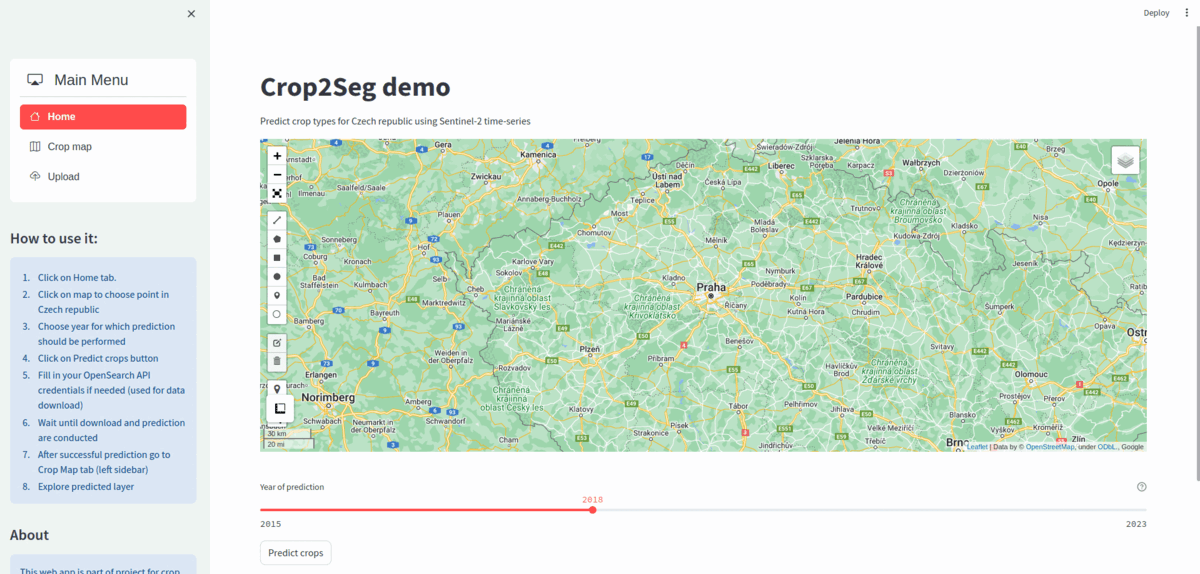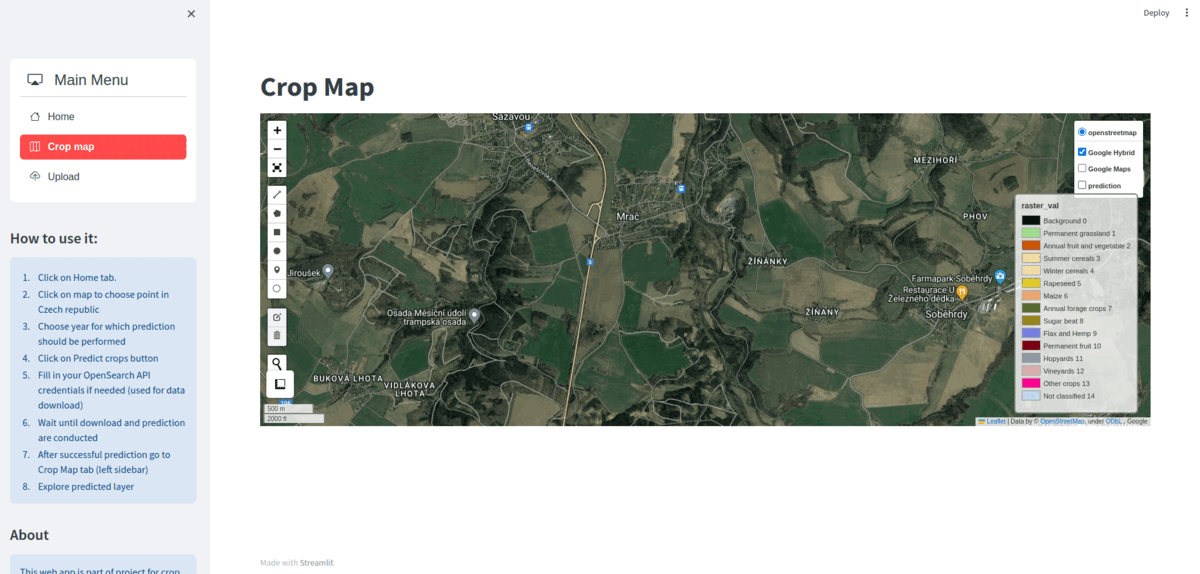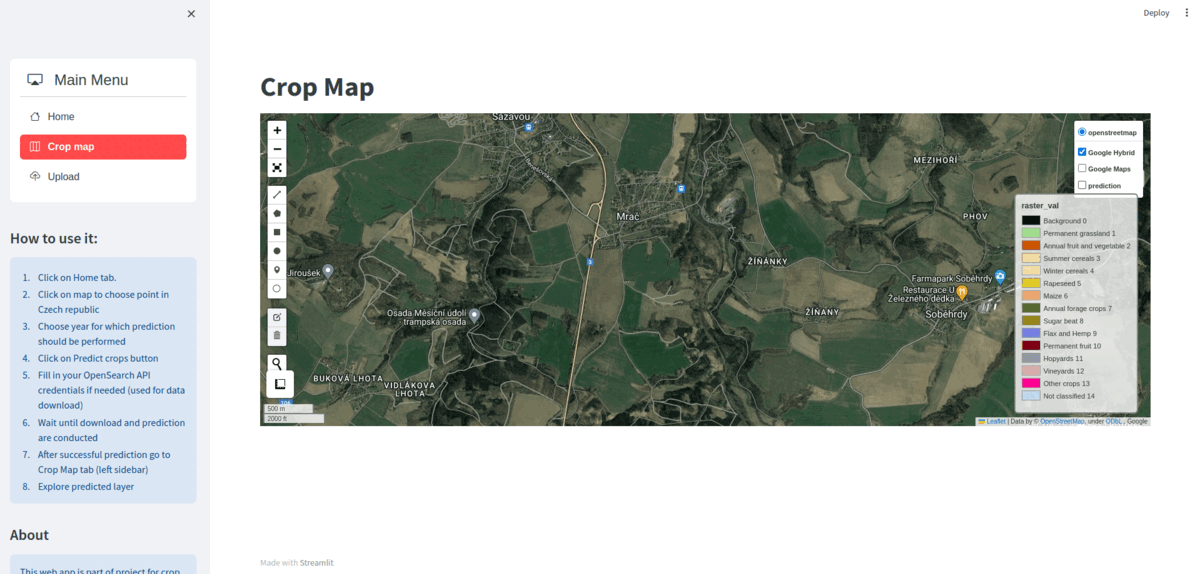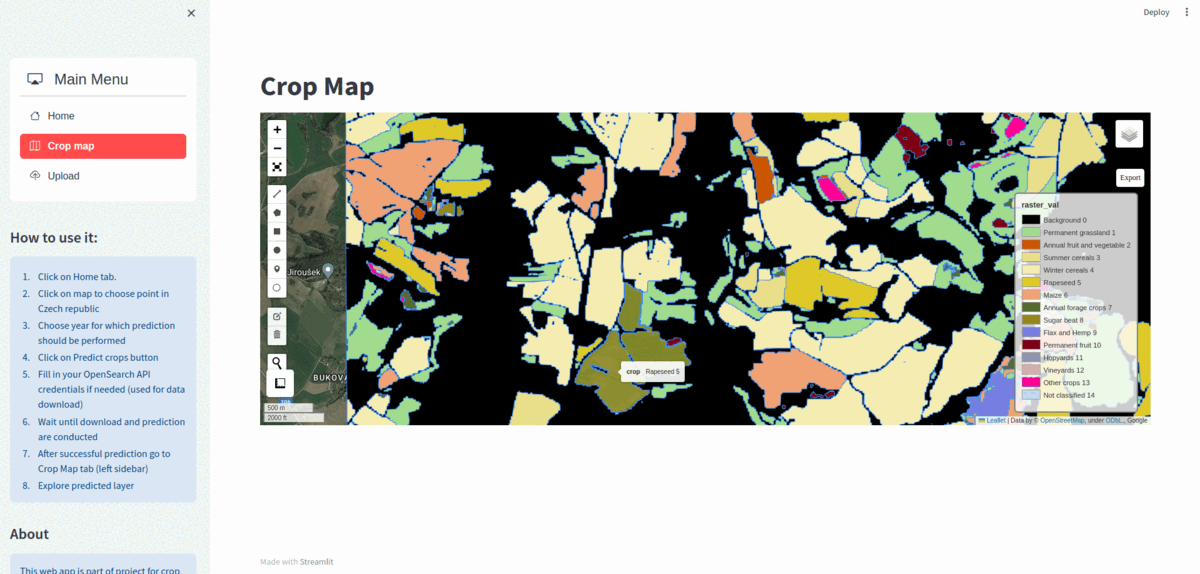
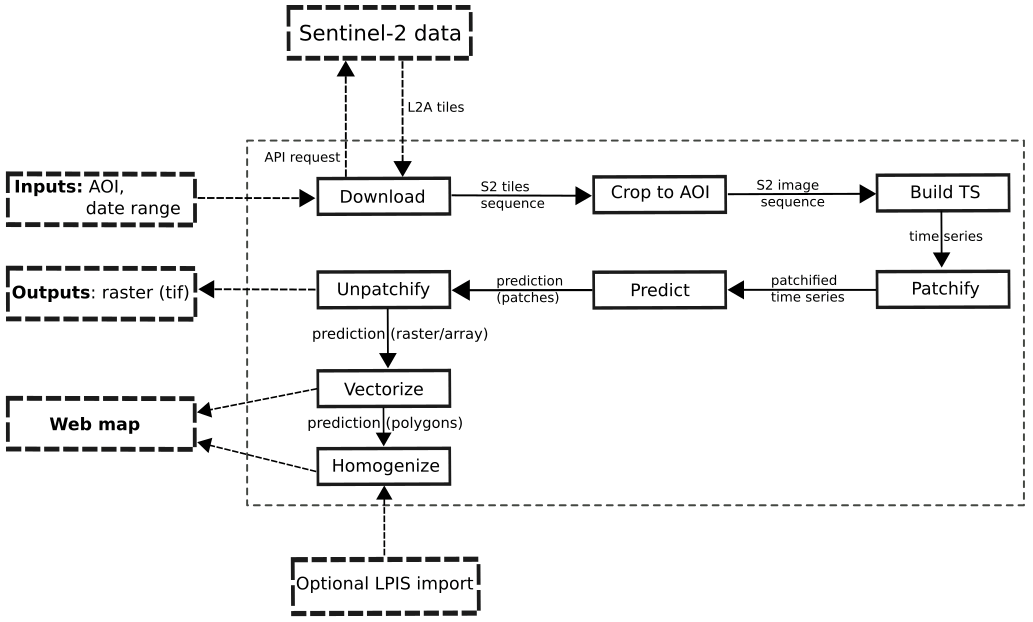
Main goal of Crop2Seg project is to explore semantic segmentation of crop types in time-series of Sentinel-2 tiles as part of author's Master's thesis. We also want to provide standalone dataset for semantic segmentation of crop types from Sentinel-2 time-series for Czech Republic in 2019.
Finally we want to provide web application which can process Sentinel-2 data and perform semantic segmentation of crop types in automatic manner.
Project was implemented in Python 3.8 therefore it is recommended to use it (it will be installed automatically within conda environment). All required dependencies are listed in requirements.txt and environment.yml files.
Note that we work with Sentinel-2 tiles and therefore all processings are very memory intensive. Especially dataset creation for training neural net requires at least 500GB of RAM. Provided web app was optimized to work with small amount of ram but it is recomended to use system with at least 6 GB of (free) RAM. On the other hand we use cached storage which is located on directory (folder) where the repo is cloned and therefore to clone this repo choose directory with enough disk space (e.g. to store time series of length of 60 one needs at least 80 GB of disk space).
