diff --git a/.github/workflows/integration-test.yml b/.github/workflows/integration-test.yml
index 9f817695..7b265ba1 100644
--- a/.github/workflows/integration-test.yml
+++ b/.github/workflows/integration-test.yml
@@ -28,6 +28,23 @@ env:
TARGET_PYTHON_VERSION: "3.9"
jobs:
+ docs-check:
+ # This job is used to check the documentation build with strict mode enabled
+ name: Docs check
+ runs-on: ubuntu-latest
+ steps:
+ - name: Checkout repo
+ uses: actions/checkout@v4
+ - name: Setup python, and load cache
+ uses: ./.github/actions/setup-env
+ with:
+ python-version: ${{ env.TARGET_PYTHON_VERSION }}
+ cache-pre-commit: false
+ cache-venv: true
+ setup-poetry: true
+ install-deps: true
+ - name: Build documentation
+ run: poetry run mkdocs build --strict
quality-test:
# This job is used to run pre-commit checks to ensure that all files are
# are formatted correctly.
diff --git a/.readthedocs.yml b/.readthedocs.yml
index 6671c7de..b273a65f 100644
--- a/.readthedocs.yml
+++ b/.readthedocs.yml
@@ -4,21 +4,27 @@ build:
os: ubuntu-22.04
tools:
python: "3.11"
- jobs:
- post_checkout:
- # Full history is required for dunamai to calculate the version
- - git fetch --unshallow || true
- post_create_environment:
- # Install poetry
- # https://python-poetry.org/docs/#installing-manually
- - pip install poetry
- # Tell poetry to not use a virtual environment
- - poetry config virtualenvs.create false
- post_install:
- # Install dependencies with 'docs' dependency group
- # https://python-poetry.org/docs/managing-dependencies/#dependency-groups
- - poetry install --with dev,docs --all-extras
+ commands:
+ # Full history is required for dunamai to calculate the version
+ - git fetch --unshallow || true
+ # Install poetry
+ # https://python-poetry.org/docs/#installing-manually
+ - pip install poetry
+ # Install poetry-dynamic-versioning plugin
+ - poetry self add "poetry-dynamic-versioning[plugin]"
+ # Build the project
+ - poetry build --format sdist
+ # Extract the built sdist
+ - mkdir -p dist/sdist && tar -xzf dist/*.tar.gz -C dist/sdist/
+ # Replace the files from the repo with the built sdist
+ - mv dist/sdist/*/pyproject.toml .
+ # Tell poetry to not use a virtual environment
+ - poetry config virtualenvs.create false
+ # Install dependencies with 'docs' dependency group
+ # https://python-poetry.org/docs/managing-dependencies/#dependency-groups
+ - poetry install --with dev,docs --all-extras
+ # Build the docs
+ - poetry run mkdocs build --clean --site-dir $READTHEDOCS_OUTPUT/html --config-file mkdocs.yml
-sphinx:
- builder: html
- configuration: docs/conf.py
+mkdocs:
+ configuration: mkdocs.yml
diff --git a/MANIFEST.in b/MANIFEST.in
deleted file mode 100644
index bda36cdf..00000000
--- a/MANIFEST.in
+++ /dev/null
@@ -1,6 +0,0 @@
-include MANIFEST.in
-include LICENSE.md
-include README.md
-include setup.py
-include pycytominer/data/*
-
diff --git a/Makefile b/Makefile
index c6fe85bd..aaf34a5c 100644
--- a/Makefile
+++ b/Makefile
@@ -18,8 +18,8 @@ test: ## Test the code with pytest
.PHONY: docs
docs: ## Build the documentation
- @echo "📚 Building documentation"
- @poetry run sphinx-build docs build
+ @echo "📚 Serving documentation"
+ @mkdocs serve
.PHONY: build
build: clean-build ## Build wheel file using poetry
diff --git a/README.md b/README.md
index 6b5d542e..a66d91d5 100644
--- a/README.md
+++ b/README.md
@@ -1,5 +1,17 @@
 +- [Data processing for image-based profiling](#data-processing-for-image-based-profiling)
+ - [Installation](##installation)
+ - [Frameworks](#frameworks)
+ - [API](#api)
+ - [Usage](#usage)
+ - [Pipeline orchestration](#pipeline-orchestration)
+ - [Other functionality](#other-functionality)
+ - [CellProfiler CSV collation](#cellprofiler-csv-collation)
+ - [Creating a cell locations lookup table](#creating-a-cell-locations-lookup-table)
+ - [Generating a GCT file for morpheus](#generating-a-gct-file-for-morpheus)
+ - [Citing pycytominer](#citing-pycytominer)
+
# Data processing for image-based profiling
[](https://github.com/cytomining/pycytominer/actions/workflows/integration-test.yml?query=branch%3Amain)
@@ -111,19 +123,7 @@ And, more specifically than that, image-based profiling readouts from [CellProfi
Therefore, we have included some custom tools in `pycytominer/cyto_utils` that provides other functionality:
-- [Data processing for image-based profiling](#data-processing-for-image-based-profiling)
- - [Installation](#installation)
- - [Frameworks](#frameworks)
- - [API](#api)
- - [Usage](#usage)
- - [Pipeline orchestration](#pipeline-orchestration)
- - [Other functionality](#other-functionality)
- - [CellProfiler CSV collation](#cellprofiler-csv-collation)
- - [Creating a cell locations lookup table](#creating-a-cell-locations-lookup-table)
- - [Generating a GCT file for morpheus](#generating-a-gct-file-for-morpheus)
- - [Citing pycytominer](#citing-pycytominer)
-
-Note, [`pycytominer.cyto_utils.cells.SingleCells()`](pycytominer/cyto_utils/cells.py) contains code to interact with single-cell SQLite files, which are output from CellProfiler.
+Note, [`pycytominer.cyto_utils.cells.SingleCells()`](./pycytominer/cyto_utils/cells.py) contains code to interact with single-cell SQLite files, which are output from CellProfiler.
Processing capabilities for SQLite files depends on SQLite file size and your available computational resources (for ex. memory and cores).
### CellProfiler CSV collation
diff --git a/docs/README.md b/docs/README.md
new file mode 100644
index 00000000..7f5067ed
--- /dev/null
+++ b/docs/README.md
@@ -0,0 +1,204 @@
+
+- [Data processing for image-based profiling](#data-processing-for-image-based-profiling)
+ - [Installation](##installation)
+ - [Frameworks](#frameworks)
+ - [API](#api)
+ - [Usage](#usage)
+ - [Pipeline orchestration](#pipeline-orchestration)
+ - [Other functionality](#other-functionality)
+ - [CellProfiler CSV collation](#cellprofiler-csv-collation)
+ - [Creating a cell locations lookup table](#creating-a-cell-locations-lookup-table)
+ - [Generating a GCT file for morpheus](#generating-a-gct-file-for-morpheus)
+ - [Citing pycytominer](#citing-pycytominer)
+
# Data processing for image-based profiling
[](https://github.com/cytomining/pycytominer/actions/workflows/integration-test.yml?query=branch%3Amain)
@@ -111,19 +123,7 @@ And, more specifically than that, image-based profiling readouts from [CellProfi
Therefore, we have included some custom tools in `pycytominer/cyto_utils` that provides other functionality:
-- [Data processing for image-based profiling](#data-processing-for-image-based-profiling)
- - [Installation](#installation)
- - [Frameworks](#frameworks)
- - [API](#api)
- - [Usage](#usage)
- - [Pipeline orchestration](#pipeline-orchestration)
- - [Other functionality](#other-functionality)
- - [CellProfiler CSV collation](#cellprofiler-csv-collation)
- - [Creating a cell locations lookup table](#creating-a-cell-locations-lookup-table)
- - [Generating a GCT file for morpheus](#generating-a-gct-file-for-morpheus)
- - [Citing pycytominer](#citing-pycytominer)
-
-Note, [`pycytominer.cyto_utils.cells.SingleCells()`](pycytominer/cyto_utils/cells.py) contains code to interact with single-cell SQLite files, which are output from CellProfiler.
+Note, [`pycytominer.cyto_utils.cells.SingleCells()`](./pycytominer/cyto_utils/cells.py) contains code to interact with single-cell SQLite files, which are output from CellProfiler.
Processing capabilities for SQLite files depends on SQLite file size and your available computational resources (for ex. memory and cores).
### CellProfiler CSV collation
diff --git a/docs/README.md b/docs/README.md
new file mode 100644
index 00000000..7f5067ed
--- /dev/null
+++ b/docs/README.md
@@ -0,0 +1,204 @@
+ +
+# Data processing for image-based profiling
+
+[](https://github.com/cytomining/pycytominer/actions/workflows/integration-test.yml?query=branch%3Amain)
+[](https://codecov.io/github/cytomining/pycytominer?branch=main)
+[](https://github.com/astral-sh/ruff)
+[](https://pycytominer.readthedocs.io/)
+[](https://doi.org/10.48550/arXiv.2311.13417)
+
+Pycytominer is a suite of common functions used to process high dimensional readouts from high-throughput cell experiments.
+The tool is most often used for processing data through the following pipeline:
+
+
+
+# Data processing for image-based profiling
+
+[](https://github.com/cytomining/pycytominer/actions/workflows/integration-test.yml?query=branch%3Amain)
+[](https://codecov.io/github/cytomining/pycytominer?branch=main)
+[](https://github.com/astral-sh/ruff)
+[](https://pycytominer.readthedocs.io/)
+[](https://doi.org/10.48550/arXiv.2311.13417)
+
+Pycytominer is a suite of common functions used to process high dimensional readouts from high-throughput cell experiments.
+The tool is most often used for processing data through the following pipeline:
+
+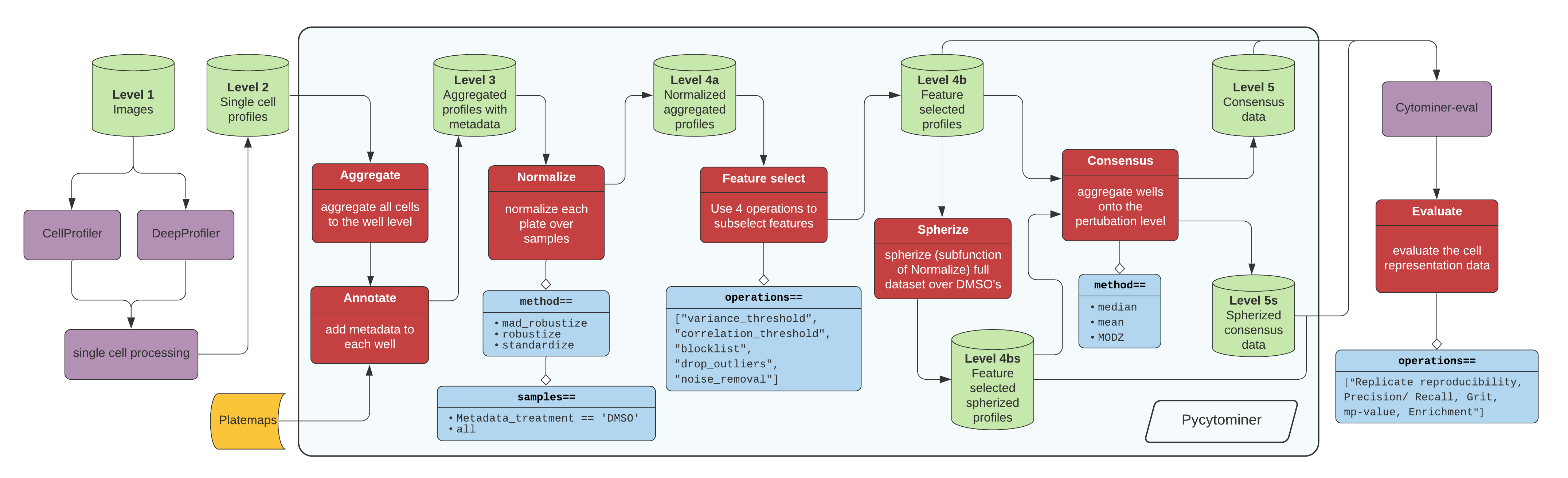 +
+[Click here for high resolution pipeline image](https://github.com/cytomining/pycytominer/blob/main/media/pipeline.png)
+
+Image data flow from a microscope to cell segmentation and feature extraction tools (e.g. CellProfiler or DeepProfiler).
+From here, additional single cell processing tools curate the single cell readouts into a form manageable for pycytominer input.
+For CellProfiler, we use [cytominer-database](https://github.com/cytomining/cytominer-database) or [CytoTable](https://github.com/cytomining/CytoTable).
+For DeepProfiler, we include single cell processing tools in [pycytominer.cyto_utils](cyto_utils.md).
+
+From the single cell output, pycytominer performs five steps using a simple API (described below), before passing along data to [cytominer-eval](https://github.com/cytomining/cytominer-eval) for quality and perturbation strength evaluation.
+
+## Installation
+
+You can install pycytominer via pip:
+
+```bash
+pip install pycytominer
+```
+
+or conda:
+
+```bash
+conda install -c conda-forge pycytominer
+```
+
+## Frameworks
+
+Pycytominer is primarily built on top of [pandas](https://pandas.pydata.org/docs/index.html), also using aspects of SQLAlchemy, sklearn, and pyarrow.
+
+Pycytominer currently supports [parquet](https://parquet.apache.org/) and compressed text file (e.g. `.csv.gz`) i/o.
+
+## API
+
+Pycytominer has five major processing functions:
+
+1. Aggregate - Average single-cell profiles based on metadata information (most often "well").
+2. Annotate - Append metadata (most often from the platemap file) to the feature profile
+3. Normalize - Transform input feature data into consistent distributions
+4. Feature select - Exclude non-informative or redundant features
+5. Consensus - Average aggregated profiles by replicates to form a "consensus signature"
+
+The API is consistent for each of these functions:
+
+```python
+# Each function takes as input a pandas DataFrame or file path
+# and transforms the input data based on the provided options and methods
+df = function(
+ profiles_or_path,
+ features,
+ samples,
+ method,
+ output_file,
+ additional_options...
+)
+```
+
+Each processing function has unique arguments, see our [documentation](https://pycytominer.readthedocs.io/) for more details.
+
+## Usage
+
+The default way to use pycytominer is within python scripts, and using pycytominer is simple and fun.
+
+```python
+# Real world example
+import pandas as pd
+import pycytominer
+
+commit = "da8ae6a3bc103346095d61b4ee02f08fc85a5d98"
+url = f"https://media.githubusercontent.com/media/broadinstitute/lincs-cell-painting/{commit}/profiles/2016_04_01_a549_48hr_batch1/SQ00014812/SQ00014812_augmented.csv.gz"
+
+df = pd.read_csv(url)
+
+normalized_df = pycytominer.normalize(
+ profiles=df,
+ method="standardize",
+ samples="Metadata_broad_sample == 'DMSO'"
+)
+```
+
+### Pipeline orchestration
+
+Pycytominer is a collection of different functions with no explicit link between steps.
+However, some options exist to use pycytominer within a pipeline framework.
+
+| Project | Format | Environment | pycytominer usage |
+| :------------------------------------------------------------------------------- | :-------- | :------------------- | :---------------------- |
+| [Profiling-recipe](https://github.com/cytomining/profiling-recipe) | yaml | agnostic | full pipeline support |
+| [CellProfiler-on-Terra](https://github.com/broadinstitute/cellprofiler-on-Terra) | WDL | google cloud / Terra | single-cell aggregation |
+| [CytoSnake](https://github.com/WayScience/CytoSnake) | snakemake | agnostic | full pipeline support |
+
+A separate project called [AuSPICES](https://github.com/broadinstitute/AuSPICEs) offers pipeline support up to image feature extraction.
+
+## Other functionality
+
+Pycytominer was written with a goal of processing any high-throughput image-based profiling data.
+However, the initial use case was developed for processing image-based profiling experiments specifically.
+And, more specifically than that, image-based profiling readouts from [CellProfiler](https://github.com/CellProfiler) measurements from [Cell Painting](https://www.nature.com/articles/nprot.2016.105) data.
+
+Therefore, we have included some custom tools in `pycytominer/cyto_utils` that provides other functionality:
+
+Note, [`pycytominer.cyto_utils.cells.SingleCells()`](cyto_utils.md##pycytominer.cyto_utils.cells) contains code to interact with single-cell SQLite files, which are output from CellProfiler.
+Processing capabilities for SQLite files depends on SQLite file size and your available computational resources (for ex. memory and cores).
+
+### CellProfiler CSV collation
+
+If running your images on a cluster, unless you have a MySQL or similar large database set up then you will likely end up with lots of different folders from the different cluster runs (often one per well or one per site), each one containing an `Image.csv`, `Nuclei.csv`, etc.
+In order to look at full plates, therefore, we first need to collate all of these CSVs into a single file (currently SQLite) per plate.
+We currently do this with a library called [cytominer-database](https://github.com/cytomining/cytominer-database).
+
+If you want to perform this data collation inside pycytominer using the `cyto_utils` function `collate` (and/or you want to be able to run the tests and have them all pass!), you will need `cytominer-database==0.3.4`; this will change your installation commands slightly:
+
+```bash
+# Example for general case commit:
+pip install "pycytominer[collate]"
+
+# Example for specific commit:
+pip install "pycytominer[collate] @ git+https://github.com/cytomining/pycytominer@77d93a3a551a438799a97ba57d49b19de0a293ab"
+```
+
+If using `pycytominer` in a conda environment, in order to run `collate.py`, you will also want to make sure to add `cytominer-database=0.3.4` to your list of dependencies.
+
+### Creating a cell locations lookup table
+
+The `CellLocation` class offers a convenient way to augment a [LoadData](https://cellprofiler-manual.s3.amazonaws.com/CPmanual/LoadData.html) file with X,Y locations of cells in each image.
+The locations information is obtained from a single cell SQLite file.
+
+To use this functionality, you will need to modify your installation command, similar to above:
+
+```bash
+# Example for general case commit:
+pip install "pycytominer[cell_locations]"
+```
+
+Example using this functionality:
+
+```bash
+metadata_input="s3://cellpainting-gallery/test-cpg0016-jump/source_4/workspace/load_data_csv/2021_08_23_Batch12/BR00126114/test_BR00126114_load_data_with_illum.parquet"
+single_single_cell_input="s3://cellpainting-gallery/test-cpg0016-jump/source_4/workspace/backend/2021_08_23_Batch12/BR00126114/test_BR00126114.sqlite"
+augmented_metadata_output="~/Desktop/load_data_with_illum_and_cell_location_subset.parquet"
+
+python \
+ -m pycytominer.cyto_utils.cell_locations_cmd \
+ --metadata_input ${metadata_input} \
+ --single_cell_input ${single_single_cell_input} \
+ --augmented_metadata_output ${augmented_metadata_output} \
+ add_cell_location
+
+# Check the output
+
+python -c "import pandas as pd; print(pd.read_parquet('${augmented_metadata_output}').head())"
+
+# It should look something like this (depends on the width of your terminal):
+
+# Metadata_Plate Metadata_Well Metadata_Site ... PathName_OrigRNA ImageNumber CellCenters
+# 0 BR00126114 A01 1 ... s3://cellpainting-gallery/cpg0016-jump/source_... 1 [{'Nuclei_Location_Center_X': 943.512129380054...
+# 1 BR00126114 A01 2 ... s3://cellpainting-gallery/cpg0016-jump/source_... 2 [{'Nuclei_Location_Center_X': 29.9516027655562...
+```
+
+### Generating a GCT file for morpheus
+
+The software [morpheus](https://software.broadinstitute.org/morpheus/) enables profile visualization in the form of interactive heatmaps.
+Pycytominer can convert profiles into a `.gct` file for drag-and-drop input into morpheus.
+
+```python
+# Real world example
+import pandas as pd
+import pycytominer
+
+commit = "da8ae6a3bc103346095d61b4ee02f08fc85a5d98"
+plate = "SQ00014812"
+url = f"https://media.githubusercontent.com/media/broadinstitute/lincs-cell-painting/{commit}/profiles/2016_04_01_a549_48hr_batch1/{plate}/{plate}_normalized_feature_select.csv.gz"
+
+df = pd.read_csv(url)
+output_file = f"{plate}.gct"
+
+pycytominer.cyto_utils.write_gct(
+ profiles=df,
+ output_file=output_file
+)
+```
+
+## Citing pycytominer
+
+If you have used `pycytominer` in your project, please use the citation below.
+You can also find the citation in the 'cite this repository' link at the top right under `about` section.
+
+APA:
+
+```text
+Serrano, E., Chandrasekaran, N., Bunten, D., Brewer, K., Tomkinson, J., Kern, R., Bornholdt, M., Fleming, S., Pei, R., Arevalo, J., Tsang, H., Rubinetti, V., Tromans-Coia, C., Becker, T., Weisbart, E., Bunne, C., Kalinin, A. A., Senft, R., Taylor, S. J., Jamali, N., Adeboye, A., Abbasi, H. S., Goodman, A., Caicedo, J., Carpenter, A. E., Cimini, B. A., Singh, S., & Way, G. P. Reproducible image-based profiling with Pycytominer. https://doi.org/10.48550/arXiv.2311.13417
+```
diff --git a/docs/conf.py b/docs/conf.py
index be8581a2..af5e0eec 100644
--- a/docs/conf.py
+++ b/docs/conf.py
@@ -1,4 +1,4 @@
-# Configuration file for the Sphinx documentation builder.
+"""Configuration file for the Sphinx documentation builder."""
#
# This file only contains a selection of the most common options. For a full
# list see the documentation:
diff --git a/docs/cyto_utils.md b/docs/cyto_utils.md
new file mode 100644
index 00000000..0251bb86
--- /dev/null
+++ b/docs/cyto_utils.md
@@ -0,0 +1,5 @@
+# Cyto utilities
+
+Functions enabling smooth interaction with CellProfiler and DeepProfiler output formats.
+
+::: pycytominer.cyto_utils
diff --git a/docs/functions.md b/docs/functions.md
new file mode 100644
index 00000000..ef4a65ab
--- /dev/null
+++ b/docs/functions.md
@@ -0,0 +1,16 @@
+# Main Functions
+
+
+
+
+::: pycytominer
+ options:
+ members:
+ - aggregate
+ - annotate
+ - consensus
+ - feature_select
+ - normalize
+
+
+
diff --git a/docs/index.rst b/docs/index.rst
deleted file mode 100644
index 460d88b8..00000000
--- a/docs/index.rst
+++ /dev/null
@@ -1,22 +0,0 @@
-.. pycytominer documentation main file, created by
- sphinx-quickstart on Fri Feb 19 14:44:29 2021.
- You can adapt this file completely to your liking, but it should at least
- contain the root `toctree` directive.
-
-.. mdinclude:: ../README.md
-
-.. toctree::
- :maxdepth: 2
- :caption: Contents:
-
- install
- walkthrough
- tutorial
- modules
-
-Indices and tables
-==================
-
-* :ref:`genindex`
-* :ref:`modindex`
-* :ref:`search`
diff --git a/docs/install.rst b/docs/install.md
similarity index 71%
rename from docs/install.rst
rename to docs/install.md
index 99213db9..9f2963ab 100644
--- a/docs/install.rst
+++ b/docs/install.md
@@ -1,14 +1,13 @@
-Install
-=======
+# Installation
To install pycytominer, use pip:
-.. code-block:: bash
-
+```bash
pip install pycytominer
+```
You can also install pycytominer with conda:
-.. code-block:: bash
-
+```bash
conda install -c conda-forge pycytominer
+```
diff --git a/docs/modules.rst b/docs/modules.rst
deleted file mode 100644
index bec89387..00000000
--- a/docs/modules.rst
+++ /dev/null
@@ -1,7 +0,0 @@
-pycytominer
-===========
-
-.. toctree::
- :maxdepth: 4
-
- pycytominer
diff --git a/docs/operations.md b/docs/operations.md
new file mode 100644
index 00000000..ba31c6ed
--- /dev/null
+++ b/docs/operations.md
@@ -0,0 +1,6 @@
+# Operations
+
+We do not recommend interacting with these functions directly.
+The core pycytominer API uses these operations internally.
+
+::: pycytominer.operations
diff --git a/docs/pycytominer.cyto_utils.rst b/docs/pycytominer.cyto_utils.rst
deleted file mode 100644
index 8618d60d..00000000
--- a/docs/pycytominer.cyto_utils.rst
+++ /dev/null
@@ -1,76 +0,0 @@
-Cyto utilities
-==============
-
-Functions enabling smooth interaction with CellProfiler and DeepProfiler output formats.
-
-pycytominer.cyto\_utils.cells module
-------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.cells
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.collate module
---------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.collate
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.features module
----------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.features
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.load module
------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.load
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.modz module
------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.modz
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.output module
--------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.output
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.util module
------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.util
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.write\_gct module
------------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.write_gct
- :members:
- :undoc-members:
- :show-inheritance:
-
-Module contents
----------------
-
-.. automodule:: pycytominer.cyto_utils
- :members:
- :undoc-members:
- :show-inheritance:
diff --git a/docs/pycytominer.operations.rst b/docs/pycytominer.operations.rst
deleted file mode 100644
index 062919d6..00000000
--- a/docs/pycytominer.operations.rst
+++ /dev/null
@@ -1,45 +0,0 @@
-Operations
-==========
-
-We do not recommend interacting with these functions directly.
-The core pycytominer API uses these operations internally.
-
-pycytominer.operations.correlation\_threshold module
-----------------------------------------------------
-
-.. automodule:: pycytominer.operations.correlation_threshold
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.operations.get\_na\_columns module
-----------------------------------------------
-
-.. automodule:: pycytominer.operations.get_na_columns
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.operations.transform module
----------------------------------------
-
-.. automodule:: pycytominer.operations.transform
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.operations.variance\_threshold module
--------------------------------------------------
-
-.. automodule:: pycytominer.operations.variance_threshold
- :members:
- :undoc-members:
- :show-inheritance:
-
-Module contents
----------------
-
-.. automodule:: pycytominer.operations
- :members:
- :undoc-members:
- :show-inheritance:
diff --git a/docs/pycytominer.rst b/docs/pycytominer.rst
deleted file mode 100644
index 01b61229..00000000
--- a/docs/pycytominer.rst
+++ /dev/null
@@ -1,59 +0,0 @@
-Core functions
-==============
-
-pycytominer.aggregate module
-----------------------------
-
-.. automodule:: pycytominer.aggregate
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.annotate module
----------------------------
-
-.. automodule:: pycytominer.annotate
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.normalize module
-----------------------------
-
-.. automodule:: pycytominer.normalize
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.feature\_select module
-----------------------------------
-
-.. automodule:: pycytominer.feature_select
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.consensus module
-----------------------------
-
-.. automodule:: pycytominer.consensus
- :members:
- :undoc-members:
- :show-inheritance:
-
-Helper functions
-----------------
-
-.. toctree::
- :maxdepth: 4
-
- pycytominer.cyto_utils
- pycytominer.operations
-
-Module contents
----------------
-
-.. automodule:: pycytominer
- :members:
- :undoc-members:
- :show-inheritance:
diff --git a/docs/tutorial.md b/docs/tutorial.md
new file mode 100644
index 00000000..492e0008
--- /dev/null
+++ b/docs/tutorial.md
@@ -0,0 +1,13 @@
+# Tutorials
+
+This [tutorial](https://github.com/cytomining/pipeline-examples#readme) shows how to run a image-based profiling pipeline using pycytominer. Using IPython notebooks, it walks through the following steps:
+
+- Downloading a dataset of single cell [CellProfiler](https://cellprofiler.org/) profiles.
+- Processing the profiles using PyCytominer. This includes the following steps:
+- Data initialization
+- Single cell aggregation to create well-level profiles
+- Addition of experiment metadata to the well-level profiles
+- Profile normalization
+- Feature selection
+- Forming consensus signatures
+- Evaluating the profile quality using [cytominer-eval](https://github.com/cytomining/cytominer-eval>).
diff --git a/docs/tutorial.rst b/docs/tutorial.rst
deleted file mode 100644
index da5d5f6f..00000000
--- a/docs/tutorial.rst
+++ /dev/null
@@ -1,14 +0,0 @@
-Tutorials
-=========
-
-`This `_ tutorial shows how to run a image-based profiling pipeline using pycytominer. Using IPython notebooks, it walks through the following steps:
-
-#. Downloading a dataset of single cell `CellProfiler `_ profiles.
-#. Processing the profiles using PyCytominer. This includes the following steps:
- #. Data initialization
- #. Single cell aggregation to create well-level profiles
- #. Addition of experiment metadata to the well-level profiles
- #. Profile normalization
- #. Feature selection
- #. Forming consensus signatures
-#. Evaluating the profile quality using `cytominer-eval `_.
\ No newline at end of file
diff --git a/docs/walkthrough.rst b/docs/walkthrough.rst
deleted file mode 100644
index 6733ed2a..00000000
--- a/docs/walkthrough.rst
+++ /dev/null
@@ -1,7 +0,0 @@
-Walkthroughs
-============
-
-.. toctree::
- :maxdepth: 1
-
- walkthroughs/single_cell_usage.ipynb
\ No newline at end of file
diff --git a/mkdocs.yml b/mkdocs.yml
new file mode 100644
index 00000000..0c264885
--- /dev/null
+++ b/mkdocs.yml
@@ -0,0 +1,67 @@
+site_name: pycytominer
+repo_url: https://github.com/cytomining/pycytominer
+site_url: https://cytomining.github.io/pycytominer
+site_description: Python package for processing image-based profiling data
+site_author: Pycytominer contributors
+edit_uri: edit/main/docs/
+repo_name: cytomining/pycytominer
+
+nav:
+ - Home: README.md
+ - Installation: install.md
+ - Main Functions: functions.md
+ - Cyto Utilities: cyto_utils.md
+ - Operations: operations.md
+ - Tutorial: tutorial.md
+ - Walkthroughs:
+ - Single Cell Usage: walkthroughs/single_cell_usage.ipynb
+plugins:
+ - search
+ - mkdocs-jupyter
+ - mkdocstrings:
+ handlers:
+ python:
+ paths:
+ - pycytominer
+ options:
+ show_source: true
+ show_submodules: true
+ show_symbol_type_heading: true
+ show_symbol_type_toc: true
+ docstring_style: numpy
+ show_root_members_full_path: true
+
+theme:
+ name: material
+ feature:
+ tabs: true
+ palette:
+ - media: "(prefers-color-scheme: light)"
+ scheme: default
+ primary: deep purple
+ accent: purple
+ toggle:
+ icon: material/brightness-7
+ name: Switch to dark mode
+ - media: "(prefers-color-scheme: dark)"
+ scheme: slate
+ primary: deep purple
+ accent: purple
+ toggle:
+ icon: material/brightness-4
+ name: Switch to light mode
+ icon:
+ repo: fontawesome/brands/github
+
+extra:
+ social:
+ - icon: fontawesome/brands/github
+ link: https://github.com/cytomining/pycytominer
+ - icon: fontawesome/brands/python
+ link: https://pypi.org/project/pycytominer
+
+markdown_extensions:
+ - toc:
+ permalink: true
+ - pymdownx.arithmatex:
+ generic: true
diff --git a/poetry.lock b/poetry.lock
index 74461f43..2048fed6 100644
--- a/poetry.lock
+++ b/poetry.lock
@@ -81,108 +81,108 @@ test = ["crate", "geojson", "pytest", "pytest-cov"]
[[package]]
name = "aiobotocore"
-version = "2.12.1"
+version = "2.12.3"
description = "Async client for aws services using botocore and aiohttp"
optional = true
python-versions = ">=3.8"
files = [
- {file = "aiobotocore-2.12.1-py3-none-any.whl", hash = "sha256:6a9a3d646cf422f45fdc1e4256e78563ebffba64733bc9b8ca9123614e8ba9af"},
- {file = "aiobotocore-2.12.1.tar.gz", hash = "sha256:8706b28f16f93c541f6ed50352115a79d8f3499539f8d0bb70aa0f7a5379c1fe"},
+ {file = "aiobotocore-2.12.3-py3-none-any.whl", hash = "sha256:86737685f4625e8f05c4e7a608a07cc97607263279f66cf6b02b640c4eafd324"},
+ {file = "aiobotocore-2.12.3.tar.gz", hash = "sha256:e2a2929207bc5d62eb556106c2224c1fd106d5c65be2eb69f15cc8c34c44c236"},
]
[package.dependencies]
aiohttp = ">=3.7.4.post0,<4.0.0"
aioitertools = ">=0.5.1,<1.0.0"
-botocore = ">=1.34.41,<1.34.52"
+botocore = ">=1.34.41,<1.34.70"
wrapt = ">=1.10.10,<2.0.0"
[package.extras]
-awscli = ["awscli (>=1.32.41,<1.32.52)"]
-boto3 = ["boto3 (>=1.34.41,<1.34.52)"]
+awscli = ["awscli (>=1.32.41,<1.32.70)"]
+boto3 = ["boto3 (>=1.34.41,<1.34.70)"]
[[package]]
name = "aiohttp"
-version = "3.9.4"
+version = "3.9.5"
description = "Async http client/server framework (asyncio)"
-optional = false
+optional = true
python-versions = ">=3.8"
files = [
- {file = "aiohttp-3.9.4-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:76d32588ef7e4a3f3adff1956a0ba96faabbdee58f2407c122dd45aa6e34f372"},
- {file = "aiohttp-3.9.4-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:56181093c10dbc6ceb8a29dfeea1e815e1dfdc020169203d87fd8d37616f73f9"},
- {file = "aiohttp-3.9.4-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:c7a5b676d3c65e88b3aca41816bf72831898fcd73f0cbb2680e9d88e819d1e4d"},
- {file = "aiohttp-3.9.4-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d1df528a85fb404899d4207a8d9934cfd6be626e30e5d3a5544a83dbae6d8a7e"},
- {file = "aiohttp-3.9.4-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:f595db1bceabd71c82e92df212dd9525a8a2c6947d39e3c994c4f27d2fe15b11"},
- {file = "aiohttp-3.9.4-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:9c0b09d76e5a4caac3d27752027fbd43dc987b95f3748fad2b924a03fe8632ad"},
- {file = "aiohttp-3.9.4-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:689eb4356649ec9535b3686200b231876fb4cab4aca54e3bece71d37f50c1d13"},
- {file = "aiohttp-3.9.4-cp310-cp310-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:a3666cf4182efdb44d73602379a66f5fdfd5da0db5e4520f0ac0dcca644a3497"},
- {file = "aiohttp-3.9.4-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:b65b0f8747b013570eea2f75726046fa54fa8e0c5db60f3b98dd5d161052004a"},
- {file = "aiohttp-3.9.4-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:a1885d2470955f70dfdd33a02e1749613c5a9c5ab855f6db38e0b9389453dce7"},
- {file = "aiohttp-3.9.4-cp310-cp310-musllinux_1_1_ppc64le.whl", hash = "sha256:0593822dcdb9483d41f12041ff7c90d4d1033ec0e880bcfaf102919b715f47f1"},
- {file = "aiohttp-3.9.4-cp310-cp310-musllinux_1_1_s390x.whl", hash = "sha256:47f6eb74e1ecb5e19a78f4a4228aa24df7fbab3b62d4a625d3f41194a08bd54f"},
- {file = "aiohttp-3.9.4-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:c8b04a3dbd54de6ccb7604242fe3ad67f2f3ca558f2d33fe19d4b08d90701a89"},
- {file = "aiohttp-3.9.4-cp310-cp310-win32.whl", hash = "sha256:8a78dfb198a328bfb38e4308ca8167028920fb747ddcf086ce706fbdd23b2926"},
- {file = "aiohttp-3.9.4-cp310-cp310-win_amd64.whl", hash = "sha256:e78da6b55275987cbc89141a1d8e75f5070e577c482dd48bd9123a76a96f0bbb"},
- {file = "aiohttp-3.9.4-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:c111b3c69060d2bafc446917534150fd049e7aedd6cbf21ba526a5a97b4402a5"},
- {file = "aiohttp-3.9.4-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:efbdd51872cf170093998c87ccdf3cb5993add3559341a8e5708bcb311934c94"},
- {file = "aiohttp-3.9.4-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:7bfdb41dc6e85d8535b00d73947548a748e9534e8e4fddd2638109ff3fb081df"},
- {file = "aiohttp-3.9.4-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:2bd9d334412961125e9f68d5b73c1d0ab9ea3f74a58a475e6b119f5293eee7ba"},
- {file = "aiohttp-3.9.4-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:35d78076736f4a668d57ade00c65d30a8ce28719d8a42471b2a06ccd1a2e3063"},
- {file = "aiohttp-3.9.4-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:824dff4f9f4d0f59d0fa3577932ee9a20e09edec8a2f813e1d6b9f89ced8293f"},
- {file = "aiohttp-3.9.4-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:52b8b4e06fc15519019e128abedaeb56412b106ab88b3c452188ca47a25c4093"},
- {file = "aiohttp-3.9.4-cp311-cp311-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:eae569fb1e7559d4f3919965617bb39f9e753967fae55ce13454bec2d1c54f09"},
- {file = "aiohttp-3.9.4-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:69b97aa5792428f321f72aeb2f118e56893371f27e0b7d05750bcad06fc42ca1"},
- {file = "aiohttp-3.9.4-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:4d79aad0ad4b980663316f26d9a492e8fab2af77c69c0f33780a56843ad2f89e"},
- {file = "aiohttp-3.9.4-cp311-cp311-musllinux_1_1_ppc64le.whl", hash = "sha256:d6577140cd7db19e430661e4b2653680194ea8c22c994bc65b7a19d8ec834403"},
- {file = "aiohttp-3.9.4-cp311-cp311-musllinux_1_1_s390x.whl", hash = "sha256:9860d455847cd98eb67897f5957b7cd69fbcb436dd3f06099230f16a66e66f79"},
- {file = "aiohttp-3.9.4-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:69ff36d3f8f5652994e08bd22f093e11cfd0444cea310f92e01b45a4e46b624e"},
- {file = "aiohttp-3.9.4-cp311-cp311-win32.whl", hash = "sha256:e27d3b5ed2c2013bce66ad67ee57cbf614288bda8cdf426c8d8fe548316f1b5f"},
- {file = "aiohttp-3.9.4-cp311-cp311-win_amd64.whl", hash = "sha256:d6a67e26daa686a6fbdb600a9af8619c80a332556245fa8e86c747d226ab1a1e"},
- {file = "aiohttp-3.9.4-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:c5ff8ff44825736a4065d8544b43b43ee4c6dd1530f3a08e6c0578a813b0aa35"},
- {file = "aiohttp-3.9.4-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:d12a244627eba4e9dc52cbf924edef905ddd6cafc6513849b4876076a6f38b0e"},
- {file = "aiohttp-3.9.4-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:dcad56c8d8348e7e468899d2fb3b309b9bc59d94e6db08710555f7436156097f"},
- {file = "aiohttp-3.9.4-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:4f7e69a7fd4b5ce419238388e55abd220336bd32212c673ceabc57ccf3d05b55"},
- {file = "aiohttp-3.9.4-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:c4870cb049f10d7680c239b55428916d84158798eb8f353e74fa2c98980dcc0b"},
- {file = "aiohttp-3.9.4-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:3b2feaf1b7031ede1bc0880cec4b0776fd347259a723d625357bb4b82f62687b"},
- {file = "aiohttp-3.9.4-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:939393e8c3f0a5bcd33ef7ace67680c318dc2ae406f15e381c0054dd658397de"},
- {file = "aiohttp-3.9.4-cp312-cp312-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:7d2334e387b2adcc944680bebcf412743f2caf4eeebd550f67249c1c3696be04"},
- {file = "aiohttp-3.9.4-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:e0198ea897680e480845ec0ffc5a14e8b694e25b3f104f63676d55bf76a82f1a"},
- {file = "aiohttp-3.9.4-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:e40d2cd22914d67c84824045861a5bb0fb46586b15dfe4f046c7495bf08306b2"},
- {file = "aiohttp-3.9.4-cp312-cp312-musllinux_1_1_ppc64le.whl", hash = "sha256:aba80e77c227f4234aa34a5ff2b6ff30c5d6a827a91d22ff6b999de9175d71bd"},
- {file = "aiohttp-3.9.4-cp312-cp312-musllinux_1_1_s390x.whl", hash = "sha256:fb68dc73bc8ac322d2e392a59a9e396c4f35cb6fdbdd749e139d1d6c985f2527"},
- {file = "aiohttp-3.9.4-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:f3460a92638dce7e47062cf088d6e7663adb135e936cb117be88d5e6c48c9d53"},
- {file = "aiohttp-3.9.4-cp312-cp312-win32.whl", hash = "sha256:32dc814ddbb254f6170bca198fe307920f6c1308a5492f049f7f63554b88ef36"},
- {file = "aiohttp-3.9.4-cp312-cp312-win_amd64.whl", hash = "sha256:63f41a909d182d2b78fe3abef557fcc14da50c7852f70ae3be60e83ff64edba5"},
- {file = "aiohttp-3.9.4-cp38-cp38-macosx_10_9_universal2.whl", hash = "sha256:c3770365675f6be220032f6609a8fbad994d6dcf3ef7dbcf295c7ee70884c9af"},
- {file = "aiohttp-3.9.4-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:305edae1dea368ce09bcb858cf5a63a064f3bff4767dec6fa60a0cc0e805a1d3"},
- {file = "aiohttp-3.9.4-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:6f121900131d116e4a93b55ab0d12ad72573f967b100e49086e496a9b24523ea"},
- {file = "aiohttp-3.9.4-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:b71e614c1ae35c3d62a293b19eface83d5e4d194e3eb2fabb10059d33e6e8cbf"},
- {file = "aiohttp-3.9.4-cp38-cp38-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:419f009fa4cfde4d16a7fc070d64f36d70a8d35a90d71aa27670bba2be4fd039"},
- {file = "aiohttp-3.9.4-cp38-cp38-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:7b39476ee69cfe64061fd77a73bf692c40021f8547cda617a3466530ef63f947"},
- {file = "aiohttp-3.9.4-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b33f34c9c7decdb2ab99c74be6443942b730b56d9c5ee48fb7df2c86492f293c"},
- {file = "aiohttp-3.9.4-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:c78700130ce2dcebb1a8103202ae795be2fa8c9351d0dd22338fe3dac74847d9"},
- {file = "aiohttp-3.9.4-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:268ba22d917655d1259af2d5659072b7dc11b4e1dc2cb9662fdd867d75afc6a4"},
- {file = "aiohttp-3.9.4-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:17e7c051f53a0d2ebf33013a9cbf020bb4e098c4bc5bce6f7b0c962108d97eab"},
- {file = "aiohttp-3.9.4-cp38-cp38-musllinux_1_1_ppc64le.whl", hash = "sha256:7be99f4abb008cb38e144f85f515598f4c2c8932bf11b65add0ff59c9c876d99"},
- {file = "aiohttp-3.9.4-cp38-cp38-musllinux_1_1_s390x.whl", hash = "sha256:d58a54d6ff08d2547656356eea8572b224e6f9bbc0cf55fa9966bcaac4ddfb10"},
- {file = "aiohttp-3.9.4-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:7673a76772bda15d0d10d1aa881b7911d0580c980dbd16e59d7ba1422b2d83cd"},
- {file = "aiohttp-3.9.4-cp38-cp38-win32.whl", hash = "sha256:e4370dda04dc8951012f30e1ce7956a0a226ac0714a7b6c389fb2f43f22a250e"},
- {file = "aiohttp-3.9.4-cp38-cp38-win_amd64.whl", hash = "sha256:eb30c4510a691bb87081192a394fb661860e75ca3896c01c6d186febe7c88530"},
- {file = "aiohttp-3.9.4-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:84e90494db7df3be5e056f91412f9fa9e611fbe8ce4aaef70647297f5943b276"},
- {file = "aiohttp-3.9.4-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:7d4845f8501ab28ebfdbeab980a50a273b415cf69e96e4e674d43d86a464df9d"},
- {file = "aiohttp-3.9.4-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:69046cd9a2a17245c4ce3c1f1a4ff8c70c7701ef222fce3d1d8435f09042bba1"},
- {file = "aiohttp-3.9.4-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:8b73a06bafc8dcc508420db43b4dd5850e41e69de99009d0351c4f3007960019"},
- {file = "aiohttp-3.9.4-cp39-cp39-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:418bb0038dfafeac923823c2e63226179976c76f981a2aaad0ad5d51f2229bca"},
- {file = "aiohttp-3.9.4-cp39-cp39-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:71a8f241456b6c2668374d5d28398f8e8cdae4cce568aaea54e0f39359cd928d"},
- {file = "aiohttp-3.9.4-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:935c369bf8acc2dc26f6eeb5222768aa7c62917c3554f7215f2ead7386b33748"},
- {file = "aiohttp-3.9.4-cp39-cp39-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:74e4e48c8752d14ecfb36d2ebb3d76d614320570e14de0a3aa7a726ff150a03c"},
- {file = "aiohttp-3.9.4-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:916b0417aeddf2c8c61291238ce25286f391a6acb6f28005dd9ce282bd6311b6"},
- {file = "aiohttp-3.9.4-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:9b6787b6d0b3518b2ee4cbeadd24a507756ee703adbac1ab6dc7c4434b8c572a"},
- {file = "aiohttp-3.9.4-cp39-cp39-musllinux_1_1_ppc64le.whl", hash = "sha256:221204dbda5ef350e8db6287937621cf75e85778b296c9c52260b522231940ed"},
- {file = "aiohttp-3.9.4-cp39-cp39-musllinux_1_1_s390x.whl", hash = "sha256:10afd99b8251022ddf81eaed1d90f5a988e349ee7d779eb429fb07b670751e8c"},
- {file = "aiohttp-3.9.4-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:2506d9f7a9b91033201be9ffe7d89c6a54150b0578803cce5cb84a943d075bc3"},
- {file = "aiohttp-3.9.4-cp39-cp39-win32.whl", hash = "sha256:e571fdd9efd65e86c6af2f332e0e95dad259bfe6beb5d15b3c3eca3a6eb5d87b"},
- {file = "aiohttp-3.9.4-cp39-cp39-win_amd64.whl", hash = "sha256:7d29dd5319d20aa3b7749719ac9685fbd926f71ac8c77b2477272725f882072d"},
- {file = "aiohttp-3.9.4.tar.gz", hash = "sha256:6ff71ede6d9a5a58cfb7b6fffc83ab5d4a63138276c771ac91ceaaddf5459644"},
+ {file = "aiohttp-3.9.5-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:fcde4c397f673fdec23e6b05ebf8d4751314fa7c24f93334bf1f1364c1c69ac7"},
+ {file = "aiohttp-3.9.5-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:5d6b3f1fabe465e819aed2c421a6743d8debbde79b6a8600739300630a01bf2c"},
+ {file = "aiohttp-3.9.5-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:6ae79c1bc12c34082d92bf9422764f799aee4746fd7a392db46b7fd357d4a17a"},
+ {file = "aiohttp-3.9.5-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:4d3ebb9e1316ec74277d19c5f482f98cc65a73ccd5430540d6d11682cd857430"},
+ {file = "aiohttp-3.9.5-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:84dabd95154f43a2ea80deffec9cb44d2e301e38a0c9d331cc4aa0166fe28ae3"},
+ {file = "aiohttp-3.9.5-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:c8a02fbeca6f63cb1f0475c799679057fc9268b77075ab7cf3f1c600e81dd46b"},
+ {file = "aiohttp-3.9.5-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:c26959ca7b75ff768e2776d8055bf9582a6267e24556bb7f7bd29e677932be72"},
+ {file = "aiohttp-3.9.5-cp310-cp310-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:714d4e5231fed4ba2762ed489b4aec07b2b9953cf4ee31e9871caac895a839c0"},
+ {file = "aiohttp-3.9.5-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:e7a6a8354f1b62e15d48e04350f13e726fa08b62c3d7b8401c0a1314f02e3558"},
+ {file = "aiohttp-3.9.5-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:c413016880e03e69d166efb5a1a95d40f83d5a3a648d16486592c49ffb76d0db"},
+ {file = "aiohttp-3.9.5-cp310-cp310-musllinux_1_1_ppc64le.whl", hash = "sha256:ff84aeb864e0fac81f676be9f4685f0527b660f1efdc40dcede3c251ef1e867f"},
+ {file = "aiohttp-3.9.5-cp310-cp310-musllinux_1_1_s390x.whl", hash = "sha256:ad7f2919d7dac062f24d6f5fe95d401597fbb015a25771f85e692d043c9d7832"},
+ {file = "aiohttp-3.9.5-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:702e2c7c187c1a498a4e2b03155d52658fdd6fda882d3d7fbb891a5cf108bb10"},
+ {file = "aiohttp-3.9.5-cp310-cp310-win32.whl", hash = "sha256:67c3119f5ddc7261d47163ed86d760ddf0e625cd6246b4ed852e82159617b5fb"},
+ {file = "aiohttp-3.9.5-cp310-cp310-win_amd64.whl", hash = "sha256:471f0ef53ccedec9995287f02caf0c068732f026455f07db3f01a46e49d76bbb"},
+ {file = "aiohttp-3.9.5-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:e0ae53e33ee7476dd3d1132f932eeb39bf6125083820049d06edcdca4381f342"},
+ {file = "aiohttp-3.9.5-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:c088c4d70d21f8ca5c0b8b5403fe84a7bc8e024161febdd4ef04575ef35d474d"},
+ {file = "aiohttp-3.9.5-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:639d0042b7670222f33b0028de6b4e2fad6451462ce7df2af8aee37dcac55424"},
+ {file = "aiohttp-3.9.5-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:f26383adb94da5e7fb388d441bf09c61e5e35f455a3217bfd790c6b6bc64b2ee"},
+ {file = "aiohttp-3.9.5-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:66331d00fb28dc90aa606d9a54304af76b335ae204d1836f65797d6fe27f1ca2"},
+ {file = "aiohttp-3.9.5-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:4ff550491f5492ab5ed3533e76b8567f4b37bd2995e780a1f46bca2024223233"},
+ {file = "aiohttp-3.9.5-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:f22eb3a6c1080d862befa0a89c380b4dafce29dc6cd56083f630073d102eb595"},
+ {file = "aiohttp-3.9.5-cp311-cp311-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:a81b1143d42b66ffc40a441379387076243ef7b51019204fd3ec36b9f69e77d6"},
+ {file = "aiohttp-3.9.5-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:f64fd07515dad67f24b6ea4a66ae2876c01031de91c93075b8093f07c0a2d93d"},
+ {file = "aiohttp-3.9.5-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:93e22add827447d2e26d67c9ac0161756007f152fdc5210277d00a85f6c92323"},
+ {file = "aiohttp-3.9.5-cp311-cp311-musllinux_1_1_ppc64le.whl", hash = "sha256:55b39c8684a46e56ef8c8d24faf02de4a2b2ac60d26cee93bc595651ff545de9"},
+ {file = "aiohttp-3.9.5-cp311-cp311-musllinux_1_1_s390x.whl", hash = "sha256:4715a9b778f4293b9f8ae7a0a7cef9829f02ff8d6277a39d7f40565c737d3771"},
+ {file = "aiohttp-3.9.5-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:afc52b8d969eff14e069a710057d15ab9ac17cd4b6753042c407dcea0e40bf75"},
+ {file = "aiohttp-3.9.5-cp311-cp311-win32.whl", hash = "sha256:b3df71da99c98534be076196791adca8819761f0bf6e08e07fd7da25127150d6"},
+ {file = "aiohttp-3.9.5-cp311-cp311-win_amd64.whl", hash = "sha256:88e311d98cc0bf45b62fc46c66753a83445f5ab20038bcc1b8a1cc05666f428a"},
+ {file = "aiohttp-3.9.5-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:c7a4b7a6cf5b6eb11e109a9755fd4fda7d57395f8c575e166d363b9fc3ec4678"},
+ {file = "aiohttp-3.9.5-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:0a158704edf0abcac8ac371fbb54044f3270bdbc93e254a82b6c82be1ef08f3c"},
+ {file = "aiohttp-3.9.5-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:d153f652a687a8e95ad367a86a61e8d53d528b0530ef382ec5aaf533140ed00f"},
+ {file = "aiohttp-3.9.5-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:82a6a97d9771cb48ae16979c3a3a9a18b600a8505b1115cfe354dfb2054468b4"},
+ {file = "aiohttp-3.9.5-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:60cdbd56f4cad9f69c35eaac0fbbdf1f77b0ff9456cebd4902f3dd1cf096464c"},
+ {file = "aiohttp-3.9.5-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:8676e8fd73141ded15ea586de0b7cda1542960a7b9ad89b2b06428e97125d4fa"},
+ {file = "aiohttp-3.9.5-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:da00da442a0e31f1c69d26d224e1efd3a1ca5bcbf210978a2ca7426dfcae9f58"},
+ {file = "aiohttp-3.9.5-cp312-cp312-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:18f634d540dd099c262e9f887c8bbacc959847cfe5da7a0e2e1cf3f14dbf2daf"},
+ {file = "aiohttp-3.9.5-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:320e8618eda64e19d11bdb3bd04ccc0a816c17eaecb7e4945d01deee2a22f95f"},
+ {file = "aiohttp-3.9.5-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:2faa61a904b83142747fc6a6d7ad8fccff898c849123030f8e75d5d967fd4a81"},
+ {file = "aiohttp-3.9.5-cp312-cp312-musllinux_1_1_ppc64le.whl", hash = "sha256:8c64a6dc3fe5db7b1b4d2b5cb84c4f677768bdc340611eca673afb7cf416ef5a"},
+ {file = "aiohttp-3.9.5-cp312-cp312-musllinux_1_1_s390x.whl", hash = "sha256:393c7aba2b55559ef7ab791c94b44f7482a07bf7640d17b341b79081f5e5cd1a"},
+ {file = "aiohttp-3.9.5-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:c671dc117c2c21a1ca10c116cfcd6e3e44da7fcde37bf83b2be485ab377b25da"},
+ {file = "aiohttp-3.9.5-cp312-cp312-win32.whl", hash = "sha256:5a7ee16aab26e76add4afc45e8f8206c95d1d75540f1039b84a03c3b3800dd59"},
+ {file = "aiohttp-3.9.5-cp312-cp312-win_amd64.whl", hash = "sha256:5ca51eadbd67045396bc92a4345d1790b7301c14d1848feaac1d6a6c9289e888"},
+ {file = "aiohttp-3.9.5-cp38-cp38-macosx_10_9_universal2.whl", hash = "sha256:694d828b5c41255e54bc2dddb51a9f5150b4eefa9886e38b52605a05d96566e8"},
+ {file = "aiohttp-3.9.5-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:0605cc2c0088fcaae79f01c913a38611ad09ba68ff482402d3410bf59039bfb8"},
+ {file = "aiohttp-3.9.5-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:4558e5012ee03d2638c681e156461d37b7a113fe13970d438d95d10173d25f78"},
+ {file = "aiohttp-3.9.5-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:9dbc053ac75ccc63dc3a3cc547b98c7258ec35a215a92bd9f983e0aac95d3d5b"},
+ {file = "aiohttp-3.9.5-cp38-cp38-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:4109adee842b90671f1b689901b948f347325045c15f46b39797ae1bf17019de"},
+ {file = "aiohttp-3.9.5-cp38-cp38-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:a6ea1a5b409a85477fd8e5ee6ad8f0e40bf2844c270955e09360418cfd09abac"},
+ {file = "aiohttp-3.9.5-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:f3c2890ca8c59ee683fd09adf32321a40fe1cf164e3387799efb2acebf090c11"},
+ {file = "aiohttp-3.9.5-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:3916c8692dbd9d55c523374a3b8213e628424d19116ac4308e434dbf6d95bbdd"},
+ {file = "aiohttp-3.9.5-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:8d1964eb7617907c792ca00b341b5ec3e01ae8c280825deadbbd678447b127e1"},
+ {file = "aiohttp-3.9.5-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:d5ab8e1f6bee051a4bf6195e38a5c13e5e161cb7bad83d8854524798bd9fcd6e"},
+ {file = "aiohttp-3.9.5-cp38-cp38-musllinux_1_1_ppc64le.whl", hash = "sha256:52c27110f3862a1afbcb2af4281fc9fdc40327fa286c4625dfee247c3ba90156"},
+ {file = "aiohttp-3.9.5-cp38-cp38-musllinux_1_1_s390x.whl", hash = "sha256:7f64cbd44443e80094309875d4f9c71d0401e966d191c3d469cde4642bc2e031"},
+ {file = "aiohttp-3.9.5-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:8b4f72fbb66279624bfe83fd5eb6aea0022dad8eec62b71e7bf63ee1caadeafe"},
+ {file = "aiohttp-3.9.5-cp38-cp38-win32.whl", hash = "sha256:6380c039ec52866c06d69b5c7aad5478b24ed11696f0e72f6b807cfb261453da"},
+ {file = "aiohttp-3.9.5-cp38-cp38-win_amd64.whl", hash = "sha256:da22dab31d7180f8c3ac7c7635f3bcd53808f374f6aa333fe0b0b9e14b01f91a"},
+ {file = "aiohttp-3.9.5-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:1732102949ff6087589408d76cd6dea656b93c896b011ecafff418c9661dc4ed"},
+ {file = "aiohttp-3.9.5-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:c6021d296318cb6f9414b48e6a439a7f5d1f665464da507e8ff640848ee2a58a"},
+ {file = "aiohttp-3.9.5-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:239f975589a944eeb1bad26b8b140a59a3a320067fb3cd10b75c3092405a1372"},
+ {file = "aiohttp-3.9.5-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3b7b30258348082826d274504fbc7c849959f1989d86c29bc355107accec6cfb"},
+ {file = "aiohttp-3.9.5-cp39-cp39-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:cd2adf5c87ff6d8b277814a28a535b59e20bfea40a101db6b3bdca7e9926bc24"},
+ {file = "aiohttp-3.9.5-cp39-cp39-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:e9a3d838441bebcf5cf442700e3963f58b5c33f015341f9ea86dcd7d503c07e2"},
+ {file = "aiohttp-3.9.5-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:9e3a1ae66e3d0c17cf65c08968a5ee3180c5a95920ec2731f53343fac9bad106"},
+ {file = "aiohttp-3.9.5-cp39-cp39-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:9c69e77370cce2d6df5d12b4e12bdcca60c47ba13d1cbbc8645dd005a20b738b"},
+ {file = "aiohttp-3.9.5-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:0cbf56238f4bbf49dab8c2dc2e6b1b68502b1e88d335bea59b3f5b9f4c001475"},
+ {file = "aiohttp-3.9.5-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:d1469f228cd9ffddd396d9948b8c9cd8022b6d1bf1e40c6f25b0fb90b4f893ed"},
+ {file = "aiohttp-3.9.5-cp39-cp39-musllinux_1_1_ppc64le.whl", hash = "sha256:45731330e754f5811c314901cebdf19dd776a44b31927fa4b4dbecab9e457b0c"},
+ {file = "aiohttp-3.9.5-cp39-cp39-musllinux_1_1_s390x.whl", hash = "sha256:3fcb4046d2904378e3aeea1df51f697b0467f2aac55d232c87ba162709478c46"},
+ {file = "aiohttp-3.9.5-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:8cf142aa6c1a751fcb364158fd710b8a9be874b81889c2bd13aa8893197455e2"},
+ {file = "aiohttp-3.9.5-cp39-cp39-win32.whl", hash = "sha256:7b179eea70833c8dee51ec42f3b4097bd6370892fa93f510f76762105568cf09"},
+ {file = "aiohttp-3.9.5-cp39-cp39-win_amd64.whl", hash = "sha256:38d80498e2e169bc61418ff36170e0aad0cd268da8b38a17c4cf29d254a8b3f1"},
+ {file = "aiohttp-3.9.5.tar.gz", hash = "sha256:edea7d15772ceeb29db4aff55e482d4bcfb6ae160ce144f2682de02f6d693551"},
]
[package.dependencies]
@@ -214,7 +214,7 @@ typing_extensions = {version = ">=4.0", markers = "python_version < \"3.10\""}
name = "aiosignal"
version = "1.3.1"
description = "aiosignal: a list of registered asynchronous callbacks"
-optional = false
+optional = true
python-versions = ">=3.7"
files = [
{file = "aiosignal-1.3.1-py3-none-any.whl", hash = "sha256:f8376fb07dd1e86a584e4fcdec80b36b7f81aac666ebc724e2c090300dd83b17"},
@@ -235,17 +235,6 @@ files = [
{file = "alabaster-0.7.13.tar.gz", hash = "sha256:a27a4a084d5e690e16e01e03ad2b2e552c61a65469419b907243193de1a84ae2"},
]
-[[package]]
-name = "anyascii"
-version = "0.3.2"
-description = "Unicode to ASCII transliteration"
-optional = false
-python-versions = ">=3.3"
-files = [
- {file = "anyascii-0.3.2-py3-none-any.whl", hash = "sha256:3b3beef6fc43d9036d3b0529050b0c48bfad8bc960e9e562d7223cfb94fe45d4"},
- {file = "anyascii-0.3.2.tar.gz", hash = "sha256:9d5d32ef844fe225b8bc7cba7f950534fae4da27a9bf3a6bea2cb0ea46ce4730"},
-]
-
[[package]]
name = "appnope"
version = "0.1.4"
@@ -259,32 +248,18 @@ files = [
[[package]]
name = "argcomplete"
-version = "3.2.3"
+version = "3.3.0"
description = "Bash tab completion for argparse"
optional = false
python-versions = ">=3.8"
files = [
- {file = "argcomplete-3.2.3-py3-none-any.whl", hash = "sha256:c12355e0494c76a2a7b73e3a59b09024ca0ba1e279fb9ed6c1b82d5b74b6a70c"},
- {file = "argcomplete-3.2.3.tar.gz", hash = "sha256:bf7900329262e481be5a15f56f19736b376df6f82ed27576fa893652c5de6c23"},
+ {file = "argcomplete-3.3.0-py3-none-any.whl", hash = "sha256:c168c3723482c031df3c207d4ba8fa702717ccb9fc0bfe4117166c1f537b4a54"},
+ {file = "argcomplete-3.3.0.tar.gz", hash = "sha256:fd03ff4a5b9e6580569d34b273f741e85cd9e072f3feeeee3eba4891c70eda62"},
]
[package.extras]
test = ["coverage", "mypy", "pexpect", "ruff", "wheel"]
-[[package]]
-name = "astroid"
-version = "3.1.0"
-description = "An abstract syntax tree for Python with inference support."
-optional = false
-python-versions = ">=3.8.0"
-files = [
- {file = "astroid-3.1.0-py3-none-any.whl", hash = "sha256:951798f922990137ac090c53af473db7ab4e70c770e6d7fae0cec59f74411819"},
- {file = "astroid-3.1.0.tar.gz", hash = "sha256:ac248253bfa4bd924a0de213707e7ebeeb3138abeb48d798784ead1e56d419d4"},
-]
-
-[package.dependencies]
-typing-extensions = {version = ">=4.0.0", markers = "python_version < \"3.11\""}
-
[[package]]
name = "asttokens"
version = "2.4.1"
@@ -303,11 +278,26 @@ six = ">=1.12.0"
astroid = ["astroid (>=1,<2)", "astroid (>=2,<4)"]
test = ["astroid (>=1,<2)", "astroid (>=2,<4)", "pytest"]
+[[package]]
+name = "astunparse"
+version = "1.6.3"
+description = "An AST unparser for Python"
+optional = false
+python-versions = "*"
+files = [
+ {file = "astunparse-1.6.3-py2.py3-none-any.whl", hash = "sha256:c2652417f2c8b5bb325c885ae329bdf3f86424075c4fd1a128674bc6fba4b8e8"},
+ {file = "astunparse-1.6.3.tar.gz", hash = "sha256:5ad93a8456f0d084c3456d059fd9a92cce667963232cbf763eac3bc5b7940872"},
+]
+
+[package.dependencies]
+six = ">=1.6.1,<2.0"
+wheel = ">=0.23.0,<1.0"
+
[[package]]
name = "async-timeout"
version = "4.0.3"
description = "Timeout context manager for asyncio programs"
-optional = false
+optional = true
python-versions = ">=3.7"
files = [
{file = "async-timeout-4.0.3.tar.gz", hash = "sha256:4640d96be84d82d02ed59ea2b7105a0f7b33abe8703703cd0ab0bf87c427522f"},
@@ -333,20 +323,6 @@ tests = ["attrs[tests-no-zope]", "zope-interface"]
tests-mypy = ["mypy (>=1.6)", "pytest-mypy-plugins"]
tests-no-zope = ["attrs[tests-mypy]", "cloudpickle", "hypothesis", "pympler", "pytest (>=4.3.0)", "pytest-xdist[psutil]"]
-[[package]]
-name = "autodoc"
-version = "0.5.0"
-description = "Autodoc Python implementation."
