
+
Apptainer
+
+
https://apptainer.org/
+
+
Apptainer is an open-source container technology for building containers.
+Apptainer creates a single, stand-alone file that is the (container image).
+As long as you have the container image file, you can use Apptainer to run your container.
+
+
On the HTC system, you can provide the name of your Apptainer file in your submit file,
+and HTCondor will use a copy of it to create the software environment for executing your job.
+You can use Apptainer to build the container image file on CHTC servers, so there is no need to install the container software on your own computer.
+
+
Use an Existing Container
+
+
If you or a colleague have already built a container for use on CHTC, it is fairly straightforward to modify your jobs to use the container environment as discussed below.
+
+
Use a Docker container
+
+
Full Guide: Running HTC Jobs Using Docker Containers
+
+
If the container you want to use is hosted on Docker Hub, find the container “address” and provide it in your submit file.
+The address typically has the convention of user/repository:tag, though official repositories such as Python are just repository:tag.
+In your submit file, use
+
+
container_image = docker://user/repository:tag
+
+
+
If the container you want to use is hosted in a different container registry, there should still be a container “address” to use,
+but now there will be a website prefix.
+
+
container_image = docker://registry_address/user/repository:tag
+
+
+
For example, to use a container from the NVIDIA Container Registry (nvcr),
+you would have docker://nvcr.io/nvidia/repository:tag.
+
+
Back to Top
+
+
Use an Apptainer container
+
+
Full Guide: Use Apptainer Containers
+
+
For historical reasons, the Apptainer container file has the file extension .sif.
+The syntax for giving HTCondor the name of the container file depends on where it is located on the CHTC system.
+
+
If the .sif file is in a /home directory:
+
+
container_image = path/to/my-container.sif
+
+
+
If the .sif file is in a /staging directory:
+
+
container_image = file:///staging/path/to/my-container.sif
+
+
+
If the .sif file is in a /staging directory AND you are using +WantFlocking or +WantGliding:
+
+
container_image = osdf:///chtc/staging/path/to/my-container.sif
+
+
+
Back to Top
+
+
Build Your Own Container
+
+
You can build your own container with the operating system and software that you want to use.
+The general process is the same whether you are using Docker or Apptainer.
+
+
+ -
+
Consult your software’s documentation
+
+ Determine the requirements for installing the software you want to use.
+In particular you are looking for (a) the operating systems it is compatible with and (b) the prerequisite libraries or packages.
+
+ -
+
Choose a base container
+
+ The base container should at minimum use an operating system compatible with your software.
+Ideally the container you choose also has many of the prerequisite libraries/programs already installed.
+
+ -
+
Create your own definition file
+
+ The definition file contains the installation commands needed to set up your software.
+(The structure of the container “definition” file differs between Docker and Apptainer, but it is fairly straightforward to translate between the two.)
+
+ -
+
Build the container
+
+ Once the definition file has been written, you must “build” the container.
+The computer you use to build the container will run through the installation commands, almost as if you were actually installing the software on that computer,
+but will save the results into the container file(s) for later use.
+
+ -
+
Distribute the container
+
+ To use the container on CHTC servers, you’ll need to distribute the container to right location.
+For Docker containers, this means “pushing” the container to Docker Hub or similar container registry.
+For Apptainer containers, this typically means copying the container .sif file to the /staging system.
+
+
+
+
You can then use the container following the instructions above.
+
+
+ A common question is whether the software installation process is repeated each time a container is used.
+The answer is “no”.
+The software installation process only occurs when the container is actually being built.
+Once the container has been built, no changes can be made to the container when being used (on CHTC systems).
+
+
+
Build your own Docker container
+
+
Please follow the instructions in our guide Build a Docker Container Image to build your own container using Docker.
+As mentioned above, you will need to have Docker installed on your own computer.
+This is so that you can push the completed container to Docker Hub.
+
+
You are unable to push containers from CHTC to Docker Hub, so please do not build Docker containers using CHTC!
+
+
Build your own Apptainer container
+
+
Please follow the instructions in our guide Use Apptainer Containers to build your own container using Apptainer.
+You can use CHTC servers to build the container, so there is no need to install any software on your computer.
+
+
Back to Top
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Running Tensorflow Jobs
+
+
+
The contents of the guide previously at this page are not currently
+supported in CHTC, although there are plans to re-integrate them in the
+future. For questions about running Tensorflow in CHTC, email CHTC's
+Research Computing Facilitators at chtc@cs.wisc.edu
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Importance of Testing
+
+
+
Running your code in test jobs before submitting large job batches is
+CRUCIAL to effective high-throughput computing.
+
+
Why Test?
+
+
Improving your own throughput
+
+
Spending the time and effort to run test jobs will pay off in more
+effective high-throughput computing in the following ways:
+
+
+ - Better matching: Pinpointing the required amount of memory/disk
+space will allow your jobs to match to as many computers as
+possible. Requesting excessive amounts of memory and disk space will
+limit the number of slots where your jobs can run, as well as using
+unnecessary space that could be available for other users. That
+said...
+ - Fewer holds or evictions: If you don't request *enough*
+memory or disk and your job exceeds its request, it can go on hold.
+Jobs that have not been tested and run over 72 hours are liable to
+be evicted without finishing.
+ - Fewer wasted compute hours: the evictions and holds described
+above will be wasted compute hours, decreasing your priority in the
+high-throughput system while not returning any results.
+ - Making good choices: knowing how big and how long your jobs are,
+and the size of input/output files will show you how to most
+effectively use CHTC resources. Jobs under 2 hrs or so? Allow your
+jobs to flock and glide to the UW Grid and OS Pool. Input
+files of more than 5 GB? You should probably be using the CHTC large
+file staging area. Longer jobs? Include a line in your submit file
+restricting your jobs to the CHTC servers that guarantee 72 hours.