We use cesnet mirror for downloading Sentinel-2 tiles. Please create an account before using demo app.
-
To clone repo you can use just
-
cd <path to directory with enough disk space> git clone https://github.com/Many98/Crop2Seg.git
optionally (on Windows) one can use
-
Default location of repo on Windows will be C:\Users\user\GitHub\Crop2Seg
-
-
To avoid dependency conflicts when installing packages it is wise to use virtual environment e.g. venv/anaconda. We recommend to use miniconda
On Windows particularly this step can be problematic. It is needed to have properly configured PATH environment variable (check it in miniconda installation)
After miniconda is installed proceeed in terminal with following
conda env create -f environment.yml
-
Previous step created
oxminiconda environmentTo launch conda environment use terminal with following commands (on windows use anaconda prompt)
conda activate ox
Main script for application is crop2seg.py
App is currently not hosted on internet but one can run it on localhost with following:
cd <path to directory with cloned repo>
conda activate ox
streamlit run crop2seg.pyWeb interface is created using streamlit and based on this streamlit-template
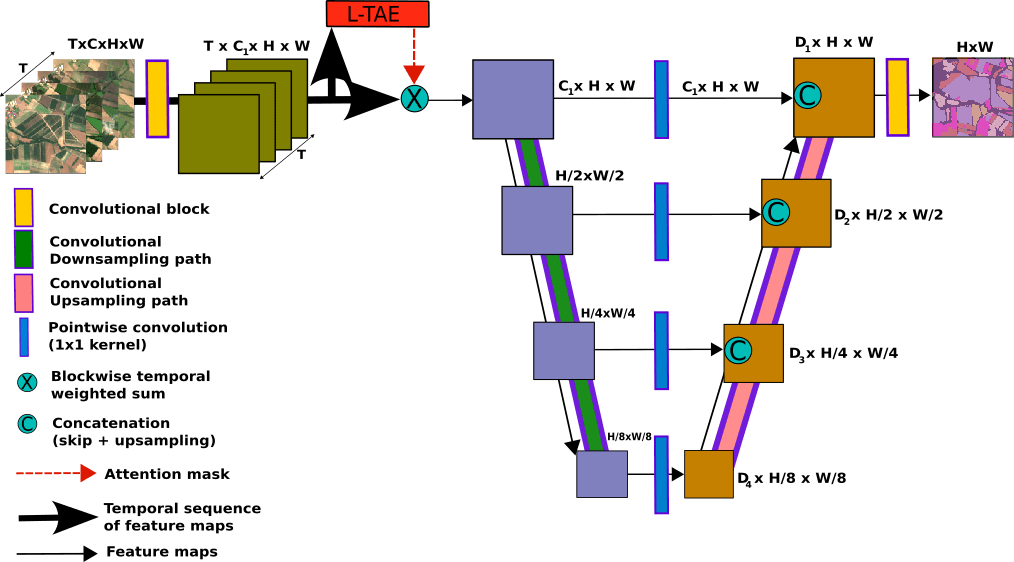
Proposed models are based on U-TAE. Modifications are mainly made within order of processing. Also few adjustments in positional encoding are proposed.
