| \n", + " | index | \n", + "
|---|---|
| Species | \n", + "\n", + " |
| Adelie Penguin (Pygoscelis adeliae) | \n", + "146 | \n", + "
| Chinstrap penguin (Pygoscelis antarctica) | \n", + "68 | \n", + "
| Gentoo penguin (Pygoscelis papua) | \n", + "120 | \n", + "
| \n", + " | index | \n", + "
|---|---|
| Species | \n", + "\n", + " |
| Adelie Penguin (Pygoscelis adeliae) | \n", + "102 | \n", + "
| Chinstrap penguin (Pygoscelis antarctica) | \n", + "47 | \n", + "
| \n", + " | index | \n", + "
|---|---|
| Species | \n", + "\n", + " |
| Adelie Penguin (Pygoscelis adeliae) | \n", + "44 | \n", + "
| Chinstrap penguin (Pygoscelis antarctica) | \n", + "21 | \n", + "
\n", " Have you run `initjs()` in this notebook? If this notebook was from another\n", @@ -879,13 +879,13 @@ "
\n", " Have you run `initjs()` in this notebook? If this notebook was from another\n", @@ -923,13 +923,13 @@ "
Machine LearningMachine LearningMachine LearningModel Training
-0.9914163090128756
+0.9871244635193133
Model Training
-1.0
+0.9900990099009901
diff --git a/rendered_notebooks/1 - Model Evaluation.html b/rendered_notebooks/1 - Model Evaluation.html
index b24fdd3..319bc9a 100644
--- a/rendered_notebooks/1 - Model Evaluation.html
+++ b/rendered_notebooks/1 - Model Evaluation.html
@@ -14896,15 +14896,32 @@ Stratification
+
In [4]:
+
+
+from matplotlib import pyplot as plt
+
+
+
+
+
+
+
+
+
+
+
+
+In [5]:
penguins.groupby("Species").Sex.count().plot(kind="bar")
+plt.show()
@@ -14919,20 +14936,6 @@ Stratification
-
-
-
- Out[4]:
-
-
-
-
-
-<AxesSubplot:xlabel='Species'>
-
-
-
-
-In [5]:
+In [6]:
y_train.reset_index().groupby(["Species"]).count()
@@ -14994,7 +14997,7 @@ Stratification
- Out[5]:
+ Out[6]:
@@ -15055,22 +15058,22 @@ Stratification
-We can address this by applying stratification.
-That is simply sampling randomly within a class (or strata) rather than randomly sampling from the entire dataframe.
+We can address this by applying stratification.
+That is simply achieved by randomly sampling *within a class** (or strata) rather than randomly sampling from the entire dataframe.
-
+
-In [6]:
+In [7]:
-X_train, X_test, y_train, y_test = train_test_split(penguins[features], penguins[target[0]], train_size=.7, random_state=42, stratify=penguins[target[0]])
-y_train.reset_index().groupby("Species").count().plot(kind="bar")
+X, y = penguins[features], penguins[target[0]]
+X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=.7, random_state=42, stratify=y)
@@ -15078,27 +15081,45 @@ Stratification
-
+
+
+
+
+
+
+To qualitatevely assess the effect of stratification, let's plot class distribution in both training and test sets:
+
+
+
+
+
+
+
+
+In [8]:
+
+
+fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 8))
-
-
-
-
-
- Out[6]:
-
-
-
+y_train.reset_index().groupby("Species").count().plot(kind="bar", ax=ax1, ylim=(0, len(y)), title="Training")
+y_test.reset_index().groupby("Species").count().plot(kind="bar", ax=ax2, ylim=(0, len(y)), title="Test")
+plt.show()
+
-
-<AxesSubplot:xlabel='Species'>
+
+
+
+
+
+
+
+
@@ -15108,7 +15129,7 @@ Stratification
- Stratification
Stratification
-In [7]:
+In [9]:
-In [8]:
+In [10]:
from sklearn.model_selection import cross_val_score
+
scores = cross_val_score(model, X_train, y_train, cv=5)
scores
@@ -15213,7 +15235,7 @@ Cross-Validation
- Out[8]:
+ Out[10]:
@@ -15233,7 +15255,7 @@ Cross-Validation
-In [9]:
+In [11]:
print(f"{scores.mean():0.2f} accuracy with a standard deviation of {scores.std():0.2f}")
@@ -15286,34 +15308,77 @@ Cross-Validation
-Time-series Validation¶
But validation can get tricky if time gets involved.
-Imagine we measured the growth of baby penguin Hank over time and wanted to us machine learning to project the development of Hank. Then our data suddenly isn't i.i.d. anymore, since it is dependent in the time dimension.
-Were we to split our data randomly for our training and test set, we would test on data points that lie in between training points, where even a simple linear interpolation can do a fairly decent job.
-Therefor, we need to split our measurements along the time axis
-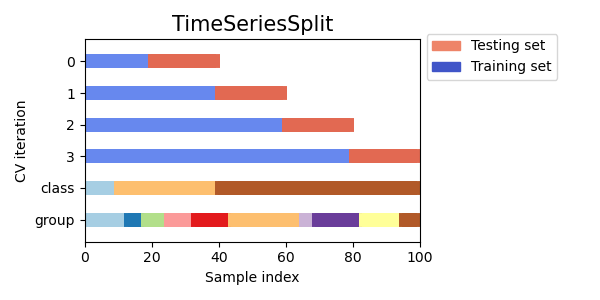 -Scikit-learn Time Series CV [Source].
-Scikit-learn Time Series CV [Source].
+Model Evaluation¶
+
+
+
+
+
+
+
+
+
+
+Brilliant! So let's recap for a moment what we have done so far, in preparation for our (final) Model evaluation.
+We have:
+
+- prepared the model pipeline:
sklearn.pipeline.Pipeline with preprocessor + model
+- generated train and test data partitions (with stratification):
(X_train, y_train) and (X_test, y_test), respectively
+- stratification guaranteed that those partitions will retain class distributions
+
+
+- assessed model performance via cross validation (i.e.
cross_val_score) on X_train(!!)
+- this had the objective of verifying model consistency on multiple data partitioning
+
+
+
+Now we need the complete our last step, namely "assess how the model we chose in CV" (we only had one model, so that was an easy choice :D ) will perform on future data!
+And we have a candidate as representative for these data: X_test.
+Please note that X_test has never been used so far (as it should have!). The take away message here is: generate test partition, and forget about it until the last step!
+Thanks to CV, We have an indication of how the SVC classifier behaves on multiple "version" of the training set. We calculated an average score of 0.99 accuracy, therefore we decided this model is to be trusted for predictions on unseen data.
+Now all we need to do, is to prove this assertion.
+To do so we need to:
+
+- train a new model on the entire **training set**
+- evaluate it's performance on **test set** (using the metric of choice - presumably the same metric we chose in CV!)
-
+
-In [10]:
+In [12]:
-import numpy as np
-from sklearn.model_selection import TimeSeriesSplit
+# training
+model = Pipeline(steps=[
+ ('preprocessor', preprocessor),
+ ('classifier', SVC()),
+])
+classifier = model.fit(X_train, y_train)
+
-X = np.array([[1, 2], [3, 4], [1, 2], [3, 4], [1, 2], [3, 4]])
-y = np.array([1, 2, 3, 4, 5, 6])
-tscv = TimeSeriesSplit(n_splits=3)
-print(tscv)
+
+
+
+
-for train, test in tscv.split(X):
- print("%s %s" % (train, test))
+
+
+
+
+
+In [13]:
+
+
+# Model evaluation
+from sklearn.metrics import accuracy_score
+
+y_pred = classifier.predict(X_test)
+print("TEST ACC: ", accuracy_score(y_true=y_test, y_pred=y_pred))
@@ -15335,10 +15400,7 @@ Time-series Validation
-TimeSeriesSplit(gap=0, max_train_size=None, n_splits=3, test_size=None)
-[0 1 2] [3]
-[0 1 2 3] [4]
-[0 1 2 3 4] [5]
+TEST ACC: 1.0
@@ -15354,9 +15416,43 @@ Time-series Validation
-Spatial Validation¶
Spatial data, like maps and satellite data has a similar problem.