+
+
+
Being a good citizen
+
+
CHTC's high-throughput system has hundreds to thousands of users,
+meaning that poor computing practices by one user can impact many other
+users. Users who submit jobs that don't finish or are evicted because
+of incorrect memory requests are using hours that could have been used
+by other people. In the worst case, untested code can cause other jobs
+running on an execute server to be evicted, directly harming someone
+else's research process. The best practices listed in these guides
+exist for a reason. Testing your code and job submissions to make sure
+they abide by CHTC recommendations will not only benefit your own
+throughput but make sure that everyone else is also getting a fair share
+of the resource.
+
+
What to Test
+
+
When running test jobs, you want to pay attention to at least the
+following five variables:
+
+
+ - disk space
+ - memory usage
+ - length of job
+ - input file size
+ - output file size
+
+
+
Memory and disk space simply make sure that your jobs have the resources
+they need to run properly. Memory is the amount of RAM needed by your
+program when it executes; disk space is how much hard drive space is
+required to store your data, executables, and any output files.
+
+
Job length has a huge impact on where your jobs can run. Within a subset
+of CHTC servers, jobs are guaranteed to run for 72 hours. Jobs that run
+for longer than 72 hours will fail, unless they have implemented a
+self-checkpointing method that allows them to resume after being
+evicted. Jobs that are shorter, around 2-4 hours, are good candidates to
+run on the UW Grid and/or OS Pool.
+
+
Input and output file size will impact how your files will be
+transferred to and from the execute nodes. Large input files will need
+to be staged on a proxy server or shared file system; small input files
+can use HTCondor's built-in file transfer system. If you have questions
+about how to handle your data, please email
+chtc@cs.wisc.edu to get in touch with a research
+computing facilitator who can advise you.
+
+
In addition to these considerations, your script/program itself should
+be thoroughly tested on your own machine until it is as bug-free and
+correct as possible. If it uses any libraries or packages, you should
+know what they are and if they have any other dependencies.
+
+
How to Test
+
+
Interactive Jobs
+
+
One of the most useful tools for testing is HTCondor's interactive job
+feature. An interactive job is essentially a job without an executable;
+you are the one running the commands instead, through a bash (shell?)
+session.
+
+
To request an interactive job:
+
+
+ -
+
Create a submit file as if you were submitting the job normally,
+with one change. Don't include an executable line; instead, list
+your executable file in the transfer_input_files line.
+
+ # sample submit file
+universe = vanilla
+log = interactive.log
+
+# executable = # delete or comment out
+should_transfer_files = YES
+when_to_transfer_output = ON_EXIT
+transfer_input_files = data_file,myprogram
+
+request_cpus = 1
+request_memory = 1GB
+request_disk = 1GB
+
+queue
+
+
+ -
+
Then, submit the job using the -i option:
+
+ $ condor_submit -i submit_file
+
+
+ You should see a message like:
+
+ Submitting job(s).
+1 job(s) submitted to cluster 4347054.
+Waiting for job to start...
+
+
+ - After a few minutes, the job should match and open an interactive
+session on an execute server, with all the files you listed in
+
transfer_input_files You are now on an execute server, much like
+one your jobs will be running on when you submit them to HTCondor.
+Here, you can try running your executable.
+ -
+
Once you are done, you can type exit to leave the interactive
+session. Note that any files you created during the session will
+be transferred back with you! Another useful tool can be to save
+your history to a file, using the following command:
+
+ $ history > history.txt
+
+
+
+
+
Scale testing
+
+
Once you know that your code works and you can successfully submit one
+job to be run by HTCondor, you should test a few jobs before submitting
+the full-size batch. After these few jobs complete, pay attention to the
+variables described above (memory, disk space, etc.) so you can edit
+your submit files before submitting your entire batch of jobs.
+
+
To find information about memory, disk space and time, look at a job's
+log file. Its name and where it is located may vary, depending on your
+submit process, but once you find it, you should see information like
+this:
+
+
001 (845638.000.000) 03/12 12:48:06 Job executing on host: <128.104.58.85:49163>
+...
+005 (845638.000.000) 03/12 12:48:06 Job terminated.
+ (1) Normal termination (return value 0)
+ Usr 0 00:00:00, Sys 0 00:00:00 - Run Remote Usage
+ Usr 0 00:00:00, Sys 0 00:00:00 - Run Local Usage
+ Usr 0 00:00:00, Sys 0 00:00:00 - Total Remote Usage
+ Usr 0 00:00:00, Sys 0 00:00:00 - Total Local Usage
+ 17 - Run Bytes Sent By Job
+ 92 - Run Bytes Received By Job
+ 17 - Total Bytes Sent By Job
+ 92 - Total Bytes Received By Job
+ Partitionable Resources : Usage Request Allocated
+ Cpus : 1 1
+ Disk (KB) : 12 1000000 26703078
+ Memory (MB) : 0 1000 1000
+
+
+
The table at the end of the log file shows how many resources you used
+and can be used to fine-tune your requests for memory and disk. If you
+didn't keep track yourself, the log file also lists when the job
+started to execute, and when it ended, thus the length of time required
+for completion.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Account Request Submitted
+
+
+
+
+
Thank you for your account request! You should get an email with “[CHTC Requests]” in the subject line within the next
+few minutes and a research
+computing facilitator should be in touch within a business day.
+
+
If you don’t get an email with the “CHTC Requests” subject or hear from a research computing facilitator within a
+day, please check your spam folder; if they aren’t in spam,
+please contact us via the email address at the bottom of this page to confirm that we received your account request.
+
+
+
+
+
+
+
+
+ Transfer Files Between CHTC and ResearchDrive
+
+
+
UW Madison provides a shared data storage for research called ResearchDrive. It
+is possible to transfer files directly between ResearchDrive and CHTC’s systems. The
+instructions in this guide may also work for accessing other data services on campus from CHTC; contact us if you
+would like to know more.
+
+
A. Pre-Requisites
+
+
In order to follow the steps in this guide, you need access to a ResearchDrive share, either as PI or member of your PI’s group, as well as a CHTC account. In what follows,
+we assume that you are transferring files to and from our HTC system, but you can
+use the same process to transfer files to and from the HPC cluster if you first log
+in to one of the HPC login nodes.
+
+
B. Transferring Files
+
+
To transfer data between ResearchDrive and CHTC, do the following:
+
+
+ - Log in:
+
+ - If you are transferring files to or from a
/staging directory, log in to transfer.chtc.wisc.edu.