| Time-Unet | W-TAE |
|---|---|
 |
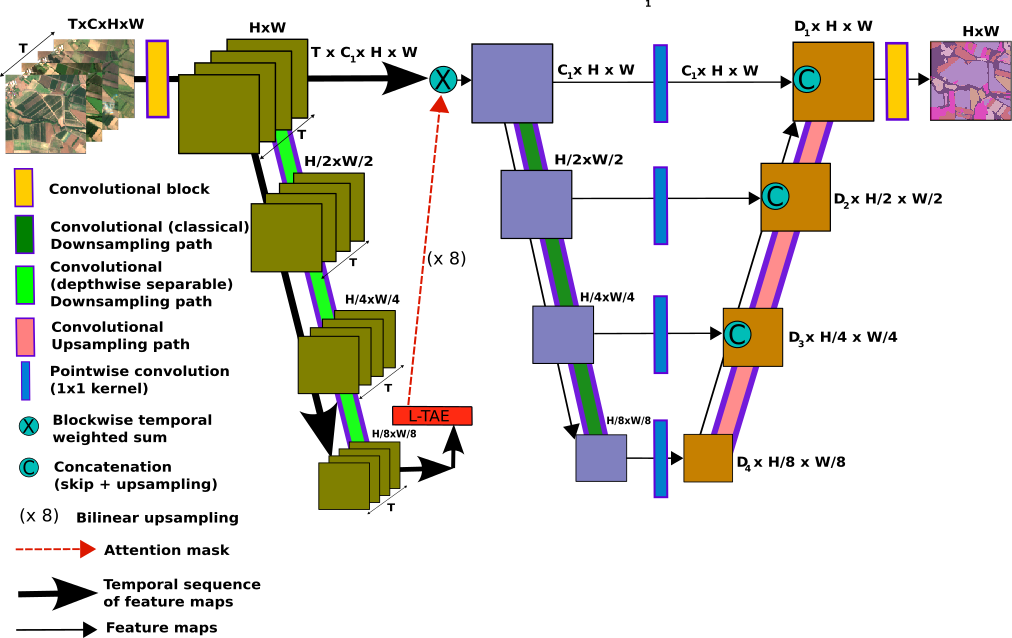 |
TODO make it publicly available
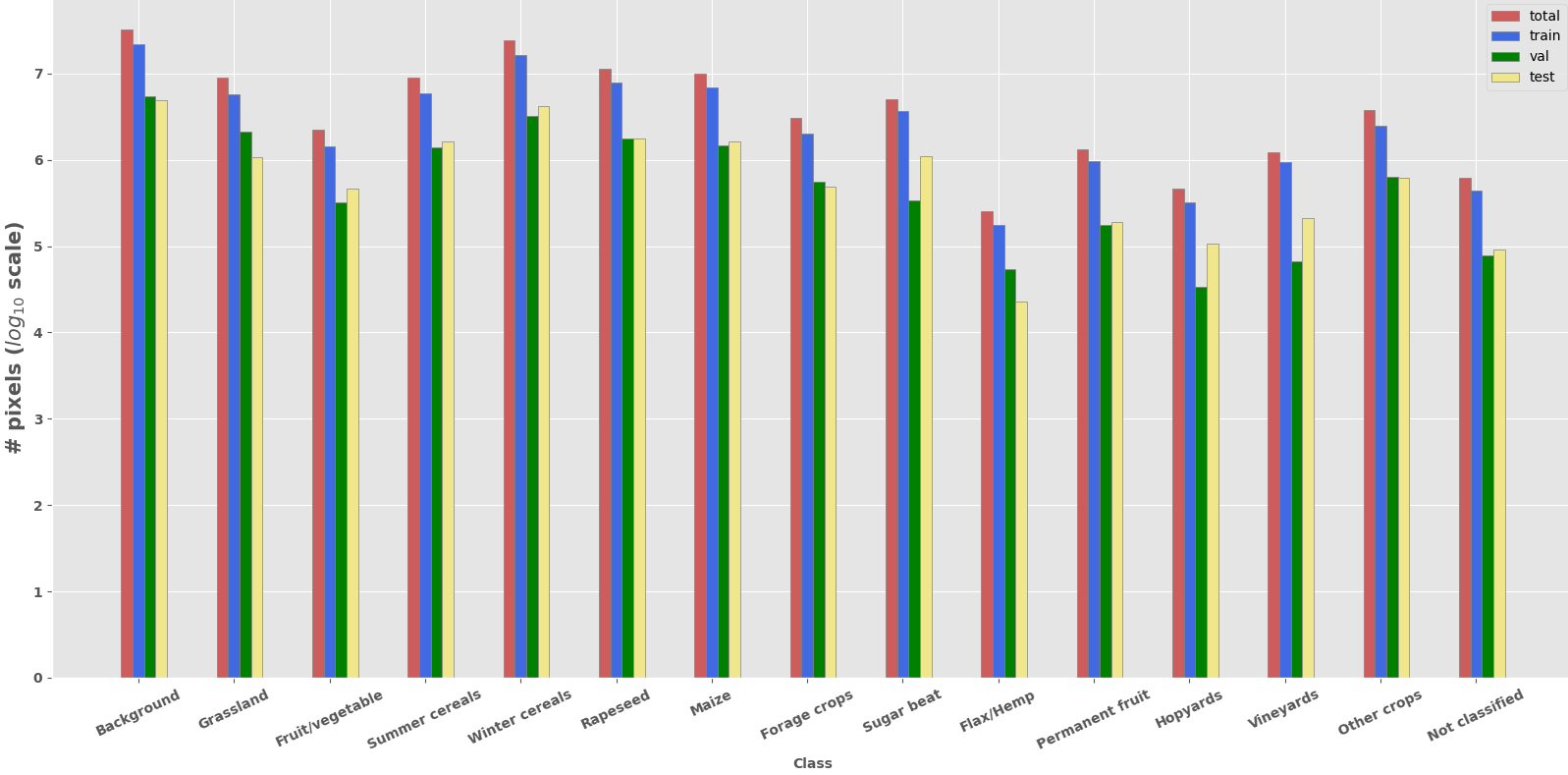
Dataset consits of 6945 Sentinel-2 time-series patches over Czech Republic for year 2019 divided into train/validation/test sub-datasets. Similarly to PASTIS we use patches of shape T x C x H x W where C=10, H=W=128. Note that lenght of time-series is irregular i.e. time-series can have from 27 to 61 acquisitions.
Dataset contains 13 classes of crops and 2 auxiliary classes (Background & Not classified). Also class representing boundary can be generated on-the-fly from ground truth (see on-the-fly boundary extraction).
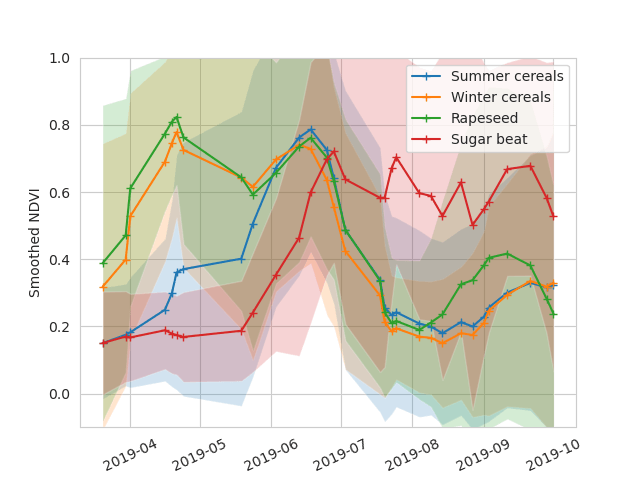
NDVI Temporal profiles of all used crop types can be found here
TODO
One can use CLI in sentinel_cli.py. It requires only subset of dependencies specified either in requirements_sentinel.txt or environment_sentinel.yml
To get help use:
$ python sentinel_cli.py --help
usage: sentinel_cli.py [-h] [--download] [--path PATH] [--platform PLATFORM] [--count COUNT] [--polygon POLYGON] [--filename FILENAME] [--product PRODUCT] [--begin BEGIN] [--polarisation POLARISATION] [--sensor SENSOR]
[--tilename TILENAME] [--cloud [CLOUD [CLOUD ...]]] [--s2timeseries]
Basic operations with Sentinel-1 and Sentinel-2. See configuration file to set specific settingsCapabilities are:
1) downloading Sentinel-1 and Sentinel-2 data (also Sentinel-2 time-series see config)
optional arguments:
-h, --help show this help message and exit
--download, -d Whether to perform download. (default: False)
--path PATH, -pa PATH
Path to the folder where Sentinel-1 and Sentinel-2 data will be downloaded (default: D:\sentinel)
--platform PLATFORM, -pl PLATFORM
Specifies the name of missionCan be: * `Sentinel-1` * Sentinel-2` (default: Sentinel-2)
