-1.6.6.dev15+gd9f20b3
+1.7.2.dev0+g8abf70a.d20240415
Why do we need a better way to visualize the gene set enrichment analysis result?¶
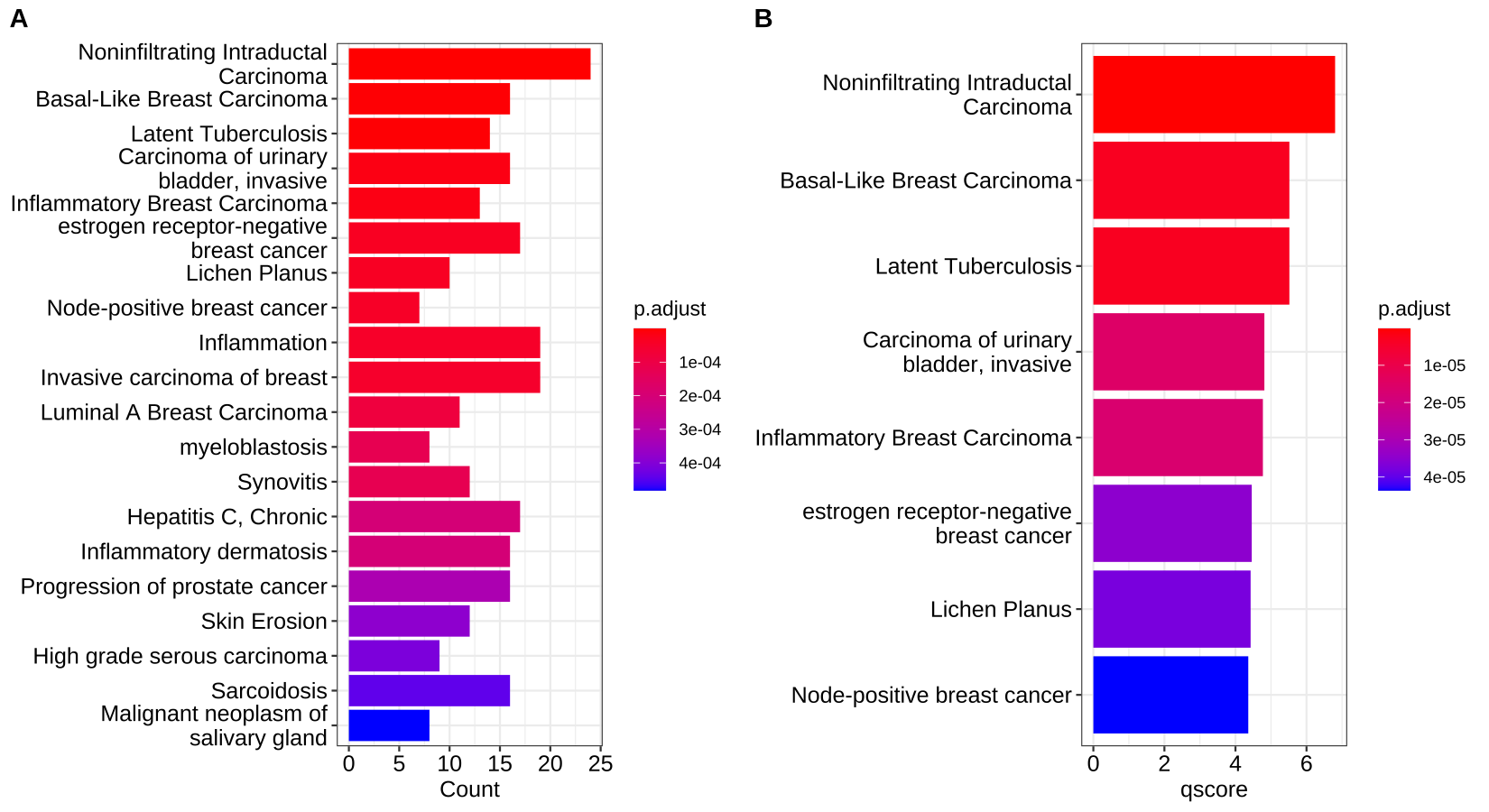
Traditionally, we have to plot lots of barplot or dotplot to visualize the gene set enrichment analysis. For example:
-
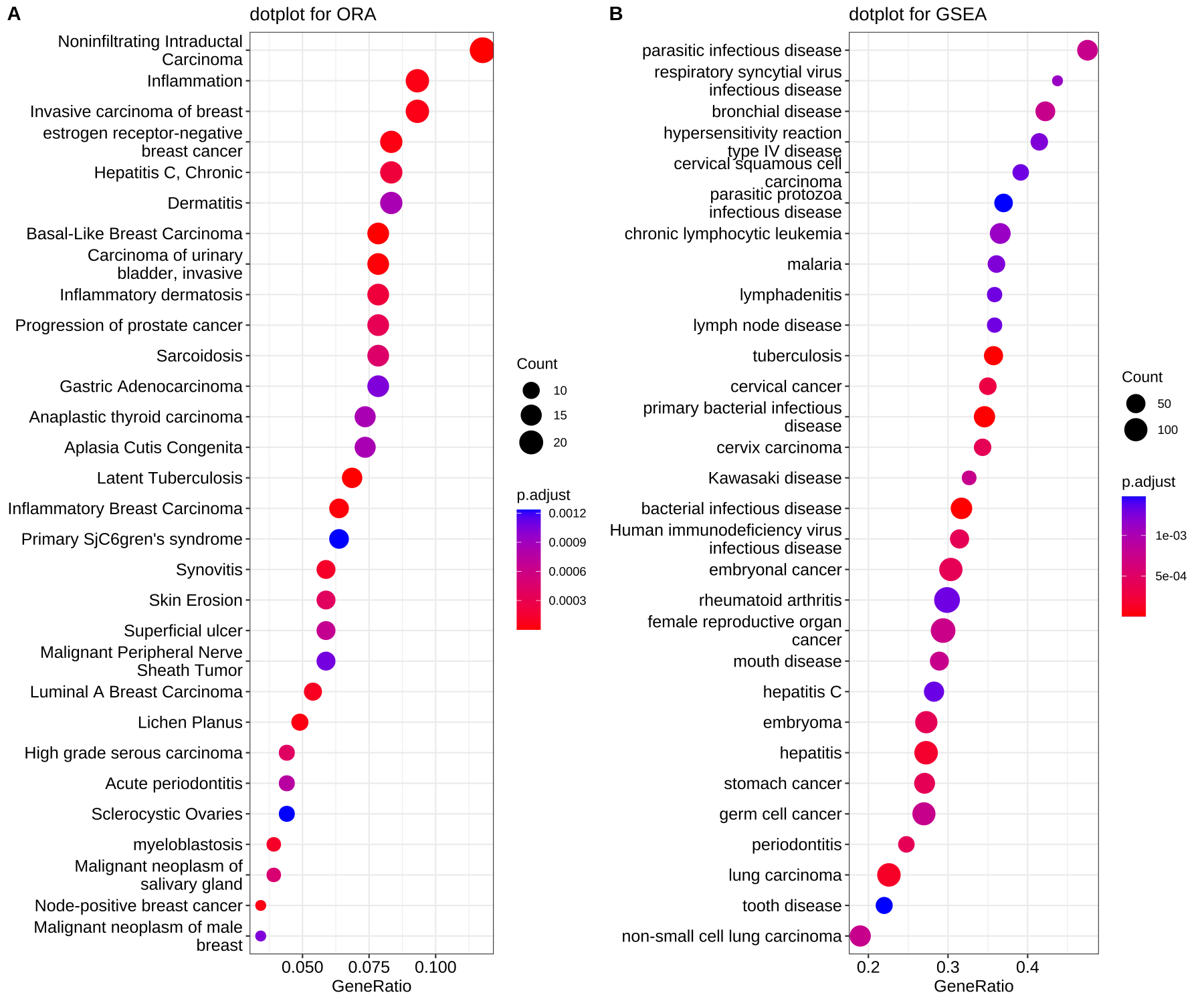


What if we had lots of cell types and performed gene set enrichment analysis for each cell types? It is not a good idea to plot lots of barplot or dotplot in the manuscript.
In this case, we can merge all the enriched result into one matrix and plot a dotHeatmap using PyComplexHeatmap
Plot¶<
-[13]:
+[9]:
left_ha = HeatmapAnnotation(
@@ -1151,7 +1151,7 @@ In this example, we used two different kinds of markers and cmap to indic
How to select marker:¶
-
+
diff --git a/docs/build/html/notebooks/gene_enrichment_analysis.ipynb b/docs/build/html/notebooks/gene_enrichment_analysis.ipynb
index 7b6fb578..b88308e1 100644
--- a/docs/build/html/notebooks/gene_enrichment_analysis.ipynb
+++ b/docs/build/html/notebooks/gene_enrichment_analysis.ipynb
@@ -20,7 +20,7 @@
"name": "stdout",
"output_type": "stream",
"text": [
- "1.6.6.dev15+gd9f20b3\n"
+ "1.7.2.dev0+g8abf70a.d20240415\n"
]
}
],
@@ -966,7 +966,7 @@
},
{
"cell_type": "code",
- "execution_count": 13,
+ "execution_count": 9,
"id": "8683f2d4-cea6-445f-ae32-cbfdb835b2ee",
"metadata": {
"tags": []
diff --git a/docs/build/html/notebooks/get_started.html b/docs/build/html/notebooks/get_started.html
index f3f8152d..7a5f4878 100644
--- a/docs/build/html/notebooks/get_started.html
+++ b/docs/build/html/notebooks/get_started.html
@@ -4,7 +4,7 @@
-
1. Import packages — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ 1. Import packages — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -18,7 +18,7 @@
-
+
diff --git a/docs/build/html/notebooks/kwargs.html b/docs/build/html/notebooks/kwargs.html
index eedb5901..4ace87e6 100644
--- a/docs/build/html/notebooks/kwargs.html
+++ b/docs/build/html/notebooks/kwargs.html
@@ -4,7 +4,7 @@
- label_kws, xticklabels_kws and yticklabels_kws: Modifying the heatmap annotations labels and xticklabels, yticklabels — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ label_kws, xticklabels_kws and yticklabels_kws: Modifying the heatmap annotations labels and xticklabels, yticklabels — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -18,7 +18,7 @@
-
+
@@ -587,7 +587,7 @@ Control gap & pad in heatmap
+
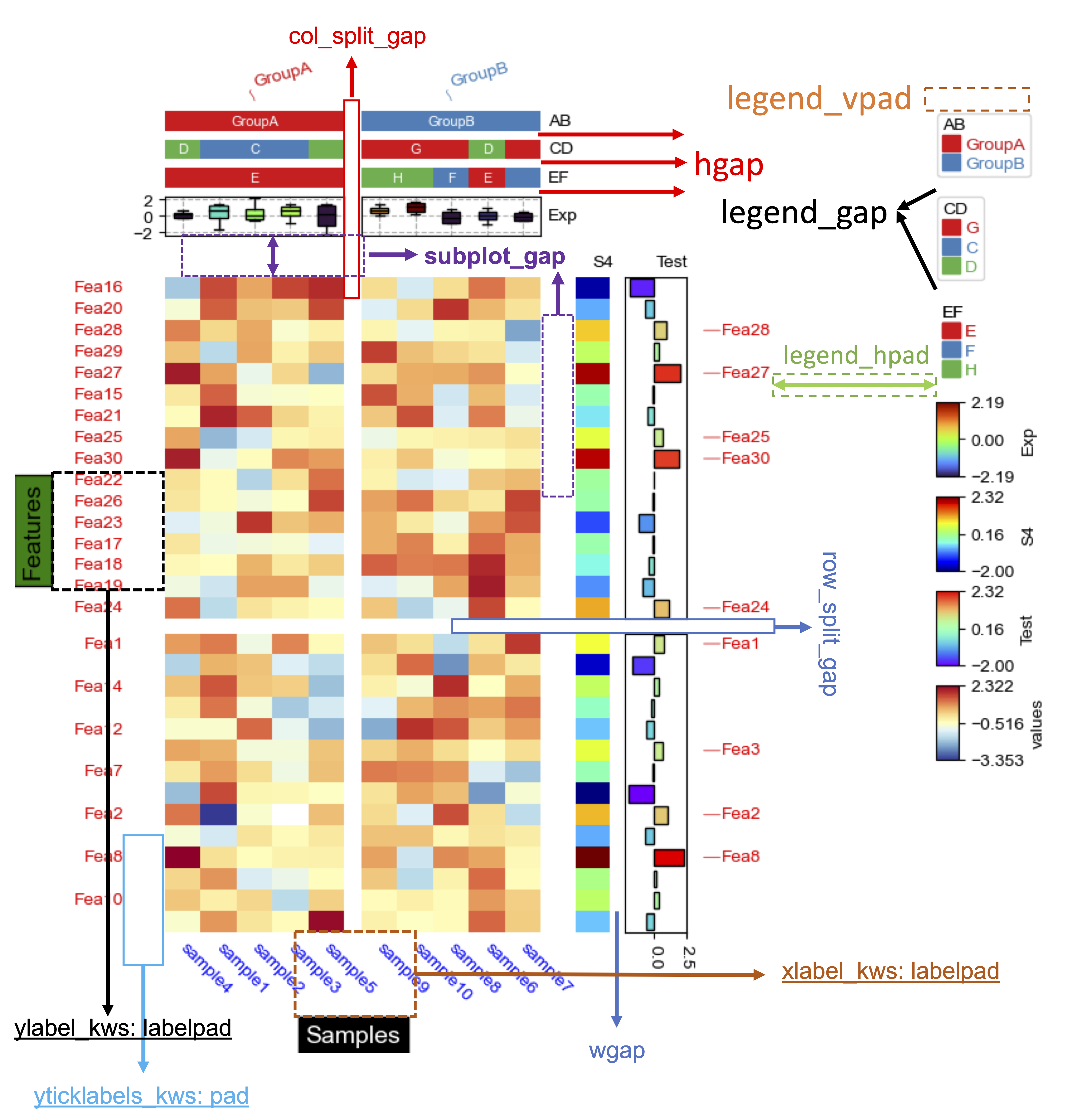
hgap and wgap for HeatmapAnnotation¶
control the gap between two rows/columns of column annotations (axis=1) or row annotations (axis=0), unit is mm
diff --git a/docs/build/html/notebooks/oncoPrint.html b/docs/build/html/notebooks/oncoPrint.html
index f365ab9a..44931732 100644
--- a/docs/build/html/notebooks/oncoPrint.html
+++ b/docs/build/html/notebooks/oncoPrint.html
@@ -4,7 +4,7 @@
- oncoPrint: visualizing TCGA Lung Adenocarcinoma Carcinoma Variants Dataset — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ oncoPrint: visualizing TCGA Lung Adenocarcinoma Carcinoma Variants Dataset — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -18,7 +18,7 @@
-
+
diff --git a/docs/build/html/notebooks/oncoPrint2.html b/docs/build/html/notebooks/oncoPrint2.html
index c1858cf2..7951afc5 100644
--- a/docs/build/html/notebooks/oncoPrint2.html
+++ b/docs/build/html/notebooks/oncoPrint2.html
@@ -4,7 +4,7 @@
- Visualizing categorical variables using oncoPrint — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ Visualizing categorical variables using oncoPrint — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -18,7 +18,7 @@
-
+
diff --git a/docs/build/html/notebooks/single_cell_methylation.html b/docs/build/html/notebooks/single_cell_methylation.html
index f3e9fcf6..c17c827c 100644
--- a/docs/build/html/notebooks/single_cell_methylation.html
+++ b/docs/build/html/notebooks/single_cell_methylation.html
@@ -4,7 +4,7 @@
- Visualizing Single Cell DNA Methylation Data (Loyfer2023) — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ Visualizing Single Cell DNA Methylation Data (Loyfer2023) — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -18,7 +18,7 @@
-
+
diff --git a/docs/build/html/notebooks/test_add_anno_img.html b/docs/build/html/notebooks/test_add_anno_img.html
index 046fc16a..f87026aa 100644
--- a/docs/build/html/notebooks/test_add_anno_img.html
+++ b/docs/build/html/notebooks/test_add_anno_img.html
@@ -4,7 +4,7 @@
- <no title> — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ <no title> — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -18,7 +18,7 @@
-
+
diff --git a/docs/build/html/oncoPrint.html b/docs/build/html/oncoPrint.html
index 71f77c50..dbc9441c 100644
--- a/docs/build/html/oncoPrint.html
+++ b/docs/build/html/oncoPrint.html
@@ -4,7 +4,7 @@
- oncoPrint — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ oncoPrint — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -17,7 +17,7 @@
-
+
diff --git a/docs/build/html/py-modindex.html b/docs/build/html/py-modindex.html
index 835d8bb0..6bfec726 100644
--- a/docs/build/html/py-modindex.html
+++ b/docs/build/html/py-modindex.html
@@ -3,7 +3,7 @@
- Python Module Index — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ Python Module Index — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -16,7 +16,7 @@
-
+
diff --git a/docs/build/html/search.html b/docs/build/html/search.html
index b239271f..c67885b0 100644
--- a/docs/build/html/search.html
+++ b/docs/build/html/search.html
@@ -3,7 +3,7 @@
- Search — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ Search — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -17,7 +17,7 @@
-
+
diff --git a/docs/build/html/searchindex.js b/docs/build/html/searchindex.js
index e391b40f..9354eff2 100644
--- a/docs/build/html/searchindex.js
+++ b/docs/build/html/searchindex.js
@@ -1 +1 @@
-Search.setIndex({"docnames": ["PyComplexHeatmap", "PyComplexHeatmap.annotations", "PyComplexHeatmap.clustermap", "PyComplexHeatmap.colors", "PyComplexHeatmap.dotHeatmap", "PyComplexHeatmap.example", "PyComplexHeatmap.oncoPrint", "PyComplexHeatmap.tools", "PyComplexHeatmap.utils", "advanced_usage", "citation", "clustermap", "dev", "dotHeatmap", "gallery", "get_started", "index", "installation", "kwargs", "modules", "more_examples", "notebooks/advanced_usage", "notebooks/bicluster", "notebooks/clustermap", "notebooks/composite_heatmaps", "notebooks/cpg_modules", "notebooks/dev", "notebooks/differential_expression_genes", "notebooks/dotHeatmap", "notebooks/gene_enrichment_analysis", "notebooks/get_started", "notebooks/kwargs", "notebooks/oncoPrint", "notebooks/oncoPrint2", "notebooks/single_cell_methylation", "notebooks/test_add_anno_img", "oncoPrint", "setup"], "filenames": ["PyComplexHeatmap.rst", "PyComplexHeatmap.annotations.rst", "PyComplexHeatmap.clustermap.rst", "PyComplexHeatmap.colors.rst", "PyComplexHeatmap.dotHeatmap.rst", "PyComplexHeatmap.example.rst", "PyComplexHeatmap.oncoPrint.rst", "PyComplexHeatmap.tools.rst", "PyComplexHeatmap.utils.rst", "advanced_usage.rst", "citation.rst", "clustermap.rst", "dev.rst", "dotHeatmap.rst", "gallery.md", "get_started.rst", "index.rst", "installation.rst", "kwargs.rst", "modules.rst", "more_examples.rst", "notebooks/advanced_usage.ipynb", "notebooks/bicluster.ipynb", "notebooks/clustermap.ipynb", "notebooks/composite_heatmaps.ipynb", "notebooks/cpg_modules.ipynb", "notebooks/dev.ipynb", "notebooks/differential_expression_genes.ipynb", "notebooks/dotHeatmap.ipynb", "notebooks/gene_enrichment_analysis.ipynb", "notebooks/get_started.ipynb", "notebooks/kwargs.ipynb", "notebooks/oncoPrint.ipynb", "notebooks/oncoPrint2.ipynb", "notebooks/single_cell_methylation.ipynb", "notebooks/test_add_anno_img.ipynb", "oncoPrint.rst", "setup.rst"], "titles": ["PyComplexHeatmap package", "PyComplexHeatmap.annotations module", "PyComplexHeatmap.clustermap module", "PyComplexHeatmap.colors module", "PyComplexHeatmap.dotHeatmap module", "PyComplexHeatmap.example module", "PyComplexHeatmap.oncoPrint module", "PyComplexHeatmap.tools module", "PyComplexHeatmap.utils module", "Advanced Usage", "Citation", "Clustermap", "For Developers", "dotHeatmap", "Gallery", "Get Started", "PyComplexHeatmap", "Installation", "Kwargs", "PyComplexHeatmap", "More Examples", "Generate dataset", "Import packages", "A quick example", "Composite two heatmaps horizontally for mouse DNA methylation array dataset", "Plot correlation matrix for CpG modules using DotClustermap", "1. Understand the layout:", "Visualizing Differential Expression Genes", "Load an example brain networks dataset from seaborn", "Visualizing gene set enrichment analysis results (clusterProfiler) using dotHeatmap", "1. Import packages", "label_kws, xticklabels_kws and yticklabels_kws: Modifying the heatmap annotations labels and xticklabels, yticklabels", "oncoPrint: visualizing TCGA Lung Adenocarcinoma Carcinoma Variants Dataset", "Visualizing categorical variables using oncoPrint", "Visualizing Single Cell DNA Methylation Data (Loyfer2023)", "<no title>", "oncoPrint", "setup module"], "terms": {"annot": [0, 2, 4, 6, 9, 11, 15, 16, 18, 19, 22, 24, 26, 28, 29, 32, 33, 34, 35], "annotationbas": [0, 1, 2, 19], "get_label_width": [0, 1], "get_max_label_width": [0, 1], "get_ticklabel_width": [0, 1], "reorder": [0, 1, 2, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "set_axes_kw": [0, 1], "set_label": [0, 1], "set_legend": [0, 1], "update_plot_kw": [0, 1], "heatmapannot": [0, 1, 2, 18, 19, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 32, 33, 34, 35], "collect_legend": [0, 1, 2, 4, 6], "plot_annot": [0, 1], "plot_legend": [0, 1, 2, 21, 23, 27, 34], "show_ticklabel": [0, 1, 21, 23, 30], "anno_barplot": [0, 1, 19, 21, 23, 25, 26, 30, 31, 32, 33], "plot": [0, 1, 2, 4, 6, 8, 9, 11, 13, 15, 16, 20, 22, 24, 27, 31, 32, 33, 34, 35], "anno_boxplot": [0, 1, 19, 21, 23, 26, 30, 31, 35], "anno_img": [0, 1, 19, 21], "anno_label": [0, 1, 16, 18, 19, 21, 22, 24, 25, 26, 27, 28, 29, 30, 32, 33, 34], "set_plot_kw": [0, 1], "set_sid": [0, 1], "anno_lineplot": [0, 1, 19, 21, 31], "anno_scatterplot": [0, 1, 19, 21, 23, 30, 31], "anno_simpl": [0, 1, 19, 21, 22, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "clustermap": [0, 1, 4, 6, 13, 16, 19, 20, 22, 23, 24], "clustermapplott": [0, 2, 4, 6, 19, 21, 22, 23, 24, 26, 27, 30, 31, 34, 35], "cal_cold_between_group": [0, 2], "cal_rowd_between_group": [0, 2], "calculate_col_dendrogram": [0, 2], "calculate_row_dendrogram": [0, 2], "format_data": [0, 2, 4, 6], "plot_dendrogram": [0, 2], "plot_matrix": [0, 2, 4, 6], "post_process": [0, 2, 4, 6], "set_axes_labels_kw": [0, 2], "set_height": [0, 2], "set_width": [0, 2], "set_xy_label": [0, 2], "standard_scal": [0, 2, 4, 6, 22], "tight_layout": [0, 2], "z_score": [0, 2, 4, 6, 21, 27], "dendrogramplott": [0, 2, 19, 22], "calculate_dendrogram": [0, 2], "calculated_linkag": [0, 2], "check_arrai": [0, 2], "get_coord": [0, 2], "reordered_ind": [0, 2], "composit": [0, 2, 16, 19, 20], "heatmap": [0, 1, 2, 4, 6, 9, 13, 15, 16, 18, 19, 20, 23, 25, 27, 32], "heatmapplott": [0, 2, 19], "plot_heatmap": [0, 2, 19], "color": [0, 1, 2, 4, 6, 8, 13, 16, 19, 21, 22, 23, 24, 25, 26, 27, 29, 30, 31, 32, 33, 34, 35], "define_cmap": [0, 3, 8, 19], "register_cmap": [0, 3, 19, 25], "dotheatmap": [0, 16, 19, 28], "dotclustermapplott": [0, 4, 13, 16, 19, 25, 29], "dotheatmap2d": [0, 4, 19], "scale": [0, 2, 4, 19, 22], "exampl": [0, 1, 2, 8, 11, 13, 16, 19, 21, 24, 25, 26, 29, 30, 31, 35], "clustermap_example0": [0, 5, 19], "clustermap_example1": [0, 5, 19], "get_kycg_example_data": [0, 5, 19], "oncoprint": [0, 16, 19], "oncoprintplott": [0, 6, 19, 32, 33], "add_default_annot": [0, 6], "get_samples_ord": [0, 6], "util": [0, 19, 21], "axis_ticklabels_overlap": [0, 8, 19], "cal_legend_width": [0, 8, 19], "cluster_label": [0, 8, 19], "despin": [0, 8, 19, 32, 33], "get_colormap": [0, 8, 19, 21], "plot_cmap_legend": [0, 4, 6, 8, 19], "plot_color_dict_legend": [0, 4, 6, 8, 19], "plot_legend_list": [0, 8, 19], "plot_marker_legend": [0, 4, 8, 19], "set_default_styl": [0, 8, 19], "to_utf8": [0, 8, 19], "class": [1, 2, 4, 6, 22], "df": [1, 21, 22, 23, 24, 26, 28, 30, 31, 35], "none": [1, 2, 4, 6, 8, 21, 22, 23, 24, 26, 27, 30, 31], "cmap": [1, 2, 4, 6, 8, 9, 13, 16, 22, 23, 24, 25, 26, 27, 29, 30, 31, 34, 35], "auto": [1, 2, 21], "height": [1, 2, 8, 21, 22, 23, 25, 26, 27, 29, 30, 31, 32, 33], "legend": [1, 2, 4, 6, 8, 21, 22, 24, 25, 27, 28, 29, 30, 31, 32, 33, 34, 35], "legend_kw": [1, 2, 4, 6, 8, 21, 23, 26, 31, 34], "plot_kw": [1, 6, 33], "sourc": [1, 2, 3, 4, 5, 6, 8, 14], "base": [1, 2, 4, 6, 8, 28], "object": [1, 2, 8, 22, 25, 27, 34], "paramet": [1, 2, 4, 6, 8, 9, 13, 16, 23, 25, 31], "datafram": [1, 2, 4, 6, 21, 26, 27, 30, 31, 32, 33, 35], "panda": [1, 2, 4, 6, 17, 22, 27, 29, 33], "seri": [1, 2], "onli": [1, 4, 6, 9, 16, 31], "one": [1, 4, 8, 23, 29], "column": [1, 2, 4, 6, 15, 16, 21, 22, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "str": [1, 2, 4, 6, 8, 21, 23, 25, 26, 30, 31, 32, 33, 35], "colormap": [1, 2, 4, 6, 9, 16, 25, 28], "set1": [1, 4, 6, 8, 21, 23, 27, 28, 29, 30, 31, 35], "dark2": [1, 8, 21, 23, 25, 27, 28, 30, 35], "bwr": [1, 2], "red": [1, 2, 4, 21, 23, 24, 25, 26, 27, 28, 30, 31, 32, 33], "jet": [1, 2, 21, 23, 25, 28, 30], "hsv": [1, 8], "rainbow": [1, 21, 26, 30, 31], "so": [1, 2], "pleas": [1, 2, 4, 10, 17, 30, 31], "see": [1, 2, 4, 21, 31], "http": [1, 2, 4, 10, 17, 21, 31, 32, 34], "matplotlib": [1, 2, 4, 8, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "org": [1, 2, 4, 10], "3": [1, 4, 15, 16, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "5": [1, 2, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "0": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "tutori": 1, "html": [1, 2, 4, 31, 32], "more": [1, 2, 16, 21, 23, 31, 32], "inform": [1, 2, 31], "run": [1, 17], "pyplot": [1, 21, 25, 27], "all": [1, 2, 4, 8, 9, 16, 23, 29, 31], "availabel": 1, "default": [1, 2, 4, 6, 8, 21, 28, 29, 31], "i": [1, 2, 4, 6, 8, 16, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 33, 35], "would": [1, 2, 21, 23], "determin": [1, 2, 4, 21, 23, 28], "dtype": [1, 2, 21, 22, 25, 26, 27, 28, 34], "each": [1, 2, 6, 23, 29, 30, 33], "dict": [1, 2, 4, 6, 8, 21, 22, 23, 25, 26, 29, 31, 34, 35], "list": [1, 2, 6, 8, 21, 22, 23, 24, 29, 33], "boxplot": 1, "barplot": [1, 29], "If": [1, 2, 4, 6, 8, 10, 23], "kei": [1, 4, 6, 8, 21], "should": [1, 2, 4, 6, 23], "exactli": 1, "same": [1, 2, 4, 23, 32, 34], "iloc": [1, 21, 23, 24, 26, 30, 31, 32, 35], "uniqu": [1, 22, 25, 28, 32, 34], "valu": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 33, 35], "name": [1, 4, 6, 21, 22, 23, 25, 26, 27, 28, 30, 31], "hex": [1, 22], "length": [1, 2, 27], "thi": [1, 4, 6, 10, 21, 23, 29, 30], "equal": 1, "nuniqu": [1, 25, 28, 32], "string": [1, 4, 6, 8], "mean": [1, 2, 4, 21, 23, 25, 28], "share": 1, "float": [1, 2, 4, 23], "axi": [1, 2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "1": [1, 2, 4, 6, 8, 12, 15, 16, 17, 21, 22, 23, 24, 25, 27, 28, 29, 31, 32, 33, 34, 35], "width": [1, 2, 6, 8, 21, 22, 23, 24, 26, 28, 29, 31, 32, 33, 34, 35], "size": [1, 2, 4, 6, 13, 16, 26, 29], "bool": [1, 2, 4], "whether": [1, 2, 4, 8, 21, 23, 31], "when": [1, 2, 4, 21, 23], "ar": [1, 2, 4, 8, 23, 24, 29, 31, 33], "vmax": [1, 2, 4, 8, 21, 24, 28, 29], "vmin": [1, 2, 4, 8, 21, 24, 28, 29], "other": [1, 2, 4, 6, 8, 23, 31, 33], "kw": [1, 2, 8], "pass": [1, 2, 4, 6, 8, 30], "plt": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "titl": [1, 2, 8], "prop": 1, "fontsiz": [1, 2, 21, 22, 23, 25, 26, 27, 29, 31, 34], "labelcolor": [1, 2, 21, 23, 26, 27, 29, 30, 31, 35], "markscal": 1, "frameon": [1, 2, 21, 23, 26, 31], "framealpha": 1, "fancybox": 1, "shadow": [1, 2], "facecolor": [1, 2, 21, 22, 23, 26, 29, 31], "edgecolor": [1, 2, 21, 26, 31], "mode": 1, "argument": 1, "pleast": 1, "type": [1, 2, 4, 6, 8, 22, 24, 29, 34], "There": 1, "an": [1, 13, 16, 21, 23], "addit": [1, 23, 31], "color_text": [1, 8, 23], "true": [1, 2, 6, 8, 9, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "which": [1, 2, 8, 28, 29, 31], "set": [1, 2, 8, 13, 16, 21, 22, 23, 25, 26, 31, 35], "text": [1, 2, 8, 21, 23, 26], "marker": [1, 4, 6, 8, 13, 16, 21, 25], "fals": [1, 2, 4, 8, 9, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "black": [1, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "user": [1, 28], "want": [1, 17, 23, 27], "us": [1, 2, 4, 6, 8, 9, 10, 13, 16, 17, 20, 22, 23, 24, 26, 31, 32, 35, 36], "custom": [1, 2, 9, 16, 25], "instead": [1, 21, 25], "blue": [1, 21, 23, 26, 27, 28, 29, 30, 31, 32, 35], "rotat": [1, 2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34], "rotation_mod": [1, 2, 21, 23, 26, 31], "ha": [1, 21], "va": [1, 21], "annotation_clip": 1, "arrowprop": [1, 21, 29, 31], "For": [1, 2, 8, 16, 21, 29, 31], "also": [1, 2, 4, 6, 23], "chang": [1, 2, 8, 9, 13, 16, 23, 27], "rang": [1, 2, 8, 21, 23, 24, 25, 26, 30, 31, 33, 35], "try": [1, 2], "df_box": [1, 21, 23, 26, 30, 31, 35], "gene1": [1, 21], "return": [1, 2, 4, 6, 8], "idx": 1, "subplot_ax": 1, "label": [1, 2, 8, 9, 16, 18, 22, 23, 24, 25, 26, 27, 28, 29, 30, 32, 33, 34, 35], "label_sid": [1, 8, 21, 23, 25, 27, 28, 29, 31, 32], "label_kw": [1, 16, 18, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 32, 34], "ticklabels_kw": [1, 31], "legend_sid": [1, 2, 8], "right": [1, 2, 8, 9, 11, 16, 25, 26, 27, 28, 29, 30, 31, 32, 33, 35], "legend_gap": [1, 2, 21, 22, 23, 25, 26, 28, 29, 30, 31, 34, 35], "legend_width": [1, 2, 8, 22, 23], "4": [1, 4, 8, 15, 16, 21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "legend_hpad": [1, 2, 21, 22, 23, 24, 25, 26, 31, 32, 34], "2": [1, 2, 4, 8, 15, 16, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "legend_vpad": [1, 2, 8, 21, 23, 24, 25, 26, 31], "orient": [1, 9, 16, 27, 29, 32], "wgap": [1, 18, 21, 26], "hgap": [1, 18, 21, 23, 26, 30], "raster": [1, 2, 22, 23, 24, 26, 31, 34, 35], "verbos": [1, 2, 8, 21, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "arg": 1, "gener": [1, 2, 9, 16, 23, 26, 30, 31, 35], "self": [1, 4], "convert": [1, 28], "int": [1, 2, 4, 21, 26, 28, 30, 31, 34], "row": [1, 2, 4, 6, 9, 15, 16, 22, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "need": 1, "specifi": [1, 2, 23], "provid": [1, 2, 8, 16, 23, 28], "have": [1, 2, 8, 17, 21, 23, 29, 34], "can": [1, 2, 4, 6, 21, 23, 27, 29, 30, 31], "from": [1, 2, 4, 6, 8, 13, 16, 17, 21, 23, 24, 25, 26, 27, 29, 31, 32, 33, 34, 35], "__subclasses__": 1, "includ": [1, 2, 8, 21], "anno_scatt": 1, "must": [1, 4], "els": [1, 21, 22, 23, 25, 26, 27, 28, 30, 31, 32, 33], "calcul": [1, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "given": [1, 4, 8, 23, 28], "invalid": 1, "top": [1, 2, 8, 9, 11, 16, 22, 25, 26, 27, 29, 31, 32, 33], "bottom": [1, 2, 8, 11, 16, 21, 25, 26, 27, 29, 30, 31, 32, 33], "left": [1, 2, 8, 9, 11, 16, 25, 26, 27, 28, 29, 30, 31, 32, 33], "alpha": [1, 2, 4, 25, 26, 28, 29, 31], "fontstyl": [1, 2, 26, 31], "horizontalalign": [1, 2, 21, 23, 25, 26, 27, 28, 29, 30, 31, 32], "verticalalign": [1, 2, 21, 26, 27, 30, 31], "visibl": [1, 2, 21, 22, 24, 25, 26, 29, 31, 32, 34], "gca": [1, 23], "yaxi": [1, 26, 31], "properti": [1, 2, 26, 31], "ax": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 28, 29, 31, 32, 33], "direct": [1, 2, 23, 31], "pad": [1, 2, 16, 18, 21, 26], "labels": [1, 2, 21, 27, 29, 31, 32, 35], "zorder": [1, 2, 26, 31], "labelbottom": [1, 2], "labeltop": [1, 2], "labelleft": [1, 2], "labelright": [1, 2], "labelrot": [1, 2, 21, 23, 26, 27, 29, 30, 31, 35], "grid_color": [1, 2], "grid_linestyl": [1, 2], "tick_param": [1, 2, 21, 31], "function": [1, 4, 8, 23, 25, 29, 30], "et": [1, 2, 22, 23], "al": [1, 2, 23], "automoti": 1, "vertic": [1, 2, 21], "gap": [1, 2, 8, 16, 18, 21, 23], "between": [1, 2, 8, 9, 16, 23, 31], "two": [1, 2, 16, 20, 23, 25, 29, 31], "mm": [1, 2, 22, 23, 24, 28, 29, 31, 32, 34, 35], "horizon": [1, 2, 23], "space": [1, 2, 8, 9, 16, 23], "up": [1, 2, 13, 16, 27, 32], "down": [1, 9, 16, 27], "show": [1, 2, 4, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "automat": [1, 2, 4, 23], "assign": [1, 29, 32], "accord": 1, "posit": [1, 26, 28], "option": [1, 4, 6, 8], "wspace": 1, "control": [1, 4, 6, 16, 18, 21, 23, 28, 33], "hspace": 1, "horizont": [1, 2, 16, 20], "pair": [1, 8, 21, 35], "collect": [1, 21, 22, 23, 24, 27, 28, 29, 30, 32, 33, 34, 35], "rtype": 1, "subplot_spec": [1, 2, 6], "figur": [1, 4, 8, 21, 22, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "add_gridspec": 1, "gridspec": 1, "gridspecfromsubplotspec": 1, "index": [1, 2, 4, 21, 22, 23, 24, 25, 26, 27, 28, 30, 31, 32, 33, 34, 35], "param": [1, 2], "kwarg": [1, 2, 4, 6, 16], "grid": [1, 21, 26, 28, 29, 31, 32, 33], "showflier": 1, "medianlinecolor": 1, "border_width": [1, 21], "border_color": [1, 21], "255": [1, 21], "merg": [1, 8, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 33, 34], "merge_width": 1, "imag": [1, 9, 16], "border": 1, "line": [1, 21, 23, 28, 30, 31], "256": [1, 21], "ignor": [1, 8, 28], "white": [1, 2, 8, 21, 22, 23, 26, 27, 28, 31, 33], "cluster": [1, 2, 8, 9, 16], "onc": 1, "extend": [1, 22, 34], "frac": 1, "major": [1, 8, 22, 29, 31], "adjust_color": [1, 34], "lumin": [1, 29, 34], "8": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 35], "relpo": [1, 21, 22, 26, 27, 29, 30, 31, 32, 34], "add": [1, 2, 9, 13, 16, 25, 27, 31], "documentatin": 1, "websit": 1, "dingwb": [1, 2, 17, 31, 34], "github": [1, 2, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "io": [1, 2, 21, 31, 32], "build": [1, 2, 9, 16, 31], "notebook": [1, 2, 31], "single_cell_methyl": 1, "distribut": 1, "fraction": 1, "arma": 1, "armb": 1, "multipl": [1, 2, 4, 9, 16], "group": [1, 2, 9, 13, 16, 24, 29, 31, 34, 35], "largest": [1, 8], "too": [1, 2, 21, 23, 28], "high": [1, 24, 25, 28], "replac": [1, 21, 26, 28, 30, 31, 32], "origin": 1, "togeth": [1, 2], "tupl": [1, 2, 21, 22], "x": [1, 4, 6, 8, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 33], "y": [1, 4, 6, 8, 21, 22, 24, 25, 26, 28, 29, 31, 32, 33], "arrow": [1, 16, 18, 21], "start": [1, 16, 21, 22, 23, 24, 27, 28, 29, 30, 32, 33, 34, 35], "point": [1, 13, 16, 21, 22, 29, 31], "rel": 1, "side": [1, 15, 16, 25, 26, 29], "about": 1, "could": [1, 2, 4, 23], "found": 1, "patch": [1, 26, 31], "fancyarrowpatch": [1, 31], "lineplot": 1, "linewidth": [1, 2, 21, 23, 25, 26, 28, 29, 31, 32, 35], "scatterplot": 1, "scatter": [1, 4, 6, 21, 23, 26, 28, 29, 30, 31, 35], "add_text": [1, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 35], "text_kw": [1, 21, 22, 23, 24, 25, 26, 27, 28, 30, 31, 35], "simpl": [1, 13, 16, 23], "categor": [1, 4, 8, 16, 23, 36], "continu": [1, 22, 25, 32], "variabl": [1, 8, 16, 36], "data": [2, 4, 6, 10, 13, 16, 20, 21, 22, 23, 24, 26, 27, 30, 31, 32, 33, 35], "top_annot": [2, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 33, 34, 35], "bottom_annot": [2, 23], "left_annot": [2, 22, 23, 24, 27, 29, 34], "right_annot": [2, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 34], "row_clust": [2, 21, 22, 26, 27, 28, 29, 30, 31, 34, 35], "col_clust": [2, 9, 22, 26, 27, 28, 29, 30, 31, 34, 35], "row_cluster_method": [2, 24, 35], "averag": [2, 21, 23, 26, 31, 35], "row_cluster_metr": [2, 24, 35], "correl": [2, 16, 20, 28, 35], "col_cluster_method": [2, 24, 35], "col_cluster_metr": [2, 24, 35], "show_rownam": [2, 21, 22, 23, 24, 26, 27, 28, 29, 30, 31, 32, 33, 35], "show_colnam": [2, 21, 22, 23, 24, 26, 27, 28, 29, 30, 31, 32, 33, 35], "row_names_sid": [2, 21, 23, 29, 30, 31, 35], "col_names_sid": [2, 23, 29], "xticklabels_kw": [2, 6, 16, 18, 21, 23, 26, 27, 29, 30, 32, 35], "yticklabels_kw": [2, 6, 16, 18, 21, 26, 27], "row_dendrogram": [2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "col_dendrogram": [2, 21, 22, 24, 27, 31, 35], "row_dendrogram_s": [2, 21], "10": [2, 10, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "col_dendrogram_s": 2, "row_split": [2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "col_split": [2, 9, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "row_dendrogram_kw": 2, "col_dendrogram_kw": [2, 21], "tree_kw": [2, 21, 23, 25, 27, 28, 30, 31, 35], "row_split_ord": [2, 21, 22, 34], "col_split_ord": [2, 9, 22, 31, 32, 34], "row_split_gap": [2, 18, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 34, 35], "col_split_gap": [2, 18, 21, 22, 23, 25, 26, 27, 28, 29, 30, 32, 33, 34, 35], "mask": [2, 4, 6], "subplot_gap": [2, 18, 21, 26, 32], "legend_anchor": [2, 23, 24, 25], "7": [2, 21, 22, 23, 25, 26, 28, 29, 30, 31, 32, 34, 35], "xlabel": [2, 16, 18, 21, 22, 23, 26, 29, 34, 35], "ylabel": [2, 16, 18, 21, 22, 23, 26, 34, 35], "xlabel_kw": [2, 16, 18, 21, 22, 23, 26, 29, 34], "ylabel_kw": [2, 16, 18, 21, 22, 23, 26, 34], "xlabel_sid": [2, 22, 29], "ylabel_sid": 2, "xlabel_bbox_kw": [2, 21, 22, 26, 29, 31], "ylabel_bbox_kw": [2, 21, 23, 26, 31], "legend_delta_x": 2, "plotter": [2, 4, 6], "numpi": [2, 4, 6, 17, 21, 22, 27, 29, 30], "arrai": [2, 4, 6, 16, 20, 22, 25, 26, 34], "perform": [2, 29], "z": [2, 25], "score": 2, "either": [2, 23], "after": 2, "method": [2, 8, 21], "linkag": [2, 9, 16, 22], "singl": [2, 16, 20], "complet": 2, "weight": [2, 22], "centroid": 2, "median": [2, 21], "ward": [2, 24, 35], "scipi": [2, 17, 35], "hierarchi": 2, "doc": 2, "refer": [2, 32], "detail": 2, "pairwis": 2, "distanc": 2, "observ": 2, "n": [2, 21, 22], "dimension": 2, "euclidean": [2, 35], "minkowski": 2, "cityblock": 2, "seuclidean": 2, "cosin": 2, "ham": 2, "jaccard": [2, 24], "chebyshev": 2, "canberra": [2, 21], "braycurti": 2, "mahalanobi": 2, "kulsinski": 2, "14": [2, 21, 22, 23, 25, 26, 27, 28, 29, 31, 33, 34], "spatial": 2, "pdist": 2, "metric": [2, 21], "ticklabel": [2, 8, 9, 16, 31], "dendrogram": [2, 22, 31], "10mm": 2, "pd": [2, 4, 6, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "number": [2, 21, 29], "hierarch": 2, "split": [2, 15, 16, 25, 28, 29, 32], "subplot": [2, 21], "my_linkag": 2, "order": [2, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "cluster_between_group": [2, 9, 31], "wa": [2, 4, 16, 23, 24, 25, 28, 34], "clsuter": [2, 9], "advanced_usag": [2, 31], "col": [2, 9, 16, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "cell": [2, 9, 16, 20, 22, 23, 29, 31, 33], "miss": 2, "infinit": 2, "1mm": 2, "asfonts": 2, "numpoint": 2, "markerscal": 2, "markerfirst": 2, "title_fonts": 2, "labelspac": 2, "alatern": 2, "we": [2, 21, 23, 28, 30, 32], "outlin": 2, "cbar": [2, 8], "cm": [2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "isinst": 2, "colorbar": [2, 8], "set_color": [2, 25, 26, 29], "set_linewidth": [2, 25, 26, 29], "divid": 2, "ax_heatmap": [2, 23, 24, 25, 26, 28, 29], "anchor": [2, 8, 21, 23, 31], "differ": [2, 8, 13, 16, 21, 29, 30], "infer": [2, 4, 28, 29], "shown": [2, 4, 6], "xticklabel": [2, 16, 18, 21, 35], "yticklabel": [2, 16, 18, 21, 35], "fontfamili": [2, 26, 31], "fontnam": [2, 21, 26, 31], "fontproperti": [2, 26, 31], "fontweight": [2, 22, 24, 26, 27, 29, 31], "xaxi": [2, 26, 31], "print": [2, 21, 22, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "sam": 2, "clip_box": [2, 26, 31], "clip_on": [2, 26, 31], "fill": [2, 4, 21, 26, 31], "in_layout": [2, 26, 31], "linestyl": [2, 26, 28, 29, 31], "get_bbox_patch": [2, 26, 31], "5000": [2, 33], "speed": 2, "center": [2, 8, 21, 24, 26, 31], "robust": [2, 8], "annot_kw": [2, 21], "fmt": [2, 21, 26, 31, 35], "linecolor": [2, 21, 23, 26, 31, 35], "na_col": [2, 8], "cbar_kwss": 2, "document": 2, "small": [2, 34], "displai": [2, 9, 16], "avoid": [2, 21], "overlap": [2, 8, 21], "To": [2, 21], "forc": [2, 9, 16], "use_linkag": 2, "row_ord": [2, 4, 6, 21, 22, 27], "col_ord": [2, 4, 6, 21, 22, 27], "fig": [2, 8, 21], "static": 2, "data2d": 2, "max": [2, 4, 22, 28, 29], "min": [2, 22, 28, 29], "normal": [2, 21, 23, 26, 30, 31, 35], "across": 2, "standard": 2, "noraml": 2, "varianc": 2, "tight_param": 2, "standar": 2, "dendrogram_kw": 2, "gap_pixel": 2, "root_x": 2, "similar": [2, 21], "upon": 2, "indic": [2, 29], "matrix": [2, 4, 6, 16, 20, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "cmlist": [2, 24], "main": [2, 15, 16, 21, 24, 31, 34], "row_gap": [2, 6], "15": [2, 21, 22, 23, 24, 26, 27, 28, 30, 31, 32], "col_gap": [2, 24], "legend_i": 2, "width_ratio": 2, "height_ratio": 2, "assembl": 2, "align": [2, 6, 33], "influenc": [2, 24], "unit": [2, 8, 31], "estim": [2, 22, 23, 24, 28, 29, 32, 34, 35], "legend_ax": [2, 8, 24], "cbar_kw": 2, "cbar_ax": 2, "squar": [2, 13, 16, 26], "xlabel_pad": 2, "ylabel_pad": 2, "xticklabels_sid": 2, "yticklabels_sid": 2, "2g": 2, "maxim": [2, 4], "minim": [2, 4], "seaborn": [2, 13, 16], "cax": [2, 8], "draw": [2, 23], "format": 2, "anno_kw": 2, "c": [3, 4, 6, 8, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 35], "hue": [4, 6, 13, 16, 25, 29], "": [4, 6, 8, 13, 16, 23, 25, 29], "o": [4, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "color_legend_kw": [4, 6], "cmap_legend_kw": [4, 6], "dot_legend_kw": 4, "dot_legend_mark": [4, 28, 29], "aggfunc": 4, "value_na": [4, 28], "hue_na": 4, "na": 4, "s_na": 4, "c_na": [4, 28], "spine": [4, 8, 21, 25, 26, 28, 29, 32, 33], "max_": [4, 28, 29], "dotclustermap": [4, 6, 16, 20], "inherit": [4, 6], "dot": [4, 6, 13, 16, 20, 29], "The": [4, 6, 16, 23, 24, 25, 29], "overwrit": [4, 6], "go": [4, 29], "stabl": 4, "api": 4, "markers_api": 4, "avail": 4, "Such": 4, "v": [4, 28], "p": [4, 25, 28, 29], "h": [4, 21, 23, 26, 27, 30, 31, 35], "d": [4, 8, 10, 21, 23, 25, 26, 28, 30, 31, 35], "_": [4, 28], "tolist": [4, 21, 22, 23, 24, 25, 28, 30, 32, 34], "It": [4, 23, 29], "pivot_t": 4, "fillna": [4, 21, 25, 28, 32], "floator": 4, "np": [4, 21, 22, 23, 26, 27, 28, 29, 30, 31, 32, 35], "aggreg": 4, "them": 4, "call": [4, 8], "pivot": [4, 28], "coeffici": 4, "valid": 4, "input": [4, 6], "paramt": [4, 6], "aspect": [6, 21], "bgcolor": 6, "whitesmok": 6, "remove_empty_row": 6, "remove_empty_column": 6, "background": [6, 23], "lightgrai": [6, 24, 25], "nvar": 6, "element": 6, "wisa": 6, "cccccc": 6, "intern": 8, "boolean": 8, "ani": 8, "legend_list": 8, "xtick": 8, "adjac": [8, 23], "tick": [8, 21], "coordin": 8, "keep": 8, "A": [8, 10, 11, 16, 20, 21, 26, 30, 31], "b": [8, 21, 26, 30, 31, 34], "len": [8, 21, 22], "new_label": 8, "plot_data": 8, "some": [8, 21], "heurist": 8, "good": [8, 29], "remov": [8, 16, 18, 25, 30, 32, 33], "current": 8, "specif": 8, "turbo": [8, 21, 23, 24, 26, 27, 30, 31, 35], "drawn": 8, "accent": 8, "tab20": [8, 25], "exp1": [8, 21, 23, 27], "exp2": [8, 21], "meth1": [8, 21], "meth2": [8, 21, 35], "legn": 8, "y0": 8, "delta_x": 8, "cmap_width": 8, "lengend": 8, "handl": 8, "legend_typ": 8, "pixel": 8, "init": 8, "2mm": 8, "5mm": 8, "boundri": 8, "obj": 8, "repres": 8, "python": [8, 10, 16, 17], "e": [8, 21, 23, 26, 30, 31, 35], "unchang": 8, "byte": 8, "utf": 8, "decod": 8, "__str__": 8, "result": [8, 13, 16, 28], "represent": 8, "dataset": [9, 13, 16, 20, 23, 26, 29, 30, 31, 33, 34, 35, 36], "select": [9, 16, 23, 26, 27, 28, 30, 31], "orent": [9, 16], "extra": [9, 16], "loop": [9, 16], "within": [9, 16], "cluster_within_group": 9, "choos": [9, 16, 30], "pycomplexheatmap": [9, 10, 17, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "diverg": 9, "qualit": 9, "how": [9, 16, 18], "you": [10, 17, 27], "softwar": [10, 28], "your": [10, 29], "work": 10, "cite": 10, "follow": [10, 21, 23, 24, 26, 31, 35], "metadata": [10, 22], "ding": [10, 16], "w": 10, "goldberg": 10, "zhou": 10, "2023": 10, "packag": [10, 15, 16, 19, 28, 29, 32], "visual": [10, 13, 16, 20, 36], "multimod": 10, "genom": 10, "imeta": 10, "e115": 10, "doi": 10, "1002": 10, "imt2": 10, "115": [10, 29], "quick": [11, 16], "understand": [12, 16], "layout": [12, 16], "load": [13, 16, 23, 24, 29], "brain": [13, 16, 23, 24], "network": [13, 16], "tradit": [13, 16], "fix": [13, 16], "five": [13, 16], "dimens": [13, 16], "gene": [13, 16, 20, 21, 23, 25, 26, 30, 31, 32, 33, 35], "enrich": [13, 16, 28], "analysi": [13, 16, 28], "clusterprofil": [13, 16], "click": 14, "picutur": 14, "view": 14, "code": [14, 21, 23, 26, 31, 35], "import": [15, 16, 21, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "along": [15, 16], "complex": 16, "biolog": [16, 29], "instal": [16, 30], "depend": [16, 29], "get": 16, "tcga": [16, 36], "lung": [16, 34, 36], "adenocarcinoma": [16, 36], "carcinoma": [16, 36], "variant": [16, 36], "advanc": 16, "usag": 16, "differenti": [16, 20, 29], "express": [16, 20], "dna": [16, 20, 28], "methyl": [16, 20, 29], "loyfer2023": [16, 20], "mous": [16, 20, 22, 23], "cpg": [16, 20, 24, 34], "modul": [16, 19, 20], "modifi": [16, 18], "galleri": [16, 28], "develop": [16, 29], "setup": [16, 17, 19], "citat": 16, "wubin": 16, "pip": 17, "upgrad": 17, "development": 17, "version": 17, "directli": 17, "com": [17, 21, 31, 34], "git": 17, "previous": 17, "updat": 17, "uninstal": 17, "again": 17, "OR": 17, "clone": [17, 27], "cd": [17, 21, 23, 26, 30, 31, 35], "py": [17, 25, 28], "submodul": 19, "content": 19, "process": [20, 22, 29], "smaller": 20, "sy": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "inlin": [21, 22, 23, 24, 25, 26, 28, 30, 31, 32, 34, 35], "pylab": [21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 34, 35], "pickl": [21, 23, 24, 26, 28, 29, 31, 32, 34, 35], "rcparam": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "dpi": [21, 22, 23, 24, 26, 28, 29, 30, 31, 32, 34, 35], "100": [21, 22, 23, 26, 28, 30, 31, 32, 33, 34, 35], "savefig": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "300": [21, 22, 23, 24, 26, 28, 29, 30, 31, 32, 34, 35], "path": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "append": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "expandus": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "project": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "__version__": [21, 22, 24, 25, 26, 29, 30, 31, 34, 35], "6": [21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 33, 34, 35], "dev15": [21, 29], "gd9f20b3": [21, 29], "font": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "arial": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "famili": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "san": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "serif": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "pdf": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "fonttyp": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "42": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "random": [21, 23, 26, 30, 31, 33, 35], "groupa": [21, 26, 30, 31, 35], "groupb": [21, 26, 30, 31, 35], "ab": [21, 23, 26, 30, 31, 35], "g": [21, 23, 24, 26, 27, 30, 31, 35], "ef": [21, 23, 26, 30, 31, 35], "f": [21, 22, 23, 24, 26, 29, 30, 31, 33, 35], "sampl": [21, 22, 23, 24, 26, 28, 30, 31, 32, 33, 35], "shape": [21, 22, 23, 25, 26, 27, 30, 31, 35], "randn": [21, 23, 26, 30, 31, 35], "df_bar": [21, 23, 26, 30, 31, 35], "uniform": [21, 23, 26, 30, 31, 35], "tmb1": [21, 23, 26, 30, 31, 35], "tmb2": [21, 23, 26, 30, 31, 35], "df_scatter": [21, 23, 26, 30, 31, 35], "df_heatmap": [21, 23, 26, 30, 31, 35], "30": [21, 23, 25, 26, 28, 29, 30, 31, 35], "11": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 33, 35], "fea": [21, 23, 26, 30, 31, 35], "nan": [21, 23, 25, 26, 30, 31, 32, 35], "df_row": [21, 22, 23, 24, 26, 28, 29, 30, 31, 34], "appli": [21, 22, 23, 25, 26, 27, 28, 30, 31, 32], "lambda": [21, 22, 23, 25, 26, 27, 28, 30, 31, 32], "sample4": [21, 26, 30, 31], "to_fram": [21, 26, 28, 30, 31], "xy": [21, 26, 30, 31], "to_seri": [21, 25, 26, 30, 31], "row_ha": [21, 22, 23, 24, 25, 26, 27, 28, 30, 31], "round": [21, 23, 26, 30, 31, 32], "12": [21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 35], "bar": [21, 23, 30, 33], "05": [21, 23, 26, 28, 29, 30, 31, 32, 35], "col_ha": [21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 34, 35], "nanmin": 21, "nanmax": [21, 28], "figsiz": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "row_cmap": [21, 23, 25, 27, 28, 30, 35], "rdylbu_r": [21, 26, 30, 31, 35], "90": [21, 22, 27, 28, 30, 34, 35], "example0": [21, 30, 35], "bbox_inch": [21, 22, 23, 24, 25, 27, 28, 29, 30, 32, 34, 35], "tight": [21, 22, 23, 24, 25, 27, 28, 29, 30, 32, 34, 35], "259323481554901": 21, "3307664641494927": 21, "32": [21, 33], "s4": [21, 26, 31], "test": [21, 26, 31], "18": [21, 26, 28, 31], "bbox": [21, 26], "boxstyl": [21, 26, 31], "circl": [21, 28], "exp": [21, 23, 26, 30, 31, 35], "turn": [21, 23, 26, 31, 35], "off": [21, 23, 26, 31, 35], "log": [21, 23, 26, 31, 35], "head": [21, 24, 25, 27, 28, 30, 32, 34], "1g": [21, 26, 31, 35], "gold": [21, 23, 26, 31, 35], "45": [21, 23, 25, 26, 27, 28, 29, 31, 32, 33, 35], "sample1": [21, 30], "404085": 21, "sample2": [21, 30], "081411": 21, "sample3": [21, 30], "594949": 21, "392642": 21, "sample5": [21, 30], "150321": 21, "229885": 21, "210195": 21, "032791": 21, "051215": 21, "261465": 21, "sample6": 21, "836030": 21, "sample7": 21, "519891": 21, "sample8": 21, "742360": 21, "sample9": 21, "617504": 21, "sample10": 21, "606197": 21, "float64": [21, 28], "fea1": 21, "941453": 21, "140686": 21, "285091": 21, "118367": 21, "214946": 21, "967923": 21, "853248": 21, "fea2": 21, "691451": 21, "561863": 21, "045189": 21, "303610": 21, "324130": 21, "240718": 21, "fea3": 21, "378372": 21, "142337": 21, "405770": 21, "274505": 21, "171325": 21, "526213": 21, "119409": 21, "fea4": 21, "212045": 21, "515978": 21, "051057": 21, "124523": 21, "378586": 21, "320444": 21, "064873": 21, "fea5": 21, "078289": 21, "476870": 21, "360113": 21, "705353": 21, "079402": 21, "284584": 21, "341059": 21, "751814": 21, "163439": 21, "038006": 21, "120816": 21, "820074": 21, "259323": 21, "757637": 21, "451777": 21, "057090": 21, "817372": 21, "082246": 21, "293083": 21, "831747": 21, "489009": 21, "258670": 21, "39": [21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 34], "fea19": 21, "fea25": 21, "fea18": 21, "fea27": 21, "fea29": 21, "fea22": 21, "fea24": 21, "fea30": 21, "fea20": 21, "fea17": 21, "fea26": 21, "fea21": 21, "fea23": 21, "fea15": 21, "fea16": 21, "fea28": 21, "fea9": 21, "fea10": 21, "fea11": 21, "fea7": 21, "fea8": 21, "fea14": 21, "fea13": 21, "fea6": 21, "fea12": 21, "37": [21, 25, 26, 31], "aaaa1": [21, 23], "bbbbb2": [21, 23], "df_bar1": [21, 30], "t1": [21, 30], "df_bar2": [21, 31], "t2": 21, "df_bar3": 21, "t3": 21, "df_bar4": [21, 30], "t4": [21, 30], "372887": 21, "061936": 21, "134849": 21, "694566": 21, "359127": 21, "132367": 21, "392230": 21, "272427": 21, "714047": 21, "625537": 21, "gene2": 21, "gene3": 21, "gene4": 21, "257733": 21, "699439": 21, "844278": 21, "608444": 21, "752454": 21, "083405": 21, "809866": 21, "431847": 21, "663268": 21, "513365": 21, "772141": 21, "387252": 21, "636622": 21, "012600": 21, "966687": 21, "476726": 21, "826088": 21, "361708": 21, "754555": 21, "343367": 21, "220518": 21, "870597": 21, "876851": 21, "9": [21, 22, 23, 24, 25, 28, 29, 31, 32, 33], "876537": 21, "395013": 21, "573748": 21, "948007": 21, "973635": 21, "386078": 21, "499509": 21, "188200": 21, "545418": 21, "328468": 21, "047464": 21, "158959": 21, "161838": 21, "163763": 21, "243494": 21, "779427": 21, "204043": 21, "542872": 21, "130472": 21, "133916": 21, "091244": 21, "788314": 21, "348602": 21, "046968": 21, "088636": 21, "612396": 21, "928272": 21, "877048": 21, "401591": 21, "980643": 21, "201984": 21, "307024": 21, "833295": 21, "660850": 21, "235867": 21, "211823": 21, "919341": 21, "290198": 21, "165467": 21, "382529": 21, "881658": 21, "073468": 21, "342374": 21, "239178": 21, "468118": 21, "962650": 21, "532729": 21, "771372": 21, "835987": 21, "436057": 21, "167528": 21, "042679": 21, "093969": 21, "936279": 21, "354470": 21, "736818": 21, "348153": 21, "335390": 21, "823424": 21, "888954": 21, "168943": 21, "042376": 21, "223754": 21, "876278": 21, "314815": 21, "541221": 21, "407723": 21, "913978": 21, "019213": 21, "885269": 21, "457418": 21, "692399": 21, "369775": 21, "256769": 21, "091064": 21, "233948": 21, "702588": 21, "314119": 21, "758320": 21, "888650": 21, "260971": 21, "891667": 21, "691901": 21, "079907": 21, "903442": 21, "754403": 21, "010756": 21, "829727": 21, "209684": 21, "739576": 21, "615991": 21, "269359": 21, "153883": 21, "386240": 21, "874027": 21, "38": [21, 28, 31], "orang": [21, 23, 24, 29], "yellowgreen": [21, 23, 24, 29], "tmb_bar": [21, 23, 31], "bar1": [21, 30], "bar4": [21, 30], "bar2": 21, "bar3": 21, "fontdict": [21, 22, 24, 30], "tab10": [21, 23], "grai": [21, 23, 24, 28], "yellow": [21, 23], "13": [21, 22, 23, 24, 25, 27, 28, 29], "here": [21, 32], "widh": 21, "40": [21, 22, 29], "20": [21, 26, 27, 28, 31], "incres": [21, 23], "ncol": [21, 23], "than": [21, 23], "support": [21, 23], "long": [21, 23, 28], "new": [21, 23], "41": [21, 29, 34], "green": [21, 23, 24, 25, 26, 27, 28, 29, 31, 33], "By": 21, "43": 21, "typic": 21, "creat": [21, 33], "annotatin": [21, 23], "df_col": [21, 22, 23, 24, 25, 28, 29, 34], "hypomethylated_sampl": [21, 22], "sample_group_color_dict": 21, "celltyp": [21, 22, 29], "ct_color_dict": 21, "ct_legend": 21, "m1": 21, "lgd": 21, "But": 21, "what": [21, 29], "mani": 21, "m2": 21, "m3": 21, "In": [21, 23, 28, 29, 31], "case": [21, 29], "col_ha_dict": 21, "sample_col": 21, "r": [21, 24, 29, 32], "jokergoo": [21, 32], "2021": 21, "03": [21, 22, 24, 29], "complexheatmap": [21, 32], "44": [21, 35], "g1": [21, 31, 35], "g2": [21, 31, 35], "g3": [21, 31, 35], "g4": [21, 31, 35], "g5": [21, 31, 35], "col_cmap": [21, 27, 31, 35], "featur": [21, 26, 31, 35], "46": 21, "loc": [21, 22, 24, 25, 28, 29, 32, 34], "47": 21, "annotaiton": [21, 31], "purpl": [21, 23, 25, 26, 28, 31, 35], "_ticklabels_kw": [21, 31], "labelpad": [21, 22, 23, 26, 29, 31, 34], "increac": [21, 22, 23, 31], "manual": [21, 22, 31], "chocol": [21, 22, 26, 31], "48": 21, "applymap": 21, "make": 21, "asterisk": 21, "locat": 21, "oper": 21, "unicod": 21, "explor": 21, "2217": 21, "courier": 21, "49": [21, 28], "fastclust": 21, "t": [21, 22, 23, 25, 27, 28, 32, 34], "nlinkag": 21, "17": [21, 25, 28], "07051523": 21, "22006417": 21, "19": [21, 23, 26, 28, 33, 35], "14461418": 21, "28842182": 21, "70552492": 21, "76359288": 21, "65679239": 21, "16": [21, 23, 24, 25, 27, 28], "21": [21, 28, 35], "91528308": 21, "22": [21, 28], "75945877": 21, "50": [21, 23, 28], "51": 21, "df_img": 21, "jpeg": 21, "52": 21, "img": 21, "diverging1": 21, "parula": [21, 22, 23, 24, 34, 35], "53": 21, "54": 21, "mpl": [21, 29], "gradient": 21, "linspac": 21, "vstack": 21, "def": 21, "plot_color_gradi": 21, "categori": [21, 28, 29], "cmap_list": 21, "adjust": [21, 29], "nrow": 21, "figh": 21, "35": [21, 29], "subplots_adjust": 21, "99": [21, 33], "set_titl": [21, 22, 24], "zip": [21, 26], "imshow": 21, "01": [21, 32], "transform": [21, 26], "transax": 21, "just": 21, "ones": 21, "set_axis_off": [21, 23], "save": 21, "later": [21, 25], "55": [21, 28], "sequenti": 21, "cmap50": 21, "56": [21, 29], "57": 21, "custom_cmap": 21, "58": 21, "59": 21, "big": [21, 23], "enough": 21, "hidden": 21, "60": 21, "matter": 21, "61": [21, 33], "home": 22, "wding2": 22, "pch": [22, 24, 25, 30, 34], "czip": 22, "palette_path": 22, "mouse_pfc": 22, "mpfc_color_palett": 22, "xlsx": 22, "cell_class_color": 22, "read_excel": 22, "sheet_nam": 22, "cellclass": [22, 29], "index_col": [22, 25, 27, 28, 32, 34, 35], "to_dict": [22, 25, 29], "multi": [22, 28], "grei": [22, 24, 29], "nonn": 22, "e06931": 22, "exc": 22, "5ec5a9": 22, "rg": 22, "6036cb": 22, "inh": 22, "e93400": 22, "delta": [22, 34], "read_csv": [22, 25, 27, 28, 32, 34, 35], "pseudo_cel": 22, "between_groups_dmr": 22, "methylpi": 22, "methylpy_rms_results_collaps": 22, "tsv": 22, "sep": [22, 25, 28, 32, 34], "lstrip": 22, "methylation_level_": 22, "startswith": 22, "renam": 22, "number_of_dm": 22, "inplac": [22, 25, 28, 32, 34], "drop": 22, "hypermethylated_sampl": 22, "sname": 22, "insert": 22, "isna": 22, "reset_index": [22, 25, 28, 32], "set_index": [22, 25, 28, 29], "read": [22, 24, 34], "major_typ": 22, "beta": [22, 24, 25, 34], "txt": [22, 28, 32], "common_row": 22, "5kb_mc_hic_clust": 22, "l2": 22, "major_type_to_cell_class": 22, "common_col": 22, "cell_type_col": 22, "258365": 22, "23": [22, 26, 28, 29], "chr": [22, 24], "end": 22, "chr14": 22, "25667890": 22, "25667923": 22, "649934": 22, "chr5": [22, 24], "70893110": 22, "70893122": 22, "333501": 22, "chr3": 22, "141598191": 22, "141598238": 22, "440745": 22, "chr1": [22, 34], "76086002": 22, "76086015": 22, "319716": 22, "chr9": 22, "16231701": 22, "16232342": 22, "444616": 22, "chr11": [22, 24], "75550115": 22, "75550656": 22, "547601": 22, "109751156": 22, "109751187": 22, "530203": 22, "68823498": 22, "68825391": 22, "29": 22, "451270": 22, "chr10": 22, "91354668": 22, "91354926": 22, "327058": 22, "74147749": 22, "74150162": 22, "31": [22, 26, 29], "541438": 22, "cellclass1": 22, "majortyp": 22, "ct": 22, "l6": 22, "cge": 22, "lamp5": 22, "IT": 22, "l23": 22, "pc": [22, 27], "mgc": 22, "odc": 22, "vip": 22, "mge": 22, "sst": 22, "opc": 22, "vlmc": 22, "l4": 22, "unknown": [22, 28], "l5": 22, "ec": 22, "pvalb": 22, "pia": 22, "asc": 22, "chrom": 22, "077450": 22, "233350": 22, "161600": 22, "733500": 22, "685500": 22, "628000": 22, "148000": 22, "468000": 22, "785000": 22, "662500": 22, "780000": 22, "092500": 22, "125000": 22, "849000": 22, "043500": 22, "839500": 22, "735500": 22, "822500": 22, "694000": 22, "630500": 22, "903500": 22, "899000": 22, "896500": 22, "637500": 22, "784500": 22, "323000": 22, "947500": 22, "743500": 22, "844000": 22, "961500": 22, "499000": 22, "926000": 22, "000000": [22, 23, 25, 27, 28, 35], "846500": 22, "954500": 22, "489500": 22, "883500": 22, "598000": 22, "884000": 22, "789500": 22, "491350": 22, "192500": 22, "623500": 22, "055500": 22, "332150": 22, "763500": 22, "464500": 22, "448000": 22, "200150": 22, "549500": 22, "377950": 22, "517000": 22, "583500": 22, "458500": 22, "419500": 22, "633500": 22, "775000": 22, "299000": 22, "082350": 22, "471000": 22, "797500": 22, "508500": 22, "773500": 22, "050000": 22, "610500": 22, "802000": 22, "869500": 22, "523500": 22, "572500": 22, "572000": 22, "887000": 22, "778000": 22, "396500": 22, "826500": 22, "697500": 22, "145450": 22, "562500": 22, "317500": 22, "807875": 22, "725875": 22, "318250": 22, "900750": 22, "807750": 22, "378837": 22, "209437": 22, "654000": 22, "526500": 22, "579750": 22, "715875": 22, "133525": 22, "605375": 22, "663125": 22, "666375": 22, "545250": 22, "307337": 22, "319063": 22, "839000": 22, "557333": 22, "631000": 22, "780667": 22, "171233": 22, "262050": 22, "886500": 22, "492667": 22, "159800": 22, "520667": 22, "726833": 22, "683833": 22, "698000": 22, "231500": 22, "830000": 22, "781667": 22, "631667": 22, "909000": 22, "222417": 22, "338917": 22, "687667": 22, "452667": 22, "457667": 22, "241333": 22, "154667": 22, "546333": 22, "629667": 22, "325667": 22, "058667": 22, "503333": 22, "321000": 22, "588000": 22, "709000": 22, "034133": 22, "859333": 22, "360333": 22, "684667": 22, "414333": 22, "081200": 22, "244667": 22, "753267": 22, "363077": 22, "607967": 22, "644540": 22, "257347": 22, "775067": 22, "890267": 22, "517487": 22, "184007": 22, "594700": 22, "720740": 22, "763000": 22, "750800": 22, "495783": 22, "471317": 22, "777000": 22, "901467": 22, "657633": 22, "362270": 22, "241253": 22, "784857": 22, "809286": 22, "508857": 22, "859286": 22, "650714": 22, "823286": 22, "763286": 22, "614143": 22, "339714": 22, "615143": 22, "859000": 22, "469000": 22, "742857": 22, "466571": 22, "851714": 22, "807286": 22, "798000": 22, "913429": 22, "423857": 22, "678143": 22, "859394": 22, "860788": 22, "766576": 22, "678030": 22, "443909": 22, "813273": 22, "867061": 22, "658548": 22, "311152": 22, "854182": 22, "765485": 22, "813455": 22, "846697": 22, "434000": 22, "893424": 22, "834667": 22, "833515": 22, "808030": 22, "302337": 22, "529615": 22, "cc": 22, "dendrogram_col": 22, "ivl": 22, "dendrogram_row": 22, "sort_valu": [22, 34], "color_dict": [22, 34], "copi": 22, "del": [22, 33], "bold": [22, 24, 27, 29], "avg": [22, 23], "n_cpg": 22, "6375": [22, 29], "exist": 22, "listdir": 22, "within_groups_dmr": 22, "join": [22, 28], "_heatmap": 22, "concat": 22, "groupbi": [22, 28, 32], "agg": 22, "to_csv": 22, "agg_beta": 22, "queri": 22, "pfc": 22, "bed": 22, "gz": 22, "outfil": 22, "3013471": 22, "3013579": 22, "3026186": 22, "3026310": 22, "3035862": 22, "3035927": 22, "3052666": 22, "3052823": 22, "3059277": 22, "3059436": 22, "chry": 22, "44290504": 22, "44290637": 22, "66427102": 22, "66427179": 22, "90367464": 22, "90367503": 22, "90732358": 22, "90732367": 22, "90792411": 22, "90792447": 22, "661968": 22, "mt": 22, "28963217": 22, "28963260": 22, "83350": 22, "923000": 22, "939000": 22, "507000": 22, "89900": 22, "833500": 22, "738500": 