-Here the data is correlated in the spatial dimension. However, we can mitigate the effect by supplying a group. In this simple example I used continents, but it's possible to group by bins on a lat-lon grid as well.
-Here especially, a cross-validation scheme is very important, as it is used to validate against every area on your map at least once.
+Now we can finally say that we have concluded our model evaluation - with a fantastic score of 0.96 Accuracy on the test set.
+
+
+
+
+
+
+
+
+
+
+
+Choosing the appropriate Evaluation Metric¶
+
+
+
+
+
+
+
+
+
+
+Ok, now for the mere sake of considering a more realistic data scenario, let's pretend our reference dataset is composed by only samples from two (out of the three) classes we have. In particular, we will crafting our dataset by choosing the most and the least represented classes, respectively.
+The very idea is to explore whether the choice of appropriate metrics could make the difference in our machine learning models evaluation.
+
+
+
+
+
+
+
+
+
+
+
+Let's recall class distributions in our dataset:
@@ -15366,7 +15462,756 @@ Spatial Validation
-In [11]:
+In [14]:
+
+
+y.reset_index().groupby(["Species"]).count()
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Out[14]:
+
+
+
+
+
+
+
+
+
+ index
+
+
+ Species
+
+
+
+
+ Adelie Penguin (Pygoscelis adeliae)
+ 146
+
+
+ Chinstrap penguin (Pygoscelis antarctica)
+ 68
+
+
+ Gentoo penguin (Pygoscelis papua)
+ 120
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+So let's select samples from the first two classes, Adelie Penguin and Chinstrap penguin:
+
+
+
+
+
+
+
+
+
+In [15]:
+
+
+samples = penguins[((penguins["Species"].str.startswith("Adelie")) | (penguins["Species"].str.startswith("Chinstrap")))]
+
+
+
+
+
+
+
+
+
+
+
+
+In [16]:
+
+
+samples.shape[0] == 146 + 68 # quick verification
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Out[16]:
+
+
+
+
+
+True
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+To make things even harder for our machine learning model, let's also see if we could get rid of clearly separating features in this toy dataset
+
+
+
+
+
+
+
+
+
+In [17]:
+
+
+import seaborn as sns
+
+pairplot_figure = sns.pairplot(samples, hue="Species")
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+OK so if we get to choose, we could definitely say that in this dataset, the Flipper Length in combination with the Culmen Depth leads to the hardest classification task for our machine learning model.
+Therefore, here is the plan:
+
+- we select only those to numerical features (iow we will get rid of the
Culmen Lenght feature)
+- we will apply an identical Model evaluation pipeline as we did in our previous example
+- Cross Validation + Evaluation on Test set
+
+
+
+The very difference this time is that we will use multiple metrics to evaluate our model to prove our point on carefully selecting evaluation metrics.
+
+
+
+
+
+
+
+
+
+In [18]:
+
+
+num_features = ["Culmen Length (mm)", "Culmen Depth (mm)", "Flipper Length (mm)"]
+selected_num_features = num_features[1:]
+cat_features = ["Sex"]
+features = selected_num_features + cat_features
+
+
+
+
+
+
+
+
+
+
+
+
+In [19]:
+
+
+num_transformer = StandardScaler()
+cat_transformer = OneHotEncoder(handle_unknown='ignore')
+
+preprocessor = ColumnTransformer(transformers=[
+ ('num', num_transformer, selected_num_features), # note here, we will only preprocess selected numerical features
+ ('cat', cat_transformer, cat_features)
+])
+
+model = Pipeline(steps=[
+ ('preprocessor', preprocessor),
+ ('classifier', SVC()),
+])
+
+
+
+
+
+
+
+
+
+
+
+
+In [20]:
+
+
+X, y = samples[features], samples[target[0]]
+X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=.7, random_state=42, stratify=y) # we also stratify on classes
+
+
+
+
+
+
+
+
+
+
+
+
+In [21]:
+
+
+y_train.reset_index().groupby("Species").count()
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Out[21]:
+
+
+
+
+
+
+
+
+
+ index
+
+
+ Species
+
+
+
+
+ Adelie Penguin (Pygoscelis adeliae)
+ 102
+
+
+ Chinstrap penguin (Pygoscelis antarctica)
+ 47
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+In [22]:
+
+
+y_test.reset_index().groupby("Species").count()
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Out[22]:
+
+
+
+
+
+
+
+
+
+ index
+
+
+ Species
+
+
+
+
+ Adelie Penguin (Pygoscelis adeliae)
+ 44
+
+
+ Chinstrap penguin (Pygoscelis antarctica)
+ 21
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+In our evaluation pipeline we will be using keep record both accuracy (ACC) and matthew correlation coefficient (MCC)
+
+
+
+
+
+
+
+
+
+In [23]:
+
+
+from sklearn.model_selection import cross_validate
+from sklearn.metrics import make_scorer
+from sklearn.metrics import matthews_corrcoef as mcc
+from sklearn.metrics import accuracy_score as acc
+
+mcc_scorer = make_scorer(mcc)
+acc_scorer = make_scorer(acc)
+scores = cross_validate(model, X_train, y_train, cv=5,
+ scoring={"MCC": mcc_scorer, "ACC": acc_scorer})
+scores
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Out[23]:
+
+
+
+
+
+{'fit_time': array([0.00225472, 0.00185323, 0.00180411, 0.00178003, 0.00177217]),
+ 'score_time': array([0.00157523, 0.00124979, 0.00123906, 0.00125003, 0.0012238 ]),
+ 'test_MCC': array([0.37796447, 0.27863911, 0.40824829, 0.02424643, 0.08625819]),
+ 'test_ACC': array([0.73333333, 0.7 , 0.76666667, 0.66666667, 0.62068966])}
+
+
+
+
+
+
+
+
+
+
+
+
+
+In [24]:
+
+
+import numpy as np
+
+print("Avg ACC in CV: ", np.average(scores["test_ACC"]))
+print("Avg MCC in CV: ", np.average(scores["test_MCC"]))
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+Avg ACC in CV: 0.697471264367816
+Avg MCC in CV: 0.2350712993854009
+
+
+
+
+
+
+
+
+
+
+
+
+
+In [25]:
+
+
+model = model.fit(X_train, y_train)
+
+print("ACC: ", acc_scorer(model, X_test, y_test))
+print("MCC: ", mcc_scorer(model, X_test, y_test))
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ACC: 0.7230769230769231
+MCC: 0.29439815585406465
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+To see exactly what happened, let's have a look at the Confusion matrix
+
+
+
+
+
+
+
+
+
+In [26]:
+
+
+from sklearn.metrics import ConfusionMatrixDisplay
+fig, ax = plt.subplots(figsize=(15, 10))
+ConfusionMatrixDisplay.from_estimator(model, X_test, y_test, ax=ax)
+plt.show()
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+As expected, the model did a pretty bad job in classifying Chinstrap Penguins and the MCC was able to catch that, whilst ACC could not as it only considers correctly classified samples!
+
+
+
+
+
+
+
+
+
+
+
+Time-series Validation¶
But validation can get tricky if time gets involved.