+ - If you are transferring files and or from your
/home directory, log into your usual submit server (typically ap2001.chtc.wisc.edu or ap2002.chtc.wisc.edu).
+
+
+ - Choose a folder: Navigate to the folder in CHTC (
/staging or /home), where you would like to transfer files.
+ - Connect to ResearchDrive: Run the following command to connect to ResearchDrive, filling in the username of
+your PI:
+
[alice@server]$ smbclient -k //research.drive.wisc.edu/PI-Name
+
+
+ Your prompt should change to look like this:
+ smb: \>
+
+
+
+ Note about NetIDs
+ If your CHTC account is not tied to your campus NetID or you are accessing a data
+storage service that doesn’t use your NetID, you’ll need to omit the -k flag above
+
+
+ - Choose a folder, part 2: If you type
ls now, you’ll see the files in ResearchDrive, not CHTC.
+Navigate through ResearchDrive (using cd) until you are at the folder where you would
+like to get or put files.
+ - Move files: To move files, you will use the
get and put commands:
+
+ - To move files from CHTC to ResearchDrive, run:
+
smb: \> put filename
+
+
+ - To move files from ResearchDrive to CHTC, run:
+
smb: \> get filename
+
+
+
+
+ - Finish: Once you are done moving files, you can type
exit to leave the connection to ResearchDrive.
+
+
+
Transferring a Batch of Files
+
+
The steps described above work well for transferring a single file, or tar archive of
+many files, at a time and is best for transferring a few files in a session. However,
+smbclient also provides options for transferring many individual files in a single command
+using the * wildcard character.
+
+
To transfer multiple files at once, first you must turn off the smbclient file transfer prompt,
+then use either mget or mput for your file transfer. For example, if you have multiple fastq.gz files
+to transfer to CHTC:
+
+
+ - Log in:
+
+ - If you are transferring files to or from a
/staging directory, log in to transfer.chtc.wisc.edu.
+ - If you are transferring files to or from your
/home directory, log into your usual submit server (typically ap2001.chtc.wisc.edu or ap2002.chtc.wisc.edu).
+
+
+ - Choose a folder: Navigate to the folder in CHTC (
/staging or /home), where you would like to put the files.
+ - Connect to ResearchDrive: Run the following command to connect to ResearchDrive, filling in the username of
+your PI:
+
[alice@server]$ smbclient -k //research.drive.wisc.edu/PI-Name
+
+
+ - Navigate to appropriate ResearchDrive directory
+
smb: \> cd path/to/files
+
+
+ - Turn of Prompting
+
smb: \> prompt
+
+
+ - Use
mget instead of get
+ This command downloads a group of files that end with “fastq.gz” to CHTC.
+ smb: \> mget *.fastq.gz
+
+
+
+
+
As another example, use smbclient to transfer multiple tar.gz output files to ResearchDrive from CHTC
+after your jobs complete:
+
+
+ - Log in:
+
+ - If you are transferring files to or from a
/staging directory, log in to transfer.chtc.wisc.edu.
+ - If you are transferring files to or from your
/home directory, log into your usual submit server (typically ap2001.chtc.wisc.edu or ap2002.chtc.wisc.edu).
+
+
+ - Choose a folder: Navigate to the folder in CHTC (
/staging or /home) where your output files are located.
+ - Connect to ResearchDrive: Run the following command to connect to ResearchDrive, filling in the username of
+your PI:
+
[alice@server]$ smbclient -k //research.drive.wisc.edu/PI-Name
+
+
+ - Navigate to appropriate ResearchDrive directory
+
smb: \> cd path/to/directory
+
+
+ - Turn off Prompting
+
smb: \> prompt
+
+
+ - Use
mput instead of put
+ smb: \> mput *.tar.gz
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Transfer Files between CHTC and your Computer
+
+
+
+
+
+
Table of Contents
+
+
+
+
+
+
+
1. Transferring Files
+
+
To transfer files to and from CHTC, you will need the same username and
+hostname information for logging in, as well as understanding
+where your files are and where you would like them to go.
+
+
+
+
A. On the command line
+
+
On Mac, Linux, or modern Windows (10+) systems, you can use the "Terminal" application and
+the scp command to copy files between your computer and CHTC servers.
+
+
Your computer to CHTC
+
+
First, open the "Terminal" application and navigate to the directory
+with the files or directories you want to transfer. Then, use this
+command to copy these files to CHTC:
+
+
$ scp file username@hostname:/home/username
+
+
+
If you would like these files to end up in a different directory inside
+your home directory, just add it to the path at the end of the command.
+
+
CHTC to your computer
+
+
Open the "Terminal" application. Do NOT log into CHTC. Instead,
+navigate to where you want the files to go on your computer. Then, use
+this command to copy these files there:
+
+
$ scp username@hostname:/home/username/file ./
+
+
+
Again, for many files, it will be easiest to create a compressed tarball
+(.tar.gz file) of your files and transfer that instead of each file
+individually.
+
+
+
+
B. Using a file transfer program (Windows/Mac)
+
+
Windows and Mac users can also use special programs to help them
+transfer files between their computers and CHTC. For Windows, we
+recommend WinSCP. It requires the
+same information as Putty (hostname, username), and once it's set up,
+looks like this:
+
+

+
+
The left window is a directory on your computer, the right window is
+your home directory in CHTC. To move files between the two, simply drag
+and drop.
+
+
There are other programs besides WinSCP that do this. Another that works
+on Mac and Windows is called Cyberduck.
+
+
+
+
C. Transferring Multiple Files
+
+
If you are transferring many files, it is advantageous to compress them
+into a single compressed file, in order to facilitate transferring them.
+Place all the files you need in a directory, and then either zip it or
+use the "tar" command to compress them:
+
+
$ tar czf data_files.tar.gz file_directory/
+
+
+
To untar or unzip files on the submit server or head nodes, you can use
+either:
+
+
[alice@submit]$ tar xzf data_files.tar.gz
+
+
+
or
+
+
[alice@submit]$ unzip data_files.zip
+
+
+
+
+
2. Creating and Editing Files in CHTC
+
+
Once you have logged in to a CHTC server, you can edit files from the
+command line, by using a command line file editor. Some common editing
+programs are:
+
+
+
+
nano is the most beginner-friendly, and emacs is the most advanced.