22, "847000": 22, "95450": 22, "966500": 22, "80500": 22, "769500": 22, "731000": 22, "752500": 22, "81300": 22, "760000": 22, "650500": 22, "772500": 22, "936500": 22, "chr4": 22, "32158998": 22, "32159443": 22, "78950": 22, "524500": 22, "791500": 22, "767750": 22, "759500": 22, "73275": 22, "791750": 22, "693250": 22, "79225": 22, "441325": 22, "69625": 22, "538750": 22, "667000": 22, "391250": 22, "46975": 22, "616500": 22, "479250": 22, "704750": 22, "549750": 22, "chr13": 22, "90011452": 22, "90011460": 22, "29100": 22, "222000": 22, "225000": 22, "228500": 22, "08115": 22, "476500": 22, "009400": 22, "329500": 22, "32350": 22, "231000": 22, "30700": 22, "390500": 22, "402500": 22, "782000": 22, "06915": 22, "314500": 22, "285500": 22, "812000": 22, "821500": 22, "126765779": 22, "126765801": 22, "81850": 22, "885000": 22, "903000": 22, "565000": 22, "87500": 22, "875000": 22, "894000": 22, "93750": 22, "906000": 22, "68550": 22, "721500": 22, "649000": 22, "783500": 22, "80050": 22, "793500": 22, "809500": 22, "764500": 22, "142126735": 22, "142126952": 22, "84475": 22, "959250": 22, "902000": 22, "706500": 22, "94350": 22, "781250": 22, "530250": 22, "97100": 22, "933250": 22, "82150": 22, "774000": 22, "728250": 22, "869750": 22, "87600": 22, "796500": 22, "863000": 22, "842750": 22, "870500": 22, "21947352": 22, "21947371": 22, "67700": 22, "261333": 22, "766667": 22, "953667": 22, "979333": 22, "93500": 22, "897333": 22, "613667": 22, "864333": 22, "89500": 22, "950667": 22, "75200": 22, "733333": 22, "855667": 22, "874333": 22, "77000": 22, "589333": 22, "506333": 22, "868333": 22, "781000": 22, "chr6": 22, "50328671": 22, "50328782": 22, "38975": 22, "316650": 22, "702250": 22, "820000": 22, "85425": 22, "458000": 22, "662250": 22, "608250": 22, "81025": 22, "889500": 22, "61000": 22, "592250": 22, "047500": 22, "632250": 22, "64975": 22, "605250": 22, "608500": 22, "305500": 22, "407325": 22, "chr12": [22, 24], "15047867": 22, "15047932": 22, "68650": 22, "750000": [22, 35], "825000": 22, "777500": 22, "266500": 22, "31450": 22, "874500": 22, "65700": 22, "867500": 22, "71350": 22, "591000": 22, "290500": 22, "740500": 22, "74400": 22, "723000": 22, "735000": 22, "755500": 22, "559500": 22, "chr8": [22, 24], "48317445": 22, "48317685": 22, "87640": 22, "938400": 22, "937200": 22, "596000": 22, "66820": 22, "844600": 22, "885600": 22, "746000": 22, "80600": 22, "880800": 22, "90840": 22, "804800": 22, "765800": 22, "848800": 22, "76380": 22, "824400": 22, "810400": 22, "916000": 22, "935600": 22, "chr2": 22, "93512302": 22, "93512369": 22, "31350": 22, "788000": 22, "581000": 22, "832500": 22, "604500": 22, "94450": 22, "937500": 22, "900500": 22, "890000": 22, "90750": 22, "956500": 22, "32550": 22, "232700": 22, "221000": 22, "799000": 22, "55550": 22, "614000": 22, "601000": 22, "435400": 22, "470000": 22, "color_palett": 22, "gc": 22, "majortype_color": 22, "silver": 22, "crimson": 22, "deepskyblu": 22, "darkseagreen": 22, "darkcyan": 22, "k": 22, "de9f00": 22, "67292f": 22, "e13f82": 22, "3f884e": 22, "78772b": 22, "4ac750": 22, "6bc22e": 22, "a8d617": 22, "314b1f": 22, "aab639": 22, "e179a2": 22, "764a71": 22, "63bc9e": 22, "cea675": 22, "a87331": 22, "9bb070": 22, "9fa600": 22, "5e461": 22, "8000": 22, "sort": [22, 34], "sum": [22, 32], "row_major_typ": 22, "cell_class_colors1": 22, "majortype_colors1": 22, "left_ha": [22, 29, 34], "right_ha": [22, 29, 34], "all_dmr_heatmap": 22, "set2": 23, "tomato": 23, "skyblu": [23, 24], "spectral_r": 23, "vertical": 23, "first": 23, "051388888888887": 23, "boarder": 23, "continnu": 23, "deep": 23, "otherwis": 23, "your_color": 23, "treat": 23, "sometim": 23, "without": 23, "do": 23, "give": 23, "anoth": [23, 25], "No": [23, 24, 28, 32], "open": [23, 24, 29], "mammal_arrai": 23, "pkl": 23, "rb": [23, 24, 29], "col_colors_dict": [23, 24, 34], "gsm4412025": 23, "gsm4412026": 23, "gsm4412027": 23, "gsm4412028": 23, "gsm4412029": 23, "gsm4412030": 23, "gsm4412031": 23, "gsm4412032": 23, "gsm4412033": 23, "gsm4412034": 23, "gsm4997945": 23, "gsm4997946": 23, "gsm4997947": 23, "gsm4997948": 23, "gsm4997949": 23, "gsm4997950": 23, "gsm4997951": 23, "gsm4997952": 23, "gsm4997953": 23, "gsm4997954": 23, "sheep": 23, "435033": 23, "432900": 23, "446626": 23, "449123": 23, "497180": 23, "515918": 23, "483706": 23, "504681": 23, "529076": 23, "446443": 23, "beluga": 23, "whale": 23, "687488": 23, "694207": 23, "706525": 23, "702734": 23, "687014": 23, "704003": 23, "705887": 23, "693806": 23, "719417": 23, "712677": 23, "560381": 23, "571552": 23, "610392": 23, "613619": 23, "675832": 23, "668502": 23, "624820": 23, "658377": 23, "702334": 23, "575034": 23, "hous": 23, "vaquita": 23, "693523": 23, "702525": 23, "716792": 23, "725095": 23, "711261": 23, "708651": 23, "717952": 23, "705486": 23, "720915": 23, "724171": 23, "581862": 23, "594443": 23, "628908": 23, "639457": 23, "680801": 23, "707493": 23, "662338": 23, "665142": 23, "751859": 23, "584952": 23, "larg": [23, 28], "fly": 23, "fox": 23, "286822": 23, "269406": 23, "296796": 23, "314719": 23, "305074": 23, "308419": 23, "268421": 23, "297236": 23, "295019": 23, "297973": 23, "281684": 23, "363926": 23, "367216": 23, "359392": 23, "289821": 23, "257351": 23, "234301": 23, "295840": 23, "249336": 23, "344848": 23, "greater": 23, "horsesho": 23, "bat": 23, "530606": 23, "517989": 23, "520719": 23, "525855": 23, "507583": 23, "511993": 23, "528107": 23, "521563": 23, "542159": 23, "509526": 23, "735151": 23, "714805": 23, "708868": 23, "738207": 23, "823604": 23, "828040": 23, "823382": 23, "796882": 23, "878783": 23, "693625": 23, "littl": 23, "brown": 23, "202525": 23, "215990": 23, "238977": 23, "241872": 23, "267750": 23, "236729": 23, "202384": 23, "240519": 23, "251148": 23, "267257": 23, "883": 23, "predictedtaxid": 23, "predictedspeci": 23, "common_nam": 23, "9940": 23, "ovis_aries_rambouillet": 23, "bovida": 23, "9749": 23, "delphinapterus_leuca": 23, "monodontida": 23, "10090": 23, "mus_musculu": 23, "murida": 23, "42100": 23, "phocoena_sinu": 23, "phocoenida": 23, "132908": 23, "pteropus_vampyru": 23, "pteropodida": 23, "59479": 23, "rhinolophus_ferrumequinum": 23, "rhinolophida": 23, "59463": 23, "myotis_lucifugu": 23, "vespertilionida": 23, "gse": 23, "basenam": 23, "ncbi_scientific_nam": 23, "taxid": 23, "tissu": [23, 24], "sex": [23, 24], "speci": 23, "successr": 23, "gse147003": 23, "ovi": 23, "ari": 23, "blood": [23, 34], "femal": [23, 24], "artiodactyla": 23, "765818": 23, "797669": 23, "759256": 23, "male": [23, 24], "749813": 23, "770433": 23, "gse164127": 23, "eptesicu": 23, "fuscu": 23, "29078": 23, "skin": 23, "chiroptera": 23, "660425": 23, "652822": 23, "664746": 23, "650848": 23, "657170": 23, "a40043": [23, 34], "00e5ff": 23, "00becc": 23, "cerebellum": 23, "b2f8ff": 23, "striatum": 23, "6cbf00": [23, 34], "liver": [23, 24, 34], "ffccee": [23, 34], "muscl": [23, 29, 34], "1e9351": 23, "put": 23, "auc": 23, "legend_pad": 23, "veri": 23, "abov": 23, "25": [23, 26, 28, 31, 32], "930555555555557": [23, 28], "last": 23, "second": 23, "worth": 23, "note": 23, "betwe": 23, "kind": [23, 29, 30], "predict": 23, "68888888888889": 23, "tranpos": 23, "columsn": 23, "obtain": 24, "pmid": [24, 34], "36617464": 24, "linearsegmentedcolormap": [24, 25], "80": [24, 29, 33, 35], "urllib": 24, "influence_of_snp_on_beta": 24, "close": 24, "snp": 24, "row_colors_dict": 24, "respect": 24, "2000": 24, "204875570030_r01c02": 24, "204875570030_r04c01": 24, "cg30848532_tc21": 24, "525089": 24, "419515": 24, "cg30147375_bc21": 24, "803776": 24, "585928": 24, "cg46239718_bc21": 24, "443958": 24, "517514": 24, "cg36100119_bc21": 24, "351977": 24, "528846": 24, "cg42738582_bc21": 24, "783958": 24, "724901": 24, "204875570030_r05c01": 24, "204875570030_r06c01": 24, "204875570035_r05c02": 24, "483276": 24, "460750": 24, "390317": 24, "510269": 24, "831463": 24, "550146": 24, "535909": 24, "450167": 24, "564107": 24, "524896": 24, "374422": 24, "551200": 24, "802178": 24, "848621": 24, "850481": 24, "target": 24, "extensionbas": 24, "probedesign": 24, "con": 24, "mapflag": 24, "ii": 24, "chr19": 24, "suboptim": 24, "hybrid": 24, "effect": 24, "artifici": 24, "low": [24, 25, 28], "meth": [24, 34], "cg": 24, "gg": 24, "ga": 24, "79760438": 24, "ca": 24, "ac": 24, "ag": 24, "109557651": 24, "gt": [24, 26], "117860829": 24, "5877949": 24, "aa": 24, "122031379": 24, "strain": 24, "molf_eij": 24, "frontal": 24, "lobe": 24, "cast_eij": 24, "darkorang": 24, "wheat": 24, "darkgrai": 24, "69": [24, 27, 34], "4986111111111": 24, "col_ha1": 24, "cm1": 24, "viridi": 24, "col_ha2": 24, "my_cmap": 24, "from_list": [24, 25], "cm2": 24, "dnam": 24, "beta_snp": 24, "df_corr": 25, "kycg_modules_correl": 25, "csv": [25, 27, 34, 35], "df_ann": 25, "kycg_modules_annot": 25, "kycg_modules_beta": 25, "cpg_std": 25, "std": [25, 28], "cpg_mean": 25, "astyp": [25, 28], "value_count": [25, 28], "28": [25, 26, 28], "33": 25, "count": [25, 27, 28, 29], "int64": 25, "isin": [25, 28], "hm": 25, "h3k4me1": 25, "h3k79me2": 25, "h3k79me3": 25, "h3f3a": 25, "h4k20me1": 25, "h2bk20ac": 25, "h3k23me2": 25, "h2afi": 25, "h2ak9ac": 25, "h2bk12ac": 25, "h2bk15ac": 25, "h3k9me1": 25, "h2bk120ub": 25, "h2azac": 25, "h3k79me1": 25, "h2a": 25, "h3_4ac": 25, "h3k27me1": 25, "h3k4me3b": 25, "h3k56ac": 25, "h3k9ac_h3k14ac": 25, "h3k9k14ac": 25, "cenpa": 25, "h1": 25, "h2afz": 25, "h2ak119": 25, "h3": 25, "h3ac": 25, "h3k27ac": 25, "h3k4me3": 25, "h3k9ac": 25, "h4": 25, "h4ac": 25, "h4k5ac": 25, "h4k5ac_h4k8ac_h4k12ac_h4k16ac": 25, "h4k8ac": 25, "histonelysineacetyl": 25, "histonelysinecrotonyl": 25, "h3k18cr": 25, "h4k16ac": 25, "h4k12ac": 25, "h3k27me3b": 25, "h2bub": 25, "h2ak5ac": 25, "h3k36me3b": 25, "h2bk120ac": 25, "h3k36ac": 25, "h4k91ac": 25, "h4k20me3": 25, "h2afy2": 25, "h3k36me2": 25, "h3k27me3": 25, "h2ak119ub": 25, "cg04735237": 25, "cg00643814": 25, "cg24865495": 25, "cg25376651": 25, "cg27485084": 25, "cg12609052": 25, "cg07354679": 25, "cg02592525": 25, "cg12747056": 25, "cg17718960": 25, "cg19987665": 25, "cg18406033": 25, "cg09678971": 25, "cg00461612": 25, "cg18689454": 25, "cg11923631": 25, "cg27251412": 25, "cg08089567": 25, "cg16717549": 25, "cg09510531": 25, "559916": 25, "686845": 25, "561415": 25, "699960": 25, "711086": 25, "563690": 25, "527953": 25, "621757": 25, "623319": 25, "048462": 25, "103904": 25, "002533": 25, "074093": 25, "114526": 25, "151763": 25, "140823": 25, "117356": 25, "100510": 25, "126291": 25, "538034": 25, "568960": 25, "687787": 25, "593650": 25, "676068": 25, "606604": 25, "589549": 25, "747987": 25, "185989": 25, "087952": 25, "242909": 25, "065101": 25, "054958": 25, "096098": 25, "089246": 25, "078234": 25, "025965": 25, "069120": 25, "614696": 25, "646452": 25, "786480": 25, "602911": 25, "519926": 25, "642344": 25, "642081": 25, "022499": 25, "108524": 25, "015151": 25, "028957": 25, "274560": 25, "302464": 25, "284823": 25, "265835": 25, "262931": 25, "268134": 25, "564983": 25, "727579": 25, "624186": 25, "416198": 25, "505130": 25, "596338": 25, "030346": 25, "122412": 25, "063630": 25, "007048": 25, "197339": 25, "226034": 25, "218230": 25, "180297": 25, "181867": 25, "204431": 25, "655515": 25, "599935": 25, "590981": 25, "627786": 25, "674095": 25, "020895": 25, "058742": 25, "078318": 25, "055019": 25, "139691": 25, "168351": 25, "160081": 25, "142216": 25, "114554": 25, "144024": 25, "512": 25, "chromhmm": [25, 28], "chromhmm_bioc": 25, "tfb": 25, "txwk": [25, 28], "hdgf": 25, "rbfox2": 25, "srebf1": 25, "pqbp1": 25, "qui": [25, 28], "ascl1": 25, "casz1": 25, "ebf1": 25, "fezf1": 25, "foxm1": 25, "hand2": 25, "hdac3": 25, "irf2b": 25, "crx": 25, "dnmt3b": 25, "otx2": 25, "rorb": 25, "zmynd11": 25, "znf711": 25, "macrod1": 25, "atf2": 25, "batf": 25, "etv6": 25, "fo": 25, "ikzf2": 25, "irf4": 25, "junb": 25, "maf": 25, "mafg": 25, "me": 25, "fry": 25, "pdx1": 25, "keep_cpg": 25, "map": [25, 28, 29], "stack": [25, 28, 32], "keep_hm": 25, "154": 25, "245870": 25, "768918": 25, "319166": 25, "635635": 25, "270459": 25, "835897": 25, "276320": 25, "618949": 25, "276748": 25, "700196": 25, "23716": 25, "all_cmap": 25, "binar": 25, "colgroup": [25, 28], "middl": [25, 28], "heatmap_ax": [25, 29], "j": 25, "set_vis": [25, 26, 29], "var": [25, 32], "folder": 25, "3q": 25, "hwjjddlj65b030m69cfbdj4h0000gq": 25, "ipykernel_61938": 25, "92826381": 25, "matplotlibdeprecationwarn": 25, "deprec": 25, "minor": [25, 28, 29], "releas": 25, "regist": 25, "dotclustermap2": 25, "dev4": 26, "g71c725a": 26, "d20240410": 26, "ax_top": 26, "ax_right": 26, "set_axis_on": 26, "set_linestyl": [26, 29], "agg_filt": 26, "anim": 26, "bbox_patch": 26, "lt": 26, "fancybboxpatch": 26, "0x7f854d935760": 26, "children": 26, "clip_path": 26, "600x800": 26, "font_manag": 26, "0x7f854d8d56a0": 26, "fontvari": 26, "gid": 26, "math_fontfamili": 26, "dejavusan": 26, "mouseov": 26, "parse_math": 26, "path_effect": 26, "picker": 26, "000000000000007": 26, "sketch_param": 26, "snap": 26, "stretch": 26, "tightbbox": 26, "125": 26, "112": [26, 28], "875": [26, 34], "blendedaffine2d": 26, "0x7f854d8d5070": 26, "transform_rotates_text": 26, "transformed_clip_path_and_affin": 26, "unitless_posit": 26, "url": 26, "usetex": 26, "window_ext": 26, "wrap": 26, "antialias": 26, "75": [26, 28, 34], "0x7f854dded490": 26, "capstyl": 26, "butt": 26, "data_transform": 26, "affine2d": 26, "0x7f854f0bbfd0": 26, "extent": 26, "761576317947967": 26, "6388888888888813": 26, "92": 26, "78935409572574": 26, "638888888888896": 26, "hatch": 26, "joinstyl": 26, "miter": 26, "solid": 26, "mutation_aspect": 26, "mutation_scal": 26, "444444444444443": 26, "patch_transform": 26, "identitytransform": 26, "0x7f854d868730": 26, "63888889": 26, "81": [26, 33], "38888889": 26, "79": 26, "uint8": 26, "vert": 26, "76157632": 26, "7893541": 26, "expr": 27, "test_expression_dataset": 27, "selected_row": 27, "adam7": 27, "hla": 27, "doa": 27, "ptgir": 27, "opcml": 27, "it1": 27, "mageb1": 27, "label_row": 27, "c8orf34": 27, "as1": 27, "smim28": 27, "linc01114": 27, "klk3": 27, "traj45": 27, "tpmtp1": 27, "hnrnpcp4": 27, "89": [27, 33], "nci": 27, "h660": 27, "vcap": 27, "du": 27, "145": 27, "mda": 27, "pca": 27, "2b": 27, "22rv1": 27, "lncap": 27, "fgc": 27, "sup": 27, "hd1": 27, "l": 27, "540": 27, "1236": 27, "km": 27, "h2": 27, "hdlm": 27, "616": 27, "428": 27, "611": 27, "TO": 27, "175": 27, "044425": 27, "303654": 27, "604782": 27, "123861": 27, "015847": 27, "848602": 27, "215875": 27, "251406": 27, "795724": 27, "092080": 27, "070939": 27, "705117": 27, "241584": 27, "766805": 27, "132841": 27, "531804": 27, "564560": 27, "832337": 27, "442301": 27, "472336": 27, "359856": 27, "911472": 27, "795981": 27, "391838": 27, "rank2": 27, "regul": [27, 29], "top20": 27, "upregul": 27, "goi": 27, "colors_dict": 27, "row_ha_left": 27, "row_ha_right": 27, "uncom": 27, "sn": 28, "subset": 28, "collaps": 28, "load_dataset": 28, "brain_network": 28, "header": 28, "used_network": 28, "used_column": 28, "get_level_valu": 28, "comput": 28, "form": 28, "corr_mat": 28, "corr": 28, "level": [28, 34], "level_0": 28, "level_1": 28, "lh": 28, "rh": 28, "881516": 28, "049793": 28, "026902": 28, "144335": 28, "141253": 28, "119250": 28, "261589": 28, "272701": 28, "370021": 28, "366065": 28, "325680": 28, "196770": 28, "144566": 28, "366818": 28, "388756": 28, "352529": 28, "363982": 28, "341524": 28, "350452": 28, "112697": 28, "036909": 28, "144277": 28, "189683": 28, "084633": 28, "324230": 28, "332886": 28, "374322": 28, "361036": 28, "274151": 28, "142392": 28, "070452": 28, "358625": 28, "402173": 28, "302286": 28, "339989": 28, "315931": 28, "343379": 28, "343464": 28, "470239": 28, "100802": 28, "438449": 28, "339667": 28, "089811": 28, "272394": 28, "036493": 28, "171179": 28, "043298": 28, "158039": 28, "005598": 28, "060007": 28, "079078": 28, "040060": 28, "027878": 28, "075781": 28, "130549": 28, "278569": 28, "127621": 28, "014404": 28, "051249": 28, "090130": 28, "170053": 28, "124278": 28, "112148": 28, "063705": 28, "172007": 28, "040629": 28, "079687": 28, "024864": 28, "092263": 28, "068858": 28, "521377": 28, "506652": 28, "320966": 28, "141884": 28, "608392": 28, "075986": 28, "095015": 28, "012966": 28, "082816": 28, "023340": 28, "058718": 28, "034181": 28, "033355": 28, "022982": 28, "025638": 28, "431619": 28, "418708": 28, "084634": 28, "132": [28, 29], "808800071564": 28, "22361111111111": 28, "wding": 28, "miniconda3": 28, "env": 28, "lib": 28, "python3": 28, "site": 28, "189": 28, "userwarn": 28, "via": 28, "norm": 28, "465277777777779": 28, "drop_dupl": 28, "rowgroup": 28, "kycg_result": 28, "rmsk1": 28, "ensregbuild": 28, "sampleid": [28, 32], "clark2018_argelaguet2019": 28, "dataset1": 28, "luo2022": 28, "dataset2": 28, "max_p": 28, "log10": [28, 29], "pval": 28, "id": [28, 29, 34], "cpgtype": 28, "vc": 28, "term": [28, 29], "p_max": 28, "p_min": 28, "odds_ratio": 28, "pvalu": 28, "enrichtyp": 28, "het": 28, "061": 28, "neg": 28, "020000e": 28, "07": [28, 29], "26": 28, "tx": 28, "029": 28, "580000e": 28, "65": 28, "tssflnk": 28, "056": 28, "370000e": 28, "06": [28, 29], "022": 28, "660000e": 28, "346": 28, "350": 28, "760000e": 28, "02": 28, "describ": 28, "64": 28, "356918": 28, "632073": 28, "119186": 28, "deplet": 28, "srprna": 28, "satellit": 28, "simple_repeat": 28, "low_complex": 28, "reprpcwk": 28, "tssbiv": 28, "reprpc": 28, "ltr": 28, "snrna": 28, "sine": 28, "rdylgn_r": 28, "coolwarm_r": 28, "dotheatmap1": 28, "180": 28, "09263168093761": 28, "20230227_kycg": 28, "516666666666666": 28, "glob": 29, "style": 29, "tradition": 29, "lot": [29, 31], "dotplot": 29, "had": 29, "idea": 29, "manuscript": 29, "alreadi": 29, "gotten": 29, "bp": 29, "molecular": 29, "mf": 29, "kegg": 29, "pathwai": 29, "enrichment_analysis_dotheatmap": 29, "data1": 29, "descript": 29, "generatio": 29, "enrichedcategori": 29, "colid": 29, "rowid": 29, "0003779": 29, "actin": 29, "bind": 29, "044949": 29, "453212e": 29, "186": 29, "group2": 29, "ct1": 29, "610265": 29, "0030695": 29, "gtpase": 29, "activ": 29, "043016": 29, "744023e": 29, "178": 29, "171081": 29, "0060589": 29, "nucleosid": 29, "triphosphatas": 29, "0005201": 29, "extracellular": 29, "structur": 29, "constitu": 29, "021266": 29, "810239e": 29, "88": 29, "34": 29, "235806": 29, "0050839": 29, "adhes": 29, "molecul": 29, "031899": 29, "612420e": 29, "792522": 29, "840": 29, "0050905": 29, "neuromuscular": 29, "010454": 29, "224335e": 29, "122": 29, "ct2": 29, "912100": 29, "1205": 29, "0030534": 29, "adult": 29, "behavior": 29, "009426": 29, "028861e": 29, "110": 29, "219765": 29, "1558": 29, "0035019": 29, "somat": 29, "stem": 29, "popul": 29, "mainten": 29, "004799": 29, "029991e": 29, "219683": 29, "1746": 29, "0019827": 29, "009854": 29, "467128e": 29, "04": 29, "833532": 29, "2999": 29, "0021953": 29, "central": 29, "nervou": 29, "system": 29, "neuron": [29, 34], "008997": 29, "138960e": 29, "105": 29, "039103": 29, "139": 29, "metal": 29, "ion": 29, "transmembran": 29, "transport": 29, "monoatom": 29, "channel": 29, "gate": 29, "calmodulin": 29, "receptor": 29, "protein": 29, "kinas": 29, "substrat": 29, "organ": 29, "extern": 29, "encapsul": 29, "axon": 29, "guidanc": 29, "locomotori": 29, "fat": 29, "amphetamin": 29, "addict": 29, "calcium": 29, "signal": 29, "cocain": 29, "cytokin": 29, "interact": 29, "neuroact": 29, "ligand": 29, "cytoskeleton": 29, "ecm": 29, "viral": 29, "cy": 29, "focal": 29, "lupu": 29, "erythematosu": 29, "atp": 29, "chromatin": 29, "remodel": 29, "histon": 29, "modif": 29, "group1": 29, "ct3": 29, "ct4": 29, "s_min": 29, "s_max": 29, "3074": 29, "0021954": 29, "004586": 29, "027429": 29, "561784": 29, "0045444": 29, "176471": 29, "000382": 29, "417494": 29, "orrd": 29, "1e": 29, "ast": 29, "ravel": 29, "both": 29, "dashdot": 29, "cell_class_dotheatmap": 29, "106": 29, "46754159267321": 29, "ratio": 29, "total": 29, "510843": 30, "531995": 30, "188667": 30, "180811": 30, "354718": 30, "whole": 30, "separ": 30, "wiki": 31, "ital": 31, "_label_kw": 31, "tree": 31, "roundtooth": 31, "book": 32, "cbioport": 32, "tcga_lung_adenocarcinoma_provisional_ras_raf_mek_jnk_signal": 32, "dropna": 32, "mut": 32, "amp": [32, 33], "homdel": 32, "4384": 32, "kra": 32, "hra": 32, "braf": 32, "raf1": 32, "map3k1": 32, "unique_vari": 32, "v1": 32, "strip": 32, "008000": 32, "mutat": 32, "frequenc": 32, "row_vc": 32, "col_vc": 32, "kras_sampl": 32, "df_col_split": 32, "op": [32, 33], "alter": 32, "27": [32, 33], "72361111111111": 32, "ad": 32, "row_var_freq": 32, "isvar": 32, "oncoprint2": 32, "430555555555557": 32, "toi": 33, "sample_": 33, "gene_": 33, "alts_lol": 33, "amp_valu": 33, "randint": 33, "del_valu": 33, "neut_valu": 33, "alts_df": 33, "neut": 33, "prepar": 33, "annot_1_df": 33, "annot1": 33, "annot_2_df": 33, "500": 33, "annot2": 33, "annot_3_df": 33, "patient": 33, "sample_1": 33, "gene_1": 33, "gene_2": 33, "gene_3": 33, "gene_4": 33, "gene_5": 33, "95": 33, "sample_10": 33, "gene_6": 33, "96": 33, "gene_7": 33, "97": 33, "gene_8": 33, "98": 33, "gene_9": 33, "gene_10": 33, "patient2": 33, "sample_2": 33, "patient5": 33, "sample_3": 33, "patient3": 33, "sample_4": 33, "patient4": 33, "sample_5": 33, "patient1": 33, "sample_6": 33, "sample_7": 33, "sample_8": 33, "sample_9": 33, "77": 33, "78": 33, "1114": 33, "2698": 33, "4718": 33, "4633": 33, "3444": 33, "1984": 33, "2088": 33, "2316": 33, "2939": 33, "1373": 33, "a3": [33, 35], "a1": [33, 35], "a2": [33, 35], "flatten": 33, "signatur": 34, "come": 34, "36599988": 34, "raw": 34, "githubusercont": 34, "adipocyt": 34, "bladder": 34, "epithelium": 34, "mem": 34, "granulocyt": 34, "colon": 34, "macrophag": 34, "monocyt": 34, "interstiti": 34, "alveolar": 34, "pancrea": 34, "duct": 34, "skelet": 34, "endocrin": 34, "bronchu": 34, "smooth": 34, "prostat": 34, "coronari": 34, "arteri": 34, "aorta": 34, "thyroid": 34, "110426397": 34, "120667": 34, "9102": 34, "8285": 34, "918667": 34, "750667": 34, "8705": 34, "896": 34, "916333": 34, "953333": 34, "9450": 34, "61225": 34, "5485": 34, "94175": 34, "952": 34, "417": 34, "150": 34, "229": 34, "294": 34, "214": 34, "938333": 34, "110426639": 34, "420000": 34, "9304": 34, "8920": 34, "955667": 34, "964667": 34, "9285": 34, "903": 34, "924667": 34, "975333": 34, "9720": 34, "94575": 34, "8990": 34, "96225": 34, "963": 34, "606": 34, "185": 34, "348": 34, "917": 34, "484": 34, "972333": 34, "110426659": 34, "425667": 34, "9328": 34, "7635": 34, "933000": 34, "942333": 34, "8945": 34, "931": 34, "963667": 34, "899333": 34, "8895": 34, "93875": 34, "8390": 34, "92225": 34, "559": 34, "129": 34, "292": 34, "395": 34, "989667": 34, "110427109": 34, "203667": 34, "9724": 34, "9610": 34, "968667": 34, "9240": 34, "939": 34, "985000": 34, "9620": 34, "73300": 34, "7570": 34, "96525": 34, "929": 34, "225": 34, "806": 34, "951": 34, "905": 34, "974333": 34, "110427116": 34, "217333": 34, "9478": 34, "0000": 34, "976000": 34, "970667": 34, "9730": 34, "913": 34, "975000": 34, "970333": 34, "9800": 34, "65125": 34, "7955": 34, "96150": 34, "000": 34, "605": 34, "857": 34, "868": 34, "981000": 34, "clusterid": 34, "ep": 34, "granul": 34, "mono": 34, "macro": 34, "nk": 34, "bone": 34, "osteob": 34, "breast": 34, "basal": 34, "dermal": 34, "fibro": 34, "epid": 34, "kerat": 34, "eryth": 34, "prog": 34, "fallopian": 34, "gallbladd": 34, "gastric": 34, "neck": 34, "heart": 34, "cardio": 34, "kidnei": 34, "hep": 34, "alveo": 34, "bron": 34, "oligodend": 34, "acinar": 34, "musc": 34, "1e93a": 34, "ff9c00": 34, "ff9f7f": 34, "ff7f00": 34, "ff2e8d": 34, "cc0043": 34, "e5e5e5": 34, "cc4407": 34, "cc843d": 34, "663d28": 34, "1e937c": 34, "40705f": 34, "009351": 34, "e7e4bf": 34, "cca300": 34, "002929": 34, "ff99aa": 34, "ff99ff": 34, "f6ff99": 34, "ba99ff": 34, "ccccff": 34, "9e542": 34, "2ca02c": 34, "df7f00": 34, "ffd866": 34, "ffcc32": 34, "7f4c33": 34, "cc9951": 34, "b2bfff": 34, "loyfer2023_heatmap": 34, "981944444444444": 34, "download": 35, "pycomplexheatmap_test_2": 35, "c1": 35, "c2": 35, "c3": 35, "c4": 35, "c5": 35, "c6": 35, "c7": 35, "c8": 35, "c9": 35, "c10": 35, "c35": 35, "c36": 35, "c37": 35, "c38": 35, "c39": 35, "c40": 35, "c41": 35, "c42": 35, "c43": 35, "c44": 35, "00000": 35, "00": 35, "250000": 35, "111111": 35, "444444": 35, "222222": 35, "388889": 35, "611111": 35, "277778": 35, "888889": 35, "944444": 35, "555556": 35, "500000": 35, "a4": 35, "625000": 35, "375000": 35, "a5": 35, "a6": 35, "600000": 35, "100000": 35, "300000": 35, "700000": 35, "a7": 35, "a8": 35, "60000": 35, "800000": 35, "400000": 35, "200000": 35, "a9": 35, "440000": 35, "72": 35, "460000": 35, "040000": 35, "080000": 35, "560000": 35, "020000": 35, "240000": 35, "a10": 35, "a11": 35, "357143": 35, "02381": 35, "023810": 35, "071429": 35, "571429": 35, "214286": 35, "452381": 35, "166667": 35, "380952": 35, "309524": 35, "a12": 35, "062500": 35, "031250": 35, "312500": 35, "218750": 35, "531250": 35, "156250": 35, "687500": 35, "343750": 35, "437500": 35, "a13": 35, "333333": 35, "666667": 35, "a14": 35, "a15": 35, "076923": 35, "615385": 35, "384615": 35, "538462": 35, "307692": 35, "153846": 35, "a16": 35, "a17": 35, "150000": 35, "900000": 35, "550000": 35, "350000": 35, "a18": 35, "833333": 35, "a19": 35, "rdpu": 35}, "objects": {"": [[0, 0, 0, "-", "PyComplexHeatmap"]], "PyComplexHeatmap": [[1, 0, 0, "-", "annotations"], [2, 0, 0, "-", "clustermap"], [3, 0, 0, "-", "colors"], [4, 0, 0, "-", "dotHeatmap"], [5, 0, 0, "-", "example"], [6, 0, 0, "-", "oncoPrint"], [8, 0, 0, "-", "utils"]], "PyComplexHeatmap.annotations": [[1, 1, 1, "", "AnnotationBase"], [1, 1, 1, "", "HeatmapAnnotation"], [1, 1, 1, "", "anno_barplot"], [1, 1, 1, "", "anno_boxplot"], [1, 1, 1, "", "anno_img"], [1, 1, 1, "", "anno_label"], [1, 1, 1, "", "anno_lineplot"], [1, 1, 1, "", "anno_scatterplot"], [1, 1, 1, "", "anno_simple"]], "PyComplexHeatmap.annotations.AnnotationBase": [[1, 2, 1, "", "get_label_width"], [1, 2, 1, "", "get_max_label_width"], [1, 2, 1, "", "get_ticklabel_width"], [1, 2, 1, "", "reorder"], [1, 2, 1, "", "set_axes_kws"], [1, 2, 1, "", "set_label"], [1, 2, 1, "", "set_legend"], [1, 2, 1, "", "update_plot_kws"]], "PyComplexHeatmap.annotations.HeatmapAnnotation": [[1, 2, 1, "", "collect_legends"], [1, 2, 1, "", "plot_annotations"], [1, 2, 1, "", "plot_legends"], [1, 2, 1, "", "set_axes_kws"], [1, 2, 1, "", "show_ticklabels"]], "PyComplexHeatmap.annotations.anno_barplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_boxplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_img": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_label": [[1, 2, 1, "", "get_ticklabel_width"], [1, 2, 1, "", "plot"], [1, 2, 1, "", "set_plot_kws"], [1, 2, 1, "", "set_side"]], "PyComplexHeatmap.annotations.anno_lineplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_scatterplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_simple": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.clustermap": [[2, 1, 1, "", "ClusterMapPlotter"], [2, 1, 1, "", "DendrogramPlotter"], [2, 4, 1, "", "composite"], [2, 4, 1, "", "heatmap"], [2, 1, 1, "", "heatmapPlotter"], [2, 4, 1, "", "plot_heatmap"]], "PyComplexHeatmap.clustermap.ClusterMapPlotter": [[2, 2, 1, "", "cal_cold_between_groups"], [2, 2, 1, "", "cal_rowd_between_groups"], [2, 2, 1, "", "calculate_col_dendrograms"], [2, 2, 1, "", "calculate_row_dendrograms"], [2, 2, 1, "", "collect_legends"], [2, 2, 1, "", "format_data"], [2, 2, 1, "", "plot"], [2, 2, 1, "", "plot_dendrograms"], [2, 2, 1, "", "plot_legends"], [2, 2, 1, "", "plot_matrix"], [2, 2, 1, "", "post_processing"], [2, 2, 1, "", "set_axes_labels_kws"], [2, 2, 1, "", "set_height"], [2, 2, 1, "", "set_width"], [2, 2, 1, "", "set_xy_labels"], [2, 2, 1, "", "standard_scale"], [2, 2, 1, "", "tight_layout"], [2, 2, 1, "", "z_score"]], "PyComplexHeatmap.clustermap.DendrogramPlotter": [[2, 2, 1, "", "calculate_dendrogram"], [2, 3, 1, "", "calculated_linkage"], [2, 2, 1, "", "check_array"], [2, 2, 1, "", "get_coords"], [2, 2, 1, "", "plot"], [2, 3, 1, "", "reordered_ind"]], "PyComplexHeatmap.clustermap.heatmapPlotter": [[2, 2, 1, "", "plot"]], "PyComplexHeatmap.colors": [[3, 4, 1, "", "define_cmap"], [3, 4, 1, "", "register_cmap"]], "PyComplexHeatmap.dotHeatmap": [[4, 1, 1, "", "DotClustermapPlotter"], [4, 4, 1, "", "dotHeatmap2d"], [4, 4, 1, "", "scale"]], "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter": [[4, 2, 1, "", "collect_legends"], [4, 2, 1, "", "format_data"], [4, 2, 1, "", "plot_matrix"], [4, 2, 1, "", "post_processing"]], "PyComplexHeatmap.example": [[5, 4, 1, "", "clustermap_example0"], [5, 4, 1, "", "clustermap_example1"], [5, 4, 1, "", "get_kycg_example_data"]], "PyComplexHeatmap.oncoPrint": [[6, 1, 1, "", "oncoPrintPlotter"], [6, 4, 1, "", "oncoprint"]], "PyComplexHeatmap.oncoPrint.oncoPrintPlotter": [[6, 2, 1, "", "add_default_annotations"], [6, 2, 1, "", "collect_legends"], [6, 2, 1, "", "format_data"], [6, 2, 1, "", "get_samples_order"], [6, 2, 1, "", "plot_matrix"], [6, 2, 1, "", "post_processing"]], "PyComplexHeatmap.utils": [[8, 4, 1, "", "axis_ticklabels_overlap"], [8, 4, 1, "", "cal_legend_width"], [8, 4, 1, "", "cluster_labels"], [8, 4, 1, "", "define_cmap"], [8, 4, 1, "", "despine"], [8, 4, 1, "", "get_colormap"], [8, 4, 1, "", "plot_cmap_legend"], [8, 4, 1, "", "plot_color_dict_legend"], [8, 4, 1, "", "plot_legend_list"], [8, 4, 1, "", "plot_marker_legend"], [8, 4, 1, "", "set_default_style"], [8, 4, 1, "", "to_utf8"]]}, "objtypes": {"0": "py:module", "1": "py:class", "2": "py:method", "3": "py:property", "4": "py:function"}, "objnames": {"0": ["py", "module", "Python module"], "1": ["py", "class", "Python class"], "2": ["py", "method", "Python method"], "3": ["py", "property", "Python property"], "4": ["py", "function", "Python function"]}, "titleterms": {"pycomplexheatmap": [0, 1, 2, 3, 4, 5, 6, 7, 8, 16, 19, 21], "packag": [0, 22, 30], "submodul": 0, "modul": [0, 1, 2, 3, 4, 5, 6, 7, 8, 25, 37], "content": [0, 9, 11, 12, 13, 15, 16, 18, 20, 36], "annot": [1, 21, 23, 30, 31], "clustermap": [2, 11, 25, 28], "color": [3, 28], "dotheatmap": [4, 13, 29], "exampl": [5, 20, 23, 28], "oncoprint": [6, 32, 33, 36], "tool": 7, "util": 8, "advanc": 9, "usag": 9, "citat": 10, "For": 12, "develop": 12, "galleri": 14, "get": 15, "start": 15, "credit": 16, "thank": 16, "instal": 17, "depend": 17, "how": [17, 21, 29, 31], "kwarg": 18, "more": 20, "gener": 21, "dataset": [21, 24, 28, 32], "add": [21, 23, 28], "select": [21, 29], "row": [21, 23, 30], "label": [21, 31], "top": [21, 23], "heatmap": [21, 24, 28, 30, 31], "cell": [21, 34], "onli": [21, 23], "plot": [21, 23, 25, 28, 29, 30], "chang": [21, 28], "orent": 21, "down": 21, "extra": 21, "space": 21, "left": [21, 23], "right": [21, 23], "orient": 21, "us": [21, 25, 28, 29, 33], "paramet": [21, 28], "multipl": 21, "loop": 21, "cluster": 21, "between": [21, 22], "group": [21, 22, 28], "within": [21, 22], "clsuter": 21, "col_split": 21, "col_clust": 21, "true": 21, "cluster_between_group": 21, "col_split_ord": 21, "fals": 21, "cluster_within_group": 21, "custom": 21, "linkag": 21, "imag": 21, "choos": 21, "build": 21, "colormap": 21, "diverg": 21, "qualit": 21, "cmap": [21, 28], "forc": 21, "displai": 21, "all": 21, "col": 21, "ticklabel": 21, "import": [22, 30], "dmr": 22, "A": [23, 25], "quick": 23, "column": [23, 30], "anno_label": [23, 31], "anno_simpl": 23, "To": 23, "quickli": 23, "you": 23, "just": 23, "need": [23, 29], "datafram": 23, "figur": 23, "legend": 23, "separ": 23, "bottom": 23, "composit": 24, "two": 24, "horizont": 24, "mous": 24, "dna": [24, 34], "methyl": [24, 34], "arrai": 24, "correl": 25, "matrix": 25, "cpg": 25, "dotclustermap": 25, "process": 25, "data": [25, 28, 29, 34], "dot": [25, 28], "smaller": 25, "1": [26, 30], "understand": 26, "layout": 26, "visual": [27, 28, 29, 32, 33, 34], "differenti": 27, "express": 27, "gene": [27, 29], "load": 28, "an": 28, "brain": 28, "network": 28, "from": 28, "seaborn": 28, "tradit": 28, "squar": 28, "marker": [28, 29], "": 28, "simpl": 28, "fix": 28, "size": 28, "point": 28, "hue": 28, "differ": 28, "up": 28, "five": 28, "dimens": 28, "dotclustermapplott": 28, "set": 29, "enrich": 29, "analysi": 29, "result": 29, "clusterprofil": 29, "why": 29, "do": 29, "we": 29, "better": 29, "wai": 29, "read": 29, "2": 30, "3": 30, "split": 30, "4": 30, "along": 30, "side": 30, "main": 30, "label_kw": 31, "xticklabels_kw": 31, "yticklabels_kw": 31, "modifi": 31, "xticklabel": 31, "yticklabel": 31, "xlabel_kw": 31, "ylabel_kw": 31, "xlabel": 31, "ylabel": 31, "control": 31, "gap": 31, "pad": 31, "hgap": 31, "wgap": 31, "heatmapannot": 31, "subplot_gap": 31, "row_split_gap": 31, "col_split_gap": 31, "remov": 31, "arrow": 31, "tcga": 32, "lung": 32, "adenocarcinoma": 32, "carcinoma": 32, "variant": 32, "categor": 33, "variabl": 33, "singl": 34, "loyfer2023": 34, "setup": 37}, "envversion": {"sphinx.domains.c": 3, "sphinx.domains.changeset": 1, "sphinx.domains.citation": 1, "sphinx.domains.cpp": 9, "sphinx.domains.index": 1, "sphinx.domains.javascript": 3, "sphinx.domains.math": 2, "sphinx.domains.python": 4, "sphinx.domains.rst": 2, "sphinx.domains.std": 2, "sphinx.ext.todo": 2, "sphinx.ext.viewcode": 1, "nbsphinx": 4, "sphinx": 58}, "alltitles": {"PyComplexHeatmap package": [[0, "pycomplexheatmap-package"]], "Submodules": [[0, "submodules"]], "Module contents": [[0, "module-PyComplexHeatmap"]], "PyComplexHeatmap.annotations module": [[1, "module-PyComplexHeatmap.annotations"]], "PyComplexHeatmap.clustermap module": [[2, "module-PyComplexHeatmap.clustermap"]], "PyComplexHeatmap.colors module": [[3, "module-PyComplexHeatmap.colors"]], "PyComplexHeatmap.dotHeatmap module": [[4, "module-PyComplexHeatmap.dotHeatmap"]], "PyComplexHeatmap.example module": [[5, "module-PyComplexHeatmap.example"]], "PyComplexHeatmap.oncoPrint module": [[6, "module-PyComplexHeatmap.oncoPrint"]], "PyComplexHeatmap.tools module": [[7, "pycomplexheatmap-tools-module"]], "PyComplexHeatmap.utils module": [[8, "module-PyComplexHeatmap.utils"]], "Advanced Usage": [[9, "advanced-usage"]], "Contents:": [[9, null], [11, null], [12, null], [13, null], [15, null], [16, null], [18, null], [20, null], [36, null]], "Citation": [[10, "citation"]], "Clustermap": [[11, "clustermap"]], "For Developers": [[12, "for-developers"]], "dotHeatmap": [[13, "dotheatmap"]], "Gallery": [[14, "gallery"]], "Get Started": [[15, "get-started"]], "PyComplexHeatmap": [[16, "pycomplexheatmap"], [19, "pycomplexheatmap"]], "Credits and Thanks": [[16, "credits-and-thanks"]], "Installation": [[17, "installation"]], "Dependencies": [[17, "dependencies"]], "How to install?": [[17, "how-to-install"]], "Kwargs": [[18, "kwargs"]], "More Examples": [[20, "more-examples"]], "Generate dataset": [[21, "Generate-dataset"]], "Add selected rows labels": [[21, "Add-selected-rows-labels"]], "Add annotations on the top of heatmap cells": [[21, "Add-annotations-on-the-top-of-heatmap-cells"]], "Only plot the annotations": [[21, "Only-plot-the-annotations"]], "Change orentation to down and add extra space": [[21, "Change-orentation-to-down-and-add-extra-space"]], "Change orentation to the left": [[21, "Change-orentation-to-the-left"]], "Change orentation to the right": [[21, "Change-orentation-to-the-right"]], "Changing orientation using parameter orientation": [[21, "Changing-orientation-using-parameter-orientation"]], "Add multiple heatmap annotations using for loop": [[21, "Add-multiple-heatmap-annotations-using-for-loop"]], "Cluster between groups and cluster within groups": [[21, "Cluster-between-groups-and-cluster-within-groups"]], "clsuter within groups: col_split=*, col_cluster=True": [[21, "clsuter-within-groups:-col_split=*,-col_cluster=True"]], "cluster_between_groups: col_split=*, col_split_order=\"cluster_between_groups\",col_cluster=False": [[21, "cluster_between_groups:-col_split=*,-col_split_order=%22cluster_between_groups%22,col_cluster=False"]], "cluster_within_groups && cluster_between_groups: col_split=*, col_split_order=\"cluster_between_groups\",col_cluster=True": [[21, "cluster_within_groups-&&-cluster_between_groups:-col_split=*,-col_split_order=%22cluster_between_groups%22,col_cluster=True"]], "Custom annotation": [[21, "Custom-annotation"]], "Custom linkage": [[21, "Custom-linkage"]], "Image annotation": [[21, "Image-annotation"]], "Choosing Build-in colormap in PyComplexHeatmap": [[21, "Choosing-Build-in-colormap-in-PyComplexHeatmap"]], "Diverging": [[21, "Diverging"]], "Qualitative": [[21, "Qualitative"]], "How to use the Build-in cmap?": [[21, "How-to-use-the-Build-in-cmap?"]], "How to force display all row/col ticklabels?": [[21, "How-to-force-display-all-row/col-ticklabels?"]], "Import packages": [[22, "Import-packages"]], "Between groups DMRs": [[22, "Between-groups-DMRs"]], "Within groups DMRs": [[22, "Within-groups-DMRs"]], "A quick example": [[23, "A-quick-example"]], "Plotting annotations": [[23, "Plotting-annotations"]], "Only plot the row/column annotation": [[23, "Only-plot-the-row/column-annotation"]], "anno_label:": [[23, "anno_label:"]], "anno_simple:": [[23, "anno_simple:"]], "To add a annotation quickly, you just need a dataframe": [[23, "To-add-a-annotation-quickly,-you-just-need-a-dataframe"]], "Plot the figure and legend separately": [[23, "Plot-the-figure-and-legend-separately"]], "Top, bottom, left ,right annotations": [[23, "Top,-bottom,-left-,right-annotations"]], "Composite two heatmaps horizontally for mouse DNA methylation array dataset": [[24, "Composite-two-heatmaps-horizontally-for-mouse-DNA-methylation-array-dataset"]], "Plot correlation matrix for CpG modules using DotClustermap": [[25, "Plot-correlation-matrix-for-CpG-modules-using-DotClustermap"]], "Processing the data": [[25, "Processing-the-data"]], "Plotting the Dot clustermap": [[25, "Plotting-the-Dot-clustermap"]], "A smaller dot clustermap": [[25, "A-smaller-dot-clustermap"]], "1. Understand the layout:": [[26, "1.-Understand-the-layout:"]], "Visualizing Differential Expression Genes": [[27, "Visualizing-Differential-Expression-Genes"]], "Load an example brain networks dataset from seaborn": [[28, "Load-an-example-brain-networks-dataset-from-seaborn"]], "Dot Heatmap": [[28, "Dot-Heatmap"]], "Plot traditional heatmap using square marker marker='s'": [[28, "Plot-traditional-heatmap-using-square-marker-marker='s'"]], "Simple dot heatmap using fixed dot size": [[28, "Simple-dot-heatmap-using-fixed-dot-size"]], "Changing the size of point": [[28, "Changing-the-size-of-point"]], "Add parameter hue and use different colors for different groups": [[28, "Add-parameter-hue-and-use-different-colors-for-different-groups"]], "Add parameter hue and use different cmap and marker for different groups": [[28, "Add-parameter-hue-and-use-different-cmap-and-marker-for-different-groups"]], "Dot Clustermap": [[28, "Dot-Clustermap"]], "Plot clustermap using seaborn brain networks dataset": [[28, "Plot-clustermap-using-seaborn-brain-networks-dataset"]], "Visualize up to five dimension data using DotClustermapPlotter": [[28, "Visualize-up-to-five-dimension-data-using-DotClustermapPlotter"]], "Visualizing gene set enrichment analysis results (clusterProfiler) using dotHeatmap": [[29, "Visualizing-gene-set-enrichment-analysis-results-(clusterProfiler)-using-dotHeatmap"]], "Why do we need a better way to visualize the gene set enrichment analysis result?": [[29, "Why-do-we-need-a-better-way-to-visualize-the-gene-set-enrichment-analysis-result?"]], "Read data": [[29, "Read-data"]], "Plot": [[29, "Plot"]], "How to select marker:": [[29, "How-to-select-marker:"]], "1. Import packages": [[30, "1.-Import-packages"]], "2. Plot heatmap annotations": [[30, "2.-Plot-heatmap-annotations"]], "3. Plot the heatmap with rows and columns split": [[30, "3.-Plot-the-heatmap-with-rows-and-columns-split"]], "4. Plot the annotations along side with main heatmap": [[30, "4.-Plot-the-annotations-along-side-with-main-heatmap"]], "label_kws, xticklabels_kws and yticklabels_kws: Modifying the heatmap annotations labels and xticklabels, yticklabels": [[31, "label_kws,-xticklabels_kws-and-yticklabels_kws:-Modifying-the-heatmap-annotations-labels-and-xticklabels,-yticklabels"]], "xlabel_kws and ylabel_kws: Modifying xlabel and ylabel": [[31, "xlabel_kws-and-ylabel_kws:-Modifying-xlabel-and-ylabel"]], "Control gap & pad in heatmap": [[31, "Control-gap-&-pad-in-heatmap"]], "hgap and wgap for HeatmapAnnotation": [[31, "hgap-and-wgap-for-HeatmapAnnotation"]], "subplot_gap": [[31, "subplot_gap"]], "row_split_gap and col_split_gap": [[31, "row_split_gap-and-col_split_gap"]], "How to remove arrow in anno_label": [[31, "How-to-remove-arrow-in-anno_label"]], "oncoPrint: visualizing TCGA Lung Adenocarcinoma Carcinoma Variants Dataset": [[32, "oncoPrint:-visualizing-TCGA-Lung-Adenocarcinoma-Carcinoma-Variants-Dataset"]], "Visualizing categorical variables using oncoPrint": [[33, "Visualizing-categorical-variables-using-oncoPrint"]], "Visualizing Single Cell DNA Methylation Data (Loyfer2023)": [[34, "Visualizing-Single-Cell-DNA-Methylation-Data-(Loyfer2023)"]], "oncoPrint": [[36, "oncoprint"]], "setup module": [[37, "setup-module"]]}, "indexentries": {"pycomplexheatmap": [[0, "module-PyComplexHeatmap"]], "module": [[0, "module-PyComplexHeatmap"], [1, "module-PyComplexHeatmap.annotations"], [2, "module-PyComplexHeatmap.clustermap"], [3, "module-PyComplexHeatmap.colors"], [4, "module-PyComplexHeatmap.dotHeatmap"], [5, "module-PyComplexHeatmap.example"], [6, "module-PyComplexHeatmap.oncoPrint"], [8, "module-PyComplexHeatmap.utils"]], "annotationbase (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.AnnotationBase"]], "heatmapannotation (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation"]], "pycomplexheatmap.annotations": [[1, "module-PyComplexHeatmap.annotations"]], "anno_barplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_barplot"]], "anno_boxplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_boxplot"]], "anno_img (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_img"]], "anno_label (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_label"]], "anno_lineplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_lineplot"]], "anno_scatterplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_scatterplot"]], "anno_simple (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_simple"]], "collect_legends() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.collect_legends"]], "get_label_width() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.get_label_width"]], "get_max_label_width() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.get_max_label_width"]], "get_ticklabel_width() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.get_ticklabel_width"]], "get_ticklabel_width() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.get_ticklabel_width"]], "plot() (pycomplexheatmap.annotations.anno_barplot method)": [[1, "PyComplexHeatmap.annotations.anno_barplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_boxplot method)": [[1, "PyComplexHeatmap.annotations.anno_boxplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_img method)": [[1, "PyComplexHeatmap.annotations.anno_img.plot"]], "plot() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.plot"]], "plot() (pycomplexheatmap.annotations.anno_lineplot method)": [[1, "PyComplexHeatmap.annotations.anno_lineplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_scatterplot method)": [[1, "PyComplexHeatmap.annotations.anno_scatterplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_simple method)": [[1, "PyComplexHeatmap.annotations.anno_simple.plot"]], "plot_annotations() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.plot_annotations"]], "plot_legends() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.plot_legends"]], "reorder() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.reorder"]], "set_axes_kws() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.set_axes_kws"]], "set_axes_kws() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.set_axes_kws"]], "set_label() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.set_label"]], "set_legend() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.set_legend"]], "set_plot_kws() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.set_plot_kws"]], "set_side() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.set_side"]], "show_ticklabels() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.show_ticklabels"]], "update_plot_kws() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.update_plot_kws"]], "clustermapplotter (class in pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter"]], "dendrogramplotter (class in pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter"]], "pycomplexheatmap.clustermap": [[2, "module-PyComplexHeatmap.clustermap"]], "cal_cold_between_groups() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.cal_cold_between_groups"]], "cal_rowd_between_groups() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.cal_rowd_between_groups"]], "calculate_col_dendrograms() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.calculate_col_dendrograms"]], "calculate_dendrogram() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.calculate_dendrogram"]], "calculate_row_dendrograms() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.calculate_row_dendrograms"]], "calculated_linkage (pycomplexheatmap.clustermap.dendrogramplotter property)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.calculated_linkage"]], "check_array() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.check_array"]], "collect_legends() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.collect_legends"]], "composite() (in module pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.composite"]], "format_data() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.format_data"]], "get_coords() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.get_coords"]], "heatmap() (in module pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.heatmap"]], "heatmapplotter (class in pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.heatmapPlotter"]], "plot() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot"]], "plot() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.plot"]], "plot() (pycomplexheatmap.clustermap.heatmapplotter method)": [[2, "PyComplexHeatmap.clustermap.heatmapPlotter.plot"]], "plot_dendrograms() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot_dendrograms"]], "plot_heatmap() (in module pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.plot_heatmap"]], "plot_legends() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot_legends"]], "plot_matrix() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot_matrix"]], "post_processing() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.post_processing"]], "reordered_ind (pycomplexheatmap.clustermap.dendrogramplotter property)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.reordered_ind"]], "set_axes_labels_kws() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_axes_labels_kws"]], "set_height() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_height"]], "set_width() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_width"]], "set_xy_labels() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_xy_labels"]], "standard_scale() (pycomplexheatmap.clustermap.clustermapplotter static method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.standard_scale"]], "tight_layout() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.tight_layout"]], "z_score() (pycomplexheatmap.clustermap.clustermapplotter static method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.z_score"]], "pycomplexheatmap.colors": [[3, "module-PyComplexHeatmap.colors"]], "define_cmap() (in module pycomplexheatmap.colors)": [[3, "PyComplexHeatmap.colors.define_cmap"]], "register_cmap() (in module pycomplexheatmap.colors)": [[3, "PyComplexHeatmap.colors.register_cmap"]], "dotclustermapplotter (class in pycomplexheatmap.dotheatmap)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter"]], "pycomplexheatmap.dotheatmap": [[4, "module-PyComplexHeatmap.dotHeatmap"]], "collect_legends() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.collect_legends"]], "dotheatmap2d() (in module pycomplexheatmap.dotheatmap)": [[4, "PyComplexHeatmap.dotHeatmap.dotHeatmap2d"]], "format_data() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.format_data"]], "plot_matrix() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.plot_matrix"]], "post_processing() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.post_processing"]], "scale() (in module pycomplexheatmap.dotheatmap)": [[4, "PyComplexHeatmap.dotHeatmap.scale"]], "pycomplexheatmap.example": [[5, "module-PyComplexHeatmap.example"]], "clustermap_example0() (in module pycomplexheatmap.example)": [[5, "PyComplexHeatmap.example.clustermap_example0"]], "clustermap_example1() (in module pycomplexheatmap.example)": [[5, "PyComplexHeatmap.example.clustermap_example1"]], "get_kycg_example_data() (in module pycomplexheatmap.example)": [[5, "PyComplexHeatmap.example.get_kycg_example_data"]], "pycomplexheatmap.oncoprint": [[6, "module-PyComplexHeatmap.oncoPrint"]], "add_default_annotations() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.add_default_annotations"]], "collect_legends() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.collect_legends"]], "format_data() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.format_data"]], "get_samples_order() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.get_samples_order"]], "oncoprintplotter (class in pycomplexheatmap.oncoprint)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter"]], "oncoprint() (in module pycomplexheatmap.oncoprint)": [[6, "PyComplexHeatmap.oncoPrint.oncoprint"]], "plot_matrix() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.plot_matrix"]], "post_processing() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.post_processing"]], "pycomplexheatmap.utils": [[8, "module-PyComplexHeatmap.utils"]], "axis_ticklabels_overlap() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.axis_ticklabels_overlap"]], "cal_legend_width() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.cal_legend_width"]], "cluster_labels() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.cluster_labels"]], "define_cmap() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.define_cmap"]], "despine() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.despine"]], "get_colormap() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.get_colormap"]], "plot_cmap_legend() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_cmap_legend"]], "plot_color_dict_legend() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_color_dict_legend"]], "plot_legend_list() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_legend_list"]], "plot_marker_legend() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_marker_legend"]], "set_default_style() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.set_default_style"]], "to_utf8() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.to_utf8"]]}})