+Imagine we measured the growth of baby penguin Hank over time and wanted to us machine learning to project the development of Hank. Then our data suddenly isn't i.i.d. anymore, since it is dependent in the time dimension.
+Were we to split our data randomly for our training and test set, we would test on data points that lie in between training points, where even a simple linear interpolation can do a fairly decent job.
+Therefor, we need to split our measurements along the time axis
+ +Scikit-learn Time Series CV [Source].
+Scikit-learn Time Series CV [Source].
+
+
+
+
+
+
+
+
+
+In [27]:
+
+
+import numpy as np
+from sklearn.model_selection import TimeSeriesSplit
+
+X = np.array([[1, 2], [3, 4], [1, 2], [3, 4], [1, 2], [3, 4]])
+y = np.array([1, 2, 3, 4, 5, 6])
+tscv = TimeSeriesSplit(n_splits=3)
+print(tscv)
+
+for train, test in tscv.split(X):
+ print("%s %s" % (train, test))
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+TimeSeriesSplit(gap=0, max_train_size=None, n_splits=3, test_size=None)
+[0 1 2] [3]
+[0 1 2 3] [4]
+[0 1 2 3 4] [5]
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+Spatial Validation¶
Spatial data, like maps and satellite data has a similar problem.
+Here the data is correlated in the spatial dimension. However, we can mitigate the effect by supplying a group. In this simple example I used continents, but it's possible to group by bins on a lat-lon grid as well.
+Here especially, a cross-validation scheme is very important, as it is used to validate against every area on your map at least once.
+
+
+
+
+
+
+
+
+
+In [28]:
@@ -15348,7 +15348,7 @@ Tree importance vs Permutatio
- Tree importance vs Permutatio
@@ -15639,7 +15639,7 @@
Tree importance vs Permutatio
@@ -15639,7 +15639,7 @@ Shap Inspection
-
+
Visualization omitted, Javascript library not loaded!
Have you run `initjs()` in this notebook? If this notebook was from another
@@ -15649,8 +15649,8 @@ Shap Inspection
@@ -15693,7 +15693,7 @@ Shap Inspection
-
+
Visualization omitted, Javascript library not loaded!
Have you run `initjs()` in this notebook? If this notebook was from another
@@ -15703,8 +15703,8 @@ Shap Inspection
@@ -15834,7 +15834,7 @@ Model Inspection
-tensor([[0.3527, 0.3850]], grad_fn=<SigmoidBackward0>)
+tensor([[0.5887, 0.6065]], grad_fn=<SigmoidBackward0>)
from matplotlib import pyplot as plt
+penguins.groupby("Species").Sex.count().plot(kind="bar")
+plt.show()
Stratification
-
-
-
- Out[4]:
-
-
-
-
-
-<AxesSubplot:xlabel='Species'>
-
-
-
-
-In [5]:
+In [6]:
y_train.reset_index().groupby(["Species"]).count()
@@ -14994,7 +14997,7 @@ Stratification
- Out[5]:
+ Out[6]:
@@ -15055,22 +15058,22 @@ Stratification
-We can address this by applying stratification.
-That is simply sampling randomly within a class (or strata) rather than randomly sampling from the entire dataframe.
+We can address this by applying stratification.
+That is simply achieved by randomly sampling *within a class** (or strata) rather than randomly sampling from the entire dataframe.
-
+
-In [6]:
+In [7]:
-X_train, X_test, y_train, y_test = train_test_split(penguins[features], penguins[target[0]], train_size=.7, random_state=42, stratify=penguins[target[0]])
-y_train.reset_index().groupby("Species").count().plot(kind="bar")
+X, y = penguins[features], penguins[target[0]]
+X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=.7, random_state=42, stratify=y)
@@ -15078,27 +15081,45 @@ Stratification
-
+
+
+
+
+
+
+To qualitatevely assess the effect of stratification, let's plot class distribution in both training and test sets:
+
+
+
+
+
+
+
+
+In [8]:
+
+
+fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 8))
-
-
-
-
-
- Out[6]:
-
-
-
+y_train.reset_index().groupby("Species").count().plot(kind="bar", ax=ax1, ylim=(0, len(y)), title="Training")
+y_test.reset_index().groupby("Species").count().plot(kind="bar", ax=ax2, ylim=(0, len(y)), title="Test")
+plt.show()
+
-
-<AxesSubplot:xlabel='Species'>
+
+
+
+
+
+
+
+
@@ -15108,7 +15129,7 @@ Stratification
- Stratification
Stratification
-In [7]:
+In [9]:
-In [8]:
+In [10]:
from sklearn.model_selection import cross_val_score
+
scores = cross_val_score(model, X_train, y_train, cv=5)
scores
@@ -15213,7 +15235,7 @@ Cross-Validation
- Out[8]:
+ Out[10]:
@@ -15233,7 +15255,7 @@ Cross-Validation
-In [9]:
+In [11]:
print(f"{scores.mean():0.2f} accuracy with a standard deviation of {scores.std():0.2f}")
@@ -15286,34 +15308,77 @@ Cross-Validation
-Time-series Validation¶
But validation can get tricky if time gets involved.
-Imagine we measured the growth of baby penguin Hank over time and wanted to us machine learning to project the development of Hank. Then our data suddenly isn't i.i.d. anymore, since it is dependent in the time dimension.
-Were we to split our data randomly for our training and test set, we would test on data points that lie in between training points, where even a simple linear interpolation can do a fairly decent job.
-Therefor, we need to split our measurements along the time axis
- -Scikit-learn Time Series CV [Source].
-Scikit-learn Time Series CV [Source].
+Model Evaluation¶
+
+
+
+
+
+
+
+
+
+
+Brilliant! So let's recap for a moment what we have done so far, in preparation for our (final) Model evaluation.
+We have:
+
+- prepared the model pipeline:
sklearn.pipeline.Pipeline with preprocessor + model
+- generated train and test data partitions (with stratification):
(X_train, y_train) and (X_test, y_test), respectively
+- stratification guaranteed that those partitions will retain class distributions
+
+
+- assessed model performance via cross validation (i.e.
cross_val_score) on X_train(!!)
+- this had the objective of verifying model consistency on multiple data partitioning
+
+
+
+Now we need the complete our last step, namely "assess how the model we chose in CV" (we only had one model, so that was an easy choice :D ) will perform on future data!
+And we have a candidate as representative for these data: X_test.
+Please note that X_test has never been used so far (as it should have!). The take away message here is: generate test partition, and forget about it until the last step!
+Thanks to CV, We have an indication of how the SVC classifier behaves on multiple "version" of the training set. We calculated an average score of 0.99 accuracy, therefore we decided this model is to be trusted for predictions on unseen data.
+Now all we need to do, is to prove this assertion.
+To do so we need to:
+
+- train a new model on the entire **training set**
+- evaluate it's performance on **test set** (using the metric of choice - presumably the same metric we chose in CV!)
-
+
-In [10]:
+In [12]:
-import numpy as np
-from sklearn.model_selection import TimeSeriesSplit
+# training
+model = Pipeline(steps=[
+ ('preprocessor', preprocessor),
+ ('classifier', SVC()),
+])
+classifier = model.fit(X_train, y_train)
+
-X = np.array([[1, 2], [3, 4], [1, 2], [3, 4], [1, 2], [3, 4]])
-y = np.array([1, 2, 3, 4, 5, 6])
-tscv = TimeSeriesSplit(n_splits=3)
-print(tscv)
+
+
+
+
-for train, test in tscv.split(X):
- print("%s %s" % (train, test))
+
+
+
+
+
+In [13]:
+
+
+# Model evaluation
+from sklearn.metrics import accuracy_score
+
+y_pred = classifier.predict(X_test)
+print("TEST ACC: ", accuracy_score(y_true=y_test, y_pred=y_pred))
@@ -15335,10 +15400,7 @@ Time-series Validation
-TimeSeriesSplit(gap=0, max_train_size=None, n_splits=3, test_size=None)
-[0 1 2] [3]
-[0 1 2 3] [4]
-[0 1 2 3 4] [5]
+TEST ACC: 1.0
@@ -15354,9 +15416,43 @@ Time-series Validation
-Spatial Validation¶
Spatial data, like maps and satellite data has a similar problem.