-optional = false
-python-versions = "*"
-files = [
- {file = "autodoc-0.5.0.tar.gz", hash = "sha256:c4387c5a0f1c09b055bb2e384542ee1e016542f313b2a33d904ca77f0460ded3"},
-]
-
-[package.dependencies]
-decorator = "*"
-webtest = "*"
-
[[package]]
name = "babel"
version = "2.14.0"
@@ -441,17 +417,17 @@ css = ["tinycss2 (>=1.1.0,<1.3)"]
[[package]]
name = "boto3"
-version = "1.34.51"
+version = "1.34.69"
description = "The AWS SDK for Python"
optional = true
-python-versions = ">= 3.8"
+python-versions = ">=3.8"
files = [
- {file = "boto3-1.34.51-py3-none-any.whl", hash = "sha256:67732634dc7d0afda879bd9a5e2d0818a2c14a98bef766b95a3e253ea5104cb9"},
- {file = "boto3-1.34.51.tar.gz", hash = "sha256:2cd9463e738a184cbce8a6824027c22163c5f73e277a35ff5aa0fb0e845b4301"},
+ {file = "boto3-1.34.69-py3-none-any.whl", hash = "sha256:2e25ef6bd325217c2da329829478be063155897d8d3b29f31f7f23ab548519b1"},
+ {file = "boto3-1.34.69.tar.gz", hash = "sha256:898a5fed26b1351352703421d1a8b886ef2a74be6c97d5ecc92432ae01fda203"},
]
[package.dependencies]
-botocore = ">=1.34.51,<1.35.0"
+botocore = ">=1.34.69,<1.35.0"
jmespath = ">=0.7.1,<2.0.0"
s3transfer = ">=0.10.0,<0.11.0"
@@ -460,13 +436,13 @@ crt = ["botocore[crt] (>=1.21.0,<2.0a0)"]
[[package]]
name = "botocore"
-version = "1.34.51"
+version = "1.34.69"
description = "Low-level, data-driven core of boto 3."
optional = true
-python-versions = ">= 3.8"
+python-versions = ">=3.8"
files = [
- {file = "botocore-1.34.51-py3-none-any.whl", hash = "sha256:01d5156247f991b3466a8404e3d7460a9ecbd9b214f9992d6ba797d9ddc6f120"},
- {file = "botocore-1.34.51.tar.gz", hash = "sha256:5086217442e67dd9de36ec7e87a0c663f76b7790d5fb6a12de565af95e87e319"},
+ {file = "botocore-1.34.69-py3-none-any.whl", hash = "sha256:d3802d076d4d507bf506f9845a6970ce43adc3d819dd57c2791f5c19ed6e5950"},
+ {file = "botocore-1.34.69.tar.gz", hash = "sha256:d1ab2bff3c2fd51719c2021d9fa2f30fbb9ed0a308f69e9a774ac92c8091380a"},
]
[package.dependencies]
@@ -474,12 +450,23 @@ jmespath = ">=0.7.1,<2.0.0"
python-dateutil = ">=2.1,<3.0.0"
urllib3 = [
{version = ">=1.25.4,<1.27", markers = "python_version < \"3.10\""},
- {version = ">=1.25.4,<2.1", markers = "python_version >= \"3.10\""},
+ {version = ">=1.25.4,<2.2.0 || >2.2.0,<3", markers = "python_version >= \"3.10\""},
]
[package.extras]
crt = ["awscrt (==0.19.19)"]
+[[package]]
+name = "bracex"
+version = "2.4"
+description = "Bash style brace expander."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "bracex-2.4-py3-none-any.whl", hash = "sha256:efdc71eff95eaff5e0f8cfebe7d01adf2c8637c8c92edaf63ef348c241a82418"},
+ {file = "bracex-2.4.tar.gz", hash = "sha256:a27eaf1df42cf561fed58b7a8f3fdf129d1ea16a81e1fadd1d17989bc6384beb"},
+]
+
[[package]]
name = "certifi"
version = "2024.2.2"
@@ -705,19 +692,36 @@ files = [
{file = "colorama-0.4.6.tar.gz", hash = "sha256:08695f5cb7ed6e0531a20572697297273c47b8cae5a63ffc6d6ed5c201be6e44"},
]
+[[package]]
+name = "comm"
+version = "0.2.2"
+description = "Jupyter Python Comm implementation, for usage in ipykernel, xeus-python etc."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "comm-0.2.2-py3-none-any.whl", hash = "sha256:e6fb86cb70ff661ee8c9c14e7d36d6de3b4066f1441be4063df9c5009f0a64d3"},
+ {file = "comm-0.2.2.tar.gz", hash = "sha256:3fd7a84065306e07bea1773df6eb8282de51ba82f77c72f9c85716ab11fe980e"},
+]
+
+[package.dependencies]
+traitlets = ">=4"
+
+[package.extras]
+test = ["pytest"]
+
[[package]]
name = "commitizen"
-version = "3.20.0"
+version = "3.24.0"
description = "Python commitizen client tool"
optional = false
python-versions = ">=3.8"
files = [
- {file = "commitizen-3.20.0-py3-none-any.whl", hash = "sha256:f079d9642347d314afae75664d289b0de80cf0343c99c3dcfa85782f164333f3"},
- {file = "commitizen-3.20.0.tar.gz", hash = "sha256:17aebc8f36326fa3e65dcc08195303579e356be84b0d65c3c4bfed85b8822411"},
+ {file = "commitizen-3.24.0-py3-none-any.whl", hash = "sha256:d9e28b1dcd97cea64dcb50be25292ceb730470d933f1da37131f9540f762df36"},
+ {file = "commitizen-3.24.0.tar.gz", hash = "sha256:088e01ae8265f1d6fa5a4d11a05e4fd7092d958c881837c35f6c65aad27331a9"},
]
[package.dependencies]
-argcomplete = ">=1.12.1,<3.3"
+argcomplete = ">=1.12.1,<3.4"
charset-normalizer = ">=2.1.0,<4"
colorama = ">=0.4.1,<0.5.0"
decli = ">=0.6.0,<0.7.0"
@@ -731,78 +735,78 @@ tomlkit = ">=0.5.3,<1.0.0"
[[package]]
name = "configparser"
-version = "6.0.1"
+version = "7.0.0"
description = "Updated configparser from stdlib for earlier Pythons."
optional = true
python-versions = ">=3.8"
files = [
- {file = "configparser-6.0.1-py3-none-any.whl", hash = "sha256:5a0da275bea56f871abaa9e0806331791e9d8ae2938e8b8797b99ab3e8e192c4"},
- {file = "configparser-6.0.1.tar.gz", hash = "sha256:db45513e971e509496b150be31bd67b0e14ab20b78a383b677e4b158e2c682d8"},
+ {file = "configparser-7.0.0-py3-none-any.whl", hash = "sha256:f46d52a12811c637104c6bb8eb33693be0038ab6bf01d69aae009c39ec8c2017"},
+ {file = "configparser-7.0.0.tar.gz", hash = "sha256:af3c618a67aaaedc4d689fd7317d238f566b9aa03cae50102e92d7f0dfe78ba0"},
]
[package.extras]
-docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "rst.linker (>=1.9)", "sphinx (<7.2.5)", "sphinx (>=3.5)", "sphinx-lint"]
-testing = ["pytest (>=6)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-mypy", "pytest-ruff (>=0.2.1)", "types-backports"]
+docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "rst.linker (>=1.9)", "sphinx (>=3.5)", "sphinx-lint"]
+testing = ["pytest (>=6,!=8.1.1)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-mypy", "pytest-ruff (>=0.2.1)", "types-backports"]
[[package]]
name = "coverage"
-version = "7.4.4"
+version = "7.5.0"
description = "Code coverage measurement for Python"
optional = false
python-versions = ">=3.8"
files = [
- {file = "coverage-7.4.4-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:e0be5efd5127542ef31f165de269f77560d6cdef525fffa446de6f7e9186cfb2"},
- {file = "coverage-7.4.4-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:ccd341521be3d1b3daeb41960ae94a5e87abe2f46f17224ba5d6f2b8398016cf"},
- {file = "coverage-7.4.4-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:09fa497a8ab37784fbb20ab699c246053ac294d13fc7eb40ec007a5043ec91f8"},
- {file = "coverage-7.4.4-cp310-cp310-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:b1a93009cb80730c9bca5d6d4665494b725b6e8e157c1cb7f2db5b4b122ea562"},
- {file = "coverage-7.4.4-cp310-cp310-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:690db6517f09336559dc0b5f55342df62370a48f5469fabf502db2c6d1cffcd2"},
- {file = "coverage-7.4.4-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:09c3255458533cb76ef55da8cc49ffab9e33f083739c8bd4f58e79fecfe288f7"},
- {file = "coverage-7.4.4-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:8ce1415194b4a6bd0cdcc3a1dfbf58b63f910dcb7330fe15bdff542c56949f87"},
- {file = "coverage-7.4.4-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:b91cbc4b195444e7e258ba27ac33769c41b94967919f10037e6355e998af255c"},
- {file = "coverage-7.4.4-cp310-cp310-win32.whl", hash = "sha256:598825b51b81c808cb6f078dcb972f96af96b078faa47af7dfcdf282835baa8d"},
- {file = "coverage-7.4.4-cp310-cp310-win_amd64.whl", hash = "sha256:09ef9199ed6653989ebbcaacc9b62b514bb63ea2f90256e71fea3ed74bd8ff6f"},
- {file = "coverage-7.4.4-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:0f9f50e7ef2a71e2fae92774c99170eb8304e3fdf9c8c3c7ae9bab3e7229c5cf"},
- {file = "coverage-7.4.4-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:623512f8ba53c422fcfb2ce68362c97945095b864cda94a92edbaf5994201083"},
- {file = "coverage-7.4.4-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:0513b9508b93da4e1716744ef6ebc507aff016ba115ffe8ecff744d1322a7b63"},
- {file = "coverage-7.4.4-cp311-cp311-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:40209e141059b9370a2657c9b15607815359ab3ef9918f0196b6fccce8d3230f"},
- {file = "coverage-7.4.4-cp311-cp311-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:8a2b2b78c78293782fd3767d53e6474582f62443d0504b1554370bde86cc8227"},
- {file = "coverage-7.4.4-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:73bfb9c09951125d06ee473bed216e2c3742f530fc5acc1383883125de76d9cd"},
- {file = "coverage-7.4.4-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:1f384c3cc76aeedce208643697fb3e8437604b512255de6d18dae3f27655a384"},
- {file = "coverage-7.4.4-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:54eb8d1bf7cacfbf2a3186019bcf01d11c666bd495ed18717162f7eb1e9dd00b"},
- {file = "coverage-7.4.4-cp311-cp311-win32.whl", hash = "sha256:cac99918c7bba15302a2d81f0312c08054a3359eaa1929c7e4b26ebe41e9b286"},
- {file = "coverage-7.4.4-cp311-cp311-win_amd64.whl", hash = "sha256:b14706df8b2de49869ae03a5ccbc211f4041750cd4a66f698df89d44f4bd30ec"},
- {file = "coverage-7.4.4-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:201bef2eea65e0e9c56343115ba3814e896afe6d36ffd37bab783261db430f76"},
- {file = "coverage-7.4.4-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:41c9c5f3de16b903b610d09650e5e27adbfa7f500302718c9ffd1c12cf9d6818"},
- {file = "coverage-7.4.4-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d898fe162d26929b5960e4e138651f7427048e72c853607f2b200909794ed978"},
- {file = "coverage-7.4.4-cp312-cp312-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:3ea79bb50e805cd6ac058dfa3b5c8f6c040cb87fe83de10845857f5535d1db70"},
- {file = "coverage-7.4.4-cp312-cp312-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:ce4b94265ca988c3f8e479e741693d143026632672e3ff924f25fab50518dd51"},
- {file = "coverage-7.4.4-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:00838a35b882694afda09f85e469c96367daa3f3f2b097d846a7216993d37f4c"},
- {file = "coverage-7.4.4-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:fdfafb32984684eb03c2d83e1e51f64f0906b11e64482df3c5db936ce3839d48"},
- {file = "coverage-7.4.4-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:69eb372f7e2ece89f14751fbcbe470295d73ed41ecd37ca36ed2eb47512a6ab9"},
- {file = "coverage-7.4.4-cp312-cp312-win32.whl", hash = "sha256:137eb07173141545e07403cca94ab625cc1cc6bc4c1e97b6e3846270e7e1fea0"},
- {file = "coverage-7.4.4-cp312-cp312-win_amd64.whl", hash = "sha256:d71eec7d83298f1af3326ce0ff1d0ea83c7cb98f72b577097f9083b20bdaf05e"},
- {file = "coverage-7.4.4-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:d5ae728ff3b5401cc320d792866987e7e7e880e6ebd24433b70a33b643bb0384"},
- {file = "coverage-7.4.4-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:cc4f1358cb0c78edef3ed237ef2c86056206bb8d9140e73b6b89fbcfcbdd40e1"},
- {file = "coverage-7.4.4-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:8130a2aa2acb8788e0b56938786c33c7c98562697bf9f4c7d6e8e5e3a0501e4a"},
- {file = "coverage-7.4.4-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:cf271892d13e43bc2b51e6908ec9a6a5094a4df1d8af0bfc360088ee6c684409"},
- {file = "coverage-7.4.4-cp38-cp38-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:a4cdc86d54b5da0df6d3d3a2f0b710949286094c3a6700c21e9015932b81447e"},
- {file = "coverage-7.4.4-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:ae71e7ddb7a413dd60052e90528f2f65270aad4b509563af6d03d53e979feafd"},
- {file = "coverage-7.4.4-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:38dd60d7bf242c4ed5b38e094baf6401faa114fc09e9e6632374388a404f98e7"},
- {file = "coverage-7.4.4-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:aa5b1c1bfc28384f1f53b69a023d789f72b2e0ab1b3787aae16992a7ca21056c"},
- {file = "coverage-7.4.4-cp38-cp38-win32.whl", hash = "sha256:dfa8fe35a0bb90382837b238fff375de15f0dcdb9ae68ff85f7a63649c98527e"},
- {file = "coverage-7.4.4-cp38-cp38-win_amd64.whl", hash = "sha256:b2991665420a803495e0b90a79233c1433d6ed77ef282e8e152a324bbbc5e0c8"},
- {file = "coverage-7.4.4-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:3b799445b9f7ee8bf299cfaed6f5b226c0037b74886a4e11515e569b36fe310d"},
- {file = "coverage-7.4.4-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:b4d33f418f46362995f1e9d4f3a35a1b6322cb959c31d88ae56b0298e1c22357"},
- {file = "coverage-7.4.4-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:aadacf9a2f407a4688d700e4ebab33a7e2e408f2ca04dbf4aef17585389eff3e"},
- {file = "coverage-7.4.4-cp39-cp39-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:7c95949560050d04d46b919301826525597f07b33beba6187d04fa64d47ac82e"},
- {file = "coverage-7.4.4-cp39-cp39-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:ff7687ca3d7028d8a5f0ebae95a6e4827c5616b31a4ee1192bdfde697db110d4"},
- {file = "coverage-7.4.4-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:5fc1de20b2d4a061b3df27ab9b7c7111e9a710f10dc2b84d33a4ab25065994ec"},
- {file = "coverage-7.4.4-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:c74880fc64d4958159fbd537a091d2a585448a8f8508bf248d72112723974cbd"},
- {file = "coverage-7.4.4-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:742a76a12aa45b44d236815d282b03cfb1de3b4323f3e4ec933acfae08e54ade"},
- {file = "coverage-7.4.4-cp39-cp39-win32.whl", hash = "sha256:d89d7b2974cae412400e88f35d86af72208e1ede1a541954af5d944a8ba46c57"},
- {file = "coverage-7.4.4-cp39-cp39-win_amd64.whl", hash = "sha256:9ca28a302acb19b6af89e90f33ee3e1906961f94b54ea37de6737b7ca9d8827c"},
- {file = "coverage-7.4.4-pp38.pp39.pp310-none-any.whl", hash = "sha256:b2c5edc4ac10a7ef6605a966c58929ec6c1bd0917fb8c15cb3363f65aa40e677"},
- {file = "coverage-7.4.4.tar.gz", hash = "sha256:c901df83d097649e257e803be22592aedfd5182f07b3cc87d640bbb9afd50f49"},
+ {file = "coverage-7.5.0-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:432949a32c3e3f820af808db1833d6d1631664d53dd3ce487aa25d574e18ad1c"},
+ {file = "coverage-7.5.0-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:2bd7065249703cbeb6d4ce679c734bef0ee69baa7bff9724361ada04a15b7e3b"},
+ {file = "coverage-7.5.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:bbfe6389c5522b99768a93d89aca52ef92310a96b99782973b9d11e80511f932"},
+ {file = "coverage-7.5.0-cp310-cp310-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:39793731182c4be939b4be0cdecde074b833f6171313cf53481f869937129ed3"},
+ {file = "coverage-7.5.0-cp310-cp310-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:85a5dbe1ba1bf38d6c63b6d2c42132d45cbee6d9f0c51b52c59aa4afba057517"},
+ {file = "coverage-7.5.0-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:357754dcdfd811462a725e7501a9b4556388e8ecf66e79df6f4b988fa3d0b39a"},
+ {file = "coverage-7.5.0-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:a81eb64feded34f40c8986869a2f764f0fe2db58c0530d3a4afbcde50f314880"},
+ {file = "coverage-7.5.0-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:51431d0abbed3a868e967f8257c5faf283d41ec882f58413cf295a389bb22e58"},
+ {file = "coverage-7.5.0-cp310-cp310-win32.whl", hash = "sha256:f609ebcb0242d84b7adeee2b06c11a2ddaec5464d21888b2c8255f5fd6a98ae4"},
+ {file = "coverage-7.5.0-cp310-cp310-win_amd64.whl", hash = "sha256:6782cd6216fab5a83216cc39f13ebe30adfac2fa72688c5a4d8d180cd52e8f6a"},
+ {file = "coverage-7.5.0-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:e768d870801f68c74c2b669fc909839660180c366501d4cc4b87efd6b0eee375"},
+ {file = "coverage-7.5.0-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:84921b10aeb2dd453247fd10de22907984eaf80901b578a5cf0bb1e279a587cb"},
+ {file = "coverage-7.5.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:710c62b6e35a9a766b99b15cdc56d5aeda0914edae8bb467e9c355f75d14ee95"},
+ {file = "coverage-7.5.0-cp311-cp311-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:c379cdd3efc0658e652a14112d51a7668f6bfca7445c5a10dee7eabecabba19d"},
+ {file = "coverage-7.5.0-cp311-cp311-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:fea9d3ca80bcf17edb2c08a4704259dadac196fe5e9274067e7a20511fad1743"},
+ {file = "coverage-7.5.0-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:41327143c5b1d715f5f98a397608f90ab9ebba606ae4e6f3389c2145410c52b1"},
+ {file = "coverage-7.5.0-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:565b2e82d0968c977e0b0f7cbf25fd06d78d4856289abc79694c8edcce6eb2de"},
+ {file = "coverage-7.5.0-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:cf3539007202ebfe03923128fedfdd245db5860a36810136ad95a564a2fdffff"},
+ {file = "coverage-7.5.0-cp311-cp311-win32.whl", hash = "sha256:bf0b4b8d9caa8d64df838e0f8dcf68fb570c5733b726d1494b87f3da85db3a2d"},
+ {file = "coverage-7.5.0-cp311-cp311-win_amd64.whl", hash = "sha256:9c6384cc90e37cfb60435bbbe0488444e54b98700f727f16f64d8bfda0b84656"},
+ {file = "coverage-7.5.0-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:fed7a72d54bd52f4aeb6c6e951f363903bd7d70bc1cad64dd1f087980d309ab9"},
+ {file = "coverage-7.5.0-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:cbe6581fcff7c8e262eb574244f81f5faaea539e712a058e6707a9d272fe5b64"},
+ {file = "coverage-7.5.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ad97ec0da94b378e593ef532b980c15e377df9b9608c7c6da3506953182398af"},
+ {file = "coverage-7.5.0-cp312-cp312-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:bd4bacd62aa2f1a1627352fe68885d6ee694bdaebb16038b6e680f2924a9b2cc"},
+ {file = "coverage-7.5.0-cp312-cp312-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:adf032b6c105881f9d77fa17d9eebe0ad1f9bfb2ad25777811f97c5362aa07f2"},
+ {file = "coverage-7.5.0-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:4ba01d9ba112b55bfa4b24808ec431197bb34f09f66f7cb4fd0258ff9d3711b1"},
+ {file = "coverage-7.5.0-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:f0bfe42523893c188e9616d853c47685e1c575fe25f737adf473d0405dcfa7eb"},
+ {file = "coverage-7.5.0-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:a9a7ef30a1b02547c1b23fa9a5564f03c9982fc71eb2ecb7f98c96d7a0db5cf2"},
+ {file = "coverage-7.5.0-cp312-cp312-win32.whl", hash = "sha256:3c2b77f295edb9fcdb6a250f83e6481c679335ca7e6e4a955e4290350f2d22a4"},
+ {file = "coverage-7.5.0-cp312-cp312-win_amd64.whl", hash = "sha256:427e1e627b0963ac02d7c8730ca6d935df10280d230508c0ba059505e9233475"},
+ {file = "coverage-7.5.0-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:9dd88fce54abbdbf4c42fb1fea0e498973d07816f24c0e27a1ecaf91883ce69e"},
+ {file = "coverage-7.5.0-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:a898c11dca8f8c97b467138004a30133974aacd572818c383596f8d5b2eb04a9"},
+ {file = "coverage-7.5.0-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:07dfdd492d645eea1bd70fb1d6febdcf47db178b0d99161d8e4eed18e7f62fe7"},
+ {file = "coverage-7.5.0-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:d3d117890b6eee85887b1eed41eefe2e598ad6e40523d9f94c4c4b213258e4a4"},
+ {file = "coverage-7.5.0-cp38-cp38-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:6afd2e84e7da40fe23ca588379f815fb6dbbb1b757c883935ed11647205111cb"},
+ {file = "coverage-7.5.0-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:a9960dd1891b2ddf13a7fe45339cd59ecee3abb6b8326d8b932d0c5da208104f"},
+ {file = "coverage-7.5.0-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:ced268e82af993d7801a9db2dbc1d2322e786c5dc76295d8e89473d46c6b84d4"},
+ {file = "coverage-7.5.0-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:e7c211f25777746d468d76f11719e64acb40eed410d81c26cefac641975beb88"},
+ {file = "coverage-7.5.0-cp38-cp38-win32.whl", hash = "sha256:262fffc1f6c1a26125d5d573e1ec379285a3723363f3bd9c83923c9593a2ac25"},
+ {file = "coverage-7.5.0-cp38-cp38-win_amd64.whl", hash = "sha256:eed462b4541c540d63ab57b3fc69e7d8c84d5957668854ee4e408b50e92ce26a"},
+ {file = "coverage-7.5.0-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:d0194d654e360b3e6cc9b774e83235bae6b9b2cac3be09040880bb0e8a88f4a1"},
+ {file = "coverage-7.5.0-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:33c020d3322662e74bc507fb11488773a96894aa82a622c35a5a28673c0c26f5"},
+ {file = "coverage-7.5.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:0cbdf2cae14a06827bec50bd58e49249452d211d9caddd8bd80e35b53cb04631"},
+ {file = "coverage-7.5.0-cp39-cp39-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:3235d7c781232e525b0761730e052388a01548bd7f67d0067a253887c6e8df46"},
+ {file = "coverage-7.5.0-cp39-cp39-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:db2de4e546f0ec4b2787d625e0b16b78e99c3e21bc1722b4977c0dddf11ca84e"},
+ {file = "coverage-7.5.0-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:4d0e206259b73af35c4ec1319fd04003776e11e859936658cb6ceffdeba0f5be"},
+ {file = "coverage-7.5.0-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:2055c4fb9a6ff624253d432aa471a37202cd8f458c033d6d989be4499aed037b"},
+ {file = "coverage-7.5.0-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:075299460948cd12722a970c7eae43d25d37989da682997687b34ae6b87c0ef0"},
+ {file = "coverage-7.5.0-cp39-cp39-win32.whl", hash = "sha256:280132aada3bc2f0fac939a5771db4fbb84f245cb35b94fae4994d4c1f80dae7"},
+ {file = "coverage-7.5.0-cp39-cp39-win_amd64.whl", hash = "sha256:c58536f6892559e030e6924896a44098bc1290663ea12532c78cef71d0df8493"},
+ {file = "coverage-7.5.0-pp38.pp39.pp310-none-any.whl", hash = "sha256:2b57780b51084d5223eee7b59f0d4911c31c16ee5aa12737c7a02455829ff067"},
+ {file = "coverage-7.5.0.tar.gz", hash = "sha256:cf62d17310f34084c59c01e027259076479128d11e4661bb6c9acb38c5e19bb8"},
]
[package.dependencies]
@@ -865,6 +869,37 @@ files = [
{file = "dbfread-2.0.7.tar.gz", hash = "sha256:07c8a9af06ffad3f6f03e8fe91ad7d2733e31a26d2b72c4dd4cfbae07ee3b73d"},
]
+[[package]]
+name = "debugpy"
+version = "1.8.1"
+description = "An implementation of the Debug Adapter Protocol for Python"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "debugpy-1.8.1-cp310-cp310-macosx_11_0_x86_64.whl", hash = "sha256:3bda0f1e943d386cc7a0e71bfa59f4137909e2ed947fb3946c506e113000f741"},
+ {file = "debugpy-1.8.1-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:dda73bf69ea479c8577a0448f8c707691152e6c4de7f0c4dec5a4bc11dee516e"},
+ {file = "debugpy-1.8.1-cp310-cp310-win32.whl", hash = "sha256:3a79c6f62adef994b2dbe9fc2cc9cc3864a23575b6e387339ab739873bea53d0"},
+ {file = "debugpy-1.8.1-cp310-cp310-win_amd64.whl", hash = "sha256:7eb7bd2b56ea3bedb009616d9e2f64aab8fc7000d481faec3cd26c98a964bcdd"},
+ {file = "debugpy-1.8.1-cp311-cp311-macosx_11_0_universal2.whl", hash = "sha256:016a9fcfc2c6b57f939673c874310d8581d51a0fe0858e7fac4e240c5eb743cb"},
+ {file = "debugpy-1.8.1-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:fd97ed11a4c7f6d042d320ce03d83b20c3fb40da892f994bc041bbc415d7a099"},
+ {file = "debugpy-1.8.1-cp311-cp311-win32.whl", hash = "sha256:0de56aba8249c28a300bdb0672a9b94785074eb82eb672db66c8144fff673146"},
+ {file = "debugpy-1.8.1-cp311-cp311-win_amd64.whl", hash = "sha256:1a9fe0829c2b854757b4fd0a338d93bc17249a3bf69ecf765c61d4c522bb92a8"},
+ {file = "debugpy-1.8.1-cp312-cp312-macosx_11_0_universal2.whl", hash = "sha256:3ebb70ba1a6524d19fa7bb122f44b74170c447d5746a503e36adc244a20ac539"},
+ {file = "debugpy-1.8.1-cp312-cp312-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:a2e658a9630f27534e63922ebf655a6ab60c370f4d2fc5c02a5b19baf4410ace"},
+ {file = "debugpy-1.8.1-cp312-cp312-win32.whl", hash = "sha256:caad2846e21188797a1f17fc09c31b84c7c3c23baf2516fed5b40b378515bbf0"},
+ {file = "debugpy-1.8.1-cp312-cp312-win_amd64.whl", hash = "sha256:edcc9f58ec0fd121a25bc950d4578df47428d72e1a0d66c07403b04eb93bcf98"},
+ {file = "debugpy-1.8.1-cp38-cp38-macosx_11_0_x86_64.whl", hash = "sha256:7a3afa222f6fd3d9dfecd52729bc2e12c93e22a7491405a0ecbf9e1d32d45b39"},
+ {file = "debugpy-1.8.1-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:d915a18f0597ef685e88bb35e5d7ab968964b7befefe1aaea1eb5b2640b586c7"},
+ {file = "debugpy-1.8.1-cp38-cp38-win32.whl", hash = "sha256:92116039b5500633cc8d44ecc187abe2dfa9b90f7a82bbf81d079fcdd506bae9"},
+ {file = "debugpy-1.8.1-cp38-cp38-win_amd64.whl", hash = "sha256:e38beb7992b5afd9d5244e96ad5fa9135e94993b0c551ceebf3fe1a5d9beb234"},
+ {file = "debugpy-1.8.1-cp39-cp39-macosx_11_0_x86_64.whl", hash = "sha256:bfb20cb57486c8e4793d41996652e5a6a885b4d9175dd369045dad59eaacea42"},
+ {file = "debugpy-1.8.1-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:efd3fdd3f67a7e576dd869c184c5dd71d9aaa36ded271939da352880c012e703"},
+ {file = "debugpy-1.8.1-cp39-cp39-win32.whl", hash = "sha256:58911e8521ca0c785ac7a0539f1e77e0ce2df753f786188f382229278b4cdf23"},
+ {file = "debugpy-1.8.1-cp39-cp39-win_amd64.whl", hash = "sha256:6df9aa9599eb05ca179fb0b810282255202a66835c6efb1d112d21ecb830ddd3"},
+ {file = "debugpy-1.8.1-py2.py3-none-any.whl", hash = "sha256:28acbe2241222b87e255260c76741e1fbf04fdc3b6d094fcf57b6c6f75ce1242"},
+ {file = "debugpy-1.8.1.zip", hash = "sha256:f696d6be15be87aef621917585f9bb94b1dc9e8aced570db1b8a6fc14e8f9b42"},
+]
+
[[package]]
name = "decli"
version = "0.6.1"
@@ -922,29 +957,18 @@ files = [
[[package]]
name = "dunamai"
-version = "1.19.2"
+version = "1.20.0"
description = "Dynamic version generation"
optional = false
python-versions = ">=3.5"
files = [
- {file = "dunamai-1.19.2-py3-none-any.whl", hash = "sha256:bc126b17571a44d68ed826cec596e0f61dc01edca8b21486f70014936a5d44f2"},
- {file = "dunamai-1.19.2.tar.gz", hash = "sha256:3be4049890763e19b8df1d52960dbea60b3e263eb0c96144a677ae0633734d2e"},
+ {file = "dunamai-1.20.0-py3-none-any.whl", hash = "sha256:a2185c227351a52a013c7d7a695d3f3cb6625c3eed14a5295adbbcc7e2f7f8d4"},
+ {file = "dunamai-1.20.0.tar.gz", hash = "sha256:c3f1ee64a1e6cc9ebc98adafa944efaccd0db32482d2177e59c1ff6bdf23cd70"},
]
[package.dependencies]
packaging = ">=20.9"
-[[package]]
-name = "entrypoints"
-version = "0.4"
-description = "Discover and load entry points from installed packages."
-optional = false
-python-versions = ">=3.6"
-files = [
- {file = "entrypoints-0.4-py3-none-any.whl", hash = "sha256:f174b5ff827504fd3cd97cc3f8649f3693f51538c7e4bdf3ef002c8429d42f9f"},
- {file = "entrypoints-0.4.tar.gz", hash = "sha256:b706eddaa9218a19ebcd67b56818f05bb27589b1ca9e8d797b74affad4ccacd4"},
-]
-
[[package]]
name = "et-xmlfile"
version = "1.1.0"
@@ -958,13 +982,13 @@ files = [
[[package]]
name = "exceptiongroup"
-version = "1.2.0"
+version = "1.2.1"
description = "Backport of PEP 654 (exception groups)"
optional = false
python-versions = ">=3.7"
files = [
- {file = "exceptiongroup-1.2.0-py3-none-any.whl", hash = "sha256:4bfd3996ac73b41e9b9628b04e079f193850720ea5945fc96a08633c66912f14"},
- {file = "exceptiongroup-1.2.0.tar.gz", hash = "sha256:91f5c769735f051a4290d52edd0858999b57e5876e9f85937691bd4c9fa3ed68"},
+ {file = "exceptiongroup-1.2.1-py3-none-any.whl", hash = "sha256:5258b9ed329c5bbdd31a309f53cbfb0b155341807f6ff7606a1e801a891b29ad"},
+ {file = "exceptiongroup-1.2.1.tar.gz", hash = "sha256:a4785e48b045528f5bfe627b6ad554ff32def154f42372786903b7abcfe1aa16"},
]
[package.extras]
@@ -1000,13 +1024,13 @@ devel = ["colorama", "json-spec", "jsonschema", "pylint", "pytest", "pytest-benc
[[package]]
name = "filelock"
-version = "3.13.3"
+version = "3.13.4"
description = "A platform independent file lock."
optional = false
python-versions = ">=3.8"
files = [
- {file = "filelock-3.13.3-py3-none-any.whl", hash = "sha256:5ffa845303983e7a0b7ae17636509bc97997d58afeafa72fb141a17b152284cb"},
- {file = "filelock-3.13.3.tar.gz", hash = "sha256:a79895a25bbefdf55d1a2a0a80968f7dbb28edcd6d4234a0afb3f37ecde4b546"},
+ {file = "filelock-3.13.4-py3-none-any.whl", hash = "sha256:404e5e9253aa60ad457cae1be07c0f0ca90a63931200a47d9b6a6af84fd7b45f"},
+ {file = "filelock-3.13.4.tar.gz", hash = "sha256:d13f466618bfde72bd2c18255e269f72542c6e70e7bac83a0232d6b1cc5c8cf4"},
]
[package.extras]
@@ -1032,7 +1056,7 @@ termcolor = "*"
name = "frozenlist"
version = "1.4.1"
description = "A list-like structure which implements collections.abc.MutableSequence"
-optional = false
+optional = true
python-versions = ">=3.8"
files = [
{file = "frozenlist-1.4.1-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:f9aa1878d1083b276b0196f2dfbe00c9b7e752475ed3b682025ff20c1c1f51ac"},
@@ -1149,23 +1173,6 @@ smb = ["smbprotocol"]
ssh = ["paramiko"]
tqdm = ["tqdm"]
-[[package]]
-name = "furo"
-version = "2023.9.10"
-description = "A clean customisable Sphinx documentation theme."
-optional = false
-python-versions = ">=3.8"
-files = [
- {file = "furo-2023.9.10-py3-none-any.whl", hash = "sha256:513092538537dc5c596691da06e3c370714ec99bc438680edc1debffb73e5bfc"},
- {file = "furo-2023.9.10.tar.gz", hash = "sha256:5707530a476d2a63b8cad83b4f961f3739a69f4b058bcf38a03a39fa537195b2"},
-]
-
-[package.dependencies]
-beautifulsoup4 = "*"
-pygments = ">=2.7"
-sphinx = ">=6.0,<8.0"
-sphinx-basic-ng = "*"
-
[[package]]
name = "ghp-import"
version = "2.1.0"
@@ -1255,28 +1262,29 @@ docs = ["Sphinx", "furo"]
test = ["objgraph", "psutil"]
[[package]]
-name = "groundwork-sphinx-theme"
-version = "1.1.1"
-description = "Sphinx theme for groundwork projects (Based on flask_theme)"
+name = "griffe"
+version = "0.44.0"
+description = "Signatures for entire Python programs. Extract the structure, the frame, the skeleton of your project, to generate API documentation or find breaking changes in your API."
optional = false
-python-versions = "*"
+python-versions = ">=3.8"
files = [
- {file = "groundwork-sphinx-theme-1.1.1.tar.gz", hash = "sha256:5f1217bf34daba5fcd93e8a2a04ef5a6c2b144ec43754aec7ce14de501e23fe4"},
- {file = "groundwork_sphinx_theme-1.1.1-py2.py3-none-any.whl", hash = "sha256:b4796989ce3cd6d5b5a3c7ebe52a8c9cf848110e9f12c2c88e2b3c0e4e8f0e24"},
+ {file = "griffe-0.44.0-py3-none-any.whl", hash = "sha256:8a4471c469ba980b87c843f1168850ce39d0c1d0c7be140dca2480f76c8e5446"},
+ {file = "griffe-0.44.0.tar.gz", hash = "sha256:34aee1571042f9bf00529bc715de4516fb6f482b164e90d030300601009e0223"},
]
[package.dependencies]
-Sphinx = ">=1.6.1"
+astunparse = {version = ">=1.6", markers = "python_version < \"3.9\""}
+colorama = ">=0.4"
[[package]]
name = "identify"
-version = "2.5.35"
+version = "2.5.36"
description = "File identification library for Python"
optional = false
python-versions = ">=3.8"
files = [
- {file = "identify-2.5.35-py2.py3-none-any.whl", hash = "sha256:c4de0081837b211594f8e877a6b4fad7ca32bbfc1a9307fdd61c28bfe923f13e"},
- {file = "identify-2.5.35.tar.gz", hash = "sha256:10a7ca245cfcd756a554a7288159f72ff105ad233c7c4b9c6f0f4d108f5f6791"},
+ {file = "identify-2.5.36-py2.py3-none-any.whl", hash = "sha256:37d93f380f4de590500d9dba7db359d0d3da95ffe7f9de1753faa159e71e7dfa"},
+ {file = "identify-2.5.36.tar.gz", hash = "sha256:e5e00f54165f9047fbebeb4a560f9acfb8af4c88232be60a488e9b68d122745d"},
]
[package.extras]
@@ -1352,6 +1360,39 @@ files = [
{file = "iniconfig-2.0.0.tar.gz", hash = "sha256:2d91e135bf72d31a410b17c16da610a82cb55f6b0477d1a902134b24a455b8b3"},
]
+[[package]]
+name = "ipykernel"
+version = "6.29.4"
+description = "IPython Kernel for Jupyter"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "ipykernel-6.29.4-py3-none-any.whl", hash = "sha256:1181e653d95c6808039c509ef8e67c4126b3b3af7781496c7cbfb5ed938a27da"},
+ {file = "ipykernel-6.29.4.tar.gz", hash = "sha256:3d44070060f9475ac2092b760123fadf105d2e2493c24848b6691a7c4f42af5c"},
+]
+
+[package.dependencies]
+appnope = {version = "*", markers = "platform_system == \"Darwin\""}
+comm = ">=0.1.1"
+debugpy = ">=1.6.5"
+ipython = ">=7.23.1"
+jupyter-client = ">=6.1.12"
+jupyter-core = ">=4.12,<5.0.dev0 || >=5.1.dev0"
+matplotlib-inline = ">=0.1"
+nest-asyncio = "*"
+packaging = "*"
+psutil = "*"
+pyzmq = ">=24"
+tornado = ">=6.1"
+traitlets = ">=5.4.0"
+
+[package.extras]
+cov = ["coverage[toml]", "curio", "matplotlib", "pytest-cov", "trio"]
+docs = ["myst-parser", "pydata-sphinx-theme", "sphinx", "sphinx-autodoc-typehints", "sphinxcontrib-github-alt", "sphinxcontrib-spelling", "trio"]
+pyqt5 = ["pyqt5"]
+pyside6 = ["pyside6"]
+test = ["flaky", "ipyparallel", "pre-commit", "pytest (>=7.0)", "pytest-asyncio (>=0.23.5)", "pytest-cov", "pytest-timeout"]
+
[[package]]
name = "ipython"
version = "8.12.3"
@@ -1454,13 +1495,13 @@ files = [
[[package]]
name = "joblib"
-version = "1.3.2"
+version = "1.4.0"
description = "Lightweight pipelining with Python functions"
optional = false
-python-versions = ">=3.7"
+python-versions = ">=3.8"
files = [
- {file = "joblib-1.3.2-py3-none-any.whl", hash = "sha256:ef4331c65f239985f3f2220ecc87db222f08fd22097a3dd5698f693875f8cbb9"},
- {file = "joblib-1.3.2.tar.gz", hash = "sha256:92f865e621e17784e7955080b6d042489e3b8e294949cc44c6eac304f59772b1"},
+ {file = "joblib-1.4.0-py3-none-any.whl", hash = "sha256:42942470d4062537be4d54c83511186da1fc14ba354961a2114da91efa9a4ed7"},
+ {file = "joblib-1.4.0.tar.gz", hash = "sha256:1eb0dc091919cd384490de890cb5dfd538410a6d4b3b54eef09fb8c50b409b1c"},
]
[[package]]
@@ -1555,6 +1596,35 @@ files = [
{file = "jupyterlab_pygments-0.3.0.tar.gz", hash = "sha256:721aca4d9029252b11cfa9d185e5b5af4d54772bb8072f9b7036f4170054d35d"},
]
+[[package]]
+name = "jupytext"
+version = "1.16.1"
+description = "Jupyter notebooks as Markdown documents, Julia, Python or R scripts"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "jupytext-1.16.1-py3-none-any.whl", hash = "sha256:796ec4f68ada663569e5d38d4ef03738a01284bfe21c943c485bc36433898bd0"},
+ {file = "jupytext-1.16.1.tar.gz", hash = "sha256:68c7b68685e870e80e60fda8286fbd6269e9c74dc1df4316df6fe46eabc94c99"},
+]
+
+[package.dependencies]
+markdown-it-py = ">=1.0"
+mdit-py-plugins = "*"
+nbformat = "*"
+packaging = "*"
+pyyaml = "*"
+toml = "*"
+
+[package.extras]
+dev = ["jupytext[test-cov,test-external]"]
+docs = ["myst-parser", "sphinx", "sphinx-copybutton", "sphinx-rtd-theme"]
+test = ["pytest", "pytest-randomly", "pytest-xdist"]
+test-cov = ["jupytext[test-integration]", "pytest-cov (>=2.6.1)"]
+test-external = ["autopep8", "black", "flake8", "gitpython", "isort", "jupyter-fs (<0.4.0)", "jupytext[test-integration]", "pre-commit", "sphinx-gallery (<0.8)"]
+test-functional = ["jupytext[test]"]
+test-integration = ["ipykernel", "jupyter-server (!=2.11)", "jupytext[test-functional]", "nbconvert"]
+test-ui = ["calysto-bash"]
+
[[package]]
name = "leather"
version = "0.4.0"
@@ -1569,124 +1639,6 @@ files = [
[package.extras]
test = ["cssselect (>=0.9.1)", "lxml (>=3.6.0)", "pytest", "pytest-cov"]
-[[package]]
-name = "livereload"
-version = "2.6.3"
-description = "Python LiveReload is an awesome tool for web developers"
-optional = false
-python-versions = "*"
-files = [
- {file = "livereload-2.6.3-py2.py3-none-any.whl", hash = "sha256:ad4ac6f53b2d62bb6ce1a5e6e96f1f00976a32348afedcb4b6d68df2a1d346e4"},
- {file = "livereload-2.6.3.tar.gz", hash = "sha256:776f2f865e59fde56490a56bcc6773b6917366bce0c267c60ee8aaf1a0959869"},
-]
-
-[package.dependencies]
-six = "*"
-tornado = {version = "*", markers = "python_version > \"2.7\""}
-
-[[package]]
-name = "lxml"
-version = "5.1.1"
-description = "Powerful and Pythonic XML processing library combining libxml2/libxslt with the ElementTree API."
-optional = false
-python-versions = ">=3.6"
-files = [
- {file = "lxml-5.1.1-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:9cc30dc3c49ea914fa62ea73b57198b541cf2cd522fcf2b9559f99a24df769bb"},
- {file = "lxml-5.1.1-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:f1d0824e3ddb969fe1337b1bc45cf0bec8095b342f36903f41a74b7769cc8c73"},
- {file = "lxml-5.1.1-cp310-cp310-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:4958c378d9387c45ef8c4859495cf6be76f863e4e3b31494f6ec7f2c48d3b8e3"},
- {file = "lxml-5.1.1-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:aff34295a6c87638a1f1905355cf3a97e4026c45c0cf3bb6ed6bc35b885b4a33"},
- {file = "lxml-5.1.1-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b174885fd2cabd1ad48585296f495e25d607f02db99668c08b2afaceb668e21b"},
- {file = "lxml-5.1.1-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:1b0611bba10d6f5467b86673e8f6bba4de0d00f7d111eea843bc872abfe11b5c"},
- {file = "lxml-5.1.1-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:adff469b7dbfe9f3babc9e4479449ee97085ba70ac492fbe5f0f7217940c6731"},
- {file = "lxml-5.1.1-cp310-cp310-win32.whl", hash = "sha256:99bcdf665576a26b44c7ce767d76b769a4418b0a13cda8300b26fb7b2647bd5b"},
- {file = "lxml-5.1.1-cp310-cp310-win_amd64.whl", hash = "sha256:3da8db291568c27b2bb248dcfc8838ca3149f373a24e204bcd1c2c89e2813d14"},
- {file = "lxml-5.1.1-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:906966babd374fdfe46e130fc656488003f0d0d63b7cba612aa5a796c8804283"},
- {file = "lxml-5.1.1-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:9c03f3715c68fc707d9383d56e482d95d198ba07cb3dad4aee9e5a5ca06b2536"},
- {file = "lxml-5.1.1-cp311-cp311-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:d26243d994d4077a50056e9008848e5b421be0c6f0fd4e932a9463e1d89fc42b"},
- {file = "lxml-5.1.1-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:2de00750318ae6869b9dfa6429a4f82b8ecad043049414547474d09db549c2ee"},
- {file = "lxml-5.1.1-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:29b2771b4eec4e85063f10294facdd9829d010e6cc9668040d0cf936dc56733a"},
- {file = "lxml-5.1.1-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:d9358f7268c161dc0a1c3216018f26c04954b5dd47ba6dead79da6598f4725d4"},
- {file = "lxml-5.1.1-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:8a943826e7a9254eed661a7134fcde3c832a9fecd989d0f47c6e08c7b769cb2c"},
- {file = "lxml-5.1.1-cp311-cp311-win32.whl", hash = "sha256:74d0967c6f91eec6fe91159f9e8ccb3720fa0fbf9f462109c7bef62550df397c"},
- {file = "lxml-5.1.1-cp311-cp311-win_amd64.whl", hash = "sha256:26974096654241df08a30dc2eb0e139c1ad5653660aa4b2ced66000230e96c14"},
- {file = "lxml-5.1.1-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:55e13a19829dcdbf0c5233062977aeb6daf72e65124909128045976f659164e8"},
- {file = "lxml-5.1.1-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:adedfb61be862f48907218e3a24bf051fd2ecca53358f3958b0bdb17d7881c20"},
- {file = "lxml-5.1.1-cp312-cp312-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:77425482e4311d1cff119a2b5ab26c52ec209d2a3d728a54db3223ab91995e20"},
- {file = "lxml-5.1.1-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:1d380f183bd03ab827899753ea96dabe27d2025eb0bfd4f2ac0eee4afa0f351d"},
- {file = "lxml-5.1.1-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:f8682af96b5ad5093aab9eee5e4ff24cb7a9796c78699d914dd456ebfe7484a6"},
- {file = "lxml-5.1.1-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:68eed33377a9925aed7ba56c8611d50aaa1e45638c07a92b4b4b0a0436cc2dd2"},
- {file = "lxml-5.1.1-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:c7c1d2f6e9c7a1c4478146ee38d16dbe0eb3be998424bc0f01346c671c38b86d"},
- {file = "lxml-5.1.1-cp312-cp312-win32.whl", hash = "sha256:81107c8de3e463052ae8fd05fd31b97c371c7a9ce4a189b8bb5f45b0b3545fb9"},
- {file = "lxml-5.1.1-cp312-cp312-win_amd64.whl", hash = "sha256:0e46181d15fae102c53621bed9356b7a599a1e837b978c934a350dd00842b1d9"},
- {file = "lxml-5.1.1-cp36-cp36m-macosx_10_9_x86_64.whl", hash = "sha256:48dd28b9f410329de709a4bb6169c58f2cd8bff25f5a48d647678ec9b8a40c65"},
- {file = "lxml-5.1.1-cp36-cp36m-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:bf7e57dbe7b3c605e63849d9c8dae246a6ab9002223c57cd3f3dec7c3a0a8e6d"},
- {file = "lxml-5.1.1-cp36-cp36m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:5020b3081030b5cfc8149eee231167aea4ff68df73a610e1d542809e1f11fde7"},
- {file = "lxml-5.1.1-cp36-cp36m-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:77842b79b63c83c04dcfe2f045c78e15e4d97c86838eabd2e6518c1ed97e3900"},
- {file = "lxml-5.1.1-cp36-cp36m-musllinux_1_1_aarch64.whl", hash = "sha256:666432274881cb2535e71dbe745e08ef10fe25c81fbb1a6b1e3c973177823b0c"},
- {file = "lxml-5.1.1-cp36-cp36m-musllinux_1_1_x86_64.whl", hash = "sha256:a103426e809640a2d985062d2f4b28db2f0fe4469ff72a67cb31fa70613158f1"},
- {file = "lxml-5.1.1-cp36-cp36m-win32.whl", hash = "sha256:95a51324a55000c55f4ab79e1f7f1e0bc42b7a24e39633f79542753023a9d4b7"},
- {file = "lxml-5.1.1-cp36-cp36m-win_amd64.whl", hash = "sha256:bd46b5b19ac969de8e87fb3d04414641d12ee489e2ea6cc75344087829b31c63"},
- {file = "lxml-5.1.1-cp37-cp37m-macosx_10_9_x86_64.whl", hash = "sha256:59ca75cfcf646ff64aa19ca4e7fd2a0fde77268d5a87856525d9e0b69b77d0c4"},
- {file = "lxml-5.1.1-cp37-cp37m-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:d55ddc73dec971277b181a6d1a6abdd34f50e4511e1e60f6b4ebe22cbaad05bb"},
- {file = "lxml-5.1.1-cp37-cp37m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:56f1e813ff660d031c77edba90a068d57e47ae93a9e811330fc88946fa68af9a"},
- {file = "lxml-5.1.1-cp37-cp37m-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:43f21b5929185fa4560836942020bb00a0fcdec9f67be98cac1a4b99501757c1"},
- {file = "lxml-5.1.1-cp37-cp37m-musllinux_1_1_aarch64.whl", hash = "sha256:1528b37e83c3aeecb438e76e5be6279b353275560125a9c3f4d74642c5f110f9"},
- {file = "lxml-5.1.1-cp37-cp37m-musllinux_1_1_x86_64.whl", hash = "sha256:2992480a25434d2df31413136ef87effab14d43b07f1f54c5012c4f6c7530144"},
- {file = "lxml-5.1.1-cp37-cp37m-win32.whl", hash = "sha256:1d0270d33fbde6e1c6758ff58e2e284144f5331aa05dfe7f44ceafdf4e9d31aa"},
- {file = "lxml-5.1.1-cp37-cp37m-win_amd64.whl", hash = "sha256:dec3491aa69a91ed07f5e6bc033e2b1a9424447ad5312ee69ac973e94d79083a"},
- {file = "lxml-5.1.1-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:5bd2595ebe95214446e00a1ab94571f778b126e17736ea222c07505c4e092289"},
- {file = "lxml-5.1.1-cp38-cp38-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:bfbdadc3cfe552331ecb0bbdcabf148d1697c73aa4321151e0e6c1704eeb76a7"},
- {file = "lxml-5.1.1-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:52358249292bc155af681a9240ec3d944c1195f0124aa10ec4e3635adc1e10a1"},
- {file = "lxml-5.1.1-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:036b36c48cd775e4fd2084b34ae62ffeefa7a01f955f5a5b816f9257c308cfc0"},
- {file = "lxml-5.1.1-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:f05ab8cea65363d0cc7ce818f42407504b6d94ca885b4cde0270f021e2f4ef61"},
- {file = "lxml-5.1.1-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:d94a28c16cc430b68c374b37b8bb536ba5f0a4a080be0e1daa8310c44a00a75c"},
- {file = "lxml-5.1.1-cp38-cp38-win32.whl", hash = "sha256:9113fe65a62f834b8e994c8f48e7b2179bf81878c0ec80ad7feba51ab9417663"},
- {file = "lxml-5.1.1-cp38-cp38-win_amd64.whl", hash = "sha256:acff17e0cd5344677757a152631d8411efac6a84e4476d60123a9b33f5d6c511"},
- {file = "lxml-5.1.1-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:a94a97380ad689d751eb0a1e1ccd2a0622c5141771a31abe9a16075f80027e95"},
- {file = "lxml-5.1.1-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:d1f4d37b3f8d2d44493edce3d65ac987127bababd8ae208a6f0d7d260852346e"},
- {file = "lxml-5.1.1-cp39-cp39-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:3c5940f188189956ccb3d1adb413001ada79f2d2b81087d2612a0cc4a1197eed"},
- {file = "lxml-5.1.1-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:50007f4e94dc4e38030487a8b6c4af87a2d51ed059c7b74b29e3dd937cb1dfe1"},
- {file = "lxml-5.1.1-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:a305d0469177fd78a0a9aa2231c60218266bb85d4b7955f9b67dab628c9267fd"},
- {file = "lxml-5.1.1-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:13b73d78a8023203722cf98e9ea0b222da83110d1d5ef437ef8782a7755b4586"},
- {file = "lxml-5.1.1-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:bc6904519dd1f92eb82f7d49814a33bbc444d0b66b1438e76daf3f79ef4aa38f"},
- {file = "lxml-5.1.1-cp39-cp39-win32.whl", hash = "sha256:04ef231dde88294a5499f61a74cdc42af97d8d5ecec1b0a645d1c7d436942789"},
- {file = "lxml-5.1.1-cp39-cp39-win_amd64.whl", hash = "sha256:071e5123d1eca861708c4be5b54e4d88923fa33fab3aa02722e907518b07071c"},
- {file = "lxml-5.1.1-pp310-pypy310_pp73-macosx_10_9_x86_64.whl", hash = "sha256:19c6bc7476eeac4598ff925ae98597610109e21af4cd7ab1e060efcfc4b1c6e2"},
- {file = "lxml-5.1.1-pp310-pypy310_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:20cd17eb21f5ae54da96791c49e1fbd3327bf66b2c00556cdf8d0552c2270f92"},
- {file = "lxml-5.1.1-pp310-pypy310_pp73-win_amd64.whl", hash = "sha256:a02ed1ebc469734dbfed5b688f709334de19e7a333cba7ae187b17d2b2c1d4ff"},
- {file = "lxml-5.1.1-pp37-pypy37_pp73-macosx_10_9_x86_64.whl", hash = "sha256:161838cb95c97e8d76d01e544a3570b52ab6b863f4897a90e1f073bb110a75ba"},
- {file = "lxml-5.1.1-pp37-pypy37_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:d1abbf2249467a37da45fb2d7ff37e578dfc9813f142800e58db9da761cb7899"},
- {file = "lxml-5.1.1-pp37-pypy37_pp73-win_amd64.whl", hash = "sha256:6c49eb5deaed1990fde5b5d80d6800aec1b5fd6113346b5f11068d988f68f2c4"},
- {file = "lxml-5.1.1-pp38-pypy38_pp73-macosx_10_9_x86_64.whl", hash = "sha256:60ceffdca5d637fe8ee95c7f06733a6c9646e07da80997efe3af2d4b4f366e36"},
- {file = "lxml-5.1.1-pp38-pypy38_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:a76a7b35e7660c74eb3f943c19f5f78c882dceab890cf8017027b6100b79ad8e"},
- {file = "lxml-5.1.1-pp38-pypy38_pp73-win_amd64.whl", hash = "sha256:5dcb373720b70aa05419e508265dd86f06886ca0388967f6f024fbc4d551379f"},
- {file = "lxml-5.1.1-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:3641bc124b037921de4220538a5ebb52354fd2799fc2bbfb335d28096063c7d6"},
- {file = "lxml-5.1.1-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:3a6e9b34f59c9755aa279c652e1c48c333c665d05a88afcd8e5ff0bde86f3b14"},
- {file = "lxml-5.1.1-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:318847c165063549c8fda6b162a0d068689b10deb825cb3859caef69fddaaaff"},
- {file = "lxml-5.1.1.tar.gz", hash = "sha256:42a8aa957e98bd8b884a8142175ec24ce4ef0a57760e8879f193bfe64b757ca9"},
-]
-
-[package.extras]
-cssselect = ["cssselect (>=0.7)"]
-html5 = ["html5lib"]
-htmlsoup = ["BeautifulSoup4"]
-source = ["Cython (>=3.0.9)"]
-
-[[package]]
-name = "m2r2"
-version = "0.3.3.post2"
-description = "Markdown and reStructuredText in a single file."
-optional = false
-python-versions = ">=3.7"
-files = [
- {file = "m2r2-0.3.3.post2-py3-none-any.whl", hash = "sha256:86157721eb6eabcd54d4eea7195890cc58fa6188b8d0abea633383cfbb5e11e3"},
- {file = "m2r2-0.3.3.post2.tar.gz", hash = "sha256:e62bcb0e74b3ce19cda0737a0556b04cf4a43b785072fcef474558f2c1482ca8"},
-]
-
-[package.dependencies]
-docutils = ">=0.19"
-mistune = "0.8.4"
-
[[package]]
name = "markdown"
version = "3.6"
@@ -1705,6 +1657,30 @@ importlib-metadata = {version = ">=4.4", markers = "python_version < \"3.10\""}
docs = ["mdx-gh-links (>=0.2)", "mkdocs (>=1.5)", "mkdocs-gen-files", "mkdocs-literate-nav", "mkdocs-nature (>=0.6)", "mkdocs-section-index", "mkdocstrings[python]"]
testing = ["coverage", "pyyaml"]
+[[package]]
+name = "markdown-it-py"
+version = "3.0.0"
+description = "Python port of markdown-it. Markdown parsing, done right!"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "markdown-it-py-3.0.0.tar.gz", hash = "sha256:e3f60a94fa066dc52ec76661e37c851cb232d92f9886b15cb560aaada2df8feb"},
+ {file = "markdown_it_py-3.0.0-py3-none-any.whl", hash = "sha256:355216845c60bd96232cd8d8c40e8f9765cc86f46880e43a8fd22dc1a1a8cab1"},
+]
+
+[package.dependencies]
+mdurl = ">=0.1,<1.0"
+
+[package.extras]
+benchmarking = ["psutil", "pytest", "pytest-benchmark"]
+code-style = ["pre-commit (>=3.0,<4.0)"]
+compare = ["commonmark (>=0.9,<1.0)", "markdown (>=3.4,<4.0)", "mistletoe (>=1.0,<2.0)", "mistune (>=2.0,<3.0)", "panflute (>=2.3,<3.0)"]
+linkify = ["linkify-it-py (>=1,<3)"]
+plugins = ["mdit-py-plugins"]
+profiling = ["gprof2dot"]
+rtd = ["jupyter_sphinx", "mdit-py-plugins", "myst-parser", "pyyaml", "sphinx", "sphinx-copybutton", "sphinx-design", "sphinx_book_theme"]
+testing = ["coverage", "pytest", "pytest-cov", "pytest-regressions"]
+
[[package]]
name = "markupsafe"
version = "2.1.5"
@@ -1776,18 +1752,48 @@ files = [
[[package]]
name = "matplotlib-inline"
-version = "0.1.6"
+version = "0.1.7"
description = "Inline Matplotlib backend for Jupyter"
optional = false
-python-versions = ">=3.5"
+python-versions = ">=3.8"
files = [
- {file = "matplotlib-inline-0.1.6.tar.gz", hash = "sha256:f887e5f10ba98e8d2b150ddcf4702c1e5f8b3a20005eb0f74bfdbd360ee6f304"},
- {file = "matplotlib_inline-0.1.6-py3-none-any.whl", hash = "sha256:f1f41aab5328aa5aaea9b16d083b128102f8712542f819fe7e6a420ff581b311"},
+ {file = "matplotlib_inline-0.1.7-py3-none-any.whl", hash = "sha256:df192d39a4ff8f21b1895d72e6a13f5fcc5099f00fa84384e0ea28c2cc0653ca"},
+ {file = "matplotlib_inline-0.1.7.tar.gz", hash = "sha256:8423b23ec666be3d16e16b60bdd8ac4e86e840ebd1dd11a30b9f117f2fa0ab90"},
]
[package.dependencies]
traitlets = "*"
+[[package]]
+name = "mdit-py-plugins"
+version = "0.4.0"
+description = "Collection of plugins for markdown-it-py"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mdit_py_plugins-0.4.0-py3-none-any.whl", hash = "sha256:b51b3bb70691f57f974e257e367107857a93b36f322a9e6d44ca5bf28ec2def9"},
+ {file = "mdit_py_plugins-0.4.0.tar.gz", hash = "sha256:d8ab27e9aed6c38aa716819fedfde15ca275715955f8a185a8e1cf90fb1d2c1b"},
+]
+
+[package.dependencies]
+markdown-it-py = ">=1.0.0,<4.0.0"
+
+[package.extras]
+code-style = ["pre-commit"]
+rtd = ["myst-parser", "sphinx-book-theme"]
+testing = ["coverage", "pytest", "pytest-cov", "pytest-regressions"]
+
+[[package]]
+name = "mdurl"
+version = "0.1.2"
+description = "Markdown URL utilities"
+optional = false
+python-versions = ">=3.7"
+files = [
+ {file = "mdurl-0.1.2-py3-none-any.whl", hash = "sha256:84008a41e51615a49fc9966191ff91509e3c40b939176e643fd50a5c2196b8f8"},
+ {file = "mdurl-0.1.2.tar.gz", hash = "sha256:bb413d29f5eea38f31dd4754dd7377d4465116fb207585f97bf925588687c1ba"},
+]
+
[[package]]
name = "mergedeep"
version = "1.3.4"
@@ -1801,45 +1807,196 @@ files = [
[[package]]
name = "mistune"
-version = "0.8.4"
-description = "The fastest markdown parser in pure Python"
+version = "3.0.2"
+description = "A sane and fast Markdown parser with useful plugins and renderers"
optional = false
-python-versions = "*"
+python-versions = ">=3.7"
files = [
- {file = "mistune-0.8.4-py2.py3-none-any.whl", hash = "sha256:88a1051873018da288eee8538d476dffe1262495144b33ecb586c4ab266bb8d4"},
- {file = "mistune-0.8.4.tar.gz", hash = "sha256:59a3429db53c50b5c6bcc8a07f8848cb00d7dc8bdb431a4ab41920d201d4756e"},
+ {file = "mistune-3.0.2-py3-none-any.whl", hash = "sha256:71481854c30fdbc938963d3605b72501f5c10a9320ecd412c121c163a1c7d205"},
+ {file = "mistune-3.0.2.tar.gz", hash = "sha256:fc7f93ded930c92394ef2cb6f04a8aabab4117a91449e72dcc8dfa646a508be8"},
]
[[package]]
name = "mkdocs"
-version = "1.5.3"
+version = "1.6.0"
description = "Project documentation with Markdown."
optional = false
-python-versions = ">=3.7"
+python-versions = ">=3.8"
files = [
- {file = "mkdocs-1.5.3-py3-none-any.whl", hash = "sha256:3b3a78e736b31158d64dbb2f8ba29bd46a379d0c6e324c2246c3bc3d2189cfc1"},
- {file = "mkdocs-1.5.3.tar.gz", hash = "sha256:eb7c99214dcb945313ba30426c2451b735992c73c2e10838f76d09e39ff4d0e2"},
+ {file = "mkdocs-1.6.0-py3-none-any.whl", hash = "sha256:1eb5cb7676b7d89323e62b56235010216319217d4af5ddc543a91beb8d125ea7"},
+ {file = "mkdocs-1.6.0.tar.gz", hash = "sha256:a73f735824ef83a4f3bcb7a231dcab23f5a838f88b7efc54a0eef5fbdbc3c512"},
]
[package.dependencies]
click = ">=7.0"
colorama = {version = ">=0.4", markers = "platform_system == \"Windows\""}
ghp-import = ">=1.0"
-importlib-metadata = {version = ">=4.3", markers = "python_version < \"3.10\""}
+importlib-metadata = {version = ">=4.4", markers = "python_version < \"3.10\""}
jinja2 = ">=2.11.1"
-markdown = ">=3.2.1"
+markdown = ">=3.3.6"
markupsafe = ">=2.0.1"
mergedeep = ">=1.3.4"
+mkdocs-get-deps = ">=0.2.0"
packaging = ">=20.5"
pathspec = ">=0.11.1"
-platformdirs = ">=2.2.0"
pyyaml = ">=5.1"
pyyaml-env-tag = ">=0.1"
watchdog = ">=2.0"
[package.extras]
i18n = ["babel (>=2.9.0)"]
-min-versions = ["babel (==2.9.0)", "click (==7.0)", "colorama (==0.4)", "ghp-import (==1.0)", "importlib-metadata (==4.3)", "jinja2 (==2.11.1)", "markdown (==3.2.1)", "markupsafe (==2.0.1)", "mergedeep (==1.3.4)", "packaging (==20.5)", "pathspec (==0.11.1)", "platformdirs (==2.2.0)", "pyyaml (==5.1)", "pyyaml-env-tag (==0.1)", "typing-extensions (==3.10)", "watchdog (==2.0)"]
+min-versions = ["babel (==2.9.0)", "click (==7.0)", "colorama (==0.4)", "ghp-import (==1.0)", "importlib-metadata (==4.4)", "jinja2 (==2.11.1)", "markdown (==3.3.6)", "markupsafe (==2.0.1)", "mergedeep (==1.3.4)", "mkdocs-get-deps (==0.2.0)", "packaging (==20.5)", "pathspec (==0.11.1)", "pyyaml (==5.1)", "pyyaml-env-tag (==0.1)", "watchdog (==2.0)"]
+
+[[package]]
+name = "mkdocs-autorefs"
+version = "1.0.1"
+description = "Automatically link across pages in MkDocs."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocs_autorefs-1.0.1-py3-none-any.whl", hash = "sha256:aacdfae1ab197780fb7a2dac92ad8a3d8f7ca8049a9cbe56a4218cd52e8da570"},
+ {file = "mkdocs_autorefs-1.0.1.tar.gz", hash = "sha256:f684edf847eced40b570b57846b15f0bf57fb93ac2c510450775dcf16accb971"},
+]
+
+[package.dependencies]
+Markdown = ">=3.3"
+markupsafe = ">=2.0.1"
+mkdocs = ">=1.1"
+
+[[package]]
+name = "mkdocs-get-deps"
+version = "0.2.0"
+description = "MkDocs extension that lists all dependencies according to a mkdocs.yml file"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocs_get_deps-0.2.0-py3-none-any.whl", hash = "sha256:2bf11d0b133e77a0dd036abeeb06dec8775e46efa526dc70667d8863eefc6134"},
+ {file = "mkdocs_get_deps-0.2.0.tar.gz", hash = "sha256:162b3d129c7fad9b19abfdcb9c1458a651628e4b1dea628ac68790fb3061c60c"},
+]
+
+[package.dependencies]
+importlib-metadata = {version = ">=4.3", markers = "python_version < \"3.10\""}
+mergedeep = ">=1.3.4"
+platformdirs = ">=2.2.0"
+pyyaml = ">=5.1"
+
+[[package]]
+name = "mkdocs-include-markdown-plugin"
+version = "6.0.5"
+description = "Mkdocs Markdown includer plugin."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocs_include_markdown_plugin-6.0.5-py3-none-any.whl", hash = "sha256:db41aa1937a618afa3497616f457d4e51d9123b13b2034bb15505ff9ce061f86"},
+ {file = "mkdocs_include_markdown_plugin-6.0.5.tar.gz", hash = "sha256:ad10779cf0dc4ff180aaa0079163271877b3c2fd31e36d5579854fe1d4b0d1ae"},
+]
+
+[package.dependencies]
+mkdocs = ">=1.4"
+wcmatch = ">=8,<9"
+
+[package.extras]
+cache = ["platformdirs"]
+
+[[package]]
+name = "mkdocs-jupyter"
+version = "0.24.7"
+description = "Use Jupyter in mkdocs websites"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocs_jupyter-0.24.7-py3-none-any.whl", hash = "sha256:893d04bea1e007479a46e4e72852cd4d280c4d358ce4a0445250f3f80c639723"},
+]
+
+[package.dependencies]
+ipykernel = ">6.0.0,<7.0.0"
+jupytext = ">1.13.8,<2"
+mkdocs = ">=1.4.0,<2"
+mkdocs-material = ">9.0.0"
+nbconvert = ">=7.2.9,<8"
+pygments = ">2.12.0"
+
+[[package]]
+name = "mkdocs-material"
+version = "9.5.19"
+description = "Documentation that simply works"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocs_material-9.5.19-py3-none-any.whl", hash = "sha256:ea96e150b6c95f5e4ffe47d78bb712c7bacdd91d2a0bec47f46b6fa0705a86ec"},
+ {file = "mkdocs_material-9.5.19.tar.gz", hash = "sha256:7473e06e17e23af608a30ef583fdde8f36389dd3ef56b1d503eed54c89c9618c"},
+]
+
+[package.dependencies]
+babel = ">=2.10,<3.0"
+colorama = ">=0.4,<1.0"
+jinja2 = ">=3.0,<4.0"
+markdown = ">=3.2,<4.0"
+mkdocs = ">=1.6,<2.0"
+mkdocs-material-extensions = ">=1.3,<2.0"
+paginate = ">=0.5,<1.0"
+pygments = ">=2.16,<3.0"
+pymdown-extensions = ">=10.2,<11.0"
+regex = ">=2022.4"
+requests = ">=2.26,<3.0"
+
+[package.extras]
+git = ["mkdocs-git-committers-plugin-2 (>=1.1,<2.0)", "mkdocs-git-revision-date-localized-plugin (>=1.2.4,<2.0)"]
+imaging = ["cairosvg (>=2.6,<3.0)", "pillow (>=10.2,<11.0)"]
+recommended = ["mkdocs-minify-plugin (>=0.7,<1.0)", "mkdocs-redirects (>=1.2,<2.0)", "mkdocs-rss-plugin (>=1.6,<2.0)"]
+
+[[package]]
+name = "mkdocs-material-extensions"
+version = "1.3.1"
+description = "Extension pack for Python Markdown and MkDocs Material."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocs_material_extensions-1.3.1-py3-none-any.whl", hash = "sha256:adff8b62700b25cb77b53358dad940f3ef973dd6db797907c49e3c2ef3ab4e31"},
+ {file = "mkdocs_material_extensions-1.3.1.tar.gz", hash = "sha256:10c9511cea88f568257f960358a467d12b970e1f7b2c0e5fb2bb48cab1928443"},
+]
+
+[[package]]
+name = "mkdocstrings"
+version = "0.23.0"
+description = "Automatic documentation from sources, for MkDocs."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocstrings-0.23.0-py3-none-any.whl", hash = "sha256:051fa4014dfcd9ed90254ae91de2dbb4f24e166347dae7be9a997fe16316c65e"},
+ {file = "mkdocstrings-0.23.0.tar.gz", hash = "sha256:d9c6a37ffbe7c14a7a54ef1258c70b8d394e6a33a1c80832bce40b9567138d1c"},
+]
+
+[package.dependencies]
+importlib-metadata = {version = ">=4.6", markers = "python_version < \"3.10\""}
+Jinja2 = ">=2.11.1"
+Markdown = ">=3.3"
+MarkupSafe = ">=1.1"
+mkdocs = ">=1.2"
+mkdocs-autorefs = ">=0.3.1"
+mkdocstrings-python = {version = ">=0.5.2", optional = true, markers = "extra == \"python\""}
+pymdown-extensions = ">=6.3"
+typing-extensions = {version = ">=4.1", markers = "python_version < \"3.10\""}
+
+[package.extras]
+crystal = ["mkdocstrings-crystal (>=0.3.4)"]
+python = ["mkdocstrings-python (>=0.5.2)"]
+python-legacy = ["mkdocstrings-python-legacy (>=0.2.1)"]
+
+[[package]]
+name = "mkdocstrings-python"
+version = "1.8.0"
+description = "A Python handler for mkdocstrings."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocstrings_python-1.8.0-py3-none-any.whl", hash = "sha256:4209970cc90bec194568682a535848a8d8489516c6ed4adbe58bbc67b699ca9d"},
+ {file = "mkdocstrings_python-1.8.0.tar.gz", hash = "sha256:1488bddf50ee42c07d9a488dddc197f8e8999c2899687043ec5dd1643d057192"},
+]
+
+[package.dependencies]
+griffe = ">=0.37"
+mkdocstrings = ">=0.20"
[[package]]
name = "mock"
@@ -1861,7 +2018,7 @@ test = ["pytest", "pytest-cov"]
name = "multidict"
version = "6.0.5"
description = "multidict implementation"
-optional = false
+optional = true
python-versions = ">=3.7"
files = [
{file = "multidict-6.0.5-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:228b644ae063c10e7f324ab1ab6b548bdf6f8b47f3ec234fef1093bc2735e5f9"},
@@ -1980,56 +2137,57 @@ test = ["flaky", "ipykernel (>=6.19.3)", "ipython", "ipywidgets", "nbconvert (>=
[[package]]
name = "nbconvert"
-version = "6.5.4"
-description = "Converting Jupyter Notebooks"
+version = "7.16.3"
+description = "Converting Jupyter Notebooks (.ipynb files) to other formats. Output formats include asciidoc, html, latex, markdown, pdf, py, rst, script. nbconvert can be used both as a Python library (`import nbconvert`) or as a command line tool (invoked as `jupyter nbconvert ...`)."