--count COUNT, -c COUNT
Number of products to be downloaded (default: 4)
--polygon POLYGON, -p POLYGON
Polygon defining Area of interest. E.g. `--polygon "[[14.31, 49.44], [13.89, 50.28], [15.55, 50.28]]"` (default: [[14.31, 49.44], [13.89, 50.28], [15.55, 50.28]])
--filename FILENAME, -f FILENAME
Filename of product to be downloaded (default: None)
--product PRODUCT, -pr PRODUCT
Specifies the type of product of particular Sentinel mission.For Sentinel_1 : * 'SLC' * 'GRD'For Sentinel_2 : * 'S2MSI2A' (in other parts code this is referred as 'L2A') * 'S2MSI1C' (in other parts code this is referred as 'L1C')If None then both (all)
product types will be downloaded (but only if 'filename' parameter is not used (set to None)) (default: S2MSI2A)
--begin BEGIN, -b BEGIN
Specifies sensing start date (Specifies interval e.g. `--begin "[NOW-30DAYS TO NOW]"`)To download Sentinel-2 time series use --s2_time_series argument (default: [NOW-30DAYS TO NOW])
--polarisation POLARISATION, -pol POLARISATION
Specifies the polarisation mode of Sentinel 1 radar.Can be: * 'HH' * 'VV' * 'HV' * 'VH' * 'HH HV' * 'VV VH' (default: VV VH)
--sensor SENSOR, -s SENSOR
Specifies the sensor operational mode of Sentinel 1 radar.Can be: * 'SM' * 'IW' (usually used) * 'EW' * 'WV' (default: IW)
--tilename TILENAME, -t TILENAME
Specifies name of particular tile. e.g. `--tilename T33UWQ``This can be done instead of performing search based on 'polygon' parameter. (default: None)
--cloud [CLOUD [CLOUD ...]], -cl [CLOUD [CLOUD ...]]
Specifies interval of allowed overall cloud coverage percentage of Sentinel-2 tile.E.g. `--cloud "[0 TO 5.5]"` is translated as [0 TO 5.5] for API used (default: ['[0 TO 5]'])
--s2timeseries, -ts Whether to download whole time series of Sentinel-2 data.Works only with Sentinel-2.See configuration to set tiles, dates and cloud cover percentages (default: False)Main script for model training/testing/finetuning etc. is train.py