-Here the data is correlated in the spatial dimension. However, we can mitigate the effect by supplying a group. In this simple example I used continents, but it's possible to group by bins on a lat-lon grid as well.
-Here especially, a cross-validation scheme is very important, as it is used to validate against every area on your map at least once.
+Now we can finally say that we have concluded our model evaluation - with a fantastic score of 0.96 Accuracy on the test set.
+
+
+
+
+
+
+
+
+
+
+
+Choosing the appropriate Evaluation Metric¶
+
+
+
+
+
+
+
+
+
+
+Ok, now for the mere sake of considering a more realistic data scenario, let's pretend our reference dataset is composed by only samples from two (out of the three) classes we have. In particular, we will crafting our dataset by choosing the most and the least represented classes, respectively.
+The very idea is to explore whether the choice of appropriate metrics could make the difference in our machine learning models evaluation.
+
+
+
+
+
+
+
+
+
+
+
+Let's recall class distributions in our dataset:
@@ -15366,7 +15462,756 @@ Spatial Validation
-In [11]:
+In [14]:
+
+
+y.reset_index().groupby(["Species"]).count()
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Out[14]:
+
+
+
+
+
+
+
+
+
+ index
+
+
+ Species
+
+
+
+
+ Adelie Penguin (Pygoscelis adeliae)
+ 146
+
+
+ Chinstrap penguin (Pygoscelis antarctica)
+ 68
+
+
+ Gentoo penguin (Pygoscelis papua)
+ 120
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+So let's select samples from the first two classes, Adelie Penguin and Chinstrap penguin:
+
+
+
+
+
+
+
+
+
+In [15]:
+
+
+samples = penguins[((penguins["Species"].str.startswith("Adelie")) | (penguins["Species"].str.startswith("Chinstrap")))]
+
+
+
+
+
+
+
+
+
+
+
+
+In [16]:
+
+
+samples.shape[0] == 146 + 68 # quick verification
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Out[16]:
+
+
+
+
+
+True
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+To make things even harder for our machine learning model, let's also see if we could get rid of clearly separating features in this toy dataset
+
+
+
+
+
+
+
+
+
+In [17]:
+
+
+import seaborn as sns
+
+pairplot_figure = sns.pairplot(samples, hue="Species")
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+OK so if we get to choose, we could definitely say that in this dataset, the Flipper Length in combination with the Culmen Depth leads to the hardest classification task for our machine learning model.
+Therefore, here is the plan:
+
+- we select only those to numerical features (iow we will get rid of the
Culmen Lenght feature)
+- we will apply an identical Model evaluation pipeline as we did in our previous example
+- Cross Validation + Evaluation on Test set
+
+
+
+The very difference this time is that we will use multiple metrics to evaluate our model to prove our point on carefully selecting evaluation metrics.
+
+
+
+
+
+
+
+
+
+In [18]:
+
+
+num_features = ["Culmen Length (mm)", "Culmen Depth (mm)", "Flipper Length (mm)"]
+selected_num_features = num_features[1:]
+cat_features = ["Sex"]
+features = selected_num_features + cat_features
+
+
+
+
+
+
+
+
+
+
+
+
+In [19]:
+
+
+num_transformer = StandardScaler()
+cat_transformer = OneHotEncoder(handle_unknown='ignore')
+
+preprocessor = ColumnTransformer(transformers=[
+ ('num', num_transformer, selected_num_features), # note here, we will only preprocess selected numerical features
+ ('cat', cat_transformer, cat_features)
+])
+
+model = Pipeline(steps=[
+ ('preprocessor', preprocessor),
+ ('classifier', SVC()),
+])
+
+
+
+
+
+
+
+
+
+
+
+
+In [20]:
+
+
+X, y = samples[features], samples[target[0]]
+X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=.7, random_state=42, stratify=y) # we also stratify on classes
+
+
+
+
+
+
+
+
+
+
+
+
+In [21]:
+
+
+y_train.reset_index().groupby("Species").count()
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Out[21]:
+
+
+
+
+
+
+
+
+
+ index
+
+
+ Species
+
+
+
+
+ Adelie Penguin (Pygoscelis adeliae)
+ 102
+
+
+ Chinstrap penguin (Pygoscelis antarctica)
+ 47
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+In [22]:
+
+
+y_test.reset_index().groupby("Species").count()
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Out[22]:
+
+
+
+
+
+
+
+
+
+ index
+
+
+ Species
+
+
+
+
+ Adelie Penguin (Pygoscelis adeliae)
+ 44
+
+
+ Chinstrap penguin (Pygoscelis antarctica)
+ 21
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+In our evaluation pipeline we will be using keep record both accuracy (ACC) and matthew correlation coefficient (MCC)
+
+
+
+
+
+
+
+
+
+In [23]:
+
+
+from sklearn.model_selection import cross_validate
+from sklearn.metrics import make_scorer
+from sklearn.metrics import matthews_corrcoef as mcc
+from sklearn.metrics import accuracy_score as acc
+
+mcc_scorer = make_scorer(mcc)
+acc_scorer = make_scorer(acc)
+scores = cross_validate(model, X_train, y_train, cv=5,
+ scoring={"MCC": mcc_scorer, "ACC": acc_scorer})
+scores
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Out[23]:
+
+
+
+
+
+{'fit_time': array([0.00225472, 0.00185323, 0.00180411, 0.00178003, 0.00177217]),
+ 'score_time': array([0.00157523, 0.00124979, 0.00123906, 0.00125003, 0.0012238 ]),
+ 'test_MCC': array([0.37796447, 0.27863911, 0.40824829, 0.02424643, 0.08625819]),
+ 'test_ACC': array([0.73333333, 0.7 , 0.76666667, 0.66666667, 0.62068966])}
+
+
+
+
+
+
+
+
+
+
+
+
+
+In [24]:
+
+
+import numpy as np
+
+print("Avg ACC in CV: ", np.average(scores["test_ACC"]))
+print("Avg MCC in CV: ", np.average(scores["test_MCC"]))
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+Avg ACC in CV: 0.697471264367816
+Avg MCC in CV: 0.2350712993854009
+
+
+
+
+
+
+
+
+
+
+
+
+
+In [25]:
+
+
+model = model.fit(X_train, y_train)
+
+print("ACC: ", acc_scorer(model, X_test, y_test))
+print("MCC: ", mcc_scorer(model, X_test, y_test))
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ACC: 0.7230769230769231
+MCC: 0.29439815585406465
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+To see exactly what happened, let's have a look at the Confusion matrix
+
+
+
+
+
+
+
+
+
+In [26]:
+
+
+from sklearn.metrics import ConfusionMatrixDisplay
+fig, ax = plt.subplots(figsize=(15, 10))
+ConfusionMatrixDisplay.from_estimator(model, X_test, y_test, ax=ax)
+plt.show()
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+As expected, the model did a pretty bad job in classifying Chinstrap Penguins and the MCC was able to catch that, whilst ACC could not as it only considers correctly classified samples!
+
+
+
+
+
+
+
+
+
+
+
+Time-series Validation¶
But validation can get tricky if time gets involved.