optional = false
-python-versions = ">=3.7"
+python-versions = ">=3.8"
files = [
- {file = "nbconvert-6.5.4-py3-none-any.whl", hash = "sha256:d679a947f849a966cbbd0bf6e7fedcfdb64be3b20ce7cef11ad55c13f5820e19"},
- {file = "nbconvert-6.5.4.tar.gz", hash = "sha256:9e3c7c6d491374cbdd5f35d268c05809357716d346f4573186bbeab32ee50bc1"},
+ {file = "nbconvert-7.16.3-py3-none-any.whl", hash = "sha256:ddeff14beeeedf3dd0bc506623e41e4507e551736de59df69a91f86700292b3b"},
+ {file = "nbconvert-7.16.3.tar.gz", hash = "sha256:a6733b78ce3d47c3f85e504998495b07e6ea9cf9bf6ec1c98dda63ec6ad19142"},
]
[package.dependencies]
beautifulsoup4 = "*"
-bleach = "*"
+bleach = "!=5.0.0"
defusedxml = "*"
-entrypoints = ">=0.2.2"
+importlib-metadata = {version = ">=3.6", markers = "python_version < \"3.10\""}
jinja2 = ">=3.0"
jupyter-core = ">=4.7"
jupyterlab-pygments = "*"
-lxml = "*"
-MarkupSafe = ">=2.0"
-mistune = ">=0.8.1,<2"
+markupsafe = ">=2.0"
+mistune = ">=2.0.3,<4"
nbclient = ">=0.5.0"
-nbformat = ">=5.1"
+nbformat = ">=5.7"
packaging = "*"
pandocfilters = ">=1.4.1"
pygments = ">=2.4.1"
tinycss2 = "*"
-traitlets = ">=5.0"
+traitlets = ">=5.1"
[package.extras]
-all = ["ipykernel", "ipython", "ipywidgets (>=7)", "nbsphinx (>=0.2.12)", "pre-commit", "pyppeteer (>=1,<1.1)", "pytest", "pytest-cov", "pytest-dependency", "sphinx (>=1.5.1)", "sphinx-rtd-theme", "tornado (>=6.1)"]
-docs = ["ipython", "nbsphinx (>=0.2.12)", "sphinx (>=1.5.1)", "sphinx-rtd-theme"]
+all = ["nbconvert[docs,qtpdf,serve,test,webpdf]"]
+docs = ["ipykernel", "ipython", "myst-parser", "nbsphinx (>=0.2.12)", "pydata-sphinx-theme", "sphinx (==5.0.2)", "sphinxcontrib-spelling"]
+qtpdf = ["nbconvert[qtpng]"]
+qtpng = ["pyqtwebengine (>=5.15)"]
serve = ["tornado (>=6.1)"]
-test = ["ipykernel", "ipywidgets (>=7)", "pre-commit", "pyppeteer (>=1,<1.1)", "pytest", "pytest-cov", "pytest-dependency"]
-webpdf = ["pyppeteer (>=1,<1.1)"]
+test = ["flaky", "ipykernel", "ipywidgets (>=7.5)", "pytest (>=7)"]
+webpdf = ["playwright"]
[[package]]
name = "nbformat"
-version = "5.10.3"
+version = "5.10.4"
description = "The Jupyter Notebook format"
optional = false
python-versions = ">=3.8"
files = [
- {file = "nbformat-5.10.3-py3-none-any.whl", hash = "sha256:d9476ca28676799af85385f409b49d95e199951477a159a576ef2a675151e5e8"},
- {file = "nbformat-5.10.3.tar.gz", hash = "sha256:60ed5e910ef7c6264b87d644f276b1b49e24011930deef54605188ddeb211685"},
+ {file = "nbformat-5.10.4-py3-none-any.whl", hash = "sha256:3b48d6c8fbca4b299bf3982ea7db1af21580e4fec269ad087b9e81588891200b"},
+ {file = "nbformat-5.10.4.tar.gz", hash = "sha256:322168b14f937a5d11362988ecac2a4952d3d8e3a2cbeb2319584631226d5b3a"},
]
[package.dependencies]
-fastjsonschema = "*"
+fastjsonschema = ">=2.15"
jsonschema = ">=2.6"
-jupyter-core = "*"
+jupyter-core = ">=4.12,<5.0.dev0 || >=5.1.dev0"
traitlets = ">=5.1"
[package.extras]
@@ -2055,6 +2213,17 @@ nbformat = "*"
sphinx = ">=1.8"
traitlets = ">=5"
+[[package]]
+name = "nest-asyncio"
+version = "1.6.0"
+description = "Patch asyncio to allow nested event loops"
+optional = false
+python-versions = ">=3.5"
+files = [
+ {file = "nest_asyncio-1.6.0-py3-none-any.whl", hash = "sha256:87af6efd6b5e897c81050477ef65c62e2b2f35d51703cae01aff2905b1852e1c"},
+ {file = "nest_asyncio-1.6.0.tar.gz", hash = "sha256:6f172d5449aca15afd6c646851f4e31e02c598d553a667e38cafa997cfec55fe"},
+]
+
[[package]]
name = "nodeenv"
version = "1.8.0"
@@ -2145,6 +2314,16 @@ files = [
{file = "packaging-24.0.tar.gz", hash = "sha256:eb82c5e3e56209074766e6885bb04b8c38a0c015d0a30036ebe7ece34c9989e9"},
]
+[[package]]
+name = "paginate"
+version = "0.5.6"
+description = "Divides large result sets into pages for easier browsing"
+optional = false
+python-versions = "*"
+files = [
+ {file = "paginate-0.5.6.tar.gz", hash = "sha256:5e6007b6a9398177a7e1648d04fdd9f8c9766a1a945bceac82f1929e8c78af2d"},
+]
+
[[package]]
name = "pandas"
version = "2.0.3"
@@ -2236,18 +2415,18 @@ files = [
[[package]]
name = "parso"
-version = "0.8.3"
+version = "0.8.4"
description = "A Python Parser"
optional = false
python-versions = ">=3.6"
files = [
- {file = "parso-0.8.3-py2.py3-none-any.whl", hash = "sha256:c001d4636cd3aecdaf33cbb40aebb59b094be2a74c556778ef5576c175e19e75"},
- {file = "parso-0.8.3.tar.gz", hash = "sha256:8c07be290bb59f03588915921e29e8a50002acaf2cdc5fa0e0114f91709fafa0"},
+ {file = "parso-0.8.4-py2.py3-none-any.whl", hash = "sha256:a418670a20291dacd2dddc80c377c5c3791378ee1e8d12bffc35420643d43f18"},
+ {file = "parso-0.8.4.tar.gz", hash = "sha256:eb3a7b58240fb99099a345571deecc0f9540ea5f4dd2fe14c2a99d6b281ab92d"},
]
[package.extras]
-qa = ["flake8 (==3.8.3)", "mypy (==0.782)"]
-testing = ["docopt", "pytest (<6.0.0)"]
+qa = ["flake8 (==5.0.4)", "mypy (==0.971)", "types-setuptools (==67.2.0.1)"]
+testing = ["docopt", "pytest"]
[[package]]
name = "pathspec"
@@ -2298,28 +2477,29 @@ files = [
[[package]]
name = "platformdirs"
-version = "4.2.0"
-description = "A small Python package for determining appropriate platform-specific dirs, e.g. a \"user data dir\"."
+version = "4.2.1"
+description = "A small Python package for determining appropriate platform-specific dirs, e.g. a `user data dir`."
optional = false
python-versions = ">=3.8"
files = [
- {file = "platformdirs-4.2.0-py3-none-any.whl", hash = "sha256:0614df2a2f37e1a662acbd8e2b25b92ccf8632929bc6d43467e17fe89c75e068"},
- {file = "platformdirs-4.2.0.tar.gz", hash = "sha256:ef0cc731df711022c174543cb70a9b5bd22e5a9337c8624ef2c2ceb8ddad8768"},
+ {file = "platformdirs-4.2.1-py3-none-any.whl", hash = "sha256:17d5a1161b3fd67b390023cb2d3b026bbd40abde6fdb052dfbd3a29c3ba22ee1"},
+ {file = "platformdirs-4.2.1.tar.gz", hash = "sha256:031cd18d4ec63ec53e82dceaac0417d218a6863f7745dfcc9efe7793b7039bdf"},
]
[package.extras]
docs = ["furo (>=2023.9.10)", "proselint (>=0.13)", "sphinx (>=7.2.6)", "sphinx-autodoc-typehints (>=1.25.2)"]
test = ["appdirs (==1.4.4)", "covdefaults (>=2.3)", "pytest (>=7.4.3)", "pytest-cov (>=4.1)", "pytest-mock (>=3.12)"]
+type = ["mypy (>=1.8)"]
[[package]]
name = "pluggy"
-version = "1.4.0"
+version = "1.5.0"
description = "plugin and hook calling mechanisms for python"
optional = false
python-versions = ">=3.8"
files = [
- {file = "pluggy-1.4.0-py3-none-any.whl", hash = "sha256:7db9f7b503d67d1c5b95f59773ebb58a8c1c288129a88665838012cfb07b8981"},
- {file = "pluggy-1.4.0.tar.gz", hash = "sha256:8c85c2876142a764e5b7548e7d9a0e0ddb46f5185161049a79b7e974454223be"},
+ {file = "pluggy-1.5.0-py3-none-any.whl", hash = "sha256:44e1ad92c8ca002de6377e165f3e0f1be63266ab4d554740532335b9d75ea669"},
+ {file = "pluggy-1.5.0.tar.gz", hash = "sha256:2cffa88e94fdc978c4c574f15f9e59b7f4201d439195c3715ca9e2486f1d0cf1"},
]
[package.extras]
@@ -2358,6 +2538,34 @@ files = [
[package.dependencies]
wcwidth = "*"
+[[package]]
+name = "psutil"
+version = "5.9.8"
+description = "Cross-platform lib for process and system monitoring in Python."
+optional = false
+python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*, !=3.3.*, !=3.4.*, !=3.5.*"
+files = [
+ {file = "psutil-5.9.8-cp27-cp27m-macosx_10_9_x86_64.whl", hash = "sha256:26bd09967ae00920df88e0352a91cff1a78f8d69b3ecabbfe733610c0af486c8"},
+ {file = "psutil-5.9.8-cp27-cp27m-manylinux2010_i686.whl", hash = "sha256:05806de88103b25903dff19bb6692bd2e714ccf9e668d050d144012055cbca73"},
+ {file = "psutil-5.9.8-cp27-cp27m-manylinux2010_x86_64.whl", hash = "sha256:611052c4bc70432ec770d5d54f64206aa7203a101ec273a0cd82418c86503bb7"},
+ {file = "psutil-5.9.8-cp27-cp27mu-manylinux2010_i686.whl", hash = "sha256:50187900d73c1381ba1454cf40308c2bf6f34268518b3f36a9b663ca87e65e36"},
+ {file = "psutil-5.9.8-cp27-cp27mu-manylinux2010_x86_64.whl", hash = "sha256:02615ed8c5ea222323408ceba16c60e99c3f91639b07da6373fb7e6539abc56d"},
+ {file = "psutil-5.9.8-cp27-none-win32.whl", hash = "sha256:36f435891adb138ed3c9e58c6af3e2e6ca9ac2f365efe1f9cfef2794e6c93b4e"},
+ {file = "psutil-5.9.8-cp27-none-win_amd64.whl", hash = "sha256:bd1184ceb3f87651a67b2708d4c3338e9b10c5df903f2e3776b62303b26cb631"},
+ {file = "psutil-5.9.8-cp36-abi3-macosx_10_9_x86_64.whl", hash = "sha256:aee678c8720623dc456fa20659af736241f575d79429a0e5e9cf88ae0605cc81"},
+ {file = "psutil-5.9.8-cp36-abi3-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:8cb6403ce6d8e047495a701dc7c5bd788add903f8986d523e3e20b98b733e421"},
+ {file = "psutil-5.9.8-cp36-abi3-manylinux_2_12_x86_64.manylinux2010_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:d06016f7f8625a1825ba3732081d77c94589dca78b7a3fc072194851e88461a4"},
+ {file = "psutil-5.9.8-cp36-cp36m-win32.whl", hash = "sha256:7d79560ad97af658a0f6adfef8b834b53f64746d45b403f225b85c5c2c140eee"},
+ {file = "psutil-5.9.8-cp36-cp36m-win_amd64.whl", hash = "sha256:27cc40c3493bb10de1be4b3f07cae4c010ce715290a5be22b98493509c6299e2"},
+ {file = "psutil-5.9.8-cp37-abi3-win32.whl", hash = "sha256:bc56c2a1b0d15aa3eaa5a60c9f3f8e3e565303b465dbf57a1b730e7a2b9844e0"},
+ {file = "psutil-5.9.8-cp37-abi3-win_amd64.whl", hash = "sha256:8db4c1b57507eef143a15a6884ca10f7c73876cdf5d51e713151c1236a0e68cf"},
+ {file = "psutil-5.9.8-cp38-abi3-macosx_11_0_arm64.whl", hash = "sha256:d16bbddf0693323b8c6123dd804100241da461e41d6e332fb0ba6058f630f8c8"},
+ {file = "psutil-5.9.8.tar.gz", hash = "sha256:6be126e3225486dff286a8fb9a06246a5253f4c7c53b475ea5f5ac934e64194c"},
+]
+
+[package.extras]
+test = ["enum34", "ipaddress", "mock", "pywin32", "wmi"]
+
[[package]]
name = "ptyprocess"
version = "0.7.0"
@@ -2385,61 +2593,61 @@ tests = ["pytest"]
[[package]]
name = "pyarrow"
-version = "15.0.2"
+version = "16.0.0"
description = "Python library for Apache Arrow"
optional = false
python-versions = ">=3.8"
files = [
- {file = "pyarrow-15.0.2-cp310-cp310-macosx_10_15_x86_64.whl", hash = "sha256:88b340f0a1d05b5ccc3d2d986279045655b1fe8e41aba6ca44ea28da0d1455d8"},
- {file = "pyarrow-15.0.2-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:eaa8f96cecf32da508e6c7f69bb8401f03745c050c1dd42ec2596f2e98deecac"},
- {file = "pyarrow-15.0.2-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:23c6753ed4f6adb8461e7c383e418391b8d8453c5d67e17f416c3a5d5709afbd"},
- {file = "pyarrow-15.0.2-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:f639c059035011db8c0497e541a8a45d98a58dbe34dc8fadd0ef128f2cee46e5"},
- {file = "pyarrow-15.0.2-cp310-cp310-manylinux_2_28_aarch64.whl", hash = "sha256:290e36a59a0993e9a5224ed2fb3e53375770f07379a0ea03ee2fce2e6d30b423"},
- {file = "pyarrow-15.0.2-cp310-cp310-manylinux_2_28_x86_64.whl", hash = "sha256:06c2bb2a98bc792f040bef31ad3e9be6a63d0cb39189227c08a7d955db96816e"},
- {file = "pyarrow-15.0.2-cp310-cp310-win_amd64.whl", hash = "sha256:f7a197f3670606a960ddc12adbe8075cea5f707ad7bf0dffa09637fdbb89f76c"},
- {file = "pyarrow-15.0.2-cp311-cp311-macosx_10_15_x86_64.whl", hash = "sha256:5f8bc839ea36b1f99984c78e06e7a06054693dc2af8920f6fb416b5bca9944e4"},
- {file = "pyarrow-15.0.2-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:f5e81dfb4e519baa6b4c80410421528c214427e77ca0ea9461eb4097c328fa33"},
- {file = "pyarrow-15.0.2-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3a4f240852b302a7af4646c8bfe9950c4691a419847001178662a98915fd7ee7"},
- {file = "pyarrow-15.0.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:4e7d9cfb5a1e648e172428c7a42b744610956f3b70f524aa3a6c02a448ba853e"},
- {file = "pyarrow-15.0.2-cp311-cp311-manylinux_2_28_aarch64.whl", hash = "sha256:2d4f905209de70c0eb5b2de6763104d5a9a37430f137678edfb9a675bac9cd98"},
- {file = "pyarrow-15.0.2-cp311-cp311-manylinux_2_28_x86_64.whl", hash = "sha256:90adb99e8ce5f36fbecbbc422e7dcbcbed07d985eed6062e459e23f9e71fd197"},
- {file = "pyarrow-15.0.2-cp311-cp311-win_amd64.whl", hash = "sha256:b116e7fd7889294cbd24eb90cd9bdd3850be3738d61297855a71ac3b8124ee38"},
- {file = "pyarrow-15.0.2-cp312-cp312-macosx_10_15_x86_64.whl", hash = "sha256:25335e6f1f07fdaa026a61c758ee7d19ce824a866b27bba744348fa73bb5a440"},
- {file = "pyarrow-15.0.2-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:90f19e976d9c3d8e73c80be84ddbe2f830b6304e4c576349d9360e335cd627fc"},
- {file = "pyarrow-15.0.2-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:a22366249bf5fd40ddacc4f03cd3160f2d7c247692945afb1899bab8a140ddfb"},
- {file = "pyarrow-15.0.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:c2a335198f886b07e4b5ea16d08ee06557e07db54a8400cc0d03c7f6a22f785f"},
- {file = "pyarrow-15.0.2-cp312-cp312-manylinux_2_28_aarch64.whl", hash = "sha256:3e6d459c0c22f0b9c810a3917a1de3ee704b021a5fb8b3bacf968eece6df098f"},
- {file = "pyarrow-15.0.2-cp312-cp312-manylinux_2_28_x86_64.whl", hash = "sha256:033b7cad32198754d93465dcfb71d0ba7cb7cd5c9afd7052cab7214676eec38b"},
- {file = "pyarrow-15.0.2-cp312-cp312-win_amd64.whl", hash = "sha256:29850d050379d6e8b5a693098f4de7fd6a2bea4365bfd073d7c57c57b95041ee"},
- {file = "pyarrow-15.0.2-cp38-cp38-macosx_10_15_x86_64.whl", hash = "sha256:7167107d7fb6dcadb375b4b691b7e316f4368f39f6f45405a05535d7ad5e5058"},
- {file = "pyarrow-15.0.2-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:e85241b44cc3d365ef950432a1b3bd44ac54626f37b2e3a0cc89c20e45dfd8bf"},
- {file = "pyarrow-15.0.2-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:248723e4ed3255fcd73edcecc209744d58a9ca852e4cf3d2577811b6d4b59818"},
- {file = "pyarrow-15.0.2-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:3ff3bdfe6f1b81ca5b73b70a8d482d37a766433823e0c21e22d1d7dde76ca33f"},
- {file = "pyarrow-15.0.2-cp38-cp38-manylinux_2_28_aarch64.whl", hash = "sha256:f3d77463dee7e9f284ef42d341689b459a63ff2e75cee2b9302058d0d98fe142"},
- {file = "pyarrow-15.0.2-cp38-cp38-manylinux_2_28_x86_64.whl", hash = "sha256:8c1faf2482fb89766e79745670cbca04e7018497d85be9242d5350cba21357e1"},
- {file = "pyarrow-15.0.2-cp38-cp38-win_amd64.whl", hash = "sha256:28f3016958a8e45a1069303a4a4f6a7d4910643fc08adb1e2e4a7ff056272ad3"},
- {file = "pyarrow-15.0.2-cp39-cp39-macosx_10_15_x86_64.whl", hash = "sha256:89722cb64286ab3d4daf168386f6968c126057b8c7ec3ef96302e81d8cdb8ae4"},
- {file = "pyarrow-15.0.2-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:cd0ba387705044b3ac77b1b317165c0498299b08261d8122c96051024f953cd5"},
- {file = "pyarrow-15.0.2-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ad2459bf1f22b6a5cdcc27ebfd99307d5526b62d217b984b9f5c974651398832"},
- {file = "pyarrow-15.0.2-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:58922e4bfece8b02abf7159f1f53a8f4d9f8e08f2d988109126c17c3bb261f22"},
- {file = "pyarrow-15.0.2-cp39-cp39-manylinux_2_28_aarch64.whl", hash = "sha256:adccc81d3dc0478ea0b498807b39a8d41628fa9210729b2f718b78cb997c7c91"},
- {file = "pyarrow-15.0.2-cp39-cp39-manylinux_2_28_x86_64.whl", hash = "sha256:8bd2baa5fe531571847983f36a30ddbf65261ef23e496862ece83bdceb70420d"},
- {file = "pyarrow-15.0.2-cp39-cp39-win_amd64.whl", hash = "sha256:6669799a1d4ca9da9c7e06ef48368320f5856f36f9a4dd31a11839dda3f6cc8c"},
- {file = "pyarrow-15.0.2.tar.gz", hash = "sha256:9c9bc803cb3b7bfacc1e96ffbfd923601065d9d3f911179d81e72d99fd74a3d9"},
-]
-
-[package.dependencies]
-numpy = ">=1.16.6,<2"
+ {file = "pyarrow-16.0.0-cp310-cp310-macosx_10_15_x86_64.whl", hash = "sha256:22a1fdb1254e5095d629e29cd1ea98ed04b4bbfd8e42cc670a6b639ccc208b60"},
+ {file = "pyarrow-16.0.0-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:574a00260a4ed9d118a14770edbd440b848fcae5a3024128be9d0274dbcaf858"},
+ {file = "pyarrow-16.0.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:c0815d0ddb733b8c1b53a05827a91f1b8bde6240f3b20bf9ba5d650eb9b89cdf"},
+ {file = "pyarrow-16.0.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:df0080339387b5d30de31e0a149c0c11a827a10c82f0c67d9afae3981d1aabb7"},
+ {file = "pyarrow-16.0.0-cp310-cp310-manylinux_2_28_aarch64.whl", hash = "sha256:edf38cce0bf0dcf726e074159c60516447e4474904c0033f018c1f33d7dac6c5"},
+ {file = "pyarrow-16.0.0-cp310-cp310-manylinux_2_28_x86_64.whl", hash = "sha256:91d28f9a40f1264eab2af7905a4d95320ac2f287891e9c8b0035f264fe3c3a4b"},
+ {file = "pyarrow-16.0.0-cp310-cp310-win_amd64.whl", hash = "sha256:99af421ee451a78884d7faea23816c429e263bd3618b22d38e7992c9ce2a7ad9"},
+ {file = "pyarrow-16.0.0-cp311-cp311-macosx_10_15_x86_64.whl", hash = "sha256:d22d0941e6c7bafddf5f4c0662e46f2075850f1c044bf1a03150dd9e189427ce"},
+ {file = "pyarrow-16.0.0-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:266ddb7e823f03733c15adc8b5078db2df6980f9aa93d6bb57ece615df4e0ba7"},
+ {file = "pyarrow-16.0.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:5cc23090224b6594f5a92d26ad47465af47c1d9c079dd4a0061ae39551889efe"},
+ {file = "pyarrow-16.0.0-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:56850a0afe9ef37249d5387355449c0f94d12ff7994af88f16803a26d38f2016"},
+ {file = "pyarrow-16.0.0-cp311-cp311-manylinux_2_28_aarch64.whl", hash = "sha256:705db70d3e2293c2f6f8e84874b5b775f690465798f66e94bb2c07bab0a6bb55"},
+ {file = "pyarrow-16.0.0-cp311-cp311-manylinux_2_28_x86_64.whl", hash = "sha256:5448564754c154997bc09e95a44b81b9e31ae918a86c0fcb35c4aa4922756f55"},
+ {file = "pyarrow-16.0.0-cp311-cp311-win_amd64.whl", hash = "sha256:729f7b262aa620c9df8b9967db96c1575e4cfc8c25d078a06968e527b8d6ec05"},
+ {file = "pyarrow-16.0.0-cp312-cp312-macosx_10_15_x86_64.whl", hash = "sha256:fb8065dbc0d051bf2ae2453af0484d99a43135cadabacf0af588a3be81fbbb9b"},
+ {file = "pyarrow-16.0.0-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:20ce707d9aa390593ea93218b19d0eadab56390311cb87aad32c9a869b0e958c"},
+ {file = "pyarrow-16.0.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:5823275c8addbbb50cd4e6a6839952682a33255b447277e37a6f518d6972f4e1"},
+ {file = "pyarrow-16.0.0-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:1ab8b9050752b16a8b53fcd9853bf07d8daf19093533e990085168f40c64d978"},
+ {file = "pyarrow-16.0.0-cp312-cp312-manylinux_2_28_aarch64.whl", hash = "sha256:42e56557bc7c5c10d3e42c3b32f6cff649a29d637e8f4e8b311d334cc4326730"},
+ {file = "pyarrow-16.0.0-cp312-cp312-manylinux_2_28_x86_64.whl", hash = "sha256:2a7abdee4a4a7cfa239e2e8d721224c4b34ffe69a0ca7981354fe03c1328789b"},
+ {file = "pyarrow-16.0.0-cp312-cp312-win_amd64.whl", hash = "sha256:ef2f309b68396bcc5a354106741d333494d6a0d3e1951271849787109f0229a6"},
+ {file = "pyarrow-16.0.0-cp38-cp38-macosx_10_15_x86_64.whl", hash = "sha256:ed66e5217b4526fa3585b5e39b0b82f501b88a10d36bd0d2a4d8aa7b5a48e2df"},
+ {file = "pyarrow-16.0.0-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:cc8814310486f2a73c661ba8354540f17eef51e1b6dd090b93e3419d3a097b3a"},
+ {file = "pyarrow-16.0.0-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3c2f5e239db7ed43e0ad2baf46a6465f89c824cc703f38ef0fde927d8e0955f7"},
+ {file = "pyarrow-16.0.0-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:f293e92d1db251447cb028ae12f7bc47526e4649c3a9924c8376cab4ad6b98bd"},
+ {file = "pyarrow-16.0.0-cp38-cp38-manylinux_2_28_aarch64.whl", hash = "sha256:dd9334a07b6dc21afe0857aa31842365a62eca664e415a3f9536e3a8bb832c07"},
+ {file = "pyarrow-16.0.0-cp38-cp38-manylinux_2_28_x86_64.whl", hash = "sha256:d91073d1e2fef2c121154680e2ba7e35ecf8d4969cc0af1fa6f14a8675858159"},
+ {file = "pyarrow-16.0.0-cp38-cp38-win_amd64.whl", hash = "sha256:71d52561cd7aefd22cf52538f262850b0cc9e4ec50af2aaa601da3a16ef48877"},
+ {file = "pyarrow-16.0.0-cp39-cp39-macosx_10_15_x86_64.whl", hash = "sha256:b93c9a50b965ee0bf4fef65e53b758a7e8dcc0c2d86cebcc037aaaf1b306ecc0"},
+ {file = "pyarrow-16.0.0-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:d831690844706e374c455fba2fb8cfcb7b797bfe53ceda4b54334316e1ac4fa4"},
+ {file = "pyarrow-16.0.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:35692ce8ad0b8c666aa60f83950957096d92f2a9d8d7deda93fb835e6053307e"},
+ {file = "pyarrow-16.0.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:9dd3151d098e56f16a8389c1247137f9e4c22720b01c6f3aa6dec29a99b74d80"},
+ {file = "pyarrow-16.0.0-cp39-cp39-manylinux_2_28_aarch64.whl", hash = "sha256:bd40467bdb3cbaf2044ed7a6f7f251c8f941c8b31275aaaf88e746c4f3ca4a7a"},
+ {file = "pyarrow-16.0.0-cp39-cp39-manylinux_2_28_x86_64.whl", hash = "sha256:00a1dcb22ad4ceb8af87f7bd30cc3354788776c417f493089e0a0af981bc8d80"},
+ {file = "pyarrow-16.0.0-cp39-cp39-win_amd64.whl", hash = "sha256:fda9a7cebd1b1d46c97b511f60f73a5b766a6de4c5236f144f41a5d5afec1f35"},
+ {file = "pyarrow-16.0.0.tar.gz", hash = "sha256:59bb1f1edbbf4114c72415f039f1359f1a57d166a331c3229788ccbfbb31689a"},
+]
+
+[package.dependencies]
+numpy = ">=1.16.6"
[[package]]
name = "pycparser"
-version = "2.21"
+version = "2.22"
description = "C parser in Python"
optional = false
-python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*, !=3.3.*"
+python-versions = ">=3.8"
files = [
- {file = "pycparser-2.21-py2.py3-none-any.whl", hash = "sha256:8ee45429555515e1f6b185e78100aea234072576aa43ab53aefcae078162fca9"},
- {file = "pycparser-2.21.tar.gz", hash = "sha256:e644fdec12f7872f86c58ff790da456218b10f863970249516d60a5eaca77206"},
+ {file = "pycparser-2.22-py3-none-any.whl", hash = "sha256:c3702b6d3dd8c7abc1afa565d7e63d53a1d0bd86cdc24edd75470f4de499cfcc"},
+ {file = "pycparser-2.22.tar.gz", hash = "sha256:491c8be9c040f5390f5bf44a5b07752bd07f56edf992381b05c701439eec10f6"},
]
[[package]]
@@ -2457,6 +2665,24 @@ files = [
plugins = ["importlib-metadata"]
windows-terminal = ["colorama (>=0.4.6)"]
+[[package]]
+name = "pymdown-extensions"
+version = "10.8"
+description = "Extension pack for Python Markdown."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "pymdown_extensions-10.8-py3-none-any.whl", hash = "sha256:3539003ff0d5e219ba979d2dc961d18fcad5ac259e66c764482e8347b4c0503c"},
+ {file = "pymdown_extensions-10.8.tar.gz", hash = "sha256:91ca336caf414e1e5e0626feca86e145de9f85a3921a7bcbd32890b51738c428"},
+]
+
+[package.dependencies]
+markdown = ">=3.6"
+pyyaml = "*"
+
+[package.extras]
+extra = ["pygments (>=2.12)"]
+
[[package]]
name = "pytest"
version = "8.1.1"
@@ -2649,104 +2875,99 @@ pyyaml = "*"
[[package]]
name = "pyzmq"
-version = "25.1.2"
+version = "26.0.2"
description = "Python bindings for 0MQ"
optional = false
-python-versions = ">=3.6"
+python-versions = ">=3.7"
files = [
- {file = "pyzmq-25.1.2-cp310-cp310-macosx_10_15_universal2.whl", hash = "sha256:e624c789359f1a16f83f35e2c705d07663ff2b4d4479bad35621178d8f0f6ea4"},
- {file = "pyzmq-25.1.2-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:49151b0efece79f6a79d41a461d78535356136ee70084a1c22532fc6383f4ad0"},
- {file = "pyzmq-25.1.2-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d9a5f194cf730f2b24d6af1f833c14c10f41023da46a7f736f48b6d35061e76e"},
- {file = "pyzmq-25.1.2-cp310-cp310-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:faf79a302f834d9e8304fafdc11d0d042266667ac45209afa57e5efc998e3872"},
- {file = "pyzmq-25.1.2-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:7f51a7b4ead28d3fca8dda53216314a553b0f7a91ee8fc46a72b402a78c3e43d"},
- {file = "pyzmq-25.1.2-cp310-cp310-manylinux_2_28_x86_64.whl", hash = "sha256:0ddd6d71d4ef17ba5a87becf7ddf01b371eaba553c603477679ae817a8d84d75"},
- {file = "pyzmq-25.1.2-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:246747b88917e4867e2367b005fc8eefbb4a54b7db363d6c92f89d69abfff4b6"},
- {file = "pyzmq-25.1.2-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:00c48ae2fd81e2a50c3485de1b9d5c7c57cd85dc8ec55683eac16846e57ac979"},
- {file = "pyzmq-25.1.2-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:5a68d491fc20762b630e5db2191dd07ff89834086740f70e978bb2ef2668be08"},
- {file = "pyzmq-25.1.2-cp310-cp310-win32.whl", hash = "sha256:09dfe949e83087da88c4a76767df04b22304a682d6154de2c572625c62ad6886"},
- {file = "pyzmq-25.1.2-cp310-cp310-win_amd64.whl", hash = "sha256:fa99973d2ed20417744fca0073390ad65ce225b546febb0580358e36aa90dba6"},
- {file = "pyzmq-25.1.2-cp311-cp311-macosx_10_15_universal2.whl", hash = "sha256:82544e0e2d0c1811482d37eef297020a040c32e0687c1f6fc23a75b75db8062c"},
- {file = "pyzmq-25.1.2-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:01171fc48542348cd1a360a4b6c3e7d8f46cdcf53a8d40f84db6707a6768acc1"},
- {file = "pyzmq-25.1.2-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:bc69c96735ab501419c432110016329bf0dea8898ce16fab97c6d9106dc0b348"},
- {file = "pyzmq-25.1.2-cp311-cp311-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:3e124e6b1dd3dfbeb695435dff0e383256655bb18082e094a8dd1f6293114642"},
- {file = "pyzmq-25.1.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:7598d2ba821caa37a0f9d54c25164a4fa351ce019d64d0b44b45540950458840"},
- {file = "pyzmq-25.1.2-cp311-cp311-manylinux_2_28_x86_64.whl", hash = "sha256:d1299d7e964c13607efd148ca1f07dcbf27c3ab9e125d1d0ae1d580a1682399d"},
- {file = "pyzmq-25.1.2-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:4e6f689880d5ad87918430957297c975203a082d9a036cc426648fcbedae769b"},
- {file = "pyzmq-25.1.2-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:cc69949484171cc961e6ecd4a8911b9ce7a0d1f738fcae717177c231bf77437b"},
- {file = "pyzmq-25.1.2-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:9880078f683466b7f567b8624bfc16cad65077be046b6e8abb53bed4eeb82dd3"},
- {file = "pyzmq-25.1.2-cp311-cp311-win32.whl", hash = "sha256:4e5837af3e5aaa99a091302df5ee001149baff06ad22b722d34e30df5f0d9097"},
- {file = "pyzmq-25.1.2-cp311-cp311-win_amd64.whl", hash = "sha256:25c2dbb97d38b5ac9fd15586e048ec5eb1e38f3d47fe7d92167b0c77bb3584e9"},
- {file = "pyzmq-25.1.2-cp312-cp312-macosx_10_15_universal2.whl", hash = "sha256:11e70516688190e9c2db14fcf93c04192b02d457b582a1f6190b154691b4c93a"},
- {file = "pyzmq-25.1.2-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:313c3794d650d1fccaaab2df942af9f2c01d6217c846177cfcbc693c7410839e"},
- {file = "pyzmq-25.1.2-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:1b3cbba2f47062b85fe0ef9de5b987612140a9ba3a9c6d2543c6dec9f7c2ab27"},
- {file = "pyzmq-25.1.2-cp312-cp312-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:fc31baa0c32a2ca660784d5af3b9487e13b61b3032cb01a115fce6588e1bed30"},
- {file = "pyzmq-25.1.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:02c9087b109070c5ab0b383079fa1b5f797f8d43e9a66c07a4b8b8bdecfd88ee"},
- {file = "pyzmq-25.1.2-cp312-cp312-manylinux_2_28_x86_64.whl", hash = "sha256:f8429b17cbb746c3e043cb986328da023657e79d5ed258b711c06a70c2ea7537"},
- {file = "pyzmq-25.1.2-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:5074adeacede5f810b7ef39607ee59d94e948b4fd954495bdb072f8c54558181"},
- {file = "pyzmq-25.1.2-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:7ae8f354b895cbd85212da245f1a5ad8159e7840e37d78b476bb4f4c3f32a9fe"},
- {file = "pyzmq-25.1.2-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:b264bf2cc96b5bc43ce0e852be995e400376bd87ceb363822e2cb1964fcdc737"},
- {file = "pyzmq-25.1.2-cp312-cp312-win32.whl", hash = "sha256:02bbc1a87b76e04fd780b45e7f695471ae6de747769e540da909173d50ff8e2d"},
- {file = "pyzmq-25.1.2-cp312-cp312-win_amd64.whl", hash = "sha256:ced111c2e81506abd1dc142e6cd7b68dd53747b3b7ae5edbea4578c5eeff96b7"},
- {file = "pyzmq-25.1.2-cp36-cp36m-macosx_10_9_x86_64.whl", hash = "sha256:7b6d09a8962a91151f0976008eb7b29b433a560fde056ec7a3db9ec8f1075438"},
- {file = "pyzmq-25.1.2-cp36-cp36m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:967668420f36878a3c9ecb5ab33c9d0ff8d054f9c0233d995a6d25b0e95e1b6b"},
- {file = "pyzmq-25.1.2-cp36-cp36m-manylinux_2_5_i686.manylinux1_i686.whl", hash = "sha256:5edac3f57c7ddaacdb4d40f6ef2f9e299471fc38d112f4bc6d60ab9365445fb0"},
- {file = "pyzmq-25.1.2-cp36-cp36m-manylinux_2_5_x86_64.manylinux1_x86_64.whl", hash = "sha256:0dabfb10ef897f3b7e101cacba1437bd3a5032ee667b7ead32bbcdd1a8422fe7"},
- {file = "pyzmq-25.1.2-cp36-cp36m-musllinux_1_1_aarch64.whl", hash = "sha256:2c6441e0398c2baacfe5ba30c937d274cfc2dc5b55e82e3749e333aabffde561"},
- {file = "pyzmq-25.1.2-cp36-cp36m-musllinux_1_1_i686.whl", hash = "sha256:16b726c1f6c2e7625706549f9dbe9b06004dfbec30dbed4bf50cbdfc73e5b32a"},
- {file = "pyzmq-25.1.2-cp36-cp36m-musllinux_1_1_x86_64.whl", hash = "sha256:a86c2dd76ef71a773e70551a07318b8e52379f58dafa7ae1e0a4be78efd1ff16"},
- {file = "pyzmq-25.1.2-cp36-cp36m-win32.whl", hash = "sha256:359f7f74b5d3c65dae137f33eb2bcfa7ad9ebefd1cab85c935f063f1dbb245cc"},
- {file = "pyzmq-25.1.2-cp36-cp36m-win_amd64.whl", hash = "sha256:55875492f820d0eb3417b51d96fea549cde77893ae3790fd25491c5754ea2f68"},
- {file = "pyzmq-25.1.2-cp37-cp37m-macosx_10_9_x86_64.whl", hash = "sha256:b8c8a419dfb02e91b453615c69568442e897aaf77561ee0064d789705ff37a92"},
- {file = "pyzmq-25.1.2-cp37-cp37m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:8807c87fa893527ae8a524c15fc505d9950d5e856f03dae5921b5e9aa3b8783b"},
- {file = "pyzmq-25.1.2-cp37-cp37m-manylinux_2_5_i686.manylinux1_i686.whl", hash = "sha256:5e319ed7d6b8f5fad9b76daa0a68497bc6f129858ad956331a5835785761e003"},
- {file = "pyzmq-25.1.2-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.whl", hash = "sha256:3c53687dde4d9d473c587ae80cc328e5b102b517447456184b485587ebd18b62"},
- {file = "pyzmq-25.1.2-cp37-cp37m-musllinux_1_1_aarch64.whl", hash = "sha256:9add2e5b33d2cd765ad96d5eb734a5e795a0755f7fc49aa04f76d7ddda73fd70"},
- {file = "pyzmq-25.1.2-cp37-cp37m-musllinux_1_1_i686.whl", hash = "sha256:e690145a8c0c273c28d3b89d6fb32c45e0d9605b2293c10e650265bf5c11cfec"},
- {file = "pyzmq-25.1.2-cp37-cp37m-musllinux_1_1_x86_64.whl", hash = "sha256:00a06faa7165634f0cac1abb27e54d7a0b3b44eb9994530b8ec73cf52e15353b"},
- {file = "pyzmq-25.1.2-cp37-cp37m-win32.whl", hash = "sha256:0f97bc2f1f13cb16905a5f3e1fbdf100e712d841482b2237484360f8bc4cb3d7"},
- {file = "pyzmq-25.1.2-cp37-cp37m-win_amd64.whl", hash = "sha256:6cc0020b74b2e410287e5942e1e10886ff81ac77789eb20bec13f7ae681f0fdd"},
- {file = "pyzmq-25.1.2-cp38-cp38-macosx_10_15_universal2.whl", hash = "sha256:bef02cfcbded83473bdd86dd8d3729cd82b2e569b75844fb4ea08fee3c26ae41"},
- {file = "pyzmq-25.1.2-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:e10a4b5a4b1192d74853cc71a5e9fd022594573926c2a3a4802020360aa719d8"},
- {file = "pyzmq-25.1.2-cp38-cp38-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:8c5f80e578427d4695adac6fdf4370c14a2feafdc8cb35549c219b90652536ae"},
- {file = "pyzmq-25.1.2-cp38-cp38-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:5dde6751e857910c1339890f3524de74007958557593b9e7e8c5f01cd919f8a7"},
- {file = "pyzmq-25.1.2-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ea1608dd169da230a0ad602d5b1ebd39807ac96cae1845c3ceed39af08a5c6df"},
- {file = "pyzmq-25.1.2-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:0f513130c4c361201da9bc69df25a086487250e16b5571ead521b31ff6b02220"},
- {file = "pyzmq-25.1.2-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:019744b99da30330798bb37df33549d59d380c78e516e3bab9c9b84f87a9592f"},
- {file = "pyzmq-25.1.2-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:2e2713ef44be5d52dd8b8e2023d706bf66cb22072e97fc71b168e01d25192755"},
- {file = "pyzmq-25.1.2-cp38-cp38-win32.whl", hash = "sha256:07cd61a20a535524906595e09344505a9bd46f1da7a07e504b315d41cd42eb07"},
- {file = "pyzmq-25.1.2-cp38-cp38-win_amd64.whl", hash = "sha256:eb7e49a17fb8c77d3119d41a4523e432eb0c6932187c37deb6fbb00cc3028088"},
- {file = "pyzmq-25.1.2-cp39-cp39-macosx_10_15_universal2.whl", hash = "sha256:94504ff66f278ab4b7e03e4cba7e7e400cb73bfa9d3d71f58d8972a8dc67e7a6"},
- {file = "pyzmq-25.1.2-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:6dd0d50bbf9dca1d0bdea219ae6b40f713a3fb477c06ca3714f208fd69e16fd8"},
- {file = "pyzmq-25.1.2-cp39-cp39-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:004ff469d21e86f0ef0369717351073e0e577428e514c47c8480770d5e24a565"},
- {file = "pyzmq-25.1.2-cp39-cp39-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:c0b5ca88a8928147b7b1e2dfa09f3b6c256bc1135a1338536cbc9ea13d3b7add"},
- {file = "pyzmq-25.1.2-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:2c9a79f1d2495b167119d02be7448bfba57fad2a4207c4f68abc0bab4b92925b"},
- {file = "pyzmq-25.1.2-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:518efd91c3d8ac9f9b4f7dd0e2b7b8bf1a4fe82a308009016b07eaa48681af82"},
- {file = "pyzmq-25.1.2-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:1ec23bd7b3a893ae676d0e54ad47d18064e6c5ae1fadc2f195143fb27373f7f6"},
- {file = "pyzmq-25.1.2-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:db36c27baed588a5a8346b971477b718fdc66cf5b80cbfbd914b4d6d355e44e2"},
- {file = "pyzmq-25.1.2-cp39-cp39-win32.whl", hash = "sha256:39b1067f13aba39d794a24761e385e2eddc26295826530a8c7b6c6c341584289"},
- {file = "pyzmq-25.1.2-cp39-cp39-win_amd64.whl", hash = "sha256:8e9f3fabc445d0ce320ea2c59a75fe3ea591fdbdeebec5db6de530dd4b09412e"},
- {file = "pyzmq-25.1.2-pp310-pypy310_pp73-macosx_10_9_x86_64.whl", hash = "sha256:a8c1d566344aee826b74e472e16edae0a02e2a044f14f7c24e123002dcff1c05"},
- {file = "pyzmq-25.1.2-pp310-pypy310_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:759cfd391a0996345ba94b6a5110fca9c557ad4166d86a6e81ea526c376a01e8"},
- {file = "pyzmq-25.1.2-pp310-pypy310_pp73-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:7c61e346ac34b74028ede1c6b4bcecf649d69b707b3ff9dc0fab453821b04d1e"},
- {file = "pyzmq-25.1.2-pp310-pypy310_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:4cb8fc1f8d69b411b8ec0b5f1ffbcaf14c1db95b6bccea21d83610987435f1a4"},
- {file = "pyzmq-25.1.2-pp310-pypy310_pp73-win_amd64.whl", hash = "sha256:3c00c9b7d1ca8165c610437ca0c92e7b5607b2f9076f4eb4b095c85d6e680a1d"},
- {file = "pyzmq-25.1.2-pp37-pypy37_pp73-macosx_10_9_x86_64.whl", hash = "sha256:df0c7a16ebb94452d2909b9a7b3337940e9a87a824c4fc1c7c36bb4404cb0cde"},
- {file = "pyzmq-25.1.2-pp37-pypy37_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:45999e7f7ed5c390f2e87ece7f6c56bf979fb213550229e711e45ecc7d42ccb8"},
- {file = "pyzmq-25.1.2-pp37-pypy37_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:ac170e9e048b40c605358667aca3d94e98f604a18c44bdb4c102e67070f3ac9b"},
- {file = "pyzmq-25.1.2-pp37-pypy37_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d1b604734bec94f05f81b360a272fc824334267426ae9905ff32dc2be433ab96"},
- {file = "pyzmq-25.1.2-pp37-pypy37_pp73-win_amd64.whl", hash = "sha256:a793ac733e3d895d96f865f1806f160696422554e46d30105807fdc9841b9f7d"},
- {file = "pyzmq-25.1.2-pp38-pypy38_pp73-macosx_10_9_x86_64.whl", hash = "sha256:0806175f2ae5ad4b835ecd87f5f85583316b69f17e97786f7443baaf54b9bb98"},
- {file = "pyzmq-25.1.2-pp38-pypy38_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:ef12e259e7bc317c7597d4f6ef59b97b913e162d83b421dd0db3d6410f17a244"},
- {file = "pyzmq-25.1.2-pp38-pypy38_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:ea253b368eb41116011add00f8d5726762320b1bda892f744c91997b65754d73"},
- {file = "pyzmq-25.1.2-pp38-pypy38_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:1b9b1f2ad6498445a941d9a4fee096d387fee436e45cc660e72e768d3d8ee611"},
- {file = "pyzmq-25.1.2-pp38-pypy38_pp73-win_amd64.whl", hash = "sha256:8b14c75979ce932c53b79976a395cb2a8cd3aaf14aef75e8c2cb55a330b9b49d"},
- {file = "pyzmq-25.1.2-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:889370d5174a741a62566c003ee8ddba4b04c3f09a97b8000092b7ca83ec9c49"},
- {file = "pyzmq-25.1.2-pp39-pypy39_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:9a18fff090441a40ffda8a7f4f18f03dc56ae73f148f1832e109f9bffa85df15"},
- {file = "pyzmq-25.1.2-pp39-pypy39_pp73-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:99a6b36f95c98839ad98f8c553d8507644c880cf1e0a57fe5e3a3f3969040882"},
- {file = "pyzmq-25.1.2-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:4345c9a27f4310afbb9c01750e9461ff33d6fb74cd2456b107525bbeebcb5be3"},
- {file = "pyzmq-25.1.2-pp39-pypy39_pp73-manylinux_2_28_x86_64.whl", hash = "sha256:3516e0b6224cf6e43e341d56da15fd33bdc37fa0c06af4f029f7d7dfceceabbc"},
- {file = "pyzmq-25.1.2-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:146b9b1f29ead41255387fb07be56dc29639262c0f7344f570eecdcd8d683314"},
- {file = "pyzmq-25.1.2.tar.gz", hash = "sha256:93f1aa311e8bb912e34f004cf186407a4e90eec4f0ecc0efd26056bf7eda0226"},
+ {file = "pyzmq-26.0.2-cp310-cp310-macosx_10_15_universal2.whl", hash = "sha256:1a60a03b01e8c9c58932ec0cca15b1712d911c2800eb82d4281bc1ae5b6dad50"},
+ {file = "pyzmq-26.0.2-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:949067079e14ea1973bd740255e0840118c163d4bce8837f539d749f145cf5c3"},
+ {file = "pyzmq-26.0.2-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:37e7edfa6cf96d036a403775c96afa25058d1bb940a79786a9a2fc94a783abe3"},
+ {file = "pyzmq-26.0.2-cp310-cp310-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:903cc7a84a7d4326b43755c368780800e035aa3d711deae84a533fdffa8755b0"},
+ {file = "pyzmq-26.0.2-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:6cb2e41af165e5f327d06fbdd79a42a4e930267fade4e9f92d17f3ccce03f3a7"},
+ {file = "pyzmq-26.0.2-cp310-cp310-manylinux_2_28_x86_64.whl", hash = "sha256:55353b8189adcfc4c125fc4ce59d477744118e9c0ec379dd0999c5fa120ac4f5"},
+ {file = "pyzmq-26.0.2-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:f961423ff6236a752ced80057a20e623044df95924ed1009f844cde8b3a595f9"},
+ {file = "pyzmq-26.0.2-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:ba77fe84fe4f5f3dc0ef681a6d366685c8ffe1c8439c1d7530997b05ac06a04b"},
+ {file = "pyzmq-26.0.2-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:52589f0a745ef61b9c75c872cf91f8c1f7c0668eb3dd99d7abd639d8c0fb9ca7"},
+ {file = "pyzmq-26.0.2-cp310-cp310-win32.whl", hash = "sha256:b7b6d2a46c7afe2ad03ec8faf9967090c8ceae85c4d8934d17d7cae6f9062b64"},
+ {file = "pyzmq-26.0.2-cp310-cp310-win_amd64.whl", hash = "sha256:86531e20de249d9204cc6d8b13d5a30537748c78820215161d8a3b9ea58ca111"},
+ {file = "pyzmq-26.0.2-cp310-cp310-win_arm64.whl", hash = "sha256:f26a05029ecd2bd306b941ff8cb80f7620b7901421052bc429d238305b1cbf2f"},
+ {file = "pyzmq-26.0.2-cp311-cp311-macosx_10_15_universal2.whl", hash = "sha256:70770e296a9cb03d955540c99360aab861cbb3cba29516abbd106a15dbd91268"},
+ {file = "pyzmq-26.0.2-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:2740fd7161b39e178554ebf21aa5667a1c9ef0cd2cb74298fd4ef017dae7aec4"},
+ {file = "pyzmq-26.0.2-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:f5e3706c32dea077faa42b1c92d825b7f86c866f72532d342e0be5e64d14d858"},
+ {file = "pyzmq-26.0.2-cp311-cp311-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:0fa1416876194927f7723d6b7171b95e1115602967fc6bfccbc0d2d51d8ebae1"},
+ {file = "pyzmq-26.0.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:4ef9a79a48794099c57dc2df00340b5d47c5caa1792f9ddb8c7a26b1280bd575"},
+ {file = "pyzmq-26.0.2-cp311-cp311-manylinux_2_28_x86_64.whl", hash = "sha256:1c60fcdfa3229aeee4291c5d60faed3a813b18bdadb86299c4bf49e8e51e8605"},
+ {file = "pyzmq-26.0.2-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:e943c39c206b04df2eb5d71305761d7c3ca75fd49452115ea92db1b5b98dbdef"},
+ {file = "pyzmq-26.0.2-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:8da0ed8a598693731c76659880a668f4748b59158f26ed283a93f7f04d47447e"},
+ {file = "pyzmq-26.0.2-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:7bf51970b11d67096bede97cdbad0f4333f7664f4708b9b2acb352bf4faa3140"},
+ {file = "pyzmq-26.0.2-cp311-cp311-win32.whl", hash = "sha256:6f8e6bd5d066be605faa9fe5ec10aa1a46ad9f18fc8646f2b9aaefc8fb575742"},
+ {file = "pyzmq-26.0.2-cp311-cp311-win_amd64.whl", hash = "sha256:6d03da3a0ae691b361edcb39530075461202f699ce05adbb15055a0e1c9bcaa4"},
+ {file = "pyzmq-26.0.2-cp311-cp311-win_arm64.whl", hash = "sha256:f84e33321b68ff00b60e9dbd1a483e31ab6022c577c8de525b8e771bd274ce68"},
+ {file = "pyzmq-26.0.2-cp312-cp312-macosx_10_15_universal2.whl", hash = "sha256:44c33ebd1c62a01db7fbc24e18bdda569d6639217d13d5929e986a2b0f69070d"},
+ {file = "pyzmq-26.0.2-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:ac04f904b4fce4afea9cdccbb78e24d468cb610a839d5a698853e14e2a3f9ecf"},
+ {file = "pyzmq-26.0.2-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:f2133de5ba9adc5f481884ccb699eac9ce789708292945c05746880f95b241c0"},
+ {file = "pyzmq-26.0.2-cp312-cp312-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:7753c67c570d7fc80c2dc59b90ca1196f1224e0e2e29a548980c95fe0fe27fc1"},
+ {file = "pyzmq-26.0.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:8d4e51632e6b12e65e8d9d7612446ecda2eda637a868afa7bce16270194650dd"},
+ {file = "pyzmq-26.0.2-cp312-cp312-manylinux_2_28_x86_64.whl", hash = "sha256:d6c38806f6ecd0acf3104b8d7e76a206bcf56dadd6ce03720d2fa9d9157d5718"},
+ {file = "pyzmq-26.0.2-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:48f496bbe14686b51cec15406323ae6942851e14022efd7fc0e2ecd092c5982c"},
+ {file = "pyzmq-26.0.2-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:e84a3161149c75bb7a7dc8646384186c34033e286a67fec1ad1bdedea165e7f4"},
+ {file = "pyzmq-26.0.2-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:dabf796c67aa9f5a4fcc956d47f0d48b5c1ed288d628cf53aa1cf08e88654343"},
+ {file = "pyzmq-26.0.2-cp312-cp312-win32.whl", hash = "sha256:3eee4c676af1b109f708d80ef0cf57ecb8aaa5900d1edaf90406aea7e0e20e37"},
+ {file = "pyzmq-26.0.2-cp312-cp312-win_amd64.whl", hash = "sha256:26721fec65846b3e4450dad050d67d31b017f97e67f7e0647b5f98aa47f828cf"},
+ {file = "pyzmq-26.0.2-cp312-cp312-win_arm64.whl", hash = "sha256:653955c6c233e90de128a1b8e882abc7216f41f44218056bd519969c8c413a15"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-macosx_10_9_x86_64.whl", hash = "sha256:becd8d8fb068fbb5a52096efd83a2d8e54354383f691781f53a4c26aee944542"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:7a15e5465e7083c12517209c9dd24722b25e9b63c49a563922922fc03554eb35"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:e8158ac8616941f874841f9fa0f6d2f1466178c2ff91ea08353fdc19de0d40c2"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ea2c6a53e28c7066ea7db86fcc0b71d78d01b818bb11d4a4341ec35059885295"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-musllinux_1_1_aarch64.whl", hash = "sha256:bdbc7dab0b0e9c62c97b732899c4242e3282ba803bad668e03650b59b165466e"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-musllinux_1_1_i686.whl", hash = "sha256:e74b6d5ef57bb65bf1b4a37453d8d86d88550dde3fb0f23b1f1a24e60c70af5b"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-musllinux_1_1_x86_64.whl", hash = "sha256:ed4c6ee624ecbc77b18aeeb07bf0700d26571ab95b8f723f0d02e056b5bce438"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-win32.whl", hash = "sha256:8a98b3cb0484b83c19d8fb5524c8a469cd9f10e743f5904ac285d92678ee761f"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-win_amd64.whl", hash = "sha256:aa5f95d71b6eca9cec28aa0a2f8310ea53dea313b63db74932879ff860c1fb8d"},
+ {file = "pyzmq-26.0.2-cp38-cp38-macosx_10_15_universal2.whl", hash = "sha256:5ff56c76ce77b9805378a7a73032c17cbdb1a5b84faa1df03c5d3e306e5616df"},
+ {file = "pyzmq-26.0.2-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:bab697fc1574fee4b81da955678708567c43c813c84c91074e452bda5346c921"},
+ {file = "pyzmq-26.0.2-cp38-cp38-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:0c0fed8aa9ba0488ee1cbdaa304deea92d52fab43d373297002cfcc69c0a20c5"},
+ {file = "pyzmq-26.0.2-cp38-cp38-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:606b922699fcec472ed814dda4dc3ff7c748254e0b26762a0ba21a726eb1c107"},
+ {file = "pyzmq-26.0.2-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:45f0fd82bad4d199fa993fbf0ac586a7ac5879addbe436a35a389df7e0eb4c91"},
+ {file = "pyzmq-26.0.2-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:166c5e41045939a52c01e6f374e493d9a6a45dfe677360d3e7026e38c42e8906"},
+ {file = "pyzmq-26.0.2-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:d566e859e8b8d5bca08467c093061774924b3d78a5ba290e82735b2569edc84b"},
+ {file = "pyzmq-26.0.2-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:264ee0e72b72ca59279dc320deab5ae0fac0d97881aed1875ce4bde2e56ffde0"},
+ {file = "pyzmq-26.0.2-cp38-cp38-win32.whl", hash = "sha256:3152bbd3a4744cbdd83dfb210ed701838b8b0c9065cef14671d6d91df12197d0"},
+ {file = "pyzmq-26.0.2-cp38-cp38-win_amd64.whl", hash = "sha256:bf77601d75ca692c179154b7e5943c286a4aaffec02c491afe05e60493ce95f2"},
+ {file = "pyzmq-26.0.2-cp39-cp39-macosx_10_15_universal2.whl", hash = "sha256:c770a7545b3deca2db185b59175e710a820dd4ed43619f4c02e90b0e227c6252"},
+ {file = "pyzmq-26.0.2-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:d47175f0a380bfd051726bc5c0054036ae4a5d8caf922c62c8a172ccd95c1a2a"},
+ {file = "pyzmq-26.0.2-cp39-cp39-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:9bce298c1ce077837e110367c321285dc4246b531cde1abfc27e4a5bbe2bed4d"},
+ {file = "pyzmq-26.0.2-cp39-cp39-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:c40b09b7e184d6e3e1be1c8af2cc320c0f9f610d8a5df3dd866e6e6e4e32b235"},
+ {file = "pyzmq-26.0.2-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d420d856bf728713874cefb911398efe69e1577835851dd297a308a78c14c249"},
+ {file = "pyzmq-26.0.2-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:d792d3cab987058451e55c70c5926e93e2ceb68ca5a2334863bb903eb860c9cb"},
+ {file = "pyzmq-26.0.2-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:83ec17729cf6d3464dab98a11e98294fcd50e6b17eaabd3d841515c23f6dbd3a"},
+ {file = "pyzmq-26.0.2-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:47c17d5ebfa88ae90f08960c97b49917098665b8cd8be31f2c24e177bcf37a0f"},
+ {file = "pyzmq-26.0.2-cp39-cp39-win32.whl", hash = "sha256:d509685d1cd1d018705a811c5f9d5bc237790936ead6d06f6558b77e16cc7235"},
+ {file = "pyzmq-26.0.2-cp39-cp39-win_amd64.whl", hash = "sha256:c7cc8cc009e8f6989a6d86c96f87dae5f5fb07d6c96916cdc7719d546152c7db"},
+ {file = "pyzmq-26.0.2-cp39-cp39-win_arm64.whl", hash = "sha256:3ada31cb879cd7532f4a85b501f4255c747d4813ab76b35c49ed510ce4865b45"},
+ {file = "pyzmq-26.0.2-pp310-pypy310_pp73-macosx_10_9_x86_64.whl", hash = "sha256:0a6ceaddc830dd3ca86cb8451cf373d1f05215368e11834538c2902ed5205139"},
+ {file = "pyzmq-26.0.2-pp310-pypy310_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:6a967681463aa7a99eb9a62bb18229b653b45c10ff0947b31cc0837a83dfb86f"},
+ {file = "pyzmq-26.0.2-pp310-pypy310_pp73-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:6472a73bc115bc40a2076609a90894775abe6faf19a78375675a2f889a613071"},
+ {file = "pyzmq-26.0.2-pp310-pypy310_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:5d6aea92bcccfe5e5524d3c70a6f16ffdae548390ddad26f4207d55c55a40593"},
+ {file = "pyzmq-26.0.2-pp310-pypy310_pp73-win_amd64.whl", hash = "sha256:e025f6351e49d48a5aa2f5a09293aa769b0ee7369c25bed551647234b7fa0c75"},
+ {file = "pyzmq-26.0.2-pp37-pypy37_pp73-macosx_10_9_x86_64.whl", hash = "sha256:40bd7ebe4dbb37d27f0c56e2a844f360239343a99be422085e13e97da13f73f9"},
+ {file = "pyzmq-26.0.2-pp37-pypy37_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:1dd40d586ad6f53764104df6e01810fe1b4e88fd353774629a5e6fe253813f79"},
+ {file = "pyzmq-26.0.2-pp37-pypy37_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:f2aca15e9ad8c8657b5b3d7ae3d1724dc8c1c1059c06b4b674c3aa36305f4930"},
+ {file = "pyzmq-26.0.2-pp37-pypy37_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:450ec234736732eb0ebeffdb95a352450d4592f12c3e087e2a9183386d22c8bf"},
+ {file = "pyzmq-26.0.2-pp37-pypy37_pp73-win_amd64.whl", hash = "sha256:f43be2bebbd09360a2f23af83b243dc25ffe7b583ea8c722e6df03e03a55f02f"},
+ {file = "pyzmq-26.0.2-pp38-pypy38_pp73-macosx_10_9_x86_64.whl", hash = "sha256:867f55e54aff254940bcec5eec068e7c0ac1e6bf360ab91479394a8bf356b0e6"},
+ {file = "pyzmq-26.0.2-pp38-pypy38_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:b4dbc033c5ad46f8c429bf238c25a889b8c1d86bfe23a74e1031a991cb3f0000"},
+ {file = "pyzmq-26.0.2-pp38-pypy38_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:6e8dd2961462e337e21092ec2da0c69d814dcb1b6e892955a37444a425e9cfb8"},
+ {file = "pyzmq-26.0.2-pp38-pypy38_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:35391e72df6c14a09b697c7b94384947c1dd326aca883ff98ff137acdf586c33"},
+ {file = "pyzmq-26.0.2-pp38-pypy38_pp73-win_amd64.whl", hash = "sha256:1c3d3c92fa54eda94ab369ca5b8d35059987c326ba5e55326eb068862f64b1fc"},
+ {file = "pyzmq-26.0.2-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:e7aa61a9cc4f0523373e31fc9255bf4567185a099f85ca3598e64de484da3ab2"},
+ {file = "pyzmq-26.0.2-pp39-pypy39_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ee53a8191271f144cc20b12c19daa9f1546adc84a2f33839e3338039b55c373c"},
+ {file = "pyzmq-26.0.2-pp39-pypy39_pp73-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:ac60a980f07fa988983f7bfe6404ef3f1e4303f5288a01713bc1266df6d18783"},
+ {file = "pyzmq-26.0.2-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:88896b1b4817d7b2fe1ec7205c4bbe07bf5d92fb249bf2d226ddea8761996068"},
+ {file = "pyzmq-26.0.2-pp39-pypy39_pp73-manylinux_2_28_x86_64.whl", hash = "sha256:18dfffe23751edee917764ffa133d5d3fef28dfd1cf3adebef8c90bc854c74c4"},
+ {file = "pyzmq-26.0.2-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:6926dd14cfe6967d3322640b6d5c3c3039db71716a5e43cca6e3b474e73e0b36"},
+ {file = "pyzmq-26.0.2.tar.gz", hash = "sha256:f0f9bb370449158359bb72a3e12c658327670c0ffe6fbcd1af083152b64f9df0"},
]
[package.dependencies]
@@ -2768,19 +2989,121 @@ prompt_toolkit = ">=2.0,<=3.0.36"
[[package]]
name = "referencing"
-version = "0.34.0"
+version = "0.35.0"
description = "JSON Referencing + Python"
optional = false
python-versions = ">=3.8"
files = [
- {file = "referencing-0.34.0-py3-none-any.whl", hash = "sha256:d53ae300ceddd3169f1ffa9caf2cb7b769e92657e4fafb23d34b93679116dfd4"},
- {file = "referencing-0.34.0.tar.gz", hash = "sha256:5773bd84ef41799a5a8ca72dc34590c041eb01bf9aa02632b4a973fb0181a844"},
+ {file = "referencing-0.35.0-py3-none-any.whl", hash = "sha256:8080727b30e364e5783152903672df9b6b091c926a146a759080b62ca3126cd6"},
+ {file = "referencing-0.35.0.tar.gz", hash = "sha256:191e936b0c696d0af17ad7430a3dc68e88bc11be6514f4757dc890f04ab05889"},
]
[package.dependencies]
attrs = ">=22.2.0"
rpds-py = ">=0.7.0"
+[[package]]
+name = "regex"
+version = "2024.4.16"
+description = "Alternative regular expression module, to replace re."
