@@ -94,80 +85,80 @@ Functions and dataExamples
The following example creates an area source for São Paulo State (Brasil). The vehicles function creates a data.frame with information about the São Paulo Fleet using data from [@detran2016], the emissionFactors creates a data.frame with emission factors for CO and PM [@cetesbEV2015]. The totalEmission calculates the total emissions of CO for these vehicles and this emission factors. The next 3 lines opens different data: wrf file, a raster and the area shapefiles. These data are the input for areaSouce that creates an area source based on an image of persistent lights of the Defense Meteorological Satellite Program (DMSP) for São Paulo and Minas Gerais (Brasil) states and finally the function emission calculates the CO emissions.
-library(EmissV)
-
-fleet <- vehicles(example = T)
-# using a example of vehicles (DETRAN 2016 data and SP vahicle distribution):
-# Category Type Fuel Use SP ...
-# Light Duty Vehicles Gasohol LDV_E25 LDV E25 41 km/d 11624342 ...
-# Light Duty Vehicles Ethanol LDV_E100 LDV E100 41 km/d 874627 ...
-# Light Duty Vehicles Flex LDV_F LDV FLEX 41 km/d 9845022 ...
-# Diesel Trucks TRUCKS_B5 TRUCKS B5 110 km/d 710634 ...
-# Diesel Urban Busses CBUS_B5 BUS B5 165 km/d 792630 ...
-# Diesel Intercity Busses MBUS_B5 BUS B5 165 km/d 21865 ...
-# Gasohol Motorcycles MOTO_E25 MOTO E25 140 km/d 3227921 ...
-# Flex Motorcycles MOTO_F MOTO FLEX 140 km/d 235056 ...
-
-# dropping the fleet from Rio de Janeiro (RJ), Parana (PR) and Santa Catarina (SC)
-fleet <- fleet[,c(-6,-8,-9)]
-
-EF <- emissionFactor(example = T)
-# using a example emission factor (values calculated from CETESB 2015):
-# CO PM
-# Light Duty Vehicles Gasohol 1.75 g/km 0.0013 g/km
-# Light Duty Vehicles Ethanol 10.04 g/km 0.0000 g/km
-# Light Duty Vehicles Flex 0.39 g/km 0.0010 g/km
-# Diesel Trucks 0.45 g/km 0.0612 g/km
-# Diesel Urban Busses 0.77 g/km 0.1052 g/km
-# Diesel Intercity Busses 1.48 g/km 0.1693 g/km
-# Gasohol Motorcycles 1.61 g/km 0.0000 g/km
-# Flex Motorcycles 0.75 g/km 0.0000 g/km
-
-TOTAL <- totalEmission(fleet,EF,pol = c("CO"),verbose = T)
-# Total of CO : 1128297.0993334 t year-1
-
-grid <- gridInfo(paste(system.file("extdata", package = "EmissV"),
- "/wrfinput_d02",sep=""))
-# Grid information from: .../EmissV/extdata/wrfinput_d02
-
-raster <- raster::raster(paste(system.file("extdata", package = "EmissV"),
- "/dmsp_hi-res.tiff",sep=""))
-
-shape <- raster::shapefile(paste(system.file("extdata", package = "EmissV"),
- "/BR.shp",sep=""),verbose = F)[12,1]
-
-Minas_Gerais <- areaSource(shape,raster,grid,name = "Minas Gerais")
-# processing Minas Gerais area ...
-# fraction of Minas Gerais area inside the domain = 0.0147607845622591
-
-shape <- raster::shapefile(paste(system.file("extdata", package = "EmissV"),
- "/BR.shp",sep=""),verbose = F)[22,1]
-
-Sao_Paulo <- areaSource(shape,raster,grid,name = "Sao Paulo")
-# processing Sao Paulo area ...
-# fraction of Sao Paulo area inside the domain = 0.473260323300595
-
-sp::spplot(raster::merge(drop_units(TOTAL$CO[[1]]) * Sao_Paulo,
- drop_units(TOTAL$CO[[2]]) * Minas_Gerais),
- scales = list(draw=TRUE),ylab="Lat",xlab="Lon",
- # main=list(label="Emissions of CO [g/d]"),
- col.regions = c("#031638","#001E48","#002756","#003062",
- "#003A6E","#004579","#005084","#005C8E",
- "#006897","#0074A1","#0081AA","#008FB3",
- "#009EBD","#00AFC8","#00C2D6","#00E3F0"))
-
-CO_emissions <- emission(total = TOTAL,
- pol = "CO",
- area = list(SP = Sao_Paulo, MG = Minas_Gerais),
- grid = grid,
- mm = 28,
- plot = T)
-# calculating emissions for CO using molar mass = 28 ...
+
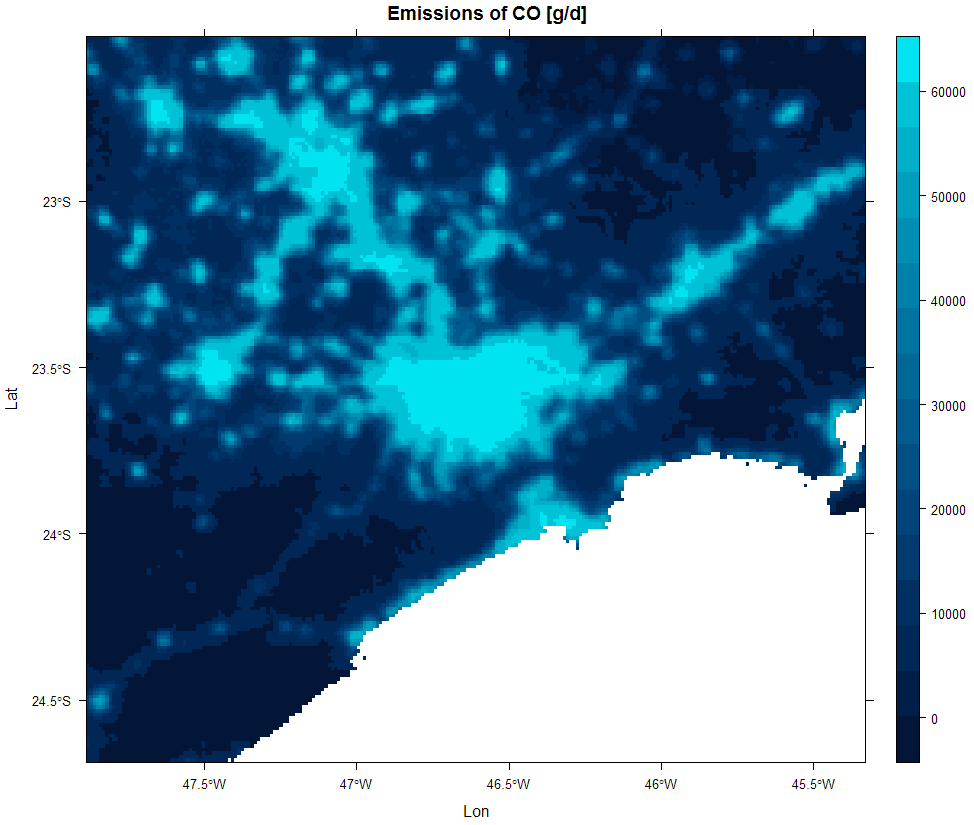
Emissions of CO using nocturnal lights.