\ No newline at end of file
+Search.setIndex({"docnames": ["PyComplexHeatmap", "PyComplexHeatmap.annotations", "PyComplexHeatmap.clustermap", "PyComplexHeatmap.colors", "PyComplexHeatmap.dotHeatmap", "PyComplexHeatmap.example", "PyComplexHeatmap.oncoPrint", "PyComplexHeatmap.tools", "PyComplexHeatmap.utils", "advanced_usage", "citation", "clustermap", "dev", "dotHeatmap", "gallery", "get_started", "index", "installation", "kwargs", "modules", "more_examples", "notebooks/advanced_usage", "notebooks/bicluster", "notebooks/clustermap", "notebooks/composite_heatmaps", "notebooks/cpg_modules", "notebooks/dev", "notebooks/differential_expression_genes", "notebooks/dotHeatmap", "notebooks/gene_enrichment_analysis", "notebooks/get_started", "notebooks/kwargs", "notebooks/oncoPrint", "notebooks/oncoPrint2", "notebooks/single_cell_methylation", "notebooks/test_add_anno_img", "oncoPrint", "setup"], "filenames": ["PyComplexHeatmap.rst", "PyComplexHeatmap.annotations.rst", "PyComplexHeatmap.clustermap.rst", "PyComplexHeatmap.colors.rst", "PyComplexHeatmap.dotHeatmap.rst", "PyComplexHeatmap.example.rst", "PyComplexHeatmap.oncoPrint.rst", "PyComplexHeatmap.tools.rst", "PyComplexHeatmap.utils.rst", "advanced_usage.rst", "citation.rst", "clustermap.rst", "dev.rst", "dotHeatmap.rst", "gallery.md", "get_started.rst", "index.rst", "installation.rst", "kwargs.rst", "modules.rst", "more_examples.rst", "notebooks/advanced_usage.ipynb", "notebooks/bicluster.ipynb", "notebooks/clustermap.ipynb", "notebooks/composite_heatmaps.ipynb", "notebooks/cpg_modules.ipynb", "notebooks/dev.ipynb", "notebooks/differential_expression_genes.ipynb", "notebooks/dotHeatmap.ipynb", "notebooks/gene_enrichment_analysis.ipynb", "notebooks/get_started.ipynb", "notebooks/kwargs.ipynb", "notebooks/oncoPrint.ipynb", "notebooks/oncoPrint2.ipynb", "notebooks/single_cell_methylation.ipynb", "notebooks/test_add_anno_img.ipynb", "oncoPrint.rst", "setup.rst"], "titles": ["PyComplexHeatmap package", "PyComplexHeatmap.annotations module", "PyComplexHeatmap.clustermap module", "PyComplexHeatmap.colors module", "PyComplexHeatmap.dotHeatmap module", "PyComplexHeatmap.example module", "PyComplexHeatmap.oncoPrint module", "PyComplexHeatmap.tools module", "PyComplexHeatmap.utils module", "Advanced Usage", "Citation", "Clustermap", "For Developers", "dotHeatmap", "Gallery", "Get Started", "PyComplexHeatmap", "Installation", "Kwargs", "PyComplexHeatmap", "More Examples", "Generate dataset", "Import packages", "A quick example", "Composite two heatmaps horizontally for mouse DNA methylation array dataset", "Plot correlation matrix for CpG modules using DotClustermap", "1. Understand the layout:", "Visualizing Differential Expression Genes", "Load an example brain networks dataset from seaborn", "Visualizing gene set enrichment analysis results (clusterProfiler) using dotHeatmap", "1. Import packages", "label_kws, xticklabels_kws and yticklabels_kws: Modifying the heatmap annotations labels and xticklabels, yticklabels", "oncoPrint: visualizing TCGA Lung Adenocarcinoma Carcinoma Variants Dataset", "Visualizing categorical variables using oncoPrint", "Visualizing Single Cell DNA Methylation Data (Loyfer2023)", "<no title>", "oncoPrint", "setup module"], "terms": {"annot": [0, 2, 4, 6, 9, 11, 15, 16, 18, 19, 22, 24, 26, 28, 29, 32, 33, 34, 35], "annotationbas": [0, 1, 2, 19], "get_label_width": [0, 1], "get_max_label_width": [0, 1], "get_ticklabel_width": [0, 1], "reorder": [0, 1, 2, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "set_axes_kw": [0, 1], "set_label": [0, 1], "set_legend": [0, 1], "update_plot_kw": [0, 1], "heatmapannot": [0, 1, 2, 18, 19, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 32, 33, 34, 35], "collect_legend": [0, 1, 2, 4, 6], "plot_annot": [0, 1], "plot_legend": [0, 1, 2, 21, 23, 27, 34], "show_ticklabel": [0, 1, 21, 23, 30], "anno_barplot": [0, 1, 19, 21, 23, 25, 26, 30, 31, 32, 33], "plot": [0, 1, 2, 4, 6, 8, 9, 11, 13, 15, 16, 20, 22, 24, 27, 31, 32, 33, 34, 35], "anno_boxplot": [0, 1, 19, 21, 23, 26, 30, 31, 35], "anno_img": [0, 1, 19, 21], "anno_label": [0, 1, 16, 18, 19, 21, 22, 24, 25, 26, 27, 28, 29, 30, 32, 33, 34], "set_plot_kw": [0, 1], "set_sid": [0, 1], "anno_lineplot": [0, 1, 19, 21, 31], "anno_scatterplot": [0, 1, 19, 21, 23, 30, 31], "anno_simpl": [0, 1, 19, 21, 22, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "clustermap": [0, 1, 4, 6, 13, 16, 19, 20, 22, 23, 24], "clustermapplott": [0, 2, 4, 6, 19, 21, 22, 23, 24, 26, 27, 30, 31, 34, 35], "cal_cold_between_group": [0, 2], "cal_rowd_between_group": [0, 2], "calculate_col_dendrogram": [0, 2], "calculate_row_dendrogram": [0, 2], "format_data": [0, 2, 4, 6], "plot_dendrogram": [0, 2], "plot_matrix": [0, 2, 4, 6], "post_process": [0, 2, 4, 6], "set_axes_labels_kw": [0, 2], "set_height": [0, 2], "set_width": [0, 2], "set_xy_label": [0, 2], "standard_scal": [0, 2, 4, 6, 22], "tight_layout": [0, 2], "z_score": [0, 2, 4, 6, 21, 27], "dendrogramplott": [0, 2, 19, 22], "calculate_dendrogram": [0, 2], "calculated_linkag": [0, 2], "check_arrai": [0, 2], "get_coord": [0, 2], "reordered_ind": [0, 2], "composit": [0, 2, 16, 19, 20], "heatmap": [0, 1, 2, 4, 6, 9, 13, 15, 16, 18, 19, 20, 23, 25, 27, 32], "heatmapplott": [0, 2, 19], "plot_heatmap": [0, 2, 19], "color": [0, 1, 2, 4, 6, 8, 13, 16, 19, 21, 22, 23, 24, 25, 26, 27, 29, 30, 31, 32, 33, 34, 35], "define_cmap": [0, 3, 8, 19], "register_cmap": [0, 3, 19, 25], "dotheatmap": [0, 16, 19, 28], "dotclustermapplott": [0, 4, 13, 16, 19, 25, 29], "dotheatmap2d": [0, 4, 19], "scale": [0, 2, 4, 19, 22], "exampl": [0, 1, 2, 8, 11, 13, 16, 19, 21, 24, 25, 26, 29, 30, 31, 35], "clustermap_example0": [0, 5, 19], "clustermap_example1": [0, 5, 19], "get_kycg_example_data": [0, 5, 19], "oncoprint": [0, 16, 19], "oncoprintplott": [0, 6, 19, 32, 33], "add_default_annot": [0, 6], "get_samples_ord": [0, 6], "util": [0, 19, 21], "axis_ticklabels_overlap": [0, 8, 19], "cal_legend_width": [0, 8, 19], "cluster_label": [0, 8, 19], "despin": [0, 8, 19, 32, 33], "get_colormap": [0, 8, 19, 21], "plot_cmap_legend": [0, 4, 6, 8, 19], "plot_color_dict_legend": [0, 4, 6, 8, 19], "plot_legend_list": [0, 8, 19], "plot_marker_legend": [0, 4, 8, 19], "set_default_styl": [0, 8, 19], "to_utf8": [0, 8, 19], "class": [1, 2, 4, 6, 22], "df": [1, 21, 22, 23, 24, 26, 28, 30, 31, 35], "none": [1, 2, 4, 6, 8, 21, 22, 23, 24, 26, 27, 30, 31], "cmap": [1, 2, 4, 6, 8, 9, 13, 16, 22, 23, 24, 25, 26, 27, 29, 30, 31, 34, 35], "auto": [1, 2, 21], "height": [1, 2, 8, 21, 22, 23, 25, 26, 27, 29, 30, 31, 32, 33], "legend": [1, 2, 4, 6, 8, 21, 22, 24, 25, 27, 28, 29, 30, 31, 32, 33, 34, 35], "legend_kw": [1, 2, 4, 6, 8, 21, 23, 26, 31, 34], "plot_kw": [1, 6, 33], "sourc": [1, 2, 3, 4, 5, 6, 8, 14], "base": [1, 2, 4, 6, 8, 28], "object": [1, 2, 8, 22, 25, 27, 34], "paramet": [1, 2, 4, 6, 8, 9, 13, 16, 23, 25, 31], "datafram": [1, 2, 4, 6, 21, 26, 27, 30, 31, 32, 33, 35], "panda": [1, 2, 4, 6, 17, 22, 27, 29, 33], "seri": [1, 2], "onli": [1, 4, 6, 9, 16, 31], "one": [1, 4, 8, 23, 29], "column": [1, 2, 4, 6, 15, 16, 21, 22, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "str": [1, 2, 4, 6, 8, 21, 23, 25, 26, 30, 31, 32, 33, 35], "colormap": [1, 2, 4, 6, 9, 16, 25, 28], "set1": [1, 4, 6, 8, 21, 23, 27, 28, 29, 30, 31, 35], "dark2": [1, 8, 21, 23, 25, 27, 28, 30, 35], "bwr": [1, 2], "red": [1, 2, 4, 21, 23, 24, 25, 26, 27, 28, 30, 31, 32, 33], "jet": [1, 2, 21, 23, 25, 28, 30], "hsv": [1, 8], "rainbow": [1, 21, 26, 30, 31], "so": [1, 2], "pleas": [1, 2, 4, 10, 17, 30, 31], "see": [1, 2, 4, 21, 31], "http": [1, 2, 4, 10, 17, 21, 31, 32, 34], "matplotlib": [1, 2, 4, 8, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "org": [1, 2, 4, 10], "3": [1, 4, 15, 16, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "5": [1, 2, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "0": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "tutori": 1, "html": [1, 2, 4, 31, 32], "more": [1, 2, 16, 21, 23, 31, 32], "inform": [1, 2, 31], "run": [1, 17], "pyplot": [1, 21, 25, 27], "all": [1, 2, 4, 8, 9, 16, 23, 29, 31], "availabel": 1, "default": [1, 2, 4, 6, 8, 21, 28, 29, 31], "i": [1, 2, 4, 6, 8, 16, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 33, 35], "would": [1, 2, 21, 23], "determin": [1, 2, 4, 21, 23, 28], "dtype": [1, 2, 21, 22, 25, 26, 27, 28, 34], "each": [1, 2, 6, 23, 29, 30, 33], "dict": [1, 2, 4, 6, 8, 21, 22, 23, 25, 26, 29, 31, 34, 35], "list": [1, 2, 6, 8, 21, 22, 23, 24, 29, 33], "boxplot": 1, "barplot": [1, 29], "If": [1, 2, 4, 6, 8, 10, 23], "kei": [1, 4, 6, 8, 21], "should": [1, 2, 4, 6, 23], "exactli": 1, "same": [1, 2, 4, 23, 32, 34], "iloc": [1, 21, 23, 24, 26, 30, 31, 32, 35], "uniqu": [1, 22, 25, 28, 32, 34], "valu": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 33, 35], "name": [1, 4, 6, 21, 22, 23, 25, 26, 27, 28, 30, 31], "hex": [1, 22], "length": [1, 2, 27], "thi": [1, 4, 6, 10, 21, 23, 29, 30], "equal": 1, "nuniqu": [1, 25, 28, 32], "string": [1, 4, 6, 8], "mean": [1, 2, 4, 21, 23, 25, 28], "share": 1, "float": [1, 2, 4, 23], "axi": [1, 2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "1": [1, 2, 4, 6, 8, 12, 15, 16, 17, 21, 22, 23, 24, 25, 27, 28, 29, 31, 32, 33, 34, 35], "width": [1, 2, 6, 8, 21, 22, 23, 24, 26, 28, 29, 31, 32, 33, 34, 35], "size": [1, 2, 4, 6, 13, 16, 26, 29], "bool": [1, 2, 4], "whether": [1, 2, 4, 8, 21, 23, 31], "when": [1, 2, 4, 21, 23], "ar": [1, 2, 4, 8, 23, 24, 29, 31, 33], "vmax": [1, 2, 4, 8, 21, 24, 28, 29], "vmin": [1, 2, 4, 8, 21, 24, 28, 29], "other": [1, 2, 4, 6, 8, 23, 31, 33], "kw": [1, 2, 8], "pass": [1, 2, 4, 6, 8, 30], "plt": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "titl": [1, 2, 8], "prop": 1, "fontsiz": [1, 2, 21, 22, 23, 25, 26, 27, 29, 31, 34], "labelcolor": [1, 2, 21, 23, 26, 27, 29, 30, 31, 35], "markscal": 1, "frameon": [1, 2, 21, 23, 26, 31], "framealpha": 1, "fancybox": 1, "shadow": [1, 2], "facecolor": [1, 2, 21, 22, 23, 26, 29, 31], "edgecolor": [1, 2, 21, 26, 31], "mode": 1, "argument": 1, "pleast": 1, "type": [1, 2, 4, 6, 8, 22, 24, 29, 34], "There": 1, "an": [1, 13, 16, 21, 23], "addit": [1, 23, 31], "color_text": [1, 8, 23], "true": [1, 2, 6, 8, 9, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "which": [1, 2, 8, 28, 29, 31], "set": [1, 2, 8, 13, 16, 21, 22, 23, 25, 26, 31, 35], "text": [1, 2, 8, 21, 23, 26], "marker": [1, 4, 6, 8, 13, 16, 21, 25], "fals": [1, 2, 4, 8, 9, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "black": [1, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "user": [1, 28], "want": [1, 17, 23, 27], "us": [1, 2, 4, 6, 8, 9, 10, 13, 16, 17, 20, 22, 23, 24, 26, 31, 32, 35, 36], "custom": [1, 2, 9, 16, 25], "instead": [1, 21, 25], "blue": [1, 21, 23, 26, 27, 28, 29, 30, 31, 32, 35], "rotat": [1, 2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34], "rotation_mod": [1, 2, 21, 23, 26, 31], "ha": [1, 21], "va": [1, 21], "annotation_clip": 1, "arrowprop": [1, 21, 29, 31], "For": [1, 2, 8, 16, 21, 29, 31], "also": [1, 2, 4, 6, 23], "chang": [1, 2, 8, 9, 13, 16, 23, 27], "rang": [1, 2, 8, 21, 23, 24, 25, 26, 30, 31, 33, 35], "try": [1, 2], "df_box": [1, 21, 23, 26, 30, 31, 35], "gene1": [1, 21], "return": [1, 2, 4, 6, 8], "idx": 1, "subplot_ax": 1, "label": [1, 2, 8, 9, 16, 18, 22, 23, 24, 25, 26, 27, 28, 29, 30, 32, 33, 34, 35], "label_sid": [1, 8, 21, 23, 25, 27, 28, 29, 31, 32], "label_kw": [1, 16, 18, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 32, 34], "ticklabels_kw": [1, 31], "legend_sid": [1, 2, 8], "right": [1, 2, 8, 9, 11, 16, 25, 26, 27, 28, 29, 30, 31, 32, 33, 35], "legend_gap": [1, 2, 21, 22, 23, 25, 26, 28, 29, 30, 31, 34, 35], "legend_width": [1, 2, 8, 22, 23], "4": [1, 4, 8, 15, 16, 21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "legend_hpad": [1, 2, 21, 22, 23, 24, 25, 26, 31, 32, 34], "2": [1, 2, 4, 8, 15, 16, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "legend_vpad": [1, 2, 8, 21, 23, 24, 25, 26, 31], "orient": [1, 9, 16, 27, 29, 32], "wgap": [1, 18, 21, 26], "hgap": [1, 18, 21, 23, 26, 30], "raster": [1, 2, 22, 23, 24, 26, 31, 34, 35], "verbos": [1, 2, 8, 21, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "arg": 1, "gener": [1, 2, 9, 16, 23, 26, 30, 31, 35], "self": [1, 4], "convert": [1, 28], "int": [1, 2, 4, 21, 26, 28, 30, 31, 34], "row": [1, 2, 4, 6, 9, 15, 16, 22, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "need": 1, "specifi": [1, 2, 23], "provid": [1, 2, 8, 16, 23, 28], "have": [1, 2, 8, 17, 21, 23, 29, 34], "can": [1, 2, 4, 6, 21, 23, 27, 29, 30, 31], "from": [1, 2, 4, 6, 8, 13, 16, 17, 21, 23, 24, 25, 26, 27, 29, 31, 32, 33, 34, 35], "__subclasses__": 1, "includ": [1, 2, 8, 21], "anno_scatt": 1, "must": [1, 4], "els": [1, 21, 22, 23, 25, 26, 27, 28, 30, 31, 32, 33], "calcul": [1, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "given": [1, 4, 8, 23, 28], "invalid": 1, "top": [1, 2, 8, 9, 11, 16, 22, 25, 26, 27, 29, 31, 32, 33], "bottom": [1, 2, 8, 11, 16, 21, 25, 26, 27, 29, 30, 31, 32, 33], "left": [1, 2, 8, 9, 11, 16, 25, 26, 27, 28, 29, 30, 31, 32, 33], "alpha": [1, 2, 4, 25, 26, 28, 29, 31], "fontstyl": [1, 2, 26, 31], "horizontalalign": [1, 2, 21, 23, 25, 26, 27, 28, 29, 30, 31, 32], "verticalalign": [1, 2, 21, 26, 27, 30, 31], "visibl": [1, 2, 21, 22, 24, 25, 26, 29, 31, 32, 34], "gca": [1, 23], "yaxi": [1, 26, 31], "properti": [1, 2, 26, 31], "ax": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 28, 29, 31, 32, 33], "direct": [1, 2, 23, 31], "pad": [1, 2, 16, 18, 21, 26], "labels": [1, 2, 21, 27, 29, 31, 32, 35], "zorder": [1, 2, 26, 31], "labelbottom": [1, 2], "labeltop": [1, 2], "labelleft": [1, 2], "labelright": [1, 2], "labelrot": [1, 2, 21, 23, 26, 27, 29, 30, 31, 35], "grid_color": [1, 2], "grid_linestyl": [1, 2], "tick_param": [1, 2, 21, 31], "function": [1, 4, 8, 23, 25, 29, 30], "et": [1, 2, 22, 23], "al": [1, 2, 23], "automoti": 1, "vertic": [1, 2, 21], "gap": [1, 2, 8, 16, 18, 21, 23], "between": [1, 2, 8, 9, 16, 23, 31], "two": [1, 2, 16, 20, 23, 25, 29, 31], "mm": [1, 2, 22, 23, 24, 28, 29, 31, 32, 34, 35], "horizon": [1, 2, 23], "space": [1, 2, 8, 9, 16, 23], "up": [1, 2, 13, 16, 27, 32], "down": [1, 9, 16, 27], "show": [1, 2, 4, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "automat": [1, 2, 4, 23], "assign": [1, 29, 32], "accord": 1, "posit": [1, 26, 28], "option": [1, 4, 6, 8], "wspace": 1, "control": [1, 4, 6, 16, 18, 21, 23, 28, 33], "hspace": 1, "horizont": [1, 2, 16, 20], "pair": [1, 8, 21, 35], "collect": [1, 21, 22, 23, 24, 27, 28, 29, 30, 32, 33, 34, 35], "rtype": 1, "subplot_spec": [1, 2, 6], "figur": [1, 4, 8, 21, 22, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "add_gridspec": 1, "gridspec": 1, "gridspecfromsubplotspec": 1, "index": [1, 2, 4, 21, 22, 23, 24, 25, 26, 27, 28, 30, 31, 32, 33, 34, 35], "param": [1, 2], "kwarg": [1, 2, 4, 6, 16], "grid": [1, 21, 26, 28, 29, 31, 32, 33], "showflier": 1, "medianlinecolor": 1, "border_width": [1, 21], "border_color": [1, 21], "255": [1, 21], "merg": [1, 8, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 33, 34], "merge_width": 1, "imag": [1, 9, 16], "border": 1, "line": [1, 21, 23, 28, 30, 31], "256": [1, 21], "ignor": [1, 8, 28], "white": [1, 2, 8, 21, 22, 23, 26, 27, 28, 31, 33], "cluster": [1, 2, 8, 9, 16], "onc": 1, "extend": [1, 22, 34], "frac": 1, "major": [1, 8, 22, 29, 31], "adjust_color": [1, 34], "lumin": [1, 29, 34], "8": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 35], "relpo": [1, 21, 22, 26, 27, 29, 30, 31, 32, 34], "add": [1, 2, 9, 13, 16, 25, 27, 31], "documentatin": 1, "websit": 1, "dingwb": [1, 2, 17, 31, 34], "github": [1, 2, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "io": [1, 2, 21, 31, 32], "build": [1, 2, 9, 16, 31], "notebook": [1, 2, 31], "single_cell_methyl": 1, "distribut": 1, "fraction": 1, "arma": 1, "armb": 1, "multipl": [1, 2, 4, 9, 16], "group": [1, 2, 9, 13, 16, 24, 29, 31, 34, 35], "largest": [1, 8], "too": [1, 2, 21, 23, 28], "high": [1, 24, 25, 28], "replac": [1, 21, 26, 28, 30, 31, 32], "origin": 1, "togeth": [1, 2], "tupl": [1, 2, 21, 22], "x": [1, 4, 6, 8, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 33], "y": [1, 4, 6, 8, 21, 22, 24, 25, 26, 28, 29, 31, 32, 33], "arrow": [1, 16, 18, 21], "start": [1, 16, 21, 22, 23, 24, 27, 28, 29, 30, 32, 33, 34, 35], "point": [1, 13, 16, 21, 22, 29, 31], "rel": 1, "side": [1, 15, 16, 25, 26, 29], "about": 1, "could": [1, 2, 4, 23], "found": 1, "patch": [1, 26, 31], "fancyarrowpatch": [1, 31], "lineplot": 1, "linewidth": [1, 2, 21, 23, 25, 26, 28, 29, 31, 32, 35], "scatterplot": 1, "scatter": [1, 4, 6, 21, 23, 26, 28, 29, 30, 31, 35], "add_text": [1, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 35], "text_kw": [1, 21, 22, 23, 24, 25, 26, 27, 28, 30, 31, 35], "simpl": [1, 13, 16, 23], "categor": [1, 4, 8, 16, 23, 36], "continu": [1, 22, 25, 32], "variabl": [1, 8, 16, 36], "data": [2, 4, 6, 10, 13, 16, 20, 21, 22, 23, 24, 26, 27, 30, 31, 32, 33, 35], "top_annot": [2, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 33, 34, 35], "bottom_annot": [2, 23], "left_annot": [2, 22, 23, 24, 27, 29, 34], "right_annot": [2, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 34], "row_clust": [2, 21, 22, 26, 27, 28, 29, 30, 31, 34, 35], "col_clust": [2, 9, 22, 26, 27, 28, 29, 30, 31, 34, 35], "row_cluster_method": [2, 24, 35], "averag": [2, 21, 23, 26, 31, 35], "row_cluster_metr": [2, 24, 35], "correl": [2, 16, 20, 28, 35], "col_cluster_method": [2, 24, 35], "col_cluster_metr": [2, 24, 35], "show_rownam": [2, 21, 22, 23, 24, 26, 27, 28, 29, 30, 31, 32, 33, 35], "show_colnam": [2, 21, 22, 23, 24, 26, 27, 28, 29, 30, 31, 32, 33, 35], "row_names_sid": [2, 21, 23, 29, 30, 31, 35], "col_names_sid": [2, 23, 29], "xticklabels_kw": [2, 6, 16, 18, 21, 23, 26, 27, 29, 30, 32, 35], "yticklabels_kw": [2, 6, 16, 18, 21, 26, 27], "row_dendrogram": [2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "col_dendrogram": [2, 21, 22, 24, 27, 31, 35], "row_dendrogram_s": [2, 21], "10": [2, 10, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "col_dendrogram_s": 2, "row_split": [2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "col_split": [2, 9, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "row_dendrogram_kw": 2, "col_dendrogram_kw": [2, 21], "tree_kw": [2, 21, 23, 25, 27, 28, 30, 31, 35], "row_split_ord": [2, 21, 22, 34], "col_split_ord": [2, 9, 22, 31, 32, 34], "row_split_gap": [2, 18, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 34, 35], "col_split_gap": [2, 18, 21, 22, 23, 25, 26, 27, 28, 29, 30, 32, 33, 34, 35], "mask": [2, 4, 6], "subplot_gap": [2, 18, 21, 26, 32], "legend_anchor": [2, 23, 24, 25], "7": [2, 21, 22, 23, 25, 26, 28, 29, 30, 31, 32, 34, 35], "xlabel": [2, 16, 18, 21, 22, 23, 26, 29, 34, 35], "ylabel": [2, 16, 18, 21, 22, 23, 26, 34, 35], "xlabel_kw": [2, 16, 18, 21, 22, 23, 26, 29, 34], "ylabel_kw": [2, 16, 18, 21, 22, 23, 26, 34], "xlabel_sid": [2, 22, 29], "ylabel_sid": 2, "xlabel_bbox_kw": [2, 21, 22, 26, 29, 31], "ylabel_bbox_kw": [2, 21, 23, 26, 31], "legend_delta_x": 2, "plotter": [2, 4, 6], "numpi": [2, 4, 6, 17, 21, 22, 27, 29, 30], "arrai": [2, 4, 6, 16, 20, 22, 25, 26, 34], "perform": [2, 29], "z": [2, 25], "score": 2, "either": [2, 23], "after": 2, "method": [2, 8, 21], "linkag": [2, 9, 16, 22], "singl": [2, 16, 20], "complet": 2, "weight": [2, 22], "centroid": 2, "median": [2, 21], "ward": [2, 24, 35], "scipi": [2, 17, 35], "hierarchi": 2, "doc": 2, "refer": [2, 32], "detail": 2, "pairwis": 2, "distanc": 2, "observ": 2, "n": [2, 21, 22], "dimension": 2, "euclidean": [2, 35], "minkowski": 2, "cityblock": 2, "seuclidean": 2, "cosin": 2, "ham": 2, "jaccard": [2, 24], "chebyshev": 2, "canberra": [2, 21], "braycurti": 2, "mahalanobi": 2, "kulsinski": 2, "14": [2, 21, 22, 23, 25, 26, 27, 28, 29, 31, 33, 34], "spatial": 2, "pdist": 2, "metric": [2, 21], "ticklabel": [2, 8, 9, 16, 31], "dendrogram": [2, 22, 31], "10mm": 2, "pd": [2, 4, 6, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "number": [2, 21, 29], "hierarch": 2, "split": [2, 15, 16, 25, 28, 29, 32], "subplot": [2, 21], "my_linkag": 2, "order": [2, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "cluster_between_group": [2, 9, 31], "wa": [2, 4, 16, 23, 24, 25, 28, 34], "clsuter": [2, 9], "advanced_usag": [2, 31], "col": [2, 9, 16, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "cell": [2, 9, 16, 20, 22, 23, 29, 31, 33], "miss": 2, "infinit": 2, "1mm": 2, "asfonts": 2, "numpoint": 2, "markerscal": 2, "markerfirst": 2, "title_fonts": 2, "labelspac": 2, "alatern": 2, "we": [2, 21, 23, 28, 30, 32], "outlin": 2, "cbar": [2, 8], "cm": [2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "isinst": 2, "colorbar": [2, 8], "set_color": [2, 25, 26, 29], "set_linewidth": [2, 25, 26, 29], "divid": 2, "ax_heatmap": [2, 23, 24, 25, 26, 28, 29], "anchor": [2, 8, 21, 23, 31], "differ": [2, 8, 13, 16, 21, 29, 30], "infer": [2, 4, 28, 29], "shown": [2, 4, 6], "xticklabel": [2, 16, 18, 21, 35], "yticklabel": [2, 16, 18, 21, 35], "fontfamili": [2, 26, 31], "fontnam": [2, 21, 26, 31], "fontproperti": [2, 26, 31], "fontweight": [2, 22, 24, 26, 27, 29, 31], "xaxi": [2, 26, 31], "print": [2, 21, 22, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "sam": 2, "clip_box": [2, 26, 31], "clip_on": [2, 26, 31], "fill": [2, 4, 21, 26, 31], "in_layout": [2, 26, 31], "linestyl": [2, 26, 28, 29, 31], "get_bbox_patch": [2, 26, 31], "5000": [2, 33], "speed": 2, "center": [2, 8, 21, 24, 26, 31], "robust": [2, 8], "annot_kw": [2, 21], "fmt": [2, 21, 26, 31, 35], "linecolor": [2, 21, 23, 26, 31, 35], "na_col": [2, 8], "cbar_kwss": 2, "document": 2, "small": [2, 34], "displai": [2, 9, 16], "avoid": [2, 21], "overlap": [2, 8, 21], "To": [2, 21], "forc": [2, 9, 16], "use_linkag": 2, "row_ord": [2, 4, 6, 21, 22, 27], "col_ord": [2, 4, 6, 21, 22, 27], "fig": [2, 8, 21], "static": 2, "data2d": 2, "max": [2, 4, 22, 28, 29], "min": [2, 22, 28, 29], "normal": [2, 21, 23, 26, 30, 31, 35], "across": 2, "standard": 2, "noraml": 2, "varianc": 2, "tight_param": 2, "standar": 2, "dendrogram_kw": 2, "gap_pixel": 2, "root_x": 2, "similar": [2, 21], "upon": 2, "indic": [2, 29], "matrix": [2, 4, 6, 16, 20, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "cmlist": [2, 24], "main": [2, 15, 16, 21, 24, 31, 34], "row_gap": [2, 6], "15": [2, 21, 22, 23, 24, 26, 27, 28, 30, 31, 32], "col_gap": [2, 24], "legend_i": 2, "width_ratio": 2, "height_ratio": 2, "assembl": 2, "align": [2, 6, 33], "influenc": [2, 24], "unit": [2, 8, 31], "estim": [2, 22, 23, 24, 28, 29, 32, 34, 35], "legend_ax": [2, 8, 24], "cbar_kw": 2, "cbar_ax": 2, "squar": [2, 13, 16, 26], "xlabel_pad": 2, "ylabel_pad": 2, "xticklabels_sid": 2, "yticklabels_sid": 2, "2g": 2, "maxim": [2, 4], "minim": [2, 4], "seaborn": [2, 13, 16], "cax": [2, 8], "draw": [2, 23], "format": 2, "anno_kw": 2, "c": [3, 4, 6, 8, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 35], "hue": [4, 6, 13, 16, 25, 29], "": [4, 6, 8, 13, 16, 23, 25, 29], "o": [4, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "color_legend_kw": [4, 6], "cmap_legend_kw": [4, 6], "dot_legend_kw": 4, "dot_legend_mark": [4, 28, 29], "aggfunc": 4, "value_na": [4, 28], "hue_na": 4, "na": 4, "s_na": 4, "c_na": [4, 28], "spine": [4, 8, 21, 25, 26, 28, 29, 32, 33], "max_": [4, 28, 29], "dotclustermap": [4, 6, 16, 20], "inherit": [4, 6], "dot": [4, 6, 13, 16, 20, 29], "The": [4, 6, 16, 23, 24, 25, 29], "overwrit": [4, 6], "go": [4, 29], "stabl": 4, "api": 4, "markers_api": 4, "avail": 4, "Such": 4, "v": [4, 28], "p": [4, 25, 28, 29], "h": [4, 21, 23, 26, 27, 30, 31, 35], "d": [4, 8, 10, 21, 23, 25, 26, 28, 30, 31, 35], "_": [4, 28], "tolist": [4, 21, 22, 23, 24, 25, 28, 30, 32, 34], "It": [4, 23, 29], "pivot_t": 4, "fillna": [4, 21, 25, 28, 32], "floator": 4, "np": [4, 21, 22, 23, 26, 27, 28, 29, 30, 31, 32, 35], "aggreg": 4, "them": 4, "call": [4, 8], "pivot": [4, 28], "coeffici": 4, "valid": 4, "input": [4, 6], "paramt": [4, 6], "aspect": [6, 21], "bgcolor": 6, "whitesmok": 6, "remove_empty_row": 6, "remove_empty_column": 6, "background": [6, 23], "lightgrai": [6, 24, 25], "nvar": 6, "element": 6, "wisa": 6, "cccccc": 6, "intern": 8, "boolean": 8, "ani": 8, "legend_list": 8, "xtick": 8, "adjac": [8, 23], "tick": [8, 21], "coordin": 8, "keep": 8, "A": [8, 10, 11, 16, 20, 21, 26, 30, 31], "b": [8, 21, 26, 30, 31, 34], "len": [8, 21, 22], "new_label": 8, "plot_data": 8, "some": [8, 21], "heurist": 8, "good": [8, 29], "remov": [8, 16, 18, 25, 30, 32, 33], "current": 8, "specif": 8, "turbo": [8, 21, 23, 24, 26, 27, 30, 31, 35], "drawn": 8, "accent": 8, "tab20": [8, 25], "exp1": [8, 21, 23, 27], "exp2": [8, 21], "meth1": [8, 21], "meth2": [8, 21, 35], "legn": 8, "y0": 8, "delta_x": 8, "cmap_width": 8, "lengend": 8, "handl": 8, "legend_typ": 8, "pixel": 8, "init": 8, "2mm": 8, "5mm": 8, "boundri": 8, "obj": 8, "repres": 8, "python": [8, 10, 16, 17], "e": [8, 21, 23, 26, 30, 31, 35], "unchang": 8, "byte": 8, "utf": 8, "decod": 8, "__str__": 8, "result": [8, 13, 16, 28], "represent": 8, "dataset": [9, 13, 16, 20, 23, 26, 29, 30, 31, 33, 34, 35, 36], "select": [9, 16, 23, 26, 27, 28, 30, 31], "orent": [9, 16], "extra": [9, 16], "loop": [9, 16], "within": [9, 16], "cluster_within_group": 9, "choos": [9, 16, 30], "pycomplexheatmap": [9, 10, 17, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "diverg": 9, "qualit": 9, "how": [9, 16, 18], "you": [10, 17, 27], "softwar": [10, 28], "your": [10, 29], "work": 10, "cite": 10, "follow": [10, 21, 23, 24, 26, 31, 35], "metadata": [10, 22], "ding": [10, 16], "w": 10, "goldberg": 10, "zhou": 10, "2023": 10, "packag": [10, 15, 16, 19, 28, 29, 32], "visual": [10, 13, 16, 20, 36], "multimod": 10, "genom": 10, "imeta": 10, "e115": 10, "doi": 10, "1002": 10, "imt2": 10, "115": [10, 29], "quick": [11, 16], "understand": [12, 16], "layout": [12, 16], "load": [13, 16, 23, 24, 29], "brain": [13, 16, 23, 24], "network": [13, 16], "tradit": [13, 16], "fix": [13, 16], "five": [13, 16], "dimens": [13, 16], "gene": [13, 16, 20, 21, 23, 25, 26, 30, 31, 32, 33, 35], "enrich": [13, 16, 28], "analysi": [13, 16, 28], "clusterprofil": [13, 16], "click": 14, "picutur": 14, "view": 14, "code": [14, 21, 23, 26, 31, 35], "import": [15, 16, 21, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "along": [15, 16], "complex": 16, "biolog": [16, 29], "instal": [16, 30], "depend": [16, 29], "get": 16, "tcga": [16, 36], "lung": [16, 34, 36], "adenocarcinoma": [16, 36], "carcinoma": [16, 36], "variant": [16, 36], "advanc": 16, "usag": 16, "differenti": [16, 20, 29], "express": [16, 20], "dna": [16, 20, 28], "methyl": [16, 20, 29], "loyfer2023": [16, 20], "mous": [16, 20, 22, 23], "cpg": [16, 20, 24, 34], "modul": [16, 19, 20], "modifi": [16, 18], "galleri": [16, 28], "develop": [16, 29], "setup": [16, 17, 19], "citat": 16, "wubin": 16, "pip": 17, "upgrad": 17, "development": 17, "version": 17, "directli": 17, "com": [17, 21, 31, 34], "git": 17, "previous": 17, "updat": 17, "uninstal": 17, "again": 17, "OR": 17, "clone": [17, 27], "cd": [17, 21, 23, 26, 30, 31, 35], "py": [17, 25, 28], "submodul": 19, "content": 19, "process": [20, 22, 29], "smaller": 20, "sy": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "inlin": [21, 22, 23, 24, 25, 26, 28, 30, 31, 32, 34, 35], "pylab": [21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 34, 35], "pickl": [21, 23, 24, 26, 28, 29, 31, 32, 34, 35], "rcparam": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "dpi": [21, 22, 23, 24, 26, 28, 29, 30, 31, 32, 34, 35], "100": [21, 22, 23, 26, 28, 30, 31, 32, 33, 34, 35], "savefig": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "300": [21, 22, 23, 24, 26, 28, 29, 30, 31, 32, 34, 35], "path": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "append": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "expandus": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "project": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "__version__": [21, 22, 24, 25, 26, 29, 30, 31, 34, 35], "6": [21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 33, 34, 35], "dev15": 21, "gd9f20b3": 21, "font": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "arial": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "famili": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "san": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "serif": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "pdf": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "fonttyp": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "42": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "random": [21, 23, 26, 30, 31, 33, 35], "groupa": [21, 26, 30, 31, 35], "groupb": [21, 26, 30, 31, 35], "ab": [21, 23, 26, 30, 31, 35], "g": [21, 23, 24, 26, 27, 30, 31, 35], "ef": [21, 23, 26, 30, 31, 35], "f": [21, 22, 23, 24, 26, 29, 30, 31, 33, 35], "sampl": [21, 22, 23, 24, 26, 28, 30, 31, 32, 33, 35], "shape": [21, 22, 23, 25, 26, 27, 30, 31, 35], "randn": [21, 23, 26, 30, 31, 35], "df_bar": [21, 23, 26, 30, 31, 35], "uniform": [21, 23, 26, 30, 31, 35], "tmb1": [21, 23, 26, 30, 31, 35], "tmb2": [21, 23, 26, 30, 31, 35], "df_scatter": [21, 23, 26, 30, 31, 35], "df_heatmap": [21, 23, 26, 30, 31, 35], "30": [21, 23, 25, 26, 28, 29, 30, 31, 35], "11": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 33, 35], "fea": [21, 23, 26, 30, 31, 35], "nan": [21, 23, 25, 26, 30, 31, 32, 35], "df_row": [21, 22, 23, 24, 26, 28, 29, 30, 31, 34], "appli": [21, 22, 23, 25, 26, 27, 28, 30, 31, 32], "lambda": [21, 22, 23, 25, 26, 27, 28, 30, 31, 32], "sample4": [21, 26, 30, 31], "to_fram": [21, 26, 28, 30, 31], "xy": [21, 26, 30, 31], "to_seri": [21, 25, 26, 30, 31], "row_ha": [21, 22, 23, 24, 25, 26, 27, 28, 30, 31], "round": [21, 23, 26, 30, 31, 32], "12": [21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 35], "bar": [21, 23, 30, 33], "05": [21, 23, 26, 28, 29, 30, 31, 32, 35], "col_ha": [21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 34, 35], "nanmin": 21, "nanmax": [21, 28], "figsiz": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "row_cmap": [21, 23, 25, 27, 28, 30, 35], "rdylbu_r": [21, 26, 30, 31, 35], "90": [21, 22, 27, 28, 30, 34, 35], "example0": [21, 30, 35], "bbox_inch": [21, 22, 23, 24, 25, 27, 28, 29, 30, 32, 34, 35], "tight": [21, 22, 23, 24, 25, 27, 28, 29, 30, 32, 34, 35], "259323481554901": 21, "3307664641494927": 21, "32": [21, 33], "s4": [21, 26, 31], "test": [21, 26, 31], "18": [21, 26, 28, 31], "bbox": [21, 26], "boxstyl": [21, 26, 31], "circl": [21, 28], "exp": [21, 23, 26, 30, 31, 35], "turn": [21, 23, 26, 31, 35], "off": [21, 23, 26, 31, 35], "log": [21, 23, 26, 31, 35], "head": [21, 24, 25, 27, 28, 30, 32, 34], "1g": [21, 26, 31, 35], "gold": [21, 23, 26, 31, 35], "45": [21, 23, 25, 26, 27, 28, 29, 31, 32, 33, 35], "sample1": [21, 30], "404085": 21, "sample2": [21, 30], "081411": 21, "sample3": [21, 30], "594949": 21, "392642": 21, "sample5": [21, 30], "150321": 21, "229885": 21, "210195": 21, "032791": 21, "051215": 21, "261465": 21, "sample6": 21, "836030": 21, "sample7": 21, "519891": 21, "sample8": 21, "742360": 21, "sample9": 21, "617504": 21, "sample10": 21, "606197": 21, "float64": [21, 28], "fea1": 21, "941453": 21, "140686": 21, "285091": 21, "118367": 21, "214946": 21, "967923": 21, "853248": 21, "fea2": 21, "691451": 21, "561863": 21, "045189": 21, "303610": 21, "324130": 21, "240718": 21, "fea3": 21, "378372": 21, "142337": 21, "405770": 21, "274505": 21, "171325": 21, "526213": 21, "119409": 21, "fea4": 21, "212045": 21, "515978": 21, "051057": 21, "124523": 21, "378586": 21, "320444": 21, "064873": 21, "fea5": 21, "078289": 21, "476870": 21, "360113": 21, "705353": 21, "079402": 21, "284584": 21, "341059": 21, "751814": 21, "163439": 21, "038006": 21, "120816": 21, "820074": 21, "259323": 21, "757637": 21, "451777": 21, "057090": 21, "817372": 21, "082246": 21, "293083": 21, "831747": 21, "489009": 21, "258670": 21, "39": [21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 34], "fea19": 21, "fea25": 21, "fea18": 21, "fea27": 21, "fea29": 21, "fea22": 21, "fea24": 21, "fea30": 21, "fea20": 21, "fea17": 21, "fea26": 21, "fea21": 21, "fea23": 21, "fea15": 21, "fea16": 21, "fea28": 21, "fea9": 21, "fea10": 21, "fea11": 21, "fea7": 21, "fea8": 21, "fea14": 21, "fea13": 21, "fea6": 21, "fea12": 21, "37": [21, 25, 26, 31], "aaaa1": [21, 23], "bbbbb2": [21, 23], "df_bar1": [21, 30], "t1": [21, 30], "df_bar2": [21, 31], "t2": 21, "df_bar3": 21, "t3": 21, "df_bar4": [21, 30], "t4": [21, 30], "372887": 21, "061936": 21, "134849": 21, "694566": 21, "359127": 21, "132367": 21, "392230": 21, "272427": 21, "714047": 21, "625537": 21, "gene2": 21, "gene3": 21, "gene4": 21, "257733": 21, "699439": 21, "844278": 21, "608444": 21, "752454": 21, "083405": 21, "809866": 21, "431847": 21, "663268": 21, "513365": 21, "772141": 21, "387252": 21, "636622": 21, "012600": 21, "966687": 21, "476726": 21, "826088": 21, "361708": 21, "754555": 21, "343367": 21, "220518": 21, "870597": 21, "876851": 21, "9": [21, 22, 23, 24, 25, 28, 29, 31, 32, 33], "876537": 21, "395013": 21, "573748": 21, "948007": 21, "973635": 21, "386078": 21, "499509": 21, "188200": 21, "545418": 21, "328468": 21, "047464": 21, "158959": 21, "161838": 21, "163763": 21, "243494": 21, "779427": 21, "204043": 21, "542872": 21, "130472": 21, "133916": 21, "091244": 21, "788314": 21, "348602": 21, "046968": 21, "088636": 21, "612396": 21, "928272": 21, "877048": 21, "401591": 21, "980643": 21, "201984": 21, "307024": 21, "833295": 21, "660850": 21, "235867": 21, "211823": 21, "919341": 21, "290198": 21, "165467": 21, "382529": 21, "881658": 21, "073468": 21, "342374": 21, "239178": 21, "468118": 21, "962650": 21, "532729": 21, "771372": 21, "835987": 21, "436057": 21, "167528": 21, "042679": 21, "093969": 21, "936279": 21, "354470": 21, "736818": 21, "348153": 21, "335390": 21, "823424": 21, "888954": 21, "168943": 21, "042376": 21, "223754": 21, "876278": 21, "314815": 21, "541221": 21, "407723": 21, "913978": 21, "019213": 21, "885269": 21, "457418": 21, "692399": 21, "369775": 21, "256769": 21, "091064": 21, "233948": 21, "702588": 21, "314119": 21, "758320": 21, "888650": 21, "260971": 21, "891667": 21, "691901": 21, "079907": 21, "903442": 21, "754403": 21, "010756": 21, "829727": 21, "209684": 21, "739576": 21, "615991": 21, "269359": 21, "153883": 21, "386240": 21, "874027": 21, "38": [21, 28, 31], "orang": [21, 23, 24, 29], "yellowgreen": [21, 23, 24, 29], "tmb_bar": [21, 23, 31], "bar1": [21, 30], "bar4": [21, 30], "bar2": 21, "bar3": 21, "fontdict": [21, 22, 24, 30], "tab10": [21, 23], "grai": [21, 23, 24, 28], "yellow": [21, 23], "13": [21, 22, 23, 24, 25, 27, 28], "here": [21, 32], "widh": 21, "40": [21, 22, 29], "20": [21, 26, 27, 28, 31], "incres": [21, 23], "ncol": [21, 23], "than": [21, 23], "support": [21, 23], "long": [21, 23, 28], "new": [21, 23], "41": [21, 29, 34], "green": [21, 23, 24, 25, 26, 27, 28, 29, 31, 33], "By": 21, "43": 21, "typic": 21, "creat": [21, 33], "annotatin": [21, 23], "df_col": [21, 22, 23, 24, 25, 28, 29, 34], "hypomethylated_sampl": [21, 22], "sample_group_color_dict": 21, "celltyp": [21, 22, 29], "ct_color_dict": 21, "ct_legend": 21, "m1": 21, "lgd": 21, "But": 21, "what": [21, 29], "mani": 21, "m2": 21, "m3": 21, "In": [21, 23, 28, 29, 31], "case": [21, 29], "col_ha_dict": 21, "sample_col": 21, "r": [21, 24, 29, 32], "jokergoo": [21, 32], "2021": 21, "03": [21, 22, 24, 29], "complexheatmap": [21, 32], "44": [21, 35], "g1": [21, 31, 35], "g2": [21, 31, 35], "g3": [21, 31, 35], "g4": [21, 31, 35], "g5": [21, 31, 35], "col_cmap": [21, 27, 31, 35], "featur": [21, 26, 31, 35], "46": 21, "loc": [21, 22, 24, 25, 28, 29, 32, 34], "47": 21, "annotaiton": [21, 31], "purpl": [21, 23, 25, 26, 28, 31, 35], "_ticklabels_kw": [21, 31], "labelpad": [21, 22, 23, 26, 29, 31, 34], "increac": [21, 22, 23, 31], "manual": [21, 22, 31], "chocol": [21, 22, 26, 31], "48": 21, "applymap": 21, "make": 21, "asterisk": 21, "locat": 21, "oper": 21, "unicod": 21, "explor": 21, "2217": 21, "courier": 21, "49": [21, 28], "fastclust": 21, "t": [21, 22, 23, 25, 27, 28, 32, 34], "nlinkag": 21, "17": [21, 25, 28], "07051523": 21, "22006417": 21, "19": [21, 23, 26, 28, 33, 35], "14461418": 21, "28842182": 21, "70552492": 21, "76359288": 21, "65679239": 21, "16": [21, 23, 24, 25, 27, 28], "21": [21, 28, 35], "91528308": 21, "22": [21, 28], "75945877": 21, "50": [21, 23, 28], "51": 21, "df_img": 21, "jpeg": 21, "52": 21, "img": 21, "diverging1": 21, "parula": [21, 22, 23, 24, 34, 35], "53": 21, "54": 21, "mpl": [21, 29], "gradient": 21, "linspac": 21, "vstack": 21, "def": 21, "plot_color_gradi": 21, "categori": [21, 28, 29], "cmap_list": 21, "adjust": [21, 29], "nrow": 21, "figh": 21, "35": [21, 29], "subplots_adjust": 21, "99": [21, 33], "set_titl": [21, 22, 24], "zip": [21, 26], "imshow": 21, "01": [21, 32], "transform": [21, 26], "transax": 21, "just": 21, "ones": 21, "set_axis_off": [21, 23], "save": 21, "later": [21, 25], "55": [21, 28], "sequenti": 21, "cmap50": 21, "56": [21, 29], "57": 21, "custom_cmap": 21, "58": 21, "59": 21, "big": [21, 23], "enough": 21, "hidden": 21, "60": 21, "matter": 21, "61": [21, 33], "home": 22, "wding2": 22, "pch": [22, 24, 25, 30, 34], "czip": 22, "palette_path": 22, "mouse_pfc": 22, "mpfc_color_palett": 22, "xlsx": 22, "cell_class_color": 22, "read_excel": 22, "sheet_nam": 22, "cellclass": [22, 29], "index_col": [22, 25, 27, 28, 32, 34, 35], "to_dict": [22, 25, 29], "multi": [22, 28], "grei": [22, 24, 29], "nonn": 22, "e06931": 22, "exc": 22, "5ec5a9": 22, "rg": 22, "6036cb": 22, "inh": 22, "e93400": 22, "delta": [22, 34], "read_csv": [22, 25, 27, 28, 32, 34, 35], "pseudo_cel": 22, "between_groups_dmr": 22, "methylpi": 22, "methylpy_rms_results_collaps": 22, "tsv": 22, "sep": [22, 25, 28, 32, 34], "lstrip": 22, "methylation_level_": 22, "startswith": 22, "renam": 22, "number_of_dm": 22, "inplac": [22, 25, 28, 32, 34], "drop": 22, "hypermethylated_sampl": 22, "sname": 22, "insert": 22, "isna": 22, "reset_index": [22, 25, 28, 32], "set_index": [22, 25, 28, 29], "read": [22, 24, 34], "major_typ": 22, "beta": [22, 24, 25, 34], "txt": [22, 28, 32], "common_row": 22, "5kb_mc_hic_clust": 22, "l2": 22, "major_type_to_cell_class": 22, "common_col": 22, "cell_type_col": 22, "258365": 22, "23": [22, 26, 28, 29], "chr": [22, 24], "end": 22, "chr14": 22, "25667890": 22, "25667923": 22, "649934": 22, "chr5": [22, 24], "70893110": 22, "70893122": 22, "333501": 22, "chr3": 22, "141598191": 22, "141598238": 22, "440745": 22, "chr1": [22, 34], "76086002": 22, "76086015": 22, "319716": 22, "chr9": 22, "16231701": 22, "16232342": 22, "444616": 22, "chr11": [22, 24], "75550115": 22, "75550656": 22, "547601": 22, "109751156": 22, "109751187": 22, "530203": 22, "68823498": 22, "68825391": 22, "29": 22, "451270": 22, "chr10": 22, "91354668": 22, "91354926": 22, "327058": 22, "74147749": 22, "74150162": 22, "31": [22, 26, 29], "541438": 22, "cellclass1": 22, "majortyp": 22, "ct": 22, "l6": 22, "cge": 22, "lamp5": 22, "IT": 22, "l23": 22, "pc": [22, 27], "mgc": 22, "odc": 22, "vip": 22, "mge": 22, "sst": 22, "opc": 22, "vlmc": 22, "l4": 22, "unknown": [22, 28], "l5": 22, "ec": 22, "pvalb": 22, "pia": 22, "asc": 22, "chrom": 22, "077450": 22, "233350": 22, "161600": 22, "733500": 22, "685500": 22, "628000": 22, "148000": 22, "468000": 22, "785000": 22, "662500": 22, "780000": 22, "092500": 22, "125000": 22, "849000": 22, "043500": 22, "839500": 22, "735500": 22, "822500": 22, "694000": 22, "630500": 22, "903500": 22, "899000": 22, "896500": 22, "637500": 22, "784500": 22, "323000": 22, "947500": 22, "743500": 22, "844000": 22, "961500": 22, "499000": 22, "926000": 22, "000000": [22, 23, 25, 27, 28, 35], "846500": 22, "954500": 22, "489500": 22, "883500": 22, "598000": 22, "884000": 22, "789500": 22, "491350": 22, "192500": 22, "623500": 22, "055500": 22, "332150": 22, "763500": 22, "464500": 22, "448000": 22, "200150": 22, "549500": 22, "377950": 22, "517000": 22, "583500": 22, "458500": 22, "419500": 22, "633500": 22, "775000": 22, "299000": 22, "082350": 22, "471000": 22, "797500": 22, "508500": 22, "773500": 22, "050000": 22, "610500": 22, "802000": 22, "869500": 22, "523500": 22, "572500": 22, "572000": 22, "887000": 22, "778000": 22, "396500": 22, "826500": 22, "697500": 22, "145450": 22, "562500": 22, "317500": 22, "807875": 22, "725875": 22, "318250": 22, "900750": 22, "807750": 22, "378837": 22, "209437": 22, "654000": 22, "526500": 22, "579750": 22, "715875": 22, "133525": 22, "605375": 22, "663125": 22, "666375": 22, "545250": 22, "307337": 22, "319063": 22, "839000": 22, "557333": 22, "631000": 22, "780667": 22, "171233": 22, "262050": 22, "886500": 22, "492667": 22, "159800": 22, "520667": 22, "726833": 22, "683833": 22, "698000": 22, "231500": 22, "830000": 22, "781667": 22, "631667": 22, "909000": 22, "222417": 22, "338917": 22, "687667": 22, "452667": 22, "457667": 22, "241333": 22, "154667": 22, "546333": 22, "629667": 22, "325667": 22, "058667": 22, "503333": 22, "321000": 22, "588000": 22, "709000": 22, "034133": 22, "859333": 22, "360333": 22, "684667": 22, "414333": 22, "081200": 22, "244667": 22, "753267": 22, "363077": 22, "607967": 22, "644540": 22, "257347": 22, "775067": 22, "890267": 22, "517487": 22, "184007": 22, "594700": 22, "720740": 22, "763000": 22, "750800": 22, "495783": 22, "471317": 22, "777000": 22, "901467": 22, "657633": 22, "362270": 22, "241253": 22, "784857": 22, "809286": 22, "508857": 22, "859286": 22, "650714": 22, "823286": 22, "763286": 22, "614143": 22, "339714": 22, "615143": 22, "859000": 22, "469000": 22, "742857": 22, "466571": 22, "851714": 22, "807286": 22, "798000": 22, "913429": 22, "423857": 22, "678143": 22, "859394": 22, "860788": 22, "766576": 22, "678030": 22, "443909": 22, "813273": 22, "867061": 22, "658548": 22, "311152": 22, "854182": 22, "765485": 22, "813455": 22, "846697": 22, "434000": 22, "893424": 22, "834667": 22, "833515": 22, "808030": 22, "302337": 22, "529615": 22, "cc": 22, "dendrogram_col": 22, "ivl": 22, "dendrogram_row": 22, "sort_valu": [22, 34], "color_dict": [22, 34], "copi": 22, "del": [22, 33], "bold": [22, 24, 27, 29], "avg": [22, 23], "n_cpg": 22, "6375": [22, 29], "exist": 22, "listdir": 22, "within_groups_dmr": 22, "join": [22, 28], "_heatmap": 22, "concat": 22, "groupbi": [22, 28, 32], "agg": 22, "to_csv": 22, "agg_beta": 22, "queri": 22, "pfc": 22, "bed": 22, "gz": 22, "outfil": 22, "3013471": 22, "3013579": 22, "3026186": 22, "3026310": 22, "3035862": 22, "3035927": 22, "3052666": 22, "3052823": 22, "3059277": 22, "3059436": 22, "chry": 22, "44290504": 22, "44290637": 22, "66427102": 22, "66427179": 22, "90367464": 22, "90367503": 22, "90732358": 22, "90732367": 22, "90792411": 22, "90792447": 22, "661968": 22, "mt": 22, "28963217": 22, "28963260": 22, "83350": 22, "923000": 22, "939000": 22, "507000": 22, "89900": 22, "833500": 22, "738500": 22, "847000": 22, "95450": 22, "966500": 22, "80500": 22, "769500": 22, "731000": 22, "752500": 22, "81300": 22, "760000": 22, "650500": 22, "772500": 22, "936500": 22, "chr4": 22, "32158998": 22, "32159443": 22, "78950": 22, "524500": 22, "791500": 22, "767750": 22, "759500": 22, "73275": 22, "791750": 22, "693250": 22, "79225": 22, "441325": 22, "69625": 22, "538750": 22, "667000": 22, "391250": 22, "46975": 22, "616500": 22, "479250": 22, "704750": 22, "549750": 22, "chr13": 22, "90011452": 22, "90011460": 22, "29100": 22, "222000": 22, "225000": 22, "228500": 22, "08115": 22, "476500": 22, "009400": 22, "329500": 22, "32350": 22, "231000": 22, "30700": 22, "390500": 22, "402500": 22, "782000": 22, "06915": 22, "314500": 22, "285500": 22, "812000": 22, "821500": 22, "126765779": 22, "126765801": 22, "81850": 22, "885000": 22, "903000": 22, "565000": 22, "87500": 22, "875000": 22, "894000": 22, "93750": 22, "906000": 22, "68550": 22, "721500": 22, "649000": 22, "783500": 22, "80050": 22, "793500": 22, "809500": 22, "764500": 22, "142126735": 22, "142126952": 22, "84475": 22, "959250": 22, "902000": 22, "706500": 22, "94350": 22, "781250": 22, "530250": 22, "97100": 22, "933250": 22, "82150": 22, "774000": 22, "728250": 22, "869750": 22, "87600": 22, "796500": 22, "863000": 22, "842750": 22, "870500": 22, "21947352": 22, "21947371": 22, "67700": 22, "261333": 22, "766667": 22, "953667": 22, "979333": 22, "93500": 22, "897333": 22, "613667": 22, "864333": 22, "89500": 22, "950667": 22, "75200": 22, "733333": 22, "855667": 22, "874333": 22, "77000": 22, "589333": 22, "506333": 22, "868333": 22, "781000": 22, "chr6": 22, "50328671": 22, "50328782": 22, "38975": 22, "316650": 22, "702250": 22, "820000": 22, "85425": 22, "458000": 22, "662250": 22, "608250": 22, "81025": 22, "889500": 22, "61000": 22, "592250": 22, "047500": 22, "632250": 22, "64975": 22, "605250": 22, "608500": 22, "305500": 22, "407325": 22, "chr12": [22, 24], "15047867": 22, "15047932": 22, "68650": 22, "750000": [22, 35], "825000": 22, "777500": 22, "266500": 22, "31450": 22, "874500": 22, "65700": 22, "867500": 22, "71350": 22, "591000": 22, "290500": 22, "740500": 22, "74400": 22, "723000": 22, "735000": 22, "755500": 22, "559500": 22, "chr8": [22, 24], "48317445": 22, "48317685": 22, "87640": 22, "938400": 22, "937200": 22, "596000": 22, "66820": 22, "844600": 22, "885600": 22, "746000": 22, "80600": 22, "880800": 22, "90840": 22, "804800": 22, "765800": 22, "848800": 22, "76380": 22, "824400": 22, "810400": 22, "916000": 22, "935600": 22, "chr2": 22, "93512302": 22, "93512369": 22, "31350": 22, "788000": 22, "581000": 22, "832500": 22, "604500": 22, "94450": 22, "937500": 22, "900500": 22, "890000": 22, "90750": 22, "956500": 22, "32550": 22, "232700": 22, "221000": 22, "799000": 22, "55550": 22, "614000": 22, "601000": 22, "435400": 22, "470000": 22, "color_palett": 22, "gc": 22, "majortype_color": 22, "silver": 22, "crimson": 22, "deepskyblu": 22, "darkseagreen": 22, "darkcyan": 22, "k": 22, "de9f00": 22, "67292f": 22, "e13f82": 22, "3f884e": 22, "78772b": 22, "4ac750": 22, "6bc22e": 22, "a8d617": 22, "314b1f": 22, "aab639": 22, "e179a2": 22, "764a71": 22, "63bc9e": 22, "cea675": 22, "a87331": 22, "9bb070": 22, "9fa600": 22, "5e461": 22, "8000": 22, "sort": [22, 34], "sum": [22, 32], "row_major_typ": 