+optional = false
+python-versions = ">=3.7"
+files = [
+ {file = "regex-2024.4.16-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:fb83cc090eac63c006871fd24db5e30a1f282faa46328572661c0a24a2323a08"},
+ {file = "regex-2024.4.16-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:8c91e1763696c0eb66340c4df98623c2d4e77d0746b8f8f2bee2c6883fd1fe18"},
+ {file = "regex-2024.4.16-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:10188fe732dec829c7acca7422cdd1bf57d853c7199d5a9e96bb4d40db239c73"},
+ {file = "regex-2024.4.16-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:956b58d692f235cfbf5b4f3abd6d99bf102f161ccfe20d2fd0904f51c72c4c66"},
+ {file = "regex-2024.4.16-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:a70b51f55fd954d1f194271695821dd62054d949efd6368d8be64edd37f55c86"},
+ {file = "regex-2024.4.16-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:5c02fcd2bf45162280613d2e4a1ca3ac558ff921ae4e308ecb307650d3a6ee51"},
+ {file = "regex-2024.4.16-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:c4ed75ea6892a56896d78f11006161eea52c45a14994794bcfa1654430984b22"},
+ {file = "regex-2024.4.16-cp310-cp310-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:bd727ad276bb91928879f3aa6396c9a1d34e5e180dce40578421a691eeb77f47"},
+ {file = "regex-2024.4.16-cp310-cp310-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:7cbc5d9e8a1781e7be17da67b92580d6ce4dcef5819c1b1b89f49d9678cc278c"},
+ {file = "regex-2024.4.16-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:78fddb22b9ef810b63ef341c9fcf6455232d97cfe03938cbc29e2672c436670e"},
+ {file = "regex-2024.4.16-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:445ca8d3c5a01309633a0c9db57150312a181146315693273e35d936472df912"},
+ {file = "regex-2024.4.16-cp310-cp310-musllinux_1_1_ppc64le.whl", hash = "sha256:95399831a206211d6bc40224af1c635cb8790ddd5c7493e0bd03b85711076a53"},
+ {file = "regex-2024.4.16-cp310-cp310-musllinux_1_1_s390x.whl", hash = "sha256:7731728b6568fc286d86745f27f07266de49603a6fdc4d19c87e8c247be452af"},
+ {file = "regex-2024.4.16-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:4facc913e10bdba42ec0aee76d029aedda628161a7ce4116b16680a0413f658a"},
+ {file = "regex-2024.4.16-cp310-cp310-win32.whl", hash = "sha256:911742856ce98d879acbea33fcc03c1d8dc1106234c5e7d068932c945db209c0"},
+ {file = "regex-2024.4.16-cp310-cp310-win_amd64.whl", hash = "sha256:e0a2df336d1135a0b3a67f3bbf78a75f69562c1199ed9935372b82215cddd6e2"},
+ {file = "regex-2024.4.16-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:1210365faba7c2150451eb78ec5687871c796b0f1fa701bfd2a4a25420482d26"},
+ {file = "regex-2024.4.16-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:9ab40412f8cd6f615bfedea40c8bf0407d41bf83b96f6fc9ff34976d6b7037fd"},
+ {file = "regex-2024.4.16-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:fd80d1280d473500d8086d104962a82d77bfbf2b118053824b7be28cd5a79ea5"},
+ {file = "regex-2024.4.16-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:7bb966fdd9217e53abf824f437a5a2d643a38d4fd5fd0ca711b9da683d452969"},
+ {file = "regex-2024.4.16-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:20b7a68444f536365af42a75ccecb7ab41a896a04acf58432db9e206f4e525d6"},
+ {file = "regex-2024.4.16-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:b74586dd0b039c62416034f811d7ee62810174bb70dffcca6439f5236249eb09"},
+ {file = "regex-2024.4.16-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:0c8290b44d8b0af4e77048646c10c6e3aa583c1ca67f3b5ffb6e06cf0c6f0f89"},
+ {file = "regex-2024.4.16-cp311-cp311-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:f2d80a6749724b37853ece57988b39c4e79d2b5fe2869a86e8aeae3bbeef9eb0"},
+ {file = "regex-2024.4.16-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:3a1018e97aeb24e4f939afcd88211ace472ba566efc5bdf53fd8fd7f41fa7170"},
+ {file = "regex-2024.4.16-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:8d015604ee6204e76569d2f44e5a210728fa917115bef0d102f4107e622b08d5"},
+ {file = "regex-2024.4.16-cp311-cp311-musllinux_1_1_ppc64le.whl", hash = "sha256:3d5ac5234fb5053850d79dd8eb1015cb0d7d9ed951fa37aa9e6249a19aa4f336"},
+ {file = "regex-2024.4.16-cp311-cp311-musllinux_1_1_s390x.whl", hash = "sha256:0a38d151e2cdd66d16dab550c22f9521ba79761423b87c01dae0a6e9add79c0d"},
+ {file = "regex-2024.4.16-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:159dc4e59a159cb8e4e8f8961eb1fa5d58f93cb1acd1701d8aff38d45e1a84a6"},
+ {file = "regex-2024.4.16-cp311-cp311-win32.whl", hash = "sha256:ba2336d6548dee3117520545cfe44dc28a250aa091f8281d28804aa8d707d93d"},
+ {file = "regex-2024.4.16-cp311-cp311-win_amd64.whl", hash = "sha256:8f83b6fd3dc3ba94d2b22717f9c8b8512354fd95221ac661784df2769ea9bba9"},
+ {file = "regex-2024.4.16-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:80b696e8972b81edf0af2a259e1b2a4a661f818fae22e5fa4fa1a995fb4a40fd"},
+ {file = "regex-2024.4.16-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:d61ae114d2a2311f61d90c2ef1358518e8f05eafda76eaf9c772a077e0b465ec"},
+ {file = "regex-2024.4.16-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:8ba6745440b9a27336443b0c285d705ce73adb9ec90e2f2004c64d95ab5a7598"},
+ {file = "regex-2024.4.16-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:6295004b2dd37b0835ea5c14a33e00e8cfa3c4add4d587b77287825f3418d310"},
+ {file = "regex-2024.4.16-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:4aba818dcc7263852aabb172ec27b71d2abca02a593b95fa79351b2774eb1d2b"},
+ {file = "regex-2024.4.16-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:d0800631e565c47520aaa04ae38b96abc5196fe8b4aa9bd864445bd2b5848a7a"},
+ {file = "regex-2024.4.16-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:08dea89f859c3df48a440dbdcd7b7155bc675f2fa2ec8c521d02dc69e877db70"},
+ {file = "regex-2024.4.16-cp312-cp312-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:eeaa0b5328b785abc344acc6241cffde50dc394a0644a968add75fcefe15b9d4"},
+ {file = "regex-2024.4.16-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:4e819a806420bc010489f4e741b3036071aba209f2e0989d4750b08b12a9343f"},
+ {file = "regex-2024.4.16-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:c2d0e7cbb6341e830adcbfa2479fdeebbfbb328f11edd6b5675674e7a1e37730"},
+ {file = "regex-2024.4.16-cp312-cp312-musllinux_1_1_ppc64le.whl", hash = "sha256:91797b98f5e34b6a49f54be33f72e2fb658018ae532be2f79f7c63b4ae225145"},
+ {file = "regex-2024.4.16-cp312-cp312-musllinux_1_1_s390x.whl", hash = "sha256:d2da13568eff02b30fd54fccd1e042a70fe920d816616fda4bf54ec705668d81"},
+ {file = "regex-2024.4.16-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:370c68dc5570b394cbaadff50e64d705f64debed30573e5c313c360689b6aadc"},
+ {file = "regex-2024.4.16-cp312-cp312-win32.whl", hash = "sha256:904c883cf10a975b02ab3478bce652f0f5346a2c28d0a8521d97bb23c323cc8b"},
+ {file = "regex-2024.4.16-cp312-cp312-win_amd64.whl", hash = "sha256:785c071c982dce54d44ea0b79cd6dfafddeccdd98cfa5f7b86ef69b381b457d9"},
+ {file = "regex-2024.4.16-cp37-cp37m-macosx_10_9_x86_64.whl", hash = "sha256:e2f142b45c6fed48166faeb4303b4b58c9fcd827da63f4cf0a123c3480ae11fb"},
+ {file = "regex-2024.4.16-cp37-cp37m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:e87ab229332ceb127a165612d839ab87795972102cb9830e5f12b8c9a5c1b508"},
+ {file = "regex-2024.4.16-cp37-cp37m-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:81500ed5af2090b4a9157a59dbc89873a25c33db1bb9a8cf123837dcc9765047"},
+ {file = "regex-2024.4.16-cp37-cp37m-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:b340cccad138ecb363324aa26893963dcabb02bb25e440ebdf42e30963f1a4e0"},
+ {file = "regex-2024.4.16-cp37-cp37m-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:2c72608e70f053643437bd2be0608f7f1c46d4022e4104d76826f0839199347a"},
+ {file = "regex-2024.4.16-cp37-cp37m-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:a01fe2305e6232ef3e8f40bfc0f0f3a04def9aab514910fa4203bafbc0bb4682"},
+ {file = "regex-2024.4.16-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:03576e3a423d19dda13e55598f0fd507b5d660d42c51b02df4e0d97824fdcae3"},
+ {file = "regex-2024.4.16-cp37-cp37m-musllinux_1_1_aarch64.whl", hash = "sha256:549c3584993772e25f02d0656ac48abdda73169fe347263948cf2b1cead622f3"},
+ {file = "regex-2024.4.16-cp37-cp37m-musllinux_1_1_i686.whl", hash = "sha256:34422d5a69a60b7e9a07a690094e824b66f5ddc662a5fc600d65b7c174a05f04"},
+ {file = "regex-2024.4.16-cp37-cp37m-musllinux_1_1_ppc64le.whl", hash = "sha256:5f580c651a72b75c39e311343fe6875d6f58cf51c471a97f15a938d9fe4e0d37"},
+ {file = "regex-2024.4.16-cp37-cp37m-musllinux_1_1_s390x.whl", hash = "sha256:3399dd8a7495bbb2bacd59b84840eef9057826c664472e86c91d675d007137f5"},
+ {file = "regex-2024.4.16-cp37-cp37m-musllinux_1_1_x86_64.whl", hash = "sha256:8d1f86f3f4e2388aa3310b50694ac44daefbd1681def26b4519bd050a398dc5a"},
+ {file = "regex-2024.4.16-cp37-cp37m-win32.whl", hash = "sha256:dd5acc0a7d38fdc7a3a6fd3ad14c880819008ecb3379626e56b163165162cc46"},
+ {file = "regex-2024.4.16-cp37-cp37m-win_amd64.whl", hash = "sha256:ba8122e3bb94ecda29a8de4cf889f600171424ea586847aa92c334772d200331"},
+ {file = "regex-2024.4.16-cp38-cp38-macosx_10_9_universal2.whl", hash = "sha256:743deffdf3b3481da32e8a96887e2aa945ec6685af1cfe2bcc292638c9ba2f48"},
+ {file = "regex-2024.4.16-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:7571f19f4a3fd00af9341c7801d1ad1967fc9c3f5e62402683047e7166b9f2b4"},
+ {file = "regex-2024.4.16-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:df79012ebf6f4efb8d307b1328226aef24ca446b3ff8d0e30202d7ebcb977a8c"},
+ {file = "regex-2024.4.16-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:e757d475953269fbf4b441207bb7dbdd1c43180711b6208e129b637792ac0b93"},
+ {file = "regex-2024.4.16-cp38-cp38-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:4313ab9bf6a81206c8ac28fdfcddc0435299dc88cad12cc6305fd0e78b81f9e4"},
+ {file = "regex-2024.4.16-cp38-cp38-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:d83c2bc678453646f1a18f8db1e927a2d3f4935031b9ad8a76e56760461105dd"},
+ {file = "regex-2024.4.16-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:9df1bfef97db938469ef0a7354b2d591a2d438bc497b2c489471bec0e6baf7c4"},
+ {file = "regex-2024.4.16-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:62120ed0de69b3649cc68e2965376048793f466c5a6c4370fb27c16c1beac22d"},
+ {file = "regex-2024.4.16-cp38-cp38-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:c2ef6f7990b6e8758fe48ad08f7e2f66c8f11dc66e24093304b87cae9037bb4a"},
+ {file = "regex-2024.4.16-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:8fc6976a3395fe4d1fbeb984adaa8ec652a1e12f36b56ec8c236e5117b585427"},
+ {file = "regex-2024.4.16-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:03e68f44340528111067cecf12721c3df4811c67268b897fbe695c95f860ac42"},
+ {file = "regex-2024.4.16-cp38-cp38-musllinux_1_1_ppc64le.whl", hash = "sha256:ec7e0043b91115f427998febaa2beb82c82df708168b35ece3accb610b91fac1"},
+ {file = "regex-2024.4.16-cp38-cp38-musllinux_1_1_s390x.whl", hash = "sha256:c21fc21a4c7480479d12fd8e679b699f744f76bb05f53a1d14182b31f55aac76"},
+ {file = "regex-2024.4.16-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:12f6a3f2f58bb7344751919a1876ee1b976fe08b9ffccb4bbea66f26af6017b9"},
+ {file = "regex-2024.4.16-cp38-cp38-win32.whl", hash = "sha256:479595a4fbe9ed8f8f72c59717e8cf222da2e4c07b6ae5b65411e6302af9708e"},
+ {file = "regex-2024.4.16-cp38-cp38-win_amd64.whl", hash = "sha256:0534b034fba6101611968fae8e856c1698da97ce2efb5c2b895fc8b9e23a5834"},
+ {file = "regex-2024.4.16-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:a7ccdd1c4a3472a7533b0a7aa9ee34c9a2bef859ba86deec07aff2ad7e0c3b94"},
+ {file = "regex-2024.4.16-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:6f2f017c5be19984fbbf55f8af6caba25e62c71293213f044da3ada7091a4455"},
+ {file = "regex-2024.4.16-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:803b8905b52de78b173d3c1e83df0efb929621e7b7c5766c0843704d5332682f"},
+ {file = "regex-2024.4.16-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:684008ec44ad275832a5a152f6e764bbe1914bea10968017b6feaecdad5736e0"},
+ {file = "regex-2024.4.16-cp39-cp39-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:65436dce9fdc0aeeb0a0effe0839cb3d6a05f45aa45a4d9f9c60989beca78b9c"},
+ {file = "regex-2024.4.16-cp39-cp39-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:ea355eb43b11764cf799dda62c658c4d2fdb16af41f59bb1ccfec517b60bcb07"},
+ {file = "regex-2024.4.16-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:98c1165f3809ce7774f05cb74e5408cd3aa93ee8573ae959a97a53db3ca3180d"},
+ {file = "regex-2024.4.16-cp39-cp39-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:cccc79a9be9b64c881f18305a7c715ba199e471a3973faeb7ba84172abb3f317"},
+ {file = "regex-2024.4.16-cp39-cp39-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:00169caa125f35d1bca6045d65a662af0202704489fada95346cfa092ec23f39"},
+ {file = "regex-2024.4.16-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:6cc38067209354e16c5609b66285af17a2863a47585bcf75285cab33d4c3b8df"},
+ {file = "regex-2024.4.16-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:23cff1b267038501b179ccbbd74a821ac4a7192a1852d1d558e562b507d46013"},
+ {file = "regex-2024.4.16-cp39-cp39-musllinux_1_1_ppc64le.whl", hash = "sha256:b9d320b3bf82a39f248769fc7f188e00f93526cc0fe739cfa197868633d44701"},
+ {file = "regex-2024.4.16-cp39-cp39-musllinux_1_1_s390x.whl", hash = "sha256:89ec7f2c08937421bbbb8b48c54096fa4f88347946d4747021ad85f1b3021b3c"},
+ {file = "regex-2024.4.16-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:4918fd5f8b43aa7ec031e0fef1ee02deb80b6afd49c85f0790be1dc4ce34cb50"},
+ {file = "regex-2024.4.16-cp39-cp39-win32.whl", hash = "sha256:684e52023aec43bdf0250e843e1fdd6febbe831bd9d52da72333fa201aaa2335"},
+ {file = "regex-2024.4.16-cp39-cp39-win_amd64.whl", hash = "sha256:e697e1c0238133589e00c244a8b676bc2cfc3ab4961318d902040d099fec7483"},
+ {file = "regex-2024.4.16.tar.gz", hash = "sha256:fa454d26f2e87ad661c4f0c5a5fe4cf6aab1e307d1b94f16ffdfcb089ba685c0"},
+]
+
[[package]]
name = "requests"
version = "2.31.0"
@@ -2912,28 +3235,28 @@ files = [
[[package]]
name = "ruff"
-version = "0.3.4"
+version = "0.3.7"
description = "An extremely fast Python linter and code formatter, written in Rust."
optional = false
python-versions = ">=3.7"
files = [
- {file = "ruff-0.3.4-py3-none-macosx_10_12_x86_64.macosx_11_0_arm64.macosx_10_12_universal2.whl", hash = "sha256:60c870a7d46efcbc8385d27ec07fe534ac32f3b251e4fc44b3cbfd9e09609ef4"},
- {file = "ruff-0.3.4-py3-none-macosx_10_12_x86_64.whl", hash = "sha256:6fc14fa742e1d8f24910e1fff0bd5e26d395b0e0e04cc1b15c7c5e5fe5b4af91"},
- {file = "ruff-0.3.4-py3-none-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d3ee7880f653cc03749a3bfea720cf2a192e4f884925b0cf7eecce82f0ce5854"},
- {file = "ruff-0.3.4-py3-none-manylinux_2_17_armv7l.manylinux2014_armv7l.whl", hash = "sha256:cf133dd744f2470b347f602452a88e70dadfbe0fcfb5fd46e093d55da65f82f7"},
- {file = "ruff-0.3.4-py3-none-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:3f3860057590e810c7ffea75669bdc6927bfd91e29b4baa9258fd48b540a4365"},
- {file = "ruff-0.3.4-py3-none-manylinux_2_17_ppc64.manylinux2014_ppc64.whl", hash = "sha256:986f2377f7cf12efac1f515fc1a5b753c000ed1e0a6de96747cdf2da20a1b369"},
- {file = "ruff-0.3.4-py3-none-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:c4fd98e85869603e65f554fdc5cddf0712e352fe6e61d29d5a6fe087ec82b76c"},
- {file = "ruff-0.3.4-py3-none-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:64abeed785dad51801b423fa51840b1764b35d6c461ea8caef9cf9e5e5ab34d9"},
- {file = "ruff-0.3.4-py3-none-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:df52972138318bc7546d92348a1ee58449bc3f9eaf0db278906eb511889c4b50"},
- {file = "ruff-0.3.4-py3-none-musllinux_1_2_aarch64.whl", hash = "sha256:98e98300056445ba2cc27d0b325fd044dc17fcc38e4e4d2c7711585bd0a958ed"},
- {file = "ruff-0.3.4-py3-none-musllinux_1_2_armv7l.whl", hash = "sha256:519cf6a0ebed244dce1dc8aecd3dc99add7a2ee15bb68cf19588bb5bf58e0488"},
- {file = "ruff-0.3.4-py3-none-musllinux_1_2_i686.whl", hash = "sha256:bb0acfb921030d00070539c038cd24bb1df73a2981e9f55942514af8b17be94e"},
- {file = "ruff-0.3.4-py3-none-musllinux_1_2_x86_64.whl", hash = "sha256:cf187a7e7098233d0d0c71175375c5162f880126c4c716fa28a8ac418dcf3378"},
- {file = "ruff-0.3.4-py3-none-win32.whl", hash = "sha256:af27ac187c0a331e8ef91d84bf1c3c6a5dea97e912a7560ac0cef25c526a4102"},
- {file = "ruff-0.3.4-py3-none-win_amd64.whl", hash = "sha256:de0d5069b165e5a32b3c6ffbb81c350b1e3d3483347196ffdf86dc0ef9e37dd6"},
- {file = "ruff-0.3.4-py3-none-win_arm64.whl", hash = "sha256:6810563cc08ad0096b57c717bd78aeac888a1bfd38654d9113cb3dc4d3f74232"},
- {file = "ruff-0.3.4.tar.gz", hash = "sha256:f0f4484c6541a99862b693e13a151435a279b271cff20e37101116a21e2a1ad1"},
+ {file = "ruff-0.3.7-py3-none-macosx_10_12_x86_64.macosx_11_0_arm64.macosx_10_12_universal2.whl", hash = "sha256:0e8377cccb2f07abd25e84fc5b2cbe48eeb0fea9f1719cad7caedb061d70e5ce"},
+ {file = "ruff-0.3.7-py3-none-macosx_10_12_x86_64.whl", hash = "sha256:15a4d1cc1e64e556fa0d67bfd388fed416b7f3b26d5d1c3e7d192c897e39ba4b"},
+ {file = "ruff-0.3.7-py3-none-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d28bdf3d7dc71dd46929fafeec98ba89b7c3550c3f0978e36389b5631b793663"},
+ {file = "ruff-0.3.7-py3-none-manylinux_2_17_armv7l.manylinux2014_armv7l.whl", hash = "sha256:379b67d4f49774ba679593b232dcd90d9e10f04d96e3c8ce4a28037ae473f7bb"},
+ {file = "ruff-0.3.7-py3-none-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:c060aea8ad5ef21cdfbbe05475ab5104ce7827b639a78dd55383a6e9895b7c51"},
+ {file = "ruff-0.3.7-py3-none-manylinux_2_17_ppc64.manylinux2014_ppc64.whl", hash = "sha256:ebf8f615dde968272d70502c083ebf963b6781aacd3079081e03b32adfe4d58a"},
+ {file = "ruff-0.3.7-py3-none-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:d48098bd8f5c38897b03604f5428901b65e3c97d40b3952e38637b5404b739a2"},
+ {file = "ruff-0.3.7-py3-none-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:da8a4fda219bf9024692b1bc68c9cff4b80507879ada8769dc7e985755d662ea"},
+ {file = "ruff-0.3.7-py3-none-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:6c44e0149f1d8b48c4d5c33d88c677a4aa22fd09b1683d6a7ff55b816b5d074f"},
+ {file = "ruff-0.3.7-py3-none-musllinux_1_2_aarch64.whl", hash = "sha256:3050ec0af72b709a62ecc2aca941b9cd479a7bf2b36cc4562f0033d688e44fa1"},
+ {file = "ruff-0.3.7-py3-none-musllinux_1_2_armv7l.whl", hash = "sha256:a29cc38e4c1ab00da18a3f6777f8b50099d73326981bb7d182e54a9a21bb4ff7"},
+ {file = "ruff-0.3.7-py3-none-musllinux_1_2_i686.whl", hash = "sha256:5b15cc59c19edca917f51b1956637db47e200b0fc5e6e1878233d3a938384b0b"},
+ {file = "ruff-0.3.7-py3-none-musllinux_1_2_x86_64.whl", hash = "sha256:e491045781b1e38b72c91247cf4634f040f8d0cb3e6d3d64d38dcf43616650b4"},
+ {file = "ruff-0.3.7-py3-none-win32.whl", hash = "sha256:bc931de87593d64fad3a22e201e55ad76271f1d5bfc44e1a1887edd0903c7d9f"},
+ {file = "ruff-0.3.7-py3-none-win_amd64.whl", hash = "sha256:5ef0e501e1e39f35e03c2acb1d1238c595b8bb36cf7a170e7c1df1b73da00e74"},
+ {file = "ruff-0.3.7-py3-none-win_arm64.whl", hash = "sha256:789e144f6dc7019d1f92a812891c645274ed08af6037d11fc65fcbc183b7d59f"},
+ {file = "ruff-0.3.7.tar.gz", hash = "sha256:d5c1aebee5162c2226784800ae031f660c350e7a3402c4d1f8ea4e97e232e3ba"},
]
[[package]]
@@ -3060,18 +3383,18 @@ test = ["asv", "gmpy2", "mpmath", "pytest", "pytest-cov", "pytest-xdist", "sciki
[[package]]
name = "setuptools"
-version = "69.2.0"
+version = "69.5.1"
description = "Easily download, build, install, upgrade, and uninstall Python packages"
optional = false
python-versions = ">=3.8"
files = [
- {file = "setuptools-69.2.0-py3-none-any.whl", hash = "sha256:c21c49fb1042386df081cb5d86759792ab89efca84cf114889191cd09aacc80c"},
- {file = "setuptools-69.2.0.tar.gz", hash = "sha256:0ff4183f8f42cd8fa3acea16c45205521a4ef28f73c6391d8a25e92893134f2e"},
+ {file = "setuptools-69.5.1-py3-none-any.whl", hash = "sha256:c636ac361bc47580504644275c9ad802c50415c7522212252c033bd15f301f32"},
+ {file = "setuptools-69.5.1.tar.gz", hash = "sha256:6c1fccdac05a97e598fb0ae3bbed5904ccb317337a51139dcd51453611bbb987"},
]
[package.extras]
-docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "pygments-github-lexers (==0.0.5)", "rst.linker (>=1.9)", "sphinx (<7.2.5)", "sphinx (>=3.5)", "sphinx-favicon", "sphinx-inline-tabs", "sphinx-lint", "sphinx-notfound-page (>=1,<2)", "sphinx-reredirects", "sphinxcontrib-towncrier"]
-testing = ["build[virtualenv]", "filelock (>=3.4.0)", "importlib-metadata", "ini2toml[lite] (>=0.9)", "jaraco.develop (>=7.21)", "jaraco.envs (>=2.2)", "jaraco.path (>=3.2.0)", "mypy (==1.9)", "packaging (>=23.2)", "pip (>=19.1)", "pytest (>=6)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-home (>=0.5)", "pytest-mypy (>=0.9.1)", "pytest-perf", "pytest-ruff (>=0.2.1)", "pytest-timeout", "pytest-xdist (>=3)", "tomli", "tomli-w (>=1.0.0)", "virtualenv (>=13.0.0)", "wheel"]
+docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "pygments-github-lexers (==0.0.5)", "rst.linker (>=1.9)", "sphinx (>=3.5)", "sphinx-favicon", "sphinx-inline-tabs", "sphinx-lint", "sphinx-notfound-page (>=1,<2)", "sphinx-reredirects", "sphinxcontrib-towncrier"]
+testing = ["build[virtualenv]", "filelock (>=3.4.0)", "importlib-metadata", "ini2toml[lite] (>=0.9)", "jaraco.develop (>=7.21)", "jaraco.envs (>=2.2)", "jaraco.path (>=3.2.0)", "mypy (==1.9)", "packaging (>=23.2)", "pip (>=19.1)", "pytest (>=6,!=8.1.1)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-home (>=0.5)", "pytest-mypy", "pytest-perf", "pytest-ruff (>=0.2.1)", "pytest-timeout", "pytest-xdist (>=3)", "tomli", "tomli-w (>=1.0.0)", "virtualenv (>=13.0.0)", "wheel"]
testing-integration = ["build[virtualenv] (>=1.0.3)", "filelock (>=3.4.0)", "jaraco.envs (>=2.2)", "jaraco.path (>=3.2.0)", "packaging (>=23.2)", "pytest", "pytest-enabler", "pytest-xdist", "tomli", "virtualenv (>=13.0.0)", "wheel"]
[[package]]
@@ -3142,84 +3465,6 @@ docs = ["sphinxcontrib-websupport"]
lint = ["docutils-stubs", "flake8 (>=3.5.0)", "flake8-simplify", "isort", "mypy (>=0.990)", "ruff", "sphinx-lint", "types-requests"]
test = ["cython", "filelock", "html5lib", "pytest (>=4.6)"]
-[[package]]
-name = "sphinx-autoapi"
-version = "3.0.0"
-description = "Sphinx API documentation generator"
-optional = false
-python-versions = ">=3.8"
-files = [
- {file = "sphinx-autoapi-3.0.0.tar.gz", hash = "sha256:09ebd674a32b44467222b0fb8a917b97c89523f20dbf05b52cb8a3f0e15714de"},
- {file = "sphinx_autoapi-3.0.0-py2.py3-none-any.whl", hash = "sha256:ea207793cba1feff7b2ded0e29364f2995a4d157303a98603cee0ce94cea2688"},
-]
-
-[package.dependencies]
-anyascii = "*"
-astroid = [
- {version = ">=2.7", markers = "python_version < \"3.12\""},
- {version = ">=3.0.0a1", markers = "python_version >= \"3.12\""},
-]
-Jinja2 = "*"
-PyYAML = "*"
-sphinx = ">=6.1.0"
-
-[package.extras]
-docs = ["furo", "sphinx", "sphinx-design"]
-
-[[package]]
-name = "sphinx-autobuild"
-version = "2021.3.14"
-description = "Rebuild Sphinx documentation on changes, with live-reload in the browser."
-optional = false
-python-versions = ">=3.6"
-files = [
- {file = "sphinx-autobuild-2021.3.14.tar.gz", hash = "sha256:de1ca3b66e271d2b5b5140c35034c89e47f263f2cd5db302c9217065f7443f05"},
- {file = "sphinx_autobuild-2021.3.14-py3-none-any.whl", hash = "sha256:8fe8cbfdb75db04475232f05187c776f46f6e9e04cacf1e49ce81bdac649ccac"},
-]
-
-[package.dependencies]
-colorama = "*"
-livereload = "*"
-sphinx = "*"
-
-[package.extras]
-test = ["pytest", "pytest-cov"]
-
-[[package]]
-name = "sphinx-basic-ng"
-version = "1.0.0b2"
-description = "A modern skeleton for Sphinx themes."
-optional = false
-python-versions = ">=3.7"
-files = [
- {file = "sphinx_basic_ng-1.0.0b2-py3-none-any.whl", hash = "sha256:eb09aedbabfb650607e9b4b68c9d240b90b1e1be221d6ad71d61c52e29f7932b"},
- {file = "sphinx_basic_ng-1.0.0b2.tar.gz", hash = "sha256:9ec55a47c90c8c002b5960c57492ec3021f5193cb26cebc2dc4ea226848651c9"},
-]
-
-[package.dependencies]
-sphinx = ">=4.0"
-
-[package.extras]
-docs = ["furo", "ipython", "myst-parser", "sphinx-copybutton", "sphinx-inline-tabs"]
-
-[[package]]
-name = "sphinx-copybutton"
-version = "0.5.2"
-description = "Add a copy button to each of your code cells."
-optional = false
-python-versions = ">=3.7"
-files = [
- {file = "sphinx-copybutton-0.5.2.tar.gz", hash = "sha256:4cf17c82fb9646d1bc9ca92ac280813a3b605d8c421225fd9913154103ee1fbd"},
- {file = "sphinx_copybutton-0.5.2-py3-none-any.whl", hash = "sha256:fb543fd386d917746c9a2c50360c7905b605726b9355cd26e9974857afeae06e"},
-]
-
-[package.dependencies]
-sphinx = ">=1.8"
-
-[package.extras]
-code-style = ["pre-commit (==2.12.1)"]
-rtd = ["ipython", "myst-nb", "sphinx", "sphinx-book-theme", "sphinx-examples"]
-
[[package]]
name = "sphinxcontrib-applehelp"
version = "1.0.4"
@@ -3445,13 +3690,13 @@ files = [
[[package]]
name = "tinycss2"
-version = "1.2.1"
+version = "1.3.0"
description = "A tiny CSS parser"
optional = false
-python-versions = ">=3.7"
+python-versions = ">=3.8"
files = [
- {file = "tinycss2-1.2.1-py3-none-any.whl", hash = "sha256:2b80a96d41e7c3914b8cda8bc7f705a4d9c49275616e886103dd839dfc847847"},
- {file = "tinycss2-1.2.1.tar.gz", hash = "sha256:8cff3a8f066c2ec677c06dbc7b45619804a6938478d9d73c284b29d14ecb0627"},
+ {file = "tinycss2-1.3.0-py3-none-any.whl", hash = "sha256:54a8dbdffb334d536851be0226030e9505965bb2f30f21a4a82c55fb2a80fae7"},
+ {file = "tinycss2-1.3.0.tar.gz", hash = "sha256:152f9acabd296a8375fbca5b84c961ff95971fcfc32e79550c8df8e29118c54d"},
]
[package.dependencies]
@@ -3459,7 +3704,18 @@ webencodings = ">=0.4"
[package.extras]
doc = ["sphinx", "sphinx_rtd_theme"]
-test = ["flake8", "isort", "pytest"]
+test = ["pytest", "ruff"]
+
+[[package]]
+name = "toml"
+version = "0.10.2"
+description = "Python Library for Tom's Obvious, Minimal Language"
+optional = false
+python-versions = ">=2.6, !=3.0.*, !=3.1.*, !=3.2.*"
+files = [
+ {file = "toml-0.10.2-py2.py3-none-any.whl", hash = "sha256:806143ae5bfb6a3c6e736a764057db0e6a0e05e338b5630894a5f779cabb4f9b"},
+ {file = "toml-0.10.2.tar.gz", hash = "sha256:b3bda1d108d5dd99f4a20d24d9c348e91c4db7ab1b749200bded2f839ccbe68f"},
+]
[[package]]
name = "tomli"
@@ -3505,28 +3761,28 @@ files = [
[[package]]
name = "traitlets"
-version = "5.14.2"
+version = "5.14.3"
description = "Traitlets Python configuration system"
optional = false
python-versions = ">=3.8"
files = [
- {file = "traitlets-5.14.2-py3-none-any.whl", hash = "sha256:fcdf85684a772ddeba87db2f398ce00b40ff550d1528c03c14dbf6a02003cd80"},
- {file = "traitlets-5.14.2.tar.gz", hash = "sha256:8cdd83c040dab7d1dee822678e5f5d100b514f7b72b01615b26fc5718916fdf9"},
+ {file = "traitlets-5.14.3-py3-none-any.whl", hash = "sha256:b74e89e397b1ed28cc831db7aea759ba6640cb3de13090ca145426688ff1ac4f"},
+ {file = "traitlets-5.14.3.tar.gz", hash = "sha256:9ed0579d3502c94b4b3732ac120375cda96f923114522847de4b3bb98b96b6b7"},
]
[package.extras]
docs = ["myst-parser", "pydata-sphinx-theme", "sphinx"]
-test = ["argcomplete (>=3.0.3)", "mypy (>=1.7.0)", "pre-commit", "pytest (>=7.0,<8.1)", "pytest-mock", "pytest-mypy-testing"]
+test = ["argcomplete (>=3.0.3)", "mypy (>=1.7.0)", "pre-commit", "pytest (>=7.0,<8.2)", "pytest-mock", "pytest-mypy-testing"]
[[package]]
name = "typing-extensions"
-version = "4.10.0"
+version = "4.11.0"
description = "Backported and Experimental Type Hints for Python 3.8+"
optional = false
python-versions = ">=3.8"
files = [
- {file = "typing_extensions-4.10.0-py3-none-any.whl", hash = "sha256:69b1a937c3a517342112fb4c6df7e72fc39a38e7891a5730ed4985b5214b5475"},
- {file = "typing_extensions-4.10.0.tar.gz", hash = "sha256:b0abd7c89e8fb96f98db18d86106ff1d90ab692004eb746cf6eda2682f91b3cb"},
+ {file = "typing_extensions-4.11.0-py3-none-any.whl", hash = "sha256:c1f94d72897edaf4ce775bb7558d5b79d8126906a14ea5ed1635921406c0387a"},
+ {file = "typing_extensions-4.11.0.tar.gz", hash = "sha256:83f085bd5ca59c80295fc2a82ab5dac679cbe02b9f33f7d83af68e241bea51b0"},
]
[[package]]
@@ -3558,30 +3814,30 @@ socks = ["PySocks (>=1.5.6,!=1.5.7,<2.0)"]
[[package]]
name = "urllib3"
-version = "2.0.7"
+version = "2.2.1"
description = "HTTP library with thread-safe connection pooling, file post, and more."
optional = false
-python-versions = ">=3.7"
+python-versions = ">=3.8"
files = [
- {file = "urllib3-2.0.7-py3-none-any.whl", hash = "sha256:fdb6d215c776278489906c2f8916e6e7d4f5a9b602ccbcfdf7f016fc8da0596e"},
- {file = "urllib3-2.0.7.tar.gz", hash = "sha256:c97dfde1f7bd43a71c8d2a58e369e9b2bf692d1334ea9f9cae55add7d0dd0f84"},
+ {file = "urllib3-2.2.1-py3-none-any.whl", hash = "sha256:450b20ec296a467077128bff42b73080516e71b56ff59a60a02bef2232c4fa9d"},
+ {file = "urllib3-2.2.1.tar.gz", hash = "sha256:d0570876c61ab9e520d776c38acbbb5b05a776d3f9ff98a5c8fd5162a444cf19"},
]
[package.extras]
brotli = ["brotli (>=1.0.9)", "brotlicffi (>=0.8.0)"]
-secure = ["certifi", "cryptography (>=1.9)", "idna (>=2.0.0)", "pyopenssl (>=17.1.0)", "urllib3-secure-extra"]
+h2 = ["h2 (>=4,<5)"]
socks = ["pysocks (>=1.5.6,!=1.5.7,<2.0)"]
zstd = ["zstandard (>=0.18.0)"]
[[package]]
name = "virtualenv"
-version = "20.25.1"
+version = "20.26.0"
description = "Virtual Python Environment builder"
optional = false
python-versions = ">=3.7"
files = [
- {file = "virtualenv-20.25.1-py3-none-any.whl", hash = "sha256:961c026ac520bac5f69acb8ea063e8a4f071bcc9457b9c1f28f6b085c511583a"},
- {file = "virtualenv-20.25.1.tar.gz", hash = "sha256:e08e13ecdca7a0bd53798f356d5831434afa5b07b93f0abdf0797b7a06ffe197"},
+ {file = "virtualenv-20.26.0-py3-none-any.whl", hash = "sha256:0846377ea76e818daaa3e00a4365c018bc3ac9760cbb3544de542885aad61fb3"},
+ {file = "virtualenv-20.26.0.tar.gz", hash = "sha256:ec25a9671a5102c8d2657f62792a27b48f016664c6873f6beed3800008577210"},
]
[package.dependencies]
@@ -3590,24 +3846,9 @@ filelock = ">=3.12.2,<4"
platformdirs = ">=3.9.1,<5"
[package.extras]
-docs = ["furo (>=2023.7.26)", "proselint (>=0.13)", "sphinx (>=7.1.2)", "sphinx-argparse (>=0.4)", "sphinxcontrib-towncrier (>=0.2.1a0)", "towncrier (>=23.6)"]
+docs = ["furo (>=2023.7.26)", "proselint (>=0.13)", "sphinx (>=7.1.2,!=7.3)", "sphinx-argparse (>=0.4)", "sphinxcontrib-towncrier (>=0.2.1a0)", "towncrier (>=23.6)"]
test = ["covdefaults (>=2.3)", "coverage (>=7.2.7)", "coverage-enable-subprocess (>=1)", "flaky (>=3.7)", "packaging (>=23.1)", "pytest (>=7.4)", "pytest-env (>=0.8.2)", "pytest-freezer (>=0.4.8)", "pytest-mock (>=3.11.1)", "pytest-randomly (>=3.12)", "pytest-timeout (>=2.1)", "setuptools (>=68)", "time-machine (>=2.10)"]
-[[package]]
-name = "waitress"
-version = "3.0.0"
-description = "Waitress WSGI server"
-optional = false
-python-versions = ">=3.8.0"
-files = [
- {file = "waitress-3.0.0-py3-none-any.whl", hash = "sha256:2a06f242f4ba0cc563444ca3d1998959447477363a2d7e9b8b4d75d35cfd1669"},
- {file = "waitress-3.0.0.tar.gz", hash = "sha256:005da479b04134cdd9dd602d1ee7c49d79de0537610d653674cc6cbde222b8a1"},
-]
-
-[package.extras]
-docs = ["Sphinx (>=1.8.1)", "docutils", "pylons-sphinx-themes (>=1.0.9)"]
-testing = ["coverage (>=5.0)", "pytest", "pytest-cov"]
-
[[package]]
name = "watchdog"
version = "4.0.0"
@@ -3649,6 +3890,20 @@ files = [
[package.extras]
watchmedo = ["PyYAML (>=3.10)"]
+[[package]]
+name = "wcmatch"
+version = "8.5.1"
+description = "Wildcard/glob file name matcher."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "wcmatch-8.5.1-py3-none-any.whl", hash = "sha256:24c19cedc92bc9c9e27f39db4e1824d72f95bd2cea32b254a47a45b1a1b227ed"},
+ {file = "wcmatch-8.5.1.tar.gz", hash = "sha256:c0088c7f6426cf6bf27e530e2b7b734031905f7e490475fd83c7c5008ab581b3"},
+]
+
+[package.dependencies]
+bracex = ">=2.1.1"
+
[[package]]
name = "wcwidth"
version = "0.2.13"
@@ -3672,39 +3927,18 @@ files = [
]
[[package]]
-name = "webob"
-version = "1.8.7"
-description = "WSGI request and response object"
-optional = false
-python-versions = ">=2.7,!=3.0.*,!=3.1.*,!=3.2.*"
-files = [
- {file = "WebOb-1.8.7-py2.py3-none-any.whl", hash = "sha256:73aae30359291c14fa3b956f8b5ca31960e420c28c1bec002547fb04928cf89b"},
- {file = "WebOb-1.8.7.tar.gz", hash = "sha256:b64ef5141be559cfade448f044fa45c2260351edcb6a8ef6b7e00c7dcef0c323"},
-]
-
-[package.extras]
-docs = ["Sphinx (>=1.7.5)", "pylons-sphinx-themes"]
-testing = ["coverage", "pytest (>=3.1.0)", "pytest-cov", "pytest-xdist"]
-
-[[package]]
-name = "webtest"
-version = "3.0.0"
-description = "Helper to test WSGI applications"
+name = "wheel"
+version = "0.43.0"
+description = "A built-package format for Python"
optional = false
-python-versions = ">=3.6, <4"
+python-versions = ">=3.8"
files = [
- {file = "WebTest-3.0.0-py3-none-any.whl", hash = "sha256:2a001a9efa40d2a7e5d9cd8d1527c75f41814eb6afce2c3d207402547b1e5ead"},
- {file = "WebTest-3.0.0.tar.gz", hash = "sha256:54bd969725838d9861a9fa27f8d971f79d275d94ae255f5c501f53bb6d9929eb"},
+ {file = "wheel-0.43.0-py3-none-any.whl", hash = "sha256:55c570405f142630c6b9f72fe09d9b67cf1477fcf543ae5b8dcb1f5b7377da81"},
+ {file = "wheel-0.43.0.tar.gz", hash = "sha256:465ef92c69fa5c5da2d1cf8ac40559a8c940886afcef87dcf14b9470862f1d85"},
]
-[package.dependencies]
-beautifulsoup4 = "*"
-waitress = ">=0.8.5"
-WebOb = ">=1.2"
-
[package.extras]
-docs = ["Sphinx (>=1.8.1)", "docutils", "pylons-sphinx-themes (>=1.0.8)"]
-tests = ["PasteDeploy", "WSGIProxy2", "coverage", "pyquery", "pytest", "pytest-cov"]
+test = ["pytest (>=6.0.0)", "setuptools (>=65)"]
[[package]]
name = "wrapt"
@@ -3805,7 +4039,7 @@ test = ["pytest", "pytest-cov"]
name = "yarl"
version = "1.9.4"
description = "Yet another URL library"
-optional = false
+optional = true
python-versions = ">=3.7"
files = [
{file = "yarl-1.9.4-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:a8c1df72eb746f4136fe9a2e72b0c9dc1da1cbd23b5372f94b5820ff8ae30e0e"},
@@ -3926,4 +4160,4 @@ collate = ["cytominer-database"]
[metadata]
lock-version = "2.0"
python-versions = ">=3.8,<4.0"
-content-hash = "8cd2e790b6c426bf02ddd240f8cd3d34bc72aa79d23cb012ab51bd48bff15a38"
+content-hash = "70a56ee15e4d18ef704d2dde4ac560fd4089a00a75df0ef9650126aec0e75f27"
diff --git a/pycytominer/__about__.py b/pycytominer/__about__.py
index c76a4b74..cbc13087 100644
--- a/pycytominer/__about__.py
+++ b/pycytominer/__about__.py
@@ -1,6 +1,8 @@
+"""Pycytominer package metadata."""
+
__project__ = "pycytominer"
# These version placeholders are updated during build by poetry-dynamic-versioning
-__version__ = "0.0.0"
-__version_tuple__ = (0, 0, 0)
+__version__ = "1.1.0.post29.dev0+9615c0d"
+__version_tuple__ = (1, 1, 0, "post29", "dev0", "9615c0d")
__license__ = "BSD 3-Clause License"
__author__ = "Pycytominer Contributors"
diff --git a/pycytominer/__config__.py b/pycytominer/__config__.py
index 4ccc6df3..dd6cb0f4 100644
--- a/pycytominer/__config__.py
+++ b/pycytominer/__config__.py
@@ -1,6 +1,4 @@
-"""
-Module used for pycytominer configuration details.
-"""
+"""Module used for pycytominer configuration details."""
import pandas as pd
diff --git a/pycytominer/__init__.py b/pycytominer/__init__.py
index 88937371..843b014d 100644
--- a/pycytominer/__init__.py
+++ b/pycytominer/__init__.py
@@ -1,3 +1,5 @@
+"""Pycytominer is a suite of common functions used to process high dimensional readouts from high-throughput cell experiments."""
+
from pycytominer import __about__, __config__
from .aggregate import aggregate
diff --git a/pycytominer/aggregate.py b/pycytominer/aggregate.py
index bf8403ba..b8e06a78 100644
--- a/pycytominer/aggregate.py
+++ b/pycytominer/aggregate.py
@@ -1,6 +1,4 @@
-"""
-Aggregate profiles based on given grouping variables.
-"""
+"""Aggregate profiles based on given grouping variables."""
from typing import Any, Dict, List, Optional, Union
@@ -67,7 +65,6 @@ def aggregate(
data.
"""
-
# Check that the operation is supported
operation = check_aggregate_operation(operation)
diff --git a/pycytominer/annotate.py b/pycytominer/annotate.py
index d5eb2bcb..e7eca779 100644
--- a/pycytominer/annotate.py
+++ b/pycytominer/annotate.py
@@ -1,6 +1,4 @@
-"""
-Annotates profiles with metadata information
-"""
+"""Annotates profiles with metadata information."""
import os
@@ -77,7 +75,6 @@ def annotate(
DataFrame. If you specify output_file, then write to file and do not return
data.
"""
-
# Load Data
profiles = load_profiles(profiles)
platemap = load_platemap(platemap, add_metadata_id_to_platemap)
diff --git a/pycytominer/consensus.py b/pycytominer/consensus.py
index 7ea19714..933df97d 100644
--- a/pycytominer/consensus.py
+++ b/pycytominer/consensus.py
@@ -1,6 +1,4 @@
-"""
-Acquire consensus signatures for input samples
-"""
+"""Acquire consensus signatures for input samples."""
from pycytominer import aggregate
from pycytominer.cyto_utils import (
diff --git a/pycytominer/cyto_utils/DeepProfiler_processing.py b/pycytominer/cyto_utils/DeepProfiler_processing.py
index e33f1036..254f4291 100644
--- a/pycytominer/cyto_utils/DeepProfiler_processing.py
+++ b/pycytominer/cyto_utils/DeepProfiler_processing.py
@@ -1,6 +1,4 @@
-"""
-Utility function to load and process the output files of a DeepProfiler run.
-"""
+"""Utility function to load and process the output files of a DeepProfiler run."""
import os
import pathlib
@@ -17,7 +15,7 @@
class DeepProfilerData:
- """This class holds all functions needed to load and annotate the DeepProfiler (DP) run.
+ """Class that holds all functions needed to load and annotate the DeepProfiler (DP) run.
Attributes
----------
@@ -58,7 +56,6 @@ def __init__(
See above for all other parameters.
"""
-
self.index_df = pd.read_csv(index_file, dtype=str)
self.profile_dir = profile_dir
self.filename_delimiter = filename_delimiter
@@ -67,9 +64,7 @@ def __init__(
self.file_extension = f".{self.file_extension}"
def build_filenames(self):
- """
- Create file names indicated by plate, well, and site information
- """
+ """Create file names indicated by plate, well, and site information."""
self.filenames = self.index_df.apply(
self.build_filename_from_index, axis="columns"
)
@@ -78,9 +73,7 @@ def build_filenames(self):
]
def build_filename_from_index(self, row):
- """
- Builds the name of the profile files
- """
+ """Build the name of the profile files."""
plate = row["Metadata_Plate"]
well = row["Metadata_Well"]
site = row["Metadata_Site"]
@@ -89,9 +82,10 @@ def build_filename_from_index(self, row):
return filename
def extract_filename_metadata(self, npz_file, delimiter="_"):
- """
- Extract metadata (site, well and plate) from the filename.
- The input format of the file: path/plate/well{delimiter}site.npz
+ """Extract metadata (site, well and plate) from the filename.
+
+ This function is used to extract the metadata from the filename of the npz files.
+ It expects a naming convetion of path/plate/well{delimiter}site.npz.
Arguments
---------
@@ -120,7 +114,7 @@ def extract_filename_metadata(self, npz_file, delimiter="_"):
class AggregateDeepProfiler:
- """This class holds all functions needed to aggregate the DeepProfiler (DP) run.
+ """Class that holds all functions needed to aggregate the DeepProfiler (DP) run.
Attributes
----------
@@ -183,10 +177,9 @@ def __init__(
self.output_file = output_file
def setup_aggregate(self):
- """
- Sets up the file_aggregate attribute. This is a helper function to aggregate_deep().
+ """Set up the file_aggregate attribute.
- the file_aggregate dictionary contains the file locations and metadata for each grouping.
+ A helper function to `aggregate_deep` that aggregate the file_aggregate dictionary contains the file locations and metadata for each grouping.
If for example we are grouping by well then the keys of self.file_aggregate would be:
plate1/well1, plate1/well2, plate2/well1, etc.
"""
@@ -218,7 +211,7 @@ def setup_aggregate(self):
def aggregate_deep(self):
"""
- Main function of this class. Aggregates the profiles into a pandas dataframe.
+ Aggregate the DeepProfiler profiles into a pandas dataframe.
For each key in file_aggregate, the profiles are loaded, concatenated and then aggregated.
If files are missing, we throw a warning but continue the code.
@@ -310,7 +303,9 @@ def aggregate_deep(self):
class SingleCellDeepProfiler:
- """This class holds functions needed to analyze single cells from the DeepProfiler (DP) run. Only pycytominer.normalization() is implemented.
+ """Class that holds functions needed to analyze single cells from the DeepProfiler (DP) run.
+
+ Only pycytominer.normalization() is implemented.
Attributes
----------
@@ -354,15 +349,14 @@ def __init__(
---------
See above for all parameters.
"""
-
self.deep_data = deep_data
def get_single_cells(
self, output=False, location_x_col_index=0, location_y_col_index=1
):
- """
- Sets up the single_cells attribute or output as a variable. This is a helper function to normalize_deep_single_cells().
- single_cells is a pandas dataframe in the format expected by pycytominer.normalize().
+ """Set up a single_cells dataframe in the format expected by pycytominer.normalize().
+
+ Helper function to normalize_deep_single_cells() that sets up the single_cells attribute or outputs it as a dataframe.
Arguments
-----------
@@ -417,7 +411,7 @@ def normalize_deep_single_cells(
spherize_epsilon=1e-6,
):
"""
- Normalizes all cells into a pandas dataframe.
+ Normalize all cells into a pandas dataframe.
For each file in the DP project features folder, the features from each cell are loaded.
These features are put into a profiles dataframe for use in pycytominer.normalize.
diff --git a/pycytominer/cyto_utils/__init__.py b/pycytominer/cyto_utils/__init__.py
index 25456868..f9a0407e 100644
--- a/pycytominer/cyto_utils/__init__.py
+++ b/pycytominer/cyto_utils/__init__.py
@@ -1,3 +1,5 @@
+"""A variety of utility functions for working with cytominer data."""
+
from .output import output
from .util import (
check_compartments,
diff --git a/pycytominer/cyto_utils/annotate_custom.py b/pycytominer/cyto_utils/annotate_custom.py
index a9c9f056..d06edb01 100644
--- a/pycytominer/cyto_utils/annotate_custom.py
+++ b/pycytominer/cyto_utils/annotate_custom.py
@@ -1,10 +1,12 @@
+"""Functions to annotate data frames with custom options according to CMAP specifications."""
+
import numpy as np
def annotate_cmap(
annotated, annotate_join_on, cell_id="unknown", perturbation_mode="none"
):
- """Annotates data frame with custom options according to CMAP specifications
+ """Annotates data frame with custom options according to CMAP specifications.
Parameters
----------
@@ -103,7 +105,7 @@ def annotate_cmap(
def cp_clean(profiles):
- """Specifically clean certain column names derived from different CellProfiler versions
+ """Specifically clean certain column names derived from different CellProfiler versions.
Parameters
----------
@@ -115,7 +117,6 @@ def cp_clean(profiles):
profiles
Renamed to standard metadata
"""
-
profiles = profiles.rename(
{
"Image_Metadata_Plate": "Metadata_Plate",
diff --git a/pycytominer/cyto_utils/cell_locations.py b/pycytominer/cyto_utils/cell_locations.py
index 612ae873..04ee9077 100644
--- a/pycytominer/cyto_utils/cell_locations.py
+++ b/pycytominer/cyto_utils/cell_locations.py
@@ -1,6 +1,4 @@
-"""
-Utility function to augment a metadata file with X,Y locations of cells in each image
-"""
+"""Utility function to augment a metadata file with X,Y locations of cells in each image."""
import pathlib
import pandas as pd
@@ -14,8 +12,7 @@
class CellLocation:
- """This class holds all the functions augment a metadata file with X,Y
- locations of cells in each image.
+ """Class holding all the functions augment a metadata file with X,Y locations of cells in each image.
In the metadata file, which is either a CSV or a Parquet file,
- Each row is single multi-channel image
@@ -107,13 +104,13 @@ def __init__(
)
def _expanduser(self, obj: Union[str, None]):
- """Expand the user home directory in a path"""
+ """Expand the user home directory in a path."""
if obj is not None and isinstance(obj, str) and not obj.startswith("s3://"):
return pathlib.Path(obj).expanduser().as_posix()
return obj
def _parse_s3_path(self, s3_path: str):
- """Parse an S3 path into a bucket and key
+ """Parse an S3 path into a bucket and key.
Parameters
----------
@@ -127,7 +124,6 @@ def _parse_s3_path(self, s3_path: str):
str
The key
"""
-
s3_path = s3_path.replace("s3://", "")
bucket = s3_path.split("/")[0]
@@ -137,7 +133,7 @@ def _parse_s3_path(self, s3_path: str):
return bucket, key
def _s3_file_exists(self, s3_path: str):
- """Check if a file exists on S3
+ """Check if a file exists on S3.
Parameters
----------
@@ -149,7 +145,6 @@ def _s3_file_exists(self, s3_path: str):
bool
True if the file exists on S3, False otherwise
"""
-
bucket, key = self._parse_s3_path(s3_path)
try:
@@ -163,10 +158,7 @@ def _s3_file_exists(self, s3_path: str):
return True
def _download_s3(self, uri: str):
- """
- Download a file from S3 to a temporary file and return the temporary path
- """
-
+ """Download a file from S3 to a temporary file and return the temporary path."""
bucket, key = self._parse_s3_path(uri)
tmp_file = tempfile.NamedTemporaryFile(
@@ -178,14 +170,13 @@ def _download_s3(self, uri: str):
return tmp_file.name
def _load_metadata(self):
- """Load the metadata into a Pandas DataFrame
+ """Load the metadata into a Pandas DataFrame.
Returns
-------
Pandas DataFrame
The metadata loaded into a Pandas DataFrame
"""
-
if not isinstance(self.metadata_input, pd.DataFrame):
# verify that the metadata file is a CSV or a Parquet file
@@ -224,7 +215,7 @@ def _load_metadata(self):
return df
def _create_nested_df(self, df: pd.DataFrame):
- """Create a new column `CellCenters` by nesting the X and Y locations of cell from an image into the row of the image
+ """Create a new column `CellCenters` by nesting the X and Y locations of cell from an image into the row of the image.
Parameters
----------
@@ -235,7 +226,6 @@ def _create_nested_df(self, df: pd.DataFrame):
-------
Pandas DataFrame
"""
-
# define a dictionary to store the output
output_df_list = collections.defaultdict(list)
@@ -271,10 +261,7 @@ def _create_nested_df(self, df: pd.DataFrame):
return pd.DataFrame(output_df_list)
def _get_single_cell_engine(self):
- """
- Get the sqlalchemy.engine.Engine object for the single_cell file
- """
-
+ """Get the sqlalchemy.engine.Engine object for the single_cell file."""
if isinstance(self.single_cell_input, str):
# check if the single_cell file is a SQLite file
if not self.single_cell_input.endswith(".sqlite"):
@@ -298,10 +285,7 @@ def _get_single_cell_engine(self):
return temp_single_cell_input, engine
def _check_single_cell_correctness(self, engine: sqlalchemy.engine.Engine):
- """
- Check that the single_cell file has the required tables and columns
- """
-
+ """Check that the single_cell file has the required tables and columns."""
inspector = sqlalchemy.inspect(engine)
if not all(
@@ -342,9 +326,7 @@ def _check_single_cell_correctness(self, engine: sqlalchemy.engine.Engine):
)
def _get_joined_image_nuclei_tables(self):
- """
- Merge the Image and Nuclei tables in SQL
- """
+ """Merge the Image and Nuclei tables in SQL."""
# get the sqlalchemy.engine.Engine object for the single_cell file
temp_single_cell_input, engine = self._get_single_cell_engine()
@@ -382,18 +364,18 @@ def _get_joined_image_nuclei_tables(self):
return joined_df
def _load_single_cell(self):
- """Load the required columns from the `Image` and `Nuclei` tables in the single_cell file or sqlalchemy.engine.Engine object into a Pandas DataFrame
+ """Load the required columns from the `Image` and `Nuclei` tables in the single_cell file or sqlalchemy.engine.Engine object into a Pandas DataFrame.
Returns
-------
Pandas DataFrame
The required columns from the `Image` and `Nuclei` tables loaded into a Pandas DataFrame
"""
-
return self._create_nested_df(self._get_joined_image_nuclei_tables())
def add_cell_location(self):
"""Add the X,Y locations of all cells to the metadata file in the corresponding row, packed into a single column.
+
Optionally, save the augmented metadata file as a Parquet file.
Returns
@@ -401,7 +383,6 @@ def add_cell_location(self):
Pandas DataFrame
Either a data frame or the path to a Parquet file with the X,Y locations of all cells packed into a single column
"""
-
# If self.augmented_metadata_output is not None and it is a str and the file already exists, there is nothing to do
if (
self.augmented_metadata_output is not None
diff --git a/pycytominer/cyto_utils/cell_locations_cmd.py b/pycytominer/cyto_utils/cell_locations_cmd.py
index f0e08891..3b96965f 100644
--- a/pycytominer/cyto_utils/cell_locations_cmd.py
+++ b/pycytominer/cyto_utils/cell_locations_cmd.py
@@ -1,3 +1,5 @@
+"""CLI for cell location calculations."""
+
from pycytominer.cyto_utils.cell_locations import CellLocation
import fire
diff --git a/pycytominer/cyto_utils/cells.py b/pycytominer/cyto_utils/cells.py
index 4e0a80f9..0e07ae61 100644
--- a/pycytominer/cyto_utils/cells.py
+++ b/pycytominer/cyto_utils/cells.py
@@ -1,3 +1,5 @@
+"""Module containing the SingleCells class, which is used to interact with single cell morphological profiles."""
+
from typing import Dict, Union, Optional
import numpy as np
@@ -25,8 +27,7 @@
class SingleCells:
- """This is a class to interact with single cell morphological profiles. Interaction
- includes aggregation, normalization, and output.
+ """Class to interact with single cell morphological profiles including aggregation, normalization, and output.
Attributes
----------
@@ -115,7 +116,7 @@ def __init__(
object_feature="Metadata_ObjectNumber",
default_datatype_float=np.float64,
):
- """Constructor method"""
+ """Construct a SingleCells object."""
# Check compartments specified
check_compartments(compartments)
@@ -179,21 +180,20 @@ def __init__(
self.load_image(image_table_name=self.image_table_name)
def _check_subsampling(self):
- """Internal method checking if subsampling options were specified correctly.
+ """Check if subsampling options were specified correctly.
Returns
-------
None
Nothing is returned.
"""
-
# Check that the user didn't specify both subset frac and subsample all
assert ( # noqa: S101
self.subsample_frac == 1 or self.subsample_n == "all"
), "Do not set both subsample_frac and subsample_n"
def set_output_file(self, output_file):
- """Setting operation to conveniently rename output file.
+ """Set or modify output file.
Parameters
----------
@@ -205,11 +205,10 @@ def set_output_file(self, output_file):
None
Nothing is returned.
"""
-
self.output_file = output_file
def set_subsample_frac(self, subsample_frac):
- """Setting operation to conveniently update the subsample fraction.
+ """Set or update the subsample fraction.
Parameters
----------
@@ -221,12 +220,11 @@ def set_subsample_frac(self, subsample_frac):
None
Nothing is returned.
"""
-
self.subsample_frac = subsample_frac
self._check_subsampling()
def set_subsample_n(self, subsample_n):
- """Setting operation to conveniently update the subsample n.
+ """Set or update the subsample n.
Parameters
----------
@@ -238,7 +236,6 @@ def set_subsample_n(self, subsample_n):
None
Nothing is returned.
"""
-
try:
self.subsample_n = int(subsample_n)
except ValueError:
@@ -246,7 +243,7 @@ def set_subsample_n(self, subsample_n):
self._check_subsampling()
def set_subsample_random_state(self, random_state):
- """Setting operation to conveniently update the subsample random state.
+ """Set or update the subsample random state.
Parameters
----------
@@ -258,11 +255,10 @@ def set_subsample_random_state(self, random_state):
None
Nothing is returned.
"""
-
self.subsampling_random_state = random_state
def load_image(self, image_table_name=None):
- """Load image table from sqlite file
+ """Load image table from sqlite file.
Returns
-------
@@ -317,7 +313,6 @@ def count_cells(self, compartment="cells", count_subset=False):
pandas.core.frame.DataFrame
DataFrame of cell counts in the experiment.
"""
-
check_compartments(compartment)
if count_subset:
@@ -359,7 +354,6 @@ def subsample_profiles(self, df, rename_col=True):
pandas.core.frame.DataFrame
A subsampled pandas dataframe of single cell profiles.
"""
-
if self.subsampling_random_state is None:
random_state = np.random.randint(0, 10000, size=1)[0]
self.set_subsample_random_state(random_state)
@@ -398,7 +392,6 @@ def get_subsample(self, df=None, compartment="cells", rename_col=True):
None
Nothing is returned.
"""
-
check_compartments(compartment)
query_cols = "TableNumber, ImageNumber, ObjectNumber"
@@ -443,7 +436,7 @@ def split_column_categories(self, col_names):
return meta_cols, feat_cols
def load_compartment(self, compartment):
- """Creates the compartment dataframe.