+Imagine we measured the growth of baby penguin Hank over time and wanted to us machine learning to project the development of Hank. Then our data suddenly isn't i.i.d. anymore, since it is dependent in the time dimension.
+Were we to split our data randomly for our training and test set, we would test on data points that lie in between training points, where even a simple linear interpolation can do a fairly decent job.
+Therefor, we need to split our measurements along the time axis
+ +Scikit-learn Time Series CV [Source].
+Scikit-learn Time Series CV [Source].
+
+
+
+
+
+
+
+
+
+In [27]:
+
+
+import numpy as np
+from sklearn.model_selection import TimeSeriesSplit
+
+X = np.array([[1, 2], [3, 4], [1, 2], [3, 4], [1, 2], [3, 4]])
+y = np.array([1, 2, 3, 4, 5, 6])
+tscv = TimeSeriesSplit(n_splits=3)
+print(tscv)
+
+for train, test in tscv.split(X):
+ print("%s %s" % (train, test))
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+TimeSeriesSplit(gap=0, max_train_size=None, n_splits=3, test_size=None)
+[0 1 2] [3]
+[0 1 2 3] [4]
+[0 1 2 3 4] [5]
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+Spatial Validation¶
Spatial data, like maps and satellite data has a similar problem.
+Here the data is correlated in the spatial dimension. However, we can mitigate the effect by supplying a group. In this simple example I used continents, but it's possible to group by bins on a lat-lon grid as well.
+Here especially, a cross-validation scheme is very important, as it is used to validate against every area on your map at least once.
+
+
+
+
+
+
+
+
+
+In [28]:
@@ -15348,7 +15348,7 @@ Tree importance vs Permutatio
- Tree importance vs Permutatio
@@ -15639,7 +15639,7 @@
Tree importance vs Permutatio
@@ -15639,7 +15639,7 @@ Shap Inspection
-
+
Visualization omitted, Javascript library not loaded!
Have you run `initjs()` in this notebook? If this notebook was from another
@@ -15649,8 +15649,8 @@ Shap Inspection
@@ -15693,7 +15693,7 @@ Shap Inspection
-
+
Visualization omitted, Javascript library not loaded!
Have you run `initjs()` in this notebook? If this notebook was from another
@@ -15703,8 +15703,8 @@ Shap Inspection
@@ -15834,7 +15834,7 @@ Model Inspection
-tensor([[0.3527, 0.3850]], grad_fn=<SigmoidBackward0>)
+tensor([[0.5887, 0.6065]], grad_fn=<SigmoidBackward0>)
<AxesSubplot:xlabel='Species'>-
y_train.reset_index().groupby(["Species"]).count()
@@ -14994,7 +14997,7 @@ Stratification
- Out[5]:
+ Out[6]:
@@ -15055,22 +15058,22 @@ Stratification
-We can address this by applying stratification.
-That is simply sampling randomly within a class (or strata) rather than randomly sampling from the entire dataframe.
+We can address this by applying stratification.
+That is simply achieved by randomly sampling *within a class** (or strata) rather than randomly sampling from the entire dataframe.
X_train, X_test, y_train, y_test = train_test_split(penguins[features], penguins[target[0]], train_size=.7, random_state=42, stratify=penguins[target[0]])
-y_train.reset_index().groupby("Species").count().plot(kind="bar")
+X, y = penguins[features], penguins[target[0]]
+X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=.7, random_state=42, stratify=y)
Stratification
-
+
+
+
+
+
+
+To qualitatevely assess the effect of stratification, let's plot class distribution in both training and test sets:
+
+
+
+
+
+
+
+
+In [8]:
+
+
+fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 8))
-
-
-
-
-
- Out[6]:
-
-
-
+y_train.reset_index().groupby("Species").count().plot(kind="bar", ax=ax1, ylim=(0, len(y)), title="Training")
+y_test.reset_index().groupby("Species").count().plot(kind="bar", ax=ax2, ylim=(0, len(y)), title="Test")
+plt.show()
+
-
-<AxesSubplot:xlabel='Species'>
+
+
+
+
+
+
+
+
@@ -15108,7 +15129,7 @@ Stratification
- Stratification
Stratification
-In [7]:
+In [9]:
-In [8]:
+In [10]:
from sklearn.model_selection import cross_val_score
+
scores = cross_val_score(model, X_train, y_train, cv=5)
scores
@@ -15213,7 +15235,7 @@ Cross-Validation
- Out[8]:
+ Out[10]:
@@ -15233,7 +15255,7 @@ Cross-Validation
-In [9]:
+In [11]:
print(f"{scores.mean():0.2f} accuracy with a standard deviation of {scores.std():0.2f}")
@@ -15286,34 +15308,77 @@ Cross-Validation
-Time-series Validation¶
But validation can get tricky if time gets involved.
-Imagine we measured the growth of baby penguin Hank over time and wanted to us machine learning to project the development of Hank. Then our data suddenly isn't i.i.d. anymore, since it is dependent in the time dimension.
-Were we to split our data randomly for our training and test set, we would test on data points that lie in between training points, where even a simple linear interpolation can do a fairly decent job.
-Therefor, we need to split our measurements along the time axis
- -Scikit-learn Time Series CV [Source].
-Scikit-learn Time Series CV [Source].
+Model Evaluation¶
+
+
+
+
+
+
+
+
+
+
+Brilliant! So let's recap for a moment what we have done so far, in preparation for our (final) Model evaluation.
+We have:
+
+- prepared the model pipeline:
sklearn.pipeline.Pipeline with preprocessor + model
+- generated train and test data partitions (with stratification):
(X_train, y_train) and (X_test, y_test), respectively
+- stratification guaranteed that those partitions will retain class distributions
+
+
+- assessed model performance via cross validation (i.e.
cross_val_score) on X_train(!!)
+- this had the objective of verifying model consistency on multiple data partitioning
+
+
+
+Now we need the complete our last step, namely "assess how the model we chose in CV" (we only had one model, so that was an easy choice :D ) will perform on future data!
+And we have a candidate as representative for these data: X_test.
+Please note that X_test has never been used so far (as it should have!). The take away message here is: generate test partition, and forget about it until the last step!
+Thanks to CV, We have an indication of how the SVC classifier behaves on multiple "version" of the training set. We calculated an average score of 0.99 accuracy, therefore we decided this model is to be trusted for predictions on unseen data.
+Now all we need to do, is to prove this assertion.
+To do so we need to:
+
+- train a new model on the entire **training set**
+- evaluate it's performance on **test set** (using the metric of choice - presumably the same metric we chose in CV!)
-
+
-In [10]:
+In [12]:
-import numpy as np
-from sklearn.model_selection import TimeSeriesSplit
+# training
+model = Pipeline(steps=[
+ ('preprocessor', preprocessor),
+ ('classifier', SVC()),
+])
+classifier = model.fit(X_train, y_train)
+
-X = np.array([[1, 2], [3, 4], [1, 2], [3, 4], [1, 2], [3, 4]])
-y = np.array([1, 2, 3, 4, 5, 6])
-tscv = TimeSeriesSplit(n_splits=3)
-print(tscv)
+
+
+
+
-for train, test in tscv.split(X):
- print("%s %s" % (train, test))
+
+
+
+
+
+In [13]:
+
+
+# Model evaluation
+from sklearn.metrics import accuracy_score
+
+y_pred = classifier.predict(X_test)
+print("TEST ACC: ", accuracy_score(y_true=y_test, y_pred=y_pred))
@@ -15335,10 +15400,7 @@ Time-series Validation
-TimeSeriesSplit(gap=0, max_train_size=None, n_splits=3, test_size=None)
-[0 1 2] [3]
-[0 1 2 3] [4]
-[0 1 2 3 4] [5]
+TEST ACC: 1.0
@@ -15354,9 +15416,43 @@ Time-series Validation
-Spatial Validation¶
Spatial data, like maps and satellite data has a similar problem.