+This Software Carpentry
+lesson describes
+how to use nano, and there are many other resources online with
+instructions for these text editors.
+
+
Some of the file transfer programs mentioned above
+allow you to edit files on CHTC servers through the interface.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+

+
+
+
+
+
+
+
+
+ Policies and Expectations for Using CHTC
+
+
+
This page lists important policies and expectations for using CHTC computing and
+data services. Our goal is to support a community of users and a variety of
+research. If an individual user is taking
+action that negatively impacts our services, we reserve the right to
+deactivate their account or remove files without notice.
+
+
Access and Use
+
+
Use of CHTC services are free to use in support of UW - Madison’s research and
+teaching mission.
+
+
Accounts are linked to individuals and should NOT be shared. We are happy to make new
+accounts for individuals or group-owned spaces for sharing files. Accounts that we
+notice being shared will be immediately disabled and a meeting with the PI
+(faculty advisor) may be necessary to reinstate the account.
+
+
For more information on the process for obtaining an account, see our
+How to Request an Account guide.
+
+
Data Policies
+
+
CHTC data locations are not backed up, and users should
+treat CHTC compute systems as temporary storage locations for active,
+currently-queued computational work. Users should remove data from CHTC
+systems upon completion of a batch of computational work and keep copies of
+all essential files in a non-CHTC location. CHTC staff reserve the right
+to delete data from any CHTC data location at at any time, to preserve
+systems performance, and are not responsible for data loss or file system
+corruption, which are possible in the absence of back-ups.
+
+
CHTC is not HIPAA-compliant and users should not bring HIPAA data into
+CHTC services. If you have data security concerns or any questions about
+data security in CHTC, please get in touch!
+
+
To request a change in the quotas for a storage location, please see
+our Request a Quota Change guide.
+
+
Export Control
+
+
Users agree not to access, utilize, store, or in any way run export controlled data, information,
+programs, etc. on CHTC software, equipment, or computing resources without prior review by the
+UW-Madison Export Control Office.
+
+
Export controlled information is subject to federal government rules on handling and viewing and has
+restrictions on who and where it may be accessed. A license can be required for access by foreign
+persons and in foreign jurisdictions so it’s important to ensure that all legal requirements are
+followed.
+If you have export controlled information that you would like to use on the CHTC, or you are unsure
+if the information you have is export controlled, please contact the Export Control Office at
+exportcontrol@grad.wisc.edu for guidance.
+
+
Note: The CHTC is not compliant with Controlled Unclassified Information (CUI) requirements.
+
+
User Expectations
+
+
Because our systems are shared by many CHTC users, everyone contributes to
+helping the systems run smoothly. The following are some best practices
+to get the most out of CHTC without harming other users. Our goal
+is always to help you get your work done - if you think the following recommendations
+limit your capacity to run work, please contact us to discuss alternatives.
+
+
Never run computationally intensive tasks on the login nodes for either
+system. As a rule of thumb, anything that runs for more than a few seconds, or
+is known to use a lot of cores or memory should not be run directly, but as a job.
+Small scripts and commands (to compress data, create directories,
+etc.) that run within a few minutes on the submit server are okay,
+but their use should be minimized when possible. If you have questions about this,
+please contact the facilitation team. CHTC staff reserve the right to kill any long-running or problematic processes on the
+head nodes and/or disable user accounts that violate this policy
+
+
Avoid unsupervised scripts on the login nodes. Automating tasks via tools like
+cron, watch, or using a workflow manager (not including HTCondor’s DAGMan) on the login node is not allowed without prior
+discussion with the facilitation or infrastructure team.
+
+
+ (HTC system specific): Since use of watch with condor_q is prohibited,
+we recommend using condor_watch_q as an alternative for live updates on your jobs
+in the queue. condor_watch_q is more efficient and will not impair system performance.
+
+
+
Test your jobs. We recommend testing a small version of your overall workflow
+before submitting your full workflow. By testing a smaller version of your jobs,
+you can determine resource requests, runtimes, and whether you may need an increase
+in your user quota. Both our HTC and HPC systems use a fair shair policy and each
+researcher has a user priority. Submitting many jobs that fail or do not produce
+the unexpected output will decrease your user priority without helping you complete
+your research. User priorities naturally reset over time.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
 +
+
+
+
+
+Join Hundreds of UW-Madison Researchers using CHTC Resources for:
+ ✓ RNA/DNA Sequencing Analyses ✓ Machine Learning Workflows
+ ✓ Economic Simulations & Predictions ✓ Weather Modeling Analyses
+ ✓ Chemical Reaction Predictions ✓ Computer Vision & Artificial Intelligence Decision Making
+ … and much more!
+
+Who We Are
+
+We are the University of Wisconsin-Madison’s core computational resource provider for large scale computing. UW-Madison staff, students, faculty, and external collaborators are welcome to use the Center for High Throughput Computing's (CHTC) resources to carry out their computationally-intensive research.
+
+CHTC provides a variety of resources and tools to meet the demands of the University’s research community.
+
+We provide no-cost compute resources (CPUs/GPUs, high-memory servers, etc.), as well as no-cost personalized consultations and classroom support.
+
+
+Your Home for Research Computing is Here
+
+As a leading research institution, UW-Madison needs a leading research computing center. Established in 2006, the Center for High Throughput Computing (CHTC), aims to bring the power of High Throughput Computing (HTC) to all fields of research and to allow the future of HTC to be shaped by insight from all fields. To advance this mission, the CHTC provides researchers access to state-of-the-art High Throughput Computing and High Performance Computing systems, as well as tools for data management and specalized hardware.
+
+Beyond CHTC's compute resources, CHTC’s Research Facilitation team helps researchers of all backgrounds identify their needs for large scale computing, practice skills needed to do their work, and provide support for implementing workflows.