+ """Create the compartment dataframe.
Note: makes use of default_datatype_float attribute
for setting a default floating point datatype.
@@ -458,7 +451,6 @@ def load_compartment(self, compartment):
pandas.core.frame.DataFrame
Compartment dataframe.
"""
-
# Get data useful to pre-alloc memory
num_cells = self.count_sql_table_rows(compartment)
col_names = self.get_sql_table_col_names(compartment)
@@ -495,7 +487,7 @@ def aggregate_compartment(
add_image_features=False,
n_aggregation_memory_strata=1,
):
- """Aggregate morphological profiles. Uses pycytominer.aggregate()
+ """Aggregate morphological profiles. Uses pycytominer.aggregate().
Parameters
----------
@@ -520,7 +512,6 @@ def aggregate_compartment(
pandas.core.frame.DataFrame
DataFrame of aggregated profiles.
"""
-
check_compartments(compartment)
if (self.subsample_frac < 1 or self.subsample_n != "all") and compute_subsample:
@@ -600,8 +591,7 @@ def _compartment_df_generator(
compartment,
n_aggregation_memory_strata=1,
):
- """A generator function that returns chunks of the entire compartment
- table from disk.
+ """Yield chunks of the entire compartment table from disk.
We want to return dataframes with all compartment entries within unique
combinations of self.merge_cols when aggregated by self.strata
@@ -622,7 +612,6 @@ def _compartment_df_generator(
between chunks, and thus groupby aggregations are valid
"""
-
assert ( # noqa: S101
n_aggregation_memory_strata > 0
), "Number of strata to pull into memory at once (n_aggregation_memory_strata) must be > 0"
@@ -712,7 +701,6 @@ def merge_single_cells(
if output_file=None returns a Pandas dataframe
else will write to file and return the filepath of the file
"""
-
# Load the single cell dataframe by merging on the specific linking columns
sc_df = ""
linking_check_cols = []
@@ -856,7 +844,6 @@ def aggregate_profiles(
if output_file=None) returns a Pandas dataframe
else will write to file and return the filepath of the file
"""
-
if output_file is not None:
self.set_output_file(output_file)
@@ -894,9 +881,7 @@ def aggregate_profiles(
def _sqlite_strata_conditions(df, dtypes, n=1):
- """Given a dataframe where columns are merge_cols and rows are unique
- value combinations that appear as aggregation strata, return a list
- of strings which constitute valid SQLite conditional statements.
+ """Construct a list of strings which constitute valid SQLite conditional statements.
Parameters
----------
@@ -927,13 +912,17 @@ def _sqlite_strata_conditions(df, dtypes, n=1):
[3] | [1, 2]
[4] | [1]
- >>> _sqlite_strata_conditions(df, dtypes={'TableNumber': 'integer', 'ImageNumber': 'integer'}, n=1)
+ >>> _sqlite_strata_conditions(
+ ... df, dtypes={"TableNumber": "integer", "ImageNumber": "integer"}, n=1
+ ... )
["(TableNumber in (1) and ImageNumber in (1))",
"(TableNumber in (2) and ImageNumber in (1, 2, 3))",
"(TableNumber in (3) and ImageNumber in (1, 2))",
"(TableNumber in (4) and ImageNumber in (1))"]
- >>> _sqlite_strata_conditions(df, dtypes={'TableNumber': 'text', 'ImageNumber': 'integer'}, n=2)
+ >>> _sqlite_strata_conditions(
+ ... df, dtypes={"TableNumber": "text", "ImageNumber": "integer"}, n=2
+ ... )
["(TableNumber in ('1') and ImageNumber in (1))
or (TableNumber in ('2') and ImageNumber in (1, 2, 3))",
"(TableNumber in ('3') and ImageNumber in (1, 2))
diff --git a/pycytominer/cyto_utils/collate.py b/pycytominer/cyto_utils/collate.py
index a21752fd..39b3e12b 100644
--- a/pycytominer/cyto_utils/collate.py
+++ b/pycytominer/cyto_utils/collate.py
@@ -1,3 +1,5 @@
+"""Module that provides functions for collating CellProfiler-created CSVs into a single SQLite file."""
+
import os
import pathlib
import subprocess
@@ -6,7 +8,7 @@
def run_check_errors(cmd):
- """Run a system command, and exit if an error occurred, otherwise continue"""
+ """Run a system command, and exit if an error occurred, otherwise continue."""
if isinstance(cmd, str):
cmd = cmd.split()
output = subprocess.run(cmd, capture_output=True, text=True) # noqa: S603
@@ -34,7 +36,7 @@ def collate(
image_feature_categories=["Granularity", "Texture", "ImageQuality", "Threshold"],
printtoscreen=True,
):
- """Collate the CellProfiler-created CSVs into a single SQLite file by calling cytominer-database
+ """Collate the CellProfiler-created CSVs into a single SQLite file by calling cytominer-database.
Parameters
----------
@@ -67,7 +69,6 @@ def collate(
printtoscreen: bool, optional, default True
Whether or not to print output to the terminal
"""
-
from pycytominer.cyto_utils.cells import SingleCells
# Check if optional dependency cytominer-database is installed
diff --git a/pycytominer/cyto_utils/collate_cmd.py b/pycytominer/cyto_utils/collate_cmd.py
index 7819d4bd..6f012a01 100644
--- a/pycytominer/cyto_utils/collate_cmd.py
+++ b/pycytominer/cyto_utils/collate_cmd.py
@@ -1,3 +1,5 @@
+"""Command line interface for collate function in pycytominer.cyto_utils.collate."""
+
import argparse
from pycytominer.cyto_utils.collate import collate
diff --git a/pycytominer/cyto_utils/cp_image_features.py b/pycytominer/cyto_utils/cp_image_features.py
index 1e9f29b2..b11db2aa 100644
--- a/pycytominer/cyto_utils/cp_image_features.py
+++ b/pycytominer/cyto_utils/cp_image_features.py
@@ -1,13 +1,11 @@
-"""
-Functions for counting the number of fields and aggregating other images features
-"""
+"""Functions for counting the number of fields and aggregating other images features."""
import numpy as np
from pycytominer import aggregate
def aggregate_fields_count(image_df, strata, fields_of_view_feature):
- """Compute the number of fields per well and create a new column called Metadata_Site_Count
+ """Compute the number of fields per well and create a new column called Metadata_Site_Count.
Parameters
----------
@@ -24,7 +22,6 @@ def aggregate_fields_count(image_df, strata, fields_of_view_feature):
DataFrame with the Metadata_Site_Count column.
"""
-
fields_count_df = image_df.loc[:, list(np.union1d(strata, fields_of_view_feature))]
fields_count_df = (
@@ -62,7 +59,6 @@ def aggregate_image_count_features(
remove_cols : list of str
Columns to remove from the image table before aggregating using aggregate_image_features()
"""
-
count_features = list(
image_features_df.columns[
image_features_df.columns.str.startswith("Metadata_" + str(count_prefix))
@@ -112,7 +108,6 @@ def aggregate_image_features(
DataFrame of aggregated image features.
"""
-
# Aggregate image count features
if count_prefix in image_feature_categories:
df, remove_cols = aggregate_image_count_features(
diff --git a/pycytominer/cyto_utils/features.py b/pycytominer/cyto_utils/features.py
index 449873df..304e71f9 100644
--- a/pycytominer/cyto_utils/features.py
+++ b/pycytominer/cyto_utils/features.py
@@ -1,6 +1,4 @@
-"""
-Utility function to manipulate cell profiler features
-"""
+"""Utility function to manipulate cell profiler features."""
import os
import pandas as pd
@@ -25,7 +23,6 @@ def get_blocklist_features(blocklist_file=blocklist_file, population_df=None):
blocklist_features : list of str
Features to exclude from downstream analysis.
"""
-
blocklist = pd.read_csv(blocklist_file)
assert any( # noqa: S101
@@ -48,7 +45,7 @@ def label_compartment(cp_features, compartment, metadata_cols):
cp_features : list of str
All features being used.
compartment : str
- Measured compartment.
+ Measured compartment.
metadata_cols : list
Columns that should be considered metadata.
@@ -57,7 +54,6 @@ def label_compartment(cp_features, compartment, metadata_cols):
cp_features: list of str
Recoded column names with appropriate metadata and compartment labels.
"""
-
compartment = compartment.Title()
avail_compartments = ["Cells", "Cytoplasm", "Nuceli", "Image", "Barcode"]
@@ -97,7 +93,6 @@ def infer_cp_features(
features: list of str
List of Cell Painting features.
"""
-
compartments = convert_compartment_format_to_list(compartments)
compartments = [x.title() for x in compartments]
@@ -135,7 +130,6 @@ def count_na_features(population_df, features):
-------
Dataframe of NA counts per feature
"""
-
return pd.DataFrame(population_df.loc[:, features].isna().sum(), columns=["num_na"])
@@ -156,7 +150,7 @@ def drop_outlier_features(
"Metadata_treatment == 'control'" (include all quotes).
If "all", use all samples to calculate.
outlier_cutoff : int or float, default 500
- see https://github.com/cytomining/pycytominer/issues/237 for details.
+ see https://github.com/cytomining/pycytominer/issues/237 for details.
Threshold to remove features if absolute values is greater
Returns
@@ -164,7 +158,6 @@ def drop_outlier_features(
outlier_features: list of str
Features greater than the threshold.
"""
-
# Subset dataframe
if samples != "all":
population_df.query(samples, inplace=True)
@@ -186,7 +179,7 @@ def drop_outlier_features(
def convert_compartment_format_to_list(compartments):
- """Converts compartment to a list.
+ """Convert cell painting compartments to a list.
Parameters
----------
@@ -198,7 +191,6 @@ def convert_compartment_format_to_list(compartments):
compartments : list of str
List of Cell Painting compartments.
"""
-
if isinstance(compartments, list):
compartments = [x.lower() for x in compartments]
elif isinstance(compartments, str):
diff --git a/pycytominer/cyto_utils/load.py b/pycytominer/cyto_utils/load.py
index 7abdd06d..198fd990 100644
--- a/pycytominer/cyto_utils/load.py
+++ b/pycytominer/cyto_utils/load.py
@@ -1,3 +1,5 @@
+"""Module for loading data from various file formats."""
+
import csv
import gzip
import pathlib
@@ -7,7 +9,7 @@
def is_path_a_parquet_file(file: Union[str, pathlib.PurePath]) -> bool:
- """Checks if the provided file path is a parquet file.
+ """Check if the provided file path is a parquet file.
Identify parquet files by inspecting the file extensions.
If the file does not end with `parquet`, this will return False, else True.
@@ -30,7 +32,6 @@ def is_path_a_parquet_file(file: Union[str, pathlib.PurePath]) -> bool:
FileNotFoundError
Raised if the provided path in the `file` does not exist
"""
-
file = pathlib.PurePath(file)
try:
# strict=true tests if path exists
@@ -47,7 +48,7 @@ def is_path_a_parquet_file(file: Union[str, pathlib.PurePath]) -> bool:
def infer_delim(file: str):
"""
- Sniff the delimiter in the given file
+ Sniff the delimiter in the given file.
Parameters
----------
@@ -72,7 +73,7 @@ def infer_delim(file: str):
def load_profiles(profiles):
"""
- Unless a dataframe is provided, load the given profile dataframe from path or string
+ Unless a dataframe is provided, load the given profile dataframe from path or string.
Parameters
----------
@@ -83,8 +84,8 @@ def load_profiles(profiles):
------
pandas DataFrame of profiles
- Raises:
- -------
+ Raises
+ ------
FileNotFoundError
Raised if the provided profile does not exists
"""
@@ -102,7 +103,7 @@ def load_profiles(profiles):
def load_platemap(platemap, add_metadata_id=True):
"""
- Unless a dataframe is provided, load the given platemap dataframe from path or string
+ Unless a dataframe is provided, load the given platemap dataframe from path or string.
Parameters
----------
diff --git a/pycytominer/cyto_utils/modz.py b/pycytominer/cyto_utils/modz.py
index 6e598ed5..5f719f0b 100644
--- a/pycytominer/cyto_utils/modz.py
+++ b/pycytominer/cyto_utils/modz.py
@@ -1,3 +1,5 @@
+"""Module for performing a modified z score transformation."""
+
import numpy as np
import pandas as pd
from pycytominer.cyto_utils.util import (
@@ -88,7 +90,7 @@ def modz(
min_weight=0.01,
precision=4,
):
- """Collapse replicates into a consensus signature using a weighted transformation
+ """Collapse replicates into a consensus signature using a weighted transformation.
Parameters
----------
diff --git a/pycytominer/cyto_utils/output.py b/pycytominer/cyto_utils/output.py
index aa5290a0..d0cd4f4d 100644
--- a/pycytominer/cyto_utils/output.py
+++ b/pycytominer/cyto_utils/output.py
@@ -1,6 +1,4 @@
-"""
-Utility function to compress output data
-"""
+"""Utility function to compress output data."""
from typing import Dict, Union, Optional
@@ -18,7 +16,7 @@ def output(
compression_options: Union[str, Dict] = {"method": "gzip", "mtime": 1},
**kwargs,
):
- """Given an output file and compression options, write file to disk
+ """Given an output file and compression options, write file to disk.
Parameters
----------
@@ -78,7 +76,6 @@ def output(
float_format=None,
)
"""
-
if output_type == "csv":
compression_options = set_compression_method(compression=compression_options)
@@ -99,7 +96,7 @@ def output(
def set_compression_method(compression: Union[str, Dict]):
- """Set the compression options
+ """Set the compression options.
Parameters
----------
@@ -112,7 +109,6 @@ def set_compression_method(compression: Union[str, Dict]):
compression, dict
A formated dictionary expected by output()
"""
-
if compression is None:
compression = {"method": None}
@@ -124,7 +120,7 @@ def set_compression_method(compression: Union[str, Dict]):
def check_compression_method(compression: str):
- """Ensure compression options are set properly
+ """Ensure compression options are set properly.
Parameters
----------
diff --git a/pycytominer/cyto_utils/single_cell_ingest_utils.py b/pycytominer/cyto_utils/single_cell_ingest_utils.py
index 2a5a5308..d9dcc070 100644
--- a/pycytominer/cyto_utils/single_cell_ingest_utils.py
+++ b/pycytominer/cyto_utils/single_cell_ingest_utils.py
@@ -1,9 +1,11 @@
+"""Utility functions for single cell ingest."""
+
from collections import Counter
from pycytominer.cyto_utils import get_default_compartments
def get_default_linking_cols():
- """Define the standard experiment linking columns between tables
+ """Define the standard experiment linking columns between tables.
Returns
-------
@@ -26,7 +28,7 @@ def get_default_linking_cols():
def assert_linking_cols_complete(linking_cols="default", compartments="default"):
- """Confirm that the linking cols and compartments are compatible
+ """Confirm that the linking cols and compartments are compatible.
Parameters
----------
@@ -75,8 +77,7 @@ def assert_linking_cols_complete(linking_cols="default", compartments="default")
def provide_linking_cols_feature_name_update(linking_cols="default"):
- """Output a dictionary to use to update pandas dataframe column names. The linking
- cols must be Metadata.
+ """Output a dictionary to use to update pandas dataframe column names from linking cols in the Metadata.
Parameters
----------
diff --git a/pycytominer/cyto_utils/util.py b/pycytominer/cyto_utils/util.py
index c550e61f..3a7302ad 100644
--- a/pycytominer/cyto_utils/util.py
+++ b/pycytominer/cyto_utils/util.py
@@ -1,6 +1,4 @@
-"""
-Miscellaneous utility functions
-"""
+"""Miscellaneous utility functions."""
import os
import warnings
@@ -16,7 +14,7 @@
def get_default_compartments():
- """Returns default compartments.
+ """Return default compartments.
Returns
-------
@@ -24,12 +22,11 @@ def get_default_compartments():
Default compartments.
"""
-
return ["cells", "cytoplasm", "nuclei"]
def check_compartments(compartments):
- """Checks if the input compartments are noncanonical compartments.
+ """Check if the input compartments are noncanonical compartments.
Parameters
----------
@@ -42,7 +39,6 @@ def check_compartments(compartments):
Nothing is returned.
"""
-
default_compartments = get_default_compartments()
compartments = convert_compartment_format_to_list(compartments)
@@ -60,13 +56,13 @@ def check_compartments(compartments):
def load_known_metadata_dictionary(metadata_file=default_metadata_file):
- """From a tab separated text file (two columns: ["compartment", "feature"]), load
- previously known metadata columns per compartment.
+ """Load previously known metadata columns per compartment from metadata text file.
Parameters
----------
metadata_file : str, optional
- File location of the metadata text file. Uses a default dictionary if you do not specify.
+ File location of the metadata text file which should be a tab-separated file with two columns: ["compartment", "feature"].
+ If not provided, the default metadata file will be used.
Returns
-------
@@ -74,7 +70,6 @@ def load_known_metadata_dictionary(metadata_file=default_metadata_file):
Compartment (keys) mappings to previously known metadata (values).
"""
-
metadata_dict = {}
with open(metadata_file) as meta_fh:
next(meta_fh)
@@ -103,7 +98,6 @@ def check_correlation_method(method):
Correctly formatted correlation method.
"""
-
method = method.lower()
avail_methods = ["pearson", "spearman", "kendall"]
assert ( # noqa: S101
@@ -127,7 +121,6 @@ def check_aggregate_operation(operation):
Correctly formatted operation method.
"""
-
operation = operation.lower()
avail_ops = ["mean", "median"]
assert ( # noqa: S101
@@ -151,7 +144,6 @@ def check_consensus_operation(operation):
Correctly formatted operation method.
"""
-
operation = operation.lower()
avail_ops = ["modz"] # All aggregation operations are also supported
try:
@@ -178,7 +170,6 @@ def check_fields_of_view_format(fields_of_view):
Correctly formatted fields_of_view variable.
"""
-
if fields_of_view != "all":
if isinstance(fields_of_view, list):
if all(isinstance(x, int) for x in fields_of_view):
@@ -214,7 +205,6 @@ def check_fields_of_view(data_fields_of_view, input_fields_of_view):
Nothing is returned.
"""
-
try:
assert len( # noqa: S101
list(np.intersect1d(data_fields_of_view, input_fields_of_view))
@@ -226,7 +216,7 @@ def check_fields_of_view(data_fields_of_view, input_fields_of_view):
def check_image_features(image_features, image_columns):
- """Confirm that the input list of image features are present in the image table
+ """Confirm that the input list of image features are present in the image table.
Parameters
----------
@@ -240,7 +230,6 @@ def check_image_features(image_features, image_columns):
None
Nothing is returned.
"""
-
if "Image" in list({img_col.split("_")[0] for img_col in image_columns}):
# Image has already been prepended to most, but not all, columns
level = 1
@@ -281,7 +270,6 @@ def extract_image_features(image_feature_categories, image_df, image_cols, strat
Correctly formatted image feature categories.
"""
-
# Check if the input image feature groups are valid.
check_image_features(image_feature_categories, list(image_df.columns))
@@ -320,13 +308,13 @@ def get_pairwise_correlation(population_df, method="pearson"):
Includes metadata and observation features.
method : str, default "pearson"
Which correlation matrix to use to test cutoff.
+
Returns
-------
list of str
Features to exclude from the population_df.
"""
-
# Check that the input method is supported
method = check_correlation_method(method)
diff --git a/pycytominer/cyto_utils/write_gct.py b/pycytominer/cyto_utils/write_gct.py
index 812811bd..02bba65b 100644
--- a/pycytominer/cyto_utils/write_gct.py
+++ b/pycytominer/cyto_utils/write_gct.py
@@ -1,7 +1,8 @@
-"""
+"""Module to write a gct file from a pandas DataFrame.
+
Transform profiles into a gct (Gene Cluster Text) file
A gct is a tab deliminted text file that traditionally stores gene expression data
-File Format Description: https://clue.io/connectopedia/gct_format
+File Format Description: https://clue.io/connectopedia/gct_format.
Modified from cytominer_scripts "write_gcg" written in R
https://github.com/broadinstitute/cytominer_scripts/blob/master/write_gct.R
@@ -21,7 +22,7 @@ def write_gct(
feature_metadata=None,
version="#1.3",
):
- """Convert profiles to a .gct file
+ """Convert profiles to a .gct file.
Parameters
----------
@@ -47,7 +48,6 @@ def write_gct(
None
Writes gct to file
"""
-
# Note, only version 1.3 is currently supported
assert version == "#1.3", "Only version #1.3 is currently supported." # noqa: S101
diff --git a/pycytominer/feature_select.py b/pycytominer/feature_select.py
index c1fd87d1..9d164e7b 100644
--- a/pycytominer/feature_select.py
+++ b/pycytominer/feature_select.py
@@ -1,6 +1,4 @@
-"""
-Select features to use in downstream analysis based on specified selection method
-"""
+"""Select features to use in downstream analysis based on specified selection method."""
from pycytominer.cyto_utils import (
drop_outlier_features,
@@ -37,7 +35,7 @@ def feature_select(
noise_removal_perturb_groups=None,
noise_removal_stdev_cutoff=None,
):
- """Performs feature selection based on the given operation.
+ """Perform feature selection based on the given operation.
Parameters
----------
@@ -100,7 +98,6 @@ def feature_select(
data.
"""
-
all_ops = [
"variance_threshold",
"correlation_threshold",
diff --git a/pycytominer/normalize.py b/pycytominer/normalize.py
index 7a83ca5f..28d4b8bc 100644
--- a/pycytominer/normalize.py
+++ b/pycytominer/normalize.py
@@ -1,6 +1,4 @@
-"""
-Normalize observation features based on specified normalization method
-"""
+"""Normalize observation features based on specified normalization method."""
import pandas as pd
from sklearn.preprocessing import RobustScaler, StandardScaler
@@ -25,7 +23,7 @@ def normalize(
spherize_method="ZCA-cor",
spherize_epsilon=1e-6,
):
- """Normalize profiling features
+ """Normalize profiling features.
Parameters
----------
@@ -58,15 +56,13 @@ def normalize(
If provided, will write normalized profiles as a specified file type (either CSV or parquet).
If not specified and output_file is provided, then the file will be outputed as CSV as default.
compression_options : str or dict, optional
- Contains compression options as input to
- pd.DataFrame.to_csv(compression=compression_options). pandas version >= 1.2.
+ Contains compression options as input to `pd.DataFrame.to_csv(compression=compression_options)`.
float_format : str, optional
Decimal precision to use in writing output file as input to
pd.DataFrame.to_csv(float_format=float_format). For example, use "%.3g" for 3
decimal precision.
mad_robustize_epsilon: float, optional
- The mad_robustize fudge factor parameter. The function only uses
- this variable if method = "mad_robustize". Set this to 0 if
+ The mad_robustize fudge factor parameter. The function only uses this variable if method = "mad_robustize". Set this to 0 if
mad_robustize generates features with large values.
spherize_center : bool
If the function should center data before sphering (aka whitening). The
@@ -88,38 +84,37 @@ def normalize(
Examples
--------
+ ```python
import pandas as pd
from pycytominer import normalize
- data_df = pd.DataFrame(
- {
- "Metadata_plate": ["a", "a", "a", "a", "b", "b", "b", "b"],
- "Metadata_treatment": [
- "drug",
- "drug",
- "control",
- "control",
- "drug",
- "drug",
- "control",
- "control",
- ],
- "x": [1, 2, 8, 2, 5, 5, 5, 1],
- "y": [3, 1, 7, 4, 5, 9, 6, 1],
- "z": [1, 8, 2, 5, 6, 22, 2, 2],
- "zz": [14, 46, 1, 6, 30, 100, 2, 2],
- }
- ).reset_index(drop=True)
+ data_df = pd.DataFrame({
+ "Metadata_plate": ["a", "a", "a", "a", "b", "b", "b", "b"],
+ "Metadata_treatment": [
+ "drug",
+ "drug",
+ "control",
+ "control",
+ "drug",
+ "drug",
+ "control",
+ "control",
+ ],
+ "x": [1, 2, 8, 2, 5, 5, 5, 1],
+ "y": [3, 1, 7, 4, 5, 9, 6, 1],
+ "z": [1, 8, 2, 5, 6, 22, 2, 2],
+ "zz": [14, 46, 1, 6, 30, 100, 2, 2],
+ }).reset_index(drop=True)
normalized_df = normalize(
profiles=data_df,
features=["x", "y", "z", "zz"],
meta_features="infer",
samples="Metadata_treatment == 'control'",
- method="standardize"
+ method="standardize",
)
+ ```
"""
-
# Load Data
profiles = load_profiles(profiles)
diff --git a/pycytominer/operations/__init__.py b/pycytominer/operations/__init__.py
index 39a59b30..07f5bba8 100644
--- a/pycytominer/operations/__init__.py
+++ b/pycytominer/operations/__init__.py
@@ -1,3 +1,5 @@
+"""Module containing statistical operations for data processing."""
+
from .correlation_threshold import correlation_threshold
from .get_na_columns import get_na_columns
from .noise_removal import noise_removal
diff --git a/pycytominer/operations/correlation_threshold.py b/pycytominer/operations/correlation_threshold.py
index 7c4522ba..d6526949 100644
--- a/pycytominer/operations/correlation_threshold.py
+++ b/pycytominer/operations/correlation_threshold.py
@@ -1,6 +1,7 @@
-"""
-Returns list of features such that no two features have a correlation greater than a
-specified threshold
+"""Module for correlation threshold operation.
+
+The correlation threshold operation list of features such that no two features have a correlation greater than a
+specified threshold.
"""
from pycytominer.cyto_utils import (
@@ -13,7 +14,7 @@
def correlation_threshold(
population_df, features="infer", samples="all", threshold=0.9, method="pearson"
):
- """Exclude features that have correlations above a certain threshold
+ """Exclude features that have correlations above a certain threshold.
Parameters
----------
@@ -38,7 +39,6 @@ def correlation_threshold(
excluded_features : list of str
List of features to exclude from the population_df.
"""
-
# Check that the input method is supported
method = check_correlation_method(method)
@@ -80,8 +80,9 @@ def correlation_threshold(
def determine_high_cor_pair(correlation_row, sorted_correlation_pairs):
- """Select highest correlated variable given a correlation row with columns:
- ["pair_a", "pair_b", "correlation"]. For use in a pandas.apply().
+ """Select highest correlated variable given a correlation row.
+
+ From a row with columns: ["pair_a", "pair_b", "correlation"]. For use in a pandas.apply().
Parameters
----------
@@ -94,7 +95,6 @@ def determine_high_cor_pair(correlation_row, sorted_correlation_pairs):
-------
The feature that has a lower total correlation sum with all other features
"""
-
pair_a = correlation_row["pair_a"]
pair_b = correlation_row["pair_b"]
diff --git a/pycytominer/operations/get_na_columns.py b/pycytominer/operations/get_na_columns.py
index ad36c377..69393318 100644
--- a/pycytominer/operations/get_na_columns.py
+++ b/pycytominer/operations/get_na_columns.py
@@ -1,13 +1,14 @@
-"""
+"""Function to get columns with NA values above a certain threshold.
+
Remove variables with specified threshold of NA values
-Note: This was called `drop_na_columns` in cytominer for R
+Note: This was called `drop_na_columns` in cytominer for R.
"""
from pycytominer.cyto_utils.features import infer_cp_features
def get_na_columns(population_df, features="infer", samples="all", cutoff=0.05):
- """Get features that have more NA values than cutoff defined
+ """Get features that have more NA values than cutoff defined.
Parameters
----------
@@ -30,7 +31,6 @@ def get_na_columns(population_df, features="infer", samples="all", cutoff=0.05):
excluded_features : list of str
List of features to exclude from the population_df.
"""
-
if samples != "all":
population_df.query(samples, inplace=True)
diff --git a/pycytominer/operations/noise_removal.py b/pycytominer/operations/noise_removal.py
index aba1a29e..f52c03ff 100644
--- a/pycytominer/operations/noise_removal.py
+++ b/pycytominer/operations/noise_removal.py
@@ -1,6 +1,4 @@
-"""
-Remove noisy features, as defined by features with excessive standard deviation within the same perturbation group.
-"""
+"""Remove noisy features, as defined by features with excessive standard deviation within the same perturbation group."""
from pycytominer.cyto_utils import infer_cp_features
@@ -12,7 +10,7 @@ def noise_removal(
samples="all",
noise_removal_stdev_cutoff=0.8,
):
- """
+ """Remove features with excessive standard deviation within the same perturbation group.
Parameters
----------
diff --git a/pycytominer/operations/transform.py b/pycytominer/operations/transform.py
index 83a889a4..5077d977 100644
--- a/pycytominer/operations/transform.py
+++ b/pycytominer/operations/transform.py
@@ -13,14 +13,14 @@
class Spherize(BaseEstimator, TransformerMixin):
- """Class to apply a sphering transform (aka whitening) data in the base sklearn
- transform API. Note, this implementation is modified/inspired from the following
- sources:
+ """Class to apply a sphering transform (aka whitening) data in the base sklearn transform API.
+
+ This implementation is modified/inspired from the following sources:
1) A custom function written by Juan C. Caicedo
2) A custom ZCA function at https://github.com/mwv/zca
3) Notes from Niranj Chandrasekaran (https://github.com/cytomining/pycytominer/issues/90)
4) The R package "whitening" written by Strimmer et al (http://strimmerlab.org/software/whitening/)
- 5) Kessy et al. 2016 "Optimal Whitening and Decorrelation" [1]_
+ 5) Kessy et al. 2016 "Optimal Whitening and Decorrelation" [1]_.
Attributes
----------
@@ -33,7 +33,8 @@ class Spherize(BaseEstimator, TransformerMixin):
"""
def __init__(self, epsilon=1e-6, center=True, method="ZCA", return_numpy=False):
- """
+ """Construct a Spherize object.
+
Parameters
----------
epsilon : float, default 1e-6
@@ -65,7 +66,7 @@ def __init__(self, epsilon=1e-6, center=True, method="ZCA", return_numpy=False):
raise ValueError("PCA-cor and ZCA-cor require center=True")
def fit(self, X, y=None):
- """Identify the sphering transform given self.X
+ """Identify the sphering transform given self.X.
Parameters
----------
@@ -221,7 +222,7 @@ def fit(self, X, y=None):
return self
def transform(self, X, y=None):
- """Perform the sphering transform
+ """Perform the sphering transform.
Parameters
----------
@@ -235,7 +236,6 @@ def transform(self, X, y=None):
pandas.core.frame.DataFrame
Spherized dataframe
"""
-
columns = X.columns
# Get Numpy representation of the DataFrame
@@ -261,7 +261,7 @@ def transform(self, X, y=None):
class RobustMAD(BaseEstimator, TransformerMixin):
- """Class to perform a "Robust" normalization with respect to median and mad
+ """Class to perform a "Robust" normalization with respect to median and mad.
scaled = (x - median) / mad
@@ -298,7 +298,7 @@ def fit(self, X, y=None):
return self
def transform(self, X, copy=None):
- """Apply the RobustMAD calculation
+ """Apply the RobustMAD calculation.
Parameters
----------
diff --git a/pycytominer/operations/variance_threshold.py b/pycytominer/operations/variance_threshold.py
index 67d3b767..d4b999fc 100644
--- a/pycytominer/operations/variance_threshold.py
+++ b/pycytominer/operations/variance_threshold.py
@@ -1,6 +1,6 @@
-"""
-Remove variables with near-zero variance.
-Modified from caret::nearZeroVar()
+"""Remove variables with near-zero variance.
+
+Modified from caret::nearZeroVar().
"""
import numpy as np
@@ -11,7 +11,7 @@
def variance_threshold(
population_df, features="infer", samples="all", freq_cut=0.05, unique_cut=0.01
):
- """Exclude features that have low variance (low information content)
+ """Exclude features that have low variance (low information content).
Parameters
----------
@@ -42,7 +42,6 @@ def variance_threshold(
List of features to exclude from the population_df.
"""
-
if not 0 <= freq_cut <= 1:
raise ValueError("freq_cut variable must be between (0 and 1)")
if not 0 <= unique_cut <= 1:
@@ -80,7 +79,6 @@ def variance_threshold(
def calculate_frequency(feature_column, freq_cut):
"""Calculate frequency of second most common to most common feature.
- Used in pandas.apply()
Parameters
----------
@@ -97,7 +95,6 @@ def calculate_frequency(feature_column, freq_cut):
Feature name if it passes threshold, "NA" otherwise
"""
-
val_count = feature_column.value_counts()
try:
max_count = val_count.iloc[0]
diff --git a/pyproject.toml b/pyproject.toml
index a8ea5428..7452a30b 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -1,7 +1,7 @@
[tool.poetry]
name = "pycytominer"
# This version is a placeholder updated during build by poetry-dynamic-versioning
-version = "0.0.0"
+version = "1.1.0.post29.dev0+9615c0d"
description = "Python package for processing image-based profiling data"
authors = [
"Erik Serrano",
@@ -39,7 +39,9 @@ license = "BSD-3-Clause"
readme = "README.md"
homepage = "https://pycytominer.readthedocs.io/"
repository = "https://github.com/cytomining/pycytominer"
-include = [{ path = "poetry.lock", format = "sdist" }]
+include = [
+ { path = "poetry.lock", format = "sdist" },
+]
[tool.poetry.dependencies]
python = ">=3.8,<4.0"
@@ -78,22 +80,18 @@ ruff = "^0.3.4"
optional = true
[tool.poetry.group.docs.dependencies]
-mkdocs = "*"
-sphinx = "~7.1"
-sphinx-autobuild = "^2021.3.14"
-sphinx-autoapi = "^3.0.0"
+mkdocs = "^1.6.0"
ipython = "^8.12"
-groundwork-sphinx-theme = "^1.1.1"
-sphinx-copybutton = "^0.5.2"
nbsphinx = "^0.9.3"
-m2r2 = "^0.3.3.post2"
-furo = "^2023.9.10"
mock = "^5.1.0"
-autodoc = "^0.5.0"
dunamai = "^1.19.0"
+mkdocs-material = "^9.5.19"
+mkdocstrings = {extras = ["python"], version = "^0.23.0"}
+mkdocs-include-markdown-plugin = "^6.0.5"
+mkdocs-jupyter = "^0.24.7"
[tool.poetry-dynamic-versioning]
-enable = true
+enable = false
style = "pep440"
vcs = "git"
@@ -117,6 +115,10 @@ version_files = [
target-version = "py38"
line-length = 88
fix = true
+extend-include = ["*.ipynb"]
+exclude = [
+ "walkthroughs/nbconverted/*",
+]
[tool.ruff.lint]
select = [
@@ -144,6 +146,8 @@ select = [
"S",
# isort
"I",
+ # pydocstyle
+ "D",
# The following checks are disabled, but we are working towards enabling them in the future.
# flake8-bugbear
# "B",
@@ -158,12 +162,21 @@ ignore = [
# DoNotAssignLambda
"E731",
]
+exclude = ["*.ipynb"]
+
+[tool.ruff.lint.pydocstyle]
+# Use Google-style docstrings.
+convention = "numpy"
[tool.ruff.lint.per-file-ignores]
# Ignore `E402` and `F401` (unusued imports) in all `__init__.py` files
"__init__.py" = ["E402", "F401"]
-# Ignore assert statements in tests
-"tests/*" = ["S101"]
+"tests/*" = [
+ # Allow assert statements in tests
+ "S101",
+ # Disable docstring checks in tests
+ "D",
+]
"pycytominer/cyto_utils/*" = [
# I (isort) is ignored due to circular dependencies in the cyto_utils module
"I",
@@ -173,6 +186,8 @@ ignore = [
[tool.ruff.format]
preview = true
+# Enable reformatting of code snippets in docstrings.
+docstring-code-format = true
[tool.pytest.ini_options]
testpaths = "tests"
diff --git a/tests/test_aggregate.py b/tests/test_aggregate.py
index d1a08889..694b7f16 100644
--- a/tests/test_aggregate.py
+++ b/tests/test_aggregate.py
@@ -49,9 +49,7 @@
def test_aggregate_median_allvar():
- """
- Testing aggregate pycytominer function
- """
+ """Testing aggregate pycytominer function."""
aggregate_result = aggregate(
population_df=data_df, strata=["g"], features="infer", operation="median"
)
@@ -78,9 +76,7 @@ def test_aggregate_median_allvar():
def test_aggregate_mean_allvar():
- """
- Testing aggregate pycytominer function
- """
+ """Testing aggregate pycytominer function."""
aggregate_result = aggregate(
population_df=data_df, strata=["g"], features="infer", operation="mean"
)
@@ -95,9 +91,7 @@ def test_aggregate_mean_allvar():
def test_aggregate_median_subsetvar():
- """
- Testing aggregate pycytominer function
- """
+ """Testing aggregate pycytominer function."""
aggregate_result = aggregate(
population_df=data_df, strata=["g"], features=["Cells_x"], operation="median"
)
@@ -109,9 +103,7 @@ def test_aggregate_median_subsetvar():
def test_aggregate_mean_subsetvar():
- """
- Testing aggregate pycytominer function
- """
+ """Testing aggregate pycytominer function."""
aggregate_result = aggregate(
population_df=data_df, strata=["g"], features=["Cells_x"], operation="mean"
)
@@ -123,10 +115,7 @@ def test_aggregate_mean_subsetvar():
def test_aggregate_median_dtype_confirm():
- """
- Testing aggregate pycytominer function
- """
-
+ """Testing aggregate pycytominer function."""
# Convert dtype of one variable to object
data_dtype_df = data_df.copy()
data_dtype_df.Cells_x = data_dtype_df.Cells_x.astype(str)
@@ -145,10 +134,7 @@ def test_aggregate_median_dtype_confirm():
def test_aggregate_median_with_missing_values():
- """
- Testing aggregate pycytominer function
- """
-
+ """Testing aggregate pycytominer function."""
# Convert dtype of one variable to object
data_dtype_df = data_missing_df.copy()
data_dtype_df.Cells_x = data_dtype_df.Cells_x.astype(str)
@@ -167,10 +153,7 @@ def test_aggregate_median_with_missing_values():
def test_aggregate_compute_object_count():
- """
- Testing aggregate pycytominer function
- """
-
+ """Testing aggregate pycytominer function."""
aggregate_result = aggregate(
population_df=data_df,
strata=["g"],
@@ -212,10 +195,7 @@ def test_aggregate_compute_object_count():
def test_aggregate_incorrect_object_feature():
- """
- Testing aggregate pycytominer function
- """
-
+ """Testing aggregate pycytominer function."""
incorrect_object_feature = "DOES NOT EXIST"
with pytest.raises(KeyError) as err:
@@ -250,10 +230,7 @@ def test_aggregate_incorrect_object_feature():
def test_custom_objectnumber_feature():
- """
- Testing aggregate pycytominer function
- """
-
+ """Testing aggregate pycytominer function."""
data_df_copy = data_df.copy().rename(
columns={"Metadata_ObjectNumber": "Custom_ObjectNumber_Feature"}
)
@@ -287,9 +264,7 @@ def test_custom_objectnumber_feature():
def test_output_type():
- """
- Testing aggregate pycytominer function
- """
+ """Testing aggregate pycytominer function."""
# dictionary with the output name associated with the file type
output_dict = {"csv": test_output_file_csv, "parquet": test_output_file_parquet}
diff --git a/tests/test_cyto_utils/conftest.py b/tests/test_cyto_utils/conftest.py
index 20ac6cc9..78422f70 100644
--- a/tests/test_cyto_utils/conftest.py
+++ b/tests/test_cyto_utils/conftest.py
@@ -1,6 +1,4 @@
-"""
-conftest.py for pytest
-"""
+"""conftest.py for pytest."""
import os
import pathlib
@@ -14,10 +12,7 @@
@pytest.fixture(name="data_dir_cell_locations")
def fixture_data_dir_cell_locations() -> str:
- """
- Provide a data directory for cell_locations test data
- """
-
+ """Provide a data directory for cell_locations test data."""
return (
f"{pathlib.Path(__file__).parent.parent}/test_data/cell_locations_example_data"
)
@@ -25,9 +20,7 @@ def fixture_data_dir_cell_locations() -> str:
@pytest.fixture(name="metadata_input_file")
def fixture_metadata_input_file(data_dir_cell_locations: str) -> str:
- """
- Provide a metadata input file for cell_locations test data
- """
+ """Provide a metadata input file for cell_locations test data."""
return os.path.join(
data_dir_cell_locations, "test_BR00126114_load_data_with_illum.parquet"
)
@@ -35,33 +28,25 @@ def fixture_metadata_input_file(data_dir_cell_locations: str) -> str:
@pytest.fixture(name="single_cell_input_file")
def fixture_single_cell_input_file(data_dir_cell_locations: str) -> str:
- """
- Provide a single cell input file for cell_locations test data
- """
+ """Provide a single cell input file for cell_locations test data."""
return os.path.join(data_dir_cell_locations, "test_BR00126114.sqlite")
@pytest.fixture(name="metadata_input_file_s3")
def fixture_metadata_input_file_s3() -> str:
- """
- Provide a metadata input file for cell_locations test data
- """
+ """Provide a metadata input file for cell_locations test data."""
return "s3://cellpainting-gallery/cpg0016-jump/source_4/workspace/load_data_csv/2021_08_23_Batch12/BR00126114/load_data_with_illum.parquet"
@pytest.fixture(name="single_cell_input_file_s3")
def fixture_single_cell_input_file_s3() -> str:
- """
- Provide a single cell input file for cell_locations test data
- """
+ """Provide a single cell input file for cell_locations test data."""
return "s3://cellpainting-gallery/cpg0016-jump/source_4/workspace/backend/2021_08_23_Batch12/BR00126114/BR00126114.sqlite"
@pytest.fixture(name="metadata_input_dataframe")
def fixture_metadata_input_dataframe(metadata_input_file: str) -> pd.DataFrame:
- """
- Provide a metadata input file for cell_locations test data
- """
+ """Provide a metadata input file for cell_locations test data."""
return pd.read_parquet(metadata_input_file)
@@ -69,9 +54,7 @@ def fixture_metadata_input_dataframe(metadata_input_file: str) -> pd.DataFrame:
def fixture_single_cell_input_engine(
single_cell_input_file: str,
) -> sqlalchemy.engine.Engine:
- """
- Provide a single cell input file for cell_locations test data
- """
+ """Provide a single cell input file for cell_locations test data."""
return sqlalchemy.create_engine(f"sqlite:///{single_cell_input_file}")
@@ -80,9 +63,7 @@ def fixture_cell_loc_obj1(
metadata_input_file: str,
single_cell_input_file: str,
) -> CellLocation:
- """
- Provide a CellLocation object with file inputs
- """
+ """Provide a CellLocation object with file inputs."""
return CellLocation(
metadata_input=metadata_input_file,
single_cell_input=single_cell_input_file,
@@ -94,9 +75,7 @@ def fixture_cell_loc_obj2(
metadata_input_dataframe: pd.DataFrame,
single_cell_input_engine: sqlalchemy.engine.Engine,
) -> CellLocation:
- """
- Provide a CellLocation object with in-memory inputs
- """
+ """Provide a CellLocation object with in-memory inputs."""
return CellLocation(
metadata_input=metadata_input_dataframe,
single_cell_input=single_cell_input_engine,
@@ -108,9 +87,7 @@ def fixture_cell_loc_obj3(
metadata_input_file_s3: str,
single_cell_input_file_s3: str,
) -> CellLocation:
- """
- Provide a CellLocation object with s3 inputs
- """
+ """Provide a CellLocation object with s3 inputs."""
return CellLocation(
metadata_input=metadata_input_file_s3,
single_cell_input=single_cell_input_file_s3,
diff --git a/tests/test_cyto_utils/test_DeepProfiler_processing.py b/tests/test_cyto_utils/test_DeepProfiler_processing.py
index 600006d3..cbf51c3b 100644
--- a/tests/test_cyto_utils/test_DeepProfiler_processing.py
+++ b/tests/test_cyto_utils/test_DeepProfiler_processing.py
@@ -1,6 +1,4 @@
-"""
-This tests the output from a DeepProfiler run (May 2021)
-"""
+"""This tests the output from a DeepProfiler run (May 2021)."""
import os
import pathlib
@@ -24,7 +22,7 @@
@pytest.fixture(scope="session")
def deep_profiler_data(tmp_path_factory):
- """This fixture returns the DeepProfilerData object and the output folder"""
+ """This fixture returns the DeepProfilerData object and the output folder."""
example_project_dir = ROOT_DIR / "tests" / "test_data" / "DeepProfiler_example_data"
profile_dir = example_project_dir / "outputs" / "results" / "features"
index_file = example_project_dir / "inputs" / "metadata" / "test_index.csv"
@@ -41,7 +39,7 @@ def deep_profiler_data(tmp_path_factory):
@pytest.fixture(scope="session")
def single_cell_deep_profiler(deep_profiler_data):
- """This fixture returns the single cell data and the SingleCellDeepProfiler object"""
+ """This fixture returns the single cell data and the SingleCellDeepProfiler object."""
deep_data, output_folder = deep_profiler_data
# compile single cell data from DP run
@@ -52,7 +50,7 @@ def single_cell_deep_profiler(deep_profiler_data):
def test_single_cell(single_cell_deep_profiler):
- """Test output from SingleCellDeepProfiler.get_single_cells()"""
+ """Test output from SingleCellDeepProfiler.get_single_cells()."""
single_cells, single_cells_DP, output_folder = single_cell_deep_profiler
meta_cols = [x for x in single_cells.columns if x.startswith("Location_")]
@@ -66,8 +64,7 @@ def test_single_cell(single_cell_deep_profiler):
def test_single_cell_normalize(single_cell_deep_profiler):
- """Test output from SingleCellDeepProfiler.normalize_deep_single_cells()"""
-
+ """Test output from SingleCellDeepProfiler.normalize_deep_single_cells()."""
single_cells, single_cells_DP, output_folder = single_cell_deep_profiler
# normalize single cell data with DP processing
diff --git a/tests/test_cyto_utils/test_cell_locations.py b/tests/test_cyto_utils/test_cell_locations.py
index e8100598..622c4121 100644
--- a/tests/test_cyto_utils/test_cell_locations.py
+++ b/tests/test_cyto_utils/test_cell_locations.py
@@ -1,4 +1,4 @@
-"""This tests the output from CellLocation class"""
+"""This tests the output from CellLocation class."""
from typing import List, Type
@@ -14,7 +14,6 @@ def get_metadata_input_dataframe(cell_loc: CellLocation) -> pd.DataFrame:
Gathers the metadata input dataframe given various conditions
from a CellLocation object.
"""
-
return (
pd.read_parquet(
cell_loc.metadata_input,
@@ -41,10 +40,7 @@ def test_output_shape_and_required_columns(
cell_loc_param: List[str],
request: Type[FixtureRequest],
):
- """
- This tests the shape of the output from CellLocation class and verifies that the required columns are present
- """
-
+ """This tests the shape of the output from CellLocation class and verifies that the required columns are present."""
cls_cell_loc = request.getfixturevalue(cell_loc_param)
cell_loc = cls_cell_loc.add_cell_location()
metadata_input_dataframe = get_metadata_input_dataframe(cell_loc=cls_cell_loc)
@@ -74,10 +70,7 @@ def test_output_value_correctness(
cell_loc_param: List[str],
request: Type[FixtureRequest],
):
- """
- This tests the correctness of the values in the output from CellLocation class by comparing the values in the output to the values in the input
- """
-
+ """This tests the correctness of the values in the output from CellLocation class by comparing the values in the output to the values in the input."""
cls_cell_loc = request.getfixturevalue(cell_loc_param)
cell_loc = cls_cell_loc.add_cell_location()
metadata_input_dataframe = get_metadata_input_dataframe(cell_loc=cls_cell_loc)
diff --git a/tests/test_cyto_utils/test_cells.py b/tests/test_cyto_utils/test_cells.py
index 395be7df..ac63fe01 100644
--- a/tests/test_cyto_utils/test_cells.py
+++ b/tests/test_cyto_utils/test_cells.py
@@ -205,9 +205,7 @@ def build_random_data(
def test_SingleCells_init():
- """
- Testing initialization of SingleCells
- """
+ """Testing initialization of SingleCells."""
assert AP.sql_file == TMP_SQLITE_FILE
assert AP.strata == ["Metadata_Plate", "Metadata_Well"]
assert AP.merge_cols == ["TableNumber", "ImageNumber"]
@@ -234,9 +232,7 @@ def test_SingleCells_init():
def test_SingleCells_reset_variables():
- """
- Testing initialization of SingleCells
- """
+ """Testing initialization of SingleCells."""
ap_switch = SingleCells(sql_file=TMP_SQLITE_FILE)
assert ap_switch.subsample_frac == 1
assert ap_switch.subsample_n == "all"
@@ -529,10 +525,7 @@ def test_merge_single_cells_subsample():
def test_merge_single_cells_annotate():
- """
- Tests SingleCells.merge_single_cells using optional annotate functionality
- """
-
+ """Tests SingleCells.merge_single_cells using optional annotate functionality."""
expected_sc_merged_df = annotate(
profiles=AP.merge_single_cells(),
platemap=PLATEMAP_DF,
@@ -546,10 +539,7 @@ def test_merge_single_cells_annotate():
def test_merge_single_cells_cytominer_database_test_file():
- """
- Tests SingleCells.merge_single_cells using cytominer-database test file
- """
-
+ """Tests SingleCells.merge_single_cells using cytominer-database test file."""
# read test file based on cytominer-database exports
sql_path = pathlib.Path(
f"{os.path.dirname(__file__)}/../test_data/cytominer_database_example_data/test_SQ00014613.sqlite",
@@ -1007,9 +997,7 @@ def test_add_image_features():
def test_load_non_canonical_image_table():
- """
- Loading an image table with non-canonical image table name
- """
+ """Loading an image table with non-canonical image table name."""
# test for exception loading image table with default table name "image"
with pytest.raises(OperationalError):
AP_IMAGE_DIFF_NAME.load_image()
diff --git a/tests/test_cyto_utils/test_features_count_na.py b/tests/test_cyto_utils/test_features_count_na.py
index 2232ece6..4de9be20 100644
--- a/tests/test_cyto_utils/test_features_count_na.py
+++ b/tests/test_cyto_utils/test_features_count_na.py
@@ -12,9 +12,7 @@
def test_count_na_features():
- """
- Testing count_na_features pycytominer function
- """
+ """Testing count_na_features pycytominer function."""
count_na_features_result = count_na_features(
population_df=data_df, features=["x", "zz"]
)
diff --git a/tests/test_cyto_utils/test_output.py b/tests/test_cyto_utils/test_output.py
index 72d5a9cf..bf8aec40 100644
--- a/tests/test_cyto_utils/test_output.py
+++ b/tests/test_cyto_utils/test_output.py
@@ -75,10 +75,7 @@ def test_output_tsv():
def test_output_parquet():
- """
- Tests using output function with parquet type
- """
-
+ """Tests using output function with parquet type."""
output_filename = pathlib.Path(f"{TMPDIR}/test_output.parquet")
# test with base output arguments and
diff --git a/tests/test_feature_select.py b/tests/test_feature_select.py
index 8cc6a205..b4f8e7c1 100644
--- a/tests/test_feature_select.py
+++ b/tests/test_feature_select.py
@@ -74,9 +74,7 @@
def test_feature_select_noise_removal():
- """
- Testing noise_removal feature selection operation
- """
+ """Testing noise_removal feature selection operation."""
# Set perturbation groups for the test dataframes
data_df_groups = ["a", "a", "a", "b", "b", "b"]
@@ -223,9 +221,7 @@ def test_feature_select_noise_removal():
def test_feature_select_get_na_columns():
- """
- Testing feature_select and get_na_columns pycytominer function
- """
+ """Testing feature_select and get_na_columns pycytominer function."""
result = feature_select(
data_na_df, features=data_na_df.columns.tolist(), operation="drop_na_columns"
)
@@ -256,9 +252,7 @@ def test_feature_select_get_na_columns():
def test_feature_select_get_na_columns_feature_infer():
- """
- Testing feature_select and get_na_columns pycytominer function
- """
+ """Testing feature_select and get_na_columns pycytominer function."""
result = feature_select(
data_feature_infer_df,
features="infer",
@@ -288,9 +282,7 @@ def test_feature_select_get_na_columns_feature_infer():
def test_feature_select_variance_threshold():
- """
- Testing feature_select and variance_threshold pycytominer function
- """
+ """Testing feature_select and variance_threshold pycytominer function."""
result = feature_select(
data_unique_test_df,
features=data_unique_test_df.columns.tolist(),
@@ -331,10 +323,7 @@ def test_feature_select_variance_threshold():
def test_feature_select_correlation_threshold():
- """
- Testing feature_select and correlation_threshold pycytominer function
- """
-
+ """Testing feature_select and correlation_threshold pycytominer function."""
result = feature_select(
data_df, features=data_df.columns.tolist(), operation="correlation_threshold"
)
@@ -417,10 +406,7 @@ def test_feature_select_compress():
def test_feature_select_blocklist():
- """
- Testing feature_select and get_na_columns pycytominer function
- """
-
+ """Testing feature_select and get_na_columns pycytominer function."""
data_blocklist_df = pd.DataFrame({
"Nuclei_Correlation_Manders_AGP_DNA": [1, 3, 8, 5, 2, 2],
"y": [1, 2, 8, 5, 2, 1],
@@ -442,9 +428,7 @@ def test_feature_select_blocklist():
def test_feature_select_drop_outlier():
- """
- Testing feature_select and get_na_columns pycytominer function
- """
+ """Testing feature_select and get_na_columns pycytominer function."""
result = feature_select(
data_outlier_df, features="infer", operation="drop_outliers"
)
@@ -467,9 +451,7 @@ def test_feature_select_drop_outlier():
def test_output_type():
- """
- Testing feature_select pycytominer function
- """
+ """Testing feature_select pycytominer function."""
# dictionary with the output name associated with the file type
output_dict = {"csv": output_test_file_csv, "parquet": output_test_file_parquet}
diff --git a/tests/test_normalize.py b/tests/test_normalize.py
index 223a0992..b7e8082f 100644
--- a/tests/test_normalize.py
+++ b/tests/test_normalize.py
@@ -84,7 +84,7 @@ def test_normalize_standardize_allsamples():
Testing normalize pycytominer function
method = "standardize"
meta_features = None
- samples="all"
+ samples="all".
"""
normalize_result = normalize(
profiles=data_df.copy(),
@@ -120,7 +120,7 @@ def test_normalize_standardize_ctrlsamples():
Testing normalize pycytominer function
method = "standardize"
meta_features = None
- samples="Metadata_treatment == 'control'"
+ samples="Metadata_treatment == 'control'".
"""
normalize_result = normalize(
profiles=data_df.copy(),
@@ -156,7 +156,7 @@ def test_normalize_robustize_allsamples():
Testing normalize pycytominer function
method = "robustize"
meta_features = None
- samples="all"
+ samples="all".
"""
normalize_result = normalize(
profiles=data_df.copy(),
@@ -192,7 +192,7 @@ def test_normalize_robustize_ctrlsamples():
Testing normalize pycytominer function
method = "robustize"
meta_features = None
- samples="Metadata_treatment == 'control'"
+ samples="Metadata_treatment == 'control'".
"""
normalize_result = normalize(
profiles=data_df.copy(),
@@ -228,7 +228,7 @@ def test_normalize_robustize_mad_allsamples():
Testing normalize pycytominer function
method = "mad_robustize"
meta_features = None
- samples="all"
+ samples="all".
"""
normalize_result = normalize(
profiles=data_df.copy(),
@@ -264,7 +264,7 @@ def test_normalize_robustize_mad_allsamples_novar():
Testing normalize pycytominer function
method = "mad_robustize"
meta_features = None
- samples="all"
+ samples="all".
"""
features = ["x", "y", "z", "zz", "yy"]
@@ -308,7 +308,7 @@ def test_normalize_standardize_allsamples_fromfile():
data_file provided
method = "standardize"
meta_features = None
- samples="all"
+ samples="all".
"""
normalize_result = normalize(
profiles=data_file,
@@ -356,7 +356,7 @@ def test_normalize_standardize_allsamples_output():
data_file provided
method = "standardize"
meta_features = None
- samples="all"
+ samples="all".
"""
out_normalize_file = os.path.join(tmpdir, "test_normalize_output.csv")
@@ -501,9 +501,7 @@ def test_normalize_spherize():
def test_spherize_epsilon():
- """
- Test that epsilon is successfully passed to the spherize transform method
- """
+ """Test that epsilon is successfully passed to the spherize transform method."""
sphere_norm_df = normalize(
data_spherize_df, features=["a", "b", "c", "d"], meta_features=["id"]
)
@@ -532,9 +530,7 @@ def test_spherize_epsilon():
def test_output_type():
- """
- Testing normalize pycytominer function with output
- """
+ """Testing normalize pycytominer function with output."""
# dictionary with the output name associated with the file type
output_dict = {"csv": output_test_file_csv, "parquet": output_test_file_parquet}
diff --git a/tests/test_operations/test_get_na_columns.py b/tests/test_operations/test_get_na_columns.py
index 9c8bd557..555d9c01 100644
--- a/tests/test_operations/test_get_na_columns.py
+++ b/tests/test_operations/test_get_na_columns.py
@@ -13,9 +13,7 @@
def test_get_na_columns():
- """
- Testing get_na_columns pycytominer function
- """
+ """Testing get_na_columns pycytominer function."""
get_na_columns_result = get_na_columns(
population_df=data_df, features=["x", "y", "zz"], cutoff=0.4
)
@@ -41,9 +39,7 @@ def test_get_na_columns():
def test_get_na_columns_sample():
- """
- Testing get_na_columns pycyominer function with samples option
- """
+ """Testing get_na_columns pycyominer function with samples option."""
data_sample_id_df = data_df.assign(
Metadata_sample=[f"sample_{x}" for x in range(0, data_df.shape[0])]
)
diff --git a/tests/test_operations/test_transform.py b/tests/test_operations/test_transform.py
index d66e1615..85a9f6e0 100644
--- a/tests/test_operations/test_transform.py
+++ b/tests/test_operations/test_transform.py
@@ -79,9 +79,7 @@ def test_spherize_precenter():
def test_robust_mad():
- """
- Testing the RobustMAD class
- """
+ """Testing the RobustMAD class."""
scaler = RobustMAD()
scaler = scaler.fit(data_df)
transform_df = scaler.transform(data_df)
diff --git a/tests/test_operations/test_variance_threshold.py b/tests/test_operations/test_variance_threshold.py
index a1a19764..ceb4e97f 100644
--- a/tests/test_operations/test_variance_threshold.py
+++ b/tests/test_operations/test_variance_threshold.py
@@ -28,9 +28,7 @@
def test_calculate_frequency():
- """
- Testing calculate_frequency pycytominer function for variance threshold calculation
- """
+ """Testing calculate_frequency pycytominer function for variance threshold calculation."""
freq_cut = 0.05
excluded_features_freq = data_df.apply(
diff --git a/walkthroughs/nbconverted/single_cell_usage.py b/walkthroughs/nbconverted/single_cell_usage.py
deleted file mode 100644
index c21c8691..00000000
--- a/walkthroughs/nbconverted/single_cell_usage.py
+++ /dev/null
@@ -1,242 +0,0 @@
-#!/usr/bin/env python
-
-# # Single-cell Profiling Walkthrough
-#
-# Welcome to this walkthrough where we will guide you through the process of extracting single cell morphology features using the [pycytominer](https://github.com/cytomining/pycytominer) API.
-#
-# For this walkthrough, we will be working with the NF1-Schwann cell morphology dataset.
-# If you would like the more information about this dataset, you can refer to this [repository](https://github.com/WayScience/nf1_cellpainting_data)
-#
-# From the mentioned repo, we specifically used this [dataset](https://github.com/WayScience/nf1_cellpainting_data/tree/main/2.cellprofiler_analysis/analysis_output) and the associated [metadata](https://github.com/WayScience/nf1_cellpainting_data/tree/main/0.download_data/metadata) to generate the walkthrough.
-#
-#
-# Let's get started with the walkthrough below!
-
-# In[1]:
-
-
-import pathlib
-
-# ignore mix type warnings from pandas
-import warnings
-
-import pandas as pd
-
-from pycytominer import annotate, feature_select, normalize
-
-# pycytominer imports
-from pycytominer.cyto_utils.cells import SingleCells
-
-warnings.filterwarnings("ignore")
-
-
-# ## About the inputs
-#
-#
-# In this section, we will set up the expected input and output paths that will be generated throughout this walkthrough. Let's take a look at the explanation of these inputs and outputs.
-#
-# For this workflow, we have two main inputs:
-#
-# - **plate_data** (SQLite file): This contains the quantified single-cell morphology features that we'll be working with.
-# - **plate_map** (CSV file): This contains additional information related to the cells, providing valuable context of our single-cell morphology dataset.
-#
-# Now, let's explore the outputs generated in this workflow. In this walkthrough, we will be generating four profiles:
-#
-# - **sc_profiles_path**: This refers to the single-cell morphology profile that will be generated.
-# - **anno_profiles_path**: This corresponds to the annotated single-cell morphology profile, meaning the information from the **plate_map** is added to each single-cell
-# - **norm_profiles_path**: This represents the normalized single-cell morphology profile.
-# - **feat_profiles_path**: Lastly, this refers to the selected features from the single-cell morphology profile.
-#
-# **Note**: All profiles are outputted as `.csv` files, however, users can output these files as `parquet` or other file file formats.
-# For more information about output formats, please refer the documentation [here](https://pycytominer.readthedocs.io/en/latest/pycytominer.cyto_utils.html#pycytominer.cyto_utils.output.output)
-#
-# These profiles will serve as important outputs that will help us analyze and interpret the single-cell morphology data effectively. Now that we have a clear understanding of the inputs and outputs, let's proceed further in our walkthrough.
-
-# In[2]:
-
-
-# Setting file paths
-data_dir = pathlib.Path("./data/").resolve(strict=True)
-metadata_dir = (data_dir / "metadata").resolve(strict=True)
-out_dir = pathlib.Path("results")
-out_dir.mkdir(exist_ok=True)
-
-# input file paths
-plate_data = pathlib.Path("./data/nf1_data.sqlite").resolve(strict=True)
-plate_map = (metadata_dir / "platemap_NF1_CP.csv").resolve(strict=True)
-
-# setting output paths
-sc_profiles_path = out_dir / "nf1_single_cell_profile.csv.gz"
-anno_profiles_path = out_dir / "nf1_annotated_profile.csv.gz"
-norm_profiles_path = out_dir / "nf1_normalized_profile.csv.gz"
-feat_profiles_path = out_dir / "nf1_features_profile.csv.gz"
-
-
-# ## Generating Merged Single-cell Morphology Dataset
-#
-# In this section of the walkthrough, our goal is to load the NF1 dataset and create a merged single-cell morphology dataset.
-#
-# Currently, the NF1 morphology `SQLite` dataset was generated by using [CellProfiler's](https://github.com/CellProfiler/CellProfiler) [ExportToDatabase](https://cellprofiler-manual.s3.amazonaws.com/CellProfiler-4.2.5/modules/fileprocessing.html?highlight=exporttodatabase#module-cellprofiler.modules.exporttodatabase) function, where each table represents a different compartment, such as Image, Cell, Nucleus, and Cytoplasm.
-#
-# To achieve this, we will utilize the `SingleCells` class, which offers a range of functionalities specifically designed for single-cell analysis. You can find detailed documentation on these functionalities [here](https://pycytominer.readthedocs.io/en/latest/pycytominer.cyto_utils.html#pycytominer.cyto_utils.cells.SingleCells).
-#
-# However, for our purpose in this walkthrough, we will focus on using the `SingleCells` class to merge all the tables within the NF1 sqlite file into a merged single-cell morphology dataset.
-#
-# ### Updating defaults
-# Before we proceed further, it is important to update the default parameters in the `SingleCells`class to accommodate the table name changes in our NF1 dataset.
-#
-# Since the table names in our NF1 dataset differ from the default table names recognized by the `SingleCells` class, we need to make adjustments to ensure proper recognition of these table name changes.
-
-# In[3]:
-
-
-# update compartment names and strata
-strata = ["Image_Metadata_Well", "Image_Metadata_Plate"]
-compartments = ["Per_Cells", "Per_Cytoplasm", "Per_Nuclei"]
-
-# Updating linking columns for merging all compartments
-linking_cols = {
- "Per_Cytoplasm": {
- "Per_Cells": "Cytoplasm_Parent_Cells",
- "Per_Nuclei": "Cytoplasm_Parent_Nuclei",
- },
- "Per_Cells": {"Per_Cytoplasm": "Cells_Number_Object_Number"},
- "Per_Nuclei": {"Per_Cytoplasm": "Nuclei_Number_Object_Number"},
-}
-
-
-# Now that we have stored the updated parameters, we can use them as inputs for `SingleCells` class to merge all the NF1 sqlite tables into a single consolidated dataset.
-#
-# This is done through the `merge_single_cells` method. For more infromation about `merge_single_cells` please refer to the documentation [here](https://pycytominer.readthedocs.io/en/latest/pycytominer.cyto_utils.html#pycytominer.cyto_utils.cells.SingleCells.merge_single_cells)
-
-# In[4]:
-
-
-# setting up sqlite address
-sqlite_address = f"sqlite:///{plate_data!s}"
-
-# loading single cell morphology data into pycyotminer's SingleCells Object
-single_cell_profile = SingleCells(
- sql_file=sqlite_address,
- compartments=compartments,
- compartment_linking_cols=linking_cols,
- image_table_name="Per_Image",
- strata=strata,
- merge_cols=["ImageNumber"],
- image_cols="ImageNumber",
- load_image_data=True,
-)
-
-# merging all sqlite table into a single tabular dataset (csv) and save as
-# compressed csv file
-single_cell_profile.merge_single_cells(
- sc_output_file=sc_profiles_path, compression_options="gzip"
-)
-
-
-# Now that we have created our merged single-cell profile, let's move on to the next step: loading our `platemaps`.
-#
-# `Platemaps` provide us with additional information that is crucial for our analysis. They contain details such as well positions, genotypes, gene names, perturbation types, and more. In other words, platemaps serve as a valuable source of metadata for our single-cell morphology profile.