-Here the data is correlated in the spatial dimension. However, we can mitigate the effect by supplying a group. In this simple example I used continents, but it's possible to group by bins on a lat-lon grid as well.
-Here especially, a cross-validation scheme is very important, as it is used to validate against every area on your map at least once.
+Now we can finally say that we have concluded our model evaluation - with a fantastic score of 0.96 Accuracy on the test set.
+
+
+
+
+
+
+
+
+
+
+
+Choosing the appropriate Evaluation Metric¶
+
+
+
+
+
+
+
+
+
+
+Ok, now for the mere sake of considering a more realistic data scenario, let's pretend our reference dataset is composed by only samples from two (out of the three) classes we have. In particular, we will crafting our dataset by choosing the most and the least represented classes, respectively.
+The very idea is to explore whether the choice of appropriate metrics could make the difference in our machine learning models evaluation.
+
+
+
+
+
+
+
+
+
+
+
+Let's recall class distributions in our dataset:
@@ -15366,7 +15462,756 @@ Spatial Validation
-In [11]:
+In [14]:
+
+
+y.reset_index().groupby(["Species"]).count()
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Out[14]:
+
+
+
+
+
+
+
+
+
+ index
+
+
+ Species
+
+
+
+
+ Adelie Penguin (Pygoscelis adeliae)
+ 146
+
+
+ Chinstrap penguin (Pygoscelis antarctica)
+ 68
+
+
+ Gentoo penguin (Pygoscelis papua)
+ 120
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+So let's select samples from the first two classes, Adelie Penguin and Chinstrap penguin:
+
+
+
+
+
+
+
+
+
+In [15]:
+
+
+samples = penguins[((penguins["Species"].str.startswith("Adelie")) | (penguins["Species"].str.startswith("Chinstrap")))]
+
+
+
+
+
+
+
+
+
+
+
+
+In [16]:
+
+
+samples.shape[0] == 146 + 68 # quick verification
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Out[16]:
+
+
+
+
+
+True
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+To make things even harder for our machine learning model, let's also see if we could get rid of clearly separating features in this toy dataset
+
+
+
+
+
+
+
+
+
+In [17]:
+
+
+import seaborn as sns
+
+pairplot_figure = sns.pairplot(samples, hue="Species")
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+OK so if we get to choose, we could definitely say that in this dataset, the Flipper Length in combination with the Culmen Depth leads to the hardest classification task for our machine learning model.
+Therefore, here is the plan:
+
+- we select only those to numerical features (iow we will get rid of the
Culmen Lenght feature)
+- we will apply an identical Model evaluation pipeline as we did in our previous example
+- Cross Validation + Evaluation on Test set
+
+
+
+The very difference this time is that we will use multiple metrics to evaluate our model to prove our point on carefully selecting evaluation metrics.
+
+
+
+
+
+
+
+
+
+In [18]:
+
+
+num_features = ["Culmen Length (mm)", "Culmen Depth (mm)", "Flipper Length (mm)"]
+selected_num_features = num_features[1:]
+cat_features = ["Sex"]
+features = selected_num_features + cat_features
+
+
+
+
+
+
+
+
+
+
+
+
+In [19]:
+
+
+num_transformer = StandardScaler()
+cat_transformer = OneHotEncoder(handle_unknown='ignore')
+
+preprocessor = ColumnTransformer(transformers=[
+ ('num', num_transformer, selected_num_features), # note here, we will only preprocess selected numerical features
+ ('cat', cat_transformer, cat_features)
+])
+
+model = Pipeline(steps=[
+ ('preprocessor', preprocessor),
+ ('classifier', SVC()),
+])
+
+
+
+
+
+
+
+
+
+
+
+
+In [20]:
+
+
+X, y = samples[features], samples[target[0]]
+X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=.7, random_state=42, stratify=y) # we also stratify on classes
+
+
+
+
+
+
+
+
+
+
+
+
+In [21]:
+
+
+y_train.reset_index().groupby("Species").count()
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Out[21]:
+
+
+
+
+
+
+
+
+
+ index
+
+
+ Species
+
+
+
+
+ Adelie Penguin (Pygoscelis adeliae)
+ 102
+
+
+ Chinstrap penguin (Pygoscelis antarctica)
+ 47
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+In [22]:
+
+
+y_test.reset_index().groupby("Species").count()
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Out[22]:
+
+
+
+
+
+
+
+
+
+ index
+
+
+ Species
+
+
+
+
+ Adelie Penguin (Pygoscelis adeliae)
+ 44
+
+
+ Chinstrap penguin (Pygoscelis antarctica)
+ 21
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+In our evaluation pipeline we will be using keep record both accuracy (ACC) and matthew correlation coefficient (MCC)
+
+
+
+
+
+
+
+
+
+In [23]:
+
+
+from sklearn.model_selection import cross_validate
+from sklearn.metrics import make_scorer
+from sklearn.metrics import matthews_corrcoef as mcc
+from sklearn.metrics import accuracy_score as acc
+
+mcc_scorer = make_scorer(mcc)
+acc_scorer = make_scorer(acc)
+scores = cross_validate(model, X_train, y_train, cv=5,
+ scoring={"MCC": mcc_scorer, "ACC": acc_scorer})
+scores
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ Out[23]:
+
+
+
+
+
+{'fit_time': array([0.00225472, 0.00185323, 0.00180411, 0.00178003, 0.00177217]),
+ 'score_time': array([0.00157523, 0.00124979, 0.00123906, 0.00125003, 0.0012238 ]),
+ 'test_MCC': array([0.37796447, 0.27863911, 0.40824829, 0.02424643, 0.08625819]),
+ 'test_ACC': array([0.73333333, 0.7 , 0.76666667, 0.66666667, 0.62068966])}
+
+
+
+
+
+
+
+
+
+
+
+
+
+In [24]:
+
+
+import numpy as np
+
+print("Avg ACC in CV: ", np.average(scores["test_ACC"]))
+print("Avg MCC in CV: ", np.average(scores["test_MCC"]))
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+Avg ACC in CV: 0.697471264367816
+Avg MCC in CV: 0.2350712993854009
+
+
+
+
+
+
+
+
+
+
+
+
+
+In [25]:
+
+
+model = model.fit(X_train, y_train)
+
+print("ACC: ", acc_scorer(model, X_test, y_test))
+print("MCC: ", mcc_scorer(model, X_test, y_test))
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ACC: 0.7230769230769231
+MCC: 0.29439815585406465
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+To see exactly what happened, let's have a look at the Confusion matrix
+
+
+
+
+
+
+
+
+
+In [26]:
+
+
+from sklearn.metrics import ConfusionMatrixDisplay
+fig, ax = plt.subplots(figsize=(15, 10))
+ConfusionMatrixDisplay.from_estimator(model, X_test, y_test, ax=ax)
+plt.show()
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+As expected, the model did a pretty bad job in classifying Chinstrap Penguins and the MCC was able to catch that, whilst ACC could not as it only considers correctly classified samples!
+
+
+
+
+
+
+
+
+
+
+
+Time-series Validation¶
But validation can get tricky if time gets involved.
+Imagine we measured the growth of baby penguin Hank over time and wanted to us machine learning to project the development of Hank. Then our data suddenly isn't i.i.d. anymore, since it is dependent in the time dimension.
+Were we to split our data randomly for our training and test set, we would test on data points that lie in between training points, where even a simple linear interpolation can do a fairly decent job.
+Therefor, we need to split our measurements along the time axis
+ +Scikit-learn Time Series CV [Source].