22, "cell_class_colors1": 22, "majortype_colors1": 22, "left_ha": [22, 29, 34], "right_ha": [22, 29, 34], "all_dmr_heatmap": 22, "set2": 23, "tomato": 23, "skyblu": [23, 24], "spectral_r": 23, "vertical": 23, "first": 23, "051388888888887": 23, "boarder": 23, "continnu": 23, "deep": 23, "otherwis": 23, "your_color": 23, "treat": 23, "sometim": 23, "without": 23, "do": 23, "give": 23, "anoth": [23, 25], "No": [23, 24, 28, 32], "open": [23, 24, 29], "mammal_arrai": 23, "pkl": 23, "rb": [23, 24, 29], "col_colors_dict": [23, 24, 34], "gsm4412025": 23, "gsm4412026": 23, "gsm4412027": 23, "gsm4412028": 23, "gsm4412029": 23, "gsm4412030": 23, "gsm4412031": 23, "gsm4412032": 23, "gsm4412033": 23, "gsm4412034": 23, "gsm4997945": 23, "gsm4997946": 23, "gsm4997947": 23, "gsm4997948": 23, "gsm4997949": 23, "gsm4997950": 23, "gsm4997951": 23, "gsm4997952": 23, "gsm4997953": 23, "gsm4997954": 23, "sheep": 23, "435033": 23, "432900": 23, "446626": 23, "449123": 23, "497180": 23, "515918": 23, "483706": 23, "504681": 23, "529076": 23, "446443": 23, "beluga": 23, "whale": 23, "687488": 23, "694207": 23, "706525": 23, "702734": 23, "687014": 23, "704003": 23, "705887": 23, "693806": 23, "719417": 23, "712677": 23, "560381": 23, "571552": 23, "610392": 23, "613619": 23, "675832": 23, "668502": 23, "624820": 23, "658377": 23, "702334": 23, "575034": 23, "hous": 23, "vaquita": 23, "693523": 23, "702525": 23, "716792": 23, "725095": 23, "711261": 23, "708651": 23, "717952": 23, "705486": 23, "720915": 23, "724171": 23, "581862": 23, "594443": 23, "628908": 23, "639457": 23, "680801": 23, "707493": 23, "662338": 23, "665142": 23, "751859": 23, "584952": 23, "larg": [23, 28], "fly": 23, "fox": 23, "286822": 23, "269406": 23, "296796": 23, "314719": 23, "305074": 23, "308419": 23, "268421": 23, "297236": 23, "295019": 23, "297973": 23, "281684": 23, "363926": 23, "367216": 23, "359392": 23, "289821": 23, "257351": 23, "234301": 23, "295840": 23, "249336": 23, "344848": 23, "greater": 23, "horsesho": 23, "bat": 23, "530606": 23, "517989": 23, "520719": 23, "525855": 23, "507583": 23, "511993": 23, "528107": 23, "521563": 23, "542159": 23, "509526": 23, "735151": 23, "714805": 23, "708868": 23, "738207": 23, "823604": 23, "828040": 23, "823382": 23, "796882": 23, "878783": 23, "693625": 23, "littl": 23, "brown": 23, "202525": 23, "215990": 23, "238977": 23, "241872": 23, "267750": 23, "236729": 23, "202384": 23, "240519": 23, "251148": 23, "267257": 23, "883": 23, "predictedtaxid": 23, "predictedspeci": 23, "common_nam": 23, "9940": 23, "ovis_aries_rambouillet": 23, "bovida": 23, "9749": 23, "delphinapterus_leuca": 23, "monodontida": 23, "10090": 23, "mus_musculu": 23, "murida": 23, "42100": 23, "phocoena_sinu": 23, "phocoenida": 23, "132908": 23, "pteropus_vampyru": 23, "pteropodida": 23, "59479": 23, "rhinolophus_ferrumequinum": 23, "rhinolophida": 23, "59463": 23, "myotis_lucifugu": 23, "vespertilionida": 23, "gse": 23, "basenam": 23, "ncbi_scientific_nam": 23, "taxid": 23, "tissu": [23, 24], "sex": [23, 24], "speci": 23, "successr": 23, "gse147003": 23, "ovi": 23, "ari": 23, "blood": [23, 34], "femal": [23, 24], "artiodactyla": 23, "765818": 23, "797669": 23, "759256": 23, "male": [23, 24], "749813": 23, "770433": 23, "gse164127": 23, "eptesicu": 23, "fuscu": 23, "29078": 23, "skin": 23, "chiroptera": 23, "660425": 23, "652822": 23, "664746": 23, "650848": 23, "657170": 23, "a40043": [23, 34], "00e5ff": 23, "00becc": 23, "cerebellum": 23, "b2f8ff": 23, "striatum": 23, "6cbf00": [23, 34], "liver": [23, 24, 34], "ffccee": [23, 34], "muscl": [23, 29, 34], "1e9351": 23, "put": 23, "auc": 23, "legend_pad": 23, "veri": 23, "abov": 23, "25": [23, 26, 28, 31, 32], "930555555555557": [23, 28], "last": 23, "second": 23, "worth": 23, "note": 23, "betwe": 23, "kind": [23, 29, 30], "predict": 23, "68888888888889": 23, "tranpos": 23, "columsn": 23, "obtain": 24, "pmid": [24, 34], "36617464": 24, "linearsegmentedcolormap": [24, 25], "80": [24, 29, 33, 35], "urllib": 24, "influence_of_snp_on_beta": 24, "close": 24, "snp": 24, "row_colors_dict": 24, "respect": 24, "2000": 24, "204875570030_r01c02": 24, "204875570030_r04c01": 24, "cg30848532_tc21": 24, "525089": 24, "419515": 24, "cg30147375_bc21": 24, "803776": 24, "585928": 24, "cg46239718_bc21": 24, "443958": 24, "517514": 24, "cg36100119_bc21": 24, "351977": 24, "528846": 24, "cg42738582_bc21": 24, "783958": 24, "724901": 24, "204875570030_r05c01": 24, "204875570030_r06c01": 24, "204875570035_r05c02": 24, "483276": 24, "460750": 24, "390317": 24, "510269": 24, "831463": 24, "550146": 24, "535909": 24, "450167": 24, "564107": 24, "524896": 24, "374422": 24, "551200": 24, "802178": 24, "848621": 24, "850481": 24, "target": 24, "extensionbas": 24, "probedesign": 24, "con": 24, "mapflag": 24, "ii": 24, "chr19": 24, "suboptim": 24, "hybrid": 24, "effect": 24, "artifici": 24, "low": [24, 25, 28], "meth": [24, 34], "cg": 24, "gg": 24, "ga": 24, "79760438": 24, "ca": 24, "ac": 24, "ag": 24, "109557651": 24, "gt": [24, 26], "117860829": 24, "5877949": 24, "aa": 24, "122031379": 24, "strain": 24, "molf_eij": 24, "frontal": 24, "lobe": 24, "cast_eij": 24, "darkorang": 24, "wheat": 24, "darkgrai": 24, "69": [24, 27, 34], "4986111111111": 24, "col_ha1": 24, "cm1": 24, "viridi": 24, "col_ha2": 24, "my_cmap": 24, "from_list": [24, 25], "cm2": 24, "dnam": 24, "beta_snp": 24, "df_corr": 25, "kycg_modules_correl": 25, "csv": [25, 27, 34, 35], "df_ann": 25, "kycg_modules_annot": 25, "kycg_modules_beta": 25, "cpg_std": 25, "std": [25, 28], "cpg_mean": 25, "astyp": [25, 28], "value_count": [25, 28], "28": [25, 26, 28], "33": 25, "count": [25, 27, 28, 29], "int64": 25, "isin": [25, 28], "hm": 25, "h3k4me1": 25, "h3k79me2": 25, "h3k79me3": 25, "h3f3a": 25, "h4k20me1": 25, "h2bk20ac": 25, "h3k23me2": 25, "h2afi": 25, "h2ak9ac": 25, "h2bk12ac": 25, "h2bk15ac": 25, "h3k9me1": 25, "h2bk120ub": 25, "h2azac": 25, "h3k79me1": 25, "h2a": 25, "h3_4ac": 25, "h3k27me1": 25, "h3k4me3b": 25, "h3k56ac": 25, "h3k9ac_h3k14ac": 25, "h3k9k14ac": 25, "cenpa": 25, "h1": 25, "h2afz": 25, "h2ak119": 25, "h3": 25, "h3ac": 25, "h3k27ac": 25, "h3k4me3": 25, "h3k9ac": 25, "h4": 25, "h4ac": 25, "h4k5ac": 25, "h4k5ac_h4k8ac_h4k12ac_h4k16ac": 25, "h4k8ac": 25, "histonelysineacetyl": 25, "histonelysinecrotonyl": 25, "h3k18cr": 25, "h4k16ac": 25, "h4k12ac": 25, "h3k27me3b": 25, "h2bub": 25, "h2ak5ac": 25, "h3k36me3b": 25, "h2bk120ac": 25, "h3k36ac": 25, "h4k91ac": 25, "h4k20me3": 25, "h2afy2": 25, "h3k36me2": 25, "h3k27me3": 25, "h2ak119ub": 25, "cg04735237": 25, "cg00643814": 25, "cg24865495": 25, "cg25376651": 25, "cg27485084": 25, "cg12609052": 25, "cg07354679": 25, "cg02592525": 25, "cg12747056": 25, "cg17718960": 25, "cg19987665": 25, "cg18406033": 25, "cg09678971": 25, "cg00461612": 25, "cg18689454": 25, "cg11923631": 25, "cg27251412": 25, "cg08089567": 25, "cg16717549": 25, "cg09510531": 25, "559916": 25, "686845": 25, "561415": 25, "699960": 25, "711086": 25, "563690": 25, "527953": 25, "621757": 25, "623319": 25, "048462": 25, "103904": 25, "002533": 25, "074093": 25, "114526": 25, "151763": 25, "140823": 25, "117356": 25, "100510": 25, "126291": 25, "538034": 25, "568960": 25, "687787": 25, "593650": 25, "676068": 25, "606604": 25, "589549": 25, "747987": 25, "185989": 25, "087952": 25, "242909": 25, "065101": 25, "054958": 25, "096098": 25, "089246": 25, "078234": 25, "025965": 25, "069120": 25, "614696": 25, "646452": 25, "786480": 25, "602911": 25, "519926": 25, "642344": 25, "642081": 25, "022499": 25, "108524": 25, "015151": 25, "028957": 25, "274560": 25, "302464": 25, "284823": 25, "265835": 25, "262931": 25, "268134": 25, "564983": 25, "727579": 25, "624186": 25, "416198": 25, "505130": 25, "596338": 25, "030346": 25, "122412": 25, "063630": 25, "007048": 25, "197339": 25, "226034": 25, "218230": 25, "180297": 25, "181867": 25, "204431": 25, "655515": 25, "599935": 25, "590981": 25, "627786": 25, "674095": 25, "020895": 25, "058742": 25, "078318": 25, "055019": 25, "139691": 25, "168351": 25, "160081": 25, "142216": 25, "114554": 25, "144024": 25, "512": 25, "chromhmm": [25, 28], "chromhmm_bioc": 25, "tfb": 25, "txwk": [25, 28], "hdgf": 25, "rbfox2": 25, "srebf1": 25, "pqbp1": 25, "qui": [25, 28], "ascl1": 25, "casz1": 25, "ebf1": 25, "fezf1": 25, "foxm1": 25, "hand2": 25, "hdac3": 25, "irf2b": 25, "crx": 25, "dnmt3b": 25, "otx2": 25, "rorb": 25, "zmynd11": 25, "znf711": 25, "macrod1": 25, "atf2": 25, "batf": 25, "etv6": 25, "fo": 25, "ikzf2": 25, "irf4": 25, "junb": 25, "maf": 25, "mafg": 25, "me": 25, "fry": 25, "pdx1": 25, "keep_cpg": 25, "map": [25, 28, 29], "stack": [25, 28, 32], "keep_hm": 25, "154": 25, "245870": 25, "768918": 25, "319166": 25, "635635": 25, "270459": 25, "835897": 25, "276320": 25, "618949": 25, "276748": 25, "700196": 25, "23716": 25, "all_cmap": 25, "binar": 25, "colgroup": [25, 28], "middl": [25, 28], "heatmap_ax": [25, 29], "j": 25, "set_vis": [25, 26, 29], "var": [25, 32], "folder": 25, "3q": 25, "hwjjddlj65b030m69cfbdj4h0000gq": 25, "ipykernel_61938": 25, "92826381": 25, "matplotlibdeprecationwarn": 25, "deprec": 25, "minor": [25, 28, 29], "releas": 25, "regist": 25, "dotclustermap2": 25, "dev4": 26, "g71c725a": 26, "d20240410": 26, "ax_top": 26, "ax_right": 26, "set_axis_on": 26, "set_linestyl": [26, 29], "agg_filt": 26, "anim": 26, "bbox_patch": 26, "lt": 26, "fancybboxpatch": 26, "0x7f854d935760": 26, "children": 26, "clip_path": 26, "600x800": 26, "font_manag": 26, "0x7f854d8d56a0": 26, "fontvari": 26, "gid": 26, "math_fontfamili": 26, "dejavusan": 26, "mouseov": 26, "parse_math": 26, "path_effect": 26, "picker": 26, "000000000000007": 26, "sketch_param": 26, "snap": 26, "stretch": 26, "tightbbox": 26, "125": 26, "112": [26, 28], "875": [26, 34], "blendedaffine2d": 26, "0x7f854d8d5070": 26, "transform_rotates_text": 26, "transformed_clip_path_and_affin": 26, "unitless_posit": 26, "url": 26, "usetex": 26, "window_ext": 26, "wrap": 26, "antialias": 26, "75": [26, 28, 34], "0x7f854dded490": 26, "capstyl": 26, "butt": 26, "data_transform": 26, "affine2d": 26, "0x7f854f0bbfd0": 26, "extent": 26, "761576317947967": 26, "6388888888888813": 26, "92": 26, "78935409572574": 26, "638888888888896": 26, "hatch": 26, "joinstyl": 26, "miter": 26, "solid": 26, "mutation_aspect": 26, "mutation_scal": 26, "444444444444443": 26, "patch_transform": 26, "identitytransform": 26, "0x7f854d868730": 26, "63888889": 26, "81": [26, 33], "38888889": 26, "79": 26, "uint8": 26, "vert": 26, "76157632": 26, "7893541": 26, "expr": 27, "test_expression_dataset": 27, "selected_row": 27, "adam7": 27, "hla": 27, "doa": 27, "ptgir": 27, "opcml": 27, "it1": 27, "mageb1": 27, "label_row": 27, "c8orf34": 27, "as1": 27, "smim28": 27, "linc01114": 27, "klk3": 27, "traj45": 27, "tpmtp1": 27, "hnrnpcp4": 27, "89": [27, 33], "nci": 27, "h660": 27, "vcap": 27, "du": 27, "145": 27, "mda": 27, "pca": 27, "2b": 27, "22rv1": 27, "lncap": 27, "fgc": 27, "sup": 27, "hd1": 27, "l": 27, "540": 27, "1236": 27, "km": 27, "h2": 27, "hdlm": 27, "616": 27, "428": 27, "611": 27, "TO": 27, "175": 27, "044425": 27, "303654": 27, "604782": 27, "123861": 27, "015847": 27, "848602": 27, "215875": 27, "251406": 27, "795724": 27, "092080": 27, "070939": 27, "705117": 27, "241584": 27, "766805": 27, "132841": 27, "531804": 27, "564560": 27, "832337": 27, "442301": 27, "472336": 27, "359856": 27, "911472": 27, "795981": 27, "391838": 27, "rank2": 27, "regul": [27, 29], "top20": 27, "upregul": 27, "goi": 27, "colors_dict": 27, "row_ha_left": 27, "row_ha_right": 27, "uncom": 27, "sn": 28, "subset": 28, "collaps": 28, "load_dataset": 28, "brain_network": 28, "header": 28, "used_network": 28, "used_column": 28, "get_level_valu": 28, "comput": 28, "form": 28, "corr_mat": 28, "corr": 28, "level": [28, 34], "level_0": 28, "level_1": 28, "lh": 28, "rh": 28, "881516": 28, "049793": 28, "026902": 28, "144335": 28, "141253": 28, "119250": 28, "261589": 28, "272701": 28, "370021": 28, "366065": 28, "325680": 28, "196770": 28, "144566": 28, "366818": 28, "388756": 28, "352529": 28, "363982": 28, "341524": 28, "350452": 28, "112697": 28, "036909": 28, "144277": 28, "189683": 28, "084633": 28, "324230": 28, "332886": 28, "374322": 28, "361036": 28, "274151": 28, "142392": 28, "070452": 28, "358625": 28, "402173": 28, "302286": 28, "339989": 28, "315931": 28, "343379": 28, "343464": 28, "470239": 28, "100802": 28, "438449": 28, "339667": 28, "089811": 28, "272394": 28, "036493": 28, "171179": 28, "043298": 28, "158039": 28, "005598": 28, "060007": 28, "079078": 28, "040060": 28, "027878": 28, "075781": 28, "130549": 28, "278569": 28, "127621": 28, "014404": 28, "051249": 28, "090130": 28, "170053": 28, "124278": 28, "112148": 28, "063705": 28, "172007": 28, "040629": 28, "079687": 28, "024864": 28, "092263": 28, "068858": 28, "521377": 28, "506652": 28, "320966": 28, "141884": 28, "608392": 28, "075986": 28, "095015": 28, "012966": 28, "082816": 28, "023340": 28, "058718": 28, "034181": 28, "033355": 28, "022982": 28, "025638": 28, "431619": 28, "418708": 28, "084634": 28, "132": [28, 29], "808800071564": 28, "22361111111111": 28, "wding": 28, "miniconda3": 28, "env": 28, "lib": 28, "python3": 28, "site": 28, "189": 28, "userwarn": 28, "via": 28, "norm": 28, "465277777777779": 28, "drop_dupl": 28, "rowgroup": 28, "kycg_result": 28, "rmsk1": 28, "ensregbuild": 28, "sampleid": [28, 32], "clark2018_argelaguet2019": 28, "dataset1": 28, "luo2022": 28, "dataset2": 28, "max_p": 28, "log10": [28, 29], "pval": 28, "id": [28, 29, 34], "cpgtype": 28, "vc": 28, "term": [28, 29], "p_max": 28, "p_min": 28, "odds_ratio": 28, "pvalu": 28, "enrichtyp": 28, "het": 28, "061": 28, "neg": 28, "020000e": 28, "07": [28, 29], "26": 28, "tx": 28, "029": 28, "580000e": 28, "65": 28, "tssflnk": 28, "056": 28, "370000e": 28, "06": [28, 29], "022": 28, "660000e": 28, "346": 28, "350": 28, "760000e": 28, "02": 28, "describ": 28, "64": 28, "356918": 28, "632073": 28, "119186": 28, "deplet": 28, "srprna": 28, "satellit": 28, "simple_repeat": 28, "low_complex": 28, "reprpcwk": 28, "tssbiv": 28, "reprpc": 28, "ltr": 28, "snrna": 28, "sine": 28, "rdylgn_r": 28, "coolwarm_r": 28, "dotheatmap1": 28, "180": 28, "09263168093761": 28, "20230227_kycg": 28, "516666666666666": 28, "glob": 29, "style": 29, "dev0": 29, "g8abf70a": 29, "d20240415": 29, "tradition": 29, "lot": [29, 31], "dotplot": 29, "had": 29, "idea": 29, "manuscript": 29, "alreadi": 29, "gotten": 29, "bp": 29, "molecular": 29, "mf": 29, "kegg": 29, "pathwai": 29, "enrichment_analysis_dotheatmap": 29, "data1": 29, "descript": 29, "generatio": 29, "enrichedcategori": 29, "colid": 29, "rowid": 29, "0003779": 29, "actin": 29, "bind": 29, "044949": 29, "453212e": 29, "186": 29, "group2": 29, "ct1": 29, "610265": 29, "0030695": 29, "gtpase": 29, "activ": 29, "043016": 29, "744023e": 29, "178": 29, "171081": 29, "0060589": 29, "nucleosid": 29, "triphosphatas": 29, "0005201": 29, "extracellular": 29, "structur": 29, "constitu": 29, "021266": 29, "810239e": 29, "88": 29, "34": 29, "235806": 29, "0050839": 29, "adhes": 29, "molecul": 29, "031899": 29, "612420e": 29, "792522": 29, "840": 29, "0050905": 29, "neuromuscular": 29, "010454": 29, "224335e": 29, "122": 29, "ct2": 29, "912100": 29, "1205": 29, "0030534": 29, "adult": 29, "behavior": 29, "009426": 29, "028861e": 29, "110": 29, "219765": 29, "1558": 29, "0035019": 29, "somat": 29, "stem": 29, "popul": 29, "mainten": 29, "004799": 29, "029991e": 29, "219683": 29, "1746": 29, "0019827": 29, "009854": 29, "467128e": 29, "04": 29, "833532": 29, "2999": 29, "0021953": 29, "central": 29, "nervou": 29, "system": 29, "neuron": [29, 34], "008997": 29, "138960e": 29, "105": 29, "039103": 29, "139": 29, "metal": 29, "ion": 29, "transmembran": 29, "transport": 29, "monoatom": 29, "channel": 29, "gate": 29, "calmodulin": 29, "receptor": 29, "protein": 29, "kinas": 29, "substrat": 29, "organ": 29, "extern": 29, "encapsul": 29, "axon": 29, "guidanc": 29, "locomotori": 29, "fat": 29, "amphetamin": 29, "addict": 29, "calcium": 29, "signal": 29, "cocain": 29, "cytokin": 29, "interact": 29, "neuroact": 29, "ligand": 29, "cytoskeleton": 29, "ecm": 29, "viral": 29, "cy": 29, "focal": 29, "lupu": 29, "erythematosu": 29, "atp": 29, "chromatin": 29, "remodel": 29, "histon": 29, "modif": 29, "group1": 29, "ct3": 29, "ct4": 29, "s_min": 29, "s_max": 29, "3074": 29, "0021954": 29, "004586": 29, "027429": 29, "561784": 29, "0045444": 29, "176471": 29, "000382": 29, "417494": 29, "orrd": 29, "1e": 29, "ast": 29, "ravel": 29, "both": 29, "dashdot": 29, "cell_class_dotheatmap": 29, "106": 29, "46754159267321": 29, "ratio": 29, "total": 29, "510843": 30, "531995": 30, "188667": 30, "180811": 30, "354718": 30, "whole": 30, "separ": 30, "wiki": 31, "ital": 31, "_label_kw": 31, "tree": 31, "roundtooth": 31, "book": 32, "cbioport": 32, "tcga_lung_adenocarcinoma_provisional_ras_raf_mek_jnk_signal": 32, "dropna": 32, "mut": 32, "amp": [32, 33], "homdel": 32, "4384": 32, "kra": 32, "hra": 32, "braf": 32, "raf1": 32, "map3k1": 32, "unique_vari": 32, "v1": 32, "strip": 32, "008000": 32, "mutat": 32, "frequenc": 32, "row_vc": 32, "col_vc": 32, "kras_sampl": 32, "df_col_split": 32, "op": [32, 33], "alter": 32, "27": [32, 33], "72361111111111": 32, "ad": 32, "row_var_freq": 32, "isvar": 32, "oncoprint2": 32, "430555555555557": 32, "toi": 33, "sample_": 33, "gene_": 33, "alts_lol": 33, "amp_valu": 33, "randint": 33, "del_valu": 33, "neut_valu": 33, "alts_df": 33, "neut": 33, "prepar": 33, "annot_1_df": 33, "annot1": 33, "annot_2_df": 33, "500": 33, "annot2": 33, "annot_3_df": 33, "patient": 33, "sample_1": 33, "gene_1": 33, "gene_2": 33, "gene_3": 33, "gene_4": 33, "gene_5": 33, "95": 33, "sample_10": 33, "gene_6": 33, "96": 33, "gene_7": 33, "97": 33, "gene_8": 33, "98": 33, "gene_9": 33, "gene_10": 33, "patient2": 33, "sample_2": 33, "patient5": 33, "sample_3": 33, "patient3": 33, "sample_4": 33, "patient4": 33, "sample_5": 33, "patient1": 33, "sample_6": 33, "sample_7": 33, "sample_8": 33, "sample_9": 33, "77": 33, "78": 33, "1114": 33, "2698": 33, "4718": 33, "4633": 33, "3444": 33, "1984": 33, "2088": 33, "2316": 33, "2939": 33, "1373": 33, "a3": [33, 35], "a1": [33, 35], "a2": [33, 35], "flatten": 33, "signatur": 34, "come": 34, "36599988": 34, "raw": 34, "githubusercont": 34, "adipocyt": 34, "bladder": 34, "epithelium": 34, "mem": 34, "granulocyt": 34, "colon": 34, "macrophag": 34, "monocyt": 34, "interstiti": 34, "alveolar": 34, "pancrea": 34, "duct": 34, "skelet": 34, "endocrin": 34, "bronchu": 34, "smooth": 34, "prostat": 34, "coronari": 34, "arteri": 34, "aorta": 34, "thyroid": 34, "110426397": 34, "120667": 34, "9102": 34, "8285": 34, "918667": 34, "750667": 34, "8705": 34, "896": 34, "916333": 34, "953333": 34, "9450": 34, "61225": 34, "5485": 34, "94175": 34, "952": 34, "417": 34, "150": 34, "229": 34, "294": 34, "214": 34, "938333": 34, "110426639": 34, "420000": 34, "9304": 34, "8920": 34, "955667": 34, "964667": 34, "9285": 34, "903": 34, "924667": 34, "975333": 34, "9720": 34, "94575": 34, "8990": 34, "96225": 34, "963": 34, "606": 34, "185": 34, "348": 34, "917": 34, "484": 34, "972333": 34, "110426659": 34, "425667": 34, "9328": 34, "7635": 34, "933000": 34, "942333": 34, "8945": 34, "931": 34, "963667": 34, "899333": 34, "8895": 34, "93875": 34, "8390": 34, "92225": 34, "559": 34, "129": 34, "292": 34, "395": 34, "989667": 34, "110427109": 34, "203667": 34, "9724": 34, "9610": 34, "968667": 34, "9240": 34, "939": 34, "985000": 34, "9620": 34, "73300": 34, "7570": 34, "96525": 34, "929": 34, "225": 34, "806": 34, "951": 34, "905": 34, "974333": 34, "110427116": 34, "217333": 34, "9478": 34, "0000": 34, "976000": 34, "970667": 34, "9730": 34, "913": 34, "975000": 34, "970333": 34, "9800": 34, "65125": 34, "7955": 34, "96150": 34, "000": 34, "605": 34, "857": 34, "868": 34, "981000": 34, "clusterid": 34, "ep": 34, "granul": 34, "mono": 34, "macro": 34, "nk": 34, "bone": 34, "osteob": 34, "breast": 34, "basal": 34, "dermal": 34, "fibro": 34, "epid": 34, "kerat": 34, "eryth": 34, "prog": 34, "fallopian": 34, "gallbladd": 34, "gastric": 34, "neck": 34, "heart": 34, "cardio": 34, "kidnei": 34, "hep": 34, "alveo": 34, "bron": 34, "oligodend": 34, "acinar": 34, "musc": 34, "1e93a": 34, "ff9c00": 34, "ff9f7f": 34, "ff7f00": 34, "ff2e8d": 34, "cc0043": 34, "e5e5e5": 34, "cc4407": 34, "cc843d": 34, "663d28": 34, "1e937c": 34, "40705f": 34, "009351": 34, "e7e4bf": 34, "cca300": 34, "002929": 34, "ff99aa": 34, "ff99ff": 34, "f6ff99": 34, "ba99ff": 34, "ccccff": 34, "9e542": 34, "2ca02c": 34, "df7f00": 34, "ffd866": 34, "ffcc32": 34, "7f4c33": 34, "cc9951": 34, "b2bfff": 34, "loyfer2023_heatmap": 34, "981944444444444": 34, "download": 35, "pycomplexheatmap_test_2": 35, "c1": 35, "c2": 35, "c3": 35, "c4": 35, "c5": 35, "c6": 35, "c7": 35, "c8": 35, "c9": 35, "c10": 35, "c35": 35, "c36": 35, "c37": 35, "c38": 35, "c39": 35, "c40": 35, "c41": 35, "c42": 35, "c43": 35, "c44": 35, "00000": 35, "00": 35, "250000": 35, "111111": 35, "444444": 35, "222222": 35, "388889": 35, "611111": 35, "277778": 35, "888889": 35, "944444": 35, "555556": 35, "500000": 35, "a4": 35, "625000": 35, "375000": 35, "a5": 35, "a6": 35, "600000": 35, "100000": 35, "300000": 35, "700000": 35, "a7": 35, "a8": 35, "60000": 35, "800000": 35, "400000": 35, "200000": 35, "a9": 35, "440000": 35, "72": 35, "460000": 35, "040000": 35, "080000": 35, "560000": 35, "020000": 35, "240000": 35, "a10": 35, "a11": 35, "357143": 35, "02381": 35, "023810": 35, "071429": 35, "571429": 35, "214286": 35, "452381": 35, "166667": 35, "380952": 35, "309524": 35, "a12": 35, "062500": 35, "031250": 35, "312500": 35, "218750": 35, "531250": 35, "156250": 35, "687500": 35, "343750": 35, "437500": 35, "a13": 35, "333333": 35, "666667": 35, "a14": 35, "a15": 35, "076923": 35, "615385": 35, "384615": 35, "538462": 35, "307692": 35, "153846": 35, "a16": 35, "a17": 35, "150000": 35, "900000": 35, "550000": 35, "350000": 35, "a18": 35, "833333": 35, "a19": 35, "rdpu": 35}, "objects": {"": [[0, 0, 0, "-", "PyComplexHeatmap"]], "PyComplexHeatmap": [[1, 0, 0, "-", "annotations"], [2, 0, 0, "-", "clustermap"], [3, 0, 0, "-", "colors"], [4, 0, 0, "-", "dotHeatmap"], [5, 0, 0, "-", "example"], [6, 0, 0, "-", "oncoPrint"], [8, 0, 0, "-", "utils"]], "PyComplexHeatmap.annotations": [[1, 1, 1, "", "AnnotationBase"], [1, 1, 1, "", "HeatmapAnnotation"], [1, 1, 1, "", "anno_barplot"], [1, 1, 1, "", "anno_boxplot"], [1, 1, 1, "", "anno_img"], [1, 1, 1, "", "anno_label"], [1, 1, 1, "", "anno_lineplot"], [1, 1, 1, "", "anno_scatterplot"], [1, 1, 1, "", "anno_simple"]], "PyComplexHeatmap.annotations.AnnotationBase": [[1, 2, 1, "", "get_label_width"], [1, 2, 1, "", "get_max_label_width"], [1, 2, 1, "", "get_ticklabel_width"], [1, 2, 1, "", "reorder"], [1, 2, 1, "", "set_axes_kws"], [1, 2, 1, "", "set_label"], [1, 2, 1, "", "set_legend"], [1, 2, 1, "", "update_plot_kws"]], "PyComplexHeatmap.annotations.HeatmapAnnotation": [[1, 2, 1, "", "collect_legends"], [1, 2, 1, "", "plot_annotations"], [1, 2, 1, "", "plot_legends"], [1, 2, 1, "", "set_axes_kws"], [1, 2, 1, "", "show_ticklabels"]], "PyComplexHeatmap.annotations.anno_barplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_boxplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_img": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_label": [[1, 2, 1, "", "get_ticklabel_width"], [1, 2, 1, "", "plot"], [1, 2, 1, "", "set_plot_kws"], [1, 2, 1, "", "set_side"]], "PyComplexHeatmap.annotations.anno_lineplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_scatterplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_simple": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.clustermap": [[2, 1, 1, "", "ClusterMapPlotter"], [2, 1, 1, "", "DendrogramPlotter"], [2, 4, 1, "", "composite"], [2, 4, 1, "", "heatmap"], [2, 1, 1, "", "heatmapPlotter"], [2, 4, 1, "", "plot_heatmap"]], "PyComplexHeatmap.clustermap.ClusterMapPlotter": [[2, 2, 1, "", "cal_cold_between_groups"], [2, 2, 1, "", "cal_rowd_between_groups"], [2, 2, 1, "", "calculate_col_dendrograms"], [2, 2, 1, "", "calculate_row_dendrograms"], [2, 2, 1, "", "collect_legends"], [2, 2, 1, "", "format_data"], [2, 2, 1, "", "plot"], [2, 2, 1, "", "plot_dendrograms"], [2, 2, 1, "", "plot_legends"], [2, 2, 1, "", "plot_matrix"], [2, 2, 1, "", "post_processing"], [2, 2, 1, "", "set_axes_labels_kws"], [2, 2, 1, "", "set_height"], [2, 2, 1, "", "set_width"], [2, 2, 1, "", "set_xy_labels"], [2, 2, 1, "", "standard_scale"], [2, 2, 1, "", "tight_layout"], [2, 2, 1, "", "z_score"]], "PyComplexHeatmap.clustermap.DendrogramPlotter": [[2, 2, 1, "", "calculate_dendrogram"], [2, 3, 1, "", "calculated_linkage"], [2, 2, 1, "", "check_array"], [2, 2, 1, "", "get_coords"], [2, 2, 1, "", "plot"], [2, 3, 1, "", "reordered_ind"]], "PyComplexHeatmap.clustermap.heatmapPlotter": [[2, 2, 1, "", "plot"]], "PyComplexHeatmap.colors": [[3, 4, 1, "", "define_cmap"], [3, 4, 1, "", "register_cmap"]], "PyComplexHeatmap.dotHeatmap": [[4, 1, 1, "", "DotClustermapPlotter"], [4, 4, 1, "", "dotHeatmap2d"], [4, 4, 1, "", "scale"]], "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter": [[4, 2, 1, "", "collect_legends"], [4, 2, 1, "", "format_data"], [4, 2, 1, "", "plot_matrix"], [4, 2, 1, "", "post_processing"]], "PyComplexHeatmap.example": [[5, 4, 1, "", "clustermap_example0"], [5, 4, 1, "", "clustermap_example1"], [5, 4, 1, "", "get_kycg_example_data"]], "PyComplexHeatmap.oncoPrint": [[6, 1, 1, "", "oncoPrintPlotter"], [6, 4, 1, "", "oncoprint"]], "PyComplexHeatmap.oncoPrint.oncoPrintPlotter": [[6, 2, 1, "", "add_default_annotations"], [6, 2, 1, "", "collect_legends"], [6, 2, 1, "", "format_data"], [6, 2, 1, "", "get_samples_order"], [6, 2, 1, "", "plot_matrix"], [6, 2, 1, "", "post_processing"]], "PyComplexHeatmap.utils": [[8, 4, 1, "", "axis_ticklabels_overlap"], [8, 4, 1, "", "cal_legend_width"], [8, 4, 1, "", "cluster_labels"], [8, 4, 1, "", "define_cmap"], [8, 4, 1, "", "despine"], [8, 4, 1, "", "get_colormap"], [8, 4, 1, "", "plot_cmap_legend"], [8, 4, 1, "", "plot_color_dict_legend"], [8, 4, 1, "", "plot_legend_list"], [8, 4, 1, "", "plot_marker_legend"], [8, 4, 1, "", "set_default_style"], [8, 4, 1, "", "to_utf8"]]}, "objtypes": {"0": "py:module", "1": "py:class", "2": "py:method", "3": "py:property", "4": "py:function"}, "objnames": {"0": ["py", "module", "Python module"], "1": ["py", "class", "Python class"], "2": ["py", "method", "Python method"], "3": ["py", "property", "Python property"], "4": ["py", "function", "Python function"]}, "titleterms": {"pycomplexheatmap": [0, 1, 2, 3, 4, 5, 6, 7, 8, 16, 19, 21], "packag": [0, 22, 30], "submodul": 0, "modul": [0, 1, 2, 3, 4, 5, 6, 7, 8, 25, 37], "content": [0, 9, 11, 12, 13, 15, 16, 18, 20, 36], "annot": [1, 21, 23, 30, 31], "clustermap": [2, 11, 25, 28], "color": [3, 28], "dotheatmap": [4, 13, 29], "exampl": [5, 20, 23, 28], "oncoprint": [6, 32, 33, 36], "tool": 7, "util": 8, "advanc": 9, "usag": 9, "citat": 10, "For": 12, "develop": 12, "galleri": 14, "get": 15, "start": 15, "credit": 16, "thank": 16, "instal": 17, "depend": 17, "how": [17, 21, 29, 31], "kwarg": 18, "more": 20, "gener": 21, "dataset": [21, 24, 28, 32], "add": [21, 23, 28], "select": [21, 29], "row": [21, 23, 30], "label": [21, 31], "top": [21, 23], "heatmap": [21, 24, 28, 30, 31], "cell": [21, 34], "onli": [21, 23], "plot": [21, 23, 25, 28, 29, 30], "chang": [21, 28], "orent": 21, "down": 21, "extra": 21, "space": 21, "left": [21, 23], "right": [21, 23], "orient": 21, "us": [21, 25, 28, 29, 33], "paramet": [21, 28], "multipl": 21, "loop": 21, "cluster": 21, "between": [21, 22], "group": [21, 22, 28], "within": [21, 22], "clsuter": 21, "col_split": 21, "col_clust": 21, "true": 21, "cluster_between_group": 21, "col_split_ord": 21, "fals": 21, "cluster_within_group": 21, "custom": 21, "linkag": 21, "imag": 21, "choos": 21, "build": 21, "colormap": 21, "diverg": 21, "qualit": 21, "cmap": [21, 28], "forc": 21, "displai": 21, "all": 21, "col": 21, "ticklabel": 21, "import": [22, 30], "dmr": 22, "A": [23, 25], "quick": 23, "column": [23, 30], "anno_label": [23, 31], "anno_simpl": 23, "To": 23, "quickli": 23, "you": 23, "just": 23, "need": [23, 29], "datafram": 23, "figur": 23, "legend": 23, "separ": 23, "bottom": 23, "composit": 24, "two": 24, "horizont": 24, "mous": 24, "dna": [24, 34], "methyl": [24, 34], "arrai": 24, "correl": 25, "matrix": 25, "cpg": 25, "dotclustermap": 25, "process": 25, "data": [25, 28, 29, 34], "dot": [25, 28], "smaller": 25, "1": [26, 30], "understand": 26, "layout": 26, "visual": [27, 28, 29, 32, 33, 34], "differenti": 27, "express": 27, "gene": [27, 29], "load": 28, "an": 28, "brain": 28, "network": 28, "from": 28, "seaborn": 28, "tradit": 28, "squar": 28, "marker": [28, 29], "": 28, "simpl": 28, "fix": 28, "size": 28, "point": 28, "hue": 28, "differ": 28, "up": 28, "five": 28, "dimens": 28, "dotclustermapplott": 28, "set": 29, "enrich": 29, "analysi": 29, "result": 29, "clusterprofil": 29, "why": 29, "do": 29, "we": 29, "better": 29, "wai": 29, "read": 29, "2": 30, "3": 30, "split": 30, "4": 30, "along": 30, "side": 30, "main": 30, "label_kw": 31, "xticklabels_kw": 31, "yticklabels_kw": 31, "modifi": 31, "xticklabel": 31, "yticklabel": 31, "xlabel_kw": 31, "ylabel_kw": 31, "xlabel": 31, "ylabel": 31, "control": 31, "gap": 31, "pad": 31, "hgap": 31, "wgap": 31, "heatmapannot": 31, "subplot_gap": 31, "row_split_gap": 31, "col_split_gap": 31, "remov": 31, "arrow": 31, "tcga": 32, "lung": 32, "adenocarcinoma": 32, "carcinoma": 32, "variant": 32, "categor": 33, "variabl": 33, "singl": 34, "loyfer2023": 34, "setup": 37}, "envversion": {"sphinx.domains.c": 3, "sphinx.domains.changeset": 1, "sphinx.domains.citation": 1, "sphinx.domains.cpp": 9, "sphinx.domains.index": 1, "sphinx.domains.javascript": 3, "sphinx.domains.math": 2, "sphinx.domains.python": 4, "sphinx.domains.rst": 2, "sphinx.domains.std": 2, "sphinx.ext.todo": 2, "sphinx.ext.viewcode": 1, "nbsphinx": 4, "sphinx": 58}, "alltitles": {"PyComplexHeatmap package": [[0, "pycomplexheatmap-package"]], "Submodules": [[0, "submodules"]], "Module contents": [[0, "module-PyComplexHeatmap"]], "PyComplexHeatmap.annotations module": [[1, "module-PyComplexHeatmap.annotations"]], "PyComplexHeatmap.clustermap module": [[2, "module-PyComplexHeatmap.clustermap"]], "PyComplexHeatmap.colors module": [[3, "module-PyComplexHeatmap.colors"]], "PyComplexHeatmap.dotHeatmap module": [[4, "module-PyComplexHeatmap.dotHeatmap"]], "PyComplexHeatmap.example module": [[5, "module-PyComplexHeatmap.example"]], "PyComplexHeatmap.oncoPrint module": [[6, "module-PyComplexHeatmap.oncoPrint"]], "PyComplexHeatmap.tools module": [[7, "pycomplexheatmap-tools-module"]], "PyComplexHeatmap.utils module": [[8, "module-PyComplexHeatmap.utils"]], "Advanced Usage": [[9, "advanced-usage"]], "Contents:": [[9, null], [11, null], [12, null], [13, null], [15, null], [16, null], [18, null], [20, null], [36, null]], "Citation": [[10, "citation"]], "Clustermap": [[11, "clustermap"]], "For Developers": [[12, "for-developers"]], "dotHeatmap": [[13, "dotheatmap"]], "Gallery": [[14, "gallery"]], "Get Started": [[15, "get-started"]], "PyComplexHeatmap": [[16, "pycomplexheatmap"], [19, "pycomplexheatmap"]], "Credits and Thanks": [[16, "credits-and-thanks"]], "Installation": [[17, "installation"]], "Dependencies": [[17, "dependencies"]], "How to install?": [[17, "how-to-install"]], "Kwargs": [[18, "kwargs"]], "More Examples": [[20, "more-examples"]], "Generate dataset": [[21, "Generate-dataset"]], "Add selected rows labels": [[21, "Add-selected-rows-labels"]], "Add annotations on the top of heatmap cells": [[21, "Add-annotations-on-the-top-of-heatmap-cells"]], "Only plot the annotations": [[21, "Only-plot-the-annotations"]], "Change orentation to down and add extra space": [[21, "Change-orentation-to-down-and-add-extra-space"]], "Change orentation to the left": [[21, "Change-orentation-to-the-left"]], "Change orentation to the right": [[21, "Change-orentation-to-the-right"]], "Changing orientation using parameter orientation": [[21, "Changing-orientation-using-parameter-orientation"]], "Add multiple heatmap annotations using for loop": [[21, "Add-multiple-heatmap-annotations-using-for-loop"]], "Cluster between groups and cluster within groups": [[21, "Cluster-between-groups-and-cluster-within-groups"]], "clsuter within groups: col_split=*, col_cluster=True": [[21, "clsuter-within-groups:-col_split=*,-col_cluster=True"]], "cluster_between_groups: col_split=*, col_split_order=\"cluster_between_groups\",col_cluster=False": [[21, "cluster_between_groups:-col_split=*,-col_split_order=%22cluster_between_groups%22,col_cluster=False"]], "cluster_within_groups && cluster_between_groups: col_split=*, col_split_order=\"cluster_between_groups\",col_cluster=True": [[21, "cluster_within_groups-&&-cluster_between_groups:-col_split=*,-col_split_order=%22cluster_between_groups%22,col_cluster=True"]], "Custom annotation": [[21, "Custom-annotation"]], "Custom linkage": [[21, "Custom-linkage"]], "Image annotation": [[21, "Image-annotation"]], "Choosing Build-in colormap in PyComplexHeatmap": [[21, "Choosing-Build-in-colormap-in-PyComplexHeatmap"]], "Diverging": [[21, "Diverging"]], "Qualitative": [[21, "Qualitative"]], "How to use the Build-in cmap?": [[21, "How-to-use-the-Build-in-cmap?"]], "How to force display all row/col ticklabels?": [[21, "How-to-force-display-all-row/col-ticklabels?"]], "Import packages": [[22, "Import-packages"]], "Between groups DMRs": [[22, "Between-groups-DMRs"]], "Within groups DMRs": [[22, "Within-groups-DMRs"]], "A quick example": [[23, "A-quick-example"]], "Plotting annotations": [[23, "Plotting-annotations"]], "Only plot the row/column annotation": [[23, "Only-plot-the-row/column-annotation"]], "anno_label:": [[23, "anno_label:"]], "anno_simple:": [[23, "anno_simple:"]], "To add a annotation quickly, you just need a dataframe": [[23, "To-add-a-annotation-quickly,-you-just-need-a-dataframe"]], "Plot the figure and legend separately": [[23, "Plot-the-figure-and-legend-separately"]], "Top, bottom, left ,right annotations": [[23, "Top,-bottom,-left-,right-annotations"]], "Composite two heatmaps horizontally for mouse DNA methylation array dataset": [[24, "Composite-two-heatmaps-horizontally-for-mouse-DNA-methylation-array-dataset"]], "Plot correlation matrix for CpG modules using DotClustermap": [[25, "Plot-correlation-matrix-for-CpG-modules-using-DotClustermap"]], "Processing the data": [[25, "Processing-the-data"]], "Plotting the Dot clustermap": [[25, "Plotting-the-Dot-clustermap"]], "A smaller dot clustermap": [[25, "A-smaller-dot-clustermap"]], "1. Understand the layout:": [[26, "1.-Understand-the-layout:"]], "Visualizing Differential Expression Genes": [[27, "Visualizing-Differential-Expression-Genes"]], "Load an example brain networks dataset from seaborn": [[28, "Load-an-example-brain-networks-dataset-from-seaborn"]], "Dot Heatmap": [[28, "Dot-Heatmap"]], "Plot traditional heatmap using square marker marker='s'": [[28, "Plot-traditional-heatmap-using-square-marker-marker='s'"]], "Simple dot heatmap using fixed dot size": [[28, "Simple-dot-heatmap-using-fixed-dot-size"]], "Changing the size of point": [[28, "Changing-the-size-of-point"]], "Add parameter hue and use different colors for different groups": [[28, "Add-parameter-hue-and-use-different-colors-for-different-groups"]], "Add parameter hue and use different cmap and marker for different groups": [[28, "Add-parameter-hue-and-use-different-cmap-and-marker-for-different-groups"]], "Dot Clustermap": [[28, "Dot-Clustermap"]], "Plot clustermap using seaborn brain networks dataset": [[28, "Plot-clustermap-using-seaborn-brain-networks-dataset"]], "Visualize up to five dimension data using DotClustermapPlotter": [[28, "Visualize-up-to-five-dimension-data-using-DotClustermapPlotter"]], "Visualizing gene set enrichment analysis results (clusterProfiler) using dotHeatmap": [[29, "Visualizing-gene-set-enrichment-analysis-results-(clusterProfiler)-using-dotHeatmap"]], "Why do we need a better way to visualize the gene set enrichment analysis result?": [[29, "Why-do-we-need-a-better-way-to-visualize-the-gene-set-enrichment-analysis-result?"]], "Read data": [[29, "Read-data"]], "Plot": [[29, "Plot"]], "How to select marker:": [[29, "How-to-select-marker:"]], "1. Import packages": [[30, "1.-Import-packages"]], "2. Plot heatmap annotations": [[30, "2.-Plot-heatmap-annotations"]], "3. Plot the heatmap with rows and columns split": [[30, "3.-Plot-the-heatmap-with-rows-and-columns-split"]], "4. Plot the annotations along side with main heatmap": [[30, "4.-Plot-the-annotations-along-side-with-main-heatmap"]], "label_kws, xticklabels_kws and yticklabels_kws: Modifying the heatmap annotations labels and xticklabels, yticklabels": [[31, "label_kws,-xticklabels_kws-and-yticklabels_kws:-Modifying-the-heatmap-annotations-labels-and-xticklabels,-yticklabels"]], "xlabel_kws and ylabel_kws: Modifying xlabel and ylabel": [[31, "xlabel_kws-and-ylabel_kws:-Modifying-xlabel-and-ylabel"]], "Control gap & pad in heatmap": [[31, "Control-gap-&-pad-in-heatmap"]], "hgap and wgap for HeatmapAnnotation": [[31, "hgap-and-wgap-for-HeatmapAnnotation"]], "subplot_gap": [[31, "subplot_gap"]], "row_split_gap and col_split_gap": [[31, "row_split_gap-and-col_split_gap"]], "How to remove arrow in anno_label": [[31, "How-to-remove-arrow-in-anno_label"]], "oncoPrint: visualizing TCGA Lung Adenocarcinoma Carcinoma Variants Dataset": [[32, "oncoPrint:-visualizing-TCGA-Lung-Adenocarcinoma-Carcinoma-Variants-Dataset"]], "Visualizing categorical variables using oncoPrint": [[33, "Visualizing-categorical-variables-using-oncoPrint"]], "Visualizing Single Cell DNA Methylation Data (Loyfer2023)": [[34, "Visualizing-Single-Cell-DNA-Methylation-Data-(Loyfer2023)"]], "oncoPrint": [[36, "oncoprint"]], "setup module": [[37, "setup-module"]]}, "indexentries": {"pycomplexheatmap": [[0, "module-PyComplexHeatmap"]], "module": [[0, "module-PyComplexHeatmap"], [1, "module-PyComplexHeatmap.annotations"], [2, "module-PyComplexHeatmap.clustermap"], [3, "module-PyComplexHeatmap.colors"], [4, "module-PyComplexHeatmap.dotHeatmap"], [5, "module-PyComplexHeatmap.example"], [6, "module-PyComplexHeatmap.oncoPrint"], [8, "module-PyComplexHeatmap.utils"]], "annotationbase (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.AnnotationBase"]], "heatmapannotation (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation"]], "pycomplexheatmap.annotations": [[1, "module-PyComplexHeatmap.annotations"]], "anno_barplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_barplot"]], "anno_boxplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_boxplot"]], "anno_img (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_img"]], "anno_label (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_label"]], "anno_lineplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_lineplot"]], "anno_scatterplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_scatterplot"]], "anno_simple (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_simple"]], "collect_legends() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.collect_legends"]], "get_label_width() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.get_label_width"]], "get_max_label_width() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.get_max_label_width"]], "get_ticklabel_width() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.get_ticklabel_width"]], "get_ticklabel_width() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.get_ticklabel_width"]], "plot() (pycomplexheatmap.annotations.anno_barplot method)": [[1, "PyComplexHeatmap.annotations.anno_barplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_boxplot method)": [[1, "PyComplexHeatmap.annotations.anno_boxplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_img method)": [[1, "PyComplexHeatmap.annotations.anno_img.plot"]], "plot() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.plot"]], "plot() (pycomplexheatmap.annotations.anno_lineplot method)": [[1, "PyComplexHeatmap.annotations.anno_lineplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_scatterplot method)": [[1, "PyComplexHeatmap.annotations.anno_scatterplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_simple method)": [[1, "PyComplexHeatmap.annotations.anno_simple.plot"]], "plot_annotations() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.plot_annotations"]], "plot_legends() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.plot_legends"]], "reorder() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.reorder"]], "set_axes_kws() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.set_axes_kws"]], "set_axes_kws() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.set_axes_kws"]], "set_label() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.set_label"]], "set_legend() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.set_legend"]], "set_plot_kws() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.set_plot_kws"]], "set_side() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.set_side"]], "show_ticklabels() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.show_ticklabels"]], "update_plot_kws() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.update_plot_kws"]], "clustermapplotter (class in pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter"]], "dendrogramplotter (class in pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter"]], "pycomplexheatmap.clustermap": [[2, "module-PyComplexHeatmap.clustermap"]], "cal_cold_between_groups() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.cal_cold_between_groups"]], "cal_rowd_between_groups() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.cal_rowd_between_groups"]], "calculate_col_dendrograms() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.calculate_col_dendrograms"]], "calculate_dendrogram() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.calculate_dendrogram"]], "calculate_row_dendrograms() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.calculate_row_dendrograms"]], "calculated_linkage (pycomplexheatmap.clustermap.dendrogramplotter property)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.calculated_linkage"]], "check_array() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.check_array"]], "collect_legends() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.collect_legends"]], "composite() (in module pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.composite"]], "format_data() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.format_data"]], "get_coords() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.get_coords"]], "heatmap() (in module pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.heatmap"]], "heatmapplotter (class in pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.heatmapPlotter"]], "plot() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot"]], "plot() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.plot"]], "plot() (pycomplexheatmap.clustermap.heatmapplotter method)": [[2, "PyComplexHeatmap.clustermap.heatmapPlotter.plot"]], "plot_dendrograms() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot_dendrograms"]], "plot_heatmap() (in module pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.plot_heatmap"]], "plot_legends() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot_legends"]], "plot_matrix() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot_matrix"]], "post_processing() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.post_processing"]], "reordered_ind (pycomplexheatmap.clustermap.dendrogramplotter property)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.reordered_ind"]], "set_axes_labels_kws() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_axes_labels_kws"]], "set_height() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_height"]], "set_width() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_width"]], "set_xy_labels() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_xy_labels"]], "standard_scale() (pycomplexheatmap.clustermap.clustermapplotter static method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.standard_scale"]], "tight_layout() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.tight_layout"]], "z_score() (pycomplexheatmap.clustermap.clustermapplotter static method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.z_score"]], "pycomplexheatmap.colors": [[3, "module-PyComplexHeatmap.colors"]], "define_cmap() (in module pycomplexheatmap.colors)": [[3, "PyComplexHeatmap.colors.define_cmap"]], "register_cmap() (in module pycomplexheatmap.colors)": [[3, "PyComplexHeatmap.colors.register_cmap"]], "dotclustermapplotter (class in pycomplexheatmap.dotheatmap)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter"]], "pycomplexheatmap.dotheatmap": [[4, "module-PyComplexHeatmap.dotHeatmap"]], "collect_legends() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.collect_legends"]], "dotheatmap2d() (in module pycomplexheatmap.dotheatmap)": [[4, "PyComplexHeatmap.dotHeatmap.dotHeatmap2d"]], "format_data() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.format_data"]], "plot_matrix() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.plot_matrix"]], "post_processing() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.post_processing"]], "scale() (in module pycomplexheatmap.dotheatmap)": [[4, "PyComplexHeatmap.dotHeatmap.scale"]], "pycomplexheatmap.example": [[5, "module-PyComplexHeatmap.example"]], "clustermap_example0() (in module pycomplexheatmap.example)": [[5, "PyComplexHeatmap.example.clustermap_example0"]], "clustermap_example1() (in module pycomplexheatmap.example)": [[5, "PyComplexHeatmap.example.clustermap_example1"]], "get_kycg_example_data() (in module pycomplexheatmap.example)": [[5, "PyComplexHeatmap.example.get_kycg_example_data"]], "pycomplexheatmap.oncoprint": [[6, "module-PyComplexHeatmap.oncoPrint"]], "add_default_annotations() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.add_default_annotations"]], "collect_legends() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.collect_legends"]], "format_data() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.format_data"]], "get_samples_order() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.get_samples_order"]], "oncoprintplotter (class in pycomplexheatmap.oncoprint)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter"]], "oncoprint() (in module pycomplexheatmap.oncoprint)": [[6, "PyComplexHeatmap.oncoPrint.oncoprint"]], "plot_matrix() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.plot_matrix"]], "post_processing() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.post_processing"]], "pycomplexheatmap.utils": [[8, "module-PyComplexHeatmap.utils"]], "axis_ticklabels_overlap() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.axis_ticklabels_overlap"]], "cal_legend_width() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.cal_legend_width"]], "cluster_labels() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.cluster_labels"]], "define_cmap() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.define_cmap"]], "despine() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.despine"]], "get_colormap() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.get_colormap"]], "plot_cmap_legend() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_cmap_legend"]], "plot_color_dict_legend() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_color_dict_legend"]], "plot_legend_list() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_legend_list"]], "plot_marker_legend() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_marker_legend"]], "set_default_style() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.set_default_style"]], "to_utf8() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.to_utf8"]]}})
\ No newline at end of file
diff --git a/docs/build/html/setup.html b/docs/build/html/setup.html
index 7f183de5..344f5c9a 100644
--- a/docs/build/html/setup.html
+++ b/docs/build/html/setup.html
@@ -4,7 +4,7 @@
- setup module — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ setup module — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -17,7 +17,7 @@
-
+
diff --git a/notebooks/gene_enrichment_analysis.ipynb b/notebooks/gene_enrichment_analysis.ipynb
index 7b6fb578..b88308e1 100644
--- a/notebooks/gene_enrichment_analysis.ipynb
+++ b/notebooks/gene_enrichment_analysis.ipynb
@@ -20,7 +20,7 @@
"name": "stdout",
"output_type": "stream",
"text": [
- "1.6.6.dev15+gd9f20b3\n"
+ "1.7.2.dev0+g8abf70a.d20240415\n"
]
}
],
@@ -966,7 +966,7 @@
},
{
"cell_type": "code",
- "execution_count": 13,