-#
-
-# In[5]:
-
-
-# loading plate map and display it
-platemap_df = pd.read_csv(plate_map)
-
-# displaying platemap
-print(platemap_df.columns.tolist())
-
-
-# ## Annotation
-#
-# In this step of the walkthrough, we will combine the metadata with the merged single-cell morphology dataset. To accomplish this, we will utilize the `annotation` function provided by `pycytominer`.
-#
-# The `annotation` function takes two inputs: the merged single-cell morphology dataset and its associated plate map. By combining these two datasets, we will generate an annotated_profile that contains enriched information.
-#
-# More information about the `annotation` function can be found [here](https://pycytominer.readthedocs.io/en/latest/pycytominer.html#module-pycytominer.annotate)
-#
-
-# In[6]:
-
-
-# annotating merged single-cell profile with metadata
-annotate(
- profiles=sc_profiles_path,
- platemap=platemap_df,
- join_on=["Metadata_well_position", "Image_Metadata_Well"],
- output_file=anno_profiles_path,
- compression_options="gzip",
-)
-
-# save message display
-print(f"Annotated profile saved in: {anno_profiles_path}")
-
-
-# ## Normalization Step
-#
-# The next step is to normalize our dataset using the `normalize` function provided by `pycytominer`.
-# More information regards `pycytominer`'s `normalize` function can be found [here](https://pycytominer.readthedocs.io/en/latest/pycytominer.html#module-pycytominer.normalize)
-#
-# Normalization is a critical preprocessing step that improves the quality of our dataset. It addresses two key challenges: mitigating the impact of outliers and handling variations in value scales. By normalizing the data, we ensure that our downstream analysis is not heavily influenced by these factors.
-#
-# Additionally, normalization plays a crucial role in determining feature importance (which is crucial for our last step). By bringing all features to a comparable scale, it enables the identification of important features without biases caused by outliers or widely-scaled values.
-#
-# To normalize our annotated single-cell morphology profile, we will utilize the `normalize` function from `pycytominer`. This function is specifically designed to handle the normalization process for cytometry data.
-
-# In[7]:
-
-
-# normalize dataset
-normalize(
- profiles=anno_profiles_path,
- features="infer",
- image_features=False,
- meta_features="infer",
- samples="all",
- method="standardize",
- output_file=norm_profiles_path,
- compression_options="gzip",
-)
-
-# save message display
-print(f"Normalized profile saved in: {norm_profiles_path}")
-
-
-# ## Feature Selection
-#
-#
-# In the final section of our walkthrough, we will utilize the normalized dataset to extract important morphological features and generate a selected features profile.
-#
-# To accomplish this, we will make use of the `feature_select` function provided by `pycytominer`.
-# Using `pycytominer`'s `feature_select` function to our dataset, we can identify the most informative morphological features that contribute significantly to the variations observed in our data. These selected features will be utilized to create our feature profile.
-#
-# For more detailed information about the `feature_select` function, its parameters, and its capabilities, please refer to the documentation available [here](https://pycytominer.readthedocs.io/en/latest/pycytominer.html#module-pycytominer.feature_select).
-#
-
-# In[8]:
-
-
-# creating selected features profile
-feature_select(
- profiles=norm_profiles_path,
- features="infer",
- image_features=False,
- samples="all",
- operation=["variance_threshold", "correlation_threshold", "blocklist"],
- output_file=feat_profiles_path,
- compression_options="gzip",
-)
-
-# save message display
-print(f"Selected features profile saved in: {feat_profiles_path}")
-
-
-# Congratulations! You have successfully completed our walkthrough. We hope that it has provided you with a basic understanding of how to analyze cell morphology features using `pycytominer`.
-#
-# By following the steps outlined in this walkthrough, you have gained valuable insights into processing high-dimensional single-cell morphology data with ease using `pycytominer`. However, please keep in mind that `pycytominer` offers a wide range of functionalities beyond what we covered here. We encourage you to explore the documentation to discover more advanced features and techniques.
-#
-# If you have any questions or need further assistance, don't hesitate to visit the `pycytominer` repository and post your question in the [issues](https://github.com/cytomining/pycytominer/issues) section. The community is there to support you and provide guidance.
-#
-# Now that you have the knowledge and tools to analyze cell morphology features, have fun exploring and mining your data!
diff --git a/walkthroughs/single_cell_usage.ipynb b/walkthroughs/single_cell_usage.ipynb
index 440f4259..0f1c943e 100644
--- a/walkthroughs/single_cell_usage.ipynb
+++ b/walkthroughs/single_cell_usage.ipynb
@@ -35,7 +35,7 @@
"# ignore mix type warnings from pandas\n",
"import warnings\n",
"\n",
- "warnings.filterwarnings(\"ignore\")\n"
+ "warnings.filterwarnings(\"ignore\")"
]
},
{
@@ -86,7 +86,7 @@
"sc_profiles_path = out_dir / \"nf1_single_cell_profile.csv.gz\"\n",
"anno_profiles_path = out_dir / \"nf1_annotated_profile.csv.gz\"\n",
"norm_profiles_path = out_dir / \"nf1_normalized_profile.csv.gz\"\n",
- "feat_profiles_path = out_dir / \"nf1_features_profile.csv.gz\"\n"
+ "feat_profiles_path = out_dir / \"nf1_features_profile.csv.gz\""
]
},
{
@@ -128,7 +128,7 @@
" },\n",
" \"Per_Cells\": {\"Per_Cytoplasm\": \"Cells_Number_Object_Number\"},\n",
" \"Per_Nuclei\": {\"Per_Cytoplasm\": \"Nuclei_Number_Object_Number\"},\n",
- "}\n"
+ "}"
]
},
{
@@ -177,7 +177,7 @@
"# compressed csv file\n",
"single_cell_profile.merge_single_cells(\n",
" sc_output_file=sc_profiles_path, compression_options=\"gzip\"\n",
- ")\n"
+ ")"
]
},
{
@@ -208,7 +208,7 @@
"platemap_df = pd.read_csv(plate_map)\n",
"\n",
"# displaying platemap\n",
- "print(platemap_df.columns.tolist())\n"
+ "print(platemap_df.columns.tolist())"
]
},
{
@@ -249,7 +249,7 @@
")\n",
"\n",
"# save message display\n",
- "print(f\"Annotated profile saved in: {anno_profiles_path}\")\n"
+ "print(f\"Annotated profile saved in: {anno_profiles_path}\")"
]
},
{
@@ -296,7 +296,7 @@
")\n",
"\n",
"# save message display\n",
- "print(f\"Normalized profile saved in: {norm_profiles_path}\")\n"
+ "print(f\"Normalized profile saved in: {norm_profiles_path}\")"
]
},
{
@@ -341,7 +341,7 @@
")\n",
"\n",
"# save message display\n",
- "print(f\"Selected features profile saved in: {feat_profiles_path}\")\n"
+ "print(f\"Selected features profile saved in: {feat_profiles_path}\")"
]
},
{
+
+[Click here for high resolution pipeline image](https://github.com/cytomining/pycytominer/blob/main/media/pipeline.png)
+
+Image data flow from a microscope to cell segmentation and feature extraction tools (e.g. CellProfiler or DeepProfiler).
+From here, additional single cell processing tools curate the single cell readouts into a form manageable for pycytominer input.
+For CellProfiler, we use [cytominer-database](https://github.com/cytomining/cytominer-database) or [CytoTable](https://github.com/cytomining/CytoTable).
+For DeepProfiler, we include single cell processing tools in [pycytominer.cyto_utils](cyto_utils.md).
+
+From the single cell output, pycytominer performs five steps using a simple API (described below), before passing along data to [cytominer-eval](https://github.com/cytomining/cytominer-eval) for quality and perturbation strength evaluation.
+
+## Installation
+
+You can install pycytominer via pip:
+
+```bash
+pip install pycytominer
+```
+
+or conda:
+
+```bash
+conda install -c conda-forge pycytominer
+```
+
+## Frameworks
+
+Pycytominer is primarily built on top of [pandas](https://pandas.pydata.org/docs/index.html), also using aspects of SQLAlchemy, sklearn, and pyarrow.
+
+Pycytominer currently supports [parquet](https://parquet.apache.org/) and compressed text file (e.g. `.csv.gz`) i/o.
+
+## API
+
+Pycytominer has five major processing functions:
+
+1. Aggregate - Average single-cell profiles based on metadata information (most often "well").
+2. Annotate - Append metadata (most often from the platemap file) to the feature profile
+3. Normalize - Transform input feature data into consistent distributions
+4. Feature select - Exclude non-informative or redundant features
+5. Consensus - Average aggregated profiles by replicates to form a "consensus signature"
+
+The API is consistent for each of these functions:
+
+```python
+# Each function takes as input a pandas DataFrame or file path
+# and transforms the input data based on the provided options and methods
+df = function(
+ profiles_or_path,
+ features,
+ samples,
+ method,
+ output_file,
+ additional_options...
+)
+```
+
+Each processing function has unique arguments, see our [documentation](https://pycytominer.readthedocs.io/) for more details.
+
+## Usage
+
+The default way to use pycytominer is within python scripts, and using pycytominer is simple and fun.
+
+```python
+# Real world example
+import pandas as pd
+import pycytominer
+
+commit = "da8ae6a3bc103346095d61b4ee02f08fc85a5d98"
+url = f"https://media.githubusercontent.com/media/broadinstitute/lincs-cell-painting/{commit}/profiles/2016_04_01_a549_48hr_batch1/SQ00014812/SQ00014812_augmented.csv.gz"
+
+df = pd.read_csv(url)
+
+normalized_df = pycytominer.normalize(
+ profiles=df,
+ method="standardize",
+ samples="Metadata_broad_sample == 'DMSO'"
+)
+```
+
+### Pipeline orchestration
+
+Pycytominer is a collection of different functions with no explicit link between steps.
+However, some options exist to use pycytominer within a pipeline framework.
+
+| Project | Format | Environment | pycytominer usage |
+| :------------------------------------------------------------------------------- | :-------- | :------------------- | :---------------------- |
+| [Profiling-recipe](https://github.com/cytomining/profiling-recipe) | yaml | agnostic | full pipeline support |
+| [CellProfiler-on-Terra](https://github.com/broadinstitute/cellprofiler-on-Terra) | WDL | google cloud / Terra | single-cell aggregation |
+| [CytoSnake](https://github.com/WayScience/CytoSnake) | snakemake | agnostic | full pipeline support |
+
+A separate project called [AuSPICES](https://github.com/broadinstitute/AuSPICEs) offers pipeline support up to image feature extraction.
+
+## Other functionality
+
+Pycytominer was written with a goal of processing any high-throughput image-based profiling data.
+However, the initial use case was developed for processing image-based profiling experiments specifically.
+And, more specifically than that, image-based profiling readouts from [CellProfiler](https://github.com/CellProfiler) measurements from [Cell Painting](https://www.nature.com/articles/nprot.2016.105) data.
+
+Therefore, we have included some custom tools in `pycytominer/cyto_utils` that provides other functionality:
+
+Note, [`pycytominer.cyto_utils.cells.SingleCells()`](cyto_utils.md##pycytominer.cyto_utils.cells) contains code to interact with single-cell SQLite files, which are output from CellProfiler.
+Processing capabilities for SQLite files depends on SQLite file size and your available computational resources (for ex. memory and cores).
+
+### CellProfiler CSV collation
+
+If running your images on a cluster, unless you have a MySQL or similar large database set up then you will likely end up with lots of different folders from the different cluster runs (often one per well or one per site), each one containing an `Image.csv`, `Nuclei.csv`, etc.
+In order to look at full plates, therefore, we first need to collate all of these CSVs into a single file (currently SQLite) per plate.
+We currently do this with a library called [cytominer-database](https://github.com/cytomining/cytominer-database).
+
+If you want to perform this data collation inside pycytominer using the `cyto_utils` function `collate` (and/or you want to be able to run the tests and have them all pass!), you will need `cytominer-database==0.3.4`; this will change your installation commands slightly:
+
+```bash
+# Example for general case commit:
+pip install "pycytominer[collate]"
+
+# Example for specific commit:
+pip install "pycytominer[collate] @ git+https://github.com/cytomining/pycytominer@77d93a3a551a438799a97ba57d49b19de0a293ab"
+```
+
+If using `pycytominer` in a conda environment, in order to run `collate.py`, you will also want to make sure to add `cytominer-database=0.3.4` to your list of dependencies.
+
+### Creating a cell locations lookup table
+
+The `CellLocation` class offers a convenient way to augment a [LoadData](https://cellprofiler-manual.s3.amazonaws.com/CPmanual/LoadData.html) file with X,Y locations of cells in each image.
+The locations information is obtained from a single cell SQLite file.
+
+To use this functionality, you will need to modify your installation command, similar to above:
+
+```bash
+# Example for general case commit:
+pip install "pycytominer[cell_locations]"
+```
+
+Example using this functionality:
+
+```bash
+metadata_input="s3://cellpainting-gallery/test-cpg0016-jump/source_4/workspace/load_data_csv/2021_08_23_Batch12/BR00126114/test_BR00126114_load_data_with_illum.parquet"
+single_single_cell_input="s3://cellpainting-gallery/test-cpg0016-jump/source_4/workspace/backend/2021_08_23_Batch12/BR00126114/test_BR00126114.sqlite"
+augmented_metadata_output="~/Desktop/load_data_with_illum_and_cell_location_subset.parquet"
+
+python \
+ -m pycytominer.cyto_utils.cell_locations_cmd \
+ --metadata_input ${metadata_input} \
+ --single_cell_input ${single_single_cell_input} \
+ --augmented_metadata_output ${augmented_metadata_output} \
+ add_cell_location
+
+# Check the output
+
+python -c "import pandas as pd; print(pd.read_parquet('${augmented_metadata_output}').head())"
+
+# It should look something like this (depends on the width of your terminal):
+
+# Metadata_Plate Metadata_Well Metadata_Site ... PathName_OrigRNA ImageNumber CellCenters
+# 0 BR00126114 A01 1 ... s3://cellpainting-gallery/cpg0016-jump/source_... 1 [{'Nuclei_Location_Center_X': 943.512129380054...
+# 1 BR00126114 A01 2 ... s3://cellpainting-gallery/cpg0016-jump/source_... 2 [{'Nuclei_Location_Center_X': 29.9516027655562...
+```
+
+### Generating a GCT file for morpheus
+
+The software [morpheus](https://software.broadinstitute.org/morpheus/) enables profile visualization in the form of interactive heatmaps.
+Pycytominer can convert profiles into a `.gct` file for drag-and-drop input into morpheus.
+
+```python
+# Real world example
+import pandas as pd
+import pycytominer
+
+commit = "da8ae6a3bc103346095d61b4ee02f08fc85a5d98"
+plate = "SQ00014812"
+url = f"https://media.githubusercontent.com/media/broadinstitute/lincs-cell-painting/{commit}/profiles/2016_04_01_a549_48hr_batch1/{plate}/{plate}_normalized_feature_select.csv.gz"
+
+df = pd.read_csv(url)
+output_file = f"{plate}.gct"
+
+pycytominer.cyto_utils.write_gct(
+ profiles=df,
+ output_file=output_file
+)
+```
+
+## Citing pycytominer
+
+If you have used `pycytominer` in your project, please use the citation below.
+You can also find the citation in the 'cite this repository' link at the top right under `about` section.
+
+APA:
+
+```text
+Serrano, E., Chandrasekaran, N., Bunten, D., Brewer, K., Tomkinson, J., Kern, R., Bornholdt, M., Fleming, S., Pei, R., Arevalo, J., Tsang, H., Rubinetti, V., Tromans-Coia, C., Becker, T., Weisbart, E., Bunne, C., Kalinin, A. A., Senft, R., Taylor, S. J., Jamali, N., Adeboye, A., Abbasi, H. S., Goodman, A., Caicedo, J., Carpenter, A. E., Cimini, B. A., Singh, S., & Way, G. P. Reproducible image-based profiling with Pycytominer. https://doi.org/10.48550/arXiv.2311.13417
+```
diff --git a/docs/conf.py b/docs/conf.py
index be8581a2..af5e0eec 100644
--- a/docs/conf.py
+++ b/docs/conf.py
@@ -1,4 +1,4 @@
-# Configuration file for the Sphinx documentation builder.
+"""Configuration file for the Sphinx documentation builder."""
#
# This file only contains a selection of the most common options. For a full
# list see the documentation:
diff --git a/docs/cyto_utils.md b/docs/cyto_utils.md
new file mode 100644
index 00000000..0251bb86
--- /dev/null
+++ b/docs/cyto_utils.md
@@ -0,0 +1,5 @@
+# Cyto utilities
+
+Functions enabling smooth interaction with CellProfiler and DeepProfiler output formats.
+
+::: pycytominer.cyto_utils
diff --git a/docs/functions.md b/docs/functions.md
new file mode 100644
index 00000000..ef4a65ab
--- /dev/null
+++ b/docs/functions.md
@@ -0,0 +1,16 @@
+# Main Functions
+
+
+
+
+::: pycytominer
+ options:
+ members:
+ - aggregate
+ - annotate
+ - consensus
+ - feature_select
+ - normalize
+
+
+
diff --git a/docs/index.rst b/docs/index.rst
deleted file mode 100644
index 460d88b8..00000000
--- a/docs/index.rst
+++ /dev/null
@@ -1,22 +0,0 @@
-.. pycytominer documentation main file, created by
- sphinx-quickstart on Fri Feb 19 14:44:29 2021.
- You can adapt this file completely to your liking, but it should at least
- contain the root `toctree` directive.
-
-.. mdinclude:: ../README.md
-
-.. toctree::
- :maxdepth: 2
- :caption: Contents:
-
- install
- walkthrough
- tutorial
- modules
-
-Indices and tables
-==================
-
-* :ref:`genindex`
-* :ref:`modindex`
-* :ref:`search`
diff --git a/docs/install.rst b/docs/install.md
similarity index 71%
rename from docs/install.rst
rename to docs/install.md
index 99213db9..9f2963ab 100644
--- a/docs/install.rst
+++ b/docs/install.md
@@ -1,14 +1,13 @@
-Install
-=======
+# Installation
To install pycytominer, use pip:
-.. code-block:: bash
-
+```bash
pip install pycytominer
+```
You can also install pycytominer with conda:
-.. code-block:: bash
-
+```bash
conda install -c conda-forge pycytominer
+```
diff --git a/docs/modules.rst b/docs/modules.rst
deleted file mode 100644
index bec89387..00000000
--- a/docs/modules.rst
+++ /dev/null
@@ -1,7 +0,0 @@
-pycytominer
-===========
-
-.. toctree::
- :maxdepth: 4
-
- pycytominer
diff --git a/docs/operations.md b/docs/operations.md
new file mode 100644
index 00000000..ba31c6ed
--- /dev/null
+++ b/docs/operations.md
@@ -0,0 +1,6 @@
+# Operations
+
+We do not recommend interacting with these functions directly.
+The core pycytominer API uses these operations internally.
+
+::: pycytominer.operations
diff --git a/docs/pycytominer.cyto_utils.rst b/docs/pycytominer.cyto_utils.rst
deleted file mode 100644
index 8618d60d..00000000
--- a/docs/pycytominer.cyto_utils.rst
+++ /dev/null
@@ -1,76 +0,0 @@
-Cyto utilities
-==============
-
-Functions enabling smooth interaction with CellProfiler and DeepProfiler output formats.
-
-pycytominer.cyto\_utils.cells module
-------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.cells
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.collate module
---------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.collate
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.features module
----------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.features
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.load module
------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.load
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.modz module
------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.modz
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.output module
--------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.output
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.util module
------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.util
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.write\_gct module
------------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.write_gct
- :members:
- :undoc-members:
- :show-inheritance:
-
-Module contents
----------------
-
-.. automodule:: pycytominer.cyto_utils
- :members:
- :undoc-members:
- :show-inheritance:
diff --git a/docs/pycytominer.operations.rst b/docs/pycytominer.operations.rst
deleted file mode 100644
index 062919d6..00000000
--- a/docs/pycytominer.operations.rst
+++ /dev/null
@@ -1,45 +0,0 @@
-Operations
-==========
-
-We do not recommend interacting with these functions directly.
-The core pycytominer API uses these operations internally.
-
-pycytominer.operations.correlation\_threshold module
-----------------------------------------------------
-
-.. automodule:: pycytominer.operations.correlation_threshold
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.operations.get\_na\_columns module
-----------------------------------------------
-
-.. automodule:: pycytominer.operations.get_na_columns
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.operations.transform module
----------------------------------------
-
-.. automodule:: pycytominer.operations.transform
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.operations.variance\_threshold module
--------------------------------------------------
-
-.. automodule:: pycytominer.operations.variance_threshold
- :members:
- :undoc-members:
- :show-inheritance:
-
-Module contents
----------------
-
-.. automodule:: pycytominer.operations
- :members:
- :undoc-members:
- :show-inheritance:
diff --git a/docs/pycytominer.rst b/docs/pycytominer.rst
deleted file mode 100644
index 01b61229..00000000
--- a/docs/pycytominer.rst
+++ /dev/null
@@ -1,59 +0,0 @@
-Core functions
-==============
-
-pycytominer.aggregate module
-----------------------------
-
-.. automodule:: pycytominer.aggregate
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.annotate module
----------------------------
-
-.. automodule:: pycytominer.annotate
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.normalize module
-----------------------------
-
-.. automodule:: pycytominer.normalize
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.feature\_select module
-----------------------------------
-
-.. automodule:: pycytominer.feature_select
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.consensus module
-----------------------------
-
-.. automodule:: pycytominer.consensus
- :members:
- :undoc-members:
- :show-inheritance:
-
-Helper functions
-----------------
-
-.. toctree::
- :maxdepth: 4
-
- pycytominer.cyto_utils
- pycytominer.operations
-
-Module contents
----------------
-
-.. automodule:: pycytominer
- :members:
- :undoc-members:
- :show-inheritance:
diff --git a/docs/tutorial.md b/docs/tutorial.md
new file mode 100644
index 00000000..492e0008
--- /dev/null
+++ b/docs/tutorial.md
@@ -0,0 +1,13 @@
+# Tutorials
+
+This [tutorial](https://github.com/cytomining/pipeline-examples#readme) shows how to run a image-based profiling pipeline using pycytominer. Using IPython notebooks, it walks through the following steps:
+
+- Downloading a dataset of single cell [CellProfiler](https://cellprofiler.org/) profiles.
+- Processing the profiles using PyCytominer. This includes the following steps:
+- Data initialization
+- Single cell aggregation to create well-level profiles
+- Addition of experiment metadata to the well-level profiles
+- Profile normalization
+- Feature selection
+- Forming consensus signatures
+- Evaluating the profile quality using [cytominer-eval](https://github.com/cytomining/cytominer-eval>).
diff --git a/docs/tutorial.rst b/docs/tutorial.rst
deleted file mode 100644
index da5d5f6f..00000000
--- a/docs/tutorial.rst
+++ /dev/null
@@ -1,14 +0,0 @@
-Tutorials
-=========
-
-`This `_ tutorial shows how to run a image-based profiling pipeline using pycytominer. Using IPython notebooks, it walks through the following steps:
-
-#. Downloading a dataset of single cell `CellProfiler `_ profiles.
-#. Processing the profiles using PyCytominer. This includes the following steps:
- #. Data initialization
- #. Single cell aggregation to create well-level profiles
- #. Addition of experiment metadata to the well-level profiles
- #. Profile normalization
- #. Feature selection
- #. Forming consensus signatures
-#. Evaluating the profile quality using `cytominer-eval `_.
\ No newline at end of file
diff --git a/docs/walkthrough.rst b/docs/walkthrough.rst
deleted file mode 100644
index 6733ed2a..00000000
--- a/docs/walkthrough.rst
+++ /dev/null
@@ -1,7 +0,0 @@
-Walkthroughs
-============
-
-.. toctree::
- :maxdepth: 1
-
- walkthroughs/single_cell_usage.ipynb
\ No newline at end of file
diff --git a/mkdocs.yml b/mkdocs.yml
new file mode 100644
index 00000000..0c264885
--- /dev/null
+++ b/mkdocs.yml
@@ -0,0 +1,67 @@
+site_name: pycytominer
+repo_url: https://github.com/cytomining/pycytominer
+site_url: https://cytomining.github.io/pycytominer
+site_description: Python package for processing image-based profiling data
+site_author: Pycytominer contributors
+edit_uri: edit/main/docs/
+repo_name: cytomining/pycytominer
+
+nav:
+ - Home: README.md
+ - Installation: install.md
+ - Main Functions: functions.md
+ - Cyto Utilities: cyto_utils.md
+ - Operations: operations.md
+ - Tutorial: tutorial.md
+ - Walkthroughs:
+ - Single Cell Usage: walkthroughs/single_cell_usage.ipynb
+plugins:
+ - search
+ - mkdocs-jupyter
+ - mkdocstrings:
+ handlers:
+ python:
+ paths:
+ - pycytominer
+ options:
+ show_source: true
+ show_submodules: true
+ show_symbol_type_heading: true
+ show_symbol_type_toc: true
+ docstring_style: numpy
+ show_root_members_full_path: true
+
+theme:
+ name: material
+ feature:
+ tabs: true
+ palette:
+ - media: "(prefers-color-scheme: light)"
+ scheme: default
+ primary: deep purple
+ accent: purple
+ toggle:
+ icon: material/brightness-7
+ name: Switch to dark mode
+ - media: "(prefers-color-scheme: dark)"
+ scheme: slate
+ primary: deep purple
+ accent: purple
+ toggle:
+ icon: material/brightness-4
+ name: Switch to light mode
+ icon:
+ repo: fontawesome/brands/github
+
+extra:
+ social:
+ - icon: fontawesome/brands/github
+ link: https://github.com/cytomining/pycytominer
+ - icon: fontawesome/brands/python
+ link: https://pypi.org/project/pycytominer
+
+markdown_extensions:
+ - toc:
+ permalink: true
+ - pymdownx.arithmatex:
+ generic: true
diff --git a/poetry.lock b/poetry.lock
index 74461f43..2048fed6 100644
--- a/poetry.lock
+++ b/poetry.lock
@@ -81,108 +81,108 @@ test = ["crate", "geojson", "pytest", "pytest-cov"]
[[package]]
name = "aiobotocore"
-version = "2.12.1"
+version = "2.12.3"
description = "Async client for aws services using botocore and aiohttp"
optional = true
python-versions = ">=3.8"
files = [
- {file = "aiobotocore-2.12.1-py3-none-any.whl", hash = "sha256:6a9a3d646cf422f45fdc1e4256e78563ebffba64733bc9b8ca9123614e8ba9af"},
- {file = "aiobotocore-2.12.1.tar.gz", hash = "sha256:8706b28f16f93c541f6ed50352115a79d8f3499539f8d0bb70aa0f7a5379c1fe"},
+ {file = "aiobotocore-2.12.3-py3-none-any.whl", hash = "sha256:86737685f4625e8f05c4e7a608a07cc97607263279f66cf6b02b640c4eafd324"},
+ {file = "aiobotocore-2.12.3.tar.gz", hash = "sha256:e2a2929207bc5d62eb556106c2224c1fd106d5c65be2eb69f15cc8c34c44c236"},
]
[package.dependencies]
aiohttp = ">=3.7.4.post0,<4.0.0"
aioitertools = ">=0.5.1,<1.0.0"
-botocore = ">=1.34.41,<1.34.52"
+botocore = ">=1.34.41,<1.34.70"
wrapt = ">=1.10.10,<2.0.0"
[package.extras]
-awscli = ["awscli (>=1.32.41,<1.32.52)"]
-boto3 = ["boto3 (>=1.34.41,<1.34.52)"]
+awscli = ["awscli (>=1.32.41,<1.32.70)"]
+boto3 = ["boto3 (>=1.34.41,<1.34.70)"]
[[package]]
name = "aiohttp"
-version = "3.9.4"
+version = "3.9.5"
description = "Async http client/server framework (asyncio)"
-optional = false
+optional = true
python-versions = ">=3.8"
files = [
- {file = "aiohttp-3.9.4-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:76d32588ef7e4a3f3adff1956a0ba96faabbdee58f2407c122dd45aa6e34f372"},
- {file = "aiohttp-3.9.4-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:56181093c10dbc6ceb8a29dfeea1e815e1dfdc020169203d87fd8d37616f73f9"},
- {file = "aiohttp-3.9.4-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:c7a5b676d3c65e88b3aca41816bf72831898fcd73f0cbb2680e9d88e819d1e4d"},
- {file = "aiohttp-3.9.4-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d1df528a85fb404899d4207a8d9934cfd6be626e30e5d3a5544a83dbae6d8a7e"},
- {file = "aiohttp-3.9.4-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:f595db1bceabd71c82e92df212dd9525a8a2c6947d39e3c994c4f27d2fe15b11"},
- {file = "aiohttp-3.9.4-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:9c0b09d76e5a4caac3d27752027fbd43dc987b95f3748fad2b924a03fe8632ad"},
- {file = "aiohttp-3.9.4-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:689eb4356649ec9535b3686200b231876fb4cab4aca54e3bece71d37f50c1d13"},
- {file = "aiohttp-3.9.4-cp310-cp310-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:a3666cf4182efdb44d73602379a66f5fdfd5da0db5e4520f0ac0dcca644a3497"},
- {file = "aiohttp-3.9.4-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:b65b0f8747b013570eea2f75726046fa54fa8e0c5db60f3b98dd5d161052004a"},
- {file = "aiohttp-3.9.4-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:a1885d2470955f70dfdd33a02e1749613c5a9c5ab855f6db38e0b9389453dce7"},
- {file = "aiohttp-3.9.4-cp310-cp310-musllinux_1_1_ppc64le.whl", hash = "sha256:0593822dcdb9483d41f12041ff7c90d4d1033ec0e880bcfaf102919b715f47f1"},
- {file = "aiohttp-3.9.4-cp310-cp310-musllinux_1_1_s390x.whl", hash = "sha256:47f6eb74e1ecb5e19a78f4a4228aa24df7fbab3b62d4a625d3f41194a08bd54f"},
- {file = "aiohttp-3.9.4-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:c8b04a3dbd54de6ccb7604242fe3ad67f2f3ca558f2d33fe19d4b08d90701a89"},
- {file = "aiohttp-3.9.4-cp310-cp310-win32.whl", hash = "sha256:8a78dfb198a328bfb38e4308ca8167028920fb747ddcf086ce706fbdd23b2926"},
- {file = "aiohttp-3.9.4-cp310-cp310-win_amd64.whl", hash = "sha256:e78da6b55275987cbc89141a1d8e75f5070e577c482dd48bd9123a76a96f0bbb"},
- {file = "aiohttp-3.9.4-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:c111b3c69060d2bafc446917534150fd049e7aedd6cbf21ba526a5a97b4402a5"},
- {file = "aiohttp-3.9.4-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:efbdd51872cf170093998c87ccdf3cb5993add3559341a8e5708bcb311934c94"},
- {file = "aiohttp-3.9.4-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:7bfdb41dc6e85d8535b00d73947548a748e9534e8e4fddd2638109ff3fb081df"},
- {file = "aiohttp-3.9.4-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:2bd9d334412961125e9f68d5b73c1d0ab9ea3f74a58a475e6b119f5293eee7ba"},
- {file = "aiohttp-3.9.4-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:35d78076736f4a668d57ade00c65d30a8ce28719d8a42471b2a06ccd1a2e3063"},
- {file = "aiohttp-3.9.4-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:824dff4f9f4d0f59d0fa3577932ee9a20e09edec8a2f813e1d6b9f89ced8293f"},
- {file = "aiohttp-3.9.4-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:52b8b4e06fc15519019e128abedaeb56412b106ab88b3c452188ca47a25c4093"},
- {file = "aiohttp-3.9.4-cp311-cp311-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:eae569fb1e7559d4f3919965617bb39f9e753967fae55ce13454bec2d1c54f09"},
- {file = "aiohttp-3.9.4-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:69b97aa5792428f321f72aeb2f118e56893371f27e0b7d05750bcad06fc42ca1"},
- {file = "aiohttp-3.9.4-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:4d79aad0ad4b980663316f26d9a492e8fab2af77c69c0f33780a56843ad2f89e"},
- {file = "aiohttp-3.9.4-cp311-cp311-musllinux_1_1_ppc64le.whl", hash = "sha256:d6577140cd7db19e430661e4b2653680194ea8c22c994bc65b7a19d8ec834403"},
- {file = "aiohttp-3.9.4-cp311-cp311-musllinux_1_1_s390x.whl", hash = "sha256:9860d455847cd98eb67897f5957b7cd69fbcb436dd3f06099230f16a66e66f79"},
- {file = "aiohttp-3.9.4-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:69ff36d3f8f5652994e08bd22f093e11cfd0444cea310f92e01b45a4e46b624e"},
- {file = "aiohttp-3.9.4-cp311-cp311-win32.whl", hash = "sha256:e27d3b5ed2c2013bce66ad67ee57cbf614288bda8cdf426c8d8fe548316f1b5f"},
- {file = "aiohttp-3.9.4-cp311-cp311-win_amd64.whl", hash = "sha256:d6a67e26daa686a6fbdb600a9af8619c80a332556245fa8e86c747d226ab1a1e"},
- {file = "aiohttp-3.9.4-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:c5ff8ff44825736a4065d8544b43b43ee4c6dd1530f3a08e6c0578a813b0aa35"},
- {file = "aiohttp-3.9.4-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:d12a244627eba4e9dc52cbf924edef905ddd6cafc6513849b4876076a6f38b0e"},
- {file = "aiohttp-3.9.4-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:dcad56c8d8348e7e468899d2fb3b309b9bc59d94e6db08710555f7436156097f"},
- {file = "aiohttp-3.9.4-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:4f7e69a7fd4b5ce419238388e55abd220336bd32212c673ceabc57ccf3d05b55"},
- {file = "aiohttp-3.9.4-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:c4870cb049f10d7680c239b55428916d84158798eb8f353e74fa2c98980dcc0b"},
- {file = "aiohttp-3.9.4-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:3b2feaf1b7031ede1bc0880cec4b0776fd347259a723d625357bb4b82f62687b"},
- {file = "aiohttp-3.9.4-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:939393e8c3f0a5bcd33ef7ace67680c318dc2ae406f15e381c0054dd658397de"},
- {file = "aiohttp-3.9.4-cp312-cp312-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:7d2334e387b2adcc944680bebcf412743f2caf4eeebd550f67249c1c3696be04"},
- {file = "aiohttp-3.9.4-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:e0198ea897680e480845ec0ffc5a14e8b694e25b3f104f63676d55bf76a82f1a"},
- {file = "aiohttp-3.9.4-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:e40d2cd22914d67c84824045861a5bb0fb46586b15dfe4f046c7495bf08306b2"},
- {file = "aiohttp-3.9.4-cp312-cp312-musllinux_1_1_ppc64le.whl", hash = "sha256:aba80e77c227f4234aa34a5ff2b6ff30c5d6a827a91d22ff6b999de9175d71bd"},
- {file = "aiohttp-3.9.4-cp312-cp312-musllinux_1_1_s390x.whl", hash = "sha256:fb68dc73bc8ac322d2e392a59a9e396c4f35cb6fdbdd749e139d1d6c985f2527"},
- {file = "aiohttp-3.9.4-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:f3460a92638dce7e47062cf088d6e7663adb135e936cb117be88d5e6c48c9d53"},
- {file = "aiohttp-3.9.4-cp312-cp312-win32.whl", hash = "sha256:32dc814ddbb254f6170bca198fe307920f6c1308a5492f049f7f63554b88ef36"},
- {file = "aiohttp-3.9.4-cp312-cp312-win_amd64.whl", hash = "sha256:63f41a909d182d2b78fe3abef557fcc14da50c7852f70ae3be60e83ff64edba5"},
- {file = "aiohttp-3.9.4-cp38-cp38-macosx_10_9_universal2.whl", hash = "sha256:c3770365675f6be220032f6609a8fbad994d6dcf3ef7dbcf295c7ee70884c9af"},
- {file = "aiohttp-3.9.4-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:305edae1dea368ce09bcb858cf5a63a064f3bff4767dec6fa60a0cc0e805a1d3"},
- {file = "aiohttp-3.9.4-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:6f121900131d116e4a93b55ab0d12ad72573f967b100e49086e496a9b24523ea"},
- {file = "aiohttp-3.9.4-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:b71e614c1ae35c3d62a293b19eface83d5e4d194e3eb2fabb10059d33e6e8cbf"},
- {file = "aiohttp-3.9.4-cp38-cp38-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:419f009fa4cfde4d16a7fc070d64f36d70a8d35a90d71aa27670bba2be4fd039"},
- {file = "aiohttp-3.9.4-cp38-cp38-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:7b39476ee69cfe64061fd77a73bf692c40021f8547cda617a3466530ef63f947"},
- {file = "aiohttp-3.9.4-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b33f34c9c7decdb2ab99c74be6443942b730b56d9c5ee48fb7df2c86492f293c"},
- {file = "aiohttp-3.9.4-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:c78700130ce2dcebb1a8103202ae795be2fa8c9351d0dd22338fe3dac74847d9"},
- {file = "aiohttp-3.9.4-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:268ba22d917655d1259af2d5659072b7dc11b4e1dc2cb9662fdd867d75afc6a4"},
- {file = "aiohttp-3.9.4-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:17e7c051f53a0d2ebf33013a9cbf020bb4e098c4bc5bce6f7b0c962108d97eab"},
- {file = "aiohttp-3.9.4-cp38-cp38-musllinux_1_1_ppc64le.whl", hash = "sha256:7be99f4abb008cb38e144f85f515598f4c2c8932bf11b65add0ff59c9c876d99"},
- {file = "aiohttp-3.9.4-cp38-cp38-musllinux_1_1_s390x.whl", hash = "sha256:d58a54d6ff08d2547656356eea8572b224e6f9bbc0cf55fa9966bcaac4ddfb10"},
- {file = "aiohttp-3.9.4-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:7673a76772bda15d0d10d1aa881b7911d0580c980dbd16e59d7ba1422b2d83cd"},
- {file = "aiohttp-3.9.4-cp38-cp38-win32.whl", hash = "sha256:e4370dda04dc8951012f30e1ce7956a0a226ac0714a7b6c389fb2f43f22a250e"},
- {file = "aiohttp-3.9.4-cp38-cp38-win_amd64.whl", hash = "sha256:eb30c4510a691bb87081192a394fb661860e75ca3896c01c6d186febe7c88530"},
- {file = "aiohttp-3.9.4-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:84e90494db7df3be5e056f91412f9fa9e611fbe8ce4aaef70647297f5943b276"},
- {file = "aiohttp-3.9.4-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:7d4845f8501ab28ebfdbeab980a50a273b415cf69e96e4e674d43d86a464df9d"},
- {file = "aiohttp-3.9.4-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:69046cd9a2a17245c4ce3c1f1a4ff8c70c7701ef222fce3d1d8435f09042bba1"},
- {file = "aiohttp-3.9.4-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:8b73a06bafc8dcc508420db43b4dd5850e41e69de99009d0351c4f3007960019"},
- {file = "aiohttp-3.9.4-cp39-cp39-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:418bb0038dfafeac923823c2e63226179976c76f981a2aaad0ad5d51f2229bca"},
- {file = "aiohttp-3.9.4-cp39-cp39-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:71a8f241456b6c2668374d5d28398f8e8cdae4cce568aaea54e0f39359cd928d"},
- {file = "aiohttp-3.9.4-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:935c369bf8acc2dc26f6eeb5222768aa7c62917c3554f7215f2ead7386b33748"},
- {file = "aiohttp-3.9.4-cp39-cp39-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:74e4e48c8752d14ecfb36d2ebb3d76d614320570e14de0a3aa7a726ff150a03c"},
- {file = "aiohttp-3.9.4-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:916b0417aeddf2c8c61291238ce25286f391a6acb6f28005dd9ce282bd6311b6"},
- {file = "aiohttp-3.9.4-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:9b6787b6d0b3518b2ee4cbeadd24a507756ee703adbac1ab6dc7c4434b8c572a"},
- {file = "aiohttp-3.9.4-cp39-cp39-musllinux_1_1_ppc64le.whl", hash = "sha256:221204dbda5ef350e8db6287937621cf75e85778b296c9c52260b522231940ed"},
- {file = "aiohttp-3.9.4-cp39-cp39-musllinux_1_1_s390x.whl", hash = "sha256:10afd99b8251022ddf81eaed1d90f5a988e349ee7d779eb429fb07b670751e8c"},
- {file = "aiohttp-3.9.4-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:2506d9f7a9b91033201be9ffe7d89c6a54150b0578803cce5cb84a943d075bc3"},
- {file = "aiohttp-3.9.4-cp39-cp39-win32.whl", hash = "sha256:e571fdd9efd65e86c6af2f332e0e95dad259bfe6beb5d15b3c3eca3a6eb5d87b"},
- {file = "aiohttp-3.9.4-cp39-cp39-win_amd64.whl", hash = "sha256:7d29dd5319d20aa3b7749719ac9685fbd926f71ac8c77b2477272725f882072d"},
- {file = "aiohttp-3.9.4.tar.gz", hash = "sha256:6ff71ede6d9a5a58cfb7b6fffc83ab5d4a63138276c771ac91ceaaddf5459644"},
+ {file = "aiohttp-3.9.5-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:fcde4c397f673fdec23e6b05ebf8d4751314fa7c24f93334bf1f1364c1c69ac7"},
+ {file = "aiohttp-3.9.5-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:5d6b3f1fabe465e819aed2c421a6743d8debbde79b6a8600739300630a01bf2c"},
+ {file = "aiohttp-3.9.5-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:6ae79c1bc12c34082d92bf9422764f799aee4746fd7a392db46b7fd357d4a17a"},
+ {file = "aiohttp-3.9.5-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:4d3ebb9e1316ec74277d19c5f482f98cc65a73ccd5430540d6d11682cd857430"},
+ {file = "aiohttp-3.9.5-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:84dabd95154f43a2ea80deffec9cb44d2e301e38a0c9d331cc4aa0166fe28ae3"},
+ {file = "aiohttp-3.9.5-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:c8a02fbeca6f63cb1f0475c799679057fc9268b77075ab7cf3f1c600e81dd46b"},
+ {file = "aiohttp-3.9.5-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:c26959ca7b75ff768e2776d8055bf9582a6267e24556bb7f7bd29e677932be72"},
+ {file = "aiohttp-3.9.5-cp310-cp310-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:714d4e5231fed4ba2762ed489b4aec07b2b9953cf4ee31e9871caac895a839c0"},
+ {file = "aiohttp-3.9.5-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:e7a6a8354f1b62e15d48e04350f13e726fa08b62c3d7b8401c0a1314f02e3558"},
+ {file = "aiohttp-3.9.5-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:c413016880e03e69d166efb5a1a95d40f83d5a3a648d16486592c49ffb76d0db"},
+ {file = "aiohttp-3.9.5-cp310-cp310-musllinux_1_1_ppc64le.whl", hash = "sha256:ff84aeb864e0fac81f676be9f4685f0527b660f1efdc40dcede3c251ef1e867f"},
+ {file = "aiohttp-3.9.5-cp310-cp310-musllinux_1_1_s390x.whl", hash = "sha256:ad7f2919d7dac062f24d6f5fe95d401597fbb015a25771f85e692d043c9d7832"},
+ {file = "aiohttp-3.9.5-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:702e2c7c187c1a498a4e2b03155d52658fdd6fda882d3d7fbb891a5cf108bb10"},
+ {file = "aiohttp-3.9.5-cp310-cp310-win32.whl", hash = "sha256:67c3119f5ddc7261d47163ed86d760ddf0e625cd6246b4ed852e82159617b5fb"},
+ {file = "aiohttp-3.9.5-cp310-cp310-win_amd64.whl", hash = "sha256:471f0ef53ccedec9995287f02caf0c068732f026455f07db3f01a46e49d76bbb"},
+ {file = "aiohttp-3.9.5-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:e0ae53e33ee7476dd3d1132f932eeb39bf6125083820049d06edcdca4381f342"},
+ {file = "aiohttp-3.9.5-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:c088c4d70d21f8ca5c0b8b5403fe84a7bc8e024161febdd4ef04575ef35d474d"},
+ {file = "aiohttp-3.9.5-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:639d0042b7670222f33b0028de6b4e2fad6451462ce7df2af8aee37dcac55424"},
+ {file = "aiohttp-3.9.5-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:f26383adb94da5e7fb388d441bf09c61e5e35f455a3217bfd790c6b6bc64b2ee"},
+ {file = "aiohttp-3.9.5-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:66331d00fb28dc90aa606d9a54304af76b335ae204d1836f65797d6fe27f1ca2"},
+ {file = "aiohttp-3.9.5-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:4ff550491f5492ab5ed3533e76b8567f4b37bd2995e780a1f46bca2024223233"},
+ {file = "aiohttp-3.9.5-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:f22eb3a6c1080d862befa0a89c380b4dafce29dc6cd56083f630073d102eb595"},
+ {file = "aiohttp-3.9.5-cp311-cp311-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:a81b1143d42b66ffc40a441379387076243ef7b51019204fd3ec36b9f69e77d6"},
+ {file = "aiohttp-3.9.5-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:f64fd07515dad67f24b6ea4a66ae2876c01031de91c93075b8093f07c0a2d93d"},
+ {file = "aiohttp-3.9.5-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:93e22add827447d2e26d67c9ac0161756007f152fdc5210277d00a85f6c92323"},
+ {file = "aiohttp-3.9.5-cp311-cp311-musllinux_1_1_ppc64le.whl", hash = "sha256:55b39c8684a46e56ef8c8d24faf02de4a2b2ac60d26cee93bc595651ff545de9"},
+ {file = "aiohttp-3.9.5-cp311-cp311-musllinux_1_1_s390x.whl", hash = "sha256:4715a9b778f4293b9f8ae7a0a7cef9829f02ff8d6277a39d7f40565c737d3771"},
+ {file = "aiohttp-3.9.5-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:afc52b8d969eff14e069a710057d15ab9ac17cd4b6753042c407dcea0e40bf75"},
+ {file = "aiohttp-3.9.5-cp311-cp311-win32.whl", hash = "sha256:b3df71da99c98534be076196791adca8819761f0bf6e08e07fd7da25127150d6"},
+ {file = "aiohttp-3.9.5-cp311-cp311-win_amd64.whl", hash = "sha256:88e311d98cc0bf45b62fc46c66753a83445f5ab20038bcc1b8a1cc05666f428a"},
+ {file = "aiohttp-3.9.5-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:c7a4b7a6cf5b6eb11e109a9755fd4fda7d57395f8c575e166d363b9fc3ec4678"},
+ {file = "aiohttp-3.9.5-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:0a158704edf0abcac8ac371fbb54044f3270bdbc93e254a82b6c82be1ef08f3c"},
+ {file = "aiohttp-3.9.5-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:d153f652a687a8e95ad367a86a61e8d53d528b0530ef382ec5aaf533140ed00f"},
+ {file = "aiohttp-3.9.5-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:82a6a97d9771cb48ae16979c3a3a9a18b600a8505b1115cfe354dfb2054468b4"},
+ {file = "aiohttp-3.9.5-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:60cdbd56f4cad9f69c35eaac0fbbdf1f77b0ff9456cebd4902f3dd1cf096464c"},
+ {file = "aiohttp-3.9.5-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:8676e8fd73141ded15ea586de0b7cda1542960a7b9ad89b2b06428e97125d4fa"},
+ {file = "aiohttp-3.9.5-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:da00da442a0e31f1c69d26d224e1efd3a1ca5bcbf210978a2ca7426dfcae9f58"},
+ {file = "aiohttp-3.9.5-cp312-cp312-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:18f634d540dd099c262e9f887c8bbacc959847cfe5da7a0e2e1cf3f14dbf2daf"},
+ {file = "aiohttp-3.9.5-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:320e8618eda64e19d11bdb3bd04ccc0a816c17eaecb7e4945d01deee2a22f95f"},
+ {file = "aiohttp-3.9.5-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:2faa61a904b83142747fc6a6d7ad8fccff898c849123030f8e75d5d967fd4a81"},
+ {file = "aiohttp-3.9.5-cp312-cp312-musllinux_1_1_ppc64le.whl", hash = "sha256:8c64a6dc3fe5db7b1b4d2b5cb84c4f677768bdc340611eca673afb7cf416ef5a"},
+ {file = "aiohttp-3.9.5-cp312-cp312-musllinux_1_1_s390x.whl", hash = "sha256:393c7aba2b55559ef7ab791c94b44f7482a07bf7640d17b341b79081f5e5cd1a"},
+ {file = "aiohttp-3.9.5-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:c671dc117c2c21a1ca10c116cfcd6e3e44da7fcde37bf83b2be485ab377b25da"},
+ {file = "aiohttp-3.9.5-cp312-cp312-win32.whl", hash = "sha256:5a7ee16aab26e76add4afc45e8f8206c95d1d75540f1039b84a03c3b3800dd59"},
+ {file = "aiohttp-3.9.5-cp312-cp312-win_amd64.whl", hash = "sha256:5ca51eadbd67045396bc92a4345d1790b7301c14d1848feaac1d6a6c9289e888"},
+ {file = "aiohttp-3.9.5-cp38-cp38-macosx_10_9_universal2.whl", hash = "sha256:694d828b5c41255e54bc2dddb51a9f5150b4eefa9886e38b52605a05d96566e8"},
+ {file = "aiohttp-3.9.5-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:0605cc2c0088fcaae79f01c913a38611ad09ba68ff482402d3410bf59039bfb8"},
+ {file = "aiohttp-3.9.5-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:4558e5012ee03d2638c681e156461d37b7a113fe13970d438d95d10173d25f78"},
+ {file = "aiohttp-3.9.5-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:9dbc053ac75ccc63dc3a3cc547b98c7258ec35a215a92bd9f983e0aac95d3d5b"},
+ {file = "aiohttp-3.9.5-cp38-cp38-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:4109adee842b90671f1b689901b948f347325045c15f46b39797ae1bf17019de"},
+ {file = "aiohttp-3.9.5-cp38-cp38-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:a6ea1a5b409a85477fd8e5ee6ad8f0e40bf2844c270955e09360418cfd09abac"},
+ {file = "aiohttp-3.9.5-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:f3c2890ca8c59ee683fd09adf32321a40fe1cf164e3387799efb2acebf090c11"},
+ {file = "aiohttp-3.9.5-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:3916c8692dbd9d55c523374a3b8213e628424d19116ac4308e434dbf6d95bbdd"},
+ {file = "aiohttp-3.9.5-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:8d1964eb7617907c792ca00b341b5ec3e01ae8c280825deadbbd678447b127e1"},
+ {file = "aiohttp-3.9.5-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:d5ab8e1f6bee051a4bf6195e38a5c13e5e161cb7bad83d8854524798bd9fcd6e"},
+ {file = "aiohttp-3.9.5-cp38-cp38-musllinux_1_1_ppc64le.whl", hash = "sha256:52c27110f3862a1afbcb2af4281fc9fdc40327fa286c4625dfee247c3ba90156"},
+ {file = "aiohttp-3.9.5-cp38-cp38-musllinux_1_1_s390x.whl", hash = "sha256:7f64cbd44443e80094309875d4f9c71d0401e966d191c3d469cde4642bc2e031"},
+ {file = "aiohttp-3.9.5-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:8b4f72fbb66279624bfe83fd5eb6aea0022dad8eec62b71e7bf63ee1caadeafe"},
+ {file = "aiohttp-3.9.5-cp38-cp38-win32.whl", hash = "sha256:6380c039ec52866c06d69b5c7aad5478b24ed11696f0e72f6b807cfb261453da"},
+ {file = "aiohttp-3.9.5-cp38-cp38-win_amd64.whl", hash = "sha256:da22dab31d7180f8c3ac7c7635f3bcd53808f374f6aa333fe0b0b9e14b01f91a"},
+ {file = "aiohttp-3.9.5-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:1732102949ff6087589408d76cd6dea656b93c896b011ecafff418c9661dc4ed"},
+ {file = "aiohttp-3.9.5-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:c6021d296318cb6f9414b48e6a439a7f5d1f665464da507e8ff640848ee2a58a"},
+ {file = "aiohttp-3.9.5-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:239f975589a944eeb1bad26b8b140a59a3a320067fb3cd10b75c3092405a1372"},
+ {file = "aiohttp-3.9.5-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3b7b30258348082826d274504fbc7c849959f1989d86c29bc355107accec6cfb"},
+ {file = "aiohttp-3.9.5-cp39-cp39-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:cd2adf5c87ff6d8b277814a28a535b59e20bfea40a101db6b3bdca7e9926bc24"},
+ {file = "aiohttp-3.9.5-cp39-cp39-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:e9a3d838441bebcf5cf442700e3963f58b5c33f015341f9ea86dcd7d503c07e2"},
+ {file = "aiohttp-3.9.5-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:9e3a1ae66e3d0c17cf65c08968a5ee3180c5a95920ec2731f53343fac9bad106"},
+ {file = "aiohttp-3.9.5-cp39-cp39-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:9c69e77370cce2d6df5d12b4e12bdcca60c47ba13d1cbbc8645dd005a20b738b"},
+ {file = "aiohttp-3.9.5-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:0cbf56238f4bbf49dab8c2dc2e6b1b68502b1e88d335bea59b3f5b9f4c001475"},
+ {file = "aiohttp-3.9.5-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:d1469f228cd9ffddd396d9948b8c9cd8022b6d1bf1e40c6f25b0fb90b4f893ed"},
+ {file = "aiohttp-3.9.5-cp39-cp39-musllinux_1_1_ppc64le.whl", hash = "sha256:45731330e754f5811c314901cebdf19dd776a44b31927fa4b4dbecab9e457b0c"},
+ {file = "aiohttp-3.9.5-cp39-cp39-musllinux_1_1_s390x.whl", hash = "sha256:3fcb4046d2904378e3aeea1df51f697b0467f2aac55d232c87ba162709478c46"},
+ {file = "aiohttp-3.9.5-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:8cf142aa6c1a751fcb364158fd710b8a9be874b81889c2bd13aa8893197455e2"},
+ {file = "aiohttp-3.9.5-cp39-cp39-win32.whl", hash = "sha256:7b179eea70833c8dee51ec42f3b4097bd6370892fa93f510f76762105568cf09"},
+ {file = "aiohttp-3.9.5-cp39-cp39-win_amd64.whl", hash = "sha256:38d80498e2e169bc61418ff36170e0aad0cd268da8b38a17c4cf29d254a8b3f1"},
+ {file = "aiohttp-3.9.5.tar.gz", hash = "sha256:edea7d15772ceeb29db4aff55e482d4bcfb6ae160ce144f2682de02f6d693551"},
]
[package.dependencies]
@@ -214,7 +214,7 @@ typing_extensions = {version = ">=4.0", markers = "python_version < \"3.10\""}
name = "aiosignal"
version = "1.3.1"
description = "aiosignal: a list of registered asynchronous callbacks"
-optional = false
+optional = true
python-versions = ">=3.7"
files = [
{file = "aiosignal-1.3.1-py3-none-any.whl", hash = "sha256:f8376fb07dd1e86a584e4fcdec80b36b7f81aac666ebc724e2c090300dd83b17"},
@@ -235,17 +235,6 @@ files = [
{file = "alabaster-0.7.13.tar.gz", hash = "sha256:a27a4a084d5e690e16e01e03ad2b2e552c61a65469419b907243193de1a84ae2"},
]
-[[package]]
-name = "anyascii"
-version = "0.3.2"
-description = "Unicode to ASCII transliteration"
-optional = false
-python-versions = ">=3.3"
-files = [
- {file = "anyascii-0.3.2-py3-none-any.whl", hash = "sha256:3b3beef6fc43d9036d3b0529050b0c48bfad8bc960e9e562d7223cfb94fe45d4"},
- {file = "anyascii-0.3.2.tar.gz", hash = "sha256:9d5d32ef844fe225b8bc7cba7f950534fae4da27a9bf3a6bea2cb0ea46ce4730"},
-]
-
[[package]]
name = "appnope"
version = "0.1.4"
@@ -259,32 +248,18 @@ files = [
[[package]]
name = "argcomplete"
-version = "3.2.3"
+version = "3.3.0"
description = "Bash tab completion for argparse"
optional = false
python-versions = ">=3.8"
files = [
- {file = "argcomplete-3.2.3-py3-none-any.whl", hash = "sha256:c12355e0494c76a2a7b73e3a59b09024ca0ba1e279fb9ed6c1b82d5b74b6a70c"},
- {file = "argcomplete-3.2.3.tar.gz", hash = "sha256:bf7900329262e481be5a15f56f19736b376df6f82ed27576fa893652c5de6c23"},
+ {file = "argcomplete-3.3.0-py3-none-any.whl", hash = "sha256:c168c3723482c031df3c207d4ba8fa702717ccb9fc0bfe4117166c1f537b4a54"},
+ {file = "argcomplete-3.3.0.tar.gz", hash = "sha256:fd03ff4a5b9e6580569d34b273f741e85cd9e072f3feeeee3eba4891c70eda62"},
]
[package.extras]
test = ["coverage", "mypy", "pexpect", "ruff", "wheel"]
-[[package]]
-name = "astroid"
-version = "3.1.0"
-description = "An abstract syntax tree for Python with inference support."
-optional = false
-python-versions = ">=3.8.0"
-files = [
- {file = "astroid-3.1.0-py3-none-any.whl", hash = "sha256:951798f922990137ac090c53af473db7ab4e70c770e6d7fae0cec59f74411819"},
- {file = "astroid-3.1.0.tar.gz", hash = "sha256:ac248253bfa4bd924a0de213707e7ebeeb3138abeb48d798784ead1e56d419d4"},
-]
-
-[package.dependencies]
-typing-extensions = {version = ">=4.0.0", markers = "python_version < \"3.11\""}
-
[[package]]
name = "asttokens"
version = "2.4.1"
@@ -303,11 +278,26 @@ six = ">=1.12.0"
astroid = ["astroid (>=1,<2)", "astroid (>=2,<4)"]
test = ["astroid (>=1,<2)", "astroid (>=2,<4)", "pytest"]
+[[package]]
+name = "astunparse"
+version = "1.6.3"
+description = "An AST unparser for Python"
+optional = false
+python-versions = "*"
+files = [
+ {file = "astunparse-1.6.3-py2.py3-none-any.whl", hash = "sha256:c2652417f2c8b5bb325c885ae329bdf3f86424075c4fd1a128674bc6fba4b8e8"},
+ {file = "astunparse-1.6.3.tar.gz", hash = "sha256:5ad93a8456f0d084c3456d059fd9a92cce667963232cbf763eac3bc5b7940872"},
+]
+
+[package.dependencies]
+six = ">=1.6.1,<2.0"
+wheel = ">=0.23.0,<1.0"
+
[[package]]
name = "async-timeout"
version = "4.0.3"
description = "Timeout context manager for asyncio programs"
-optional = false
+optional = true
python-versions = ">=3.7"
files = [
{file = "async-timeout-4.0.3.tar.gz", hash = "sha256:4640d96be84d82d02ed59ea2b7105a0f7b33abe8703703cd0ab0bf87c427522f"},
@@ -333,20 +323,6 @@ tests = ["attrs[tests-no-zope]", "zope-interface"]
tests-mypy = ["mypy (>=1.6)", "pytest-mypy-plugins"]
tests-no-zope = ["attrs[tests-mypy]", "cloudpickle", "hypothesis", "pympler", "pytest (>=4.3.0)", "pytest-xdist[psutil]"]
-[[package]]
-name = "autodoc"
-version = "0.5.0"
-description = "Autodoc Python implementation."