+
+They offer the following services, free-of-charge:
+• Office Hours twice a week
+• Personal consultations to provide individualized guidance
+• Workshops and other informational events
+• Yearly week-long HTC summer school
+• Guest presentations for courses, seminars, and other groups
+• Email support
+
+
diff --git a/preview-fall2024-info/veritas.html b/preview-fall2024-info/veritas.html
new file mode 100644
index 000000000..b737c18de
--- /dev/null
+++ b/preview-fall2024-info/veritas.html
@@ -0,0 +1,389 @@
+
+
+
+
+
+
+VERITAS and OSG explore extreme window into the universe
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ VERITAS and OSG explore extreme window into the universe
+
+
Understanding the universe has always fascinated mankind. The VERITAS Cherenkov telescope array unravels its secrets by detecting very-high-energy gamma rays from astrophysics sources.
+
+
Gamma-ray astronomy studies the most energetic form of electromagnetic radiation, with photon energies in the GeV – TeV range (gigaelectronvolt to teraelectronvolt). The Very Energetic Radiation Imaging Telescope Array System (VERITAS) uses four 12-meter diameter imaging telescopes. The system uses the Imaging Atmospheric Cherenkov Telescope (IACT) technique to observe gamma rays that cause particle showers in Earth’s upper atmosphere.
+
+
Inside the system, there are four photo-multiplier, 499-pixel cameras, each 0.15 degree in diameter, which record images of the showers by detecting the Cherenkov light emitted by particles in the air shower. The field of view of each camera is 3.5 degrees.
+
+
There are currently three operating IACT arrays: H.E.S.S., MAGIC, and VERITAS. VERITAS is sensitive to very-high-energy (VHE) gamma rays in the energy range between ~80 GeV up to several tens of TeV. It is one of the most sensitive instruments in that energy band.
+
+
+  + The Very Energetic Radiation Imaging Telescope Array System (VERITAS) uses four 12-meter diameter imaging telescopes. The system uses the Imaging Atmospheric Cherenkov Telescope (IACT) technique to observe gamma rays that cause particle showers in Earth’s upper atmosphere. The picture shows the four telescopes at their permanent location at the Fred Lawrence Whipple Observatory (FLWO) observatory one hour south of Tucson, AZ. Courtesy Nepomuk Otte.
+
+ The Very Energetic Radiation Imaging Telescope Array System (VERITAS) uses four 12-meter diameter imaging telescopes. The system uses the Imaging Atmospheric Cherenkov Telescope (IACT) technique to observe gamma rays that cause particle showers in Earth’s upper atmosphere. The picture shows the four telescopes at their permanent location at the Fred Lawrence Whipple Observatory (FLWO) observatory one hour south of Tucson, AZ. Courtesy Nepomuk Otte.
+
+
+
A. Nepomuk Otte, Ph.D., is an assistant professor in the School of Physics at the Georgia Institute of Technology. He works in the Center for Relativistic Astrophysics and is a collaborator in VERITAS.
+
+
“We use the VERITAS telescopes in Arizona to study black holes, the remnants of exploding stars, pulsars, and other objects in the sky,” says Otte. “A high-energy gamma ray has a trillion times more energy than a light particle from the sun. When such a gamma ray hits the atmosphere, it produces millions of electrons and positrons that travel faster than the speed of light through the atmosphere.”
+
+
Otte explains that these charged particles emit a bluish flash of light as they zip through the atmosphere. The VERITAS telescopes collect that light and project it onto special cameras to take an image of the particle shower.
+
+
“In our analysis software, we compare the recorded images with simulated ones to find out if a shower was produced by an actual gamma ray or a cosmic ray, which would be a background event,” says Otte. “We also have to compare our events with simulated ones to reconstruct the energy and its origin in the sky—everything we need for a full reconstruction. For our analysis, it is crucial that we properly simulate our experiment to make sense of the data.”
+
+
Otte relies on the Open Science Pool to run the simulations. “Without simulations, we are blind because the characteristics of each recorded image depend on too many parameters to be described analytically,” says Otte. “We have to repeat every step of the experiment in the computer, from the gamma-ray interaction in the atmosphere up to the point where the digitized photon detector signals are written to disk about 100 million times. That is a very time-consuming process.” Otte then compares each recorded event with simulated ones. “The simulated events that best match the recorded event tell us what the energy of the recorded event was and whether it was a gamma ray or a cosmic ray.”
+
+
VERITAS began recording data ten years ago. Over that time span, VERITAS accumulated 10,000 hours of observations on more than 100 objects. Some objects were observed for more than 300 hours. The analysis of these large data sets is sensitive to even small differences between the experiment and the simulations, which was not important when VERITAS started. Two years ago, the VERITAS collaboration reworked the simulation models to account for these small differences by including more details about the experiment itself.
+
+
“We had to rewrite large fractions of our simulation code,” says Otte. “The added detail also meant we needed more computing power. In the past, we could do our simulations on a few hundred CPUs. Now, we need a hundred times more power because we want to simulate ten times more showers than before.”
+
+
OSG gives the VERITAS collaboration the computing power they need. “Using free cycles that others are not using is almost perfect for us,” says Otte. He and his group started using the OSG in August 2016. Initially, Otte wrote an XSEDE allocation application to use Stampede, and the XSEDE experts recommended OSG as a better fit for the project. “I knew about OSG, having used it for other experiments,” says Otte, “but the shared free cycles in this case was a huge help.”
+
+
Otte says the grand challenge in their field is to look at the air showers in the atmosphere; they see very few gamma rays and just a handful of them observed for over 100 hours. At the same time, millions of background events are recorded that are cosmic rays but look very similar to gamma-ray air showers. “So, our challenge is to dig needles out of a huge haystack,” says Otte. “This has been a huge challenge for decades.”
+
+
It was possible only after realizing the power of image analysis of air showers in the late 1980s to distinguish between gamma-ray events and background events with very high efficiency. The simulations tell what features to look for in the images to suppress the background events in the analysis.