+ "execution_count": 9,
"id": "8683f2d4-cea6-445f-ae32-cbfdb835b2ee",
"metadata": {
"tags": []
[13]:
+[9]:
left_ha = HeatmapAnnotation(
@@ -1151,7 +1151,7 @@ In this example, we used two different kinds of markers and cmap to indic
How to select marker:¶
-
+
diff --git a/docs/build/html/notebooks/gene_enrichment_analysis.ipynb b/docs/build/html/notebooks/gene_enrichment_analysis.ipynb
index 7b6fb578..b88308e1 100644
--- a/docs/build/html/notebooks/gene_enrichment_analysis.ipynb
+++ b/docs/build/html/notebooks/gene_enrichment_analysis.ipynb
@@ -20,7 +20,7 @@
"name": "stdout",
"output_type": "stream",
"text": [
- "1.6.6.dev15+gd9f20b3\n"
+ "1.7.2.dev0+g8abf70a.d20240415\n"
]
}
],
@@ -966,7 +966,7 @@
},
{
"cell_type": "code",
- "execution_count": 13,
+ "execution_count": 9,
"id": "8683f2d4-cea6-445f-ae32-cbfdb835b2ee",
"metadata": {
"tags": []
diff --git a/docs/build/html/notebooks/get_started.html b/docs/build/html/notebooks/get_started.html
index f3f8152d..7a5f4878 100644
--- a/docs/build/html/notebooks/get_started.html
+++ b/docs/build/html/notebooks/get_started.html
@@ -4,7 +4,7 @@
-
1. Import packages — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ 1. Import packages — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -18,7 +18,7 @@
-
+
diff --git a/docs/build/html/notebooks/kwargs.html b/docs/build/html/notebooks/kwargs.html
index eedb5901..4ace87e6 100644
--- a/docs/build/html/notebooks/kwargs.html
+++ b/docs/build/html/notebooks/kwargs.html
@@ -4,7 +4,7 @@
- label_kws, xticklabels_kws and yticklabels_kws: Modifying the heatmap annotations labels and xticklabels, yticklabels — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ label_kws, xticklabels_kws and yticklabels_kws: Modifying the heatmap annotations labels and xticklabels, yticklabels — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -18,7 +18,7 @@
-
+
@@ -587,7 +587,7 @@ Control gap & pad in heatmap
+

hgap and wgap for HeatmapAnnotation¶
control the gap between two rows/columns of column annotations (axis=1) or row annotations (axis=0), unit is mm
diff --git a/docs/build/html/notebooks/oncoPrint.html b/docs/build/html/notebooks/oncoPrint.html
index f365ab9a..44931732 100644
--- a/docs/build/html/notebooks/oncoPrint.html
+++ b/docs/build/html/notebooks/oncoPrint.html
@@ -4,7 +4,7 @@
- oncoPrint: visualizing TCGA Lung Adenocarcinoma Carcinoma Variants Dataset — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ oncoPrint: visualizing TCGA Lung Adenocarcinoma Carcinoma Variants Dataset — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -18,7 +18,7 @@
-
+
diff --git a/docs/build/html/notebooks/oncoPrint2.html b/docs/build/html/notebooks/oncoPrint2.html
index c1858cf2..7951afc5 100644
--- a/docs/build/html/notebooks/oncoPrint2.html
+++ b/docs/build/html/notebooks/oncoPrint2.html
@@ -4,7 +4,7 @@
- Visualizing categorical variables using oncoPrint — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ Visualizing categorical variables using oncoPrint — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -18,7 +18,7 @@
-
+
diff --git a/docs/build/html/notebooks/single_cell_methylation.html b/docs/build/html/notebooks/single_cell_methylation.html
index f3e9fcf6..c17c827c 100644
--- a/docs/build/html/notebooks/single_cell_methylation.html
+++ b/docs/build/html/notebooks/single_cell_methylation.html
@@ -4,7 +4,7 @@
- Visualizing Single Cell DNA Methylation Data (Loyfer2023) — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ Visualizing Single Cell DNA Methylation Data (Loyfer2023) — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -18,7 +18,7 @@
-
+
diff --git a/docs/build/html/notebooks/test_add_anno_img.html b/docs/build/html/notebooks/test_add_anno_img.html
index 046fc16a..f87026aa 100644
--- a/docs/build/html/notebooks/test_add_anno_img.html
+++ b/docs/build/html/notebooks/test_add_anno_img.html
@@ -4,7 +4,7 @@
- <no title> — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ <no title> — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -18,7 +18,7 @@
-
+
diff --git a/docs/build/html/oncoPrint.html b/docs/build/html/oncoPrint.html
index 71f77c50..dbc9441c 100644
--- a/docs/build/html/oncoPrint.html
+++ b/docs/build/html/oncoPrint.html
@@ -4,7 +4,7 @@
- oncoPrint — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ oncoPrint — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -17,7 +17,7 @@
-
+
diff --git a/docs/build/html/py-modindex.html b/docs/build/html/py-modindex.html
index 835d8bb0..6bfec726 100644
--- a/docs/build/html/py-modindex.html
+++ b/docs/build/html/py-modindex.html
@@ -3,7 +3,7 @@
- Python Module Index — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ Python Module Index — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -16,7 +16,7 @@
-
+
diff --git a/docs/build/html/search.html b/docs/build/html/search.html
index b239271f..c67885b0 100644
--- a/docs/build/html/search.html
+++ b/docs/build/html/search.html
@@ -3,7 +3,7 @@
- Search — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ Search — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -17,7 +17,7 @@
-
+
diff --git a/docs/build/html/searchindex.js b/docs/build/html/searchindex.js
index e391b40f..9354eff2 100644
--- a/docs/build/html/searchindex.js
+++ b/docs/build/html/searchindex.js
@@ -1 +1 @@
-Search.setIndex({"docnames": ["PyComplexHeatmap", "PyComplexHeatmap.annotations", "PyComplexHeatmap.clustermap", "PyComplexHeatmap.colors", "PyComplexHeatmap.dotHeatmap", "PyComplexHeatmap.example", "PyComplexHeatmap.oncoPrint", "PyComplexHeatmap.tools", "PyComplexHeatmap.utils", "advanced_usage", "citation", "clustermap", "dev", "dotHeatmap", "gallery", "get_started", "index", "installation", "kwargs", "modules", "more_examples", "notebooks/advanced_usage", "notebooks/bicluster", "notebooks/clustermap", "notebooks/composite_heatmaps", "notebooks/cpg_modules", "notebooks/dev", "notebooks/differential_expression_genes", "notebooks/dotHeatmap", "notebooks/gene_enrichment_analysis", "notebooks/get_started", "notebooks/kwargs", "notebooks/oncoPrint", "notebooks/oncoPrint2", "notebooks/single_cell_methylation", "notebooks/test_add_anno_img", "oncoPrint", "setup"], "filenames": ["PyComplexHeatmap.rst", "PyComplexHeatmap.annotations.rst", "PyComplexHeatmap.clustermap.rst", "PyComplexHeatmap.colors.rst", "PyComplexHeatmap.dotHeatmap.rst", "PyComplexHeatmap.example.rst", "PyComplexHeatmap.oncoPrint.rst", "PyComplexHeatmap.tools.rst", "PyComplexHeatmap.utils.rst", "advanced_usage.rst", "citation.rst", "clustermap.rst", "dev.rst", "dotHeatmap.rst", "gallery.md", "get_started.rst", "index.rst", "installation.rst", "kwargs.rst", "modules.rst", "more_examples.rst", "notebooks/advanced_usage.ipynb", "notebooks/bicluster.ipynb", "notebooks/clustermap.ipynb", "notebooks/composite_heatmaps.ipynb", "notebooks/cpg_modules.ipynb", "notebooks/dev.ipynb", "notebooks/differential_expression_genes.ipynb", "notebooks/dotHeatmap.ipynb", "notebooks/gene_enrichment_analysis.ipynb", "notebooks/get_started.ipynb", "notebooks/kwargs.ipynb", "notebooks/oncoPrint.ipynb", "notebooks/oncoPrint2.ipynb", "notebooks/single_cell_methylation.ipynb", "notebooks/test_add_anno_img.ipynb", "oncoPrint.rst", "setup.rst"], "titles": ["PyComplexHeatmap package", "PyComplexHeatmap.annotations module", "PyComplexHeatmap.clustermap module", "PyComplexHeatmap.colors module", "PyComplexHeatmap.dotHeatmap module", "PyComplexHeatmap.example module", "PyComplexHeatmap.oncoPrint module", "PyComplexHeatmap.tools module", "PyComplexHeatmap.utils module", "Advanced Usage", "Citation", "Clustermap", "For Developers", "dotHeatmap", "Gallery", "Get Started", "PyComplexHeatmap", "Installation", "Kwargs", "PyComplexHeatmap", "More Examples", "Generate dataset", "Import packages", "A quick example", "Composite two heatmaps horizontally for mouse DNA methylation array dataset", "Plot correlation matrix for CpG modules using DotClustermap", "1. Understand the layout:", "Visualizing Differential Expression Genes", "Load an example brain networks dataset from seaborn", "Visualizing gene set enrichment analysis results (clusterProfiler) using dotHeatmap", "1. Import packages", "label_kws, xticklabels_kws and yticklabels_kws: Modifying the heatmap annotations labels and xticklabels, yticklabels", "oncoPrint: visualizing TCGA Lung Adenocarcinoma Carcinoma Variants Dataset", "Visualizing categorical variables using oncoPrint", "Visualizing Single Cell DNA Methylation Data (Loyfer2023)", "<no title>", "oncoPrint", "setup module"], "terms": {"annot": [0, 2, 4, 6, 9, 11, 15, 16, 18, 19, 22, 24, 26, 28, 29, 32, 33, 34, 35], "annotationbas": [0, 1, 2, 19], "get_label_width": [0, 1], "get_max_label_width": [0, 1], "get_ticklabel_width": [0, 1], "reorder": [0, 1, 2, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "set_axes_kw": [0, 1], "set_label": [0, 1], "set_legend": [0, 1], "update_plot_kw": [0, 1], "heatmapannot": [0, 1, 2, 18, 19, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 32, 33, 34, 35], "collect_legend": [0, 1, 2, 4, 6], "plot_annot": [0, 1], "plot_legend": [0, 1, 2, 21, 23, 27, 34], "show_ticklabel": [0, 1, 21, 23, 30], "anno_barplot": [0, 1, 19, 21, 23, 25, 26, 30, 31, 32, 33], "plot": [0, 1, 2, 4, 6, 8, 9, 11, 13, 15, 16, 20, 22, 24, 27, 31, 32, 33, 34, 35], "anno_boxplot": [0, 1, 19, 21, 23, 26, 30, 31, 35], "anno_img": [0, 1, 19, 21], "anno_label": [0, 1, 16, 18, 19, 21, 22, 24, 25, 26, 27, 28, 29, 30, 32, 33, 34], "set_plot_kw": [0, 1], "set_sid": [0, 1], "anno_lineplot": [0, 1, 19, 21, 31], "anno_scatterplot": [0, 1, 19, 21, 23, 30, 31], "anno_simpl": [0, 1, 19, 21, 22, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "clustermap": [0, 1, 4, 6, 13, 16, 19, 20, 22, 23, 24], "clustermapplott": [0, 2, 4, 6, 19, 21, 22, 23, 24, 26, 27, 30, 31, 34, 35], "cal_cold_between_group": [0, 2], "cal_rowd_between_group": [0, 2], "calculate_col_dendrogram": [0, 2], "calculate_row_dendrogram": [0, 2], "format_data": [0, 2, 4, 6], "plot_dendrogram": [0, 2], "plot_matrix": [0, 2, 4, 6], "post_process": [0, 2, 4, 6], "set_axes_labels_kw": [0, 2], "set_height": [0, 2], "set_width": [0, 2], "set_xy_label": [0, 2], "standard_scal": [0, 2, 4, 6, 22], "tight_layout": [0, 2], "z_score": [0, 2, 4, 6, 21, 27], "dendrogramplott": [0, 2, 19, 22], "calculate_dendrogram": [0, 2], "calculated_linkag": [0, 2], "check_arrai": [0, 2], "get_coord": [0, 2], "reordered_ind": [0, 2], "composit": [0, 2, 16, 19, 20], "heatmap": [0, 1, 2, 4, 6, 9, 13, 15, 16, 18, 19, 20, 23, 25, 27, 32], "heatmapplott": [0, 2, 19], "plot_heatmap": [0, 2, 19], "color": [0, 1, 2, 4, 6, 8, 13, 16, 19, 21, 22, 23, 24, 25, 26, 27, 29, 30, 31, 32, 33, 34, 35], "define_cmap": [0, 3, 8, 19], "register_cmap": [0, 3, 19, 25], "dotheatmap": [0, 16, 19, 28], "dotclustermapplott": [0, 4, 13, 16, 19, 25, 29], "dotheatmap2d": [0, 4, 19], "scale": [0, 2, 4, 19, 22], "exampl": [0, 1, 2, 8, 11, 13, 16, 19, 21, 24, 25, 26, 29, 30, 31, 35], "clustermap_example0": [0, 5, 19], "clustermap_example1": [0, 5, 19], "get_kycg_example_data": [0, 5, 19], "oncoprint": [0, 16, 19], "oncoprintplott": [0, 6, 19, 32, 33], "add_default_annot": [0, 6], "get_samples_ord": [0, 6], "util": [0, 19, 21], "axis_ticklabels_overlap": [0, 8, 19], "cal_legend_width": [0, 8, 19], "cluster_label": [0, 8, 19], "despin": [0, 8, 19, 32, 33], "get_colormap": [0, 8, 19, 21], "plot_cmap_legend": [0, 4, 6, 8, 19], "plot_color_dict_legend": [0, 4, 6, 8, 19], "plot_legend_list": [0, 8, 19], "plot_marker_legend": [0, 4, 8, 19], "set_default_styl": [0, 8, 19], "to_utf8": [0, 8, 19], "class": [1, 2, 4, 6, 22], "df": [1, 21, 22, 23, 24, 26, 28, 30, 31, 35], "none": [1, 2, 4, 6, 8, 21, 22, 23, 24, 26, 27, 30, 31], "cmap": [1, 2, 4, 6, 8, 9, 13, 16, 22, 23, 24, 25, 26, 27, 29, 30, 31, 34, 35], "auto": [1, 2, 21], "height": [1, 2, 8, 21, 22, 23, 25, 26, 27, 29, 30, 31, 32, 33], "legend": [1, 2, 4, 6, 8, 21, 22, 24, 25, 27, 28, 29, 30, 31, 32, 33, 34, 35], "legend_kw": [1, 2, 4, 6, 8, 21, 23, 26, 31, 34], "plot_kw": [1, 6, 33], "sourc": [1, 2, 3, 4, 5, 6, 8, 14], "base": [1, 2, 4, 6, 8, 28], "object": [1, 2, 8, 22, 25, 27, 34], "paramet": [1, 2, 4, 6, 8, 9, 13, 16, 23, 25, 31], "datafram": [1, 2, 4, 6, 21, 26, 27, 30, 31, 32, 33, 35], "panda": [1, 2, 4, 6, 17, 22, 27, 29, 33], "seri": [1, 2], "onli": [1, 4, 6, 9, 16, 31], "one": [1, 4, 8, 23, 29], "column": [1, 2, 4, 6, 15, 16, 21, 22, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "str": [1, 2, 4, 6, 8, 21, 23, 25, 26, 30, 31, 32, 33, 35], "colormap": [1, 2, 4, 6, 9, 16, 25, 28], "set1": [1, 4, 6, 8, 21, 23, 27, 28, 29, 30, 31, 35], "dark2": [1, 8, 21, 23, 25, 27, 28, 30, 35], "bwr": [1, 2], "red": [1, 2, 4, 21, 23, 24, 25, 26, 27, 28, 30, 31, 32, 33], "jet": [1, 2, 21, 23, 25, 28, 30], "hsv": [1, 8], "rainbow": [1, 21, 26, 30, 31], "so": [1, 2], "pleas": [1, 2, 4, 10, 17, 30, 31], "see": [1, 2, 4, 21, 31], "http": [1, 2, 4, 10, 17, 21, 31, 32, 34], "matplotlib": [1, 2, 4, 8, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "org": [1, 2, 4, 10], "3": [1, 4, 15, 16, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "5": [1, 2, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "0": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "tutori": 1, "html": [1, 2, 4, 31, 32], "more": [1, 2, 16, 21, 23, 31, 32], "inform": [1, 2, 31], "run": [1, 17], "pyplot": [1, 21, 25, 27], "all": [1, 2, 4, 8, 9, 16, 23, 29, 31], "availabel": 1, "default": [1, 2, 4, 6, 8, 21, 28, 29, 31], "i": [1, 2, 4, 6, 8, 16, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 33, 35], "would": [1, 2, 21, 23], "determin": [1, 2, 4, 21, 23, 28], "dtype": [1, 2, 21, 22, 25, 26, 27, 28, 34], "each": [1, 2, 6, 23, 29, 30, 33], "dict": [1, 2, 4, 6, 8, 21, 22, 23, 25, 26, 29, 31, 34, 35], "list": [1, 2, 6, 8, 21, 22, 23, 24, 29, 33], "boxplot": 1, "barplot": [1, 29], "If": [1, 2, 4, 6, 8, 10, 23], "kei": [1, 4, 6, 8, 21], "should": [1, 2, 4, 6, 23], "exactli": 1, "same": [1, 2, 4, 23, 32, 34], "iloc": [1, 21, 23, 24, 26, 30, 31, 32, 35], "uniqu": [1, 22, 25, 28, 32, 34], "valu": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 33, 35], "name": [1, 4, 6, 21, 22, 23, 25, 26, 27, 28, 30, 31], "hex": [1, 22], "length": [1, 2, 27], "thi": [1, 4, 6, 10, 21, 23, 29, 30], "equal": 1, "nuniqu": [1, 25, 28, 32], "string": [1, 4, 6, 8], "mean": [1, 2, 4, 21, 23, 25, 28], "share": 1, "float": [1, 2, 4, 23], "axi": [1, 2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "1": [1, 2, 4, 6, 8, 12, 15, 16, 17, 21, 22, 23, 24, 25, 27, 28, 29, 31, 32, 33, 34, 35], "width": [1, 2, 6, 8, 21, 22, 23, 24, 26, 28, 29, 31, 32, 33, 34, 35], "size": [1, 2, 4, 6, 13, 16, 26, 29], "bool": [1, 2, 4], "whether": [1, 2, 4, 8, 21, 23, 31], "when": [1, 2, 4, 21, 23], "ar": [1, 2, 4, 8, 23, 24, 29, 31, 33], "vmax": [1, 2, 4, 8, 21, 24, 28, 29], "vmin": [1, 2, 4, 8, 21, 24, 28, 29], "other": [1, 2, 4, 6, 8, 23, 31, 33], "kw": [1, 2, 8], "pass": [1, 2, 4, 6, 8, 30], "plt": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "titl": [1, 2, 8], "prop": 1, "fontsiz": [1, 2, 21, 22, 23, 25, 26, 27, 29, 31, 34], "labelcolor": [1, 2, 21, 23, 26, 27, 29, 30, 31, 35], "markscal": 1, "frameon": [1, 2, 21, 23, 26, 31], "framealpha": 1, "fancybox": 1, "shadow": [1, 2], "facecolor": [1, 2, 21, 22, 23, 26, 29, 31], "edgecolor": [1, 2, 21, 26, 31], "mode": 1, "argument": 1, "pleast": 1, "type": [1, 2, 4, 6, 8, 22, 24, 29, 34], "There": 1, "an": [1, 13, 16, 21, 23], "addit": [1, 23, 31], "color_text": [1, 8, 23], "true": [1, 2, 6, 8, 9, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "which": [1, 2, 8, 28, 29, 31], "set": [1, 2, 8, 13, 16, 21, 22, 23, 25, 26, 31, 35], "text": [1, 2, 8, 21, 23, 26], "marker": [1, 4, 6, 8, 13, 16, 21, 25], "fals": [1, 2, 4, 8, 9, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "black": [1, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "user": [1, 28], "want": [1, 17, 23, 27], "us": [1, 2, 4, 6, 8, 9, 10, 13, 16, 17, 20, 22, 23, 24, 26, 31, 32, 35, 36], "custom": [1, 2, 9, 16, 25], "instead": [1, 21, 25], "blue": [1, 21, 23, 26, 27, 28, 29, 30, 31, 32, 35], "rotat": [1, 2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34], "rotation_mod": [1, 2, 21, 23, 26, 31], "ha": [1, 21], "va": [1, 21], "annotation_clip": 1, "arrowprop": [1, 21, 29, 31], "For": [1, 2, 8, 16, 21, 29, 31], "also": [1, 2, 4, 6, 23], "chang": [1, 2, 8, 9, 13, 16, 23, 27], "rang": [1, 2, 8, 21, 23, 24, 25, 26, 30, 31, 33, 35], "try": [1, 2], "df_box": [1, 21, 23, 26, 30, 31, 35], "gene1": [1, 21], "return": [1, 2, 4, 6, 8], "idx": 1, "subplot_ax": 1, "label": [1, 2, 8, 9, 16, 18, 22, 23, 24, 25, 26, 27, 28, 29, 30, 32, 33, 34, 35], "label_sid": [1, 8, 21, 23, 25, 27, 28, 29, 31, 32], "label_kw": [1, 16, 18, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 32, 34], "ticklabels_kw": [1, 31], "legend_sid": [1, 2, 8], "right": [1, 2, 8, 9, 11, 16, 25, 26, 27, 28, 29, 30, 31, 32, 33, 35], "legend_gap": [1, 2, 21, 22, 23, 25, 26, 28, 29, 30, 31, 34, 35], "legend_width": [1, 2, 8, 22, 23], "4": [1, 4, 8, 15, 16, 21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "legend_hpad": [1, 2, 21, 22, 23, 24, 25, 26, 31, 32, 34], "2": [1, 2, 4, 8, 15, 16, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "legend_vpad": [1, 2, 8, 21, 23, 24, 25, 26, 31], "orient": [1, 9, 16, 27, 29, 32], "wgap": [1, 18, 21, 26], "hgap": [1, 18, 21, 23, 26, 30], "raster": [1, 2, 22, 23, 24, 26, 31, 34, 35], "verbos": [1, 2, 8, 21, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "arg": 1, "gener": [1, 2, 9, 16, 23, 26, 30, 31, 35], "self": [1, 4], "convert": [1, 28], "int": [1, 2, 4, 21, 26, 28, 30, 31, 34], "row": [1, 2, 4, 6, 9, 15, 16, 22, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "need": 1, "specifi": [1, 2, 23], "provid": [1, 2, 8, 16, 23, 28], "have": [1, 2, 8, 17, 21, 23, 29, 34], "can": [1, 2, 4, 6, 21, 23, 27, 29, 30, 31], "from": [1, 2, 4, 6, 8, 13, 16, 17, 21, 23, 24, 25, 26, 27, 29, 31, 32, 33, 34, 35], "__subclasses__": 1, "includ": [1, 2, 8, 21], "anno_scatt": 1, "must": [1, 4], "els": [1, 21, 22, 23, 25, 26, 27, 28, 30, 31, 32, 33], "calcul": [1, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "given": [1, 4, 8, 23, 28], "invalid": 1, "top": [1, 2, 8, 9, 11, 16, 22, 25, 26, 27, 29, 31, 32, 33], "bottom": [1, 2, 8, 11, 16, 21, 25, 26, 27, 29, 30, 31, 32, 33], "left": [1, 2, 8, 9, 11, 16, 25, 26, 27, 28, 29, 30, 31, 32, 33], "alpha": [1, 2, 4, 25, 26, 28, 29, 31], "fontstyl": [1, 2, 26, 31], "horizontalalign": [1, 2, 21, 23, 25, 26, 27, 28, 29, 30, 31, 32], "verticalalign": [1, 2, 21, 26, 27, 30, 31], "visibl": [1, 2, 21, 22, 24, 25, 26, 29, 31, 32, 34], "gca": [1, 23], "yaxi": [1, 26, 31], "properti": [1, 2, 26, 31], "ax": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 28, 29, 31, 32, 33], "direct": [1, 2, 23, 31], "pad": [1, 2, 16, 18, 21, 26], "labels": [1, 2, 21, 27, 29, 31, 32, 35], "zorder": [1, 2, 26, 31], "labelbottom": [1, 2], "labeltop": [1, 2], "labelleft": [1, 2], "labelright": [1, 2], "labelrot": [1, 2, 21, 23, 26, 27, 29, 30, 31, 35], "grid_color": [1, 2], "grid_linestyl": [1, 2], "tick_param": [1, 2, 21, 31], "function": [1, 4, 8, 23, 25, 29, 30], "et": [1, 2, 22, 23], "al": [1, 2, 23], "automoti": 1, "vertic": [1, 2, 21], "gap": [1, 2, 8, 16, 18, 21, 23], "between": [1, 2, 8, 9, 16, 23, 31], "two": [1, 2, 16, 20, 23, 25, 29, 31], "mm": [1, 2, 22, 23, 24, 28, 29, 31, 32, 34, 35], "horizon": [1, 2, 23], "space": [1, 2, 8, 9, 16, 23], "up": [1, 2, 13, 16, 27, 32], "down": [1, 9, 16, 27], "show": [1, 2, 4, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "automat": [1, 2, 4, 23], "assign": [1, 29, 32], "accord": 1, "posit": [1, 26, 28], "option": [1, 4, 6, 8], "wspace": 1, "control": [1, 4, 6, 16, 18, 21, 23, 28, 33], "hspace": 1, "horizont": [1, 2, 16, 20], "pair": [1, 8, 21, 35], "collect": [1, 21, 22, 23, 24, 27, 28, 29, 30, 32, 33, 34, 35], "rtype": 1, "subplot_spec": [1, 2, 6], "figur": [1, 4, 8, 21, 22, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "add_gridspec": 1, "gridspec": 1, "gridspecfromsubplotspec": 1, "index": [1, 2, 4, 21, 22, 23, 24, 25, 26, 27, 28, 30, 31, 32, 33, 34, 35], "param": [1, 2], "kwarg": [1, 2, 4, 6, 16], "grid": [1, 21, 26, 28, 29, 31, 32, 33], "showflier": 1, "medianlinecolor": 1, "border_width": [1, 21], "border_color": [1, 21], "255": [1, 21], "merg": [1, 8, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 33, 34], "merge_width": 1, "imag": [1, 9, 16], "border": 1, "line": [1, 21, 23, 28, 30, 31], "256": [1, 21], "ignor": [1, 8, 28], "white": [1, 2, 8, 21, 22, 23, 26, 27, 28, 31, 33], "cluster": [1, 2, 8, 9, 16], "onc": 1, "extend": [1, 22, 34], "frac": 1, "major": [1, 8, 22, 29, 31], "adjust_color": [1, 34], "lumin": [1, 29, 34], "8": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 35], "relpo": [1, 21, 22, 26, 27, 29, 30, 31, 32, 34], "add": [1, 2, 9, 13, 16, 25, 27, 31], "documentatin": 1, "websit": 1, "dingwb": [1, 2, 17, 31, 34], "github": [1, 2, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "io": [1, 2, 21, 31, 32], "build": [1, 2, 9, 16, 31], "notebook": [1, 2, 31], "single_cell_methyl": 1, "distribut": 1, "fraction": 1, "arma": 1, "armb": 1, "multipl": [1, 2, 4, 9, 16], "group": [1, 2, 9, 13, 16, 24, 29, 31, 34, 35], "largest": [1, 8], "too": [1, 2, 21, 23, 28], "high": [1, 24, 25, 28], "replac": [1, 21, 26, 28, 30, 31, 32], "origin": 1, "togeth": [1, 2], "tupl": [1, 2, 21, 22], "x": [1, 4, 6, 8, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 33], "y": [1, 4, 6, 8, 21, 22, 24, 25, 26, 28, 29, 31, 32, 33], "arrow": [1, 16, 18, 21], "start": [1, 16, 21, 22, 23, 24, 27, 28, 29, 30, 32, 33, 34, 35], "point": [1, 13, 16, 21, 22, 29, 31], "rel": 1, "side": [1, 15, 16, 25, 26, 29], "about": 1, "could": [1, 2, 4, 23], "found": 1, "patch": [1, 26, 31], "fancyarrowpatch": [1, 31], "lineplot": 1, "linewidth": [1, 2, 21, 23, 25, 26, 28, 29, 31, 32, 35], "scatterplot": 1, "scatter": [1, 4, 6, 21, 23, 26, 28, 29, 30, 31, 35], "add_text": [1, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 35], "text_kw": [1, 21, 22, 23, 24, 25, 26, 27, 28, 30, 31, 35], "simpl": [1, 13, 16, 23], "categor": [1, 4, 8, 16, 23, 36], "continu": [1, 22, 25, 32], "variabl": [1, 8, 16, 36], "data": [2, 4, 6, 10, 13, 16, 20, 21, 22, 23, 24, 26, 27, 30, 31, 32, 33, 35], "top_annot": [2, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 33, 34, 35], "bottom_annot": [2, 23], "left_annot": [2, 22, 23, 24, 27, 29, 34], "right_annot": [2, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 34], "row_clust": [2, 21, 22, 26, 27, 28, 29, 30, 31, 34, 35], "col_clust": [2, 9, 22, 26, 27, 28, 29, 30, 31, 34, 35], "row_cluster_method": [2, 24, 35], "averag": [2, 21, 23, 26, 31, 35], "row_cluster_metr": [2, 24, 35], "correl": [2, 16, 20, 28, 35], "col_cluster_method": [2, 24, 35], "col_cluster_metr": [2, 24, 35], "show_rownam": [2, 21, 22, 23, 24, 26, 27, 28, 29, 30, 31, 32, 33, 35], "show_colnam": [2, 21, 22, 23, 24, 26, 27, 28, 29, 30, 31, 32, 33, 35], "row_names_sid": [2, 21, 23, 29, 30, 31, 35], "col_names_sid": [2, 23, 29], "xticklabels_kw": [2, 6, 16, 18, 21, 23, 26, 27, 29, 30, 32, 35], "yticklabels_kw": [2, 6, 16, 18, 21, 26, 27], "row_dendrogram": [2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "col_dendrogram": [2, 21, 22, 24, 27, 31, 35], "row_dendrogram_s": [2, 21], "10": [2, 10, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "col_dendrogram_s": 2, "row_split": [2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "col_split": [2, 9, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "row_dendrogram_kw": 2, "col_dendrogram_kw": [2, 21], "tree_kw": [2, 21, 23, 25, 27, 28, 30, 31, 35], "row_split_ord": [2, 21, 22, 34], "col_split_ord": [2, 9, 22, 31, 32, 34], "row_split_gap": [2, 18, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 34, 35], "col_split_gap": [2, 18, 21, 22, 23, 25, 26, 27, 28, 29, 30, 32, 33, 34, 35], "mask": [2, 4, 6], "subplot_gap": [2, 18, 21, 26, 32], "legend_anchor": [2, 23, 24, 25], "7": [2, 21, 22, 23, 25, 26, 28, 29, 30, 31, 32, 34, 35], "xlabel": [2, 16, 18, 21, 22, 23, 26, 29, 34, 35], "ylabel": [2, 16, 18, 21, 22, 23, 26, 34, 35], "xlabel_kw": [2, 16, 18, 21, 22, 23, 26, 29, 34], "ylabel_kw": [2, 16, 18, 21, 22, 23, 26, 34], "xlabel_sid": [2, 22, 29], "ylabel_sid": 2, "xlabel_bbox_kw": [2, 21, 22, 26, 29, 31], "ylabel_bbox_kw": [2, 21, 23, 26, 31], "legend_delta_x": 2, "plotter": [2, 4, 6], "numpi": [2, 4, 6, 17, 21, 22, 27, 29, 30], "arrai": [2, 4, 6, 16, 20, 22, 25, 26, 34], "perform": [2, 29], "z": [2, 25], "score": 2, "either": [2, 23], "after": 2, "method": [2, 8, 21], "linkag": [2, 9, 16, 22], "singl": [2, 16, 20], "complet": 2, "weight": [2, 22], "centroid": 2, "median": [2, 21], "ward": [2, 24, 35], "scipi": [2, 17, 35], "hierarchi": 2, "doc": 2, "refer": [2, 32], "detail": 2, "pairwis": 2, "distanc": 2, "observ": 2, "n": [2, 21, 22], "dimension": 2, "euclidean": [2, 35], "minkowski": 2, "cityblock": 2, "seuclidean": 2, "cosin": 2, "ham": 2, "jaccard": [2, 24], "chebyshev": 2, "canberra": [2, 21], "braycurti": 2, "mahalanobi": 2, "kulsinski": 2, "14": [2, 21, 22, 23, 25, 26, 27, 28, 29, 31, 33, 34], "spatial": 2, "pdist": 2, "metric": [2, 21], "ticklabel": [2, 8, 9, 16, 31], "dendrogram": [2, 22, 31], "10mm": 2, "pd": [2, 4, 6, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "number": [2, 21, 29], "hierarch": 2, "split": [2, 15, 16, 25, 28, 29, 32], "subplot": [2, 21], "my_linkag": 2, "order": [2, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "cluster_between_group": [2, 9, 31], "wa": [2, 4, 16, 23, 24, 25, 28, 34], "clsuter": [2, 9], "advanced_usag": [2, 31], "col": [2, 9, 16, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "cell": [2, 9, 16, 20, 22, 23, 29, 31, 33], "miss": 2, "infinit": 2, "1mm": 2, "asfonts": 2, "numpoint": 2, "markerscal": 2, "markerfirst": 2, "title_fonts": 2, "labelspac": 2, "alatern": 2, "we": [2, 21, 23, 28, 30, 32], "outlin": 2, "cbar": [2, 8], "cm": [2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "isinst": 2, "colorbar": [2, 8], "set_color": [2, 25, 26, 29], "set_linewidth": [2, 25, 26, 29], "divid": 2, "ax_heatmap": [2, 23, 24, 25, 26, 28, 29], "anchor": [2, 8, 21, 23, 31], "differ": [2, 8, 13, 16, 21, 29, 30], "infer": [2, 4, 28, 29], "shown": [2, 4, 6], "xticklabel": [2, 16, 18, 21, 35], "yticklabel": [2, 16, 18, 21, 35], "fontfamili": [2, 26, 31], "fontnam": [2, 21, 26, 31], "fontproperti": [2, 26, 31], "fontweight": [2, 22, 24, 26, 27, 29, 31], "xaxi": [2, 26, 31], "print": [2, 21, 22, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "sam": 2, "clip_box": [2, 26, 31], "clip_on": [2, 26, 31], "fill": [2, 4, 21, 26, 31], "in_layout": [2, 26, 31], "linestyl": [2, 26, 28, 29, 31], "get_bbox_patch": [2, 26, 31], "5000": [2, 33], "speed": 2, "center": [2, 8, 21, 24, 26, 31], "robust": [2, 8], "annot_kw": [2, 21], "fmt": [2, 21, 26, 31, 35], "linecolor": [2, 21, 23, 26, 31, 35], "na_col": [2, 8], "cbar_kwss": 2, "document": 2, "small": [2, 34], "displai": [2, 9, 16], "avoid": [2, 21], "overlap": [2, 8, 21], "To": [2, 21], "forc": [2, 9, 16], "use_linkag": 2, "row_ord": [2, 4, 6, 21, 22, 27], "col_ord": [2, 4, 6, 21, 22, 27], "fig": [2, 8, 21], "static": 2, "data2d": 2, "max": [2, 4, 22, 28, 29], "min": [2, 22, 28, 29], "normal": [2, 21, 23, 26, 30, 31, 35], "across": 2, "standard": 2, "noraml": 2, "varianc": 2, "tight_param": 2, "standar": 2, "dendrogram_kw": 2, "gap_pixel": 2, "root_x": 2, "similar": [2, 21], "upon": 2, "indic": [2, 29], "matrix": [2, 4, 6, 16, 20, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "cmlist": [2, 24], "main": [2, 15, 16, 21, 24, 31, 34], "row_gap": [2, 6], "15": [2, 21, 22, 23, 24, 26, 27, 28, 30, 31, 32], "col_gap": [2, 24], "legend_i": 2, "width_ratio": 2, "height_ratio": 2, "assembl": 2, "align": [2, 6, 33], "influenc": [2, 24], "unit": [2, 8, 31], "estim": [2, 22, 23, 24, 28, 29, 32, 34, 35], "legend_ax": [2, 8, 24], "cbar_kw": 2, "cbar_ax": 2, "squar": [2, 13, 16, 26], "xlabel_pad": 2, "ylabel_pad": 2, "xticklabels_sid": 2, "yticklabels_sid": 2, "2g": 2, "maxim": [2, 4], "minim": [2, 4], "seaborn": [2, 13, 16], "cax": [2, 8], "draw": [2, 23], "format": 2, "anno_kw": 2, "c": [3, 4, 6, 8, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 35], "hue": [4, 6, 13, 16, 25, 29], "": [4, 6, 8, 13, 16, 23, 25, 29], "o": [4, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "color_legend_kw": [4, 6], "cmap_legend_kw": [4, 6], "dot_legend_kw": 4, "dot_legend_mark": [4, 28, 29], "aggfunc": 4, "value_na": [4, 28], "hue_na": 4, "na": 4, "s_na": 4, "c_na": [4, 28], "spine": [4, 8, 21, 25, 26, 28, 29, 32, 33], "max_": [4, 28, 29], "dotclustermap": [4, 6, 16, 20], "inherit": [4, 6], "dot": [4, 6, 13, 16, 20, 29], "The": [4, 6, 16, 23, 24, 25, 29], "overwrit": [4, 6], "go": [4, 29], "stabl": 4, "api": 4, "markers_api": 4, "avail": 4, "Such": 4, "v": [4, 28], "p": [4, 25, 28, 29], "h": [4, 21, 23, 26, 27, 30, 31, 35], "d": [4, 8, 10, 21, 23, 25, 26, 28, 30, 31, 35], "_": [4, 28], "tolist": [4, 21, 22, 23, 24, 25, 28, 30, 32, 34], "It": [4, 23, 29], "pivot_t": 4, "fillna": [4, 21, 25, 28, 32], "floator": 4, "np": [4, 21, 22, 23, 26, 27, 28, 29, 30, 31, 32, 35], "aggreg": 4, "them": 4, "call": [4, 8], "pivot": [4, 28], "coeffici": 4, "valid": 4, "input": [4, 6], "paramt": [4, 6], "aspect": [6, 21], "bgcolor": 6, "whitesmok": 6, "remove_empty_row": 6, "remove_empty_column": 6, "background": [6, 23], "lightgrai": [6, 24, 25], "nvar": 6, "element": 6, "wisa": 6, "cccccc": 6, "intern": 8, "boolean": 8, "ani": 8, "legend_list": 8, "xtick": 8, "adjac": [8, 23], "tick": [8, 21], "coordin": 8, "keep": 8, "A": [8, 10, 11, 16, 20, 21, 26, 30, 31], "b": [8, 21, 26, 30, 31, 34], "len": [8, 21, 22], "new_label": 8, "plot_data": 8, "some": [8, 21], "heurist": 8, "good": [8, 29], "remov": [8, 16, 18, 25, 30, 32, 33], "current": 8, "specif": 8, "turbo": [8, 21, 23, 24, 26, 27, 30, 31, 35], "drawn": 8, "accent": 8, "tab20": [8, 25], "exp1": [8, 21, 23, 27], "exp2": [8, 21], "meth1": [8, 21], "meth2": [8, 21, 35], "legn": 8, "y0": 8, "delta_x": 8, "cmap_width": 8, "lengend": 8, "handl": 8, "legend_typ": 8, "pixel": 8, "init": 8, "2mm": 8, "5mm": 8, "boundri": 8, "obj": 8, "repres": 8, "python": [8, 10, 16, 17], "e": [8, 21, 23, 26, 30, 31, 35], "unchang": 8, "byte": 8, "utf": 8, "decod": 8, "__str__": 8, "result": [8, 13, 16, 28], "represent": 8, "dataset": [9, 13, 16, 20, 23, 26, 29, 30, 31, 33, 34, 35, 36], "select": [9, 16, 23, 26, 27, 28, 30, 31], "orent": [9, 16], "extra": [9, 16], "loop": [9, 16], "within": [9, 16], "cluster_within_group": 9, "choos": [9, 16, 30], "pycomplexheatmap": [9, 10, 17, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "diverg": 9, "qualit": 9, "how": [9, 16, 18], "you": [10, 17, 27], "softwar": [10, 28], "your": [10, 29], "work": 10, "cite": 10, "follow": [10, 21, 23, 24, 26, 31, 35], "metadata": [10, 22], "ding": [10, 16], "w": 10, "goldberg": 10, "zhou": 10, "2023": 10, "packag": [10, 15, 16, 19, 28, 29, 32], "visual": [10, 13, 16, 20, 36], "multimod": 10, "genom": 10, "imeta": 10, "e115": 10, "doi": 10, "1002": 10, "imt2": 10, "115": [10, 29], "quick": [11, 16], "understand": [12, 16], "layout": [12, 16], "load": [13, 16, 23, 24, 29], "brain": [13, 16, 23, 24], "network": [13, 16], "tradit": [13, 16], "fix": [13, 16], "five": [13, 16], "dimens": [13, 16], "gene": [13, 16, 20, 21, 23, 25, 26, 30, 31, 32, 33, 35], "enrich": [13, 16, 28], "analysi": [13, 16, 28], "clusterprofil": [13, 16], "click": 14, "picutur": 14, "view": 14, "code": [14, 21, 23, 26, 31, 35], "import": [15, 16, 21, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "along": [15, 16], "complex": 16, "biolog": [16, 29], "instal": [16, 30], "depend": [16, 29], "get": 16, "tcga": [16, 36], "lung": [16, 34, 36], "adenocarcinoma": [16, 36], "carcinoma": [16, 36], "variant": [16, 36], "advanc": 16, "usag": 16, "differenti": [16, 20, 29], "express": [16, 20], "dna": [16, 20, 28], "methyl": [16, 20, 29], "loyfer2023": [16, 20], "mous": [16, 20, 22, 23], "cpg": [16, 20, 24, 34], "modul": [16, 19, 20], "modifi": [16, 18], "galleri": [16, 28], "develop": [16, 29], "setup": [16, 17, 19], "citat": 16, "wubin": 16, "pip": 17, "upgrad": 17, "development": 17, "version": 17, "directli": 17, "com": [17, 21, 31, 34], "git": 17, "previous": 17, "updat": 17, "uninstal": 17, "again": 17, "OR": 17, "clone": [17, 27], "cd": [17, 21, 23, 26, 30, 31, 35], "py": [17, 25, 28], "submodul": 19, "content": 19, "process": [20, 22, 29], "smaller": 20, "sy": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "inlin": [21, 22, 23, 24, 25, 26, 28, 30, 31, 32, 34, 35], "pylab": [21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 34, 35], "pickl": [21, 23, 24, 26, 28, 29, 31, 32, 34, 35], "rcparam": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "dpi": [21, 22, 23, 24, 26, 28, 29, 30, 31, 32, 34, 35], "100": [21, 22, 23, 26, 28, 30, 31, 32, 33, 34, 35], "savefig": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "300": [21, 22, 23, 24, 26, 28, 29, 30, 31, 32, 34, 35], "path": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "append": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "expandus": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "project": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "__version__": [21, 22, 24, 25, 26, 29, 30, 31, 34, 35], "6": [21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 33, 34, 35], "dev15": [21, 29], "gd9f20b3": [21, 29], "font": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "arial": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "famili": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "san": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "serif": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "pdf": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "fonttyp": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "42": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "random": [21, 23, 26, 30, 31, 33, 35], "groupa": [21, 26, 30, 31, 35], "groupb": [21, 26, 30, 31, 35], "ab": [21, 23, 26, 30, 31, 35], "g": [21, 23, 24, 26, 27, 30, 31, 35], "ef": [21, 23, 26, 30, 31, 35], "f": [21, 22, 23, 24, 26, 29, 30, 31, 33, 35], "sampl": [21, 22, 23, 24, 26, 28, 30, 31, 32, 33, 35], "shape": [21, 22, 23, 25, 26, 27, 30, 31, 35], "randn": [21, 23, 26, 30, 31, 35], "df_bar": [21, 23, 26, 30, 31, 35], "uniform": [21, 23, 26, 30, 31, 35], "tmb1": [21, 23, 26, 30, 31, 35], "tmb2": [21, 23, 26, 30, 31, 35], "df_scatter": [21, 23, 26, 30, 31, 35], "df_heatmap": [21, 23, 26, 30, 31, 35], "30": [21, 23, 25, 26, 28, 29, 30, 31, 35], "11": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 33, 35], "fea": [21, 23, 26, 30, 31, 35], "nan": [21, 23, 25, 26, 30, 31, 32, 35], "df_row": [21, 22, 23, 24, 26, 28, 29, 30, 31, 34], "appli": [21, 22, 23, 25, 26, 27, 28, 30, 31, 32], "lambda": [21, 22, 23, 25, 26, 27, 28, 30, 31, 32], "sample4": [21, 26, 30, 31], "to_fram": [21, 26, 28, 30, 31], "xy": [21, 26, 30, 31], "to_seri": [21, 25, 26, 30, 31], "row_ha": [21, 22, 23, 24, 25, 26, 27, 28, 30, 31], "round": [21, 23, 26, 30, 31, 32], "12": [21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 35], "bar": [21, 23, 30, 33], "05": [21, 23, 26, 28, 29, 30, 31, 32, 35], "col_ha": [21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 34, 35], "nanmin": 21, "nanmax": [21, 28], "figsiz": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "row_cmap": [21, 23, 25, 27, 28, 30, 35], "rdylbu_r": [21, 26, 30, 31, 35], "90": [21, 22, 27, 28, 30, 34, 35], "example0": [21, 30, 35], "bbox_inch": [21, 22, 23, 24, 25, 27, 28, 29, 30, 32, 34, 35], "tight": [21, 22, 23, 24, 25, 27, 28, 29, 30, 32, 34, 35], "259323481554901": 21, "3307664641494927": 21, "32": [21, 33], "s4": [21, 26, 31], "test": [21, 26, 31], "18": [21, 26, 28, 31], "bbox": [21, 26], "boxstyl": [21, 26, 31], "circl": [21, 28], "exp": [21, 23, 26, 30, 31, 35], "turn": [21, 23, 26, 31, 35], "off": [21, 23, 26, 31, 35], "log": [21, 23, 26, 31, 35], "head": [21, 24, 25, 27, 28, 30, 32, 34], "1g": [21, 26, 31, 35], "gold": [21, 23, 26, 31, 35], "45": [21, 23, 25, 26, 27, 28, 29, 31, 32, 33, 35], "sample1": [21, 30], "404085": 21, "sample2": [21, 30], "081411": 21, "sample3": [21, 30], "594949": 21, "392642": 21, "sample5": [21, 30], "150321": 21, "229885": 21, "210195": 21, "032791": 21, "051215": 21, "261465": 21, "sample6": 21, "836030": 21, "sample7": 21, "519891": 21, "sample8": 21, "742360": 21, "sample9": 21, "617504": 21, "sample10": 21, "606197": 21, "float64": [21, 28], "fea1": 21, "941453": 21, "140686": 21, "285091": 21, "118367": 21, "214946": 21, "967923": 21, "853248": 21, "fea2": 21, "691451": 21, "561863": 21, "045189": 21, "303610": 21, "324130": 21, "240718": 21, "fea3": 21, "378372": 21, "142337": 21, "405770": 21, "274505": 21, "171325": 21, "526213": 21, "119409": 21, "fea4": 21, "212045": 21, "515978": 21, "051057": 21, "124523": 21, "378586": 21, "320444": 21, "064873": 21, "fea5": 21, "078289": 21, "476870": 21, "360113": 21, "705353": 21, "079402": 21, "284584": 21, "341059": 21, "751814": 21, "163439": 21, "038006": 21, "120816": 21, "820074": 21, "259323": 21, "757637": 21, "451777": 21, "057090": 21, "817372": 21, "082246": 21, "293083": 21, "831747": 21, "489009": 21, "258670": 21, "39": [21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 34], "fea19": 21, "fea25": 21, "fea18": 21, "fea27": 21, "fea29": 21, "fea22": 21, "fea24": 21, "fea30": 21, "fea20": 21, "fea17": 21, "fea26": 21, "fea21": 21, "fea23": 21, "fea15": 21, "fea16": 21, "fea28": 21, "fea9": 21, "fea10": 21, "fea11": 21, "fea7": 21, "fea8": 21, "fea14": 21, "fea13": 21, "fea6": 21, "fea12": 21, "37": [21, 25, 26, 31], "aaaa1": [21, 23], "bbbbb2": [21, 23], "df_bar1": [21, 30], "t1": [21, 30], "df_bar2": [21, 31], "t2": 21, "df_bar3": 21, "t3": 21, "df_bar4": [21, 30], "t4": [21, 30], "372887": 21, "061936": 21, "134849": 21, "694566": 21, "359127": 21, "132367": 21, "392230": 21, "272427": 21, "714047": 21, "625537": 21, "gene2": 21, "gene3": 21, "gene4": 21, "257733": 21, "699439": 21, "844278": 21, "608444": 21, "752454": 21, "083405": 21, "809866": 21, "431847": 21, "663268": 21, "513365": 21, "772141": 21, "387252": 21, "636622": 21, "012600": 21, "966687": 21, "476726": 21, "826088": 21, "361708": 21, "754555": 21, "343367": 21, "220518": 21, "870597": 21, "876851": 21, "9": [21, 22, 23, 24, 25, 28, 29, 31, 32, 33], "876537": 21, "395013": 21, "573748": 21, "948007": 21, "973635": 21, "386078": 21, "499509": 21, "188200": 21, "545418": 21, "328468": 21, "047464": 21, "158959": 21, "161838": 21, "163763": 21, "243494": 21, "779427": 21, "204043": 21, "542872": 21, "130472": 21, "133916": 21, "091244": 21, "788314": 21, "348602": 21, "046968": 21, "088636": 21, "612396": 21, "928272": 21, "877048": 21, "401591": 21, "980643": 21, "201984": 21, "307024": 21, "833295": 21, "660850": 21, "235867": 21, "211823": 21, "919341": 21, "290198": 21, "165467": 21, "382529": 21, "881658": 21, "073468": 21, "342374": 21, "239178": 21, "468118": 21, "962650": 21, "532729": 21, "771372": 21, "835987": 21, "436057": 21, "167528": 21, "042679": 21, "093969": 21, "936279": 21, "354470": 21, "736818": 21, "348153": 21, "335390": 21, "823424": 21, "888954": 21, "168943": 21, "042376": 21, "223754": 21, "876278": 21, "314815": 21, "541221": 21, "407723": 21, "913978": 21, "019213": 21, "885269": 21, "457418": 21, "692399": 21, "369775": 21, "256769": 21, "091064": 21, "233948": 21, "702588": 21, "314119": 21, "758320": 21, "888650": 21, "260971": 21, "891667": 21, "691901": 21, "079907": 21, "903442": 21, "754403": 21, "010756": 21, "829727": 21, "209684": 21, "739576": 21, "615991": 21, "269359": 21, "153883": 21, "386240": 21, "874027": 21, "38": [21, 28, 31], "orang": [21, 23, 24, 29], "yellowgreen": [21, 23, 24, 29], "tmb_bar": [21, 23, 31], "bar1": [21, 30], "bar4": [21, 30], "bar2": 21, "bar3": 21, "fontdict": [21, 22, 24, 30], "tab10": [21, 23], "grai": [21, 23, 24, 28], "yellow": [21, 23], "13": [21, 22, 23, 24, 25, 27, 28, 29], "here": [21, 32], "widh": 21, "40": [21, 22, 29], "20": [21, 26, 27, 28, 31], "incres": [21, 23], "ncol": [21, 23], "than": [21, 23], "support": [21, 23], "long": [21, 23, 28], "new": [21, 23], "41": [21, 29, 34], "green": [21, 23, 24, 25, 26, 27, 28, 29, 31, 33], "By": 21, "43": 21, "typic": 21, "creat": [21, 33], "annotatin": [21, 23], "df_col": [21, 22, 23, 24, 25, 28, 29, 34], "hypomethylated_sampl": [21, 22], "sample_group_color_dict": 21, "celltyp": [21, 22, 29], "ct_color_dict": 21, "ct_legend": 21, "m1": 21, "lgd": 21, "But": 21, "what": [21, 29], "mani": 21, "m2": 21, "m3": 21, "In": [21, 23, 28, 29, 31], "case": [21, 29], "col_ha_dict": 21, "sample_col": 21, "r": [21, 24, 29, 32], "jokergoo": [21, 32], "2021": 21, "03": [21, 22, 24, 29], "complexheatmap": [21, 32], "44": [21, 35], "g1": [21, 31, 35], "g2": [21, 31, 35], "g3": [21, 31, 35], "g4": [21, 31, 35], "g5": [21, 31, 35], "col_cmap": [21, 27, 31, 35], "featur": [21, 26, 31, 35], "46": 21, "loc": [21, 22, 24, 25, 28, 29, 32, 34], "47": 21, "annotaiton": [21, 31], "purpl": [21, 23, 25, 26, 28, 31, 35], "_ticklabels_kw": [21, 31], "labelpad": [21, 22, 23, 26, 29, 31, 34], "increac": [21, 22, 23, 31], "manual": [21, 22, 31], "chocol": [21, 22, 26, 31], "48": 21, "applymap": 21, "make": 21, "asterisk": 21, "locat": 21, "oper": 21, "unicod": 21, "explor": 21, "2217": 21, "courier": 21, "49": [21, 28], "fastclust": 21, "t": [21, 22, 23, 25, 27, 28, 32, 34], "nlinkag": 21, "17": [21, 25, 28], "07051523": 21, "22006417": 21, "19": [21, 23, 26, 28, 33, 35], "14461418": 21, "28842182": 21, "70552492": 21, "76359288": 21, "65679239": 21, "16": [21, 23, 24, 25, 27, 28], "21": [21, 28, 35], "91528308": 21, "22": [21, 28], "75945877": 21, "50": [21, 23, 28], "51": 21, "df_img": 21, "jpeg": 21, "52": 21, "img": 21, "diverging1": 21, "parula": [21, 22, 23, 24, 34, 35], "53": 21, "54": 21, "mpl": [21, 29], "gradient": 21, "linspac": 21, "vstack": 21, "def": 21, "plot_color_gradi": 21, "categori": [21, 28, 29], "cmap_list": 21, "adjust": [21, 29], "nrow": 21, "figh": 21, "35": [21, 29], "subplots_adjust": 21, "99": [21, 33], "set_titl": [21, 22, 24], "zip": [21, 26], "imshow": 21, "01": [21, 32], "transform": [21, 26], "transax": 21, "just": 21, "ones": 21, "set_axis_off": [21, 23], "save": 21, "later": [21, 25], "55": [21, 28], "sequenti": 21, "cmap50": 21, "56": [21, 29], "57": 21, "custom_cmap": 21, "58": 21, "59": 21, "big": [21, 23], "enough": 21, "hidden": 21, "60": 21, "matter": 21, "61": [21, 33], "home": 22, "wding2": 22, "pch": [22, 24, 25, 30, 34], "czip": 22, "palette_path": 22, "mouse_pfc": 22, "mpfc_color_palett": 22, "xlsx": 22, "cell_class_color": 22, "read_excel": 22, "sheet_nam": 22, "cellclass": [22, 29], "index_col": [22, 25, 27, 28, 32, 34, 35], "to_dict": [22, 25, 29], "multi": [22, 28], "grei": [22, 24, 29], "nonn": 22, "e06931": 22, "exc": 22, "5ec5a9": 22, "rg": 22, "6036cb": 22, "inh": 22, "e93400": 22, "delta": [22, 34], "read_csv": [22, 25, 27, 28, 32, 34, 35], "pseudo_cel": 22, "between_groups_dmr": 22, "methylpi": 22, "methylpy_rms_results_collaps": 22, "tsv": 22, "sep": [22, 25, 28, 32, 34], "lstrip": 22, "methylation_level_": 22, "startswith": 22, "renam": 22, "number_of_dm": 22, "inplac": [22, 25, 28, 32, 34], "drop": 22, "hypermethylated_sampl": 22, "sname": 22, "insert": 22, "isna": 22, "reset_index": [22, 25, 28, 32], "set_index": [22, 25, 28, 29], "read": [22, 24, 34], "major_typ": 22, "beta": [22, 24, 25, 34], "txt": [22, 28, 32], "common_row": 22, "5kb_mc_hic_clust": 22, "l2": 22, "major_type_to_cell_class": 22, "common_col": 22, "cell_type_col": 22, "258365": 22, "23": [22, 26, 28, 29], "chr": [22, 24], "end": 22, "chr14": 22, "25667890": 22, "25667923": 22, "649934": 22, "chr5": [22, 24], "70893110": 22, "70893122": 22, "333501": 22, "chr3": 22, "141598191": 22, "141598238": 22, "440745": 22, "chr1": [22, 34], "76086002": 22, "76086015": 22, "319716": 22, "chr9": 22, "16231701": 22, "16232342": 22, "444616": 22, "chr11": [22, 24], "75550115": 22, "75550656": 22, "547601": 22, "109751156": 22, "109751187": 22, "530203": 22, "68823498": 22, "68825391": 22, "29": 22, "451270": 22, "chr10": 22, "91354668": 22, "91354926": 22, "327058": 22, "74147749": 22, "74150162": 22, "31": [22, 26, 29], "541438": 22, "cellclass1": 22, "majortyp": 22, "ct": 22, "l6": 22, "cge": 22, "lamp5": 22, "IT": 22, "l23": 22, "pc": [22, 27], "mgc": 22, "odc": 22, "vip": 22, "mge": 22, "sst": 22, "opc": 22, "vlmc": 22, "l4": 22, "unknown": [22, 28], "l5": 22, "ec": 22, "pvalb": 22, "pia": 22, "asc": 22, "chrom": 22, "077450": 22, "233350": 22, "161600": 22, "733500": 22, "685500": 22, "628000": 22, "148000": 22, "468000": 22, "785000": 22, "662500": 22, "780000": 22, "092500": 22, "125000": 22, "849000": 22, "043500": 22, "839500": 22, "735500": 22, "822500": 22, "694000": 22, "630500": 22, "903500": 22, "899000": 22, "896500": 22, "637500": 22, "784500": 22, "323000": 22, "947500": 22, "743500": 22, "844000": 22, "961500": 22, "499000": 22, "926000": 22, "000000": [22, 23, 25, 27, 28, 35], "846500": 22, "954500": 22, "489500": 22, "883500": 22, "598000": 22, "884000": 22, "789500": 22, "491350": 22, "192500": 22, "623500": 22, "055500": 22, "332150": 22, "763500": 22, "464500": 22, "448000": 22, "200150": 22, "549500": 22, "377950": 22, "517000": 22, "583500": 22, "458500": 22, "419500": 22, "633500": 22, "775000": 22, "299000": 22, "082350": 22, "471000": 22, "797500": 22, "508500": 22, "773500": 22, "050000": 22, "610500": 22, "802000": 22, "869500": 22, "523500": 22, "572500": 22, "572000": 22, "887000": 22, "778000": 22, "396500": 22, "826500": 22, "697500": 22, "145450": 22, "562500": 22, "317500": 22, "807875": 22, "725875": 22, "318250": 22, "900750": 22, "807750": 22, "378837": 22, "209437": 22, "654000": 22, "526500": 22, "579750": 22, "715875": 22, "133525": 22, "605375": 22, "663125": 22, "666375": 22, "545250": 22, "307337": 22, "319063": 22, "839000": 22, "557333": 22, "631000": 22, "780667": 22, "171233": 22, "262050": 22, "886500": 22, "492667": 22, "159800": 22, "520667": 22, "726833": 22, "683833": 22, "698000": 22, "231500": 22, "830000": 22, "781667": 22, "631667": 22, "909000": 22, "222417": 22, "338917": 22, "687667": 22, "452667": 22, "457667": 22, "241333": 22, "154667": 22, "546333": 22, "629667": 22, "325667": 22, "058667": 22, "503333": 22, "321000": 22, "588000": 22, "709000": 22, "034133": 22, "859333": 22, "360333": 22, "684667": 22, "414333": 22, "081200": 22, "244667": 22, "753267": 22, "363077": 22, "607967": 22, "644540": 22, "257347": 22, "775067": 22, "890267": 22, "517487": 22, "184007": 22, "594700": 22, "720740": 22, "763000": 22, "750800": 22, "495783": 22, "471317": 22, "777000": 22, "901467": 22, "657633": 22, "362270": 22, "241253": 22, "784857": 22, "809286": 22, "508857": 22, "859286": 22, "650714": 22, "823286": 22, "763286": 22, "614143": 22, "339714": 22, "615143": 22, "859000": 22, "469000": 22, "742857": 22, "466571": 22, "851714": 22, "807286": 22, "798000": 22, "913429": 22, "423857": 22, "678143": 22, "859394": 22, "860788": 22, "766576": 22, "678030": 22, "443909": 22, "813273": 22, "867061": 22, "658548": 22, "311152": 22, "854182": 22, "765485": 22, "813455": 22, "846697": 22, "434000": 22, "893424": 22, "834667": 22, "833515": 22, "808030": 22, "302337": 22, "529615": 22, "cc": 22, "dendrogram_col": 22, "ivl": 22, "dendrogram_row": 22, "sort_valu": [22, 34], "color_dict": [22, 34], "copi": 22, "del": [22, 33], "bold": [22, 24, 27, 29], "avg": [22, 23], "n_cpg": 22, "6375": [22, 29], "exist": 22, "listdir": 22, "within_groups_dmr": 22, "join": [22, 28], "_heatmap": 22, "concat": 22, "groupbi": [22, 28, 32], "agg": 22, "to_csv": 22, "agg_beta": 22, "queri": 22, "pfc": 22, "bed": 22, "gz": 22, "outfil": 22, "3013471": 22, "3013579": 22, "3026186": 22, "3026310": 22, "3035862": 22, "3035927": 22, "3052666": 22, "3052823": 22, "3059277": 22, "3059436": 22, "chry": 22, "44290504": 22, "44290637": 22, "66427102": 22, "66427179": 22, "90367464": 22, "90367503": 22, "90732358": 22, "90732367": 22, "90792411": 22, "90792447": 22, "661968": 22, "mt": 22, "28963217": 22, "28963260": 22, "83350": 22, "923000": 22, "939000": 22, "507000": 22, "89900": 22, "833500": 22, "738500": 22, "847000": 22, "95450": 22, "966500": 22, "80500": 22, "769500": 22, "731000": 22, "752500": 22, "81300": 22, "760000": 22, "650500": 22, "772500": 22, "936500": 22, "chr4": 22, "32158998": 22, "32159443": 22, "78950": 22, "524500": 22, "791500": 22, "767750": 22, "759500": 22, "73275": 22, "791750": 22, "693250": 22, "79225": 22, "441325": 22, "69625": 22, "538750": 22, "667000": 22, "391250": 22, "46975": 22, "616500": 22, "479250": 22, "704750": 22, "549750": 22, "chr13": 22, "90011452": 22, "90011460": 22, "29100": 22, "222000": 22, "225000": 22, "228500": 22, "08115": 