+Scikit-learn Time Series CV [Source].
+
+
+
+
+
+
+
+
+
+In [27]:
+
+
+import numpy as np
+from sklearn.model_selection import TimeSeriesSplit
+
+X = np.array([[1, 2], [3, 4], [1, 2], [3, 4], [1, 2], [3, 4]])
+y = np.array([1, 2, 3, 4, 5, 6])
+tscv = TimeSeriesSplit(n_splits=3)
+print(tscv)
+
+for train, test in tscv.split(X):
+ print("%s %s" % (train, test))
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+TimeSeriesSplit(gap=0, max_train_size=None, n_splits=3, test_size=None)
+[0 1 2] [3]
+[0 1 2 3] [4]
+[0 1 2 3 4] [5]
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+Spatial Validation¶
Spatial data, like maps and satellite data has a similar problem.
+Here the data is correlated in the spatial dimension. However, we can mitigate the effect by supplying a group. In this simple example I used continents, but it's possible to group by bins on a lat-lon grid as well.
+Here especially, a cross-validation scheme is very important, as it is used to validate against every area on your map at least once.
+
+
+
+
+
+
+
+
+
+In [28]:
@@ -15348,7 +15348,7 @@ Tree importance vs Permutatio
- Tree importance vs Permutatio
@@ -15639,7 +15639,7 @@
Tree importance vs Permutatio
@@ -15639,7 +15639,7 @@ Shap Inspection
-
+
Visualization omitted, Javascript library not loaded!
Have you run `initjs()` in this notebook? If this notebook was from another
@@ -15649,8 +15649,8 @@ Shap Inspection
@@ -15693,7 +15693,7 @@ Shap Inspection
-
+
Visualization omitted, Javascript library not loaded!
Have you run `initjs()` in this notebook? If this notebook was from another
@@ -15703,8 +15703,8 @@ Shap Inspection
@@ -15834,7 +15834,7 @@ Model Inspection
-tensor([[0.3527, 0.3850]], grad_fn=<SigmoidBackward0>)
+tensor([[0.5887, 0.6065]], grad_fn=<SigmoidBackward0>)
To qualitatevely assess the effect of stratification, let's plot class distribution in both training and test sets:
+fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 8))
-
-
-
-
-
- Out[6]:
-
-
-
+y_train.reset_index().groupby("Species").count().plot(kind="bar", ax=ax1, ylim=(0, len(y)), title="Training")
+y_test.reset_index().groupby("Species").count().plot(kind="bar", ax=ax2, ylim=(0, len(y)), title="Test")
+plt.show()
+
-
-<AxesSubplot:xlabel='Species'>
+
+
+Stratification
- Stratification
Stratification
from sklearn.model_selection import cross_val_score
+
scores = cross_val_score(model, X_train, y_train, cv=5)
scores
Cross-Validation
- Out[8]:
+ Out[10]:
@@ -15233,7 +15255,7 @@ Cross-Validation
print(f"{scores.mean():0.2f} accuracy with a standard deviation of {scores.std():0.2f}")
@@ -15286,34 +15308,77 @@ Cross-Validation
-Time-series Validation¶
But validation can get tricky if time gets involved.
-Imagine we measured the growth of baby penguin Hank over time and wanted to us machine learning to project the development of Hank. Then our data suddenly isn't i.i.d. anymore, since it is dependent in the time dimension.
-Were we to split our data randomly for our training and test set, we would test on data points that lie in between training points, where even a simple linear interpolation can do a fairly decent job.
-Therefor, we need to split our measurements along the time axis
- -Scikit-learn Time Series CV [Source].
-Scikit-learn Time Series CV [Source].
+Model Evaluation¶
+
+
Brilliant! So let's recap for a moment what we have done so far, in preparation for our (final) Model evaluation.
+We have:
+-
+
- prepared the model pipeline:
sklearn.pipeline.Pipelinewithpreprocessor + model
+ - generated train and test data partitions (with stratification):
(X_train, y_train)and(X_test, y_test), respectively-
+
- stratification guaranteed that those partitions will retain class distributions +
+ - assessed model performance via cross validation (i.e.
cross_val_score) onX_train(!!)-
+
- this had the objective of verifying model consistency on multiple data partitioning +
+
Now we need the complete our last step, namely "assess how the model we chose in CV" (we only had one model, so that was an easy choice :D ) will perform on future data!
+And we have a candidate as representative for these data: X_test.
Please note that X_test has never been used so far (as it should have!). The take away message here is: generate test partition, and forget about it until the last step!
Thanks to CV, We have an indication of how the SVC classifier behaves on multiple "version" of the training set. We calculated an average score of 0.99 accuracy, therefore we decided this model is to be trusted for predictions on unseen data.
Now all we need to do, is to prove this assertion.
+To do so we need to:
+ +- train a new model on the entire **training set**
+- evaluate it's performance on **test set** (using the metric of choice - presumably the same metric we chose in CV!)import numpy as np
-from sklearn.model_selection import TimeSeriesSplit
+# training
+model = Pipeline(steps=[
+ ('preprocessor', preprocessor),
+ ('classifier', SVC()),
+])
+classifier = model.fit(X_train, y_train)
+
-X = np.array([[1, 2], [3, 4], [1, 2], [3, 4], [1, 2], [3, 4]])
-y = np.array([1, 2, 3, 4, 5, 6])
-tscv = TimeSeriesSplit(n_splits=3)
-print(tscv)
+ # Model evaluation
+from sklearn.metrics import accuracy_score
+
+y_pred = classifier.predict(X_test)
+print("TEST ACC: ", accuracy_score(y_true=y_test, y_pred=y_pred))
Time-series Validation
-TimeSeriesSplit(gap=0, max_train_size=None, n_splits=3, test_size=None)
-[0 1 2] [3]
-[0 1 2 3] [4]
-[0 1 2 3 4] [5]
+TEST ACC: 1.0
TEST ACC: 1.0
Time-series Validation
-Spatial Validation¶
Spatial data, like maps and satellite data has a similar problem.
-Here the data is correlated in the spatial dimension. However, we can mitigate the effect by supplying a group. In this simple example I used continents, but it's possible to group by bins on a lat-lon grid as well.
-Here especially, a cross-validation scheme is very important, as it is used to validate against every area on your map at least once.
+Now we can finally say that we have concluded our model evaluation - with a fantastic score of 0.96 Accuracy on the test set.
+
+
+
+
Spatial Validation¶
Spatial data, like maps and satellite data has a similar problem.
-Here the data is correlated in the spatial dimension. However, we can mitigate the effect by supplying a group. In this simple example I used continents, but it's possible to group by bins on a lat-lon grid as well.
-Here especially, a cross-validation scheme is very important, as it is used to validate against every area on your map at least once.
+Now we can finally say that we have concluded our model evaluation - with a fantastic score of 0.96 Accuracy on the test set.
Choosing the appropriate Evaluation Metric¶
+Ok, now for the mere sake of considering a more realistic data scenario, let's pretend our reference dataset is composed by only samples from two (out of the three) classes we have. In particular, we will crafting our dataset by choosing the most and the least represented classes, respectively.
+The very idea is to explore whether the choice of appropriate metrics could make the difference in our machine learning models evaluation.