+
+
“Simulations are crucial,” says Otte. “We could not make sense of the data without them. And now with bigger data sets it has become very important to also include aspects of the telescopes that did not matter before. For example, we have now recorded events with energies of several tens of TeV. These events are extremely rare, but we have them in our data. The images of these events are so bright that a lot of the camera pixels saturate. We had not included these saturation effects in our simulations before and thus made large errors in reconstructing the energy of these events.”
+
+
After the VERITAS analysis is done, the data are combined with observations in X-ray, radio, and optical and compared with models that try to explain what happens inside the source. One of the important science drivers for VERITAS is to find the origin of cosmic rays, which is a century-old puzzle. “The remnants of supernovae are prime candidates to accelerate cosmic rays,” says Otte. “In some cases, we resolve the expanding shell in gamma rays and can directly see where the cosmic rays come from.”
+
+
Otte uses the OSG mostly for the simulations. “We don’t need these massive computing resources like other experiments might,” says Otte. “Running the simulations is a single effort that takes a lot of time and a lot of computing resources. Buying resources for such a short time would not be sustainable. The sharing concept of OSG is perfect for us. We borrow the resources, do production, have the data on disk, and then do our science for the next few years on our local computing clusters at the universities.”
+
+
Without the OSG, Otte says they would be stuck with local clusters, and that would hold them back. Another important aspect for the VERITAS collaboration is they have groups across the nation with computing resources, but only the OSG can combine them all into one big virtual computing cluster. That makes them far more productive. “With tens of terabytes of data,” says Otte, “the grid makes things much easier.”
+
+
“With the help of the OSG, we are exploring a new and very exciting window into the most extreme objects in our universe. Like black holes and exploding stars, we study the origin of dark matter, which makes up 25 percent of the universe, and we don’t even understand what it is. We explore the evolution of the universe and can even test the fabric of space-time. VERITAS is a very versatile tool and a world-leading instrument.”
+
+
“As we pursue our research, we develop new technologies and algorithms. These find use in other areas as well. For example, the photon detector technology we use is also used in apparatus for cancer screening and diagnostics. And our algorithms can apply to and be used for other large data sets.”
+
+
+  + Research group of Nepomuk Otte. Courtesy Nepomuk Otte.