-optional = false
-python-versions = "*"
-files = [
- {file = "autodoc-0.5.0.tar.gz", hash = "sha256:c4387c5a0f1c09b055bb2e384542ee1e016542f313b2a33d904ca77f0460ded3"},
-]
-
-[package.dependencies]
-decorator = "*"
-webtest = "*"
-
[[package]]
name = "babel"
version = "2.14.0"
@@ -441,17 +417,17 @@ css = ["tinycss2 (>=1.1.0,<1.3)"]
[[package]]
name = "boto3"
-version = "1.34.51"
+version = "1.34.69"
description = "The AWS SDK for Python"
optional = true
-python-versions = ">= 3.8"
+python-versions = ">=3.8"
files = [
- {file = "boto3-1.34.51-py3-none-any.whl", hash = "sha256:67732634dc7d0afda879bd9a5e2d0818a2c14a98bef766b95a3e253ea5104cb9"},
- {file = "boto3-1.34.51.tar.gz", hash = "sha256:2cd9463e738a184cbce8a6824027c22163c5f73e277a35ff5aa0fb0e845b4301"},
+ {file = "boto3-1.34.69-py3-none-any.whl", hash = "sha256:2e25ef6bd325217c2da329829478be063155897d8d3b29f31f7f23ab548519b1"},
+ {file = "boto3-1.34.69.tar.gz", hash = "sha256:898a5fed26b1351352703421d1a8b886ef2a74be6c97d5ecc92432ae01fda203"},
]
[package.dependencies]
-botocore = ">=1.34.51,<1.35.0"
+botocore = ">=1.34.69,<1.35.0"
jmespath = ">=0.7.1,<2.0.0"
s3transfer = ">=0.10.0,<0.11.0"
@@ -460,13 +436,13 @@ crt = ["botocore[crt] (>=1.21.0,<2.0a0)"]
[[package]]
name = "botocore"
-version = "1.34.51"
+version = "1.34.69"
description = "Low-level, data-driven core of boto 3."
optional = true
-python-versions = ">= 3.8"
+python-versions = ">=3.8"
files = [
- {file = "botocore-1.34.51-py3-none-any.whl", hash = "sha256:01d5156247f991b3466a8404e3d7460a9ecbd9b214f9992d6ba797d9ddc6f120"},
- {file = "botocore-1.34.51.tar.gz", hash = "sha256:5086217442e67dd9de36ec7e87a0c663f76b7790d5fb6a12de565af95e87e319"},
+ {file = "botocore-1.34.69-py3-none-any.whl", hash = "sha256:d3802d076d4d507bf506f9845a6970ce43adc3d819dd57c2791f5c19ed6e5950"},
+ {file = "botocore-1.34.69.tar.gz", hash = "sha256:d1ab2bff3c2fd51719c2021d9fa2f30fbb9ed0a308f69e9a774ac92c8091380a"},
]
[package.dependencies]
@@ -474,12 +450,23 @@ jmespath = ">=0.7.1,<2.0.0"
python-dateutil = ">=2.1,<3.0.0"
urllib3 = [
{version = ">=1.25.4,<1.27", markers = "python_version < \"3.10\""},
- {version = ">=1.25.4,<2.1", markers = "python_version >= \"3.10\""},
+ {version = ">=1.25.4,<2.2.0 || >2.2.0,<3", markers = "python_version >= \"3.10\""},
]
[package.extras]
crt = ["awscrt (==0.19.19)"]
+[[package]]
+name = "bracex"
+version = "2.4"
+description = "Bash style brace expander."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "bracex-2.4-py3-none-any.whl", hash = "sha256:efdc71eff95eaff5e0f8cfebe7d01adf2c8637c8c92edaf63ef348c241a82418"},
+ {file = "bracex-2.4.tar.gz", hash = "sha256:a27eaf1df42cf561fed58b7a8f3fdf129d1ea16a81e1fadd1d17989bc6384beb"},
+]
+
[[package]]
name = "certifi"
version = "2024.2.2"
@@ -705,19 +692,36 @@ files = [
{file = "colorama-0.4.6.tar.gz", hash = "sha256:08695f5cb7ed6e0531a20572697297273c47b8cae5a63ffc6d6ed5c201be6e44"},
]
+[[package]]
+name = "comm"
+version = "0.2.2"
+description = "Jupyter Python Comm implementation, for usage in ipykernel, xeus-python etc."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "comm-0.2.2-py3-none-any.whl", hash = "sha256:e6fb86cb70ff661ee8c9c14e7d36d6de3b4066f1441be4063df9c5009f0a64d3"},
+ {file = "comm-0.2.2.tar.gz", hash = "sha256:3fd7a84065306e07bea1773df6eb8282de51ba82f77c72f9c85716ab11fe980e"},
+]
+
+[package.dependencies]
+traitlets = ">=4"
+
+[package.extras]
+test = ["pytest"]
+
[[package]]
name = "commitizen"
-version = "3.20.0"
+version = "3.24.0"
description = "Python commitizen client tool"
optional = false
python-versions = ">=3.8"
files = [
- {file = "commitizen-3.20.0-py3-none-any.whl", hash = "sha256:f079d9642347d314afae75664d289b0de80cf0343c99c3dcfa85782f164333f3"},
- {file = "commitizen-3.20.0.tar.gz", hash = "sha256:17aebc8f36326fa3e65dcc08195303579e356be84b0d65c3c4bfed85b8822411"},
+ {file = "commitizen-3.24.0-py3-none-any.whl", hash = "sha256:d9e28b1dcd97cea64dcb50be25292ceb730470d933f1da37131f9540f762df36"},
+ {file = "commitizen-3.24.0.tar.gz", hash = "sha256:088e01ae8265f1d6fa5a4d11a05e4fd7092d958c881837c35f6c65aad27331a9"},
]
[package.dependencies]
-argcomplete = ">=1.12.1,<3.3"
+argcomplete = ">=1.12.1,<3.4"
charset-normalizer = ">=2.1.0,<4"
colorama = ">=0.4.1,<0.5.0"
decli = ">=0.6.0,<0.7.0"
@@ -731,78 +735,78 @@ tomlkit = ">=0.5.3,<1.0.0"
[[package]]
name = "configparser"
-version = "6.0.1"
+version = "7.0.0"
description = "Updated configparser from stdlib for earlier Pythons."
optional = true
python-versions = ">=3.8"
files = [
- {file = "configparser-6.0.1-py3-none-any.whl", hash = "sha256:5a0da275bea56f871abaa9e0806331791e9d8ae2938e8b8797b99ab3e8e192c4"},
- {file = "configparser-6.0.1.tar.gz", hash = "sha256:db45513e971e509496b150be31bd67b0e14ab20b78a383b677e4b158e2c682d8"},
+ {file = "configparser-7.0.0-py3-none-any.whl", hash = "sha256:f46d52a12811c637104c6bb8eb33693be0038ab6bf01d69aae009c39ec8c2017"},
+ {file = "configparser-7.0.0.tar.gz", hash = "sha256:af3c618a67aaaedc4d689fd7317d238f566b9aa03cae50102e92d7f0dfe78ba0"},
]
[package.extras]
-docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "rst.linker (>=1.9)", "sphinx (<7.2.5)", "sphinx (>=3.5)", "sphinx-lint"]
-testing = ["pytest (>=6)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-mypy", "pytest-ruff (>=0.2.1)", "types-backports"]
+docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "rst.linker (>=1.9)", "sphinx (>=3.5)", "sphinx-lint"]
+testing = ["pytest (>=6,!=8.1.1)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-mypy", "pytest-ruff (>=0.2.1)", "types-backports"]
[[package]]
name = "coverage"
-version = "7.4.4"
+version = "7.5.0"
description = "Code coverage measurement for Python"
optional = false
python-versions = ">=3.8"
files = [
- {file = "coverage-7.4.4-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:e0be5efd5127542ef31f165de269f77560d6cdef525fffa446de6f7e9186cfb2"},
- {file = "coverage-7.4.4-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:ccd341521be3d1b3daeb41960ae94a5e87abe2f46f17224ba5d6f2b8398016cf"},
- {file = "coverage-7.4.4-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:09fa497a8ab37784fbb20ab699c246053ac294d13fc7eb40ec007a5043ec91f8"},
- {file = "coverage-7.4.4-cp310-cp310-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:b1a93009cb80730c9bca5d6d4665494b725b6e8e157c1cb7f2db5b4b122ea562"},
- {file = "coverage-7.4.4-cp310-cp310-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:690db6517f09336559dc0b5f55342df62370a48f5469fabf502db2c6d1cffcd2"},
- {file = "coverage-7.4.4-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:09c3255458533cb76ef55da8cc49ffab9e33f083739c8bd4f58e79fecfe288f7"},
- {file = "coverage-7.4.4-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:8ce1415194b4a6bd0cdcc3a1dfbf58b63f910dcb7330fe15bdff542c56949f87"},
- {file = "coverage-7.4.4-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:b91cbc4b195444e7e258ba27ac33769c41b94967919f10037e6355e998af255c"},
- {file = "coverage-7.4.4-cp310-cp310-win32.whl", hash = "sha256:598825b51b81c808cb6f078dcb972f96af96b078faa47af7dfcdf282835baa8d"},
- {file = "coverage-7.4.4-cp310-cp310-win_amd64.whl", hash = "sha256:09ef9199ed6653989ebbcaacc9b62b514bb63ea2f90256e71fea3ed74bd8ff6f"},
- {file = "coverage-7.4.4-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:0f9f50e7ef2a71e2fae92774c99170eb8304e3fdf9c8c3c7ae9bab3e7229c5cf"},
- {file = "coverage-7.4.4-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:623512f8ba53c422fcfb2ce68362c97945095b864cda94a92edbaf5994201083"},
- {file = "coverage-7.4.4-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:0513b9508b93da4e1716744ef6ebc507aff016ba115ffe8ecff744d1322a7b63"},
- {file = "coverage-7.4.4-cp311-cp311-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:40209e141059b9370a2657c9b15607815359ab3ef9918f0196b6fccce8d3230f"},
- {file = "coverage-7.4.4-cp311-cp311-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:8a2b2b78c78293782fd3767d53e6474582f62443d0504b1554370bde86cc8227"},
- {file = "coverage-7.4.4-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:73bfb9c09951125d06ee473bed216e2c3742f530fc5acc1383883125de76d9cd"},
- {file = "coverage-7.4.4-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:1f384c3cc76aeedce208643697fb3e8437604b512255de6d18dae3f27655a384"},
- {file = "coverage-7.4.4-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:54eb8d1bf7cacfbf2a3186019bcf01d11c666bd495ed18717162f7eb1e9dd00b"},
- {file = "coverage-7.4.4-cp311-cp311-win32.whl", hash = "sha256:cac99918c7bba15302a2d81f0312c08054a3359eaa1929c7e4b26ebe41e9b286"},
- {file = "coverage-7.4.4-cp311-cp311-win_amd64.whl", hash = "sha256:b14706df8b2de49869ae03a5ccbc211f4041750cd4a66f698df89d44f4bd30ec"},
- {file = "coverage-7.4.4-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:201bef2eea65e0e9c56343115ba3814e896afe6d36ffd37bab783261db430f76"},
- {file = "coverage-7.4.4-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:41c9c5f3de16b903b610d09650e5e27adbfa7f500302718c9ffd1c12cf9d6818"},
- {file = "coverage-7.4.4-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d898fe162d26929b5960e4e138651f7427048e72c853607f2b200909794ed978"},
- {file = "coverage-7.4.4-cp312-cp312-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:3ea79bb50e805cd6ac058dfa3b5c8f6c040cb87fe83de10845857f5535d1db70"},
- {file = "coverage-7.4.4-cp312-cp312-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:ce4b94265ca988c3f8e479e741693d143026632672e3ff924f25fab50518dd51"},
- {file = "coverage-7.4.4-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:00838a35b882694afda09f85e469c96367daa3f3f2b097d846a7216993d37f4c"},
- {file = "coverage-7.4.4-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:fdfafb32984684eb03c2d83e1e51f64f0906b11e64482df3c5db936ce3839d48"},
- {file = "coverage-7.4.4-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:69eb372f7e2ece89f14751fbcbe470295d73ed41ecd37ca36ed2eb47512a6ab9"},
- {file = "coverage-7.4.4-cp312-cp312-win32.whl", hash = "sha256:137eb07173141545e07403cca94ab625cc1cc6bc4c1e97b6e3846270e7e1fea0"},
- {file = "coverage-7.4.4-cp312-cp312-win_amd64.whl", hash = "sha256:d71eec7d83298f1af3326ce0ff1d0ea83c7cb98f72b577097f9083b20bdaf05e"},
- {file = "coverage-7.4.4-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:d5ae728ff3b5401cc320d792866987e7e7e880e6ebd24433b70a33b643bb0384"},
- {file = "coverage-7.4.4-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:cc4f1358cb0c78edef3ed237ef2c86056206bb8d9140e73b6b89fbcfcbdd40e1"},
- {file = "coverage-7.4.4-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:8130a2aa2acb8788e0b56938786c33c7c98562697bf9f4c7d6e8e5e3a0501e4a"},
- {file = "coverage-7.4.4-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:cf271892d13e43bc2b51e6908ec9a6a5094a4df1d8af0bfc360088ee6c684409"},
- {file = "coverage-7.4.4-cp38-cp38-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:a4cdc86d54b5da0df6d3d3a2f0b710949286094c3a6700c21e9015932b81447e"},
- {file = "coverage-7.4.4-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:ae71e7ddb7a413dd60052e90528f2f65270aad4b509563af6d03d53e979feafd"},
- {file = "coverage-7.4.4-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:38dd60d7bf242c4ed5b38e094baf6401faa114fc09e9e6632374388a404f98e7"},
- {file = "coverage-7.4.4-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:aa5b1c1bfc28384f1f53b69a023d789f72b2e0ab1b3787aae16992a7ca21056c"},
- {file = "coverage-7.4.4-cp38-cp38-win32.whl", hash = "sha256:dfa8fe35a0bb90382837b238fff375de15f0dcdb9ae68ff85f7a63649c98527e"},
- {file = "coverage-7.4.4-cp38-cp38-win_amd64.whl", hash = "sha256:b2991665420a803495e0b90a79233c1433d6ed77ef282e8e152a324bbbc5e0c8"},
- {file = "coverage-7.4.4-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:3b799445b9f7ee8bf299cfaed6f5b226c0037b74886a4e11515e569b36fe310d"},
- {file = "coverage-7.4.4-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:b4d33f418f46362995f1e9d4f3a35a1b6322cb959c31d88ae56b0298e1c22357"},
- {file = "coverage-7.4.4-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:aadacf9a2f407a4688d700e4ebab33a7e2e408f2ca04dbf4aef17585389eff3e"},
- {file = "coverage-7.4.4-cp39-cp39-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:7c95949560050d04d46b919301826525597f07b33beba6187d04fa64d47ac82e"},
- {file = "coverage-7.4.4-cp39-cp39-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:ff7687ca3d7028d8a5f0ebae95a6e4827c5616b31a4ee1192bdfde697db110d4"},
- {file = "coverage-7.4.4-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:5fc1de20b2d4a061b3df27ab9b7c7111e9a710f10dc2b84d33a4ab25065994ec"},
- {file = "coverage-7.4.4-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:c74880fc64d4958159fbd537a091d2a585448a8f8508bf248d72112723974cbd"},
- {file = "coverage-7.4.4-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:742a76a12aa45b44d236815d282b03cfb1de3b4323f3e4ec933acfae08e54ade"},
- {file = "coverage-7.4.4-cp39-cp39-win32.whl", hash = "sha256:d89d7b2974cae412400e88f35d86af72208e1ede1a541954af5d944a8ba46c57"},
- {file = "coverage-7.4.4-cp39-cp39-win_amd64.whl", hash = "sha256:9ca28a302acb19b6af89e90f33ee3e1906961f94b54ea37de6737b7ca9d8827c"},
- {file = "coverage-7.4.4-pp38.pp39.pp310-none-any.whl", hash = "sha256:b2c5edc4ac10a7ef6605a966c58929ec6c1bd0917fb8c15cb3363f65aa40e677"},
- {file = "coverage-7.4.4.tar.gz", hash = "sha256:c901df83d097649e257e803be22592aedfd5182f07b3cc87d640bbb9afd50f49"},
+ {file = "coverage-7.5.0-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:432949a32c3e3f820af808db1833d6d1631664d53dd3ce487aa25d574e18ad1c"},
+ {file = "coverage-7.5.0-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:2bd7065249703cbeb6d4ce679c734bef0ee69baa7bff9724361ada04a15b7e3b"},
+ {file = "coverage-7.5.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:bbfe6389c5522b99768a93d89aca52ef92310a96b99782973b9d11e80511f932"},
+ {file = "coverage-7.5.0-cp310-cp310-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:39793731182c4be939b4be0cdecde074b833f6171313cf53481f869937129ed3"},
+ {file = "coverage-7.5.0-cp310-cp310-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:85a5dbe1ba1bf38d6c63b6d2c42132d45cbee6d9f0c51b52c59aa4afba057517"},
+ {file = "coverage-7.5.0-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:357754dcdfd811462a725e7501a9b4556388e8ecf66e79df6f4b988fa3d0b39a"},
+ {file = "coverage-7.5.0-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:a81eb64feded34f40c8986869a2f764f0fe2db58c0530d3a4afbcde50f314880"},
+ {file = "coverage-7.5.0-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:51431d0abbed3a868e967f8257c5faf283d41ec882f58413cf295a389bb22e58"},
+ {file = "coverage-7.5.0-cp310-cp310-win32.whl", hash = "sha256:f609ebcb0242d84b7adeee2b06c11a2ddaec5464d21888b2c8255f5fd6a98ae4"},
+ {file = "coverage-7.5.0-cp310-cp310-win_amd64.whl", hash = "sha256:6782cd6216fab5a83216cc39f13ebe30adfac2fa72688c5a4d8d180cd52e8f6a"},
+ {file = "coverage-7.5.0-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:e768d870801f68c74c2b669fc909839660180c366501d4cc4b87efd6b0eee375"},
+ {file = "coverage-7.5.0-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:84921b10aeb2dd453247fd10de22907984eaf80901b578a5cf0bb1e279a587cb"},
+ {file = "coverage-7.5.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:710c62b6e35a9a766b99b15cdc56d5aeda0914edae8bb467e9c355f75d14ee95"},
+ {file = "coverage-7.5.0-cp311-cp311-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:c379cdd3efc0658e652a14112d51a7668f6bfca7445c5a10dee7eabecabba19d"},
+ {file = "coverage-7.5.0-cp311-cp311-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:fea9d3ca80bcf17edb2c08a4704259dadac196fe5e9274067e7a20511fad1743"},
+ {file = "coverage-7.5.0-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:41327143c5b1d715f5f98a397608f90ab9ebba606ae4e6f3389c2145410c52b1"},
+ {file = "coverage-7.5.0-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:565b2e82d0968c977e0b0f7cbf25fd06d78d4856289abc79694c8edcce6eb2de"},
+ {file = "coverage-7.5.0-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:cf3539007202ebfe03923128fedfdd245db5860a36810136ad95a564a2fdffff"},
+ {file = "coverage-7.5.0-cp311-cp311-win32.whl", hash = "sha256:bf0b4b8d9caa8d64df838e0f8dcf68fb570c5733b726d1494b87f3da85db3a2d"},
+ {file = "coverage-7.5.0-cp311-cp311-win_amd64.whl", hash = "sha256:9c6384cc90e37cfb60435bbbe0488444e54b98700f727f16f64d8bfda0b84656"},
+ {file = "coverage-7.5.0-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:fed7a72d54bd52f4aeb6c6e951f363903bd7d70bc1cad64dd1f087980d309ab9"},
+ {file = "coverage-7.5.0-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:cbe6581fcff7c8e262eb574244f81f5faaea539e712a058e6707a9d272fe5b64"},
+ {file = "coverage-7.5.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ad97ec0da94b378e593ef532b980c15e377df9b9608c7c6da3506953182398af"},
+ {file = "coverage-7.5.0-cp312-cp312-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:bd4bacd62aa2f1a1627352fe68885d6ee694bdaebb16038b6e680f2924a9b2cc"},
+ {file = "coverage-7.5.0-cp312-cp312-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:adf032b6c105881f9d77fa17d9eebe0ad1f9bfb2ad25777811f97c5362aa07f2"},
+ {file = "coverage-7.5.0-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:4ba01d9ba112b55bfa4b24808ec431197bb34f09f66f7cb4fd0258ff9d3711b1"},
+ {file = "coverage-7.5.0-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:f0bfe42523893c188e9616d853c47685e1c575fe25f737adf473d0405dcfa7eb"},
+ {file = "coverage-7.5.0-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:a9a7ef30a1b02547c1b23fa9a5564f03c9982fc71eb2ecb7f98c96d7a0db5cf2"},
+ {file = "coverage-7.5.0-cp312-cp312-win32.whl", hash = "sha256:3c2b77f295edb9fcdb6a250f83e6481c679335ca7e6e4a955e4290350f2d22a4"},
+ {file = "coverage-7.5.0-cp312-cp312-win_amd64.whl", hash = "sha256:427e1e627b0963ac02d7c8730ca6d935df10280d230508c0ba059505e9233475"},
+ {file = "coverage-7.5.0-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:9dd88fce54abbdbf4c42fb1fea0e498973d07816f24c0e27a1ecaf91883ce69e"},
+ {file = "coverage-7.5.0-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:a898c11dca8f8c97b467138004a30133974aacd572818c383596f8d5b2eb04a9"},
+ {file = "coverage-7.5.0-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:07dfdd492d645eea1bd70fb1d6febdcf47db178b0d99161d8e4eed18e7f62fe7"},
+ {file = "coverage-7.5.0-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:d3d117890b6eee85887b1eed41eefe2e598ad6e40523d9f94c4c4b213258e4a4"},
+ {file = "coverage-7.5.0-cp38-cp38-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:6afd2e84e7da40fe23ca588379f815fb6dbbb1b757c883935ed11647205111cb"},
+ {file = "coverage-7.5.0-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:a9960dd1891b2ddf13a7fe45339cd59ecee3abb6b8326d8b932d0c5da208104f"},
+ {file = "coverage-7.5.0-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:ced268e82af993d7801a9db2dbc1d2322e786c5dc76295d8e89473d46c6b84d4"},
+ {file = "coverage-7.5.0-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:e7c211f25777746d468d76f11719e64acb40eed410d81c26cefac641975beb88"},
+ {file = "coverage-7.5.0-cp38-cp38-win32.whl", hash = "sha256:262fffc1f6c1a26125d5d573e1ec379285a3723363f3bd9c83923c9593a2ac25"},
+ {file = "coverage-7.5.0-cp38-cp38-win_amd64.whl", hash = "sha256:eed462b4541c540d63ab57b3fc69e7d8c84d5957668854ee4e408b50e92ce26a"},
+ {file = "coverage-7.5.0-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:d0194d654e360b3e6cc9b774e83235bae6b9b2cac3be09040880bb0e8a88f4a1"},
+ {file = "coverage-7.5.0-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:33c020d3322662e74bc507fb11488773a96894aa82a622c35a5a28673c0c26f5"},
+ {file = "coverage-7.5.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:0cbdf2cae14a06827bec50bd58e49249452d211d9caddd8bd80e35b53cb04631"},
+ {file = "coverage-7.5.0-cp39-cp39-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:3235d7c781232e525b0761730e052388a01548bd7f67d0067a253887c6e8df46"},
+ {file = "coverage-7.5.0-cp39-cp39-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:db2de4e546f0ec4b2787d625e0b16b78e99c3e21bc1722b4977c0dddf11ca84e"},
+ {file = "coverage-7.5.0-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:4d0e206259b73af35c4ec1319fd04003776e11e859936658cb6ceffdeba0f5be"},
+ {file = "coverage-7.5.0-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:2055c4fb9a6ff624253d432aa471a37202cd8f458c033d6d989be4499aed037b"},
+ {file = "coverage-7.5.0-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:075299460948cd12722a970c7eae43d25d37989da682997687b34ae6b87c0ef0"},
+ {file = "coverage-7.5.0-cp39-cp39-win32.whl", hash = "sha256:280132aada3bc2f0fac939a5771db4fbb84f245cb35b94fae4994d4c1f80dae7"},
+ {file = "coverage-7.5.0-cp39-cp39-win_amd64.whl", hash = "sha256:c58536f6892559e030e6924896a44098bc1290663ea12532c78cef71d0df8493"},
+ {file = "coverage-7.5.0-pp38.pp39.pp310-none-any.whl", hash = "sha256:2b57780b51084d5223eee7b59f0d4911c31c16ee5aa12737c7a02455829ff067"},
+ {file = "coverage-7.5.0.tar.gz", hash = "sha256:cf62d17310f34084c59c01e027259076479128d11e4661bb6c9acb38c5e19bb8"},
]
[package.dependencies]
@@ -865,6 +869,37 @@ files = [
{file = "dbfread-2.0.7.tar.gz", hash = "sha256:07c8a9af06ffad3f6f03e8fe91ad7d2733e31a26d2b72c4dd4cfbae07ee3b73d"},
]
+[[package]]
+name = "debugpy"
+version = "1.8.1"
+description = "An implementation of the Debug Adapter Protocol for Python"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "debugpy-1.8.1-cp310-cp310-macosx_11_0_x86_64.whl", hash = "sha256:3bda0f1e943d386cc7a0e71bfa59f4137909e2ed947fb3946c506e113000f741"},
+ {file = "debugpy-1.8.1-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:dda73bf69ea479c8577a0448f8c707691152e6c4de7f0c4dec5a4bc11dee516e"},
+ {file = "debugpy-1.8.1-cp310-cp310-win32.whl", hash = "sha256:3a79c6f62adef994b2dbe9fc2cc9cc3864a23575b6e387339ab739873bea53d0"},
+ {file = "debugpy-1.8.1-cp310-cp310-win_amd64.whl", hash = "sha256:7eb7bd2b56ea3bedb009616d9e2f64aab8fc7000d481faec3cd26c98a964bcdd"},
+ {file = "debugpy-1.8.1-cp311-cp311-macosx_11_0_universal2.whl", hash = "sha256:016a9fcfc2c6b57f939673c874310d8581d51a0fe0858e7fac4e240c5eb743cb"},
+ {file = "debugpy-1.8.1-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:fd97ed11a4c7f6d042d320ce03d83b20c3fb40da892f994bc041bbc415d7a099"},
+ {file = "debugpy-1.8.1-cp311-cp311-win32.whl", hash = "sha256:0de56aba8249c28a300bdb0672a9b94785074eb82eb672db66c8144fff673146"},
+ {file = "debugpy-1.8.1-cp311-cp311-win_amd64.whl", hash = "sha256:1a9fe0829c2b854757b4fd0a338d93bc17249a3bf69ecf765c61d4c522bb92a8"},
+ {file = "debugpy-1.8.1-cp312-cp312-macosx_11_0_universal2.whl", hash = "sha256:3ebb70ba1a6524d19fa7bb122f44b74170c447d5746a503e36adc244a20ac539"},
+ {file = "debugpy-1.8.1-cp312-cp312-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:a2e658a9630f27534e63922ebf655a6ab60c370f4d2fc5c02a5b19baf4410ace"},
+ {file = "debugpy-1.8.1-cp312-cp312-win32.whl", hash = "sha256:caad2846e21188797a1f17fc09c31b84c7c3c23baf2516fed5b40b378515bbf0"},
+ {file = "debugpy-1.8.1-cp312-cp312-win_amd64.whl", hash = "sha256:edcc9f58ec0fd121a25bc950d4578df47428d72e1a0d66c07403b04eb93bcf98"},
+ {file = "debugpy-1.8.1-cp38-cp38-macosx_11_0_x86_64.whl", hash = "sha256:7a3afa222f6fd3d9dfecd52729bc2e12c93e22a7491405a0ecbf9e1d32d45b39"},
+ {file = "debugpy-1.8.1-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:d915a18f0597ef685e88bb35e5d7ab968964b7befefe1aaea1eb5b2640b586c7"},
+ {file = "debugpy-1.8.1-cp38-cp38-win32.whl", hash = "sha256:92116039b5500633cc8d44ecc187abe2dfa9b90f7a82bbf81d079fcdd506bae9"},
+ {file = "debugpy-1.8.1-cp38-cp38-win_amd64.whl", hash = "sha256:e38beb7992b5afd9d5244e96ad5fa9135e94993b0c551ceebf3fe1a5d9beb234"},
+ {file = "debugpy-1.8.1-cp39-cp39-macosx_11_0_x86_64.whl", hash = "sha256:bfb20cb57486c8e4793d41996652e5a6a885b4d9175dd369045dad59eaacea42"},
+ {file = "debugpy-1.8.1-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:efd3fdd3f67a7e576dd869c184c5dd71d9aaa36ded271939da352880c012e703"},
+ {file = "debugpy-1.8.1-cp39-cp39-win32.whl", hash = "sha256:58911e8521ca0c785ac7a0539f1e77e0ce2df753f786188f382229278b4cdf23"},
+ {file = "debugpy-1.8.1-cp39-cp39-win_amd64.whl", hash = "sha256:6df9aa9599eb05ca179fb0b810282255202a66835c6efb1d112d21ecb830ddd3"},
+ {file = "debugpy-1.8.1-py2.py3-none-any.whl", hash = "sha256:28acbe2241222b87e255260c76741e1fbf04fdc3b6d094fcf57b6c6f75ce1242"},
+ {file = "debugpy-1.8.1.zip", hash = "sha256:f696d6be15be87aef621917585f9bb94b1dc9e8aced570db1b8a6fc14e8f9b42"},
+]
+
[[package]]
name = "decli"
version = "0.6.1"
@@ -922,29 +957,18 @@ files = [
[[package]]
name = "dunamai"
-version = "1.19.2"
+version = "1.20.0"
description = "Dynamic version generation"
optional = false
python-versions = ">=3.5"
files = [
- {file = "dunamai-1.19.2-py3-none-any.whl", hash = "sha256:bc126b17571a44d68ed826cec596e0f61dc01edca8b21486f70014936a5d44f2"},
- {file = "dunamai-1.19.2.tar.gz", hash = "sha256:3be4049890763e19b8df1d52960dbea60b3e263eb0c96144a677ae0633734d2e"},
+ {file = "dunamai-1.20.0-py3-none-any.whl", hash = "sha256:a2185c227351a52a013c7d7a695d3f3cb6625c3eed14a5295adbbcc7e2f7f8d4"},
+ {file = "dunamai-1.20.0.tar.gz", hash = "sha256:c3f1ee64a1e6cc9ebc98adafa944efaccd0db32482d2177e59c1ff6bdf23cd70"},
]
[package.dependencies]
packaging = ">=20.9"
-[[package]]
-name = "entrypoints"
-version = "0.4"
-description = "Discover and load entry points from installed packages."
-optional = false
-python-versions = ">=3.6"
-files = [
- {file = "entrypoints-0.4-py3-none-any.whl", hash = "sha256:f174b5ff827504fd3cd97cc3f8649f3693f51538c7e4bdf3ef002c8429d42f9f"},
- {file = "entrypoints-0.4.tar.gz", hash = "sha256:b706eddaa9218a19ebcd67b56818f05bb27589b1ca9e8d797b74affad4ccacd4"},
-]
-
[[package]]
name = "et-xmlfile"
version = "1.1.0"
@@ -958,13 +982,13 @@ files = [
[[package]]
name = "exceptiongroup"
-version = "1.2.0"
+version = "1.2.1"
description = "Backport of PEP 654 (exception groups)"
optional = false
python-versions = ">=3.7"
files = [
- {file = "exceptiongroup-1.2.0-py3-none-any.whl", hash = "sha256:4bfd3996ac73b41e9b9628b04e079f193850720ea5945fc96a08633c66912f14"},
- {file = "exceptiongroup-1.2.0.tar.gz", hash = "sha256:91f5c769735f051a4290d52edd0858999b57e5876e9f85937691bd4c9fa3ed68"},
+ {file = "exceptiongroup-1.2.1-py3-none-any.whl", hash = "sha256:5258b9ed329c5bbdd31a309f53cbfb0b155341807f6ff7606a1e801a891b29ad"},
+ {file = "exceptiongroup-1.2.1.tar.gz", hash = "sha256:a4785e48b045528f5bfe627b6ad554ff32def154f42372786903b7abcfe1aa16"},
]
[package.extras]
@@ -1000,13 +1024,13 @@ devel = ["colorama", "json-spec", "jsonschema", "pylint", "pytest", "pytest-benc
[[package]]
name = "filelock"
-version = "3.13.3"
+version = "3.13.4"
description = "A platform independent file lock."
optional = false
python-versions = ">=3.8"
files = [
- {file = "filelock-3.13.3-py3-none-any.whl", hash = "sha256:5ffa845303983e7a0b7ae17636509bc97997d58afeafa72fb141a17b152284cb"},
- {file = "filelock-3.13.3.tar.gz", hash = "sha256:a79895a25bbefdf55d1a2a0a80968f7dbb28edcd6d4234a0afb3f37ecde4b546"},
+ {file = "filelock-3.13.4-py3-none-any.whl", hash = "sha256:404e5e9253aa60ad457cae1be07c0f0ca90a63931200a47d9b6a6af84fd7b45f"},
+ {file = "filelock-3.13.4.tar.gz", hash = "sha256:d13f466618bfde72bd2c18255e269f72542c6e70e7bac83a0232d6b1cc5c8cf4"},
]
[package.extras]
@@ -1032,7 +1056,7 @@ termcolor = "*"
name = "frozenlist"
version = "1.4.1"
description = "A list-like structure which implements collections.abc.MutableSequence"
-optional = false
+optional = true
python-versions = ">=3.8"
files = [
{file = "frozenlist-1.4.1-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:f9aa1878d1083b276b0196f2dfbe00c9b7e752475ed3b682025ff20c1c1f51ac"},
@@ -1149,23 +1173,6 @@ smb = ["smbprotocol"]
ssh = ["paramiko"]
tqdm = ["tqdm"]
-[[package]]
-name = "furo"
-version = "2023.9.10"
-description = "A clean customisable Sphinx documentation theme."
-optional = false
-python-versions = ">=3.8"
-files = [
- {file = "furo-2023.9.10-py3-none-any.whl", hash = "sha256:513092538537dc5c596691da06e3c370714ec99bc438680edc1debffb73e5bfc"},
- {file = "furo-2023.9.10.tar.gz", hash = "sha256:5707530a476d2a63b8cad83b4f961f3739a69f4b058bcf38a03a39fa537195b2"},
-]
-
-[package.dependencies]
-beautifulsoup4 = "*"
-pygments = ">=2.7"
-sphinx = ">=6.0,<8.0"
-sphinx-basic-ng = "*"
-
[[package]]
name = "ghp-import"
version = "2.1.0"
@@ -1255,28 +1262,29 @@ docs = ["Sphinx", "furo"]
test = ["objgraph", "psutil"]
[[package]]
-name = "groundwork-sphinx-theme"
-version = "1.1.1"
-description = "Sphinx theme for groundwork projects (Based on flask_theme)"
+name = "griffe"
+version = "0.44.0"
+description = "Signatures for entire Python programs. Extract the structure, the frame, the skeleton of your project, to generate API documentation or find breaking changes in your API."
optional = false
-python-versions = "*"
+python-versions = ">=3.8"
files = [
- {file = "groundwork-sphinx-theme-1.1.1.tar.gz", hash = "sha256:5f1217bf34daba5fcd93e8a2a04ef5a6c2b144ec43754aec7ce14de501e23fe4"},
- {file = "groundwork_sphinx_theme-1.1.1-py2.py3-none-any.whl", hash = "sha256:b4796989ce3cd6d5b5a3c7ebe52a8c9cf848110e9f12c2c88e2b3c0e4e8f0e24"},
+ {file = "griffe-0.44.0-py3-none-any.whl", hash = "sha256:8a4471c469ba980b87c843f1168850ce39d0c1d0c7be140dca2480f76c8e5446"},
+ {file = "griffe-0.44.0.tar.gz", hash = "sha256:34aee1571042f9bf00529bc715de4516fb6f482b164e90d030300601009e0223"},
]
[package.dependencies]
-Sphinx = ">=1.6.1"
+astunparse = {version = ">=1.6", markers = "python_version < \"3.9\""}
+colorama = ">=0.4"
[[package]]
name = "identify"
-version = "2.5.35"
+version = "2.5.36"
description = "File identification library for Python"
optional = false
python-versions = ">=3.8"
files = [
- {file = "identify-2.5.35-py2.py3-none-any.whl", hash = "sha256:c4de0081837b211594f8e877a6b4fad7ca32bbfc1a9307fdd61c28bfe923f13e"},
- {file = "identify-2.5.35.tar.gz", hash = "sha256:10a7ca245cfcd756a554a7288159f72ff105ad233c7c4b9c6f0f4d108f5f6791"},
+ {file = "identify-2.5.36-py2.py3-none-any.whl", hash = "sha256:37d93f380f4de590500d9dba7db359d0d3da95ffe7f9de1753faa159e71e7dfa"},
+ {file = "identify-2.5.36.tar.gz", hash = "sha256:e5e00f54165f9047fbebeb4a560f9acfb8af4c88232be60a488e9b68d122745d"},
]
[package.extras]
@@ -1352,6 +1360,39 @@ files = [
{file = "iniconfig-2.0.0.tar.gz", hash = "sha256:2d91e135bf72d31a410b17c16da610a82cb55f6b0477d1a902134b24a455b8b3"},
]
+[[package]]
+name = "ipykernel"
+version = "6.29.4"
+description = "IPython Kernel for Jupyter"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "ipykernel-6.29.4-py3-none-any.whl", hash = "sha256:1181e653d95c6808039c509ef8e67c4126b3b3af7781496c7cbfb5ed938a27da"},
+ {file = "ipykernel-6.29.4.tar.gz", hash = "sha256:3d44070060f9475ac2092b760123fadf105d2e2493c24848b6691a7c4f42af5c"},
+]
+
+[package.dependencies]
+appnope = {version = "*", markers = "platform_system == \"Darwin\""}
+comm = ">=0.1.1"
+debugpy = ">=1.6.5"
+ipython = ">=7.23.1"
+jupyter-client = ">=6.1.12"
+jupyter-core = ">=4.12,<5.0.dev0 || >=5.1.dev0"
+matplotlib-inline = ">=0.1"
+nest-asyncio = "*"
+packaging = "*"
+psutil = "*"
+pyzmq = ">=24"
+tornado = ">=6.1"
+traitlets = ">=5.4.0"
+
+[package.extras]
+cov = ["coverage[toml]", "curio", "matplotlib", "pytest-cov", "trio"]
+docs = ["myst-parser", "pydata-sphinx-theme", "sphinx", "sphinx-autodoc-typehints", "sphinxcontrib-github-alt", "sphinxcontrib-spelling", "trio"]
+pyqt5 = ["pyqt5"]
+pyside6 = ["pyside6"]
+test = ["flaky", "ipyparallel", "pre-commit", "pytest (>=7.0)", "pytest-asyncio (>=0.23.5)", "pytest-cov", "pytest-timeout"]
+
[[package]]
name = "ipython"
version = "8.12.3"
@@ -1454,13 +1495,13 @@ files = [
[[package]]
name = "joblib"
-version = "1.3.2"
+version = "1.4.0"
description = "Lightweight pipelining with Python functions"
optional = false
-python-versions = ">=3.7"
+python-versions = ">=3.8"
files = [
- {file = "joblib-1.3.2-py3-none-any.whl", hash = "sha256:ef4331c65f239985f3f2220ecc87db222f08fd22097a3dd5698f693875f8cbb9"},
- {file = "joblib-1.3.2.tar.gz", hash = "sha256:92f865e621e17784e7955080b6d042489e3b8e294949cc44c6eac304f59772b1"},
+ {file = "joblib-1.4.0-py3-none-any.whl", hash = "sha256:42942470d4062537be4d54c83511186da1fc14ba354961a2114da91efa9a4ed7"},
+ {file = "joblib-1.4.0.tar.gz", hash = "sha256:1eb0dc091919cd384490de890cb5dfd538410a6d4b3b54eef09fb8c50b409b1c"},
]
[[package]]
@@ -1555,6 +1596,35 @@ files = [
{file = "jupyterlab_pygments-0.3.0.tar.gz", hash = "sha256:721aca4d9029252b11cfa9d185e5b5af4d54772bb8072f9b7036f4170054d35d"},
]
+[[package]]
+name = "jupytext"
+version = "1.16.1"
+description = "Jupyter notebooks as Markdown documents, Julia, Python or R scripts"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "jupytext-1.16.1-py3-none-any.whl", hash = "sha256:796ec4f68ada663569e5d38d4ef03738a01284bfe21c943c485bc36433898bd0"},
+ {file = "jupytext-1.16.1.tar.gz", hash = "sha256:68c7b68685e870e80e60fda8286fbd6269e9c74dc1df4316df6fe46eabc94c99"},
+]
+
+[package.dependencies]
+markdown-it-py = ">=1.0"
+mdit-py-plugins = "*"
+nbformat = "*"
+packaging = "*"
+pyyaml = "*"
+toml = "*"
+
+[package.extras]
+dev = ["jupytext[test-cov,test-external]"]
+docs = ["myst-parser", "sphinx", "sphinx-copybutton", "sphinx-rtd-theme"]
+test = ["pytest", "pytest-randomly", "pytest-xdist"]
+test-cov = ["jupytext[test-integration]", "pytest-cov (>=2.6.1)"]
+test-external = ["autopep8", "black", "flake8", "gitpython", "isort", "jupyter-fs (<0.4.0)", "jupytext[test-integration]", "pre-commit", "sphinx-gallery (<0.8)"]
+test-functional = ["jupytext[test]"]
+test-integration = ["ipykernel", "jupyter-server (!=2.11)", "jupytext[test-functional]", "nbconvert"]
+test-ui = ["calysto-bash"]
+
[[package]]
name = "leather"
version = "0.4.0"
@@ -1569,124 +1639,6 @@ files = [
[package.extras]
test = ["cssselect (>=0.9.1)", "lxml (>=3.6.0)", "pytest", "pytest-cov"]
-[[package]]
-name = "livereload"
-version = "2.6.3"
-description = "Python LiveReload is an awesome tool for web developers"
-optional = false
-python-versions = "*"
-files = [
- {file = "livereload-2.6.3-py2.py3-none-any.whl", hash = "sha256:ad4ac6f53b2d62bb6ce1a5e6e96f1f00976a32348afedcb4b6d68df2a1d346e4"},
- {file = "livereload-2.6.3.tar.gz", hash = "sha256:776f2f865e59fde56490a56bcc6773b6917366bce0c267c60ee8aaf1a0959869"},
-]
-
-[package.dependencies]
-six = "*"
-tornado = {version = "*", markers = "python_version > \"2.7\""}
-
-[[package]]
-name = "lxml"
-version = "5.1.1"
-description = "Powerful and Pythonic XML processing library combining libxml2/libxslt with the ElementTree API."
-optional = false
-python-versions = ">=3.6"
-files = [
- {file = "lxml-5.1.1-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:9cc30dc3c49ea914fa62ea73b57198b541cf2cd522fcf2b9559f99a24df769bb"},
- {file = "lxml-5.1.1-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:f1d0824e3ddb969fe1337b1bc45cf0bec8095b342f36903f41a74b7769cc8c73"},
- {file = "lxml-5.1.1-cp310-cp310-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:4958c378d9387c45ef8c4859495cf6be76f863e4e3b31494f6ec7f2c48d3b8e3"},
- {file = "lxml-5.1.1-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:aff34295a6c87638a1f1905355cf3a97e4026c45c0cf3bb6ed6bc35b885b4a33"},
- {file = "lxml-5.1.1-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b174885fd2cabd1ad48585296f495e25d607f02db99668c08b2afaceb668e21b"},
- {file = "lxml-5.1.1-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:1b0611bba10d6f5467b86673e8f6bba4de0d00f7d111eea843bc872abfe11b5c"},
- {file = "lxml-5.1.1-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:adff469b7dbfe9f3babc9e4479449ee97085ba70ac492fbe5f0f7217940c6731"},
- {file = "lxml-5.1.1-cp310-cp310-win32.whl", hash = "sha256:99bcdf665576a26b44c7ce767d76b769a4418b0a13cda8300b26fb7b2647bd5b"},
- {file = "lxml-5.1.1-cp310-cp310-win_amd64.whl", hash = "sha256:3da8db291568c27b2bb248dcfc8838ca3149f373a24e204bcd1c2c89e2813d14"},
- {file = "lxml-5.1.1-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:906966babd374fdfe46e130fc656488003f0d0d63b7cba612aa5a796c8804283"},
- {file = "lxml-5.1.1-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:9c03f3715c68fc707d9383d56e482d95d198ba07cb3dad4aee9e5a5ca06b2536"},
- {file = "lxml-5.1.1-cp311-cp311-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:d26243d994d4077a50056e9008848e5b421be0c6f0fd4e932a9463e1d89fc42b"},
- {file = "lxml-5.1.1-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:2de00750318ae6869b9dfa6429a4f82b8ecad043049414547474d09db549c2ee"},
- {file = "lxml-5.1.1-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:29b2771b4eec4e85063f10294facdd9829d010e6cc9668040d0cf936dc56733a"},
- {file = "lxml-5.1.1-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:d9358f7268c161dc0a1c3216018f26c04954b5dd47ba6dead79da6598f4725d4"},
- {file = "lxml-5.1.1-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:8a943826e7a9254eed661a7134fcde3c832a9fecd989d0f47c6e08c7b769cb2c"},
- {file = "lxml-5.1.1-cp311-cp311-win32.whl", hash = "sha256:74d0967c6f91eec6fe91159f9e8ccb3720fa0fbf9f462109c7bef62550df397c"},
- {file = "lxml-5.1.1-cp311-cp311-win_amd64.whl", hash = "sha256:26974096654241df08a30dc2eb0e139c1ad5653660aa4b2ced66000230e96c14"},
- {file = "lxml-5.1.1-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:55e13a19829dcdbf0c5233062977aeb6daf72e65124909128045976f659164e8"},
- {file = "lxml-5.1.1-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:adedfb61be862f48907218e3a24bf051fd2ecca53358f3958b0bdb17d7881c20"},
- {file = "lxml-5.1.1-cp312-cp312-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:77425482e4311d1cff119a2b5ab26c52ec209d2a3d728a54db3223ab91995e20"},
- {file = "lxml-5.1.1-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:1d380f183bd03ab827899753ea96dabe27d2025eb0bfd4f2ac0eee4afa0f351d"},
- {file = "lxml-5.1.1-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:f8682af96b5ad5093aab9eee5e4ff24cb7a9796c78699d914dd456ebfe7484a6"},
- {file = "lxml-5.1.1-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:68eed33377a9925aed7ba56c8611d50aaa1e45638c07a92b4b4b0a0436cc2dd2"},
- {file = "lxml-5.1.1-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:c7c1d2f6e9c7a1c4478146ee38d16dbe0eb3be998424bc0f01346c671c38b86d"},
- {file = "lxml-5.1.1-cp312-cp312-win32.whl", hash = "sha256:81107c8de3e463052ae8fd05fd31b97c371c7a9ce4a189b8bb5f45b0b3545fb9"},
- {file = "lxml-5.1.1-cp312-cp312-win_amd64.whl", hash = "sha256:0e46181d15fae102c53621bed9356b7a599a1e837b978c934a350dd00842b1d9"},
- {file = "lxml-5.1.1-cp36-cp36m-macosx_10_9_x86_64.whl", hash = "sha256:48dd28b9f410329de709a4bb6169c58f2cd8bff25f5a48d647678ec9b8a40c65"},
- {file = "lxml-5.1.1-cp36-cp36m-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:bf7e57dbe7b3c605e63849d9c8dae246a6ab9002223c57cd3f3dec7c3a0a8e6d"},
- {file = "lxml-5.1.1-cp36-cp36m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:5020b3081030b5cfc8149eee231167aea4ff68df73a610e1d542809e1f11fde7"},
- {file = "lxml-5.1.1-cp36-cp36m-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:77842b79b63c83c04dcfe2f045c78e15e4d97c86838eabd2e6518c1ed97e3900"},
- {file = "lxml-5.1.1-cp36-cp36m-musllinux_1_1_aarch64.whl", hash = "sha256:666432274881cb2535e71dbe745e08ef10fe25c81fbb1a6b1e3c973177823b0c"},
- {file = "lxml-5.1.1-cp36-cp36m-musllinux_1_1_x86_64.whl", hash = "sha256:a103426e809640a2d985062d2f4b28db2f0fe4469ff72a67cb31fa70613158f1"},
- {file = "lxml-5.1.1-cp36-cp36m-win32.whl", hash = "sha256:95a51324a55000c55f4ab79e1f7f1e0bc42b7a24e39633f79542753023a9d4b7"},
- {file = "lxml-5.1.1-cp36-cp36m-win_amd64.whl", hash = "sha256:bd46b5b19ac969de8e87fb3d04414641d12ee489e2ea6cc75344087829b31c63"},
- {file = "lxml-5.1.1-cp37-cp37m-macosx_10_9_x86_64.whl", hash = "sha256:59ca75cfcf646ff64aa19ca4e7fd2a0fde77268d5a87856525d9e0b69b77d0c4"},
- {file = "lxml-5.1.1-cp37-cp37m-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:d55ddc73dec971277b181a6d1a6abdd34f50e4511e1e60f6b4ebe22cbaad05bb"},
- {file = "lxml-5.1.1-cp37-cp37m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:56f1e813ff660d031c77edba90a068d57e47ae93a9e811330fc88946fa68af9a"},
- {file = "lxml-5.1.1-cp37-cp37m-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:43f21b5929185fa4560836942020bb00a0fcdec9f67be98cac1a4b99501757c1"},
- {file = "lxml-5.1.1-cp37-cp37m-musllinux_1_1_aarch64.whl", hash = "sha256:1528b37e83c3aeecb438e76e5be6279b353275560125a9c3f4d74642c5f110f9"},
- {file = "lxml-5.1.1-cp37-cp37m-musllinux_1_1_x86_64.whl", hash = "sha256:2992480a25434d2df31413136ef87effab14d43b07f1f54c5012c4f6c7530144"},
- {file = "lxml-5.1.1-cp37-cp37m-win32.whl", hash = "sha256:1d0270d33fbde6e1c6758ff58e2e284144f5331aa05dfe7f44ceafdf4e9d31aa"},
- {file = "lxml-5.1.1-cp37-cp37m-win_amd64.whl", hash = "sha256:dec3491aa69a91ed07f5e6bc033e2b1a9424447ad5312ee69ac973e94d79083a"},
- {file = "lxml-5.1.1-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:5bd2595ebe95214446e00a1ab94571f778b126e17736ea222c07505c4e092289"},
- {file = "lxml-5.1.1-cp38-cp38-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:bfbdadc3cfe552331ecb0bbdcabf148d1697c73aa4321151e0e6c1704eeb76a7"},
- {file = "lxml-5.1.1-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:52358249292bc155af681a9240ec3d944c1195f0124aa10ec4e3635adc1e10a1"},
- {file = "lxml-5.1.1-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:036b36c48cd775e4fd2084b34ae62ffeefa7a01f955f5a5b816f9257c308cfc0"},
- {file = "lxml-5.1.1-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:f05ab8cea65363d0cc7ce818f42407504b6d94ca885b4cde0270f021e2f4ef61"},
- {file = "lxml-5.1.1-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:d94a28c16cc430b68c374b37b8bb536ba5f0a4a080be0e1daa8310c44a00a75c"},
- {file = "lxml-5.1.1-cp38-cp38-win32.whl", hash = "sha256:9113fe65a62f834b8e994c8f48e7b2179bf81878c0ec80ad7feba51ab9417663"},
- {file = "lxml-5.1.1-cp38-cp38-win_amd64.whl", hash = "sha256:acff17e0cd5344677757a152631d8411efac6a84e4476d60123a9b33f5d6c511"},
- {file = "lxml-5.1.1-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:a94a97380ad689d751eb0a1e1ccd2a0622c5141771a31abe9a16075f80027e95"},
- {file = "lxml-5.1.1-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:d1f4d37b3f8d2d44493edce3d65ac987127bababd8ae208a6f0d7d260852346e"},
- {file = "lxml-5.1.1-cp39-cp39-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:3c5940f188189956ccb3d1adb413001ada79f2d2b81087d2612a0cc4a1197eed"},
- {file = "lxml-5.1.1-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:50007f4e94dc4e38030487a8b6c4af87a2d51ed059c7b74b29e3dd937cb1dfe1"},
- {file = "lxml-5.1.1-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:a305d0469177fd78a0a9aa2231c60218266bb85d4b7955f9b67dab628c9267fd"},
- {file = "lxml-5.1.1-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:13b73d78a8023203722cf98e9ea0b222da83110d1d5ef437ef8782a7755b4586"},
- {file = "lxml-5.1.1-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:bc6904519dd1f92eb82f7d49814a33bbc444d0b66b1438e76daf3f79ef4aa38f"},
- {file = "lxml-5.1.1-cp39-cp39-win32.whl", hash = "sha256:04ef231dde88294a5499f61a74cdc42af97d8d5ecec1b0a645d1c7d436942789"},
- {file = "lxml-5.1.1-cp39-cp39-win_amd64.whl", hash = "sha256:071e5123d1eca861708c4be5b54e4d88923fa33fab3aa02722e907518b07071c"},
- {file = "lxml-5.1.1-pp310-pypy310_pp73-macosx_10_9_x86_64.whl", hash = "sha256:19c6bc7476eeac4598ff925ae98597610109e21af4cd7ab1e060efcfc4b1c6e2"},
- {file = "lxml-5.1.1-pp310-pypy310_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:20cd17eb21f5ae54da96791c49e1fbd3327bf66b2c00556cdf8d0552c2270f92"},
- {file = "lxml-5.1.1-pp310-pypy310_pp73-win_amd64.whl", hash = "sha256:a02ed1ebc469734dbfed5b688f709334de19e7a333cba7ae187b17d2b2c1d4ff"},
- {file = "lxml-5.1.1-pp37-pypy37_pp73-macosx_10_9_x86_64.whl", hash = "sha256:161838cb95c97e8d76d01e544a3570b52ab6b863f4897a90e1f073bb110a75ba"},
- {file = "lxml-5.1.1-pp37-pypy37_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:d1abbf2249467a37da45fb2d7ff37e578dfc9813f142800e58db9da761cb7899"},
- {file = "lxml-5.1.1-pp37-pypy37_pp73-win_amd64.whl", hash = "sha256:6c49eb5deaed1990fde5b5d80d6800aec1b5fd6113346b5f11068d988f68f2c4"},
- {file = "lxml-5.1.1-pp38-pypy38_pp73-macosx_10_9_x86_64.whl", hash = "sha256:60ceffdca5d637fe8ee95c7f06733a6c9646e07da80997efe3af2d4b4f366e36"},
- {file = "lxml-5.1.1-pp38-pypy38_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:a76a7b35e7660c74eb3f943c19f5f78c882dceab890cf8017027b6100b79ad8e"},
- {file = "lxml-5.1.1-pp38-pypy38_pp73-win_amd64.whl", hash = "sha256:5dcb373720b70aa05419e508265dd86f06886ca0388967f6f024fbc4d551379f"},
- {file = "lxml-5.1.1-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:3641bc124b037921de4220538a5ebb52354fd2799fc2bbfb335d28096063c7d6"},
- {file = "lxml-5.1.1-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:3a6e9b34f59c9755aa279c652e1c48c333c665d05a88afcd8e5ff0bde86f3b14"},
- {file = "lxml-5.1.1-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:318847c165063549c8fda6b162a0d068689b10deb825cb3859caef69fddaaaff"},
- {file = "lxml-5.1.1.tar.gz", hash = "sha256:42a8aa957e98bd8b884a8142175ec24ce4ef0a57760e8879f193bfe64b757ca9"},
-]
-
-[package.extras]
-cssselect = ["cssselect (>=0.7)"]
-html5 = ["html5lib"]
-htmlsoup = ["BeautifulSoup4"]
-source = ["Cython (>=3.0.9)"]
-
-[[package]]
-name = "m2r2"
-version = "0.3.3.post2"
-description = "Markdown and reStructuredText in a single file."
-optional = false
-python-versions = ">=3.7"
-files = [
- {file = "m2r2-0.3.3.post2-py3-none-any.whl", hash = "sha256:86157721eb6eabcd54d4eea7195890cc58fa6188b8d0abea633383cfbb5e11e3"},
- {file = "m2r2-0.3.3.post2.tar.gz", hash = "sha256:e62bcb0e74b3ce19cda0737a0556b04cf4a43b785072fcef474558f2c1482ca8"},
-]
-
-[package.dependencies]
-docutils = ">=0.19"
-mistune = "0.8.4"
-
[[package]]
name = "markdown"
version = "3.6"
@@ -1705,6 +1657,30 @@ importlib-metadata = {version = ">=4.4", markers = "python_version < \"3.10\""}
docs = ["mdx-gh-links (>=0.2)", "mkdocs (>=1.5)", "mkdocs-gen-files", "mkdocs-literate-nav", "mkdocs-nature (>=0.6)", "mkdocs-section-index", "mkdocstrings[python]"]
testing = ["coverage", "pyyaml"]
+[[package]]
+name = "markdown-it-py"
+version = "3.0.0"
+description = "Python port of markdown-it. Markdown parsing, done right!"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "markdown-it-py-3.0.0.tar.gz", hash = "sha256:e3f60a94fa066dc52ec76661e37c851cb232d92f9886b15cb560aaada2df8feb"},
+ {file = "markdown_it_py-3.0.0-py3-none-any.whl", hash = "sha256:355216845c60bd96232cd8d8c40e8f9765cc86f46880e43a8fd22dc1a1a8cab1"},
+]
+
+[package.dependencies]
+mdurl = ">=0.1,<1.0"
+
+[package.extras]
+benchmarking = ["psutil", "pytest", "pytest-benchmark"]
+code-style = ["pre-commit (>=3.0,<4.0)"]
+compare = ["commonmark (>=0.9,<1.0)", "markdown (>=3.4,<4.0)", "mistletoe (>=1.0,<2.0)", "mistune (>=2.0,<3.0)", "panflute (>=2.3,<3.0)"]
+linkify = ["linkify-it-py (>=1,<3)"]
+plugins = ["mdit-py-plugins"]
+profiling = ["gprof2dot"]
+rtd = ["jupyter_sphinx", "mdit-py-plugins", "myst-parser", "pyyaml", "sphinx", "sphinx-copybutton", "sphinx-design", "sphinx_book_theme"]
+testing = ["coverage", "pytest", "pytest-cov", "pytest-regressions"]
+
[[package]]
name = "markupsafe"
version = "2.1.5"
@@ -1776,18 +1752,48 @@ files = [
[[package]]
name = "matplotlib-inline"
-version = "0.1.6"
+version = "0.1.7"
description = "Inline Matplotlib backend for Jupyter"
optional = false
-python-versions = ">=3.5"
+python-versions = ">=3.8"
files = [
- {file = "matplotlib-inline-0.1.6.tar.gz", hash = "sha256:f887e5f10ba98e8d2b150ddcf4702c1e5f8b3a20005eb0f74bfdbd360ee6f304"},
- {file = "matplotlib_inline-0.1.6-py3-none-any.whl", hash = "sha256:f1f41aab5328aa5aaea9b16d083b128102f8712542f819fe7e6a420ff581b311"},
+ {file = "matplotlib_inline-0.1.7-py3-none-any.whl", hash = "sha256:df192d39a4ff8f21b1895d72e6a13f5fcc5099f00fa84384e0ea28c2cc0653ca"},
+ {file = "matplotlib_inline-0.1.7.tar.gz", hash = "sha256:8423b23ec666be3d16e16b60bdd8ac4e86e840ebd1dd11a30b9f117f2fa0ab90"},
]
[package.dependencies]
traitlets = "*"
+[[package]]
+name = "mdit-py-plugins"
+version = "0.4.0"
+description = "Collection of plugins for markdown-it-py"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mdit_py_plugins-0.4.0-py3-none-any.whl", hash = "sha256:b51b3bb70691f57f974e257e367107857a93b36f322a9e6d44ca5bf28ec2def9"},
+ {file = "mdit_py_plugins-0.4.0.tar.gz", hash = "sha256:d8ab27e9aed6c38aa716819fedfde15ca275715955f8a185a8e1cf90fb1d2c1b"},
+]
+
+[package.dependencies]
+markdown-it-py = ">=1.0.0,<4.0.0"
+
+[package.extras]
+code-style = ["pre-commit"]
+rtd = ["myst-parser", "sphinx-book-theme"]
+testing = ["coverage", "pytest", "pytest-cov", "pytest-regressions"]
+
+[[package]]
+name = "mdurl"
+version = "0.1.2"
+description = "Markdown URL utilities"
+optional = false
+python-versions = ">=3.7"
+files = [
+ {file = "mdurl-0.1.2-py3-none-any.whl", hash = "sha256:84008a41e51615a49fc9966191ff91509e3c40b939176e643fd50a5c2196b8f8"},
+ {file = "mdurl-0.1.2.tar.gz", hash = "sha256:bb413d29f5eea38f31dd4754dd7377d4465116fb207585f97bf925588687c1ba"},
+]
+
[[package]]
name = "mergedeep"
version = "1.3.4"
@@ -1801,45 +1807,196 @@ files = [
[[package]]
name = "mistune"
-version = "0.8.4"
-description = "The fastest markdown parser in pure Python"
+version = "3.0.2"
+description = "A sane and fast Markdown parser with useful plugins and renderers"
optional = false
-python-versions = "*"
+python-versions = ">=3.7"
files = [
- {file = "mistune-0.8.4-py2.py3-none-any.whl", hash = "sha256:88a1051873018da288eee8538d476dffe1262495144b33ecb586c4ab266bb8d4"},
- {file = "mistune-0.8.4.tar.gz", hash = "sha256:59a3429db53c50b5c6bcc8a07f8848cb00d7dc8bdb431a4ab41920d201d4756e"},
+ {file = "mistune-3.0.2-py3-none-any.whl", hash = "sha256:71481854c30fdbc938963d3605b72501f5c10a9320ecd412c121c163a1c7d205"},
+ {file = "mistune-3.0.2.tar.gz", hash = "sha256:fc7f93ded930c92394ef2cb6f04a8aabab4117a91449e72dcc8dfa646a508be8"},
]
[[package]]
name = "mkdocs"
-version = "1.5.3"
+version = "1.6.0"
description = "Project documentation with Markdown."
optional = false
-python-versions = ">=3.7"
+python-versions = ">=3.8"
files = [
- {file = "mkdocs-1.5.3-py3-none-any.whl", hash = "sha256:3b3a78e736b31158d64dbb2f8ba29bd46a379d0c6e324c2246c3bc3d2189cfc1"},
- {file = "mkdocs-1.5.3.tar.gz", hash = "sha256:eb7c99214dcb945313ba30426c2451b735992c73c2e10838f76d09e39ff4d0e2"},
+ {file = "mkdocs-1.6.0-py3-none-any.whl", hash = "sha256:1eb5cb7676b7d89323e62b56235010216319217d4af5ddc543a91beb8d125ea7"},
+ {file = "mkdocs-1.6.0.tar.gz", hash = "sha256:a73f735824ef83a4f3bcb7a231dcab23f5a838f88b7efc54a0eef5fbdbc3c512"},
]
[package.dependencies]
click = ">=7.0"
colorama = {version = ">=0.4", markers = "platform_system == \"Windows\""}
ghp-import = ">=1.0"
-importlib-metadata = {version = ">=4.3", markers = "python_version < \"3.10\""}
+importlib-metadata = {version = ">=4.4", markers = "python_version < \"3.10\""}
jinja2 = ">=2.11.1"
-markdown = ">=3.2.1"
+markdown = ">=3.3.6"
markupsafe = ">=2.0.1"
mergedeep = ">=1.3.4"
+mkdocs-get-deps = ">=0.2.0"
packaging = ">=20.5"
pathspec = ">=0.11.1"
-platformdirs = ">=2.2.0"
pyyaml = ">=5.1"
pyyaml-env-tag = ">=0.1"
watchdog = ">=2.0"
[package.extras]
i18n = ["babel (>=2.9.0)"]
-min-versions = ["babel (==2.9.0)", "click (==7.0)", "colorama (==0.4)", "ghp-import (==1.0)", "importlib-metadata (==4.3)", "jinja2 (==2.11.1)", "markdown (==3.2.1)", "markupsafe (==2.0.1)", "mergedeep (==1.3.4)", "packaging (==20.5)", "pathspec (==0.11.1)", "platformdirs (==2.2.0)", "pyyaml (==5.1)", "pyyaml-env-tag (==0.1)", "typing-extensions (==3.10)", "watchdog (==2.0)"]
+min-versions = ["babel (==2.9.0)", "click (==7.0)", "colorama (==0.4)", "ghp-import (==1.0)", "importlib-metadata (==4.4)", "jinja2 (==2.11.1)", "markdown (==3.3.6)", "markupsafe (==2.0.1)", "mergedeep (==1.3.4)", "mkdocs-get-deps (==0.2.0)", "packaging (==20.5)", "pathspec (==0.11.1)", "pyyaml (==5.1)", "pyyaml-env-tag (==0.1)", "watchdog (==2.0)"]
+
+[[package]]
+name = "mkdocs-autorefs"
+version = "1.0.1"
+description = "Automatically link across pages in MkDocs."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocs_autorefs-1.0.1-py3-none-any.whl", hash = "sha256:aacdfae1ab197780fb7a2dac92ad8a3d8f7ca8049a9cbe56a4218cd52e8da570"},
+ {file = "mkdocs_autorefs-1.0.1.tar.gz", hash = "sha256:f684edf847eced40b570b57846b15f0bf57fb93ac2c510450775dcf16accb971"},
+]
+
+[package.dependencies]
+Markdown = ">=3.3"
+markupsafe = ">=2.0.1"
+mkdocs = ">=1.1"
+
+[[package]]
+name = "mkdocs-get-deps"
+version = "0.2.0"
+description = "MkDocs extension that lists all dependencies according to a mkdocs.yml file"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocs_get_deps-0.2.0-py3-none-any.whl", hash = "sha256:2bf11d0b133e77a0dd036abeeb06dec8775e46efa526dc70667d8863eefc6134"},
+ {file = "mkdocs_get_deps-0.2.0.tar.gz", hash = "sha256:162b3d129c7fad9b19abfdcb9c1458a651628e4b1dea628ac68790fb3061c60c"},
+]
+
+[package.dependencies]
+importlib-metadata = {version = ">=4.3", markers = "python_version < \"3.10\""}
+mergedeep = ">=1.3.4"
+platformdirs = ">=2.2.0"
+pyyaml = ">=5.1"
+
+[[package]]
+name = "mkdocs-include-markdown-plugin"
+version = "6.0.5"
+description = "Mkdocs Markdown includer plugin."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocs_include_markdown_plugin-6.0.5-py3-none-any.whl", hash = "sha256:db41aa1937a618afa3497616f457d4e51d9123b13b2034bb15505ff9ce061f86"},
+ {file = "mkdocs_include_markdown_plugin-6.0.5.tar.gz", hash = "sha256:ad10779cf0dc4ff180aaa0079163271877b3c2fd31e36d5579854fe1d4b0d1ae"},
+]
+
+[package.dependencies]
+mkdocs = ">=1.4"
+wcmatch = ">=8,<9"
+
+[package.extras]
+cache = ["platformdirs"]
+
+[[package]]
+name = "mkdocs-jupyter"
+version = "0.24.7"
+description = "Use Jupyter in mkdocs websites"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocs_jupyter-0.24.7-py3-none-any.whl", hash = "sha256:893d04bea1e007479a46e4e72852cd4d280c4d358ce4a0445250f3f80c639723"},
+]
+
+[package.dependencies]
+ipykernel = ">6.0.0,<7.0.0"
+jupytext = ">1.13.8,<2"
+mkdocs = ">=1.4.0,<2"
+mkdocs-material = ">9.0.0"
+nbconvert = ">=7.2.9,<8"
+pygments = ">2.12.0"
+
+[[package]]
+name = "mkdocs-material"
+version = "9.5.19"
+description = "Documentation that simply works"
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocs_material-9.5.19-py3-none-any.whl", hash = "sha256:ea96e150b6c95f5e4ffe47d78bb712c7bacdd91d2a0bec47f46b6fa0705a86ec"},
+ {file = "mkdocs_material-9.5.19.tar.gz", hash = "sha256:7473e06e17e23af608a30ef583fdde8f36389dd3ef56b1d503eed54c89c9618c"},
+]
+
+[package.dependencies]
+babel = ">=2.10,<3.0"
+colorama = ">=0.4,<1.0"
+jinja2 = ">=3.0,<4.0"
+markdown = ">=3.2,<4.0"
+mkdocs = ">=1.6,<2.0"
+mkdocs-material-extensions = ">=1.3,<2.0"
+paginate = ">=0.5,<1.0"
+pygments = ">=2.16,<3.0"
+pymdown-extensions = ">=10.2,<11.0"
+regex = ">=2022.4"
+requests = ">=2.26,<3.0"
+
+[package.extras]
+git = ["mkdocs-git-committers-plugin-2 (>=1.1,<2.0)", "mkdocs-git-revision-date-localized-plugin (>=1.2.4,<2.0)"]
+imaging = ["cairosvg (>=2.6,<3.0)", "pillow (>=10.2,<11.0)"]
+recommended = ["mkdocs-minify-plugin (>=0.7,<1.0)", "mkdocs-redirects (>=1.2,<2.0)", "mkdocs-rss-plugin (>=1.6,<2.0)"]
+
+[[package]]
+name = "mkdocs-material-extensions"
+version = "1.3.1"
+description = "Extension pack for Python Markdown and MkDocs Material."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocs_material_extensions-1.3.1-py3-none-any.whl", hash = "sha256:adff8b62700b25cb77b53358dad940f3ef973dd6db797907c49e3c2ef3ab4e31"},
+ {file = "mkdocs_material_extensions-1.3.1.tar.gz", hash = "sha256:10c9511cea88f568257f960358a467d12b970e1f7b2c0e5fb2bb48cab1928443"},
+]
+
+[[package]]
+name = "mkdocstrings"
+version = "0.23.0"
+description = "Automatic documentation from sources, for MkDocs."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocstrings-0.23.0-py3-none-any.whl", hash = "sha256:051fa4014dfcd9ed90254ae91de2dbb4f24e166347dae7be9a997fe16316c65e"},
+ {file = "mkdocstrings-0.23.0.tar.gz", hash = "sha256:d9c6a37ffbe7c14a7a54ef1258c70b8d394e6a33a1c80832bce40b9567138d1c"},
+]
+
+[package.dependencies]
+importlib-metadata = {version = ">=4.6", markers = "python_version < \"3.10\""}
+Jinja2 = ">=2.11.1"
+Markdown = ">=3.3"
+MarkupSafe = ">=1.1"
+mkdocs = ">=1.2"
+mkdocs-autorefs = ">=0.3.1"
+mkdocstrings-python = {version = ">=0.5.2", optional = true, markers = "extra == \"python\""}
+pymdown-extensions = ">=6.3"
+typing-extensions = {version = ">=4.1", markers = "python_version < \"3.10\""}
+
+[package.extras]
+crystal = ["mkdocstrings-crystal (>=0.3.4)"]
+python = ["mkdocstrings-python (>=0.5.2)"]
+python-legacy = ["mkdocstrings-python-legacy (>=0.2.1)"]
+
+[[package]]
+name = "mkdocstrings-python"
+version = "1.8.0"
+description = "A Python handler for mkdocstrings."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "mkdocstrings_python-1.8.0-py3-none-any.whl", hash = "sha256:4209970cc90bec194568682a535848a8d8489516c6ed4adbe58bbc67b699ca9d"},
+ {file = "mkdocstrings_python-1.8.0.tar.gz", hash = "sha256:1488bddf50ee42c07d9a488dddc197f8e8999c2899687043ec5dd1643d057192"},
+]
+
+[package.dependencies]
+griffe = ">=0.37"
+mkdocstrings = ">=0.20"
[[package]]
name = "mock"
@@ -1861,7 +2018,7 @@ test = ["pytest", "pytest-cov"]
name = "multidict"
version = "6.0.5"
description = "multidict implementation"
-optional = false
+optional = true
python-versions = ">=3.7"
files = [
{file = "multidict-6.0.5-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:228b644ae063c10e7f324ab1ab6b548bdf6f8b47f3ec234fef1093bc2735e5f9"},
@@ -1980,56 +2137,57 @@ test = ["flaky", "ipykernel (>=6.19.3)", "ipython", "ipywidgets", "nbconvert (>=
[[package]]
name = "nbconvert"
-version = "6.5.4"
-description = "Converting Jupyter Notebooks"
+version = "7.16.3"
+description = "Converting Jupyter Notebooks (.ipynb files) to other formats. Output formats include asciidoc, html, latex, markdown, pdf, py, rst, script. nbconvert can be used both as a Python library (`import nbconvert`) or as a command line tool (invoked as `jupyter nbconvert ...`)."
optional = false
-python-versions = ">=3.7"
+python-versions = ">=3.8"
files = [
- {file = "nbconvert-6.5.4-py3-none-any.whl", hash = "sha256:d679a947f849a966cbbd0bf6e7fedcfdb64be3b20ce7cef11ad55c13f5820e19"},
- {file = "nbconvert-6.5.4.tar.gz", hash = "sha256:9e3c7c6d491374cbdd5f35d268c05809357716d346f4573186bbeab32ee50bc1"},
+ {file = "nbconvert-7.16.3-py3-none-any.whl", hash = "sha256:ddeff14beeeedf3dd0bc506623e41e4507e551736de59df69a91f86700292b3b"},
+ {file = "nbconvert-7.16.3.tar.gz", hash = "sha256:a6733b78ce3d47c3f85e504998495b07e6ea9cf9bf6ec1c98dda63ec6ad19142"},
]
[package.dependencies]
beautifulsoup4 = "*"
-bleach = "*"
+bleach = "!=5.0.0"
defusedxml = "*"
-entrypoints = ">=0.2.2"
+importlib-metadata = {version = ">=3.6", markers = "python_version < \"3.10\""}
jinja2 = ">=3.0"
jupyter-core = ">=4.7"
jupyterlab-pygments = "*"
-lxml = "*"
-MarkupSafe = ">=2.0"
-mistune = ">=0.8.1,<2"
+markupsafe = ">=2.0"
+mistune = ">=2.0.3,<4"
nbclient = ">=0.5.0"
-nbformat = ">=5.1"
+nbformat = ">=5.7"
packaging = "*"
pandocfilters = ">=1.4.1"
pygments = ">=2.4.1"
tinycss2 = "*"
-traitlets = ">=5.0"
+traitlets = ">=5.1"
[package.extras]
-all = ["ipykernel", "ipython", "ipywidgets (>=7)", "nbsphinx (>=0.2.12)", "pre-commit", "pyppeteer (>=1,<1.1)", "pytest", "pytest-cov", "pytest-dependency", "sphinx (>=1.5.1)", "sphinx-rtd-theme", "tornado (>=6.1)"]
-docs = ["ipython", "nbsphinx (>=0.2.12)", "sphinx (>=1.5.1)", "sphinx-rtd-theme"]
+all = ["nbconvert[docs,qtpdf,serve,test,webpdf]"]
+docs = ["ipykernel", "ipython", "myst-parser", "nbsphinx (>=0.2.12)", "pydata-sphinx-theme", "sphinx (==5.0.2)", "sphinxcontrib-spelling"]
+qtpdf = ["nbconvert[qtpng]"]
+qtpng = ["pyqtwebengine (>=5.15)"]
serve = ["tornado (>=6.1)"]
-test = ["ipykernel", "ipywidgets (>=7)", "pre-commit", "pyppeteer (>=1,<1.1)", "pytest", "pytest-cov", "pytest-dependency"]
-webpdf = ["pyppeteer (>=1,<1.1)"]
+test = ["flaky", "ipykernel", "ipywidgets (>=7.5)", "pytest (>=7)"]
+webpdf = ["playwright"]
[[package]]
name = "nbformat"
-version = "5.10.3"
+version = "5.10.4"
description = "The Jupyter Notebook format"
optional = false
python-versions = ">=3.8"
files = [
- {file = "nbformat-5.10.3-py3-none-any.whl", hash = "sha256:d9476ca28676799af85385f409b49d95e199951477a159a576ef2a675151e5e8"},
- {file = "nbformat-5.10.3.tar.gz", hash = "sha256:60ed5e910ef7c6264b87d644f276b1b49e24011930deef54605188ddeb211685"},
+ {file = "nbformat-5.10.4-py3-none-any.whl", hash = "sha256:3b48d6c8fbca4b299bf3982ea7db1af21580e4fec269ad087b9e81588891200b"},
+ {file = "nbformat-5.10.4.tar.gz", hash = "sha256:322168b14f937a5d11362988ecac2a4952d3d8e3a2cbeb2319584631226d5b3a"},
]
[package.dependencies]
-fastjsonschema = "*"
+fastjsonschema = ">=2.15"
jsonschema = ">=2.6"
-jupyter-core = "*"
+jupyter-core = ">=4.12,<5.0.dev0 || >=5.1.dev0"
traitlets = ">=5.1"
[package.extras]
@@ -2055,6 +2213,17 @@ nbformat = "*"
sphinx = ">=1.8"
traitlets = ">=5"
+[[package]]
+name = "nest-asyncio"
+version = "1.6.0"
+description = "Patch asyncio to allow nested event loops"
+optional = false
+python-versions = ">=3.5"
+files = [
+ {file = "nest_asyncio-1.6.0-py3-none-any.whl", hash = "sha256:87af6efd6b5e897c81050477ef65c62e2b2f35d51703cae01aff2905b1852e1c"},
+ {file = "nest_asyncio-1.6.0.tar.gz", hash = "sha256:6f172d5449aca15afd6c646851f4e31e02c598d553a667e38cafa997cfec55fe"},
+]
+
[[package]]
name = "nodeenv"
version = "1.8.0"
@@ -2145,6 +2314,16 @@ files = [
{file = "packaging-24.0.tar.gz", hash = "sha256:eb82c5e3e56209074766e6885bb04b8c38a0c015d0a30036ebe7ece34c9989e9"},
]
+[[package]]
+name = "paginate"
+version = "0.5.6"
+description = "Divides large result sets into pages for easier browsing"
+optional = false
+python-versions = "*"
+files = [
+ {file = "paginate-0.5.6.tar.gz", hash = "sha256:5e6007b6a9398177a7e1648d04fdd9f8c9766a1a945bceac82f1929e8c78af2d"},
+]
+
[[package]]
name = "pandas"
version = "2.0.3"
@@ -2236,18 +2415,18 @@ files = [
[[package]]
name = "parso"
-version = "0.8.3"
+version = "0.8.4"
description = "A Python Parser"
optional = false
python-versions = ">=3.6"
files = [
- {file = "parso-0.8.3-py2.py3-none-any.whl", hash = "sha256:c001d4636cd3aecdaf33cbb40aebb59b094be2a74c556778ef5576c175e19e75"},
- {file = "parso-0.8.3.tar.gz", hash = "sha256:8c07be290bb59f03588915921e29e8a50002acaf2cdc5fa0e0114f91709fafa0"},
+ {file = "parso-0.8.4-py2.py3-none-any.whl", hash = "sha256:a418670a20291dacd2dddc80c377c5c3791378ee1e8d12bffc35420643d43f18"},
+ {file = "parso-0.8.4.tar.gz", hash = "sha256:eb3a7b58240fb99099a345571deecc0f9540ea5f4dd2fe14c2a99d6b281ab92d"},
]
[package.extras]
-qa = ["flake8 (==3.8.3)", "mypy (==0.782)"]
-testing = ["docopt", "pytest (<6.0.0)"]
+qa = ["flake8 (==5.0.4)", "mypy (==0.971)", "types-setuptools (==67.2.0.1)"]
+testing = ["docopt", "pytest"]
[[package]]
name = "pathspec"
@@ -2298,28 +2477,29 @@ files = [
[[package]]
name = "platformdirs"
-version = "4.2.0"
-description = "A small Python package for determining appropriate platform-specific dirs, e.g. a \"user data dir\"."
