Sequence to sequence models convert a sequence of one type to a sequence of another type. The encoder decoder architecture is often used to achieve this. Both the encoder and decoder are built with RNNs, but the entire model is trained end-to-end (i.e. from input into encoder to output from decoder).
An encoder decoder model. Image courtesy of Towards Data Science.
At each time
The encoder inputs the data in time steps. It disregards the output at each
The context vector is passed into the decoder sequentially. The decoder calculates the next internal state using the input context vector data and the previous internal state. This information is used to calculate the output at each time step
The issue with standard encoder decoder architecture arises when input sequences increase in length, as it becomes very difficult to capture information from the entire input sequence in one vector. Since this last internal state of the encoder is the only vector passed through to the decoder, performance of the model decreases.
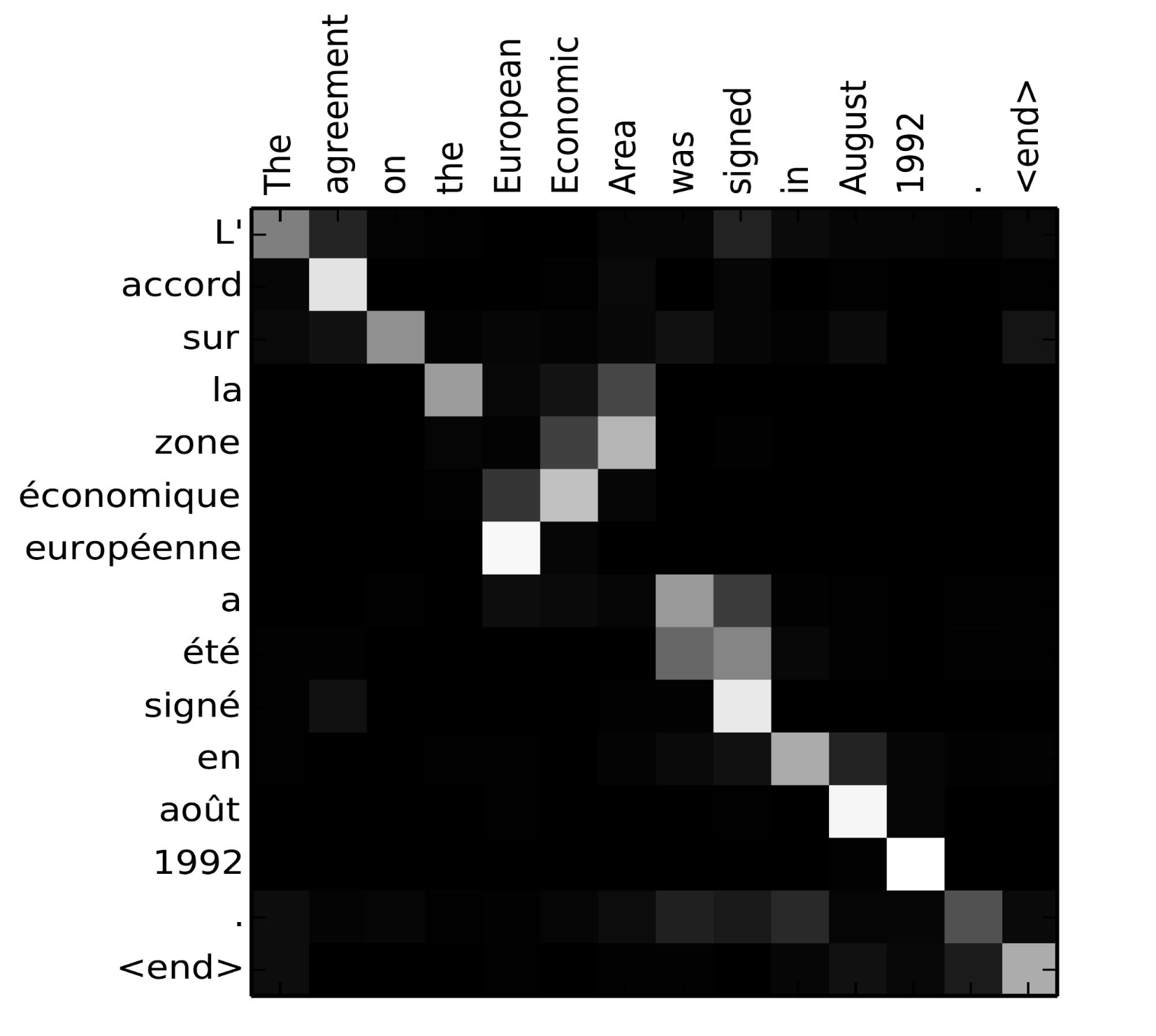
When applied to a sequence to sequence model, attention determines and focuses the model on the most relevant input features for a certain output. In general, this is done by storing all hidden states in a matrix, instead of passing only the vector representing the last hidden state to the decoder. Having access to all internal state information, it creates mappings between outputs at all time steps of the decoder and all internal states of the encoder to determine and select features most influential in producing an output at a certain time step. The "attention" given to an input feature is quantified with alignment scores.
Matrix of alignment scores of French to English translation from Bahdanau et al.
Several attention mechanisms have been introduced.
The attention mechanisms proposed by Bahdanau et al. and Luong et al. involve three aspects: the encoder model, alignment scores, and the decoder model. The main difference in approach between the two processes is when the decoder RNN is utilized.
The first step for both methods is to pass inputs into the encoder RNN and store all hidden states.
The process of calculating alignment scores differs slightly between the Bahdanau and Luong methods because the decoder RNN is used at different times.
Bahdanau et al. calculated alignment scores using the following equation:
$$ \text{score}{\text{alignment}} = W{\text{combined}} \cdot \tanh(W_{\text{decoder}} * H_{\text{decoder}} ^{(t-1)} + W_{\text{encoder}} \cdot H_{\text{encoder}}) $$
Luong et al. calculated alignment scores using three different equations:
- Dot:
$$ \text{score}{\text{alignment}} = H{\text{encoder}} \cdot H_{\text{decoder}} $$
- General:
$$ \text{score}{\text{alignment}} = W(H{\text{encoder}} \cdot H_{\text{decoder}}) $$
- Concat:
$$ \text{score}{\text{alignment}} = W \cdot \tanh(W\text{combined}(H_{\text{encoder}} + H_{\text{decoder}})) $$
Both methods applied the
For the Bahdanau attention method, the decoder RNN was used last. At time
Attention model by Bahdanau et al.
For the Luong attention method, the decoder RNN was used first, prior to calculating the alignment scores. Thus, the hidden state at time step
Attention model by Luong et al.
Attention methods highlight the most relevant input features using probability distributions; the
In soft attention, a weighted average of features and their weights is computed, and the result is fed into the RNN architecture. Generally, soft attention is parameterized by differentiable and thus continuous functions, so training can be completed via backpropagation.
However, in hard attention, the obtained weights are used to sample one feature to input into the RNN architecture. Hard attention methods are generally described by discrete variables. Because of this, other techniques must be used to train structures other than standard gradient descent, which depends on differentiable functions.
Global attention refers to when attention is calculated over the entire input sequence. Local attention refers to when a subsection of the input sequence is considered.