It has CLI interface implemented.
To find out how to use it run following command:
$ python train.py --help
usage: train.py [-h] [--model MODEL] [--encoder_widths ENCODER_WIDTHS] [--decoder_widths DECODER_WIDTHS] [--out_conv OUT_CONV] [--str_conv_k STR_CONV_K] [--str_conv_s STR_CONV_S] [--str_conv_p STR_CONV_P] [--agg_mode AGG_MODE]
[--encoder_norm ENCODER_NORM] [--n_head N_HEAD] [--d_model D_MODEL] [--d_k D_K] [--input_dim INPUT_DIM] [--num_queries NUM_QUERIES] [--temporal_dropout TEMPORAL_DROPOUT] [--augment] [--add_linear] [--add_boundary]
[--get_affine] [--dataset DATASET] [--test] [--test_region TEST_REGION] [--finetune] [--dataset_folder DATASET_FOLDER] [--norm_values_folder NORM_VALUES_FOLDER] [--weight_folder WEIGHT_FOLDER] [--res_dir RES_DIR]
[--rdm_seed RDM_SEED] [--device DEVICE] [--display_step DISPLAY_STEP] [--cache] [--epochs EPOCHS] [--batch_size BATCH_SIZE] [--lr LR] [--mono_date MONO_DATE] [--ref_date REF_DATE] [--fold FOLD]
[--num_classes NUM_CLASSES] [--ignore_index IGNORE_INDEX] [--pad_value PAD_VALUE] [--padding_mode PADDING_MODE] [--conv_type CONV_TYPE] [--use_mbconv] [--add_squeeze] [--use_doy] [--add_ndvi] [--use_abs_rel_enc]
[--seg_model SEG_MODEL] [--temp_model TEMP_MODEL] [--val_every VAL_EVERY] [--val_after VAL_AFTER]
optional arguments:
-h, --help show this help message and exit
--model MODEL Type of architecture to use. Can be one of: (utae/unet3d/timeunet/wtae)
--encoder_widths ENCODER_WIDTHS
--decoder_widths DECODER_WIDTHS
--out_conv OUT_CONV
--str_conv_k STR_CONV_K
--str_conv_s STR_CONV_S
--str_conv_p STR_CONV_P
--agg_mode AGG_MODE
--encoder_norm ENCODER_NORM
--n_head N_HEAD
--d_model D_MODEL Dimension to which map value vectors before temporalencoding.