22, "476500": 22, "009400": 22, "329500": 22, "32350": 22, "231000": 22, "30700": 22, "390500": 22, "402500": 22, "782000": 22, "06915": 22, "314500": 22, "285500": 22, "812000": 22, "821500": 22, "126765779": 22, "126765801": 22, "81850": 22, "885000": 22, "903000": 22, "565000": 22, "87500": 22, "875000": 22, "894000": 22, "93750": 22, "906000": 22, "68550": 22, "721500": 22, "649000": 22, "783500": 22, "80050": 22, "793500": 22, "809500": 22, "764500": 22, "142126735": 22, "142126952": 22, "84475": 22, "959250": 22, "902000": 22, "706500": 22, "94350": 22, "781250": 22, "530250": 22, "97100": 22, "933250": 22, "82150": 22, "774000": 22, "728250": 22, "869750": 22, "87600": 22, "796500": 22, "863000": 22, "842750": 22, "870500": 22, "21947352": 22, "21947371": 22, "67700": 22, "261333": 22, "766667": 22, "953667": 22, "979333": 22, "93500": 22, "897333": 22, "613667": 22, "864333": 22, "89500": 22, "950667": 22, "75200": 22, "733333": 22, "855667": 22, "874333": 22, "77000": 22, "589333": 22, "506333": 22, "868333": 22, "781000": 22, "chr6": 22, "50328671": 22, "50328782": 22, "38975": 22, "316650": 22, "702250": 22, "820000": 22, "85425": 22, "458000": 22, "662250": 22, "608250": 22, "81025": 22, "889500": 22, "61000": 22, "592250": 22, "047500": 22, "632250": 22, "64975": 22, "605250": 22, "608500": 22, "305500": 22, "407325": 22, "chr12": [22, 24], "15047867": 22, "15047932": 22, "68650": 22, "750000": [22, 35], "825000": 22, "777500": 22, "266500": 22, "31450": 22, "874500": 22, "65700": 22, "867500": 22, "71350": 22, "591000": 22, "290500": 22, "740500": 22, "74400": 22, "723000": 22, "735000": 22, "755500": 22, "559500": 22, "chr8": [22, 24], "48317445": 22, "48317685": 22, "87640": 22, "938400": 22, "937200": 22, "596000": 22, "66820": 22, "844600": 22, "885600": 22, "746000": 22, "80600": 22, "880800": 22, "90840": 22, "804800": 22, "765800": 22, "848800": 22, "76380": 22, "824400": 22, "810400": 22, "916000": 22, "935600": 22, "chr2": 22, "93512302": 22, "93512369": 22, "31350": 22, "788000": 22, "581000": 22, "832500": 22, "604500": 22, "94450": 22, "937500": 22, "900500": 22, "890000": 22, "90750": 22, "956500": 22, "32550": 22, "232700": 22, "221000": 22, "799000": 22, "55550": 22, "614000": 22, "601000": 22, "435400": 22, "470000": 22, "color_palett": 22, "gc": 22, "majortype_color": 22, "silver": 22, "crimson": 22, "deepskyblu": 22, "darkseagreen": 22, "darkcyan": 22, "k": 22, "de9f00": 22, "67292f": 22, "e13f82": 22, "3f884e": 22, "78772b": 22, "4ac750": 22, "6bc22e": 22, "a8d617": 22, "314b1f": 22, "aab639": 22, "e179a2": 22, "764a71": 22, "63bc9e": 22, "cea675": 22, "a87331": 22, "9bb070": 22, "9fa600": 22, "5e461": 22, "8000": 22, "sort": [22, 34], "sum": [22, 32], "row_major_typ": 22, "cell_class_colors1": 22, "majortype_colors1": 22, "left_ha": [22, 29, 34], "right_ha": [22, 29, 34], "all_dmr_heatmap": 22, "set2": 23, "tomato": 23, "skyblu": [23, 24], "spectral_r": 23, "vertical": 23, "first": 23, "051388888888887": 23, "boarder": 23, "continnu": 23, "deep": 23, "otherwis": 23, "your_color": 23, "treat": 23, "sometim": 23, "without": 23, "do": 23, "give": 23, "anoth": [23, 25], "No": [23, 24, 28, 32], "open": [23, 24, 29], "mammal_arrai": 23, "pkl": 23, "rb": [23, 24, 29], "col_colors_dict": [23, 24, 34], "gsm4412025": 23, "gsm4412026": 23, "gsm4412027": 23, "gsm4412028": 23, "gsm4412029": 23, "gsm4412030": 23, "gsm4412031": 23, "gsm4412032": 23, "gsm4412033": 23, "gsm4412034": 23, "gsm4997945": 23, "gsm4997946": 23, "gsm4997947": 23, "gsm4997948": 23, "gsm4997949": 23, "gsm4997950": 23, "gsm4997951": 23, "gsm4997952": 23, "gsm4997953": 23, "gsm4997954": 23, "sheep": 23, "435033": 23, "432900": 23, "446626": 23, "449123": 23, "497180": 23, "515918": 23, "483706": 23, "504681": 23, "529076": 23, "446443": 23, "beluga": 23, "whale": 23, "687488": 23, "694207": 23, "706525": 23, "702734": 23, "687014": 23, "704003": 23, "705887": 23, "693806": 23, "719417": 23, "712677": 23, "560381": 23, "571552": 23, "610392": 23, "613619": 23, "675832": 23, "668502": 23, "624820": 23, "658377": 23, "702334": 23, "575034": 23, "hous": 23, "vaquita": 23, "693523": 23, "702525": 23, "716792": 23, "725095": 23, "711261": 23, "708651": 23, "717952": 23, "705486": 23, "720915": 23, "724171": 23, "581862": 23, "594443": 23, "628908": 23, "639457": 23, "680801": 23, "707493": 23, "662338": 23, "665142": 23, "751859": 23, "584952": 23, "larg": [23, 28], "fly": 23, "fox": 23, "286822": 23, "269406": 23, "296796": 23, "314719": 23, "305074": 23, "308419": 23, "268421": 23, "297236": 23, "295019": 23, "297973": 23, "281684": 23, "363926": 23, "367216": 23, "359392": 23, "289821": 23, "257351": 23, "234301": 23, "295840": 23, "249336": 23, "344848": 23, "greater": 23, "horsesho": 23, "bat": 23, "530606": 23, "517989": 23, "520719": 23, "525855": 23, "507583": 23, "511993": 23, "528107": 23, "521563": 23, "542159": 23, "509526": 23, "735151": 23, "714805": 23, "708868": 23, "738207": 23, "823604": 23, "828040": 23, "823382": 23, "796882": 23, "878783": 23, "693625": 23, "littl": 23, "brown": 23, "202525": 23, "215990": 23, "238977": 23, "241872": 23, "267750": 23, "236729": 23, "202384": 23, "240519": 23, "251148": 23, "267257": 23, "883": 23, "predictedtaxid": 23, "predictedspeci": 23, "common_nam": 23, "9940": 23, "ovis_aries_rambouillet": 23, "bovida": 23, "9749": 23, "delphinapterus_leuca": 23, "monodontida": 23, "10090": 23, "mus_musculu": 23, "murida": 23, "42100": 23, "phocoena_sinu": 23, "phocoenida": 23, "132908": 23, "pteropus_vampyru": 23, "pteropodida": 23, "59479": 23, "rhinolophus_ferrumequinum": 23, "rhinolophida": 23, "59463": 23, "myotis_lucifugu": 23, "vespertilionida": 23, "gse": 23, "basenam": 23, "ncbi_scientific_nam": 23, "taxid": 23, "tissu": [23, 24], "sex": [23, 24], "speci": 23, "successr": 23, "gse147003": 23, "ovi": 23, "ari": 23, "blood": [23, 34], "femal": [23, 24], "artiodactyla": 23, "765818": 23, "797669": 23, "759256": 23, "male": [23, 24], "749813": 23, "770433": 23, "gse164127": 23, "eptesicu": 23, "fuscu": 23, "29078": 23, "skin": 23, "chiroptera": 23, "660425": 23, "652822": 23, "664746": 23, "650848": 23, "657170": 23, "a40043": [23, 34], "00e5ff": 23, "00becc": 23, "cerebellum": 23, "b2f8ff": 23, "striatum": 23, "6cbf00": [23, 34], "liver": [23, 24, 34], "ffccee": [23, 34], "muscl": [23, 29, 34], "1e9351": 23, "put": 23, "auc": 23, "legend_pad": 23, "veri": 23, "abov": 23, "25": [23, 26, 28, 31, 32], "930555555555557": [23, 28], "last": 23, "second": 23, "worth": 23, "note": 23, "betwe": 23, "kind": [23, 29, 30], "predict": 23, "68888888888889": 23, "tranpos": 23, "columsn": 23, "obtain": 24, "pmid": [24, 34], "36617464": 24, "linearsegmentedcolormap": [24, 25], "80": [24, 29, 33, 35], "urllib": 24, "influence_of_snp_on_beta": 24, "close": 24, "snp": 24, "row_colors_dict": 24, "respect": 24, "2000": 24, "204875570030_r01c02": 24, "204875570030_r04c01": 24, "cg30848532_tc21": 24, "525089": 24, "419515": 24, "cg30147375_bc21": 24, "803776": 24, "585928": 24, "cg46239718_bc21": 24, "443958": 24, "517514": 24, "cg36100119_bc21": 24, "351977": 24, "528846": 24, "cg42738582_bc21": 24, "783958": 24, "724901": 24, "204875570030_r05c01": 24, "204875570030_r06c01": 24, "204875570035_r05c02": 24, "483276": 24, "460750": 24, "390317": 24, "510269": 24, "831463": 24, "550146": 24, "535909": 24, "450167": 24, "564107": 24, "524896": 24, "374422": 24, "551200": 24, "802178": 24, "848621": 24, "850481": 24, "target": 24, "extensionbas": 24, "probedesign": 24, "con": 24, "mapflag": 24, "ii": 24, "chr19": 24, "suboptim": 24, "hybrid": 24, "effect": 24, "artifici": 24, "low": [24, 25, 28], "meth": [24, 34], "cg": 24, "gg": 24, "ga": 24, "79760438": 24, "ca": 24, "ac": 24, "ag": 24, "109557651": 24, "gt": [24, 26], "117860829": 24, "5877949": 24, "aa": 24, "122031379": 24, "strain": 24, "molf_eij": 24, "frontal": 24, "lobe": 24, "cast_eij": 24, "darkorang": 24, "wheat": 24, "darkgrai": 24, "69": [24, 27, 34], "4986111111111": 24, "col_ha1": 24, "cm1": 24, "viridi": 24, "col_ha2": 24, "my_cmap": 24, "from_list": [24, 25], "cm2": 24, "dnam": 24, "beta_snp": 24, "df_corr": 25, "kycg_modules_correl": 25, "csv": [25, 27, 34, 35], "df_ann": 25, "kycg_modules_annot": 25, "kycg_modules_beta": 25, "cpg_std": 25, "std": [25, 28], "cpg_mean": 25, "astyp": [25, 28], "value_count": [25, 28], "28": [25, 26, 28], "33": 25, "count": [25, 27, 28, 29], "int64": 25, "isin": [25, 28], "hm": 25, "h3k4me1": 25, "h3k79me2": 25, "h3k79me3": 25, "h3f3a": 25, "h4k20me1": 25, "h2bk20ac": 25, "h3k23me2": 25, "h2afi": 25, "h2ak9ac": 25, "h2bk12ac": 25, "h2bk15ac": 25, "h3k9me1": 25, "h2bk120ub": 25, "h2azac": 25, "h3k79me1": 25, "h2a": 25, "h3_4ac": 25, "h3k27me1": 25, "h3k4me3b": 25, "h3k56ac": 25, "h3k9ac_h3k14ac": 25, "h3k9k14ac": 25, "cenpa": 25, "h1": 25, "h2afz": 25, "h2ak119": 25, "h3": 25, "h3ac": 25, "h3k27ac": 25, "h3k4me3": 25, "h3k9ac": 25, "h4": 25, "h4ac": 25, "h4k5ac": 25, "h4k5ac_h4k8ac_h4k12ac_h4k16ac": 25, "h4k8ac": 25, "histonelysineacetyl": 25, "histonelysinecrotonyl": 25, "h3k18cr": 25, "h4k16ac": 25, "h4k12ac": 25, "h3k27me3b": 25, "h2bub": 25, "h2ak5ac": 25, "h3k36me3b": 25, "h2bk120ac": 25, "h3k36ac": 25, "h4k91ac": 25, "h4k20me3": 25, "h2afy2": 25, "h3k36me2": 25, "h3k27me3": 25, "h2ak119ub": 25, "cg04735237": 25, "cg00643814": 25, "cg24865495": 25, "cg25376651": 25, "cg27485084": 25, "cg12609052": 25, "cg07354679": 25, "cg02592525": 25, "cg12747056": 25, "cg17718960": 25, "cg19987665": 25, "cg18406033": 25, "cg09678971": 25, "cg00461612": 25, "cg18689454": 25, "cg11923631": 25, "cg27251412": 25, "cg08089567": 25, "cg16717549": 25, "cg09510531": 25, "559916": 25, "686845": 25, "561415": 25, "699960": 25, "711086": 25, "563690": 25, "527953": 25, "621757": 25, "623319": 25, "048462": 25, "103904": 25, "002533": 25, "074093": 25, "114526": 25, "151763": 25, "140823": 25, "117356": 25, "100510": 25, "126291": 25, "538034": 25, "568960": 25, "687787": 25, "593650": 25, "676068": 25, "606604": 25, "589549": 25, "747987": 25, "185989": 25, "087952": 25, "242909": 25, "065101": 25, "054958": 25, "096098": 25, "089246": 25, "078234": 25, "025965": 25, "069120": 25, "614696": 25, "646452": 25, "786480": 25, "602911": 25, "519926": 25, "642344": 25, "642081": 25, "022499": 25, "108524": 25, "015151": 25, "028957": 25, "274560": 25, "302464": 25, "284823": 25, "265835": 25, "262931": 25, "268134": 25, "564983": 25, "727579": 25, "624186": 25, "416198": 25, "505130": 25, "596338": 25, "030346": 25, "122412": 25, "063630": 25, "007048": 25, "197339": 25, "226034": 25, "218230": 25, "180297": 25, "181867": 25, "204431": 25, "655515": 25, "599935": 25, "590981": 25, "627786": 25, "674095": 25, "020895": 25, "058742": 25, "078318": 25, "055019": 25, "139691": 25, "168351": 25, "160081": 25, "142216": 25, "114554": 25, "144024": 25, "512": 25, "chromhmm": [25, 28], "chromhmm_bioc": 25, "tfb": 25, "txwk": [25, 28], "hdgf": 25, "rbfox2": 25, "srebf1": 25, "pqbp1": 25, "qui": [25, 28], "ascl1": 25, "casz1": 25, "ebf1": 25, "fezf1": 25, "foxm1": 25, "hand2": 25, "hdac3": 25, "irf2b": 25, "crx": 25, "dnmt3b": 25, "otx2": 25, "rorb": 25, "zmynd11": 25, "znf711": 25, "macrod1": 25, "atf2": 25, "batf": 25, "etv6": 25, "fo": 25, "ikzf2": 25, "irf4": 25, "junb": 25, "maf": 25, "mafg": 25, "me": 25, "fry": 25, "pdx1": 25, "keep_cpg": 25, "map": [25, 28, 29], "stack": [25, 28, 32], "keep_hm": 25, "154": 25, "245870": 25, "768918": 25, "319166": 25, "635635": 25, "270459": 25, "835897": 25, "276320": 25, "618949": 25, "276748": 25, "700196": 25, "23716": 25, "all_cmap": 25, "binar": 25, "colgroup": [25, 28], "middl": [25, 28], "heatmap_ax": [25, 29], "j": 25, "set_vis": [25, 26, 29], "var": [25, 32], "folder": 25, "3q": 25, "hwjjddlj65b030m69cfbdj4h0000gq": 25, "ipykernel_61938": 25, "92826381": 25, "matplotlibdeprecationwarn": 25, "deprec": 25, "minor": [25, 28, 29], "releas": 25, "regist": 25, "dotclustermap2": 25, "dev4": 26, "g71c725a": 26, "d20240410": 26, "ax_top": 26, "ax_right": 26, "set_axis_on": 26, "set_linestyl": [26, 29], "agg_filt": 26, "anim": 26, "bbox_patch": 26, "lt": 26, "fancybboxpatch": 26, "0x7f854d935760": 26, "children": 26, "clip_path": 26, "600x800": 26, "font_manag": 26, "0x7f854d8d56a0": 26, "fontvari": 26, "gid": 26, "math_fontfamili": 26, "dejavusan": 26, "mouseov": 26, "parse_math": 26, "path_effect": 26, "picker": 26, "000000000000007": 26, "sketch_param": 26, "snap": 26, "stretch": 26, "tightbbox": 26, "125": 26, "112": [26, 28], "875": [26, 34], "blendedaffine2d": 26, "0x7f854d8d5070": 26, "transform_rotates_text": 26, "transformed_clip_path_and_affin": 26, "unitless_posit": 26, "url": 26, "usetex": 26, "window_ext": 26, "wrap": 26, "antialias": 26, "75": [26, 28, 34], "0x7f854dded490": 26, "capstyl": 26, "butt": 26, "data_transform": 26, "affine2d": 26, "0x7f854f0bbfd0": 26, "extent": 26, "761576317947967": 26, "6388888888888813": 26, "92": 26, "78935409572574": 26, "638888888888896": 26, "hatch": 26, "joinstyl": 26, "miter": 26, "solid": 26, "mutation_aspect": 26, "mutation_scal": 26, "444444444444443": 26, "patch_transform": 26, "identitytransform": 26, "0x7f854d868730": 26, "63888889": 26, "81": [26, 33], "38888889": 26, "79": 26, "uint8": 26, "vert": 26, "76157632": 26, "7893541": 26, "expr": 27, "test_expression_dataset": 27, "selected_row": 27, "adam7": 27, "hla": 27, "doa": 27, "ptgir": 27, "opcml": 27, "it1": 27, "mageb1": 27, "label_row": 27, "c8orf34": 27, "as1": 27, "smim28": 27, "linc01114": 27, "klk3": 27, "traj45": 27, "tpmtp1": 27, "hnrnpcp4": 27, "89": [27, 33], "nci": 27, "h660": 27, "vcap": 27, "du": 27, "145": 27, "mda": 27, "pca": 27, "2b": 27, "22rv1": 27, "lncap": 27, "fgc": 27, "sup": 27, "hd1": 27, "l": 27, "540": 27, "1236": 27, "km": 27, "h2": 27, "hdlm": 27, "616": 27, "428": 27, "611": 27, "TO": 27, "175": 27, "044425": 27, "303654": 27, "604782": 27, "123861": 27, "015847": 27, "848602": 27, "215875": 27, "251406": 27, "795724": 27, "092080": 27, "070939": 27, "705117": 27, "241584": 27, "766805": 27, "132841": 27, "531804": 27, "564560": 27, "832337": 27, "442301": 27, "472336": 27, "359856": 27, "911472": 27, "795981": 27, "391838": 27, "rank2": 27, "regul": [27, 29], "top20": 27, "upregul": 27, "goi": 27, "colors_dict": 27, "row_ha_left": 27, "row_ha_right": 27, "uncom": 27, "sn": 28, "subset": 28, "collaps": 28, "load_dataset": 28, "brain_network": 28, "header": 28, "used_network": 28, "used_column": 28, "get_level_valu": 28, "comput": 28, "form": 28, "corr_mat": 28, "corr": 28, "level": [28, 34], "level_0": 28, "level_1": 28, "lh": 28, "rh": 28, "881516": 28, "049793": 28, "026902": 28, "144335": 28, "141253": 28, "119250": 28, "261589": 28, "272701": 28, "370021": 28, "366065": 28, "325680": 28, "196770": 28, "144566": 28, "366818": 28, "388756": 28, "352529": 28, "363982": 28, "341524": 28, "350452": 28, "112697": 28, "036909": 28, "144277": 28, "189683": 28, "084633": 28, "324230": 28, "332886": 28, "374322": 28, "361036": 28, "274151": 28, "142392": 28, "070452": 28, "358625": 28, "402173": 28, "302286": 28, "339989": 28, "315931": 28, "343379": 28, "343464": 28, "470239": 28, "100802": 28, "438449": 28, "339667": 28, "089811": 28, "272394": 28, "036493": 28, "171179": 28, "043298": 28, "158039": 28, "005598": 28, "060007": 28, "079078": 28, "040060": 28, "027878": 28, "075781": 28, "130549": 28, "278569": 28, "127621": 28, "014404": 28, "051249": 28, "090130": 28, "170053": 28, "124278": 28, "112148": 28, "063705": 28, "172007": 28, "040629": 28, "079687": 28, "024864": 28, "092263": 28, "068858": 28, "521377": 28, "506652": 28, "320966": 28, "141884": 28, "608392": 28, "075986": 28, "095015": 28, "012966": 28, "082816": 28, "023340": 28, "058718": 28, "034181": 28, "033355": 28, "022982": 28, "025638": 28, "431619": 28, "418708": 28, "084634": 28, "132": [28, 29], "808800071564": 28, "22361111111111": 28, "wding": 28, "miniconda3": 28, "env": 28, "lib": 28, "python3": 28, "site": 28, "189": 28, "userwarn": 28, "via": 28, "norm": 28, "465277777777779": 28, "drop_dupl": 28, "rowgroup": 28, "kycg_result": 28, "rmsk1": 28, "ensregbuild": 28, "sampleid": [28, 32], "clark2018_argelaguet2019": 28, "dataset1": 28, "luo2022": 28, "dataset2": 28, "max_p": 28, "log10": [28, 29], "pval": 28, "id": [28, 29, 34], "cpgtype": 28, "vc": 28, "term": [28, 29], "p_max": 28, "p_min": 28, "odds_ratio": 28, "pvalu": 28, "enrichtyp": 28, "het": 28, "061": 28, "neg": 28, "020000e": 28, "07": [28, 29], "26": 28, "tx": 28, "029": 28, "580000e": 28, "65": 28, "tssflnk": 28, "056": 28, "370000e": 28, "06": [28, 29], "022": 28, "660000e": 28, "346": 28, "350": 28, "760000e": 28, "02": 28, "describ": 28, "64": 28, "356918": 28, "632073": 28, "119186": 28, "deplet": 28, "srprna": 28, "satellit": 28, "simple_repeat": 28, "low_complex": 28, "reprpcwk": 28, "tssbiv": 28, "reprpc": 28, "ltr": 28, "snrna": 28, "sine": 28, "rdylgn_r": 28, "coolwarm_r": 28, "dotheatmap1": 28, "180": 28, "09263168093761": 28, "20230227_kycg": 28, "516666666666666": 28, "glob": 29, "style": 29, "tradition": 29, "lot": [29, 31], "dotplot": 29, "had": 29, "idea": 29, "manuscript": 29, "alreadi": 29, "gotten": 29, "bp": 29, "molecular": 29, "mf": 29, "kegg": 29, "pathwai": 29, "enrichment_analysis_dotheatmap": 29, "data1": 29, "descript": 29, "generatio": 29, "enrichedcategori": 29, "colid": 29, "rowid": 29, "0003779": 29, "actin": 29, "bind": 29, "044949": 29, "453212e": 29, "186": 29, "group2": 29, "ct1": 29, "610265": 29, "0030695": 29, "gtpase": 29, "activ": 29, "043016": 29, "744023e": 29, "178": 29, "171081": 29, "0060589": 29, "nucleosid": 29, "triphosphatas": 29, "0005201": 29, "extracellular": 29, "structur": 29, "constitu": 29, "021266": 29, "810239e": 29, "88": 29, "34": 29, "235806": 29, "0050839": 29, "adhes": 29, "molecul": 29, "031899": 29, "612420e": 29, "792522": 29, "840": 29, "0050905": 29, "neuromuscular": 29, "010454": 29, "224335e": 29, "122": 29, "ct2": 29, "912100": 29, "1205": 29, "0030534": 29, "adult": 29, "behavior": 29, "009426": 29, "028861e": 29, "110": 29, "219765": 29, "1558": 29, "0035019": 29, "somat": 29, "stem": 29, "popul": 29, "mainten": 29, "004799": 29, "029991e": 29, "219683": 29, "1746": 29, "0019827": 29, "009854": 29, "467128e": 29, "04": 29, "833532": 29, "2999": 29, "0021953": 29, "central": 29, "nervou": 29, "system": 29, "neuron": [29, 34], "008997": 29, "138960e": 29, "105": 29, "039103": 29, "139": 29, "metal": 29, "ion": 29, "transmembran": 29, "transport": 29, "monoatom": 29, "channel": 29, "gate": 29, "calmodulin": 29, "receptor": 29, "protein": 29, "kinas": 29, "substrat": 29, "organ": 29, "extern": 29, "encapsul": 29, "axon": 29, "guidanc": 29, "locomotori": 29, "fat": 29, "amphetamin": 29, "addict": 29, "calcium": 29, "signal": 29, "cocain": 29, "cytokin": 29, "interact": 29, "neuroact": 29, "ligand": 29, "cytoskeleton": 29, "ecm": 29, "viral": 29, "cy": 29, "focal": 29, "lupu": 29, "erythematosu": 29, "atp": 29, "chromatin": 29, "remodel": 29, "histon": 29, "modif": 29, "group1": 29, "ct3": 29, "ct4": 29, "s_min": 29, "s_max": 29, "3074": 29, "0021954": 29, "004586": 29, "027429": 29, "561784": 29, "0045444": 29, "176471": 29, "000382": 29, "417494": 29, "orrd": 29, "1e": 29, "ast": 29, "ravel": 29, "both": 29, "dashdot": 29, "cell_class_dotheatmap": 29, "106": 29, "46754159267321": 29, "ratio": 29, "total": 29, "510843": 30, "531995": 30, "188667": 30, "180811": 30, "354718": 30, "whole": 30, "separ": 30, "wiki": 31, "ital": 31, "_label_kw": 31, "tree": 31, "roundtooth": 31, "book": 32, "cbioport": 32, "tcga_lung_adenocarcinoma_provisional_ras_raf_mek_jnk_signal": 32, "dropna": 32, "mut": 32, "amp": [32, 33], "homdel": 32, "4384": 32, "kra": 32, "hra": 32, "braf": 32, "raf1": 32, "map3k1": 32, "unique_vari": 32, "v1": 32, "strip": 32, "008000": 32, "mutat": 32, "frequenc": 32, "row_vc": 32, "col_vc": 32, "kras_sampl": 32, "df_col_split": 32, "op": [32, 33], "alter": 32, "27": [32, 33], "72361111111111": 32, "ad": 32, "row_var_freq": 32, "isvar": 32, "oncoprint2": 32, "430555555555557": 32, "toi": 33, "sample_": 33, "gene_": 33, "alts_lol": 33, "amp_valu": 33, "randint": 33, "del_valu": 33, "neut_valu": 33, "alts_df": 33, "neut": 33, "prepar": 33, "annot_1_df": 33, "annot1": 33, "annot_2_df": 33, "500": 33, "annot2": 33, "annot_3_df": 33, "patient": 33, "sample_1": 33, "gene_1": 33, "gene_2": 33, "gene_3": 33, "gene_4": 33, "gene_5": 33, "95": 33, "sample_10": 33, "gene_6": 33, "96": 33, "gene_7": 33, "97": 33, "gene_8": 33, "98": 33, "gene_9": 33, "gene_10": 33, "patient2": 33, "sample_2": 33, "patient5": 33, "sample_3": 33, "patient3": 33, "sample_4": 33, "patient4": 33, "sample_5": 33, "patient1": 33, "sample_6": 33, "sample_7": 33, "sample_8": 33, "sample_9": 33, "77": 33, "78": 33, "1114": 33, "2698": 33, "4718": 33, "4633": 33, "3444": 33, "1984": 33, "2088": 33, "2316": 33, "2939": 33, "1373": 33, "a3": [33, 35], "a1": [33, 35], "a2": [33, 35], "flatten": 33, "signatur": 34, "come": 34, "36599988": 34, "raw": 34, "githubusercont": 34, "adipocyt": 34, "bladder": 34, "epithelium": 34, "mem": 34, "granulocyt": 34, "colon": 34, "macrophag": 34, "monocyt": 34, "interstiti": 34, "alveolar": 34, "pancrea": 34, "duct": 34, "skelet": 34, "endocrin": 34, "bronchu": 34, "smooth": 34, "prostat": 34, "coronari": 34, "arteri": 34, "aorta": 34, "thyroid": 34, "110426397": 34, "120667": 34, "9102": 34, "8285": 34, "918667": 34, "750667": 34, "8705": 34, "896": 34, "916333": 34, "953333": 34, "9450": 34, "61225": 34, "5485": 34, "94175": 34, "952": 34, "417": 34, "150": 34, "229": 34, "294": 34, "214": 34, "938333": 34, "110426639": 34, "420000": 34, "9304": 34, "8920": 34, "955667": 34, "964667": 34, "9285": 34, "903": 34, "924667": 34, "975333": 34, "9720": 34, "94575": 34, "8990": 34, "96225": 34, "963": 34, "606": 34, "185": 34, "348": 34, "917": 34, "484": 34, "972333": 34, "110426659": 34, "425667": 34, "9328": 34, "7635": 34, "933000": 34, "942333": 34, "8945": 34, "931": 34, "963667": 34, "899333": 34, "8895": 34, "93875": 34, "8390": 34, "92225": 34, "559": 34, "129": 34, "292": 34, "395": 34, "989667": 34, "110427109": 34, "203667": 34, "9724": 34, "9610": 34, "968667": 34, "9240": 34, "939": 34, "985000": 34, "9620": 34, "73300": 34, "7570": 34, "96525": 34, "929": 34, "225": 34, "806": 34, "951": 34, "905": 34, "974333": 34, "110427116": 34, "217333": 34, "9478": 34, "0000": 34, "976000": 34, "970667": 34, "9730": 34, "913": 34, "975000": 34, "970333": 34, "9800": 34, "65125": 34, "7955": 34, "96150": 34, "000": 34, "605": 34, "857": 34, "868": 34, "981000": 34, "clusterid": 34, "ep": 34, "granul": 34, "mono": 34, "macro": 34, "nk": 34, "bone": 34, "osteob": 34, "breast": 34, "basal": 34, "dermal": 34, "fibro": 34, "epid": 34, "kerat": 34, "eryth": 34, "prog": 34, "fallopian": 34, "gallbladd": 34, "gastric": 34, "neck": 34, "heart": 34, "cardio": 34, "kidnei": 34, "hep": 34, "alveo": 34, "bron": 34, "oligodend": 34, "acinar": 34, "musc": 34, "1e93a": 34, "ff9c00": 34, "ff9f7f": 34, "ff7f00": 34, "ff2e8d": 34, "cc0043": 34, "e5e5e5": 34, "cc4407": 34, "cc843d": 34, "663d28": 34, "1e937c": 34, "40705f": 34, "009351": 34, "e7e4bf": 34, "cca300": 34, "002929": 34, "ff99aa": 34, "ff99ff": 34, "f6ff99": 34, "ba99ff": 34, "ccccff": 34, "9e542": 34, "2ca02c": 34, "df7f00": 34, "ffd866": 34, "ffcc32": 34, "7f4c33": 34, "cc9951": 34, "b2bfff": 34, "loyfer2023_heatmap": 34, "981944444444444": 34, "download": 35, "pycomplexheatmap_test_2": 35, "c1": 35, "c2": 35, "c3": 35, "c4": 35, "c5": 35, "c6": 35, "c7": 35, "c8": 35, "c9": 35, "c10": 35, "c35": 35, "c36": 35, "c37": 35, "c38": 35, "c39": 35, "c40": 35, "c41": 35, "c42": 35, "c43": 35, "c44": 35, "00000": 35, "00": 35, "250000": 35, "111111": 35, "444444": 35, "222222": 35, "388889": 35, "611111": 35, "277778": 35, "888889": 35, "944444": 35, "555556": 35, "500000": 35, "a4": 35, "625000": 35, "375000": 35, "a5": 35, "a6": 35, "600000": 35, "100000": 35, "300000": 35, "700000": 35, "a7": 35, "a8": 35, "60000": 35, "800000": 35, "400000": 35, "200000": 35, "a9": 35, "440000": 35, "72": 35, "460000": 35, "040000": 35, "080000": 35, "560000": 35, "020000": 35, "240000": 35, "a10": 35, "a11": 35, "357143": 35, "02381": 35, "023810": 35, "071429": 35, "571429": 35, "214286": 35, "452381": 35, "166667": 35, "380952": 35, "309524": 35, "a12": 35, "062500": 35, "031250": 35, "312500": 35, "218750": 35, "531250": 35, "156250": 35, "687500": 35, "343750": 35, "437500": 35, "a13": 35, "333333": 35, "666667": 35, "a14": 35, "a15": 35, "076923": 35, "615385": 35, "384615": 35, "538462": 35, "307692": 35, "153846": 35, "a16": 35, "a17": 35, "150000": 35, "900000": 35, "550000": 35, "350000": 35, "a18": 35, "833333": 35, "a19": 35, "rdpu": 35}, "objects": {"": [[0, 0, 0, "-", "PyComplexHeatmap"]], "PyComplexHeatmap": [[1, 0, 0, "-", "annotations"], [2, 0, 0, "-", "clustermap"], [3, 0, 0, "-", "colors"], [4, 0, 0, "-", "dotHeatmap"], [5, 0, 0, "-", "example"], [6, 0, 0, "-", "oncoPrint"], [8, 0, 0, "-", "utils"]], "PyComplexHeatmap.annotations": [[1, 1, 1, "", "AnnotationBase"], [1, 1, 1, "", "HeatmapAnnotation"], [1, 1, 1, "", "anno_barplot"], [1, 1, 1, "", "anno_boxplot"], [1, 1, 1, "", "anno_img"], [1, 1, 1, "", "anno_label"], [1, 1, 1, "", "anno_lineplot"], [1, 1, 1, "", "anno_scatterplot"], [1, 1, 1, "", "anno_simple"]], "PyComplexHeatmap.annotations.AnnotationBase": [[1, 2, 1, "", "get_label_width"], [1, 2, 1, "", "get_max_label_width"], [1, 2, 1, "", "get_ticklabel_width"], [1, 2, 1, "", "reorder"], [1, 2, 1, "", "set_axes_kws"], [1, 2, 1, "", "set_label"], [1, 2, 1, "", "set_legend"], [1, 2, 1, "", "update_plot_kws"]], "PyComplexHeatmap.annotations.HeatmapAnnotation": [[1, 2, 1, "", "collect_legends"], [1, 2, 1, "", "plot_annotations"], [1, 2, 1, "", "plot_legends"], [1, 2, 1, "", "set_axes_kws"], [1, 2, 1, "", "show_ticklabels"]], "PyComplexHeatmap.annotations.anno_barplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_boxplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_img": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_label": [[1, 2, 1, "", "get_ticklabel_width"], [1, 2, 1, "", "plot"], [1, 2, 1, "", "set_plot_kws"], [1, 2, 1, "", "set_side"]], "PyComplexHeatmap.annotations.anno_lineplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_scatterplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_simple": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.clustermap": [[2, 1, 1, "", "ClusterMapPlotter"], [2, 1, 1, "", "DendrogramPlotter"], [2, 4, 1, "", "composite"], [2, 4, 1, "", "heatmap"], [2, 1, 1, "", "heatmapPlotter"], [2, 4, 1, "", "plot_heatmap"]], "PyComplexHeatmap.clustermap.ClusterMapPlotter": [[2, 2, 1, "", "cal_cold_between_groups"], [2, 2, 1, "", "cal_rowd_between_groups"], [2, 2, 1, "", "calculate_col_dendrograms"], [2, 2, 1, "", "calculate_row_dendrograms"], [2, 2, 1, "", "collect_legends"], [2, 2, 1, "", "format_data"], [2, 2, 1, "", "plot"], [2, 2, 1, "", "plot_dendrograms"], [2, 2, 1, "", "plot_legends"], [2, 2, 1, "", "plot_matrix"], [2, 2, 1, "", "post_processing"], [2, 2, 1, "", "set_axes_labels_kws"], [2, 2, 1, "", "set_height"], [2, 2, 1, "", "set_width"], [2, 2, 1, "", "set_xy_labels"], [2, 2, 1, "", "standard_scale"], [2, 2, 1, "", "tight_layout"], [2, 2, 1, "", "z_score"]], "PyComplexHeatmap.clustermap.DendrogramPlotter": [[2, 2, 1, "", "calculate_dendrogram"], [2, 3, 1, "", "calculated_linkage"], [2, 2, 1, "", "check_array"], [2, 2, 1, "", "get_coords"], [2, 2, 1, "", "plot"], [2, 3, 1, "", "reordered_ind"]], "PyComplexHeatmap.clustermap.heatmapPlotter": [[2, 2, 1, "", "plot"]], "PyComplexHeatmap.colors": [[3, 4, 1, "", "define_cmap"], [3, 4, 1, "", "register_cmap"]], "PyComplexHeatmap.dotHeatmap": [[4, 1, 1, "", "DotClustermapPlotter"], [4, 4, 1, "", "dotHeatmap2d"], [4, 4, 1, "", "scale"]], "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter": [[4, 2, 1, "", "collect_legends"], [4, 2, 1, "", "format_data"], [4, 2, 1, "", "plot_matrix"], [4, 2, 1, "", "post_processing"]], "PyComplexHeatmap.example": [[5, 4, 1, "", "clustermap_example0"], [5, 4, 1, "", "clustermap_example1"], [5, 4, 1, "", "get_kycg_example_data"]], "PyComplexHeatmap.oncoPrint": [[6, 1, 1, "", "oncoPrintPlotter"], [6, 4, 1, "", "oncoprint"]], "PyComplexHeatmap.oncoPrint.oncoPrintPlotter": [[6, 2, 1, "", "add_default_annotations"], [6, 2, 1, "", "collect_legends"], [6, 2, 1, "", "format_data"], [6, 2, 1, "", "get_samples_order"], [6, 2, 1, "", "plot_matrix"], [6, 2, 1, "", "post_processing"]], "PyComplexHeatmap.utils": [[8, 4, 1, "", "axis_ticklabels_overlap"], [8, 4, 1, "", "cal_legend_width"], [8, 4, 1, "", "cluster_labels"], [8, 4, 1, "", "define_cmap"], [8, 4, 1, "", "despine"], [8, 4, 1, "", "get_colormap"], [8, 4, 1, "", "plot_cmap_legend"], [8, 4, 1, "", "plot_color_dict_legend"], [8, 4, 1, "", "plot_legend_list"], [8, 4, 1, "", "plot_marker_legend"], [8, 4, 1, "", "set_default_style"], [8, 4, 1, "", "to_utf8"]]}, "objtypes": {"0": "py:module", "1": "py:class", "2": "py:method", "3": "py:property", "4": "py:function"}, "objnames": {"0": ["py", "module", "Python module"], "1": ["py", "class", "Python class"], "2": ["py", "method", "Python method"], "3": ["py", "property", "Python property"], "4": ["py", "function", "Python function"]}, "titleterms": {"pycomplexheatmap": [0, 1, 2, 3, 4, 5, 6, 7, 8, 16, 19, 21], "packag": [0, 22, 30], "submodul": 0, "modul": [0, 1, 2, 3, 4, 5, 6, 7, 8, 25, 37], "content": [0, 9, 11, 12, 13, 15, 16, 18, 20, 36], "annot": [1, 21, 23, 30, 31], "clustermap": [2, 11, 25, 28], "color": [3, 28], "dotheatmap": [4, 13, 29], "exampl": [5, 20, 23, 28], "oncoprint": [6, 32, 33, 36], "tool": 7, "util": 8, "advanc": 9, "usag": 9, "citat": 10, "For": 12, "develop": 12, "galleri": 14, "get": 15, "start": 15, "credit": 16, "thank": 16, "instal": 17, "depend": 17, "how": [17, 21, 29, 31], "kwarg": 18, "more": 20, "gener": 21, "dataset": [21, 24, 28, 32], "add": [21, 23, 28], "select": [21, 29], "row": [21, 23, 30], "label": [21, 31], "top": [21, 23], "heatmap": [21, 24, 28, 30, 31], "cell": [21, 34], "onli": [21, 23], "plot": [21, 23, 25, 28, 29, 30], "chang": [21, 28], "orent": 21, "down": 21, "extra": 21, "space": 21, "left": [21, 23], "right": [21, 23], "orient": 21, "us": [21, 25, 28, 29, 33], "paramet": [21, 28], "multipl": 21, "loop": 21, "cluster": 21, "between": [21, 22], "group": [21, 22, 28], "within": [21, 22], "clsuter": 21, "col_split": 21, "col_clust": 21, "true": 21, "cluster_between_group": 21, "col_split_ord": 21, "fals": 21, "cluster_within_group": 21, "custom": 21, "linkag": 21, "imag": 21, "choos": 21, "build": 21, "colormap": 21, "diverg": 21, "qualit": 21, "cmap": [21, 28], "forc": 21, "displai": 21, "all": 21, "col": 21, "ticklabel": 21, "import": [22, 30], "dmr": 22, "A": [23, 25], "quick": 23, "column": [23, 30], "anno_label": [23, 31], "anno_simpl": 23, "To": 23, "quickli": 23, "you": 23, "just": 23, "need": [23, 29], "datafram": 23, "figur": 23, "legend": 23, "separ": 23, "bottom": 23, "composit": 24, "two": 24, "horizont": 24, "mous": 24, "dna": [24, 34], "methyl": [24, 34], "arrai": 24, "correl": 25, "matrix": 25, "cpg": 25, "dotclustermap": 25, "process": 25, "data": [25, 28, 29, 34], "dot": [25, 28], "smaller": 25, "1": [26, 30], "understand": 26, "layout": 26, "visual": [27, 28, 29, 32, 33, 34], "differenti": 27, "express": 27, "gene": [27, 29], "load": 28, "an": 28, "brain": 28, "network": 28, "from": 28, "seaborn": 28, "tradit": 28, "squar": 28, "marker": [28, 29], "": 28, "simpl": 28, "fix": 28, "size": 28, "point": 28, "hue": 28, "differ": 28, "up": 28, "five": 28, "dimens": 28, "dotclustermapplott": 28, "set": 29, "enrich": 29, "analysi": 29, "result": 29, "clusterprofil": 29, "why": 29, "do": 29, "we": 29, "better": 29, "wai": 29, "read": 29, "2": 30, "3": 30, "split": 30, "4": 30, "along": 30, "side": 30, "main": 30, "label_kw": 31, "xticklabels_kw": 31, "yticklabels_kw": 31, "modifi": 31, "xticklabel": 31, "yticklabel": 31, "xlabel_kw": 31, "ylabel_kw": 31, "xlabel": 31, "ylabel": 31, "control": 31, "gap": 31, "pad": 31, "hgap": 31, "wgap": 31, "heatmapannot": 31, "subplot_gap": 31, "row_split_gap": 31, "col_split_gap": 31, "remov": 31, "arrow": 31, "tcga": 32, "lung": 32, "adenocarcinoma": 32, "carcinoma": 32, "variant": 32, "categor": 33, "variabl": 33, "singl": 34, "loyfer2023": 34, "setup": 37}, "envversion": {"sphinx.domains.c": 3, "sphinx.domains.changeset": 1, "sphinx.domains.citation": 1, "sphinx.domains.cpp": 9, "sphinx.domains.index": 1, "sphinx.domains.javascript": 3, "sphinx.domains.math": 2, "sphinx.domains.python": 4, "sphinx.domains.rst": 2, "sphinx.domains.std": 2, "sphinx.ext.todo": 2, "sphinx.ext.viewcode": 1, "nbsphinx": 4, "sphinx": 58}, "alltitles": {"PyComplexHeatmap package": [[0, "pycomplexheatmap-package"]], "Submodules": [[0, "submodules"]], "Module contents": [[0, "module-PyComplexHeatmap"]], "PyComplexHeatmap.annotations module": [[1, "module-PyComplexHeatmap.annotations"]], "PyComplexHeatmap.clustermap module": [[2, "module-PyComplexHeatmap.clustermap"]], "PyComplexHeatmap.colors module": [[3, "module-PyComplexHeatmap.colors"]], "PyComplexHeatmap.dotHeatmap module": [[4, "module-PyComplexHeatmap.dotHeatmap"]], "PyComplexHeatmap.example module": [[5, "module-PyComplexHeatmap.example"]], "PyComplexHeatmap.oncoPrint module": [[6, "module-PyComplexHeatmap.oncoPrint"]], "PyComplexHeatmap.tools module": [[7, "pycomplexheatmap-tools-module"]], "PyComplexHeatmap.utils module": [[8, "module-PyComplexHeatmap.utils"]], "Advanced Usage": [[9, "advanced-usage"]], "Contents:": [[9, null], [11, null], [12, null], [13, null], [15, null], [16, null], [18, null], [20, null], [36, null]], "Citation": [[10, "citation"]], "Clustermap": [[11, "clustermap"]], "For Developers": [[12, "for-developers"]], "dotHeatmap": [[13, "dotheatmap"]], "Gallery": [[14, "gallery"]], "Get Started": [[15, "get-started"]], "PyComplexHeatmap": [[16, "pycomplexheatmap"], [19, "pycomplexheatmap"]], "Credits and Thanks": [[16, "credits-and-thanks"]], "Installation": [[17, "installation"]], "Dependencies": [[17, "dependencies"]], "How to install?": [[17, "how-to-install"]], "Kwargs": [[18, "kwargs"]], "More Examples": [[20, "more-examples"]], "Generate dataset": [[21, "Generate-dataset"]], "Add selected rows labels": [[21, "Add-selected-rows-labels"]], "Add annotations on the top of heatmap cells": [[21, "Add-annotations-on-the-top-of-heatmap-cells"]], "Only plot the annotations": [[21, "Only-plot-the-annotations"]], "Change orentation to down and add extra space": [[21, "Change-orentation-to-down-and-add-extra-space"]], "Change orentation to the left": [[21, "Change-orentation-to-the-left"]], "Change orentation to the right": [[21, "Change-orentation-to-the-right"]], "Changing orientation using parameter orientation": [[21, "Changing-orientation-using-parameter-orientation"]], "Add multiple heatmap annotations using for loop": [[21, "Add-multiple-heatmap-annotations-using-for-loop"]], "Cluster between groups and cluster within groups": [[21, "Cluster-between-groups-and-cluster-within-groups"]], "clsuter within groups: col_split=*, col_cluster=True": [[21, "clsuter-within-groups:-col_split=*,-col_cluster=True"]], "cluster_between_groups: col_split=*, col_split_order=\"cluster_between_groups\",col_cluster=False": [[21, "cluster_between_groups:-col_split=*,-col_split_order=%22cluster_between_groups%22,col_cluster=False"]], "cluster_within_groups && cluster_between_groups: col_split=*, col_split_order=\"cluster_between_groups\",col_cluster=True": [[21, "cluster_within_groups-&&-cluster_between_groups:-col_split=*,-col_split_order=%22cluster_between_groups%22,col_cluster=True"]], "Custom annotation": [[21, "Custom-annotation"]], "Custom linkage": [[21, "Custom-linkage"]], "Image annotation": [[21, "Image-annotation"]], "Choosing Build-in colormap in PyComplexHeatmap": [[21, "Choosing-Build-in-colormap-in-PyComplexHeatmap"]], "Diverging": [[21, "Diverging"]], "Qualitative": [[21, "Qualitative"]], "How to use the Build-in cmap?": [[21, "How-to-use-the-Build-in-cmap?"]], "How to force display all row/col ticklabels?": [[21, "How-to-force-display-all-row/col-ticklabels?"]], "Import packages": [[22, "Import-packages"]], "Between groups DMRs": [[22, "Between-groups-DMRs"]], "Within groups DMRs": [[22, "Within-groups-DMRs"]], "A quick example": [[23, "A-quick-example"]], "Plotting annotations": [[23, "Plotting-annotations"]], "Only plot the row/column annotation": [[23, "Only-plot-the-row/column-annotation"]], "anno_label:": [[23, "anno_label:"]], "anno_simple:": [[23, "anno_simple:"]], "To add a annotation quickly, you just need a dataframe": [[23, "To-add-a-annotation-quickly,-you-just-need-a-dataframe"]], "Plot the figure and legend separately": [[23, "Plot-the-figure-and-legend-separately"]], "Top, bottom, left ,right annotations": [[23, "Top,-bottom,-left-,right-annotations"]], "Composite two heatmaps horizontally for mouse DNA methylation array dataset": [[24, "Composite-two-heatmaps-horizontally-for-mouse-DNA-methylation-array-dataset"]], "Plot correlation matrix for CpG modules using DotClustermap": [[25, "Plot-correlation-matrix-for-CpG-modules-using-DotClustermap"]], "Processing the data": [[25, "Processing-the-data"]], "Plotting the Dot clustermap": [[25, "Plotting-the-Dot-clustermap"]], "A smaller dot clustermap": [[25, "A-smaller-dot-clustermap"]], "1. Understand the layout:": [[26, "1.-Understand-the-layout:"]], "Visualizing Differential Expression Genes": [[27, "Visualizing-Differential-Expression-Genes"]], "Load an example brain networks dataset from seaborn": [[28, "Load-an-example-brain-networks-dataset-from-seaborn"]], "Dot Heatmap": [[28, "Dot-Heatmap"]], "Plot traditional heatmap using square marker marker='s'": [[28, "Plot-traditional-heatmap-using-square-marker-marker='s'"]], "Simple dot heatmap using fixed dot size": [[28, "Simple-dot-heatmap-using-fixed-dot-size"]], "Changing the size of point": [[28, "Changing-the-size-of-point"]], "Add parameter hue and use different colors for different groups": [[28, "Add-parameter-hue-and-use-different-colors-for-different-groups"]], "Add parameter hue and use different cmap and marker for different groups": [[28, "Add-parameter-hue-and-use-different-cmap-and-marker-for-different-groups"]], "Dot Clustermap": [[28, "Dot-Clustermap"]], "Plot clustermap using seaborn brain networks dataset": [[28, "Plot-clustermap-using-seaborn-brain-networks-dataset"]], "Visualize up to five dimension data using DotClustermapPlotter": [[28, "Visualize-up-to-five-dimension-data-using-DotClustermapPlotter"]], "Visualizing gene set enrichment analysis results (clusterProfiler) using dotHeatmap": [[29, "Visualizing-gene-set-enrichment-analysis-results-(clusterProfiler)-using-dotHeatmap"]], "Why do we need a better way to visualize the gene set enrichment analysis result?": [[29, "Why-do-we-need-a-better-way-to-visualize-the-gene-set-enrichment-analysis-result?"]], "Read data": [[29, "Read-data"]], "Plot": [[29, "Plot"]], "How to select marker:": [[29, "How-to-select-marker:"]], "1. Import packages": [[30, "1.-Import-packages"]], "2. Plot heatmap annotations": [[30, "2.-Plot-heatmap-annotations"]], "3. Plot the heatmap with rows and columns split": [[30, "3.-Plot-the-heatmap-with-rows-and-columns-split"]], "4. Plot the annotations along side with main heatmap": [[30, "4.-Plot-the-annotations-along-side-with-main-heatmap"]], "label_kws, xticklabels_kws and yticklabels_kws: Modifying the heatmap annotations labels and xticklabels, yticklabels": [[31, "label_kws,-xticklabels_kws-and-yticklabels_kws:-Modifying-the-heatmap-annotations-labels-and-xticklabels,-yticklabels"]], "xlabel_kws and ylabel_kws: Modifying xlabel and ylabel": [[31, "xlabel_kws-and-ylabel_kws:-Modifying-xlabel-and-ylabel"]], "Control gap & pad in heatmap": [[31, "Control-gap-&-pad-in-heatmap"]], "hgap and wgap for HeatmapAnnotation": [[31, "hgap-and-wgap-for-HeatmapAnnotation"]], "subplot_gap": [[31, "subplot_gap"]], "row_split_gap and col_split_gap": [[31, "row_split_gap-and-col_split_gap"]], "How to remove arrow in anno_label": [[31, "How-to-remove-arrow-in-anno_label"]], "oncoPrint: visualizing TCGA Lung Adenocarcinoma Carcinoma Variants Dataset": [[32, "oncoPrint:-visualizing-TCGA-Lung-Adenocarcinoma-Carcinoma-Variants-Dataset"]], "Visualizing categorical variables using oncoPrint": [[33, "Visualizing-categorical-variables-using-oncoPrint"]], "Visualizing Single Cell DNA Methylation Data (Loyfer2023)": [[34, "Visualizing-Single-Cell-DNA-Methylation-Data-(Loyfer2023)"]], "oncoPrint": [[36, "oncoprint"]], "setup module": [[37, "setup-module"]]}, "indexentries": {"pycomplexheatmap": [[0, "module-PyComplexHeatmap"]], "module": [[0, "module-PyComplexHeatmap"], [1, "module-PyComplexHeatmap.annotations"], [2, "module-PyComplexHeatmap.clustermap"], [3, "module-PyComplexHeatmap.colors"], [4, "module-PyComplexHeatmap.dotHeatmap"], [5, "module-PyComplexHeatmap.example"], [6, "module-PyComplexHeatmap.oncoPrint"], [8, "module-PyComplexHeatmap.utils"]], "annotationbase (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.AnnotationBase"]], "heatmapannotation (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation"]], "pycomplexheatmap.annotations": [[1, "module-PyComplexHeatmap.annotations"]], "anno_barplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_barplot"]], "anno_boxplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_boxplot"]], "anno_img (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_img"]], "anno_label (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_label"]], "anno_lineplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_lineplot"]], "anno_scatterplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_scatterplot"]], "anno_simple (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_simple"]], "collect_legends() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.collect_legends"]], "get_label_width() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.get_label_width"]], "get_max_label_width() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.get_max_label_width"]], "get_ticklabel_width() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.get_ticklabel_width"]], "get_ticklabel_width() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.get_ticklabel_width"]], "plot() (pycomplexheatmap.annotations.anno_barplot method)": [[1, "PyComplexHeatmap.annotations.anno_barplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_boxplot method)": [[1, "PyComplexHeatmap.annotations.anno_boxplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_img method)": [[1, "PyComplexHeatmap.annotations.anno_img.plot"]], "plot() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.plot"]], "plot() (pycomplexheatmap.annotations.anno_lineplot method)": [[1, "PyComplexHeatmap.annotations.anno_lineplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_scatterplot method)": [[1, "PyComplexHeatmap.annotations.anno_scatterplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_simple method)": [[1, "PyComplexHeatmap.annotations.anno_simple.plot"]], "plot_annotations() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.plot_annotations"]], "plot_legends() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.plot_legends"]], "reorder() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.reorder"]], "set_axes_kws() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.set_axes_kws"]], "set_axes_kws() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.set_axes_kws"]], "set_label() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.set_label"]], "set_legend() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.set_legend"]], "set_plot_kws() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.set_plot_kws"]], "set_side() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.set_side"]], "show_ticklabels() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.show_ticklabels"]], "update_plot_kws() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.update_plot_kws"]], "clustermapplotter (class in pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter"]], "dendrogramplotter (class in pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter"]], "pycomplexheatmap.clustermap": [[2, "module-PyComplexHeatmap.clustermap"]], "cal_cold_between_groups() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.cal_cold_between_groups"]], "cal_rowd_between_groups() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.cal_rowd_between_groups"]], "calculate_col_dendrograms() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.calculate_col_dendrograms"]], "calculate_dendrogram() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.calculate_dendrogram"]], "calculate_row_dendrograms() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.calculate_row_dendrograms"]], "calculated_linkage (pycomplexheatmap.clustermap.dendrogramplotter property)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.calculated_linkage"]], "check_array() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.check_array"]], "collect_legends() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.collect_legends"]], "composite() (in module pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.composite"]], "format_data() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.format_data"]], "get_coords() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.get_coords"]], "heatmap() (in module pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.heatmap"]], "heatmapplotter (class in pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.heatmapPlotter"]], "plot() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot"]], "plot() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.plot"]], "plot() (pycomplexheatmap.clustermap.heatmapplotter method)": [[2, "PyComplexHeatmap.clustermap.heatmapPlotter.plot"]], "plot_dendrograms() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot_dendrograms"]], "plot_heatmap() (in module pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.plot_heatmap"]], "plot_legends() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot_legends"]], "plot_matrix() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot_matrix"]], "post_processing() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.post_processing"]], "reordered_ind (pycomplexheatmap.clustermap.dendrogramplotter property)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.reordered_ind"]], "set_axes_labels_kws() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_axes_labels_kws"]], "set_height() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_height"]], "set_width() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_width"]], "set_xy_labels() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_xy_labels"]], "standard_scale() (pycomplexheatmap.clustermap.clustermapplotter static method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.standard_scale"]], "tight_layout() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.tight_layout"]], "z_score() (pycomplexheatmap.clustermap.clustermapplotter static method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.z_score"]], "pycomplexheatmap.colors": [[3, "module-PyComplexHeatmap.colors"]], "define_cmap() (in module pycomplexheatmap.colors)": [[3, "PyComplexHeatmap.colors.define_cmap"]], "register_cmap() (in module pycomplexheatmap.colors)": [[3, "PyComplexHeatmap.colors.register_cmap"]], "dotclustermapplotter (class in pycomplexheatmap.dotheatmap)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter"]], "pycomplexheatmap.dotheatmap": [[4, "module-PyComplexHeatmap.dotHeatmap"]], "collect_legends() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.collect_legends"]], "dotheatmap2d() (in module pycomplexheatmap.dotheatmap)": [[4, "PyComplexHeatmap.dotHeatmap.dotHeatmap2d"]], "format_data() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.format_data"]], "plot_matrix() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.plot_matrix"]], "post_processing() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.post_processing"]], "scale() (in module pycomplexheatmap.dotheatmap)": [[4, "PyComplexHeatmap.dotHeatmap.scale"]], "pycomplexheatmap.example": [[5, "module-PyComplexHeatmap.example"]], "clustermap_example0() (in module pycomplexheatmap.example)": [[5, "PyComplexHeatmap.example.clustermap_example0"]], "clustermap_example1() (in module pycomplexheatmap.example)": [[5, "PyComplexHeatmap.example.clustermap_example1"]], "get_kycg_example_data() (in module pycomplexheatmap.example)": [[5, "PyComplexHeatmap.example.get_kycg_example_data"]], "pycomplexheatmap.oncoprint": [[6, "module-PyComplexHeatmap.oncoPrint"]], "add_default_annotations() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.add_default_annotations"]], "collect_legends() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.collect_legends"]], "format_data() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.format_data"]], "get_samples_order() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.get_samples_order"]], "oncoprintplotter (class in pycomplexheatmap.oncoprint)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter"]], "oncoprint() (in module pycomplexheatmap.oncoprint)": [[6, "PyComplexHeatmap.oncoPrint.oncoprint"]], "plot_matrix() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.plot_matrix"]], "post_processing() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.post_processing"]], "pycomplexheatmap.utils": [[8, "module-PyComplexHeatmap.utils"]], "axis_ticklabels_overlap() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.axis_ticklabels_overlap"]], "cal_legend_width() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.cal_legend_width"]], "cluster_labels() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.cluster_labels"]], "define_cmap() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.define_cmap"]], "despine() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.despine"]], "get_colormap() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.get_colormap"]], "plot_cmap_legend() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_cmap_legend"]], "plot_color_dict_legend() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_color_dict_legend"]], "plot_legend_list() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_legend_list"]], "plot_marker_legend() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_marker_legend"]], "set_default_style() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.set_default_style"]], "to_utf8() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.to_utf8"]]}})