+
+ Research group of Nepomuk Otte. Courtesy Nepomuk Otte.
+
+
+
+
+
 +
+ +
+  +
+  +
+ +
+ +
+  on the African School of Physics website.](https://raw.githubusercontent.com/CHTC/Articles/main/images/asp-banner.jpeg) +
+  +
+  +
+  +
+ 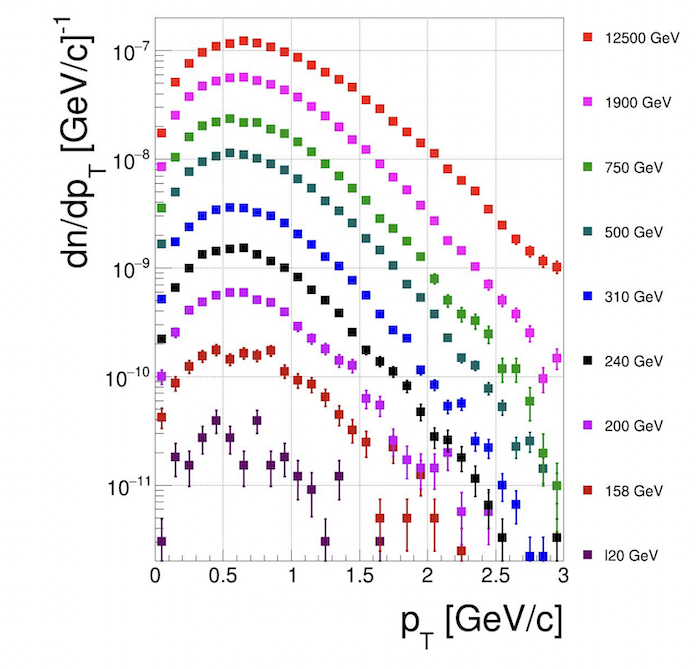 +
+  +
+  +
+ 
 +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+ 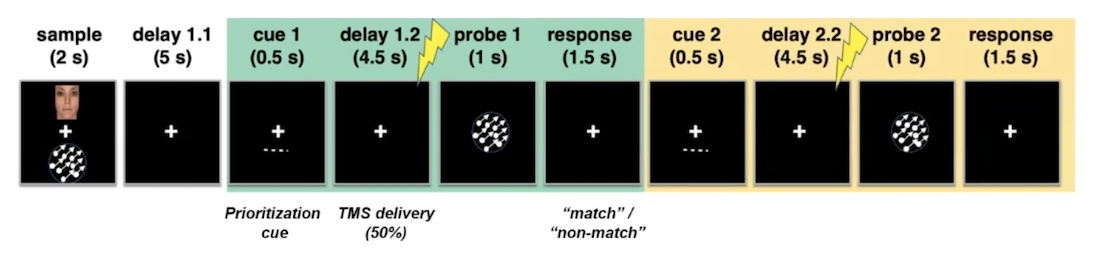 +
+  +
+  +
+  +
+  +
+ 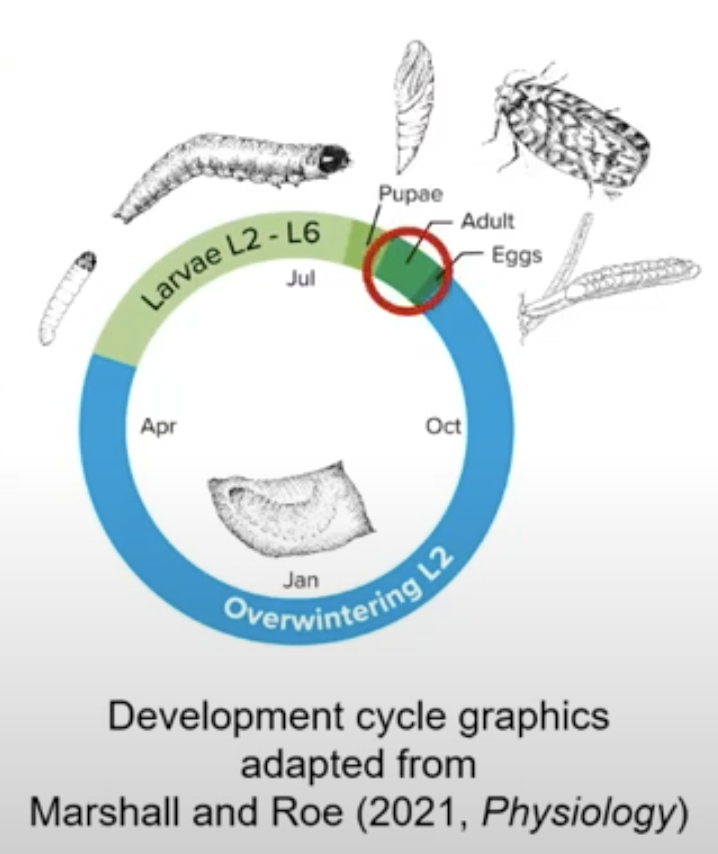 +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+ 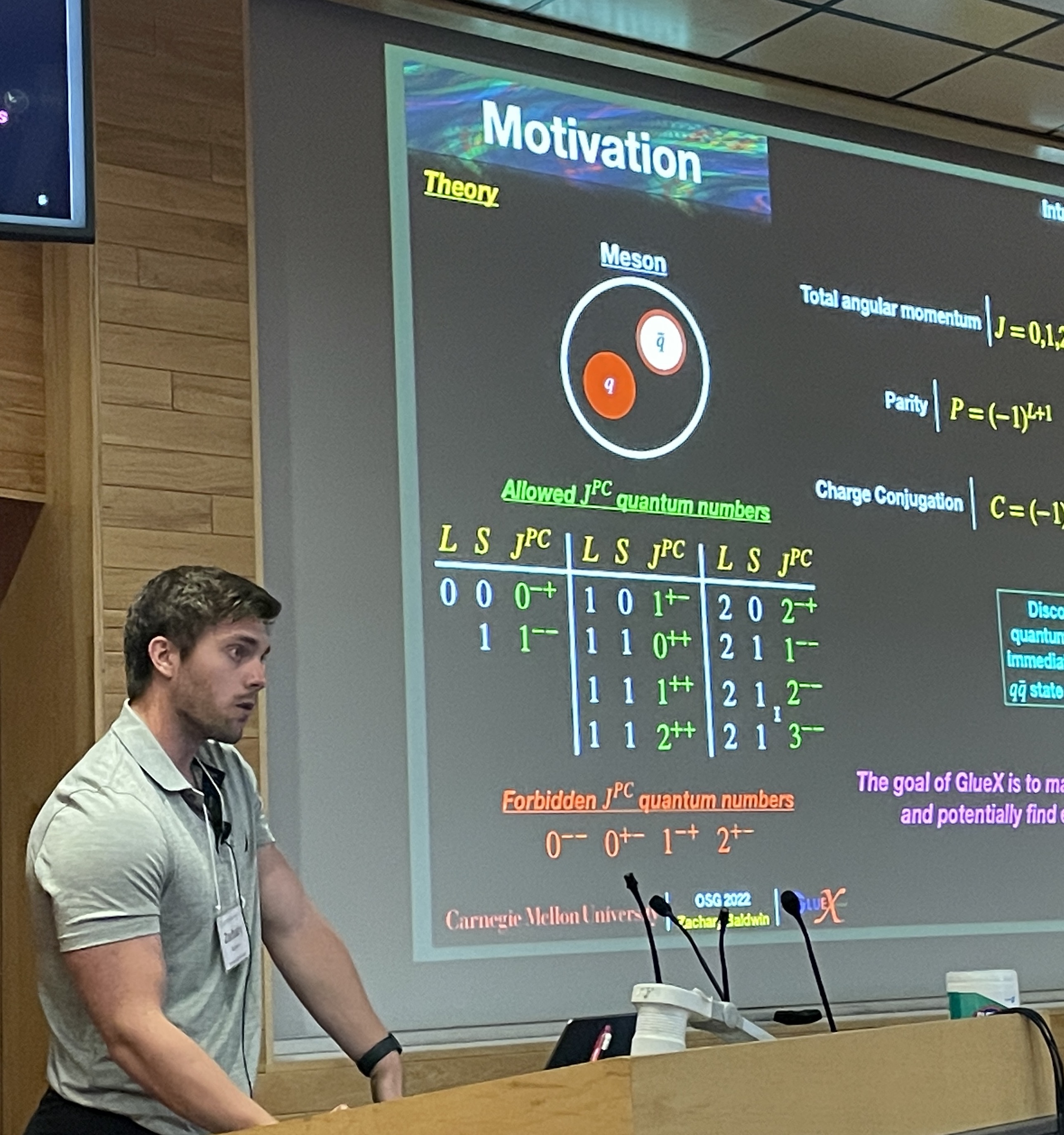 +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+ 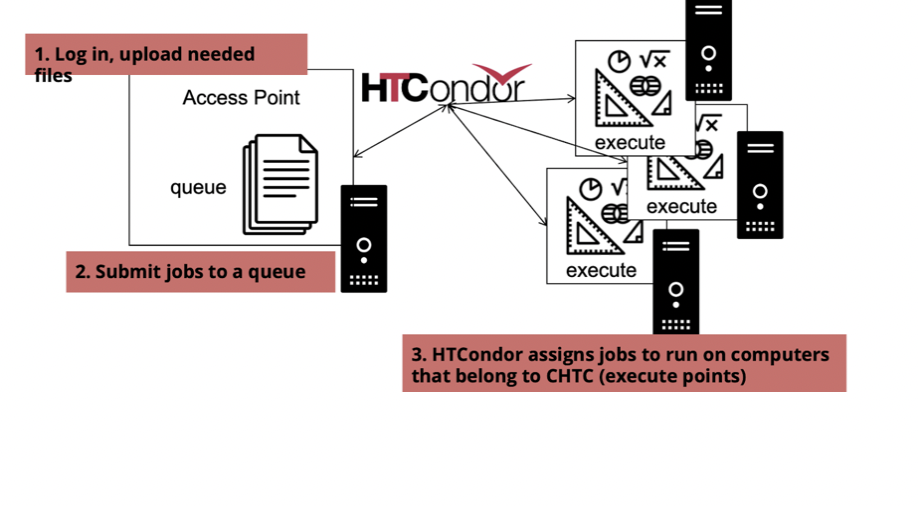 +
+ 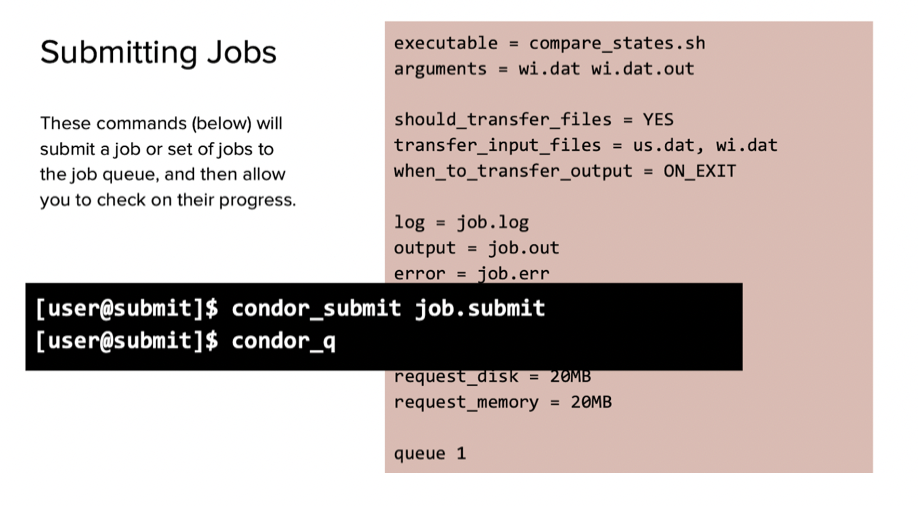 +
+ 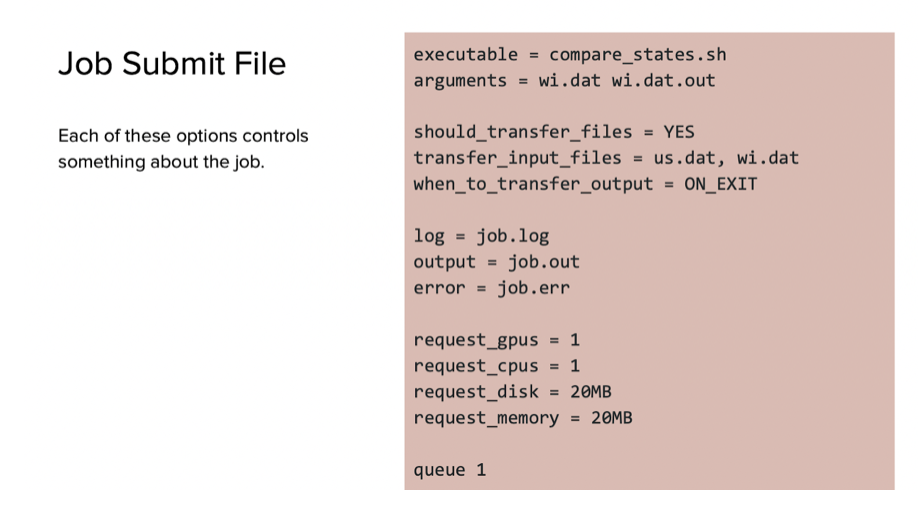 +
+  +
+ 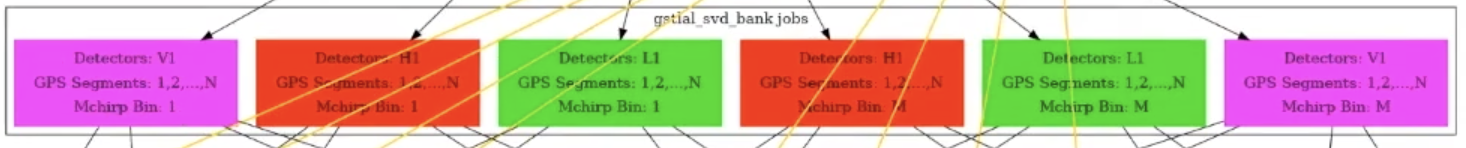 +
+  +
+  +
+  +
+  +
+  +
+  +
+ 
 +
+ 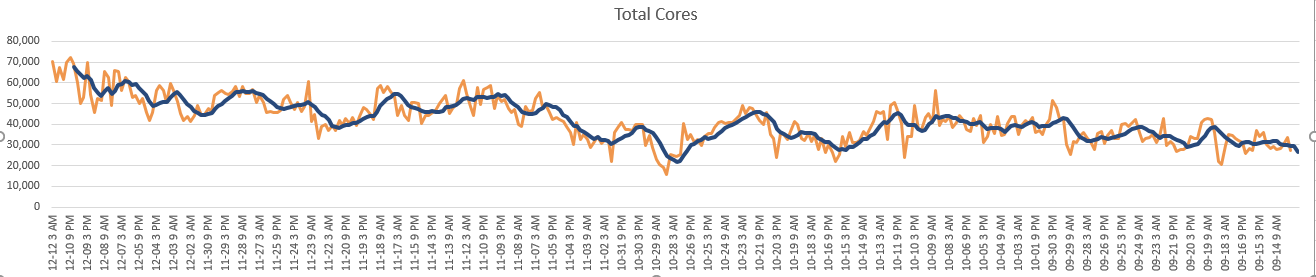 +
+  +
+  +
+  +
+  +
+ 


 +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+ 
 +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+ +
+  +
+  +
+
+
+.](https://raw.githubusercontent.com/CHTC/Articles/main/images/materials-science.jpg) +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+ 
 +
+ 





















 +
+
 +
+  +
+  +
+  +
+
+
+  +
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+
+
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+ 
 +
+ 












 +
+  +
+  +
+  +
+ 
 +
+ +
+  +
+  +
+ 

 +
+ 

 +
+  +
+