--d_k D_K Dimension of learnable query vector.
--input_dim INPUT_DIM
Number of input spectral channels
--num_queries NUM_QUERIES
Number of learnable query vectors. This vectors areaveraged.
--temporal_dropout TEMPORAL_DROPOUT
Probability of removing acquisition from time-series
--augment Whether to perform augmentation of S2TSCZCrop Dataset
--add_linear Whether to add linear transform to positional encoder
--add_boundary Whether to add boundary loss. i.e. model will segment crops and boundary
--get_affine Whether to return also affine transform
--dataset DATASET Type of dataset to use. Can be one of: (s2tsczcrop/pastis)
--test Whether to perform test run (inference)Weights stored in `--weight_folder` directory will be used
--test_region TEST_REGION
Experimental setting. Can be one of ['all', 'boundary', 'interior']
--finetune Whether to perform finetuning instead of training from scratch.Weights stored in `--weight_folder` directory will be used
--dataset_folder DATASET_FOLDER
Path to the folder where is stored dataset.
--norm_values_folder NORM_VALUES_FOLDER
Path to the folder where to look for NORM_S2_patch.json file storing normalization values
--weight_folder WEIGHT_FOLDER
Path to folder containing the network weights in model.pth.tar file and model configuration file in conf.json.If you want to resume training then this folder should also have trainlog.json file.
--res_dir RES_DIR Path to the folder where the results should be stored
--rdm_seed RDM_SEED Random seed
--device DEVICE Name of device to use for tensor computations (cuda/cpu)
--display_step DISPLAY_STEP
Interval in batches between display of training metrics
--cache If specified, the whole dataset is kept in RAM
--epochs EPOCHS Number of epochs
--batch_size BATCH_SIZE
Batch size
--lr LR Learning rate
--mono_date MONO_DATE
Whether to perform segmentation using only one element of time-series. Use integer or string in form (YYYY-MM-DD)
--ref_date REF_DATE Reference date (YYYY-MM-DD) used in relative positional encoding scheme i.e. dates are encoded as difference between actual date and reference date. If you want to use absolute encodingusing day of years
use `--use_doy` flag
--fold FOLD Specify fold. (between 1 and 5) Note that this argument is used only as legacy argument and is used only for accessing correct normalization values e.g. if using PASTIS trainednetwork for fine-tuning
--num_classes NUM_CLASSES
Number of classes used in segmentation task
--ignore_index IGNORE_INDEX
Index of class to be ignored
--pad_value PAD_VALUE
Padding value for time-series
--padding_mode PADDING_MODE
Type of padding
--conv_type CONV_TYPE
Type of convolutional layer. Must be one of '2d' or 'depthwise_separable'
--use_mbconv Whether to use MBConv module instead of classical convolutional layers
--add_squeeze Whether to add squeeze & excitation module
--use_doy Whether to use absolute positional encoding (day of year) instead of relative encoding w.r.t. reference date
--add_ndvi Whether to add NDVI channel at the end
--use_abs_rel_enc Whether to use both date representations: Relative andabsolute (DOY)
--seg_model SEG_MODEL
Model to use for segmentation
--temp_model TEMP_MODEL
Model to use for temporal encoding
--val_every VAL_EVERY
Interval in epochs between two validation steps.
--val_after VAL_AFTER
Do validation only after that many epochs.