\ No newline at end of file
+Search.setIndex({"docnames": ["PyComplexHeatmap", "PyComplexHeatmap.annotations", "PyComplexHeatmap.clustermap", "PyComplexHeatmap.colors", "PyComplexHeatmap.dotHeatmap", "PyComplexHeatmap.example", "PyComplexHeatmap.oncoPrint", "PyComplexHeatmap.tools", "PyComplexHeatmap.utils", "advanced_usage", "citation", "clustermap", "dev", "dotHeatmap", "gallery", "get_started", "index", "installation", "kwargs", "modules", "more_examples", "notebooks/advanced_usage", "notebooks/bicluster", "notebooks/clustermap", "notebooks/composite_heatmaps", "notebooks/cpg_modules", "notebooks/dev", "notebooks/differential_expression_genes", "notebooks/dotHeatmap", "notebooks/gene_enrichment_analysis", "notebooks/get_started", "notebooks/kwargs", "notebooks/oncoPrint", "notebooks/oncoPrint2", "notebooks/single_cell_methylation", "notebooks/test_add_anno_img", "oncoPrint", "setup"], "filenames": ["PyComplexHeatmap.rst", "PyComplexHeatmap.annotations.rst", "PyComplexHeatmap.clustermap.rst", "PyComplexHeatmap.colors.rst", "PyComplexHeatmap.dotHeatmap.rst", "PyComplexHeatmap.example.rst", "PyComplexHeatmap.oncoPrint.rst", "PyComplexHeatmap.tools.rst", "PyComplexHeatmap.utils.rst", "advanced_usage.rst", "citation.rst", "clustermap.rst", "dev.rst", "dotHeatmap.rst", "gallery.md", "get_started.rst", "index.rst", "installation.rst", "kwargs.rst", "modules.rst", "more_examples.rst", "notebooks/advanced_usage.ipynb", "notebooks/bicluster.ipynb", "notebooks/clustermap.ipynb", "notebooks/composite_heatmaps.ipynb", "notebooks/cpg_modules.ipynb", "notebooks/dev.ipynb", "notebooks/differential_expression_genes.ipynb", "notebooks/dotHeatmap.ipynb", "notebooks/gene_enrichment_analysis.ipynb", "notebooks/get_started.ipynb", "notebooks/kwargs.ipynb", "notebooks/oncoPrint.ipynb", "notebooks/oncoPrint2.ipynb", "notebooks/single_cell_methylation.ipynb", "notebooks/test_add_anno_img.ipynb", "oncoPrint.rst", "setup.rst"], "titles": ["PyComplexHeatmap package", "PyComplexHeatmap.annotations module", "PyComplexHeatmap.clustermap module", "PyComplexHeatmap.colors module", "PyComplexHeatmap.dotHeatmap module", "PyComplexHeatmap.example module", "PyComplexHeatmap.oncoPrint module", "PyComplexHeatmap.tools module", "PyComplexHeatmap.utils module", "Advanced Usage", "Citation", "Clustermap", "For Developers", "dotHeatmap", "Gallery", "Get Started", "PyComplexHeatmap", "Installation", "Kwargs", "PyComplexHeatmap", "More Examples", "Generate dataset", "Import packages", "A quick example", "Composite two heatmaps horizontally for mouse DNA methylation array dataset", "Plot correlation matrix for CpG modules using DotClustermap", "1. Understand the layout:", "Visualizing Differential Expression Genes", "Load an example brain networks dataset from seaborn", "Visualizing gene set enrichment analysis results (clusterProfiler) using dotHeatmap", "1. Import packages", "label_kws, xticklabels_kws and yticklabels_kws: Modifying the heatmap annotations labels and xticklabels, yticklabels", "oncoPrint: visualizing TCGA Lung Adenocarcinoma Carcinoma Variants Dataset", "Visualizing categorical variables using oncoPrint", "Visualizing Single Cell DNA Methylation Data (Loyfer2023)", "<no title>", "oncoPrint", "setup module"], "terms": {"annot": [0, 2, 4, 6, 9, 11, 15, 16, 18, 19, 22, 24, 26, 28, 29, 32, 33, 34, 35], "annotationbas": [0, 1, 2, 19], "get_label_width": [0, 1], "get_max_label_width": [0, 1], "get_ticklabel_width": [0, 1], "reorder": [0, 1, 2, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "set_axes_kw": [0, 1], "set_label": [0, 1], "set_legend": [0, 1], "update_plot_kw": [0, 1], "heatmapannot": [0, 1, 2, 18, 19, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 32, 33, 34, 35], "collect_legend": [0, 1, 2, 4, 6], "plot_annot": [0, 1], "plot_legend": [0, 1, 2, 21, 23, 27, 34], "show_ticklabel": [0, 1, 21, 23, 30], "anno_barplot": [0, 1, 19, 21, 23, 25, 26, 30, 31, 32, 33], "plot": [0, 1, 2, 4, 6, 8, 9, 11, 13, 15, 16, 20, 22, 24, 27, 31, 32, 33, 34, 35], "anno_boxplot": [0, 1, 19, 21, 23, 26, 30, 31, 35], "anno_img": [0, 1, 19, 21], "anno_label": [0, 1, 16, 18, 19, 21, 22, 24, 25, 26, 27, 28, 29, 30, 32, 33, 34], "set_plot_kw": [0, 1], "set_sid": [0, 1], "anno_lineplot": [0, 1, 19, 21, 31], "anno_scatterplot": [0, 1, 19, 21, 23, 30, 31], "anno_simpl": [0, 1, 19, 21, 22, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "clustermap": [0, 1, 4, 6, 13, 16, 19, 20, 22, 23, 24], "clustermapplott": [0, 2, 4, 6, 19, 21, 22, 23, 24, 26, 27, 30, 31, 34, 35], "cal_cold_between_group": [0, 2], "cal_rowd_between_group": [0, 2], "calculate_col_dendrogram": [0, 2], "calculate_row_dendrogram": [0, 2], "format_data": [0, 2, 4, 6], "plot_dendrogram": [0, 2], "plot_matrix": [0, 2, 4, 6], "post_process": [0, 2, 4, 6], "set_axes_labels_kw": [0, 2], "set_height": [0, 2], "set_width": [0, 2], "set_xy_label": [0, 2], "standard_scal": [0, 2, 4, 6, 22], "tight_layout": [0, 2], "z_score": [0, 2, 4, 6, 21, 27], "dendrogramplott": [0, 2, 19, 22], "calculate_dendrogram": [0, 2], "calculated_linkag": [0, 2], "check_arrai": [0, 2], "get_coord": [0, 2], "reordered_ind": [0, 2], "composit": [0, 2, 16, 19, 20], "heatmap": [0, 1, 2, 4, 6, 9, 13, 15, 16, 18, 19, 20, 23, 25, 27, 32], "heatmapplott": [0, 2, 19], "plot_heatmap": [0, 2, 19], "color": [0, 1, 2, 4, 6, 8, 13, 16, 19, 21, 22, 23, 24, 25, 26, 27, 29, 30, 31, 32, 33, 34, 35], "define_cmap": [0, 3, 8, 19], "register_cmap": [0, 3, 19, 25], "dotheatmap": [0, 16, 19, 28], "dotclustermapplott": [0, 4, 13, 16, 19, 25, 29], "dotheatmap2d": [0, 4, 19], "scale": [0, 2, 4, 19, 22], "exampl": [0, 1, 2, 8, 11, 13, 16, 19, 21, 24, 25, 26, 29, 30, 31, 35], "clustermap_example0": [0, 5, 19], "clustermap_example1": [0, 5, 19], "get_kycg_example_data": [0, 5, 19], "oncoprint": [0, 16, 19], "oncoprintplott": [0, 6, 19, 32, 33], "add_default_annot": [0, 6], "get_samples_ord": [0, 6], "util": [0, 19, 21], "axis_ticklabels_overlap": [0, 8, 19], "cal_legend_width": [0, 8, 19], "cluster_label": [0, 8, 19], "despin": [0, 8, 19, 32, 33], "get_colormap": [0, 8, 19, 21], "plot_cmap_legend": [0, 4, 6, 8, 19], "plot_color_dict_legend": [0, 4, 6, 8, 19], "plot_legend_list": [0, 8, 19], "plot_marker_legend": [0, 4, 8, 19], "set_default_styl": [0, 8, 19], "to_utf8": [0, 8, 19], "class": [1, 2, 4, 6, 22], "df": [1, 21, 22, 23, 24, 26, 28, 30, 31, 35], "none": [1, 2, 4, 6, 8, 21, 22, 23, 24, 26, 27, 30, 31], "cmap": [1, 2, 4, 6, 8, 9, 13, 16, 22, 23, 24, 25, 26, 27, 29, 30, 31, 34, 35], "auto": [1, 2, 21], "height": [1, 2, 8, 21, 22, 23, 25, 26, 27, 29, 30, 31, 32, 33], "legend": [1, 2, 4, 6, 8, 21, 22, 24, 25, 27, 28, 29, 30, 31, 32, 33, 34, 35], "legend_kw": [1, 2, 4, 6, 8, 21, 23, 26, 31, 34], "plot_kw": [1, 6, 33], "sourc": [1, 2, 3, 4, 5, 6, 8, 14], "base": [1, 2, 4, 6, 8, 28], "object": [1, 2, 8, 22, 25, 27, 34], "paramet": [1, 2, 4, 6, 8, 9, 13, 16, 23, 25, 31], "datafram": [1, 2, 4, 6, 21, 26, 27, 30, 31, 32, 33, 35], "panda": [1, 2, 4, 6, 17, 22, 27, 29, 33], "seri": [1, 2], "onli": [1, 4, 6, 9, 16, 31], "one": [1, 4, 8, 23, 29], "column": [1, 2, 4, 6, 15, 16, 21, 22, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "str": [1, 2, 4, 6, 8, 21, 23, 25, 26, 30, 31, 32, 33, 35], "colormap": [1, 2, 4, 6, 9, 16, 25, 28], "set1": [1, 4, 6, 8, 21, 23, 27, 28, 29, 30, 31, 35], "dark2": [1, 8, 21, 23, 25, 27, 28, 30, 35], "bwr": [1, 2], "red": [1, 2, 4, 21, 23, 24, 25, 26, 27, 28, 30, 31, 32, 33], "jet": [1, 2, 21, 23, 25, 28, 30], "hsv": [1, 8], "rainbow": [1, 21, 26, 30, 31], "so": [1, 2], "pleas": [1, 2, 4, 10, 17, 30, 31], "see": [1, 2, 4, 21, 31], "http": [1, 2, 4, 10, 17, 21, 31, 32, 34], "matplotlib": [1, 2, 4, 8, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "org": [1, 2, 4, 10], "3": [1, 4, 15, 16, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "5": [1, 2, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "0": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "tutori": 1, "html": [1, 2, 4, 31, 32], "more": [1, 2, 16, 21, 23, 31, 32], "inform": [1, 2, 31], "run": [1, 17], "pyplot": [1, 21, 25, 27], "all": [1, 2, 4, 8, 9, 16, 23, 29, 31], "availabel": 1, "default": [1, 2, 4, 6, 8, 21, 28, 29, 31], "i": [1, 2, 4, 6, 8, 16, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 33, 35], "would": [1, 2, 21, 23], "determin": [1, 2, 4, 21, 23, 28], "dtype": [1, 2, 21, 22, 25, 26, 27, 28, 34], "each": [1, 2, 6, 23, 29, 30, 33], "dict": [1, 2, 4, 6, 8, 21, 22, 23, 25, 26, 29, 31, 34, 35], "list": [1, 2, 6, 8, 21, 22, 23, 24, 29, 33], "boxplot": 1, "barplot": [1, 29], "If": [1, 2, 4, 6, 8, 10, 23], "kei": [1, 4, 6, 8, 21], "should": [1, 2, 4, 6, 23], "exactli": 1, "same": [1, 2, 4, 23, 32, 34], "iloc": [1, 21, 23, 24, 26, 30, 31, 32, 35], "uniqu": [1, 22, 25, 28, 32, 34], "valu": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 33, 35], "name": [1, 4, 6, 21, 22, 23, 25, 26, 27, 28, 30, 31], "hex": [1, 22], "length": [1, 2, 27], "thi": [1, 4, 6, 10, 21, 23, 29, 30], "equal": 1, "nuniqu": [1, 25, 28, 32], "string": [1, 4, 6, 8], "mean": [1, 2, 4, 21, 23, 25, 28], "share": 1, "float": [1, 2, 4, 23], "axi": [1, 2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "1": [1, 2, 4, 6, 8, 12, 15, 16, 17, 21, 22, 23, 24, 25, 27, 28, 29, 31, 32, 33, 34, 35], "width": [1, 2, 6, 8, 21, 22, 23, 24, 26, 28, 29, 31, 32, 33, 34, 35], "size": [1, 2, 4, 6, 13, 16, 26, 29], "bool": [1, 2, 4], "whether": [1, 2, 4, 8, 21, 23, 31], "when": [1, 2, 4, 21, 23], "ar": [1, 2, 4, 8, 23, 24, 29, 31, 33], "vmax": [1, 2, 4, 8, 21, 24, 28, 29], "vmin": [1, 2, 4, 8, 21, 24, 28, 29], "other": [1, 2, 4, 6, 8, 23, 31, 33], "kw": [1, 2, 8], "pass": [1, 2, 4, 6, 8, 30], "plt": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "titl": [1, 2, 8], "prop": 1, "fontsiz": [1, 2, 21, 22, 23, 25, 26, 27, 29, 31, 34], "labelcolor": [1, 2, 21, 23, 26, 27, 29, 30, 31, 35], "markscal": 1, "frameon": [1, 2, 21, 23, 26, 31], "framealpha": 1, "fancybox": 1, "shadow": [1, 2], "facecolor": [1, 2, 21, 22, 23, 26, 29, 31], "edgecolor": [1, 2, 21, 26, 31], "mode": 1, "argument": 1, "pleast": 1, "type": [1, 2, 4, 6, 8, 22, 24, 29, 34], "There": 1, "an": [1, 13, 16, 21, 23], "addit": [1, 23, 31], "color_text": [1, 8, 23], "true": [1, 2, 6, 8, 9, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "which": [1, 2, 8, 28, 29, 31], "set": [1, 2, 8, 13, 16, 21, 22, 23, 25, 26, 31, 35], "text": [1, 2, 8, 21, 23, 26], "marker": [1, 4, 6, 8, 13, 16, 21, 25], "fals": [1, 2, 4, 8, 9, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "black": [1, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "user": [1, 28], "want": [1, 17, 23, 27], "us": [1, 2, 4, 6, 8, 9, 10, 13, 16, 17, 20, 22, 23, 24, 26, 31, 32, 35, 36], "custom": [1, 2, 9, 16, 25], "instead": [1, 21, 25], "blue": [1, 21, 23, 26, 27, 28, 29, 30, 31, 32, 35], "rotat": [1, 2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34], "rotation_mod": [1, 2, 21, 23, 26, 31], "ha": [1, 21], "va": [1, 21], "annotation_clip": 1, "arrowprop": [1, 21, 29, 31], "For": [1, 2, 8, 16, 21, 29, 31], "also": [1, 2, 4, 6, 23], "chang": [1, 2, 8, 9, 13, 16, 23, 27], "rang": [1, 2, 8, 21, 23, 24, 25, 26, 30, 31, 33, 35], "try": [1, 2], "df_box": [1, 21, 23, 26, 30, 31, 35], "gene1": [1, 21], "return": [1, 2, 4, 6, 8], "idx": 1, "subplot_ax": 1, "label": [1, 2, 8, 9, 16, 18, 22, 23, 24, 25, 26, 27, 28, 29, 30, 32, 33, 34, 35], "label_sid": [1, 8, 21, 23, 25, 27, 28, 29, 31, 32], "label_kw": [1, 16, 18, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 32, 34], "ticklabels_kw": [1, 31], "legend_sid": [1, 2, 8], "right": [1, 2, 8, 9, 11, 16, 25, 26, 27, 28, 29, 30, 31, 32, 33, 35], "legend_gap": [1, 2, 21, 22, 23, 25, 26, 28, 29, 30, 31, 34, 35], "legend_width": [1, 2, 8, 22, 23], "4": [1, 4, 8, 15, 16, 21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "legend_hpad": [1, 2, 21, 22, 23, 24, 25, 26, 31, 32, 34], "2": [1, 2, 4, 8, 15, 16, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "legend_vpad": [1, 2, 8, 21, 23, 24, 25, 26, 31], "orient": [1, 9, 16, 27, 29, 32], "wgap": [1, 18, 21, 26], "hgap": [1, 18, 21, 23, 26, 30], "raster": [1, 2, 22, 23, 24, 26, 31, 34, 35], "verbos": [1, 2, 8, 21, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "arg": 1, "gener": [1, 2, 9, 16, 23, 26, 30, 31, 35], "self": [1, 4], "convert": [1, 28], "int": [1, 2, 4, 21, 26, 28, 30, 31, 34], "row": [1, 2, 4, 6, 9, 15, 16, 22, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "need": 1, "specifi": [1, 2, 23], "provid": [1, 2, 8, 16, 23, 28], "have": [1, 2, 8, 17, 21, 23, 29, 34], "can": [1, 2, 4, 6, 21, 23, 27, 29, 30, 31], "from": [1, 2, 4, 6, 8, 13, 16, 17, 21, 23, 24, 25, 26, 27, 29, 31, 32, 33, 34, 35], "__subclasses__": 1, "includ": [1, 2, 8, 21], "anno_scatt": 1, "must": [1, 4], "els": [1, 21, 22, 23, 25, 26, 27, 28, 30, 31, 32, 33], "calcul": [1, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "given": [1, 4, 8, 23, 28], "invalid": 1, "top": [1, 2, 8, 9, 11, 16, 22, 25, 26, 27, 29, 31, 32, 33], "bottom": [1, 2, 8, 11, 16, 21, 25, 26, 27, 29, 30, 31, 32, 33], "left": [1, 2, 8, 9, 11, 16, 25, 26, 27, 28, 29, 30, 31, 32, 33], "alpha": [1, 2, 4, 25, 26, 28, 29, 31], "fontstyl": [1, 2, 26, 31], "horizontalalign": [1, 2, 21, 23, 25, 26, 27, 28, 29, 30, 31, 32], "verticalalign": [1, 2, 21, 26, 27, 30, 31], "visibl": [1, 2, 21, 22, 24, 25, 26, 29, 31, 32, 34], "gca": [1, 23], "yaxi": [1, 26, 31], "properti": [1, 2, 26, 31], "ax": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 28, 29, 31, 32, 33], "direct": [1, 2, 23, 31], "pad": [1, 2, 16, 18, 21, 26], "labels": [1, 2, 21, 27, 29, 31, 32, 35], "zorder": [1, 2, 26, 31], "labelbottom": [1, 2], "labeltop": [1, 2], "labelleft": [1, 2], "labelright": [1, 2], "labelrot": [1, 2, 21, 23, 26, 27, 29, 30, 31, 35], "grid_color": [1, 2], "grid_linestyl": [1, 2], "tick_param": [1, 2, 21, 31], "function": [1, 4, 8, 23, 25, 29, 30], "et": [1, 2, 22, 23], "al": [1, 2, 23], "automoti": 1, "vertic": [1, 2, 21], "gap": [1, 2, 8, 16, 18, 21, 23], "between": [1, 2, 8, 9, 16, 23, 31], "two": [1, 2, 16, 20, 23, 25, 29, 31], "mm": [1, 2, 22, 23, 24, 28, 29, 31, 32, 34, 35], "horizon": [1, 2, 23], "space": [1, 2, 8, 9, 16, 23], "up": [1, 2, 13, 16, 27, 32], "down": [1, 9, 16, 27], "show": [1, 2, 4, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "automat": [1, 2, 4, 23], "assign": [1, 29, 32], "accord": 1, "posit": [1, 26, 28], "option": [1, 4, 6, 8], "wspace": 1, "control": [1, 4, 6, 16, 18, 21, 23, 28, 33], "hspace": 1, "horizont": [1, 2, 16, 20], "pair": [1, 8, 21, 35], "collect": [1, 21, 22, 23, 24, 27, 28, 29, 30, 32, 33, 34, 35], "rtype": 1, "subplot_spec": [1, 2, 6], "figur": [1, 4, 8, 21, 22, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "add_gridspec": 1, "gridspec": 1, "gridspecfromsubplotspec": 1, "index": [1, 2, 4, 21, 22, 23, 24, 25, 26, 27, 28, 30, 31, 32, 33, 34, 35], "param": [1, 2], "kwarg": [1, 2, 4, 6, 16], "grid": [1, 21, 26, 28, 29, 31, 32, 33], "showflier": 1, "medianlinecolor": 1, "border_width": [1, 21], "border_color": [1, 21], "255": [1, 21], "merg": [1, 8, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 33, 34], "merge_width": 1, "imag": [1, 9, 16], "border": 1, "line": [1, 21, 23, 28, 30, 31], "256": [1, 21], "ignor": [1, 8, 28], "white": [1, 2, 8, 21, 22, 23, 26, 27, 28, 31, 33], "cluster": [1, 2, 8, 9, 16], "onc": 1, "extend": [1, 22, 34], "frac": 1, "major": [1, 8, 22, 29, 31], "adjust_color": [1, 34], "lumin": [1, 29, 34], "8": [1, 2, 4, 6, 8, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 35], "relpo": [1, 21, 22, 26, 27, 29, 30, 31, 32, 34], "add": [1, 2, 9, 13, 16, 25, 27, 31], "documentatin": 1, "websit": 1, "dingwb": [1, 2, 17, 31, 34], "github": [1, 2, 17, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "io": [1, 2, 21, 31, 32], "build": [1, 2, 9, 16, 31], "notebook": [1, 2, 31], "single_cell_methyl": 1, "distribut": 1, "fraction": 1, "arma": 1, "armb": 1, "multipl": [1, 2, 4, 9, 16], "group": [1, 2, 9, 13, 16, 24, 29, 31, 34, 35], "largest": [1, 8], "too": [1, 2, 21, 23, 28], "high": [1, 24, 25, 28], "replac": [1, 21, 26, 28, 30, 31, 32], "origin": 1, "togeth": [1, 2], "tupl": [1, 2, 21, 22], "x": [1, 4, 6, 8, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 33], "y": [1, 4, 6, 8, 21, 22, 24, 25, 26, 28, 29, 31, 32, 33], "arrow": [1, 16, 18, 21], "start": [1, 16, 21, 22, 23, 24, 27, 28, 29, 30, 32, 33, 34, 35], "point": [1, 13, 16, 21, 22, 29, 31], "rel": 1, "side": [1, 15, 16, 25, 26, 29], "about": 1, "could": [1, 2, 4, 23], "found": 1, "patch": [1, 26, 31], "fancyarrowpatch": [1, 31], "lineplot": 1, "linewidth": [1, 2, 21, 23, 25, 26, 28, 29, 31, 32, 35], "scatterplot": 1, "scatter": [1, 4, 6, 21, 23, 26, 28, 29, 30, 31, 35], "add_text": [1, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 35], "text_kw": [1, 21, 22, 23, 24, 25, 26, 27, 28, 30, 31, 35], "simpl": [1, 13, 16, 23], "categor": [1, 4, 8, 16, 23, 36], "continu": [1, 22, 25, 32], "variabl": [1, 8, 16, 36], "data": [2, 4, 6, 10, 13, 16, 20, 21, 22, 23, 24, 26, 27, 30, 31, 32, 33, 35], "top_annot": [2, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 33, 34, 35], "bottom_annot": [2, 23], "left_annot": [2, 22, 23, 24, 27, 29, 34], "right_annot": [2, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 34], "row_clust": [2, 21, 22, 26, 27, 28, 29, 30, 31, 34, 35], "col_clust": [2, 9, 22, 26, 27, 28, 29, 30, 31, 34, 35], "row_cluster_method": [2, 24, 35], "averag": [2, 21, 23, 26, 31, 35], "row_cluster_metr": [2, 24, 35], "correl": [2, 16, 20, 28, 35], "col_cluster_method": [2, 24, 35], "col_cluster_metr": [2, 24, 35], "show_rownam": [2, 21, 22, 23, 24, 26, 27, 28, 29, 30, 31, 32, 33, 35], "show_colnam": [2, 21, 22, 23, 24, 26, 27, 28, 29, 30, 31, 32, 33, 35], "row_names_sid": [2, 21, 23, 29, 30, 31, 35], "col_names_sid": [2, 23, 29], "xticklabels_kw": [2, 6, 16, 18, 21, 23, 26, 27, 29, 30, 32, 35], "yticklabels_kw": [2, 6, 16, 18, 21, 26, 27], "row_dendrogram": [2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "col_dendrogram": [2, 21, 22, 24, 27, 31, 35], "row_dendrogram_s": [2, 21], "10": [2, 10, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "col_dendrogram_s": 2, "row_split": [2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "col_split": [2, 9, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "row_dendrogram_kw": 2, "col_dendrogram_kw": [2, 21], "tree_kw": [2, 21, 23, 25, 27, 28, 30, 31, 35], "row_split_ord": [2, 21, 22, 34], "col_split_ord": [2, 9, 22, 31, 32, 34], "row_split_gap": [2, 18, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 34, 35], "col_split_gap": [2, 18, 21, 22, 23, 25, 26, 27, 28, 29, 30, 32, 33, 34, 35], "mask": [2, 4, 6], "subplot_gap": [2, 18, 21, 26, 32], "legend_anchor": [2, 23, 24, 25], "7": [2, 21, 22, 23, 25, 26, 28, 29, 30, 31, 32, 34, 35], "xlabel": [2, 16, 18, 21, 22, 23, 26, 29, 34, 35], "ylabel": [2, 16, 18, 21, 22, 23, 26, 34, 35], "xlabel_kw": [2, 16, 18, 21, 22, 23, 26, 29, 34], "ylabel_kw": [2, 16, 18, 21, 22, 23, 26, 34], "xlabel_sid": [2, 22, 29], "ylabel_sid": 2, "xlabel_bbox_kw": [2, 21, 22, 26, 29, 31], "ylabel_bbox_kw": [2, 21, 23, 26, 31], "legend_delta_x": 2, "plotter": [2, 4, 6], "numpi": [2, 4, 6, 17, 21, 22, 27, 29, 30], "arrai": [2, 4, 6, 16, 20, 22, 25, 26, 34], "perform": [2, 29], "z": [2, 25], "score": 2, "either": [2, 23], "after": 2, "method": [2, 8, 21], "linkag": [2, 9, 16, 22], "singl": [2, 16, 20], "complet": 2, "weight": [2, 22], "centroid": 2, "median": [2, 21], "ward": [2, 24, 35], "scipi": [2, 17, 35], "hierarchi": 2, "doc": 2, "refer": [2, 32], "detail": 2, "pairwis": 2, "distanc": 2, "observ": 2, "n": [2, 21, 22], "dimension": 2, "euclidean": [2, 35], "minkowski": 2, "cityblock": 2, "seuclidean": 2, "cosin": 2, "ham": 2, "jaccard": [2, 24], "chebyshev": 2, "canberra": [2, 21], "braycurti": 2, "mahalanobi": 2, "kulsinski": 2, "14": [2, 21, 22, 23, 25, 26, 27, 28, 29, 31, 33, 34], "spatial": 2, "pdist": 2, "metric": [2, 21], "ticklabel": [2, 8, 9, 16, 31], "dendrogram": [2, 22, 31], "10mm": 2, "pd": [2, 4, 6, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "number": [2, 21, 29], "hierarch": 2, "split": [2, 15, 16, 25, 28, 29, 32], "subplot": [2, 21], "my_linkag": 2, "order": [2, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "cluster_between_group": [2, 9, 31], "wa": [2, 4, 16, 23, 24, 25, 28, 34], "clsuter": [2, 9], "advanced_usag": [2, 31], "col": [2, 9, 16, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "cell": [2, 9, 16, 20, 22, 23, 29, 31, 33], "miss": 2, "infinit": 2, "1mm": 2, "asfonts": 2, "numpoint": 2, "markerscal": 2, "markerfirst": 2, "title_fonts": 2, "labelspac": 2, "alatern": 2, "we": [2, 21, 23, 28, 30, 32], "outlin": 2, "cbar": [2, 8], "cm": [2, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34, 35], "isinst": 2, "colorbar": [2, 8], "set_color": [2, 25, 26, 29], "set_linewidth": [2, 25, 26, 29], "divid": 2, "ax_heatmap": [2, 23, 24, 25, 26, 28, 29], "anchor": [2, 8, 21, 23, 31], "differ": [2, 8, 13, 16, 21, 29, 30], "infer": [2, 4, 28, 29], "shown": [2, 4, 6], "xticklabel": [2, 16, 18, 21, 35], "yticklabel": [2, 16, 18, 21, 35], "fontfamili": [2, 26, 31], "fontnam": [2, 21, 26, 31], "fontproperti": [2, 26, 31], "fontweight": [2, 22, 24, 26, 27, 29, 31], "xaxi": [2, 26, 31], "print": [2, 21, 22, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "sam": 2, "clip_box": [2, 26, 31], "clip_on": [2, 26, 31], "fill": [2, 4, 21, 26, 31], "in_layout": [2, 26, 31], "linestyl": [2, 26, 28, 29, 31], "get_bbox_patch": [2, 26, 31], "5000": [2, 33], "speed": 2, "center": [2, 8, 21, 24, 26, 31], "robust": [2, 8], "annot_kw": [2, 21], "fmt": [2, 21, 26, 31, 35], "linecolor": [2, 21, 23, 26, 31, 35], "na_col": [2, 8], "cbar_kwss": 2, "document": 2, "small": [2, 34], "displai": [2, 9, 16], "avoid": [2, 21], "overlap": [2, 8, 21], "To": [2, 21], "forc": [2, 9, 16], "use_linkag": 2, "row_ord": [2, 4, 6, 21, 22, 27], "col_ord": [2, 4, 6, 21, 22, 27], "fig": [2, 8, 21], "static": 2, "data2d": 2, "max": [2, 4, 22, 28, 29], "min": [2, 22, 28, 29], "normal": [2, 21, 23, 26, 30, 31, 35], "across": 2, "standard": 2, "noraml": 2, "varianc": 2, "tight_param": 2, "standar": 2, "dendrogram_kw": 2, "gap_pixel": 2, "root_x": 2, "similar": [2, 21], "upon": 2, "indic": [2, 29], "matrix": [2, 4, 6, 16, 20, 22, 23, 24, 27, 28, 29, 32, 33, 34, 35], "cmlist": [2, 24], "main": [2, 15, 16, 21, 24, 31, 34], "row_gap": [2, 6], "15": [2, 21, 22, 23, 24, 26, 27, 28, 30, 31, 32], "col_gap": [2, 24], "legend_i": 2, "width_ratio": 2, "height_ratio": 2, "assembl": 2, "align": [2, 6, 33], "influenc": [2, 24], "unit": [2, 8, 31], "estim": [2, 22, 23, 24, 28, 29, 32, 34, 35], "legend_ax": [2, 8, 24], "cbar_kw": 2, "cbar_ax": 2, "squar": [2, 13, 16, 26], "xlabel_pad": 2, "ylabel_pad": 2, "xticklabels_sid": 2, "yticklabels_sid": 2, "2g": 2, "maxim": [2, 4], "minim": [2, 4], "seaborn": [2, 13, 16], "cax": [2, 8], "draw": [2, 23], "format": 2, "anno_kw": 2, "c": [3, 4, 6, 8, 21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 35], "hue": [4, 6, 13, 16, 25, 29], "": [4, 6, 8, 13, 16, 23, 25, 29], "o": [4, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "color_legend_kw": [4, 6], "cmap_legend_kw": [4, 6], "dot_legend_kw": 4, "dot_legend_mark": [4, 28, 29], "aggfunc": 4, "value_na": [4, 28], "hue_na": 4, "na": 4, "s_na": 4, "c_na": [4, 28], "spine": [4, 8, 21, 25, 26, 28, 29, 32, 33], "max_": [4, 28, 29], "dotclustermap": [4, 6, 16, 20], "inherit": [4, 6], "dot": [4, 6, 13, 16, 20, 29], "The": [4, 6, 16, 23, 24, 25, 29], "overwrit": [4, 6], "go": [4, 29], "stabl": 4, "api": 4, "markers_api": 4, "avail": 4, "Such": 4, "v": [4, 28], "p": [4, 25, 28, 29], "h": [4, 21, 23, 26, 27, 30, 31, 35], "d": [4, 8, 10, 21, 23, 25, 26, 28, 30, 31, 35], "_": [4, 28], "tolist": [4, 21, 22, 23, 24, 25, 28, 30, 32, 34], "It": [4, 23, 29], "pivot_t": 4, "fillna": [4, 21, 25, 28, 32], "floator": 4, "np": [4, 21, 22, 23, 26, 27, 28, 29, 30, 31, 32, 35], "aggreg": 4, "them": 4, "call": [4, 8], "pivot": [4, 28], "coeffici": 4, "valid": 4, "input": [4, 6], "paramt": [4, 6], "aspect": [6, 21], "bgcolor": 6, "whitesmok": 6, "remove_empty_row": 6, "remove_empty_column": 6, "background": [6, 23], "lightgrai": [6, 24, 25], "nvar": 6, "element": 6, "wisa": 6, "cccccc": 6, "intern": 8, "boolean": 8, "ani": 8, "legend_list": 8, "xtick": 8, "adjac": [8, 23], "tick": [8, 21], "coordin": 8, "keep": 8, "A": [8, 10, 11, 16, 20, 21, 26, 30, 31], "b": [8, 21, 26, 30, 31, 34], "len": [8, 21, 22], "new_label": 8, "plot_data": 8, "some": [8, 21], "heurist": 8, "good": [8, 29], "remov": [8, 16, 18, 25, 30, 32, 33], "current": 8, "specif": 8, "turbo": [8, 21, 23, 24, 26, 27, 30, 31, 35], "drawn": 8, "accent": 8, "tab20": [8, 25], "exp1": [8, 21, 23, 27], "exp2": [8, 21], "meth1": [8, 21], "meth2": [8, 21, 35], "legn": 8, "y0": 8, "delta_x": 8, "cmap_width": 8, "lengend": 8, "handl": 8, "legend_typ": 8, "pixel": 8, "init": 8, "2mm": 8, "5mm": 8, "boundri": 8, "obj": 8, "repres": 8, "python": [8, 10, 16, 17], "e": [8, 21, 23, 26, 30, 31, 35], "unchang": 8, "byte": 8, "utf": 8, "decod": 8, "__str__": 8, "result": [8, 13, 16, 28], "represent": 8, "dataset": [9, 13, 16, 20, 23, 26, 29, 30, 31, 33, 34, 35, 36], "select": [9, 16, 23, 26, 27, 28, 30, 31], "orent": [9, 16], "extra": [9, 16], "loop": [9, 16], "within": [9, 16], "cluster_within_group": 9, "choos": [9, 16, 30], "pycomplexheatmap": [9, 10, 17, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "diverg": 9, "qualit": 9, "how": [9, 16, 18], "you": [10, 17, 27], "softwar": [10, 28], "your": [10, 29], "work": 10, "cite": 10, "follow": [10, 21, 23, 24, 26, 31, 35], "metadata": [10, 22], "ding": [10, 16], "w": 10, "goldberg": 10, "zhou": 10, "2023": 10, "packag": [10, 15, 16, 19, 28, 29, 32], "visual": [10, 13, 16, 20, 36], "multimod": 10, "genom": 10, "imeta": 10, "e115": 10, "doi": 10, "1002": 10, "imt2": 10, "115": [10, 29], "quick": [11, 16], "understand": [12, 16], "layout": [12, 16], "load": [13, 16, 23, 24, 29], "brain": [13, 16, 23, 24], "network": [13, 16], "tradit": [13, 16], "fix": [13, 16], "five": [13, 16], "dimens": [13, 16], "gene": [13, 16, 20, 21, 23, 25, 26, 30, 31, 32, 33, 35], "enrich": [13, 16, 28], "analysi": [13, 16, 28], "clusterprofil": [13, 16], "click": 14, "picutur": 14, "view": 14, "code": [14, 21, 23, 26, 31, 35], "import": [15, 16, 21, 23, 24, 25, 26, 27, 28, 29, 31, 32, 33, 34, 35], "along": [15, 16], "complex": 16, "biolog": [16, 29], "instal": [16, 30], "depend": [16, 29], "get": 16, "tcga": [16, 36], "lung": [16, 34, 36], "adenocarcinoma": [16, 36], "carcinoma": [16, 36], "variant": [16, 36], "advanc": 16, "usag": 16, "differenti": [16, 20, 29], "express": [16, 20], "dna": [16, 20, 28], "methyl": [16, 20, 29], "loyfer2023": [16, 20], "mous": [16, 20, 22, 23], "cpg": [16, 20, 24, 34], "modul": [16, 19, 20], "modifi": [16, 18], "galleri": [16, 28], "develop": [16, 29], "setup": [16, 17, 19], "citat": 16, "wubin": 16, "pip": 17, "upgrad": 17, "development": 17, "version": 17, "directli": 17, "com": [17, 21, 31, 34], "git": 17, "previous": 17, "updat": 17, "uninstal": 17, "again": 17, "OR": 17, "clone": [17, 27], "cd": [17, 21, 23, 26, 30, 31, 35], "py": [17, 25, 28], "submodul": 19, "content": 19, "process": [20, 22, 29], "smaller": 20, "sy": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "inlin": [21, 22, 23, 24, 25, 26, 28, 30, 31, 32, 34, 35], "pylab": [21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 34, 35], "pickl": [21, 23, 24, 26, 28, 29, 31, 32, 34, 35], "rcparam": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "dpi": [21, 22, 23, 24, 26, 28, 29, 30, 31, 32, 34, 35], "100": [21, 22, 23, 26, 28, 30, 31, 32, 33, 34, 35], "savefig": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "300": [21, 22, 23, 24, 26, 28, 29, 30, 31, 32, 34, 35], "path": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "append": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "expandus": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "project": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "__version__": [21, 22, 24, 25, 26, 29, 30, 31, 34, 35], "6": [21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 32, 33, 34, 35], "dev15": 21, "gd9f20b3": 21, "font": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "arial": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "famili": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "san": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "serif": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "pdf": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "fonttyp": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "42": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35], "random": [21, 23, 26, 30, 31, 33, 35], "groupa": [21, 26, 30, 31, 35], "groupb": [21, 26, 30, 31, 35], "ab": [21, 23, 26, 30, 31, 35], "g": [21, 23, 24, 26, 27, 30, 31, 35], "ef": [21, 23, 26, 30, 31, 35], "f": [21, 22, 23, 24, 26, 29, 30, 31, 33, 35], "sampl": [21, 22, 23, 24, 26, 28, 30, 31, 32, 33, 35], "shape": [21, 22, 23, 25, 26, 27, 30, 31, 35], "randn": [21, 23, 26, 30, 31, 35], "df_bar": [21, 23, 26, 30, 31, 35], "uniform": [21, 23, 26, 30, 31, 35], "tmb1": [21, 23, 26, 30, 31, 35], "tmb2": [21, 23, 26, 30, 31, 35], "df_scatter": [21, 23, 26, 30, 31, 35], "df_heatmap": [21, 23, 26, 30, 31, 35], "30": [21, 23, 25, 26, 28, 29, 30, 31, 35], "11": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 33, 35], "fea": [21, 23, 26, 30, 31, 35], "nan": [21, 23, 25, 26, 30, 31, 32, 35], "df_row": [21, 22, 23, 24, 26, 28, 29, 30, 31, 34], "appli": [21, 22, 23, 25, 26, 27, 28, 30, 31, 32], "lambda": [21, 22, 23, 25, 26, 27, 28, 30, 31, 32], "sample4": [21, 26, 30, 31], "to_fram": [21, 26, 28, 30, 31], "xy": [21, 26, 30, 31], "to_seri": [21, 25, 26, 30, 31], "row_ha": [21, 22, 23, 24, 25, 26, 27, 28, 30, 31], "round": [21, 23, 26, 30, 31, 32], "12": [21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 35], "bar": [21, 23, 30, 33], "05": [21, 23, 26, 28, 29, 30, 31, 32, 35], "col_ha": [21, 22, 23, 24, 25, 26, 28, 29, 30, 31, 34, 35], "nanmin": 21, "nanmax": [21, 28], "figsiz": [21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35], "row_cmap": [21, 23, 25, 27, 28, 30, 35], "rdylbu_r": [21, 26, 30, 31, 35], "90": [21, 22, 27, 28, 30, 34, 35], "example0": [21, 30, 35], "bbox_inch": [21, 22, 23, 24, 25, 27, 28, 29, 30, 32, 34, 35], "tight": [21, 22, 23, 24, 25, 27, 28, 29, 30, 32, 34, 35], "259323481554901": 21, "3307664641494927": 21, "32": [21, 33], "s4": [21, 26, 31], "test": [21, 26, 31], "18": [21, 26, 28, 31], "bbox": [21, 26], "boxstyl": [21, 26, 31], "circl": [21, 28], "exp": [21, 23, 26, 30, 31, 35], "turn": [21, 23, 26, 31, 35], "off": [21, 23, 26, 31, 35], "log": [21, 23, 26, 31, 35], "head": [21, 24, 25, 27, 28, 30, 32, 34], "1g": [21, 26, 31, 35], "gold": [21, 23, 26, 31, 35], "45": [21, 23, 25, 26, 27, 28, 29, 31, 32, 33, 35], "sample1": [21, 30], "404085": 21, "sample2": [21, 30], "081411": 21, "sample3": [21, 30], "594949": 21, "392642": 21, "sample5": [21, 30], "150321": 21, "229885": 21, "210195": 21, "032791": 21, "051215": 21, "261465": 21, "sample6": 21, "836030": 21, "sample7": 21, "519891": 21, "sample8": 21, "742360": 21, "sample9": 21, "617504": 21, "sample10": 21, "606197": 21, "float64": [21, 28], "fea1": 21, "941453": 21, "140686": 21, "285091": 21, "118367": 21, "214946": 21, "967923": 21, "853248": 21, "fea2": 21, "691451": 21, "561863": 21, "045189": 21, "303610": 21, "324130": 21, "240718": 21, "fea3": 21, "378372": 21, "142337": 21, "405770": 21, "274505": 21, "171325": 21, "526213": 21, "119409": 21, "fea4": 21, "212045": 21, "515978": 21, "051057": 21, "124523": 21, "378586": 21, "320444": 21, "064873": 21, "fea5": 21, "078289": 21, "476870": 21, "360113": 21, "705353": 21, "079402": 21, "284584": 21, "341059": 21, "751814": 21, "163439": 21, "038006": 21, "120816": 21, "820074": 21, "259323": 21, "757637": 21, "451777": 21, "057090": 21, "817372": 21, "082246": 21, "293083": 21, "831747": 21, "489009": 21, "258670": 21, "39": [21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 34], "fea19": 21, "fea25": 21, "fea18": 21, "fea27": 21, "fea29": 21, "fea22": 21, "fea24": 21, "fea30": 21, "fea20": 21, "fea17": 21, "fea26": 21, "fea21": 21, "fea23": 21, "fea15": 21, "fea16": 21, "fea28": 21, "fea9": 21, "fea10": 21, "fea11": 21, "fea7": 21, "fea8": 21, "fea14": 21, "fea13": 21, "fea6": 21, "fea12": 21, "37": [21, 25, 26, 31], "aaaa1": [21, 23], "bbbbb2": [21, 23], "df_bar1": [21, 30], "t1": [21, 30], "df_bar2": [21, 31], "t2": 21, "df_bar3": 21, "t3": 21, "df_bar4": [21, 30], "t4": [21, 30], "372887": 21, "061936": 21, "134849": 21, "694566": 21, "359127": 21, "132367": 21, "392230": 21, "272427": 21, "714047": 21, "625537": 21, "gene2": 21, "gene3": 21, "gene4": 21, "257733": 21, "699439": 21, "844278": 21, "608444": 21, "752454": 21, "083405": 21, "809866": 21, "431847": 21, "663268": 21, "513365": 21, "772141": 21, "387252": 21, "636622": 21, "012600": 21, "966687": 21, "476726": 21, "826088": 21, "361708": 21, "754555": 21, "343367": 21, "220518": 21, "870597": 21, "876851": 21, "9": [21, 22, 23, 24, 25, 28, 29, 31, 32, 33], "876537": 21, "395013": 21, "573748": 21, "948007": 21, "973635": 21, "386078": 21, "499509": 21, "188200": 21, "545418": 21, "328468": 21, "047464": 21, "158959": 21, "161838": 21, "163763": 21, "243494": 21, "779427": 21, "204043": 21, "542872": 21, "130472": 21, "133916": 21, "091244": 21, "788314": 21, "348602": 21, "046968": 21, "088636": 21, "612396": 21, "928272": 21, "877048": 21, "401591": 21, "980643": 21, "201984": 21, "307024": 21, "833295": 21, "660850": 21, "235867": 21, "211823": 21, "919341": 21, "290198": 21, "165467": 21, "382529": 21, "881658": 21, "073468": 21, "342374": 21, "239178": 21, "468118": 21, "962650": 21, "532729": 21, "771372": 21, "835987": 21, "436057": 21, "167528": 21, "042679": 21, "093969": 21, "936279": 21, "354470": 21, "736818": 21, "348153": 21, "335390": 21, "823424": 21, "888954": 21, "168943": 21, "042376": 21, "223754": 21, "876278": 21, "314815": 21, "541221": 21, "407723": 21, "913978": 21, "019213": 21, "885269": 21, "457418": 21, "692399": 21, "369775": 21, "256769": 21, "091064": 21, "233948": 21, "702588": 21, "314119": 21, "758320": 21, "888650": 21, "260971": 21, "891667": 21, "691901": 21, "079907": 21, "903442": 21, "754403": 21, "010756": 21, "829727": 21, "209684": 21, "739576": 21, "615991": 21, "269359": 21, "153883": 21, "386240": 21, "874027": 21, "38": [21, 28, 31], "orang": [21, 23, 24, 29], "yellowgreen": [21, 23, 24, 29], "tmb_bar": [21, 23, 31], "bar1": [21, 30], "bar4": [21, 30], "bar2": 21, "bar3": 21, "fontdict": [21, 22, 24, 30], "tab10": [21, 23], "grai": [21, 23, 24, 28], "yellow": [21, 23], "13": [21, 22, 23, 24, 25, 27, 28], "here": [21, 32], "widh": 21, "40": [21, 22, 29], "20": [21, 26, 27, 28, 31], "incres": [21, 23], "ncol": [21, 23], "than": [21, 23], "support": [21, 23], "long": [21, 23, 28], "new": [21, 23], "41": [21, 29, 34], "green": [21, 23, 24, 25, 26, 27, 28, 29, 31, 33], "By": 21, "43": 21, "typic": 21, "creat": [21, 33], "annotatin": [21, 23], "df_col": [21, 22, 23, 24, 25, 28, 29, 34], "hypomethylated_sampl": [21, 22], "sample_group_color_dict": 21, "celltyp": [21, 22, 29], "ct_color_dict": 21, "ct_legend": 21, "m1": 21, "lgd": 21, "But": 21, "what": [21, 29], "mani": 21, "m2": 21, "m3": 21, "In": [21, 23, 28, 29, 31], "case": [21, 29], "col_ha_dict": 21, "sample_col": 21, "r": [21, 24, 29, 32], "jokergoo": [21, 32], "2021": 21, "03": [21, 22, 24, 29], "complexheatmap": [21, 32], "44": [21, 35], "g1": [21, 31, 35], "g2": [21, 31, 35], "g3": [21, 31, 35], "g4": [21, 31, 35], "g5": [21, 31, 35], "col_cmap": [21, 27, 31, 35], "featur": [21, 26, 31, 35], "46": 21, "loc": [21, 22, 24, 25, 28, 29, 32, 34], "47": 21, "annotaiton": [21, 31], "purpl": [21, 23, 25, 26, 28, 31, 35], "_ticklabels_kw": [21, 31], "labelpad": [21, 22, 23, 26, 29, 31, 34], "increac": [21, 22, 23, 31], "manual": [21, 22, 31], "chocol": [21, 22, 26, 31], "48": 21, "applymap": 21, "make": 21, "asterisk": 21, "locat": 21, "oper": 21, "unicod": 21, "explor": 21, "2217": 21, "courier": 21, "49": [21, 28], "fastclust": 21, "t": [21, 22, 23, 25, 27, 28, 32, 34], "nlinkag": 21, "17": [21, 25, 28], "07051523": 21, "22006417": 21, "19": [21, 23, 26, 28, 33, 35], "14461418": 21, "28842182": 21, "70552492": 21, "76359288": 21, "65679239": 21, "16": [21, 23, 24, 25, 27, 28], "21": [21, 28, 35], "91528308": 21, "22": [21, 28], "75945877": 21, "50": [21, 23, 28], "51": 21, "df_img": 21, "jpeg": 21, "52": 21, "img": 21, "diverging1": 21, "parula": [21, 22, 23, 24, 34, 35], "53": 21, "54": 21, "mpl": [21, 29], "gradient": 21, "linspac": 21, "vstack": 21, "def": 21, "plot_color_gradi": 21, "categori": [21, 28, 29], "cmap_list": 21, "adjust": [21, 29], "nrow": 21, "figh": 21, "35": [21, 29], "subplots_adjust": 21, "99": [21, 33], "set_titl": [21, 22, 24], "zip": [21, 26], "imshow": 21, "01": [21, 32], "transform": [21, 26], "transax": 21, "just": 21, "ones": 21, "set_axis_off": [21, 23], "save": 21, "later": [21, 25], "55": [21, 28], "sequenti": 21, "cmap50": 21, "56": [21, 29], "57": 21, "custom_cmap": 21, "58": 21, "59": 21, "big": [21, 23], "enough": 21, "hidden": 21, "60": 21, "matter": 21, "61": [21, 33], "home": 22, "wding2": 22, "pch": [22, 24, 25, 30, 34], "czip": 22, "palette_path": 22, "mouse_pfc": 22, "mpfc_color_palett": 22, "xlsx": 22, "cell_class_color": 22, "read_excel": 22, "sheet_nam": 22, "cellclass": [22, 29], "index_col": [22, 25, 27, 28, 32, 34, 35], "to_dict": [22, 25, 29], "multi": [22, 28], "grei": [22, 24, 29], "nonn": 22, "e06931": 22, "exc": 22, "5ec5a9": 22, "rg": 22, "6036cb": 22, "inh": 22, "e93400": 22, "delta": [22, 34], "read_csv": [22, 25, 27, 28, 32, 34, 35], "pseudo_cel": 22, "between_groups_dmr": 22, "methylpi": 22, "methylpy_rms_results_collaps": 22, "tsv": 22, "sep": [22, 25, 28, 32, 34], "lstrip": 22, "methylation_level_": 22, "startswith": 22, "renam": 22, "number_of_dm": 22, "inplac": [22, 25, 28, 32, 34], "drop": 22, "hypermethylated_sampl": 22, "sname": 22, "insert": 22, "isna": 22, "reset_index": [22, 25, 28, 32], "set_index": [22, 25, 28, 29], "read": [22, 24, 34], "major_typ": 22, "beta": [22, 24, 25, 34], "txt": [22, 28, 32], "common_row": 22, "5kb_mc_hic_clust": 22, "l2": 22, "major_type_to_cell_class": 22, "common_col": 22, "cell_type_col": 22, "258365": 22, "23": [22, 26, 28, 29], "chr": [22, 24], "end": 22, "chr14": 22, "25667890": 22, "25667923": 22, "649934": 22, "chr5": [22, 24], "70893110": 22, "70893122": 22, "333501": 22, "chr3": 22, "141598191": 22, "141598238": 22, "440745": 22, "chr1": [22, 34], "76086002": 22, "76086015": 22, "319716": 22, "chr9": 22, "16231701": 22, "16232342": 22, "444616": 22, "chr11": [22, 24], "75550115": 22, "75550656": 22, "547601": 22, "109751156": 22, "109751187": 22, "530203": 22, "68823498": 22, "68825391": 22, "29": 22, "451270": 22, "chr10": 22, "91354668": 22, "91354926": 22, "327058": 22, "74147749": 22, "74150162": 22, "31": [22, 26, 29], "541438": 22, "cellclass1": 22, "majortyp": 22, "ct": 22, "l6": 22, "cge": 22, "lamp5": 22, "IT": 22, "l23": 22, "pc": [22, 27], "mgc": 22, "odc": 22, "vip": 22, "mge": 22, "sst": 22, "opc": 22, "vlmc": 22, "l4": 22, "unknown": [22, 28], "l5": 22, "ec": 22, "pvalb": 22, "pia": 22, "asc": 22, "chrom": 22, "077450": 22, "233350": 22, "161600": 22, "733500": 22, "685500": 22, "628000": 22, "148000": 22, "468000": 22, "785000": 22, "662500": 22, "780000": 22, "092500": 22, "125000": 22, "849000": 22, "043500": 22, "839500": 22, "735500": 22, "822500": 22, "694000": 22, "630500": 22, "903500": 22, "899000": 22, "896500": 22, "637500": 22, "784500": 22, "323000": 22, "947500": 22, "743500": 22, "844000": 22, "961500": 22, "499000": 22, "926000": 22, "000000": [22, 23, 25, 27, 28, 35], "846500": 22, "954500": 22, "489500": 22, "883500": 22, "598000": 22, "884000": 22, "789500": 22, "491350": 22, "192500": 22, "623500": 22, "055500": 22, "332150": 22, "763500": 22, "464500": 22, "448000": 22, "200150": 22, "549500": 22, "377950": 22, "517000": 22, "583500": 22, "458500": 22, "419500": 22, "633500": 22, "775000": 22, "299000": 22, "082350": 22, "471000": 22, "797500": 22, "508500": 22, "773500": 22, "050000": 22, "610500": 22, "802000": 22, "869500": 22, "523500": 22, "572500": 22, "572000": 22, "887000": 22, "778000": 22, "396500": 22, "826500": 22, "697500": 22, "145450": 22, "562500": 22, "317500": 22, "807875": 22, "725875": 22, "318250": 22, "900750": 22, "807750": 22, "378837": 22, "209437": 22, "654000": 22, "526500": 22, "579750": 22, "715875": 22, "133525": 22, "605375": 22, "663125": 22, "666375": 22, "545250": 22, "307337": 22, "319063": 22, "839000": 22, "557333": 22, "631000": 22, "780667": 22, "171233": 22, "262050": 22, "886500": 22, "492667": 22, "159800": 22, "520667": 22, "726833": 22, "683833": 22, "698000": 22, "231500": 22, "830000": 22, "781667": 22, "631667": 22, "909000": 22, "222417": 22, "338917": 22, "687667": 22, "452667": 22, "457667": 22, "241333": 22, "154667": 22, "546333": 22, "629667": 22, "325667": 22, "058667": 22, "503333": 22, "321000": 22, "588000": 22, "709000": 22, "034133": 22, "859333": 22, "360333": 22, "684667": 22, "414333": 22, "081200": 22, "244667": 22, "753267": 22, "363077": 22, "607967": 22, "644540": 22, "257347": 22, "775067": 22, "890267": 22, "517487": 22, "184007": 22, "594700": 22, "720740": 22, "763000": 22, "750800": 22, "495783": 22, "471317": 22, "777000": 22, "901467": 22, "657633": 22, "362270": 22, "241253": 22, "784857": 22, "809286": 22, "508857": 22, "859286": 22, "650714": 22, "823286": 22, "763286": 22, "614143": 22, "339714": 22, "615143": 22, "859000": 22, "469000": 22, "742857": 22, "466571": 22, "851714": 22, "807286": 22, "798000": 22, "913429": 22, "423857": 22, "678143": 22, "859394": 22, "860788": 22, "766576": 22, "678030": 22, "443909": 22, "813273": 22, "867061": 22, "658548": 22, "311152": 22, "854182": 22, "765485": 22, "813455": 22, "846697": 22, "434000": 22, "893424": 22, "834667": 22, "833515": 22, "808030": 22, "302337": 22, "529615": 22, "cc": 22, "dendrogram_col": 22, "ivl": 22, "dendrogram_row": 22, "sort_valu": [22, 34], "color_dict": [22, 34], "copi": 22, "del": [22, 33], "bold": [22, 24, 27, 29], "avg": [22, 23], "n_cpg": 22, "6375": [22, 29], "exist": 22, "listdir": 22, "within_groups_dmr": 22, "join": [22, 28], "_heatmap": 22, "concat": 22, "groupbi": [22, 28, 32], "agg": 22, "to_csv": 22, "agg_beta": 22, "queri": 22, "pfc": 22, "bed": 22, "gz": 22, "outfil": 22, "3013471": 22, "3013579": 22, "3026186": 22, "3026310": 22, "3035862": 22, "3035927": 22, "3052666": 22, "3052823": 22, "3059277": 22, "3059436": 22, "chry": 22, "44290504": 22, "44290637": 22, "66427102": 22, "66427179": 22, "90367464": 22, "90367503": 22, "90732358": 22, "90732367": 22, "90792411": 22, "90792447": 22, "661968": 22, "mt": 22, "28963217": 22, "28963260": 22, "83350": 22, "923000": 22, "939000": 22, "507000": 22, "89900": 22, "833500": 22, "738500": 22, "847000": 22, "95450": 22, "966500": 22, "80500": 22, "769500": 22, "731000": 22, "752500": 22, "81300": 22, "760000": 22, "650500": 22, "772500": 22, "936500": 22, "chr4": 22, "32158998": 22, "32159443": 22, "78950": 22, "524500": 22, "791500": 22, "767750": 22, "759500": 22, "73275": 22, "791750": 22, "693250": 22, "79225": 22, "441325": 22, "69625": 22, "538750": 22, "667000": 22, "391250": 22, "46975": 22, "616500": 22, "479250": 22, "704750": 22, "549750": 22, "chr13": 22, "90011452": 22, "90011460": 22, "29100": 22, "222000": 22, "225000": 22, "228500": 22, "08115": 22, "476500": 22, "009400": 22, "329500": 22, "32350": 22, "231000": 22, "30700": 22, "390500": 22, "402500": 22, "782000": 22, "06915": 22, "314500": 22, "285500": 22, "812000": 22, "821500": 22, "126765779": 22, "126765801": 22, "81850": 22, "885000": 22, "903000": 22, "565000": 22, "87500": 22, "875000": 22, "894000": 22, "93750": 22, "906000": 22, "68550": 22, "721500": 22, "649000": 22, "783500": 22, "80050": 22, "793500": 22, "809500": 22, "764500": 22, "142126735": 22, "142126952": 22, "84475": 22, "959250": 22, "902000": 22, "706500": 22, "94350": 22, "781250": 22, "530250": 22, "97100": 22, "933250": 22, "82150": 22, "774000": 22, "728250": 22, "869750": 22, "87600": 22, "796500": 22, "863000": 22, "842750": 22, "870500": 22, "21947352": 22, "21947371": 22, "67700": 22, "261333": 22, "766667": 22, "953667": 22, "979333": 22, "93500": 22, "897333": 22, "613667": 22, "864333": 22, "89500": 22, "950667": 22, "75200": 22, "733333": 22, "855667": 22, "874333": 22, "77000": 22, "589333": 22, "506333": 22, "868333": 22, "781000": 22, "chr6": 22, "50328671": 22, "50328782": 22, "38975": 22, "316650": 22, "702250": 22, "820000": 22, "85425": 22, "458000": 22, "662250": 22, "608250": 22, "81025": 22, "889500": 22, "61000": 22, "592250": 22, "047500": 22, "632250": 22, "64975": 22, "605250": 22, "608500": 22, "305500": 22, "407325": 22, "chr12": [22, 24], "15047867": 22, "15047932": 22, "68650": 22, "750000": [22, 35], "825000": 22, "777500": 22, "266500": 22, "31450": 22, "874500": 22, "65700": 22, "867500": 22, "71350": 22, "591000": 22, "290500": 22, "740500": 22, "74400": 22, "723000": 22, "735000": 22, "755500": 22, "559500": 22, "chr8": [22, 24], "48317445": 22, "48317685": 22, "87640": 22, "938400": 22, "937200": 22, "596000": 22, "66820": 22, "844600": 22, "885600": 22, "746000": 22, "80600": 22, "880800": 22, "90840": 22, "804800": 22, "765800": 22, "848800": 22, "76380": 22, "824400": 22, "810400": 22, "916000": 22, "935600": 22, "chr2": 22, "93512302": 22, "93512369": 22, "31350": 22, "788000": 22, "581000": 22, "832500": 22, "604500": 22, "94450": 22, "937500": 22, "900500": 22, "890000": 22, "90750": 22, "956500": 22, "32550": 22, "232700": 22, "221000": 22, "799000": 22, "55550": 22, "614000": 22, "601000": 22, "435400": 22, "470000": 22, "color_palett": 22, "gc": 22, "majortype_color": 22, "silver": 22, "crimson": 22, "deepskyblu": 22, "darkseagreen": 22, "darkcyan": 22, "k": 22, "de9f00": 22, "67292f": 22, "e13f82": 22, "3f884e": 22, "78772b": 22, "4ac750": 22, "6bc22e": 22, "a8d617": 22, "314b1f": 22, "aab639": 22, "e179a2": 22, "764a71": 22, "63bc9e": 22, "cea675": 22, "a87331": 22, "9bb070": 22, "9fa600": 22, "5e461": 22, "8000": 22, "sort": [22, 34], "sum": [22, 32], "row_major_typ": 22, "cell_class_colors1": 22, "majortype_colors1": 22, "left_ha": [22, 29, 34], "right_ha": [22, 29, 34], "all_dmr_heatmap": 22, "set2": 23, "tomato": 23, "skyblu": [23, 24], "spectral_r": 23, "vertical": 23, "first": 23, "051388888888887": 23, "boarder": 23, "continnu": 23, "deep": 23, "otherwis": 23, "your_color": 23, "treat": 23, "sometim": 23, "without": 23, "do": 23, "give": 23, "anoth": [23, 25], "No": [23, 24, 28, 32], "open": [23, 24, 29], "mammal_arrai": 23, "pkl": 23, "rb": [23, 24, 29], "col_colors_dict": [23, 24, 34], "gsm4412025": 23, "gsm4412026": 