+ +Let's recall class distributions in our dataset:
Spatial Validation
y.reset_index().groupby(["Species"]).count()
+| + | index | +
|---|---|
| Species | ++ |
| Adelie Penguin (Pygoscelis adeliae) | +146 | +
| Chinstrap penguin (Pygoscelis antarctica) | +68 | +
| Gentoo penguin (Pygoscelis papua) | +120 | +
So let's select samples from the first two classes, Adelie Penguin and Chinstrap penguin:
samples = penguins[((penguins["Species"].str.startswith("Adelie")) | (penguins["Species"].str.startswith("Chinstrap")))]
+samples.shape[0] == 146 + 68 # quick verification
+True+
To make things even harder for our machine learning model, let's also see if we could get rid of clearly separating features in this toy dataset
+ +import seaborn as sns
+
+pairplot_figure = sns.pairplot(samples, hue="Species")
+OK so if we get to choose, we could definitely say that in this dataset, the Flipper Length in combination with the Culmen Depth leads to the hardest classification task for our machine learning model.
Therefore, here is the plan:
+-
+
- we select only those to numerical features (iow we will get rid of the
Culmen Lenghtfeature)
+ - we will apply an identical Model evaluation pipeline as we did in our previous example
-
+
- Cross Validation + Evaluation on Test set +
+
The very difference this time is that we will use multiple metrics to evaluate our model to prove our point on carefully selecting evaluation metrics.
+ +num_features = ["Culmen Length (mm)", "Culmen Depth (mm)", "Flipper Length (mm)"]
+selected_num_features = num_features[1:]
+cat_features = ["Sex"]
+features = selected_num_features + cat_features
+num_transformer = StandardScaler()
+cat_transformer = OneHotEncoder(handle_unknown='ignore')
+
+preprocessor = ColumnTransformer(transformers=[
+ ('num', num_transformer, selected_num_features), # note here, we will only preprocess selected numerical features
+ ('cat', cat_transformer, cat_features)
+])
+
+model = Pipeline(steps=[
+ ('preprocessor', preprocessor),
+ ('classifier', SVC()),
+])
+X, y = samples[features], samples[target[0]]
+X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=.7, random_state=42, stratify=y) # we also stratify on classes
+y_train.reset_index().groupby("Species").count()
+| + | index | +
|---|---|
| Species | ++ |
| Adelie Penguin (Pygoscelis adeliae) | +102 | +
| Chinstrap penguin (Pygoscelis antarctica) | +47 | +
y_test.reset_index().groupby("Species").count()
+| + | index | +
|---|---|
| Species | ++ |
| Adelie Penguin (Pygoscelis adeliae) | +44 | +
| Chinstrap penguin (Pygoscelis antarctica) | +21 | +
In our evaluation pipeline we will be using keep record both accuracy (ACC) and matthew correlation coefficient (MCC)
from sklearn.model_selection import cross_validate
+from sklearn.metrics import make_scorer
+from sklearn.metrics import matthews_corrcoef as mcc
+from sklearn.metrics import accuracy_score as acc
+
+mcc_scorer = make_scorer(mcc)
+acc_scorer = make_scorer(acc)
+scores = cross_validate(model, X_train, y_train, cv=5,
+ scoring={"MCC": mcc_scorer, "ACC": acc_scorer})
+scores
+{'fit_time': array([0.00225472, 0.00185323, 0.00180411, 0.00178003, 0.00177217]),
+ 'score_time': array([0.00157523, 0.00124979, 0.00123906, 0.00125003, 0.0012238 ]),
+ 'test_MCC': array([0.37796447, 0.27863911, 0.40824829, 0.02424643, 0.08625819]),
+ 'test_ACC': array([0.73333333, 0.7 , 0.76666667, 0.66666667, 0.62068966])}
+import numpy as np
+
+print("Avg ACC in CV: ", np.average(scores["test_ACC"]))
+print("Avg MCC in CV: ", np.average(scores["test_MCC"]))
+Avg ACC in CV: 0.697471264367816 +Avg MCC in CV: 0.2350712993854009 ++
model = model.fit(X_train, y_train)
+
+print("ACC: ", acc_scorer(model, X_test, y_test))
+print("MCC: ", mcc_scorer(model, X_test, y_test))
+ACC: 0.7230769230769231 +MCC: 0.29439815585406465 ++
To see exactly what happened, let's have a look at the Confusion matrix
+ +from sklearn.metrics import ConfusionMatrixDisplay
+fig, ax = plt.subplots(figsize=(15, 10))
+ConfusionMatrixDisplay.from_estimator(model, X_test, y_test, ax=ax)
+plt.show()
+As expected, the model did a pretty bad job in classifying Chinstrap Penguins and the MCC was able to catch that, whilst ACC could not as it only considers correctly classified samples!
Time-series Validation¶
But validation can get tricky if time gets involved.
+Imagine we measured the growth of baby penguin Hank over time and wanted to us machine learning to project the development of Hank. Then our data suddenly isn't i.i.d. anymore, since it is dependent in the time dimension.
+Were we to split our data randomly for our training and test set, we would test on data points that lie in between training points, where even a simple linear interpolation can do a fairly decent job.
+Therefor, we need to split our measurements along the time axis
+ +Scikit-learn Time Series CV [Source].
+Scikit-learn Time Series CV [Source].
import numpy as np
+from sklearn.model_selection import TimeSeriesSplit
+
+X = np.array([[1, 2], [3, 4], [1, 2], [3, 4], [1, 2], [3, 4]])
+y = np.array([1, 2, 3, 4, 5, 6])
+tscv = TimeSeriesSplit(n_splits=3)
+print(tscv)
+
+for train, test in tscv.split(X):
+ print("%s %s" % (train, test))
+TimeSeriesSplit(gap=0, max_train_size=None, n_splits=3, test_size=None) +[0 1 2] [3] +[0 1 2 3] [4] +[0 1 2 3 4] [5] ++
Spatial Validation¶
Spatial data, like maps and satellite data has a similar problem.
+Here the data is correlated in the spatial dimension. However, we can mitigate the effect by supplying a group. In this simple example I used continents, but it's possible to group by bins on a lat-lon grid as well.
+Here especially, a cross-validation scheme is very important, as it is used to validate against every area on your map at least once.
+ +Tree importance vs Permutatio
- Tree importance vs Permutatio
@@ -15639,7 +15639,7 @@
Tree importance vs Permutatio
@@ -15639,7 +15639,7 @@ Shap Inspection
-
+
Visualization omitted, Javascript library not loaded!
Have you run `initjs()` in this notebook? If this notebook was from another
@@ -15649,8 +15649,8 @@ Shap Inspection
@@ -15693,7 +15693,7 @@ Shap Inspection
-
+
Visualization omitted, Javascript library not loaded!
Have you run `initjs()` in this notebook? If this notebook was from another
@@ -15703,8 +15703,8 @@ Shap Inspection
@@ -15834,7 +15834,7 @@ Model Inspection
-tensor([[0.3527, 0.3850]], grad_fn=<SigmoidBackward0>)
+tensor([[0.5887, 0.6065]], grad_fn=<SigmoidBackward0>)
Have you run `initjs()` in this notebook? If this notebook was from another @@ -15649,8 +15649,8 @@
Shap Inspection
Shap Inspection
-
+
Visualization omitted, Javascript library not loaded!
Have you run `initjs()` in this notebook? If this notebook was from another
@@ -15703,8 +15703,8 @@ Shap Inspection
@@ -15834,7 +15834,7 @@ Model Inspection
-tensor([[0.3527, 0.3850]], grad_fn=<SigmoidBackward0>)
+tensor([[0.5887, 0.6065]], grad_fn=<SigmoidBackward0>)
Have you run `initjs()` in this notebook? If this notebook was from another @@ -15703,8 +15703,8 @@
Shap Inspection
Model Inspection
-tensor([[0.3527, 0.3850]], grad_fn=<SigmoidBackward0>)
+tensor([[0.5887, 0.6065]], grad_fn=<SigmoidBackward0>)
tensor([[0.5887, 0.6065]], grad_fn=<SigmoidBackward0>)