23, "gsm4412027": 23, "gsm4412028": 23, "gsm4412029": 23, "gsm4412030": 23, "gsm4412031": 23, "gsm4412032": 23, "gsm4412033": 23, "gsm4412034": 23, "gsm4997945": 23, "gsm4997946": 23, "gsm4997947": 23, "gsm4997948": 23, "gsm4997949": 23, "gsm4997950": 23, "gsm4997951": 23, "gsm4997952": 23, "gsm4997953": 23, "gsm4997954": 23, "sheep": 23, "435033": 23, "432900": 23, "446626": 23, "449123": 23, "497180": 23, "515918": 23, "483706": 23, "504681": 23, "529076": 23, "446443": 23, "beluga": 23, "whale": 23, "687488": 23, "694207": 23, "706525": 23, "702734": 23, "687014": 23, "704003": 23, "705887": 23, "693806": 23, "719417": 23, "712677": 23, "560381": 23, "571552": 23, "610392": 23, "613619": 23, "675832": 23, "668502": 23, "624820": 23, "658377": 23, "702334": 23, "575034": 23, "hous": 23, "vaquita": 23, "693523": 23, "702525": 23, "716792": 23, "725095": 23, "711261": 23, "708651": 23, "717952": 23, "705486": 23, "720915": 23, "724171": 23, "581862": 23, "594443": 23, "628908": 23, "639457": 23, "680801": 23, "707493": 23, "662338": 23, "665142": 23, "751859": 23, "584952": 23, "larg": [23, 28], "fly": 23, "fox": 23, "286822": 23, "269406": 23, "296796": 23, "314719": 23, "305074": 23, "308419": 23, "268421": 23, "297236": 23, "295019": 23, "297973": 23, "281684": 23, "363926": 23, "367216": 23, "359392": 23, "289821": 23, "257351": 23, "234301": 23, "295840": 23, "249336": 23, "344848": 23, "greater": 23, "horsesho": 23, "bat": 23, "530606": 23, "517989": 23, "520719": 23, "525855": 23, "507583": 23, "511993": 23, "528107": 23, "521563": 23, "542159": 23, "509526": 23, "735151": 23, "714805": 23, "708868": 23, "738207": 23, "823604": 23, "828040": 23, "823382": 23, "796882": 23, "878783": 23, "693625": 23, "littl": 23, "brown": 23, "202525": 23, "215990": 23, "238977": 23, "241872": 23, "267750": 23, "236729": 23, "202384": 23, "240519": 23, "251148": 23, "267257": 23, "883": 23, "predictedtaxid": 23, "predictedspeci": 23, "common_nam": 23, "9940": 23, "ovis_aries_rambouillet": 23, "bovida": 23, "9749": 23, "delphinapterus_leuca": 23, "monodontida": 23, "10090": 23, "mus_musculu": 23, "murida": 23, "42100": 23, "phocoena_sinu": 23, "phocoenida": 23, "132908": 23, "pteropus_vampyru": 23, "pteropodida": 23, "59479": 23, "rhinolophus_ferrumequinum": 23, "rhinolophida": 23, "59463": 23, "myotis_lucifugu": 23, "vespertilionida": 23, "gse": 23, "basenam": 23, "ncbi_scientific_nam": 23, "taxid": 23, "tissu": [23, 24], "sex": [23, 24], "speci": 23, "successr": 23, "gse147003": 23, "ovi": 23, "ari": 23, "blood": [23, 34], "femal": [23, 24], "artiodactyla": 23, "765818": 23, "797669": 23, "759256": 23, "male": [23, 24], "749813": 23, "770433": 23, "gse164127": 23, "eptesicu": 23, "fuscu": 23, "29078": 23, "skin": 23, "chiroptera": 23, "660425": 23, "652822": 23, "664746": 23, "650848": 23, "657170": 23, "a40043": [23, 34], "00e5ff": 23, "00becc": 23, "cerebellum": 23, "b2f8ff": 23, "striatum": 23, "6cbf00": [23, 34], "liver": [23, 24, 34], "ffccee": [23, 34], "muscl": [23, 29, 34], "1e9351": 23, "put": 23, "auc": 23, "legend_pad": 23, "veri": 23, "abov": 23, "25": [23, 26, 28, 31, 32], "930555555555557": [23, 28], "last": 23, "second": 23, "worth": 23, "note": 23, "betwe": 23, "kind": [23, 29, 30], "predict": 23, "68888888888889": 23, "tranpos": 23, "columsn": 23, "obtain": 24, "pmid": [24, 34], "36617464": 24, "linearsegmentedcolormap": [24, 25], "80": [24, 29, 33, 35], "urllib": 24, "influence_of_snp_on_beta": 24, "close": 24, "snp": 24, "row_colors_dict": 24, "respect": 24, "2000": 24, "204875570030_r01c02": 24, "204875570030_r04c01": 24, "cg30848532_tc21": 24, "525089": 24, "419515": 24, "cg30147375_bc21": 24, "803776": 24, "585928": 24, "cg46239718_bc21": 24, "443958": 24, "517514": 24, "cg36100119_bc21": 24, "351977": 24, "528846": 24, "cg42738582_bc21": 24, "783958": 24, "724901": 24, "204875570030_r05c01": 24, "204875570030_r06c01": 24, "204875570035_r05c02": 24, "483276": 24, "460750": 24, "390317": 24, "510269": 24, "831463": 24, "550146": 24, "535909": 24, "450167": 24, "564107": 24, "524896": 24, "374422": 24, "551200": 24, "802178": 24, "848621": 24, "850481": 24, "target": 24, "extensionbas": 24, "probedesign": 24, "con": 24, "mapflag": 24, "ii": 24, "chr19": 24, "suboptim": 24, "hybrid": 24, "effect": 24, "artifici": 24, "low": [24, 25, 28], "meth": [24, 34], "cg": 24, "gg": 24, "ga": 24, "79760438": 24, "ca": 24, "ac": 24, "ag": 24, "109557651": 24, "gt": [24, 26], "117860829": 24, "5877949": 24, "aa": 24, "122031379": 24, "strain": 24, "molf_eij": 24, "frontal": 24, "lobe": 24, "cast_eij": 24, "darkorang": 24, "wheat": 24, "darkgrai": 24, "69": [24, 27, 34], "4986111111111": 24, "col_ha1": 24, "cm1": 24, "viridi": 24, "col_ha2": 24, "my_cmap": 24, "from_list": [24, 25], "cm2": 24, "dnam": 24, "beta_snp": 24, "df_corr": 25, "kycg_modules_correl": 25, "csv": [25, 27, 34, 35], "df_ann": 25, "kycg_modules_annot": 25, "kycg_modules_beta": 25, "cpg_std": 25, "std": [25, 28], "cpg_mean": 25, "astyp": [25, 28], "value_count": [25, 28], "28": [25, 26, 28], "33": 25, "count": [25, 27, 28, 29], "int64": 25, "isin": [25, 28], "hm": 25, "h3k4me1": 25, "h3k79me2": 25, "h3k79me3": 25, "h3f3a": 25, "h4k20me1": 25, "h2bk20ac": 25, "h3k23me2": 25, "h2afi": 25, "h2ak9ac": 25, "h2bk12ac": 25, "h2bk15ac": 25, "h3k9me1": 25, "h2bk120ub": 25, "h2azac": 25, "h3k79me1": 25, "h2a": 25, "h3_4ac": 25, "h3k27me1": 25, "h3k4me3b": 25, "h3k56ac": 25, "h3k9ac_h3k14ac": 25, "h3k9k14ac": 25, "cenpa": 25, "h1": 25, "h2afz": 25, "h2ak119": 25, "h3": 25, "h3ac": 25, "h3k27ac": 25, "h3k4me3": 25, "h3k9ac": 25, "h4": 25, "h4ac": 25, "h4k5ac": 25, "h4k5ac_h4k8ac_h4k12ac_h4k16ac": 25, "h4k8ac": 25, "histonelysineacetyl": 25, "histonelysinecrotonyl": 25, "h3k18cr": 25, "h4k16ac": 25, "h4k12ac": 25, "h3k27me3b": 25, "h2bub": 25, "h2ak5ac": 25, "h3k36me3b": 25, "h2bk120ac": 25, "h3k36ac": 25, "h4k91ac": 25, "h4k20me3": 25, "h2afy2": 25, "h3k36me2": 25, "h3k27me3": 25, "h2ak119ub": 25, "cg04735237": 25, "cg00643814": 25, "cg24865495": 25, "cg25376651": 25, "cg27485084": 25, "cg12609052": 25, "cg07354679": 25, "cg02592525": 25, "cg12747056": 25, "cg17718960": 25, "cg19987665": 25, "cg18406033": 25, "cg09678971": 25, "cg00461612": 25, "cg18689454": 25, "cg11923631": 25, "cg27251412": 25, "cg08089567": 25, "cg16717549": 25, "cg09510531": 25, "559916": 25, "686845": 25, "561415": 25, "699960": 25, "711086": 25, "563690": 25, "527953": 25, "621757": 25, "623319": 25, "048462": 25, "103904": 25, "002533": 25, "074093": 25, "114526": 25, "151763": 25, "140823": 25, "117356": 25, "100510": 25, "126291": 25, "538034": 25, "568960": 25, "687787": 25, "593650": 25, "676068": 25, "606604": 25, "589549": 25, "747987": 25, "185989": 25, "087952": 25, "242909": 25, "065101": 25, "054958": 25, "096098": 25, "089246": 25, "078234": 25, "025965": 25, "069120": 25, "614696": 25, "646452": 25, "786480": 25, "602911": 25, "519926": 25, "642344": 25, "642081": 25, "022499": 25, "108524": 25, "015151": 25, "028957": 25, "274560": 25, "302464": 25, "284823": 25, "265835": 25, "262931": 25, "268134": 25, "564983": 25, "727579": 25, "624186": 25, "416198": 25, "505130": 25, "596338": 25, "030346": 25, "122412": 25, "063630": 25, "007048": 25, "197339": 25, "226034": 25, "218230": 25, "180297": 25, "181867": 25, "204431": 25, "655515": 25, "599935": 25, "590981": 25, "627786": 25, "674095": 25, "020895": 25, "058742": 25, "078318": 25, "055019": 25, "139691": 25, "168351": 25, "160081": 25, "142216": 25, "114554": 25, "144024": 25, "512": 25, "chromhmm": [25, 28], "chromhmm_bioc": 25, "tfb": 25, "txwk": [25, 28], "hdgf": 25, "rbfox2": 25, "srebf1": 25, "pqbp1": 25, "qui": [25, 28], "ascl1": 25, "casz1": 25, "ebf1": 25, "fezf1": 25, "foxm1": 25, "hand2": 25, "hdac3": 25, "irf2b": 25, "crx": 25, "dnmt3b": 25, "otx2": 25, "rorb": 25, "zmynd11": 25, "znf711": 25, "macrod1": 25, "atf2": 25, "batf": 25, "etv6": 25, "fo": 25, "ikzf2": 25, "irf4": 25, "junb": 25, "maf": 25, "mafg": 25, "me": 25, "fry": 25, "pdx1": 25, "keep_cpg": 25, "map": [25, 28, 29], "stack": [25, 28, 32], "keep_hm": 25, "154": 25, "245870": 25, "768918": 25, "319166": 25, "635635": 25, "270459": 25, "835897": 25, "276320": 25, "618949": 25, "276748": 25, "700196": 25, "23716": 25, "all_cmap": 25, "binar": 25, "colgroup": [25, 28], "middl": [25, 28], "heatmap_ax": [25, 29], "j": 25, "set_vis": [25, 26, 29], "var": [25, 32], "folder": 25, "3q": 25, "hwjjddlj65b030m69cfbdj4h0000gq": 25, "ipykernel_61938": 25, "92826381": 25, "matplotlibdeprecationwarn": 25, "deprec": 25, "minor": [25, 28, 29], "releas": 25, "regist": 25, "dotclustermap2": 25, "dev4": 26, "g71c725a": 26, "d20240410": 26, "ax_top": 26, "ax_right": 26, "set_axis_on": 26, "set_linestyl": [26, 29], "agg_filt": 26, "anim": 26, "bbox_patch": 26, "lt": 26, "fancybboxpatch": 26, "0x7f854d935760": 26, "children": 26, "clip_path": 26, "600x800": 26, "font_manag": 26, "0x7f854d8d56a0": 26, "fontvari": 26, "gid": 26, "math_fontfamili": 26, "dejavusan": 26, "mouseov": 26, "parse_math": 26, "path_effect": 26, "picker": 26, "000000000000007": 26, "sketch_param": 26, "snap": 26, "stretch": 26, "tightbbox": 26, "125": 26, "112": [26, 28], "875": [26, 34], "blendedaffine2d": 26, "0x7f854d8d5070": 26, "transform_rotates_text": 26, "transformed_clip_path_and_affin": 26, "unitless_posit": 26, "url": 26, "usetex": 26, "window_ext": 26, "wrap": 26, "antialias": 26, "75": [26, 28, 34], "0x7f854dded490": 26, "capstyl": 26, "butt": 26, "data_transform": 26, "affine2d": 26, "0x7f854f0bbfd0": 26, "extent": 26, "761576317947967": 26, "6388888888888813": 26, "92": 26, "78935409572574": 26, "638888888888896": 26, "hatch": 26, "joinstyl": 26, "miter": 26, "solid": 26, "mutation_aspect": 26, "mutation_scal": 26, "444444444444443": 26, "patch_transform": 26, "identitytransform": 26, "0x7f854d868730": 26, "63888889": 26, "81": [26, 33], "38888889": 26, "79": 26, "uint8": 26, "vert": 26, "76157632": 26, "7893541": 26, "expr": 27, "test_expression_dataset": 27, "selected_row": 27, "adam7": 27, "hla": 27, "doa": 27, "ptgir": 27, "opcml": 27, "it1": 27, "mageb1": 27, "label_row": 27, "c8orf34": 27, "as1": 27, "smim28": 27, "linc01114": 27, "klk3": 27, "traj45": 27, "tpmtp1": 27, "hnrnpcp4": 27, "89": [27, 33], "nci": 27, "h660": 27, "vcap": 27, "du": 27, "145": 27, "mda": 27, "pca": 27, "2b": 27, "22rv1": 27, "lncap": 27, "fgc": 27, "sup": 27, "hd1": 27, "l": 27, "540": 27, "1236": 27, "km": 27, "h2": 27, "hdlm": 27, "616": 27, "428": 27, "611": 27, "TO": 27, "175": 27, "044425": 27, "303654": 27, "604782": 27, "123861": 27, "015847": 27, "848602": 27, "215875": 27, "251406": 27, "795724": 27, "092080": 27, "070939": 27, "705117": 27, "241584": 27, "766805": 27, "132841": 27, "531804": 27, "564560": 27, "832337": 27, "442301": 27, "472336": 27, "359856": 27, "911472": 27, "795981": 27, "391838": 27, "rank2": 27, "regul": [27, 29], "top20": 27, "upregul": 27, "goi": 27, "colors_dict": 27, "row_ha_left": 27, "row_ha_right": 27, "uncom": 27, "sn": 28, "subset": 28, "collaps": 28, "load_dataset": 28, "brain_network": 28, "header": 28, "used_network": 28, "used_column": 28, "get_level_valu": 28, "comput": 28, "form": 28, "corr_mat": 28, "corr": 28, "level": [28, 34], "level_0": 28, "level_1": 28, "lh": 28, "rh": 28, "881516": 28, "049793": 28, "026902": 28, "144335": 28, "141253": 28, "119250": 28, "261589": 28, "272701": 28, "370021": 28, "366065": 28, "325680": 28, "196770": 28, "144566": 28, "366818": 28, "388756": 28, "352529": 28, "363982": 28, "341524": 28, "350452": 28, "112697": 28, "036909": 28, "144277": 28, "189683": 28, "084633": 28, "324230": 28, "332886": 28, "374322": 28, "361036": 28, "274151": 28, "142392": 28, "070452": 28, "358625": 28, "402173": 28, "302286": 28, "339989": 28, "315931": 28, "343379": 28, "343464": 28, "470239": 28, "100802": 28, "438449": 28, "339667": 28, "089811": 28, "272394": 28, "036493": 28, "171179": 28, "043298": 28, "158039": 28, "005598": 28, "060007": 28, "079078": 28, "040060": 28, "027878": 28, "075781": 28, "130549": 28, "278569": 28, "127621": 28, "014404": 28, "051249": 28, "090130": 28, "170053": 28, "124278": 28, "112148": 28, "063705": 28, "172007": 28, "040629": 28, "079687": 28, "024864": 28, "092263": 28, "068858": 28, "521377": 28, "506652": 28, "320966": 28, "141884": 28, "608392": 28, "075986": 28, "095015": 28, "012966": 28, "082816": 28, "023340": 28, "058718": 28, "034181": 28, "033355": 28, "022982": 28, "025638": 28, "431619": 28, "418708": 28, "084634": 28, "132": [28, 29], "808800071564": 28, "22361111111111": 28, "wding": 28, "miniconda3": 28, "env": 28, "lib": 28, "python3": 28, "site": 28, "189": 28, "userwarn": 28, "via": 28, "norm": 28, "465277777777779": 28, "drop_dupl": 28, "rowgroup": 28, "kycg_result": 28, "rmsk1": 28, "ensregbuild": 28, "sampleid": [28, 32], "clark2018_argelaguet2019": 28, "dataset1": 28, "luo2022": 28, "dataset2": 28, "max_p": 28, "log10": [28, 29], "pval": 28, "id": [28, 29, 34], "cpgtype": 28, "vc": 28, "term": [28, 29], "p_max": 28, "p_min": 28, "odds_ratio": 28, "pvalu": 28, "enrichtyp": 28, "het": 28, "061": 28, "neg": 28, "020000e": 28, "07": [28, 29], "26": 28, "tx": 28, "029": 28, "580000e": 28, "65": 28, "tssflnk": 28, "056": 28, "370000e": 28, "06": [28, 29], "022": 28, "660000e": 28, "346": 28, "350": 28, "760000e": 28, "02": 28, "describ": 28, "64": 28, "356918": 28, "632073": 28, "119186": 28, "deplet": 28, "srprna": 28, "satellit": 28, "simple_repeat": 28, "low_complex": 28, "reprpcwk": 28, "tssbiv": 28, "reprpc": 28, "ltr": 28, "snrna": 28, "sine": 28, "rdylgn_r": 28, "coolwarm_r": 28, "dotheatmap1": 28, "180": 28, "09263168093761": 28, "20230227_kycg": 28, "516666666666666": 28, "glob": 29, "style": 29, "dev0": 29, "g8abf70a": 29, "d20240415": 29, "tradition": 29, "lot": [29, 31], "dotplot": 29, "had": 29, "idea": 29, "manuscript": 29, "alreadi": 29, "gotten": 29, "bp": 29, "molecular": 29, "mf": 29, "kegg": 29, "pathwai": 29, "enrichment_analysis_dotheatmap": 29, "data1": 29, "descript": 29, "generatio": 29, "enrichedcategori": 29, "colid": 29, "rowid": 29, "0003779": 29, "actin": 29, "bind": 29, "044949": 29, "453212e": 29, "186": 29, "group2": 29, "ct1": 29, "610265": 29, "0030695": 29, "gtpase": 29, "activ": 29, "043016": 29, "744023e": 29, "178": 29, "171081": 29, "0060589": 29, "nucleosid": 29, "triphosphatas": 29, "0005201": 29, "extracellular": 29, "structur": 29, "constitu": 29, "021266": 29, "810239e": 29, "88": 29, "34": 29, "235806": 29, "0050839": 29, "adhes": 29, "molecul": 29, "031899": 29, "612420e": 29, "792522": 29, "840": 29, "0050905": 29, "neuromuscular": 29, "010454": 29, "224335e": 29, "122": 29, "ct2": 29, "912100": 29, "1205": 29, "0030534": 29, "adult": 29, "behavior": 29, "009426": 29, "028861e": 29, "110": 29, "219765": 29, "1558": 29, "0035019": 29, "somat": 29, "stem": 29, "popul": 29, "mainten": 29, "004799": 29, "029991e": 29, "219683": 29, "1746": 29, "0019827": 29, "009854": 29, "467128e": 29, "04": 29, "833532": 29, "2999": 29, "0021953": 29, "central": 29, "nervou": 29, "system": 29, "neuron": [29, 34], "008997": 29, "138960e": 29, "105": 29, "039103": 29, "139": 29, "metal": 29, "ion": 29, "transmembran": 29, "transport": 29, "monoatom": 29, "channel": 29, "gate": 29, "calmodulin": 29, "receptor": 29, "protein": 29, "kinas": 29, "substrat": 29, "organ": 29, "extern": 29, "encapsul": 29, "axon": 29, "guidanc": 29, "locomotori": 29, "fat": 29, "amphetamin": 29, "addict": 29, "calcium": 29, "signal": 29, "cocain": 29, "cytokin": 29, "interact": 29, "neuroact": 29, "ligand": 29, "cytoskeleton": 29, "ecm": 29, "viral": 29, "cy": 29, "focal": 29, "lupu": 29, "erythematosu": 29, "atp": 29, "chromatin": 29, "remodel": 29, "histon": 29, "modif": 29, "group1": 29, "ct3": 29, "ct4": 29, "s_min": 29, "s_max": 29, "3074": 29, "0021954": 29, "004586": 29, "027429": 29, "561784": 29, "0045444": 29, "176471": 29, "000382": 29, "417494": 29, "orrd": 29, "1e": 29, "ast": 29, "ravel": 29, "both": 29, "dashdot": 29, "cell_class_dotheatmap": 29, "106": 29, "46754159267321": 29, "ratio": 29, "total": 29, "510843": 30, "531995": 30, "188667": 30, "180811": 30, "354718": 30, "whole": 30, "separ": 30, "wiki": 31, "ital": 31, "_label_kw": 31, "tree": 31, "roundtooth": 31, "book": 32, "cbioport": 32, "tcga_lung_adenocarcinoma_provisional_ras_raf_mek_jnk_signal": 32, "dropna": 32, "mut": 32, "amp": [32, 33], "homdel": 32, "4384": 32, "kra": 32, "hra": 32, "braf": 32, "raf1": 32, "map3k1": 32, "unique_vari": 32, "v1": 32, "strip": 32, "008000": 32, "mutat": 32, "frequenc": 32, "row_vc": 32, "col_vc": 32, "kras_sampl": 32, "df_col_split": 32, "op": [32, 33], "alter": 32, "27": [32, 33], "72361111111111": 32, "ad": 32, "row_var_freq": 32, "isvar": 32, "oncoprint2": 32, "430555555555557": 32, "toi": 33, "sample_": 33, "gene_": 33, "alts_lol": 33, "amp_valu": 33, "randint": 33, "del_valu": 33, "neut_valu": 33, "alts_df": 33, "neut": 33, "prepar": 33, "annot_1_df": 33, "annot1": 33, "annot_2_df": 33, "500": 33, "annot2": 33, "annot_3_df": 33, "patient": 33, "sample_1": 33, "gene_1": 33, "gene_2": 33, "gene_3": 33, "gene_4": 33, "gene_5": 33, "95": 33, "sample_10": 33, "gene_6": 33, "96": 33, "gene_7": 33, "97": 33, "gene_8": 33, "98": 33, "gene_9": 33, "gene_10": 33, "patient2": 33, "sample_2": 33, "patient5": 33, "sample_3": 33, "patient3": 33, "sample_4": 33, "patient4": 33, "sample_5": 33, "patient1": 33, "sample_6": 33, "sample_7": 33, "sample_8": 33, "sample_9": 33, "77": 33, "78": 33, "1114": 33, "2698": 33, "4718": 33, "4633": 33, "3444": 33, "1984": 33, "2088": 33, "2316": 33, "2939": 33, "1373": 33, "a3": [33, 35], "a1": [33, 35], "a2": [33, 35], "flatten": 33, "signatur": 34, "come": 34, "36599988": 34, "raw": 34, "githubusercont": 34, "adipocyt": 34, "bladder": 34, "epithelium": 34, "mem": 34, "granulocyt": 34, "colon": 34, "macrophag": 34, "monocyt": 34, "interstiti": 34, "alveolar": 34, "pancrea": 34, "duct": 34, "skelet": 34, "endocrin": 34, "bronchu": 34, "smooth": 34, "prostat": 34, "coronari": 34, "arteri": 34, "aorta": 34, "thyroid": 34, "110426397": 34, "120667": 34, "9102": 34, "8285": 34, "918667": 34, "750667": 34, "8705": 34, "896": 34, "916333": 34, "953333": 34, "9450": 34, "61225": 34, "5485": 34, "94175": 34, "952": 34, "417": 34, "150": 34, "229": 34, "294": 34, "214": 34, "938333": 34, "110426639": 34, "420000": 34, "9304": 34, "8920": 34, "955667": 34, "964667": 34, "9285": 34, "903": 34, "924667": 34, "975333": 34, "9720": 34, "94575": 34, "8990": 34, "96225": 34, "963": 34, "606": 34, "185": 34, "348": 34, "917": 34, "484": 34, "972333": 34, "110426659": 34, "425667": 34, "9328": 34, "7635": 34, "933000": 34, "942333": 34, "8945": 34, "931": 34, "963667": 34, "899333": 34, "8895": 34, "93875": 34, "8390": 34, "92225": 34, "559": 34, "129": 34, "292": 34, "395": 34, "989667": 34, "110427109": 34, "203667": 34, "9724": 34, "9610": 34, "968667": 34, "9240": 34, "939": 34, "985000": 34, "9620": 34, "73300": 34, "7570": 34, "96525": 34, "929": 34, "225": 34, "806": 34, "951": 34, "905": 34, "974333": 34, "110427116": 34, "217333": 34, "9478": 34, "0000": 34, "976000": 34, "970667": 34, "9730": 34, "913": 34, "975000": 34, "970333": 34, "9800": 34, "65125": 34, "7955": 34, "96150": 34, "000": 34, "605": 34, "857": 34, "868": 34, "981000": 34, "clusterid": 34, "ep": 34, "granul": 34, "mono": 34, "macro": 34, "nk": 34, "bone": 34, "osteob": 34, "breast": 34, "basal": 34, "dermal": 34, "fibro": 34, "epid": 34, "kerat": 34, "eryth": 34, "prog": 34, "fallopian": 34, "gallbladd": 34, "gastric": 34, "neck": 34, "heart": 34, "cardio": 34, "kidnei": 34, "hep": 34, "alveo": 34, "bron": 34, "oligodend": 34, "acinar": 34, "musc": 34, "1e93a": 34, "ff9c00": 34, "ff9f7f": 34, "ff7f00": 34, "ff2e8d": 34, "cc0043": 34, "e5e5e5": 34, "cc4407": 34, "cc843d": 34, "663d28": 34, "1e937c": 34, "40705f": 34, "009351": 34, "e7e4bf": 34, "cca300": 34, "002929": 34, "ff99aa": 34, "ff99ff": 34, "f6ff99": 34, "ba99ff": 34, "ccccff": 34, "9e542": 34, "2ca02c": 34, "df7f00": 34, "ffd866": 34, "ffcc32": 34, "7f4c33": 34, "cc9951": 34, "b2bfff": 34, "loyfer2023_heatmap": 34, "981944444444444": 34, "download": 35, "pycomplexheatmap_test_2": 35, "c1": 35, "c2": 35, "c3": 35, "c4": 35, "c5": 35, "c6": 35, "c7": 35, "c8": 35, "c9": 35, "c10": 35, "c35": 35, "c36": 35, "c37": 35, "c38": 35, "c39": 35, "c40": 35, "c41": 35, "c42": 35, "c43": 35, "c44": 35, "00000": 35, "00": 35, "250000": 35, "111111": 35, "444444": 35, "222222": 35, "388889": 35, "611111": 35, "277778": 35, "888889": 35, "944444": 35, "555556": 35, "500000": 35, "a4": 35, "625000": 35, "375000": 35, "a5": 35, "a6": 35, "600000": 35, "100000": 35, "300000": 35, "700000": 35, "a7": 35, "a8": 35, "60000": 35, "800000": 35, "400000": 35, "200000": 35, "a9": 35, "440000": 35, "72": 35, "460000": 35, "040000": 35, "080000": 35, "560000": 35, "020000": 35, "240000": 35, "a10": 35, "a11": 35, "357143": 35, "02381": 35, "023810": 35, "071429": 35, "571429": 35, "214286": 35, "452381": 35, "166667": 35, "380952": 35, "309524": 35, "a12": 35, "062500": 35, "031250": 35, "312500": 35, "218750": 35, "531250": 35, "156250": 35, "687500": 35, "343750": 35, "437500": 35, "a13": 35, "333333": 35, "666667": 35, "a14": 35, "a15": 35, "076923": 35, "615385": 35, "384615": 35, "538462": 35, "307692": 35, "153846": 35, "a16": 35, "a17": 35, "150000": 35, "900000": 35, "550000": 35, "350000": 35, "a18": 35, "833333": 35, "a19": 35, "rdpu": 35}, "objects": {"": [[0, 0, 0, "-", "PyComplexHeatmap"]], "PyComplexHeatmap": [[1, 0, 0, "-", "annotations"], [2, 0, 0, "-", "clustermap"], [3, 0, 0, "-", "colors"], [4, 0, 0, "-", "dotHeatmap"], [5, 0, 0, "-", "example"], [6, 0, 0, "-", "oncoPrint"], [8, 0, 0, "-", "utils"]], "PyComplexHeatmap.annotations": [[1, 1, 1, "", "AnnotationBase"], [1, 1, 1, "", "HeatmapAnnotation"], [1, 1, 1, "", "anno_barplot"], [1, 1, 1, "", "anno_boxplot"], [1, 1, 1, "", "anno_img"], [1, 1, 1, "", "anno_label"], [1, 1, 1, "", "anno_lineplot"], [1, 1, 1, "", "anno_scatterplot"], [1, 1, 1, "", "anno_simple"]], "PyComplexHeatmap.annotations.AnnotationBase": [[1, 2, 1, "", "get_label_width"], [1, 2, 1, "", "get_max_label_width"], [1, 2, 1, "", "get_ticklabel_width"], [1, 2, 1, "", "reorder"], [1, 2, 1, "", "set_axes_kws"], [1, 2, 1, "", "set_label"], [1, 2, 1, "", "set_legend"], [1, 2, 1, "", "update_plot_kws"]], "PyComplexHeatmap.annotations.HeatmapAnnotation": [[1, 2, 1, "", "collect_legends"], [1, 2, 1, "", "plot_annotations"], [1, 2, 1, "", "plot_legends"], [1, 2, 1, "", "set_axes_kws"], [1, 2, 1, "", "show_ticklabels"]], "PyComplexHeatmap.annotations.anno_barplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_boxplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_img": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_label": [[1, 2, 1, "", "get_ticklabel_width"], [1, 2, 1, "", "plot"], [1, 2, 1, "", "set_plot_kws"], [1, 2, 1, "", "set_side"]], "PyComplexHeatmap.annotations.anno_lineplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_scatterplot": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.annotations.anno_simple": [[1, 2, 1, "", "plot"]], "PyComplexHeatmap.clustermap": [[2, 1, 1, "", "ClusterMapPlotter"], [2, 1, 1, "", "DendrogramPlotter"], [2, 4, 1, "", "composite"], [2, 4, 1, "", "heatmap"], [2, 1, 1, "", "heatmapPlotter"], [2, 4, 1, "", "plot_heatmap"]], "PyComplexHeatmap.clustermap.ClusterMapPlotter": [[2, 2, 1, "", "cal_cold_between_groups"], [2, 2, 1, "", "cal_rowd_between_groups"], [2, 2, 1, "", "calculate_col_dendrograms"], [2, 2, 1, "", "calculate_row_dendrograms"], [2, 2, 1, "", "collect_legends"], [2, 2, 1, "", "format_data"], [2, 2, 1, "", "plot"], [2, 2, 1, "", "plot_dendrograms"], [2, 2, 1, "", "plot_legends"], [2, 2, 1, "", "plot_matrix"], [2, 2, 1, "", "post_processing"], [2, 2, 1, "", "set_axes_labels_kws"], [2, 2, 1, "", "set_height"], [2, 2, 1, "", "set_width"], [2, 2, 1, "", "set_xy_labels"], [2, 2, 1, "", "standard_scale"], [2, 2, 1, "", "tight_layout"], [2, 2, 1, "", "z_score"]], "PyComplexHeatmap.clustermap.DendrogramPlotter": [[2, 2, 1, "", "calculate_dendrogram"], [2, 3, 1, "", "calculated_linkage"], [2, 2, 1, "", "check_array"], [2, 2, 1, "", "get_coords"], [2, 2, 1, "", "plot"], [2, 3, 1, "", "reordered_ind"]], "PyComplexHeatmap.clustermap.heatmapPlotter": [[2, 2, 1, "", "plot"]], "PyComplexHeatmap.colors": [[3, 4, 1, "", "define_cmap"], [3, 4, 1, "", "register_cmap"]], "PyComplexHeatmap.dotHeatmap": [[4, 1, 1, "", "DotClustermapPlotter"], [4, 4, 1, "", "dotHeatmap2d"], [4, 4, 1, "", "scale"]], "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter": [[4, 2, 1, "", "collect_legends"], [4, 2, 1, "", "format_data"], [4, 2, 1, "", "plot_matrix"], [4, 2, 1, "", "post_processing"]], "PyComplexHeatmap.example": [[5, 4, 1, "", "clustermap_example0"], [5, 4, 1, "", "clustermap_example1"], [5, 4, 1, "", "get_kycg_example_data"]], "PyComplexHeatmap.oncoPrint": [[6, 1, 1, "", "oncoPrintPlotter"], [6, 4, 1, "", "oncoprint"]], "PyComplexHeatmap.oncoPrint.oncoPrintPlotter": [[6, 2, 1, "", "add_default_annotations"], [6, 2, 1, "", "collect_legends"], [6, 2, 1, "", "format_data"], [6, 2, 1, "", "get_samples_order"], [6, 2, 1, "", "plot_matrix"], [6, 2, 1, "", "post_processing"]], "PyComplexHeatmap.utils": [[8, 4, 1, "", "axis_ticklabels_overlap"], [8, 4, 1, "", "cal_legend_width"], [8, 4, 1, "", "cluster_labels"], [8, 4, 1, "", "define_cmap"], [8, 4, 1, "", "despine"], [8, 4, 1, "", "get_colormap"], [8, 4, 1, "", "plot_cmap_legend"], [8, 4, 1, "", "plot_color_dict_legend"], [8, 4, 1, "", "plot_legend_list"], [8, 4, 1, "", "plot_marker_legend"], [8, 4, 1, "", "set_default_style"], [8, 4, 1, "", "to_utf8"]]}, "objtypes": {"0": "py:module", "1": "py:class", "2": "py:method", "3": "py:property", "4": "py:function"}, "objnames": {"0": ["py", "module", "Python module"], "1": ["py", "class", "Python class"], "2": ["py", "method", "Python method"], "3": ["py", "property", "Python property"], "4": ["py", "function", "Python function"]}, "titleterms": {"pycomplexheatmap": [0, 1, 2, 3, 4, 5, 6, 7, 8, 16, 19, 21], "packag": [0, 22, 30], "submodul": 0, "modul": [0, 1, 2, 3, 4, 5, 6, 7, 8, 25, 37], "content": [0, 9, 11, 12, 13, 15, 16, 18, 20, 36], "annot": [1, 21, 23, 30, 31], "clustermap": [2, 11, 25, 28], "color": [3, 28], "dotheatmap": [4, 13, 29], "exampl": [5, 20, 23, 28], "oncoprint": [6, 32, 33, 36], "tool": 7, "util": 8, "advanc": 9, "usag": 9, "citat": 10, "For": 12, "develop": 12, "galleri": 14, "get": 15, "start": 15, "credit": 16, "thank": 16, "instal": 17, "depend": 17, "how": [17, 21, 29, 31], "kwarg": 18, "more": 20, "gener": 21, "dataset": [21, 24, 28, 32], "add": [21, 23, 28], "select": [21, 29], "row": [21, 23, 30], "label": [21, 31], "top": [21, 23], "heatmap": [21, 24, 28, 30, 31], "cell": [21, 34], "onli": [21, 23], "plot": [21, 23, 25, 28, 29, 30], "chang": [21, 28], "orent": 21, "down": 21, "extra": 21, "space": 21, "left": [21, 23], "right": [21, 23], "orient": 21, "us": [21, 25, 28, 29, 33], "paramet": [21, 28], "multipl": 21, "loop": 21, "cluster": 21, "between": [21, 22], "group": [21, 22, 28], "within": [21, 22], "clsuter": 21, "col_split": 21, "col_clust": 21, "true": 21, "cluster_between_group": 21, "col_split_ord": 21, "fals": 21, "cluster_within_group": 21, "custom": 21, "linkag": 21, "imag": 21, "choos": 21, "build": 21, "colormap": 21, "diverg": 21, "qualit": 21, "cmap": [21, 28], "forc": 21, "displai": 21, "all": 21, "col": 21, "ticklabel": 21, "import": [22, 30], "dmr": 22, "A": [23, 25], "quick": 23, "column": [23, 30], "anno_label": [23, 31], "anno_simpl": 23, "To": 23, "quickli": 23, "you": 23, "just": 23, "need": [23, 29], "datafram": 23, "figur": 23, "legend": 23, "separ": 23, "bottom": 23, "composit": 24, "two": 24, "horizont": 24, "mous": 24, "dna": [24, 34], "methyl": [24, 34], "arrai": 24, "correl": 25, "matrix": 25, "cpg": 25, "dotclustermap": 25, "process": 25, "data": [25, 28, 29, 34], "dot": [25, 28], "smaller": 25, "1": [26, 30], "understand": 26, "layout": 26, "visual": [27, 28, 29, 32, 33, 34], "differenti": 27, "express": 27, "gene": [27, 29], "load": 28, "an": 28, "brain": 28, "network": 28, "from": 28, "seaborn": 28, "tradit": 28, "squar": 28, "marker": [28, 29], "": 28, "simpl": 28, "fix": 28, "size": 28, "point": 28, "hue": 28, "differ": 28, "up": 28, "five": 28, "dimens": 28, "dotclustermapplott": 28, "set": 29, "enrich": 29, "analysi": 29, "result": 29, "clusterprofil": 29, "why": 29, "do": 29, "we": 29, "better": 29, "wai": 29, "read": 29, "2": 30, "3": 30, "split": 30, "4": 30, "along": 30, "side": 30, "main": 30, "label_kw": 31, "xticklabels_kw": 31, "yticklabels_kw": 31, "modifi": 31, "xticklabel": 31, "yticklabel": 31, "xlabel_kw": 31, "ylabel_kw": 31, "xlabel": 31, "ylabel": 31, "control": 31, "gap": 31, "pad": 31, "hgap": 31, "wgap": 31, "heatmapannot": 31, "subplot_gap": 31, "row_split_gap": 31, "col_split_gap": 31, "remov": 31, "arrow": 31, "tcga": 32, "lung": 32, "adenocarcinoma": 32, "carcinoma": 32, "variant": 32, "categor": 33, "variabl": 33, "singl": 34, "loyfer2023": 34, "setup": 37}, "envversion": {"sphinx.domains.c": 3, "sphinx.domains.changeset": 1, "sphinx.domains.citation": 1, "sphinx.domains.cpp": 9, "sphinx.domains.index": 1, "sphinx.domains.javascript": 3, "sphinx.domains.math": 2, "sphinx.domains.python": 4, "sphinx.domains.rst": 2, "sphinx.domains.std": 2, "sphinx.ext.todo": 2, "sphinx.ext.viewcode": 1, "nbsphinx": 4, "sphinx": 58}, "alltitles": {"PyComplexHeatmap package": [[0, "pycomplexheatmap-package"]], "Submodules": [[0, "submodules"]], "Module contents": [[0, "module-PyComplexHeatmap"]], "PyComplexHeatmap.annotations module": [[1, "module-PyComplexHeatmap.annotations"]], "PyComplexHeatmap.clustermap module": [[2, "module-PyComplexHeatmap.clustermap"]], "PyComplexHeatmap.colors module": [[3, "module-PyComplexHeatmap.colors"]], "PyComplexHeatmap.dotHeatmap module": [[4, "module-PyComplexHeatmap.dotHeatmap"]], "PyComplexHeatmap.example module": [[5, "module-PyComplexHeatmap.example"]], "PyComplexHeatmap.oncoPrint module": [[6, "module-PyComplexHeatmap.oncoPrint"]], "PyComplexHeatmap.tools module": [[7, "pycomplexheatmap-tools-module"]], "PyComplexHeatmap.utils module": [[8, "module-PyComplexHeatmap.utils"]], "Advanced Usage": [[9, "advanced-usage"]], "Contents:": [[9, null], [11, null], [12, null], [13, null], [15, null], [16, null], [18, null], [20, null], [36, null]], "Citation": [[10, "citation"]], "Clustermap": [[11, "clustermap"]], "For Developers": [[12, "for-developers"]], "dotHeatmap": [[13, "dotheatmap"]], "Gallery": [[14, "gallery"]], "Get Started": [[15, "get-started"]], "PyComplexHeatmap": [[16, "pycomplexheatmap"], [19, "pycomplexheatmap"]], "Credits and Thanks": [[16, "credits-and-thanks"]], "Installation": [[17, "installation"]], "Dependencies": [[17, "dependencies"]], "How to install?": [[17, "how-to-install"]], "Kwargs": [[18, "kwargs"]], "More Examples": [[20, "more-examples"]], "Generate dataset": [[21, "Generate-dataset"]], "Add selected rows labels": [[21, "Add-selected-rows-labels"]], "Add annotations on the top of heatmap cells": [[21, "Add-annotations-on-the-top-of-heatmap-cells"]], "Only plot the annotations": [[21, "Only-plot-the-annotations"]], "Change orentation to down and add extra space": [[21, "Change-orentation-to-down-and-add-extra-space"]], "Change orentation to the left": [[21, "Change-orentation-to-the-left"]], "Change orentation to the right": [[21, "Change-orentation-to-the-right"]], "Changing orientation using parameter orientation": [[21, "Changing-orientation-using-parameter-orientation"]], "Add multiple heatmap annotations using for loop": [[21, "Add-multiple-heatmap-annotations-using-for-loop"]], "Cluster between groups and cluster within groups": [[21, "Cluster-between-groups-and-cluster-within-groups"]], "clsuter within groups: col_split=*, col_cluster=True": [[21, "clsuter-within-groups:-col_split=*,-col_cluster=True"]], "cluster_between_groups: col_split=*, col_split_order=\"cluster_between_groups\",col_cluster=False": [[21, "cluster_between_groups:-col_split=*,-col_split_order=%22cluster_between_groups%22,col_cluster=False"]], "cluster_within_groups && cluster_between_groups: col_split=*, col_split_order=\"cluster_between_groups\",col_cluster=True": [[21, "cluster_within_groups-&&-cluster_between_groups:-col_split=*,-col_split_order=%22cluster_between_groups%22,col_cluster=True"]], "Custom annotation": [[21, "Custom-annotation"]], "Custom linkage": [[21, "Custom-linkage"]], "Image annotation": [[21, "Image-annotation"]], "Choosing Build-in colormap in PyComplexHeatmap": [[21, "Choosing-Build-in-colormap-in-PyComplexHeatmap"]], "Diverging": [[21, "Diverging"]], "Qualitative": [[21, "Qualitative"]], "How to use the Build-in cmap?": [[21, "How-to-use-the-Build-in-cmap?"]], "How to force display all row/col ticklabels?": [[21, "How-to-force-display-all-row/col-ticklabels?"]], "Import packages": [[22, "Import-packages"]], "Between groups DMRs": [[22, "Between-groups-DMRs"]], "Within groups DMRs": [[22, "Within-groups-DMRs"]], "A quick example": [[23, "A-quick-example"]], "Plotting annotations": [[23, "Plotting-annotations"]], "Only plot the row/column annotation": [[23, "Only-plot-the-row/column-annotation"]], "anno_label:": [[23, "anno_label:"]], "anno_simple:": [[23, "anno_simple:"]], "To add a annotation quickly, you just need a dataframe": [[23, "To-add-a-annotation-quickly,-you-just-need-a-dataframe"]], "Plot the figure and legend separately": [[23, "Plot-the-figure-and-legend-separately"]], "Top, bottom, left ,right annotations": [[23, "Top,-bottom,-left-,right-annotations"]], "Composite two heatmaps horizontally for mouse DNA methylation array dataset": [[24, "Composite-two-heatmaps-horizontally-for-mouse-DNA-methylation-array-dataset"]], "Plot correlation matrix for CpG modules using DotClustermap": [[25, "Plot-correlation-matrix-for-CpG-modules-using-DotClustermap"]], "Processing the data": [[25, "Processing-the-data"]], "Plotting the Dot clustermap": [[25, "Plotting-the-Dot-clustermap"]], "A smaller dot clustermap": [[25, "A-smaller-dot-clustermap"]], "1. Understand the layout:": [[26, "1.-Understand-the-layout:"]], "Visualizing Differential Expression Genes": [[27, "Visualizing-Differential-Expression-Genes"]], "Load an example brain networks dataset from seaborn": [[28, "Load-an-example-brain-networks-dataset-from-seaborn"]], "Dot Heatmap": [[28, "Dot-Heatmap"]], "Plot traditional heatmap using square marker marker='s'": [[28, "Plot-traditional-heatmap-using-square-marker-marker='s'"]], "Simple dot heatmap using fixed dot size": [[28, "Simple-dot-heatmap-using-fixed-dot-size"]], "Changing the size of point": [[28, "Changing-the-size-of-point"]], "Add parameter hue and use different colors for different groups": [[28, "Add-parameter-hue-and-use-different-colors-for-different-groups"]], "Add parameter hue and use different cmap and marker for different groups": [[28, "Add-parameter-hue-and-use-different-cmap-and-marker-for-different-groups"]], "Dot Clustermap": [[28, "Dot-Clustermap"]], "Plot clustermap using seaborn brain networks dataset": [[28, "Plot-clustermap-using-seaborn-brain-networks-dataset"]], "Visualize up to five dimension data using DotClustermapPlotter": [[28, "Visualize-up-to-five-dimension-data-using-DotClustermapPlotter"]], "Visualizing gene set enrichment analysis results (clusterProfiler) using dotHeatmap": [[29, "Visualizing-gene-set-enrichment-analysis-results-(clusterProfiler)-using-dotHeatmap"]], "Why do we need a better way to visualize the gene set enrichment analysis result?": [[29, "Why-do-we-need-a-better-way-to-visualize-the-gene-set-enrichment-analysis-result?"]], "Read data": [[29, "Read-data"]], "Plot": [[29, "Plot"]], "How to select marker:": [[29, "How-to-select-marker:"]], "1. Import packages": [[30, "1.-Import-packages"]], "2. Plot heatmap annotations": [[30, "2.-Plot-heatmap-annotations"]], "3. Plot the heatmap with rows and columns split": [[30, "3.-Plot-the-heatmap-with-rows-and-columns-split"]], "4. Plot the annotations along side with main heatmap": [[30, "4.-Plot-the-annotations-along-side-with-main-heatmap"]], "label_kws, xticklabels_kws and yticklabels_kws: Modifying the heatmap annotations labels and xticklabels, yticklabels": [[31, "label_kws,-xticklabels_kws-and-yticklabels_kws:-Modifying-the-heatmap-annotations-labels-and-xticklabels,-yticklabels"]], "xlabel_kws and ylabel_kws: Modifying xlabel and ylabel": [[31, "xlabel_kws-and-ylabel_kws:-Modifying-xlabel-and-ylabel"]], "Control gap & pad in heatmap": [[31, "Control-gap-&-pad-in-heatmap"]], "hgap and wgap for HeatmapAnnotation": [[31, "hgap-and-wgap-for-HeatmapAnnotation"]], "subplot_gap": [[31, "subplot_gap"]], "row_split_gap and col_split_gap": [[31, "row_split_gap-and-col_split_gap"]], "How to remove arrow in anno_label": [[31, "How-to-remove-arrow-in-anno_label"]], "oncoPrint: visualizing TCGA Lung Adenocarcinoma Carcinoma Variants Dataset": [[32, "oncoPrint:-visualizing-TCGA-Lung-Adenocarcinoma-Carcinoma-Variants-Dataset"]], "Visualizing categorical variables using oncoPrint": [[33, "Visualizing-categorical-variables-using-oncoPrint"]], "Visualizing Single Cell DNA Methylation Data (Loyfer2023)": [[34, "Visualizing-Single-Cell-DNA-Methylation-Data-(Loyfer2023)"]], "oncoPrint": [[36, "oncoprint"]], "setup module": [[37, "setup-module"]]}, "indexentries": {"pycomplexheatmap": [[0, "module-PyComplexHeatmap"]], "module": [[0, "module-PyComplexHeatmap"], [1, "module-PyComplexHeatmap.annotations"], [2, "module-PyComplexHeatmap.clustermap"], [3, "module-PyComplexHeatmap.colors"], [4, "module-PyComplexHeatmap.dotHeatmap"], [5, "module-PyComplexHeatmap.example"], [6, "module-PyComplexHeatmap.oncoPrint"], [8, "module-PyComplexHeatmap.utils"]], "annotationbase (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.AnnotationBase"]], "heatmapannotation (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation"]], "pycomplexheatmap.annotations": [[1, "module-PyComplexHeatmap.annotations"]], "anno_barplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_barplot"]], "anno_boxplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_boxplot"]], "anno_img (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_img"]], "anno_label (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_label"]], "anno_lineplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_lineplot"]], "anno_scatterplot (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_scatterplot"]], "anno_simple (class in pycomplexheatmap.annotations)": [[1, "PyComplexHeatmap.annotations.anno_simple"]], "collect_legends() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.collect_legends"]], "get_label_width() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.get_label_width"]], "get_max_label_width() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.get_max_label_width"]], "get_ticklabel_width() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.get_ticklabel_width"]], "get_ticklabel_width() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.get_ticklabel_width"]], "plot() (pycomplexheatmap.annotations.anno_barplot method)": [[1, "PyComplexHeatmap.annotations.anno_barplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_boxplot method)": [[1, "PyComplexHeatmap.annotations.anno_boxplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_img method)": [[1, "PyComplexHeatmap.annotations.anno_img.plot"]], "plot() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.plot"]], "plot() (pycomplexheatmap.annotations.anno_lineplot method)": [[1, "PyComplexHeatmap.annotations.anno_lineplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_scatterplot method)": [[1, "PyComplexHeatmap.annotations.anno_scatterplot.plot"]], "plot() (pycomplexheatmap.annotations.anno_simple method)": [[1, "PyComplexHeatmap.annotations.anno_simple.plot"]], "plot_annotations() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.plot_annotations"]], "plot_legends() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.plot_legends"]], "reorder() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.reorder"]], "set_axes_kws() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.set_axes_kws"]], "set_axes_kws() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.set_axes_kws"]], "set_label() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.set_label"]], "set_legend() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.set_legend"]], "set_plot_kws() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.set_plot_kws"]], "set_side() (pycomplexheatmap.annotations.anno_label method)": [[1, "PyComplexHeatmap.annotations.anno_label.set_side"]], "show_ticklabels() (pycomplexheatmap.annotations.heatmapannotation method)": [[1, "PyComplexHeatmap.annotations.HeatmapAnnotation.show_ticklabels"]], "update_plot_kws() (pycomplexheatmap.annotations.annotationbase method)": [[1, "PyComplexHeatmap.annotations.AnnotationBase.update_plot_kws"]], "clustermapplotter (class in pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter"]], "dendrogramplotter (class in pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter"]], "pycomplexheatmap.clustermap": [[2, "module-PyComplexHeatmap.clustermap"]], "cal_cold_between_groups() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.cal_cold_between_groups"]], "cal_rowd_between_groups() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.cal_rowd_between_groups"]], "calculate_col_dendrograms() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.calculate_col_dendrograms"]], "calculate_dendrogram() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.calculate_dendrogram"]], "calculate_row_dendrograms() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.calculate_row_dendrograms"]], "calculated_linkage (pycomplexheatmap.clustermap.dendrogramplotter property)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.calculated_linkage"]], "check_array() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.check_array"]], "collect_legends() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.collect_legends"]], "composite() (in module pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.composite"]], "format_data() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.format_data"]], "get_coords() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.get_coords"]], "heatmap() (in module pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.heatmap"]], "heatmapplotter (class in pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.heatmapPlotter"]], "plot() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot"]], "plot() (pycomplexheatmap.clustermap.dendrogramplotter method)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.plot"]], "plot() (pycomplexheatmap.clustermap.heatmapplotter method)": [[2, "PyComplexHeatmap.clustermap.heatmapPlotter.plot"]], "plot_dendrograms() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot_dendrograms"]], "plot_heatmap() (in module pycomplexheatmap.clustermap)": [[2, "PyComplexHeatmap.clustermap.plot_heatmap"]], "plot_legends() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot_legends"]], "plot_matrix() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.plot_matrix"]], "post_processing() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.post_processing"]], "reordered_ind (pycomplexheatmap.clustermap.dendrogramplotter property)": [[2, "PyComplexHeatmap.clustermap.DendrogramPlotter.reordered_ind"]], "set_axes_labels_kws() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_axes_labels_kws"]], "set_height() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_height"]], "set_width() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_width"]], "set_xy_labels() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.set_xy_labels"]], "standard_scale() (pycomplexheatmap.clustermap.clustermapplotter static method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.standard_scale"]], "tight_layout() (pycomplexheatmap.clustermap.clustermapplotter method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.tight_layout"]], "z_score() (pycomplexheatmap.clustermap.clustermapplotter static method)": [[2, "PyComplexHeatmap.clustermap.ClusterMapPlotter.z_score"]], "pycomplexheatmap.colors": [[3, "module-PyComplexHeatmap.colors"]], "define_cmap() (in module pycomplexheatmap.colors)": [[3, "PyComplexHeatmap.colors.define_cmap"]], "register_cmap() (in module pycomplexheatmap.colors)": [[3, "PyComplexHeatmap.colors.register_cmap"]], "dotclustermapplotter (class in pycomplexheatmap.dotheatmap)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter"]], "pycomplexheatmap.dotheatmap": [[4, "module-PyComplexHeatmap.dotHeatmap"]], "collect_legends() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.collect_legends"]], "dotheatmap2d() (in module pycomplexheatmap.dotheatmap)": [[4, "PyComplexHeatmap.dotHeatmap.dotHeatmap2d"]], "format_data() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.format_data"]], "plot_matrix() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.plot_matrix"]], "post_processing() (pycomplexheatmap.dotheatmap.dotclustermapplotter method)": [[4, "PyComplexHeatmap.dotHeatmap.DotClustermapPlotter.post_processing"]], "scale() (in module pycomplexheatmap.dotheatmap)": [[4, "PyComplexHeatmap.dotHeatmap.scale"]], "pycomplexheatmap.example": [[5, "module-PyComplexHeatmap.example"]], "clustermap_example0() (in module pycomplexheatmap.example)": [[5, "PyComplexHeatmap.example.clustermap_example0"]], "clustermap_example1() (in module pycomplexheatmap.example)": [[5, "PyComplexHeatmap.example.clustermap_example1"]], "get_kycg_example_data() (in module pycomplexheatmap.example)": [[5, "PyComplexHeatmap.example.get_kycg_example_data"]], "pycomplexheatmap.oncoprint": [[6, "module-PyComplexHeatmap.oncoPrint"]], "add_default_annotations() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.add_default_annotations"]], "collect_legends() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.collect_legends"]], "format_data() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.format_data"]], "get_samples_order() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.get_samples_order"]], "oncoprintplotter (class in pycomplexheatmap.oncoprint)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter"]], "oncoprint() (in module pycomplexheatmap.oncoprint)": [[6, "PyComplexHeatmap.oncoPrint.oncoprint"]], "plot_matrix() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.plot_matrix"]], "post_processing() (pycomplexheatmap.oncoprint.oncoprintplotter method)": [[6, "PyComplexHeatmap.oncoPrint.oncoPrintPlotter.post_processing"]], "pycomplexheatmap.utils": [[8, "module-PyComplexHeatmap.utils"]], "axis_ticklabels_overlap() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.axis_ticklabels_overlap"]], "cal_legend_width() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.cal_legend_width"]], "cluster_labels() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.cluster_labels"]], "define_cmap() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.define_cmap"]], "despine() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.despine"]], "get_colormap() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.get_colormap"]], "plot_cmap_legend() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_cmap_legend"]], "plot_color_dict_legend() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_color_dict_legend"]], "plot_legend_list() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_legend_list"]], "plot_marker_legend() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.plot_marker_legend"]], "set_default_style() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.set_default_style"]], "to_utf8() (in module pycomplexheatmap.utils)": [[8, "PyComplexHeatmap.utils.to_utf8"]]}})
\ No newline at end of file
diff --git a/docs/build/html/setup.html b/docs/build/html/setup.html
index 7f183de5..344f5c9a 100644
--- a/docs/build/html/setup.html
+++ b/docs/build/html/setup.html
@@ -4,7 +4,7 @@
- setup module — PyComplexHeatmap 1.6.6.dev15+gd9f20b3 documentation
+ setup module — PyComplexHeatmap 1.7.2.dev0+g8abf70a.d20240415 documentation
@@ -17,7 +17,7 @@
-
+
diff --git a/notebooks/gene_enrichment_analysis.ipynb b/notebooks/gene_enrichment_analysis.ipynb
index 7b6fb578..b88308e1 100644
--- a/notebooks/gene_enrichment_analysis.ipynb
+++ b/notebooks/gene_enrichment_analysis.ipynb
@@ -20,7 +20,7 @@
"name": "stdout",
"output_type": "stream",
"text": [
- "1.6.6.dev15+gd9f20b3\n"
+ "1.7.2.dev0+g8abf70a.d20240415\n"
]
}
],
@@ -966,7 +966,7 @@
},
{
"cell_type": "code",
- "execution_count": 13,
+ "execution_count": 9,
"id": "8683f2d4-cea6-445f-ae32-cbfdb835b2ee",
"metadata": {
"tags": []