+version = "4.2.1"
+description = "A small Python package for determining appropriate platform-specific dirs, e.g. a `user data dir`."
optional = false
python-versions = ">=3.8"
files = [
- {file = "platformdirs-4.2.0-py3-none-any.whl", hash = "sha256:0614df2a2f37e1a662acbd8e2b25b92ccf8632929bc6d43467e17fe89c75e068"},
- {file = "platformdirs-4.2.0.tar.gz", hash = "sha256:ef0cc731df711022c174543cb70a9b5bd22e5a9337c8624ef2c2ceb8ddad8768"},
+ {file = "platformdirs-4.2.1-py3-none-any.whl", hash = "sha256:17d5a1161b3fd67b390023cb2d3b026bbd40abde6fdb052dfbd3a29c3ba22ee1"},
+ {file = "platformdirs-4.2.1.tar.gz", hash = "sha256:031cd18d4ec63ec53e82dceaac0417d218a6863f7745dfcc9efe7793b7039bdf"},
]
[package.extras]
docs = ["furo (>=2023.9.10)", "proselint (>=0.13)", "sphinx (>=7.2.6)", "sphinx-autodoc-typehints (>=1.25.2)"]
test = ["appdirs (==1.4.4)", "covdefaults (>=2.3)", "pytest (>=7.4.3)", "pytest-cov (>=4.1)", "pytest-mock (>=3.12)"]
+type = ["mypy (>=1.8)"]
[[package]]
name = "pluggy"
-version = "1.4.0"
+version = "1.5.0"
description = "plugin and hook calling mechanisms for python"
optional = false
python-versions = ">=3.8"
files = [
- {file = "pluggy-1.4.0-py3-none-any.whl", hash = "sha256:7db9f7b503d67d1c5b95f59773ebb58a8c1c288129a88665838012cfb07b8981"},
- {file = "pluggy-1.4.0.tar.gz", hash = "sha256:8c85c2876142a764e5b7548e7d9a0e0ddb46f5185161049a79b7e974454223be"},
+ {file = "pluggy-1.5.0-py3-none-any.whl", hash = "sha256:44e1ad92c8ca002de6377e165f3e0f1be63266ab4d554740532335b9d75ea669"},
+ {file = "pluggy-1.5.0.tar.gz", hash = "sha256:2cffa88e94fdc978c4c574f15f9e59b7f4201d439195c3715ca9e2486f1d0cf1"},
]
[package.extras]
@@ -2358,6 +2538,34 @@ files = [
[package.dependencies]
wcwidth = "*"
+[[package]]
+name = "psutil"
+version = "5.9.8"
+description = "Cross-platform lib for process and system monitoring in Python."
+optional = false
+python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*, !=3.3.*, !=3.4.*, !=3.5.*"
+files = [
+ {file = "psutil-5.9.8-cp27-cp27m-macosx_10_9_x86_64.whl", hash = "sha256:26bd09967ae00920df88e0352a91cff1a78f8d69b3ecabbfe733610c0af486c8"},
+ {file = "psutil-5.9.8-cp27-cp27m-manylinux2010_i686.whl", hash = "sha256:05806de88103b25903dff19bb6692bd2e714ccf9e668d050d144012055cbca73"},
+ {file = "psutil-5.9.8-cp27-cp27m-manylinux2010_x86_64.whl", hash = "sha256:611052c4bc70432ec770d5d54f64206aa7203a101ec273a0cd82418c86503bb7"},
+ {file = "psutil-5.9.8-cp27-cp27mu-manylinux2010_i686.whl", hash = "sha256:50187900d73c1381ba1454cf40308c2bf6f34268518b3f36a9b663ca87e65e36"},
+ {file = "psutil-5.9.8-cp27-cp27mu-manylinux2010_x86_64.whl", hash = "sha256:02615ed8c5ea222323408ceba16c60e99c3f91639b07da6373fb7e6539abc56d"},
+ {file = "psutil-5.9.8-cp27-none-win32.whl", hash = "sha256:36f435891adb138ed3c9e58c6af3e2e6ca9ac2f365efe1f9cfef2794e6c93b4e"},
+ {file = "psutil-5.9.8-cp27-none-win_amd64.whl", hash = "sha256:bd1184ceb3f87651a67b2708d4c3338e9b10c5df903f2e3776b62303b26cb631"},
+ {file = "psutil-5.9.8-cp36-abi3-macosx_10_9_x86_64.whl", hash = "sha256:aee678c8720623dc456fa20659af736241f575d79429a0e5e9cf88ae0605cc81"},
+ {file = "psutil-5.9.8-cp36-abi3-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:8cb6403ce6d8e047495a701dc7c5bd788add903f8986d523e3e20b98b733e421"},
+ {file = "psutil-5.9.8-cp36-abi3-manylinux_2_12_x86_64.manylinux2010_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:d06016f7f8625a1825ba3732081d77c94589dca78b7a3fc072194851e88461a4"},
+ {file = "psutil-5.9.8-cp36-cp36m-win32.whl", hash = "sha256:7d79560ad97af658a0f6adfef8b834b53f64746d45b403f225b85c5c2c140eee"},
+ {file = "psutil-5.9.8-cp36-cp36m-win_amd64.whl", hash = "sha256:27cc40c3493bb10de1be4b3f07cae4c010ce715290a5be22b98493509c6299e2"},
+ {file = "psutil-5.9.8-cp37-abi3-win32.whl", hash = "sha256:bc56c2a1b0d15aa3eaa5a60c9f3f8e3e565303b465dbf57a1b730e7a2b9844e0"},
+ {file = "psutil-5.9.8-cp37-abi3-win_amd64.whl", hash = "sha256:8db4c1b57507eef143a15a6884ca10f7c73876cdf5d51e713151c1236a0e68cf"},
+ {file = "psutil-5.9.8-cp38-abi3-macosx_11_0_arm64.whl", hash = "sha256:d16bbddf0693323b8c6123dd804100241da461e41d6e332fb0ba6058f630f8c8"},
+ {file = "psutil-5.9.8.tar.gz", hash = "sha256:6be126e3225486dff286a8fb9a06246a5253f4c7c53b475ea5f5ac934e64194c"},
+]
+
+[package.extras]
+test = ["enum34", "ipaddress", "mock", "pywin32", "wmi"]
+
[[package]]
name = "ptyprocess"
version = "0.7.0"
@@ -2385,61 +2593,61 @@ tests = ["pytest"]
[[package]]
name = "pyarrow"
-version = "15.0.2"
+version = "16.0.0"
description = "Python library for Apache Arrow"
optional = false
python-versions = ">=3.8"
files = [
- {file = "pyarrow-15.0.2-cp310-cp310-macosx_10_15_x86_64.whl", hash = "sha256:88b340f0a1d05b5ccc3d2d986279045655b1fe8e41aba6ca44ea28da0d1455d8"},
- {file = "pyarrow-15.0.2-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:eaa8f96cecf32da508e6c7f69bb8401f03745c050c1dd42ec2596f2e98deecac"},
- {file = "pyarrow-15.0.2-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:23c6753ed4f6adb8461e7c383e418391b8d8453c5d67e17f416c3a5d5709afbd"},
- {file = "pyarrow-15.0.2-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:f639c059035011db8c0497e541a8a45d98a58dbe34dc8fadd0ef128f2cee46e5"},
- {file = "pyarrow-15.0.2-cp310-cp310-manylinux_2_28_aarch64.whl", hash = "sha256:290e36a59a0993e9a5224ed2fb3e53375770f07379a0ea03ee2fce2e6d30b423"},
- {file = "pyarrow-15.0.2-cp310-cp310-manylinux_2_28_x86_64.whl", hash = "sha256:06c2bb2a98bc792f040bef31ad3e9be6a63d0cb39189227c08a7d955db96816e"},
- {file = "pyarrow-15.0.2-cp310-cp310-win_amd64.whl", hash = "sha256:f7a197f3670606a960ddc12adbe8075cea5f707ad7bf0dffa09637fdbb89f76c"},
- {file = "pyarrow-15.0.2-cp311-cp311-macosx_10_15_x86_64.whl", hash = "sha256:5f8bc839ea36b1f99984c78e06e7a06054693dc2af8920f6fb416b5bca9944e4"},
- {file = "pyarrow-15.0.2-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:f5e81dfb4e519baa6b4c80410421528c214427e77ca0ea9461eb4097c328fa33"},
- {file = "pyarrow-15.0.2-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3a4f240852b302a7af4646c8bfe9950c4691a419847001178662a98915fd7ee7"},
- {file = "pyarrow-15.0.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:4e7d9cfb5a1e648e172428c7a42b744610956f3b70f524aa3a6c02a448ba853e"},
- {file = "pyarrow-15.0.2-cp311-cp311-manylinux_2_28_aarch64.whl", hash = "sha256:2d4f905209de70c0eb5b2de6763104d5a9a37430f137678edfb9a675bac9cd98"},
- {file = "pyarrow-15.0.2-cp311-cp311-manylinux_2_28_x86_64.whl", hash = "sha256:90adb99e8ce5f36fbecbbc422e7dcbcbed07d985eed6062e459e23f9e71fd197"},
- {file = "pyarrow-15.0.2-cp311-cp311-win_amd64.whl", hash = "sha256:b116e7fd7889294cbd24eb90cd9bdd3850be3738d61297855a71ac3b8124ee38"},
- {file = "pyarrow-15.0.2-cp312-cp312-macosx_10_15_x86_64.whl", hash = "sha256:25335e6f1f07fdaa026a61c758ee7d19ce824a866b27bba744348fa73bb5a440"},
- {file = "pyarrow-15.0.2-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:90f19e976d9c3d8e73c80be84ddbe2f830b6304e4c576349d9360e335cd627fc"},
- {file = "pyarrow-15.0.2-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:a22366249bf5fd40ddacc4f03cd3160f2d7c247692945afb1899bab8a140ddfb"},
- {file = "pyarrow-15.0.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:c2a335198f886b07e4b5ea16d08ee06557e07db54a8400cc0d03c7f6a22f785f"},
- {file = "pyarrow-15.0.2-cp312-cp312-manylinux_2_28_aarch64.whl", hash = "sha256:3e6d459c0c22f0b9c810a3917a1de3ee704b021a5fb8b3bacf968eece6df098f"},
- {file = "pyarrow-15.0.2-cp312-cp312-manylinux_2_28_x86_64.whl", hash = "sha256:033b7cad32198754d93465dcfb71d0ba7cb7cd5c9afd7052cab7214676eec38b"},
- {file = "pyarrow-15.0.2-cp312-cp312-win_amd64.whl", hash = "sha256:29850d050379d6e8b5a693098f4de7fd6a2bea4365bfd073d7c57c57b95041ee"},
- {file = "pyarrow-15.0.2-cp38-cp38-macosx_10_15_x86_64.whl", hash = "sha256:7167107d7fb6dcadb375b4b691b7e316f4368f39f6f45405a05535d7ad5e5058"},
- {file = "pyarrow-15.0.2-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:e85241b44cc3d365ef950432a1b3bd44ac54626f37b2e3a0cc89c20e45dfd8bf"},
- {file = "pyarrow-15.0.2-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:248723e4ed3255fcd73edcecc209744d58a9ca852e4cf3d2577811b6d4b59818"},
- {file = "pyarrow-15.0.2-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:3ff3bdfe6f1b81ca5b73b70a8d482d37a766433823e0c21e22d1d7dde76ca33f"},
- {file = "pyarrow-15.0.2-cp38-cp38-manylinux_2_28_aarch64.whl", hash = "sha256:f3d77463dee7e9f284ef42d341689b459a63ff2e75cee2b9302058d0d98fe142"},
- {file = "pyarrow-15.0.2-cp38-cp38-manylinux_2_28_x86_64.whl", hash = "sha256:8c1faf2482fb89766e79745670cbca04e7018497d85be9242d5350cba21357e1"},
- {file = "pyarrow-15.0.2-cp38-cp38-win_amd64.whl", hash = "sha256:28f3016958a8e45a1069303a4a4f6a7d4910643fc08adb1e2e4a7ff056272ad3"},
- {file = "pyarrow-15.0.2-cp39-cp39-macosx_10_15_x86_64.whl", hash = "sha256:89722cb64286ab3d4daf168386f6968c126057b8c7ec3ef96302e81d8cdb8ae4"},
- {file = "pyarrow-15.0.2-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:cd0ba387705044b3ac77b1b317165c0498299b08261d8122c96051024f953cd5"},
- {file = "pyarrow-15.0.2-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ad2459bf1f22b6a5cdcc27ebfd99307d5526b62d217b984b9f5c974651398832"},
- {file = "pyarrow-15.0.2-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:58922e4bfece8b02abf7159f1f53a8f4d9f8e08f2d988109126c17c3bb261f22"},
- {file = "pyarrow-15.0.2-cp39-cp39-manylinux_2_28_aarch64.whl", hash = "sha256:adccc81d3dc0478ea0b498807b39a8d41628fa9210729b2f718b78cb997c7c91"},
- {file = "pyarrow-15.0.2-cp39-cp39-manylinux_2_28_x86_64.whl", hash = "sha256:8bd2baa5fe531571847983f36a30ddbf65261ef23e496862ece83bdceb70420d"},
- {file = "pyarrow-15.0.2-cp39-cp39-win_amd64.whl", hash = "sha256:6669799a1d4ca9da9c7e06ef48368320f5856f36f9a4dd31a11839dda3f6cc8c"},
- {file = "pyarrow-15.0.2.tar.gz", hash = "sha256:9c9bc803cb3b7bfacc1e96ffbfd923601065d9d3f911179d81e72d99fd74a3d9"},
-]
-
-[package.dependencies]
-numpy = ">=1.16.6,<2"
+ {file = "pyarrow-16.0.0-cp310-cp310-macosx_10_15_x86_64.whl", hash = "sha256:22a1fdb1254e5095d629e29cd1ea98ed04b4bbfd8e42cc670a6b639ccc208b60"},
+ {file = "pyarrow-16.0.0-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:574a00260a4ed9d118a14770edbd440b848fcae5a3024128be9d0274dbcaf858"},
+ {file = "pyarrow-16.0.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:c0815d0ddb733b8c1b53a05827a91f1b8bde6240f3b20bf9ba5d650eb9b89cdf"},
+ {file = "pyarrow-16.0.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:df0080339387b5d30de31e0a149c0c11a827a10c82f0c67d9afae3981d1aabb7"},
+ {file = "pyarrow-16.0.0-cp310-cp310-manylinux_2_28_aarch64.whl", hash = "sha256:edf38cce0bf0dcf726e074159c60516447e4474904c0033f018c1f33d7dac6c5"},
+ {file = "pyarrow-16.0.0-cp310-cp310-manylinux_2_28_x86_64.whl", hash = "sha256:91d28f9a40f1264eab2af7905a4d95320ac2f287891e9c8b0035f264fe3c3a4b"},
+ {file = "pyarrow-16.0.0-cp310-cp310-win_amd64.whl", hash = "sha256:99af421ee451a78884d7faea23816c429e263bd3618b22d38e7992c9ce2a7ad9"},
+ {file = "pyarrow-16.0.0-cp311-cp311-macosx_10_15_x86_64.whl", hash = "sha256:d22d0941e6c7bafddf5f4c0662e46f2075850f1c044bf1a03150dd9e189427ce"},
+ {file = "pyarrow-16.0.0-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:266ddb7e823f03733c15adc8b5078db2df6980f9aa93d6bb57ece615df4e0ba7"},
+ {file = "pyarrow-16.0.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:5cc23090224b6594f5a92d26ad47465af47c1d9c079dd4a0061ae39551889efe"},
+ {file = "pyarrow-16.0.0-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:56850a0afe9ef37249d5387355449c0f94d12ff7994af88f16803a26d38f2016"},
+ {file = "pyarrow-16.0.0-cp311-cp311-manylinux_2_28_aarch64.whl", hash = "sha256:705db70d3e2293c2f6f8e84874b5b775f690465798f66e94bb2c07bab0a6bb55"},
+ {file = "pyarrow-16.0.0-cp311-cp311-manylinux_2_28_x86_64.whl", hash = "sha256:5448564754c154997bc09e95a44b81b9e31ae918a86c0fcb35c4aa4922756f55"},
+ {file = "pyarrow-16.0.0-cp311-cp311-win_amd64.whl", hash = "sha256:729f7b262aa620c9df8b9967db96c1575e4cfc8c25d078a06968e527b8d6ec05"},
+ {file = "pyarrow-16.0.0-cp312-cp312-macosx_10_15_x86_64.whl", hash = "sha256:fb8065dbc0d051bf2ae2453af0484d99a43135cadabacf0af588a3be81fbbb9b"},
+ {file = "pyarrow-16.0.0-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:20ce707d9aa390593ea93218b19d0eadab56390311cb87aad32c9a869b0e958c"},
+ {file = "pyarrow-16.0.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:5823275c8addbbb50cd4e6a6839952682a33255b447277e37a6f518d6972f4e1"},
+ {file = "pyarrow-16.0.0-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:1ab8b9050752b16a8b53fcd9853bf07d8daf19093533e990085168f40c64d978"},
+ {file = "pyarrow-16.0.0-cp312-cp312-manylinux_2_28_aarch64.whl", hash = "sha256:42e56557bc7c5c10d3e42c3b32f6cff649a29d637e8f4e8b311d334cc4326730"},
+ {file = "pyarrow-16.0.0-cp312-cp312-manylinux_2_28_x86_64.whl", hash = "sha256:2a7abdee4a4a7cfa239e2e8d721224c4b34ffe69a0ca7981354fe03c1328789b"},
+ {file = "pyarrow-16.0.0-cp312-cp312-win_amd64.whl", hash = "sha256:ef2f309b68396bcc5a354106741d333494d6a0d3e1951271849787109f0229a6"},
+ {file = "pyarrow-16.0.0-cp38-cp38-macosx_10_15_x86_64.whl", hash = "sha256:ed66e5217b4526fa3585b5e39b0b82f501b88a10d36bd0d2a4d8aa7b5a48e2df"},
+ {file = "pyarrow-16.0.0-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:cc8814310486f2a73c661ba8354540f17eef51e1b6dd090b93e3419d3a097b3a"},
+ {file = "pyarrow-16.0.0-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3c2f5e239db7ed43e0ad2baf46a6465f89c824cc703f38ef0fde927d8e0955f7"},
+ {file = "pyarrow-16.0.0-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:f293e92d1db251447cb028ae12f7bc47526e4649c3a9924c8376cab4ad6b98bd"},
+ {file = "pyarrow-16.0.0-cp38-cp38-manylinux_2_28_aarch64.whl", hash = "sha256:dd9334a07b6dc21afe0857aa31842365a62eca664e415a3f9536e3a8bb832c07"},
+ {file = "pyarrow-16.0.0-cp38-cp38-manylinux_2_28_x86_64.whl", hash = "sha256:d91073d1e2fef2c121154680e2ba7e35ecf8d4969cc0af1fa6f14a8675858159"},
+ {file = "pyarrow-16.0.0-cp38-cp38-win_amd64.whl", hash = "sha256:71d52561cd7aefd22cf52538f262850b0cc9e4ec50af2aaa601da3a16ef48877"},
+ {file = "pyarrow-16.0.0-cp39-cp39-macosx_10_15_x86_64.whl", hash = "sha256:b93c9a50b965ee0bf4fef65e53b758a7e8dcc0c2d86cebcc037aaaf1b306ecc0"},
+ {file = "pyarrow-16.0.0-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:d831690844706e374c455fba2fb8cfcb7b797bfe53ceda4b54334316e1ac4fa4"},
+ {file = "pyarrow-16.0.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:35692ce8ad0b8c666aa60f83950957096d92f2a9d8d7deda93fb835e6053307e"},
+ {file = "pyarrow-16.0.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:9dd3151d098e56f16a8389c1247137f9e4c22720b01c6f3aa6dec29a99b74d80"},
+ {file = "pyarrow-16.0.0-cp39-cp39-manylinux_2_28_aarch64.whl", hash = "sha256:bd40467bdb3cbaf2044ed7a6f7f251c8f941c8b31275aaaf88e746c4f3ca4a7a"},
+ {file = "pyarrow-16.0.0-cp39-cp39-manylinux_2_28_x86_64.whl", hash = "sha256:00a1dcb22ad4ceb8af87f7bd30cc3354788776c417f493089e0a0af981bc8d80"},
+ {file = "pyarrow-16.0.0-cp39-cp39-win_amd64.whl", hash = "sha256:fda9a7cebd1b1d46c97b511f60f73a5b766a6de4c5236f144f41a5d5afec1f35"},
+ {file = "pyarrow-16.0.0.tar.gz", hash = "sha256:59bb1f1edbbf4114c72415f039f1359f1a57d166a331c3229788ccbfbb31689a"},
+]
+
+[package.dependencies]
+numpy = ">=1.16.6"
[[package]]
name = "pycparser"
-version = "2.21"
+version = "2.22"
description = "C parser in Python"
optional = false
-python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*, !=3.3.*"
+python-versions = ">=3.8"
files = [
- {file = "pycparser-2.21-py2.py3-none-any.whl", hash = "sha256:8ee45429555515e1f6b185e78100aea234072576aa43ab53aefcae078162fca9"},
- {file = "pycparser-2.21.tar.gz", hash = "sha256:e644fdec12f7872f86c58ff790da456218b10f863970249516d60a5eaca77206"},
+ {file = "pycparser-2.22-py3-none-any.whl", hash = "sha256:c3702b6d3dd8c7abc1afa565d7e63d53a1d0bd86cdc24edd75470f4de499cfcc"},
+ {file = "pycparser-2.22.tar.gz", hash = "sha256:491c8be9c040f5390f5bf44a5b07752bd07f56edf992381b05c701439eec10f6"},
]
[[package]]
@@ -2457,6 +2665,24 @@ files = [
plugins = ["importlib-metadata"]
windows-terminal = ["colorama (>=0.4.6)"]
+[[package]]
+name = "pymdown-extensions"
+version = "10.8"
+description = "Extension pack for Python Markdown."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "pymdown_extensions-10.8-py3-none-any.whl", hash = "sha256:3539003ff0d5e219ba979d2dc961d18fcad5ac259e66c764482e8347b4c0503c"},
+ {file = "pymdown_extensions-10.8.tar.gz", hash = "sha256:91ca336caf414e1e5e0626feca86e145de9f85a3921a7bcbd32890b51738c428"},
+]
+
+[package.dependencies]
+markdown = ">=3.6"
+pyyaml = "*"
+
+[package.extras]
+extra = ["pygments (>=2.12)"]
+
[[package]]
name = "pytest"
version = "8.1.1"
@@ -2649,104 +2875,99 @@ pyyaml = "*"
[[package]]
name = "pyzmq"
-version = "25.1.2"
+version = "26.0.2"
description = "Python bindings for 0MQ"
optional = false
-python-versions = ">=3.6"
+python-versions = ">=3.7"
files = [
- {file = "pyzmq-25.1.2-cp310-cp310-macosx_10_15_universal2.whl", hash = "sha256:e624c789359f1a16f83f35e2c705d07663ff2b4d4479bad35621178d8f0f6ea4"},
- {file = "pyzmq-25.1.2-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:49151b0efece79f6a79d41a461d78535356136ee70084a1c22532fc6383f4ad0"},
- {file = "pyzmq-25.1.2-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d9a5f194cf730f2b24d6af1f833c14c10f41023da46a7f736f48b6d35061e76e"},
- {file = "pyzmq-25.1.2-cp310-cp310-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:faf79a302f834d9e8304fafdc11d0d042266667ac45209afa57e5efc998e3872"},
- {file = "pyzmq-25.1.2-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:7f51a7b4ead28d3fca8dda53216314a553b0f7a91ee8fc46a72b402a78c3e43d"},
- {file = "pyzmq-25.1.2-cp310-cp310-manylinux_2_28_x86_64.whl", hash = "sha256:0ddd6d71d4ef17ba5a87becf7ddf01b371eaba553c603477679ae817a8d84d75"},
- {file = "pyzmq-25.1.2-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:246747b88917e4867e2367b005fc8eefbb4a54b7db363d6c92f89d69abfff4b6"},
- {file = "pyzmq-25.1.2-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:00c48ae2fd81e2a50c3485de1b9d5c7c57cd85dc8ec55683eac16846e57ac979"},
- {file = "pyzmq-25.1.2-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:5a68d491fc20762b630e5db2191dd07ff89834086740f70e978bb2ef2668be08"},
- {file = "pyzmq-25.1.2-cp310-cp310-win32.whl", hash = "sha256:09dfe949e83087da88c4a76767df04b22304a682d6154de2c572625c62ad6886"},
- {file = "pyzmq-25.1.2-cp310-cp310-win_amd64.whl", hash = "sha256:fa99973d2ed20417744fca0073390ad65ce225b546febb0580358e36aa90dba6"},
- {file = "pyzmq-25.1.2-cp311-cp311-macosx_10_15_universal2.whl", hash = "sha256:82544e0e2d0c1811482d37eef297020a040c32e0687c1f6fc23a75b75db8062c"},
- {file = "pyzmq-25.1.2-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:01171fc48542348cd1a360a4b6c3e7d8f46cdcf53a8d40f84db6707a6768acc1"},
- {file = "pyzmq-25.1.2-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:bc69c96735ab501419c432110016329bf0dea8898ce16fab97c6d9106dc0b348"},
- {file = "pyzmq-25.1.2-cp311-cp311-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:3e124e6b1dd3dfbeb695435dff0e383256655bb18082e094a8dd1f6293114642"},
- {file = "pyzmq-25.1.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:7598d2ba821caa37a0f9d54c25164a4fa351ce019d64d0b44b45540950458840"},
- {file = "pyzmq-25.1.2-cp311-cp311-manylinux_2_28_x86_64.whl", hash = "sha256:d1299d7e964c13607efd148ca1f07dcbf27c3ab9e125d1d0ae1d580a1682399d"},
- {file = "pyzmq-25.1.2-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:4e6f689880d5ad87918430957297c975203a082d9a036cc426648fcbedae769b"},
- {file = "pyzmq-25.1.2-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:cc69949484171cc961e6ecd4a8911b9ce7a0d1f738fcae717177c231bf77437b"},
- {file = "pyzmq-25.1.2-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:9880078f683466b7f567b8624bfc16cad65077be046b6e8abb53bed4eeb82dd3"},
- {file = "pyzmq-25.1.2-cp311-cp311-win32.whl", hash = "sha256:4e5837af3e5aaa99a091302df5ee001149baff06ad22b722d34e30df5f0d9097"},
- {file = "pyzmq-25.1.2-cp311-cp311-win_amd64.whl", hash = "sha256:25c2dbb97d38b5ac9fd15586e048ec5eb1e38f3d47fe7d92167b0c77bb3584e9"},
- {file = "pyzmq-25.1.2-cp312-cp312-macosx_10_15_universal2.whl", hash = "sha256:11e70516688190e9c2db14fcf93c04192b02d457b582a1f6190b154691b4c93a"},
- {file = "pyzmq-25.1.2-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:313c3794d650d1fccaaab2df942af9f2c01d6217c846177cfcbc693c7410839e"},
- {file = "pyzmq-25.1.2-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:1b3cbba2f47062b85fe0ef9de5b987612140a9ba3a9c6d2543c6dec9f7c2ab27"},
- {file = "pyzmq-25.1.2-cp312-cp312-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:fc31baa0c32a2ca660784d5af3b9487e13b61b3032cb01a115fce6588e1bed30"},
- {file = "pyzmq-25.1.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:02c9087b109070c5ab0b383079fa1b5f797f8d43e9a66c07a4b8b8bdecfd88ee"},
- {file = "pyzmq-25.1.2-cp312-cp312-manylinux_2_28_x86_64.whl", hash = "sha256:f8429b17cbb746c3e043cb986328da023657e79d5ed258b711c06a70c2ea7537"},
- {file = "pyzmq-25.1.2-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:5074adeacede5f810b7ef39607ee59d94e948b4fd954495bdb072f8c54558181"},
- {file = "pyzmq-25.1.2-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:7ae8f354b895cbd85212da245f1a5ad8159e7840e37d78b476bb4f4c3f32a9fe"},
- {file = "pyzmq-25.1.2-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:b264bf2cc96b5bc43ce0e852be995e400376bd87ceb363822e2cb1964fcdc737"},
- {file = "pyzmq-25.1.2-cp312-cp312-win32.whl", hash = "sha256:02bbc1a87b76e04fd780b45e7f695471ae6de747769e540da909173d50ff8e2d"},
- {file = "pyzmq-25.1.2-cp312-cp312-win_amd64.whl", hash = "sha256:ced111c2e81506abd1dc142e6cd7b68dd53747b3b7ae5edbea4578c5eeff96b7"},
- {file = "pyzmq-25.1.2-cp36-cp36m-macosx_10_9_x86_64.whl", hash = "sha256:7b6d09a8962a91151f0976008eb7b29b433a560fde056ec7a3db9ec8f1075438"},
- {file = "pyzmq-25.1.2-cp36-cp36m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:967668420f36878a3c9ecb5ab33c9d0ff8d054f9c0233d995a6d25b0e95e1b6b"},
- {file = "pyzmq-25.1.2-cp36-cp36m-manylinux_2_5_i686.manylinux1_i686.whl", hash = "sha256:5edac3f57c7ddaacdb4d40f6ef2f9e299471fc38d112f4bc6d60ab9365445fb0"},
- {file = "pyzmq-25.1.2-cp36-cp36m-manylinux_2_5_x86_64.manylinux1_x86_64.whl", hash = "sha256:0dabfb10ef897f3b7e101cacba1437bd3a5032ee667b7ead32bbcdd1a8422fe7"},
- {file = "pyzmq-25.1.2-cp36-cp36m-musllinux_1_1_aarch64.whl", hash = "sha256:2c6441e0398c2baacfe5ba30c937d274cfc2dc5b55e82e3749e333aabffde561"},
- {file = "pyzmq-25.1.2-cp36-cp36m-musllinux_1_1_i686.whl", hash = "sha256:16b726c1f6c2e7625706549f9dbe9b06004dfbec30dbed4bf50cbdfc73e5b32a"},
- {file = "pyzmq-25.1.2-cp36-cp36m-musllinux_1_1_x86_64.whl", hash = "sha256:a86c2dd76ef71a773e70551a07318b8e52379f58dafa7ae1e0a4be78efd1ff16"},
- {file = "pyzmq-25.1.2-cp36-cp36m-win32.whl", hash = "sha256:359f7f74b5d3c65dae137f33eb2bcfa7ad9ebefd1cab85c935f063f1dbb245cc"},
- {file = "pyzmq-25.1.2-cp36-cp36m-win_amd64.whl", hash = "sha256:55875492f820d0eb3417b51d96fea549cde77893ae3790fd25491c5754ea2f68"},
- {file = "pyzmq-25.1.2-cp37-cp37m-macosx_10_9_x86_64.whl", hash = "sha256:b8c8a419dfb02e91b453615c69568442e897aaf77561ee0064d789705ff37a92"},
- {file = "pyzmq-25.1.2-cp37-cp37m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:8807c87fa893527ae8a524c15fc505d9950d5e856f03dae5921b5e9aa3b8783b"},
- {file = "pyzmq-25.1.2-cp37-cp37m-manylinux_2_5_i686.manylinux1_i686.whl", hash = "sha256:5e319ed7d6b8f5fad9b76daa0a68497bc6f129858ad956331a5835785761e003"},
- {file = "pyzmq-25.1.2-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.whl", hash = "sha256:3c53687dde4d9d473c587ae80cc328e5b102b517447456184b485587ebd18b62"},
- {file = "pyzmq-25.1.2-cp37-cp37m-musllinux_1_1_aarch64.whl", hash = "sha256:9add2e5b33d2cd765ad96d5eb734a5e795a0755f7fc49aa04f76d7ddda73fd70"},
- {file = "pyzmq-25.1.2-cp37-cp37m-musllinux_1_1_i686.whl", hash = "sha256:e690145a8c0c273c28d3b89d6fb32c45e0d9605b2293c10e650265bf5c11cfec"},
- {file = "pyzmq-25.1.2-cp37-cp37m-musllinux_1_1_x86_64.whl", hash = "sha256:00a06faa7165634f0cac1abb27e54d7a0b3b44eb9994530b8ec73cf52e15353b"},
- {file = "pyzmq-25.1.2-cp37-cp37m-win32.whl", hash = "sha256:0f97bc2f1f13cb16905a5f3e1fbdf100e712d841482b2237484360f8bc4cb3d7"},
- {file = "pyzmq-25.1.2-cp37-cp37m-win_amd64.whl", hash = "sha256:6cc0020b74b2e410287e5942e1e10886ff81ac77789eb20bec13f7ae681f0fdd"},
- {file = "pyzmq-25.1.2-cp38-cp38-macosx_10_15_universal2.whl", hash = "sha256:bef02cfcbded83473bdd86dd8d3729cd82b2e569b75844fb4ea08fee3c26ae41"},
- {file = "pyzmq-25.1.2-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:e10a4b5a4b1192d74853cc71a5e9fd022594573926c2a3a4802020360aa719d8"},
- {file = "pyzmq-25.1.2-cp38-cp38-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:8c5f80e578427d4695adac6fdf4370c14a2feafdc8cb35549c219b90652536ae"},
- {file = "pyzmq-25.1.2-cp38-cp38-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:5dde6751e857910c1339890f3524de74007958557593b9e7e8c5f01cd919f8a7"},
- {file = "pyzmq-25.1.2-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ea1608dd169da230a0ad602d5b1ebd39807ac96cae1845c3ceed39af08a5c6df"},
- {file = "pyzmq-25.1.2-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:0f513130c4c361201da9bc69df25a086487250e16b5571ead521b31ff6b02220"},
- {file = "pyzmq-25.1.2-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:019744b99da30330798bb37df33549d59d380c78e516e3bab9c9b84f87a9592f"},
- {file = "pyzmq-25.1.2-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:2e2713ef44be5d52dd8b8e2023d706bf66cb22072e97fc71b168e01d25192755"},
- {file = "pyzmq-25.1.2-cp38-cp38-win32.whl", hash = "sha256:07cd61a20a535524906595e09344505a9bd46f1da7a07e504b315d41cd42eb07"},
- {file = "pyzmq-25.1.2-cp38-cp38-win_amd64.whl", hash = "sha256:eb7e49a17fb8c77d3119d41a4523e432eb0c6932187c37deb6fbb00cc3028088"},
- {file = "pyzmq-25.1.2-cp39-cp39-macosx_10_15_universal2.whl", hash = "sha256:94504ff66f278ab4b7e03e4cba7e7e400cb73bfa9d3d71f58d8972a8dc67e7a6"},
- {file = "pyzmq-25.1.2-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:6dd0d50bbf9dca1d0bdea219ae6b40f713a3fb477c06ca3714f208fd69e16fd8"},
- {file = "pyzmq-25.1.2-cp39-cp39-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:004ff469d21e86f0ef0369717351073e0e577428e514c47c8480770d5e24a565"},
- {file = "pyzmq-25.1.2-cp39-cp39-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:c0b5ca88a8928147b7b1e2dfa09f3b6c256bc1135a1338536cbc9ea13d3b7add"},
- {file = "pyzmq-25.1.2-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:2c9a79f1d2495b167119d02be7448bfba57fad2a4207c4f68abc0bab4b92925b"},
- {file = "pyzmq-25.1.2-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:518efd91c3d8ac9f9b4f7dd0e2b7b8bf1a4fe82a308009016b07eaa48681af82"},
- {file = "pyzmq-25.1.2-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:1ec23bd7b3a893ae676d0e54ad47d18064e6c5ae1fadc2f195143fb27373f7f6"},
- {file = "pyzmq-25.1.2-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:db36c27baed588a5a8346b971477b718fdc66cf5b80cbfbd914b4d6d355e44e2"},
- {file = "pyzmq-25.1.2-cp39-cp39-win32.whl", hash = "sha256:39b1067f13aba39d794a24761e385e2eddc26295826530a8c7b6c6c341584289"},
- {file = "pyzmq-25.1.2-cp39-cp39-win_amd64.whl", hash = "sha256:8e9f3fabc445d0ce320ea2c59a75fe3ea591fdbdeebec5db6de530dd4b09412e"},
- {file = "pyzmq-25.1.2-pp310-pypy310_pp73-macosx_10_9_x86_64.whl", hash = "sha256:a8c1d566344aee826b74e472e16edae0a02e2a044f14f7c24e123002dcff1c05"},
- {file = "pyzmq-25.1.2-pp310-pypy310_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:759cfd391a0996345ba94b6a5110fca9c557ad4166d86a6e81ea526c376a01e8"},
- {file = "pyzmq-25.1.2-pp310-pypy310_pp73-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:7c61e346ac34b74028ede1c6b4bcecf649d69b707b3ff9dc0fab453821b04d1e"},
- {file = "pyzmq-25.1.2-pp310-pypy310_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:4cb8fc1f8d69b411b8ec0b5f1ffbcaf14c1db95b6bccea21d83610987435f1a4"},
- {file = "pyzmq-25.1.2-pp310-pypy310_pp73-win_amd64.whl", hash = "sha256:3c00c9b7d1ca8165c610437ca0c92e7b5607b2f9076f4eb4b095c85d6e680a1d"},
- {file = "pyzmq-25.1.2-pp37-pypy37_pp73-macosx_10_9_x86_64.whl", hash = "sha256:df0c7a16ebb94452d2909b9a7b3337940e9a87a824c4fc1c7c36bb4404cb0cde"},
- {file = "pyzmq-25.1.2-pp37-pypy37_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:45999e7f7ed5c390f2e87ece7f6c56bf979fb213550229e711e45ecc7d42ccb8"},
- {file = "pyzmq-25.1.2-pp37-pypy37_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:ac170e9e048b40c605358667aca3d94e98f604a18c44bdb4c102e67070f3ac9b"},
- {file = "pyzmq-25.1.2-pp37-pypy37_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d1b604734bec94f05f81b360a272fc824334267426ae9905ff32dc2be433ab96"},
- {file = "pyzmq-25.1.2-pp37-pypy37_pp73-win_amd64.whl", hash = "sha256:a793ac733e3d895d96f865f1806f160696422554e46d30105807fdc9841b9f7d"},
- {file = "pyzmq-25.1.2-pp38-pypy38_pp73-macosx_10_9_x86_64.whl", hash = "sha256:0806175f2ae5ad4b835ecd87f5f85583316b69f17e97786f7443baaf54b9bb98"},
- {file = "pyzmq-25.1.2-pp38-pypy38_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:ef12e259e7bc317c7597d4f6ef59b97b913e162d83b421dd0db3d6410f17a244"},
- {file = "pyzmq-25.1.2-pp38-pypy38_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:ea253b368eb41116011add00f8d5726762320b1bda892f744c91997b65754d73"},
- {file = "pyzmq-25.1.2-pp38-pypy38_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:1b9b1f2ad6498445a941d9a4fee096d387fee436e45cc660e72e768d3d8ee611"},
- {file = "pyzmq-25.1.2-pp38-pypy38_pp73-win_amd64.whl", hash = "sha256:8b14c75979ce932c53b79976a395cb2a8cd3aaf14aef75e8c2cb55a330b9b49d"},
- {file = "pyzmq-25.1.2-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:889370d5174a741a62566c003ee8ddba4b04c3f09a97b8000092b7ca83ec9c49"},
- {file = "pyzmq-25.1.2-pp39-pypy39_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:9a18fff090441a40ffda8a7f4f18f03dc56ae73f148f1832e109f9bffa85df15"},
- {file = "pyzmq-25.1.2-pp39-pypy39_pp73-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:99a6b36f95c98839ad98f8c553d8507644c880cf1e0a57fe5e3a3f3969040882"},
- {file = "pyzmq-25.1.2-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:4345c9a27f4310afbb9c01750e9461ff33d6fb74cd2456b107525bbeebcb5be3"},
- {file = "pyzmq-25.1.2-pp39-pypy39_pp73-manylinux_2_28_x86_64.whl", hash = "sha256:3516e0b6224cf6e43e341d56da15fd33bdc37fa0c06af4f029f7d7dfceceabbc"},
- {file = "pyzmq-25.1.2-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:146b9b1f29ead41255387fb07be56dc29639262c0f7344f570eecdcd8d683314"},
- {file = "pyzmq-25.1.2.tar.gz", hash = "sha256:93f1aa311e8bb912e34f004cf186407a4e90eec4f0ecc0efd26056bf7eda0226"},
+ {file = "pyzmq-26.0.2-cp310-cp310-macosx_10_15_universal2.whl", hash = "sha256:1a60a03b01e8c9c58932ec0cca15b1712d911c2800eb82d4281bc1ae5b6dad50"},
+ {file = "pyzmq-26.0.2-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:949067079e14ea1973bd740255e0840118c163d4bce8837f539d749f145cf5c3"},
+ {file = "pyzmq-26.0.2-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:37e7edfa6cf96d036a403775c96afa25058d1bb940a79786a9a2fc94a783abe3"},
+ {file = "pyzmq-26.0.2-cp310-cp310-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:903cc7a84a7d4326b43755c368780800e035aa3d711deae84a533fdffa8755b0"},
+ {file = "pyzmq-26.0.2-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:6cb2e41af165e5f327d06fbdd79a42a4e930267fade4e9f92d17f3ccce03f3a7"},
+ {file = "pyzmq-26.0.2-cp310-cp310-manylinux_2_28_x86_64.whl", hash = "sha256:55353b8189adcfc4c125fc4ce59d477744118e9c0ec379dd0999c5fa120ac4f5"},
+ {file = "pyzmq-26.0.2-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:f961423ff6236a752ced80057a20e623044df95924ed1009f844cde8b3a595f9"},
+ {file = "pyzmq-26.0.2-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:ba77fe84fe4f5f3dc0ef681a6d366685c8ffe1c8439c1d7530997b05ac06a04b"},
+ {file = "pyzmq-26.0.2-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:52589f0a745ef61b9c75c872cf91f8c1f7c0668eb3dd99d7abd639d8c0fb9ca7"},
+ {file = "pyzmq-26.0.2-cp310-cp310-win32.whl", hash = "sha256:b7b6d2a46c7afe2ad03ec8faf9967090c8ceae85c4d8934d17d7cae6f9062b64"},
+ {file = "pyzmq-26.0.2-cp310-cp310-win_amd64.whl", hash = "sha256:86531e20de249d9204cc6d8b13d5a30537748c78820215161d8a3b9ea58ca111"},
+ {file = "pyzmq-26.0.2-cp310-cp310-win_arm64.whl", hash = "sha256:f26a05029ecd2bd306b941ff8cb80f7620b7901421052bc429d238305b1cbf2f"},
+ {file = "pyzmq-26.0.2-cp311-cp311-macosx_10_15_universal2.whl", hash = "sha256:70770e296a9cb03d955540c99360aab861cbb3cba29516abbd106a15dbd91268"},
+ {file = "pyzmq-26.0.2-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:2740fd7161b39e178554ebf21aa5667a1c9ef0cd2cb74298fd4ef017dae7aec4"},
+ {file = "pyzmq-26.0.2-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:f5e3706c32dea077faa42b1c92d825b7f86c866f72532d342e0be5e64d14d858"},
+ {file = "pyzmq-26.0.2-cp311-cp311-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:0fa1416876194927f7723d6b7171b95e1115602967fc6bfccbc0d2d51d8ebae1"},
+ {file = "pyzmq-26.0.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:4ef9a79a48794099c57dc2df00340b5d47c5caa1792f9ddb8c7a26b1280bd575"},
+ {file = "pyzmq-26.0.2-cp311-cp311-manylinux_2_28_x86_64.whl", hash = "sha256:1c60fcdfa3229aeee4291c5d60faed3a813b18bdadb86299c4bf49e8e51e8605"},
+ {file = "pyzmq-26.0.2-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:e943c39c206b04df2eb5d71305761d7c3ca75fd49452115ea92db1b5b98dbdef"},
+ {file = "pyzmq-26.0.2-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:8da0ed8a598693731c76659880a668f4748b59158f26ed283a93f7f04d47447e"},
+ {file = "pyzmq-26.0.2-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:7bf51970b11d67096bede97cdbad0f4333f7664f4708b9b2acb352bf4faa3140"},
+ {file = "pyzmq-26.0.2-cp311-cp311-win32.whl", hash = "sha256:6f8e6bd5d066be605faa9fe5ec10aa1a46ad9f18fc8646f2b9aaefc8fb575742"},
+ {file = "pyzmq-26.0.2-cp311-cp311-win_amd64.whl", hash = "sha256:6d03da3a0ae691b361edcb39530075461202f699ce05adbb15055a0e1c9bcaa4"},
+ {file = "pyzmq-26.0.2-cp311-cp311-win_arm64.whl", hash = "sha256:f84e33321b68ff00b60e9dbd1a483e31ab6022c577c8de525b8e771bd274ce68"},
+ {file = "pyzmq-26.0.2-cp312-cp312-macosx_10_15_universal2.whl", hash = "sha256:44c33ebd1c62a01db7fbc24e18bdda569d6639217d13d5929e986a2b0f69070d"},
+ {file = "pyzmq-26.0.2-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:ac04f904b4fce4afea9cdccbb78e24d468cb610a839d5a698853e14e2a3f9ecf"},
+ {file = "pyzmq-26.0.2-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:f2133de5ba9adc5f481884ccb699eac9ce789708292945c05746880f95b241c0"},
+ {file = "pyzmq-26.0.2-cp312-cp312-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:7753c67c570d7fc80c2dc59b90ca1196f1224e0e2e29a548980c95fe0fe27fc1"},
+ {file = "pyzmq-26.0.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:8d4e51632e6b12e65e8d9d7612446ecda2eda637a868afa7bce16270194650dd"},
+ {file = "pyzmq-26.0.2-cp312-cp312-manylinux_2_28_x86_64.whl", hash = "sha256:d6c38806f6ecd0acf3104b8d7e76a206bcf56dadd6ce03720d2fa9d9157d5718"},
+ {file = "pyzmq-26.0.2-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:48f496bbe14686b51cec15406323ae6942851e14022efd7fc0e2ecd092c5982c"},
+ {file = "pyzmq-26.0.2-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:e84a3161149c75bb7a7dc8646384186c34033e286a67fec1ad1bdedea165e7f4"},
+ {file = "pyzmq-26.0.2-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:dabf796c67aa9f5a4fcc956d47f0d48b5c1ed288d628cf53aa1cf08e88654343"},
+ {file = "pyzmq-26.0.2-cp312-cp312-win32.whl", hash = "sha256:3eee4c676af1b109f708d80ef0cf57ecb8aaa5900d1edaf90406aea7e0e20e37"},
+ {file = "pyzmq-26.0.2-cp312-cp312-win_amd64.whl", hash = "sha256:26721fec65846b3e4450dad050d67d31b017f97e67f7e0647b5f98aa47f828cf"},
+ {file = "pyzmq-26.0.2-cp312-cp312-win_arm64.whl", hash = "sha256:653955c6c233e90de128a1b8e882abc7216f41f44218056bd519969c8c413a15"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-macosx_10_9_x86_64.whl", hash = "sha256:becd8d8fb068fbb5a52096efd83a2d8e54354383f691781f53a4c26aee944542"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:7a15e5465e7083c12517209c9dd24722b25e9b63c49a563922922fc03554eb35"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:e8158ac8616941f874841f9fa0f6d2f1466178c2ff91ea08353fdc19de0d40c2"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ea2c6a53e28c7066ea7db86fcc0b71d78d01b818bb11d4a4341ec35059885295"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-musllinux_1_1_aarch64.whl", hash = "sha256:bdbc7dab0b0e9c62c97b732899c4242e3282ba803bad668e03650b59b165466e"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-musllinux_1_1_i686.whl", hash = "sha256:e74b6d5ef57bb65bf1b4a37453d8d86d88550dde3fb0f23b1f1a24e60c70af5b"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-musllinux_1_1_x86_64.whl", hash = "sha256:ed4c6ee624ecbc77b18aeeb07bf0700d26571ab95b8f723f0d02e056b5bce438"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-win32.whl", hash = "sha256:8a98b3cb0484b83c19d8fb5524c8a469cd9f10e743f5904ac285d92678ee761f"},
+ {file = "pyzmq-26.0.2-cp37-cp37m-win_amd64.whl", hash = "sha256:aa5f95d71b6eca9cec28aa0a2f8310ea53dea313b63db74932879ff860c1fb8d"},
+ {file = "pyzmq-26.0.2-cp38-cp38-macosx_10_15_universal2.whl", hash = "sha256:5ff56c76ce77b9805378a7a73032c17cbdb1a5b84faa1df03c5d3e306e5616df"},
+ {file = "pyzmq-26.0.2-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:bab697fc1574fee4b81da955678708567c43c813c84c91074e452bda5346c921"},
+ {file = "pyzmq-26.0.2-cp38-cp38-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:0c0fed8aa9ba0488ee1cbdaa304deea92d52fab43d373297002cfcc69c0a20c5"},
+ {file = "pyzmq-26.0.2-cp38-cp38-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:606b922699fcec472ed814dda4dc3ff7c748254e0b26762a0ba21a726eb1c107"},
+ {file = "pyzmq-26.0.2-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:45f0fd82bad4d199fa993fbf0ac586a7ac5879addbe436a35a389df7e0eb4c91"},
+ {file = "pyzmq-26.0.2-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:166c5e41045939a52c01e6f374e493d9a6a45dfe677360d3e7026e38c42e8906"},
+ {file = "pyzmq-26.0.2-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:d566e859e8b8d5bca08467c093061774924b3d78a5ba290e82735b2569edc84b"},
+ {file = "pyzmq-26.0.2-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:264ee0e72b72ca59279dc320deab5ae0fac0d97881aed1875ce4bde2e56ffde0"},
+ {file = "pyzmq-26.0.2-cp38-cp38-win32.whl", hash = "sha256:3152bbd3a4744cbdd83dfb210ed701838b8b0c9065cef14671d6d91df12197d0"},
+ {file = "pyzmq-26.0.2-cp38-cp38-win_amd64.whl", hash = "sha256:bf77601d75ca692c179154b7e5943c286a4aaffec02c491afe05e60493ce95f2"},
+ {file = "pyzmq-26.0.2-cp39-cp39-macosx_10_15_universal2.whl", hash = "sha256:c770a7545b3deca2db185b59175e710a820dd4ed43619f4c02e90b0e227c6252"},
+ {file = "pyzmq-26.0.2-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:d47175f0a380bfd051726bc5c0054036ae4a5d8caf922c62c8a172ccd95c1a2a"},
+ {file = "pyzmq-26.0.2-cp39-cp39-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:9bce298c1ce077837e110367c321285dc4246b531cde1abfc27e4a5bbe2bed4d"},
+ {file = "pyzmq-26.0.2-cp39-cp39-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:c40b09b7e184d6e3e1be1c8af2cc320c0f9f610d8a5df3dd866e6e6e4e32b235"},
+ {file = "pyzmq-26.0.2-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d420d856bf728713874cefb911398efe69e1577835851dd297a308a78c14c249"},
+ {file = "pyzmq-26.0.2-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:d792d3cab987058451e55c70c5926e93e2ceb68ca5a2334863bb903eb860c9cb"},
+ {file = "pyzmq-26.0.2-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:83ec17729cf6d3464dab98a11e98294fcd50e6b17eaabd3d841515c23f6dbd3a"},
+ {file = "pyzmq-26.0.2-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:47c17d5ebfa88ae90f08960c97b49917098665b8cd8be31f2c24e177bcf37a0f"},
+ {file = "pyzmq-26.0.2-cp39-cp39-win32.whl", hash = "sha256:d509685d1cd1d018705a811c5f9d5bc237790936ead6d06f6558b77e16cc7235"},
+ {file = "pyzmq-26.0.2-cp39-cp39-win_amd64.whl", hash = "sha256:c7cc8cc009e8f6989a6d86c96f87dae5f5fb07d6c96916cdc7719d546152c7db"},
+ {file = "pyzmq-26.0.2-cp39-cp39-win_arm64.whl", hash = "sha256:3ada31cb879cd7532f4a85b501f4255c747d4813ab76b35c49ed510ce4865b45"},
+ {file = "pyzmq-26.0.2-pp310-pypy310_pp73-macosx_10_9_x86_64.whl", hash = "sha256:0a6ceaddc830dd3ca86cb8451cf373d1f05215368e11834538c2902ed5205139"},
+ {file = "pyzmq-26.0.2-pp310-pypy310_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:6a967681463aa7a99eb9a62bb18229b653b45c10ff0947b31cc0837a83dfb86f"},
+ {file = "pyzmq-26.0.2-pp310-pypy310_pp73-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:6472a73bc115bc40a2076609a90894775abe6faf19a78375675a2f889a613071"},
+ {file = "pyzmq-26.0.2-pp310-pypy310_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:5d6aea92bcccfe5e5524d3c70a6f16ffdae548390ddad26f4207d55c55a40593"},
+ {file = "pyzmq-26.0.2-pp310-pypy310_pp73-win_amd64.whl", hash = "sha256:e025f6351e49d48a5aa2f5a09293aa769b0ee7369c25bed551647234b7fa0c75"},
+ {file = "pyzmq-26.0.2-pp37-pypy37_pp73-macosx_10_9_x86_64.whl", hash = "sha256:40bd7ebe4dbb37d27f0c56e2a844f360239343a99be422085e13e97da13f73f9"},
+ {file = "pyzmq-26.0.2-pp37-pypy37_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:1dd40d586ad6f53764104df6e01810fe1b4e88fd353774629a5e6fe253813f79"},
+ {file = "pyzmq-26.0.2-pp37-pypy37_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:f2aca15e9ad8c8657b5b3d7ae3d1724dc8c1c1059c06b4b674c3aa36305f4930"},
+ {file = "pyzmq-26.0.2-pp37-pypy37_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:450ec234736732eb0ebeffdb95a352450d4592f12c3e087e2a9183386d22c8bf"},
+ {file = "pyzmq-26.0.2-pp37-pypy37_pp73-win_amd64.whl", hash = "sha256:f43be2bebbd09360a2f23af83b243dc25ffe7b583ea8c722e6df03e03a55f02f"},
+ {file = "pyzmq-26.0.2-pp38-pypy38_pp73-macosx_10_9_x86_64.whl", hash = "sha256:867f55e54aff254940bcec5eec068e7c0ac1e6bf360ab91479394a8bf356b0e6"},
+ {file = "pyzmq-26.0.2-pp38-pypy38_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:b4dbc033c5ad46f8c429bf238c25a889b8c1d86bfe23a74e1031a991cb3f0000"},
+ {file = "pyzmq-26.0.2-pp38-pypy38_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:6e8dd2961462e337e21092ec2da0c69d814dcb1b6e892955a37444a425e9cfb8"},
+ {file = "pyzmq-26.0.2-pp38-pypy38_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:35391e72df6c14a09b697c7b94384947c1dd326aca883ff98ff137acdf586c33"},
+ {file = "pyzmq-26.0.2-pp38-pypy38_pp73-win_amd64.whl", hash = "sha256:1c3d3c92fa54eda94ab369ca5b8d35059987c326ba5e55326eb068862f64b1fc"},
+ {file = "pyzmq-26.0.2-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:e7aa61a9cc4f0523373e31fc9255bf4567185a099f85ca3598e64de484da3ab2"},
+ {file = "pyzmq-26.0.2-pp39-pypy39_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ee53a8191271f144cc20b12c19daa9f1546adc84a2f33839e3338039b55c373c"},
+ {file = "pyzmq-26.0.2-pp39-pypy39_pp73-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:ac60a980f07fa988983f7bfe6404ef3f1e4303f5288a01713bc1266df6d18783"},
+ {file = "pyzmq-26.0.2-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:88896b1b4817d7b2fe1ec7205c4bbe07bf5d92fb249bf2d226ddea8761996068"},
+ {file = "pyzmq-26.0.2-pp39-pypy39_pp73-manylinux_2_28_x86_64.whl", hash = "sha256:18dfffe23751edee917764ffa133d5d3fef28dfd1cf3adebef8c90bc854c74c4"},
+ {file = "pyzmq-26.0.2-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:6926dd14cfe6967d3322640b6d5c3c3039db71716a5e43cca6e3b474e73e0b36"},
+ {file = "pyzmq-26.0.2.tar.gz", hash = "sha256:f0f9bb370449158359bb72a3e12c658327670c0ffe6fbcd1af083152b64f9df0"},
]
[package.dependencies]
@@ -2768,19 +2989,121 @@ prompt_toolkit = ">=2.0,<=3.0.36"
[[package]]
name = "referencing"
-version = "0.34.0"
+version = "0.35.0"
description = "JSON Referencing + Python"
optional = false
python-versions = ">=3.8"
files = [
- {file = "referencing-0.34.0-py3-none-any.whl", hash = "sha256:d53ae300ceddd3169f1ffa9caf2cb7b769e92657e4fafb23d34b93679116dfd4"},
- {file = "referencing-0.34.0.tar.gz", hash = "sha256:5773bd84ef41799a5a8ca72dc34590c041eb01bf9aa02632b4a973fb0181a844"},
+ {file = "referencing-0.35.0-py3-none-any.whl", hash = "sha256:8080727b30e364e5783152903672df9b6b091c926a146a759080b62ca3126cd6"},
+ {file = "referencing-0.35.0.tar.gz", hash = "sha256:191e936b0c696d0af17ad7430a3dc68e88bc11be6514f4757dc890f04ab05889"},
]
[package.dependencies]
attrs = ">=22.2.0"
rpds-py = ">=0.7.0"
+[[package]]
+name = "regex"
+version = "2024.4.16"
+description = "Alternative regular expression module, to replace re."
+optional = false
+python-versions = ">=3.7"
+files = [
+ {file = "regex-2024.4.16-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:fb83cc090eac63c006871fd24db5e30a1f282faa46328572661c0a24a2323a08"},
+ {file = "regex-2024.4.16-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:8c91e1763696c0eb66340c4df98623c2d4e77d0746b8f8f2bee2c6883fd1fe18"},
+ {file = "regex-2024.4.16-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:10188fe732dec829c7acca7422cdd1bf57d853c7199d5a9e96bb4d40db239c73"},
+ {file = "regex-2024.4.16-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:956b58d692f235cfbf5b4f3abd6d99bf102f161ccfe20d2fd0904f51c72c4c66"},
+ {file = "regex-2024.4.16-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:a70b51f55fd954d1f194271695821dd62054d949efd6368d8be64edd37f55c86"},
+ {file = "regex-2024.4.16-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:5c02fcd2bf45162280613d2e4a1ca3ac558ff921ae4e308ecb307650d3a6ee51"},
+ {file = "regex-2024.4.16-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:c4ed75ea6892a56896d78f11006161eea52c45a14994794bcfa1654430984b22"},
+ {file = "regex-2024.4.16-cp310-cp310-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:bd727ad276bb91928879f3aa6396c9a1d34e5e180dce40578421a691eeb77f47"},
+ {file = "regex-2024.4.16-cp310-cp310-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:7cbc5d9e8a1781e7be17da67b92580d6ce4dcef5819c1b1b89f49d9678cc278c"},
+ {file = "regex-2024.4.16-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:78fddb22b9ef810b63ef341c9fcf6455232d97cfe03938cbc29e2672c436670e"},
+ {file = "regex-2024.4.16-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:445ca8d3c5a01309633a0c9db57150312a181146315693273e35d936472df912"},
+ {file = "regex-2024.4.16-cp310-cp310-musllinux_1_1_ppc64le.whl", hash = "sha256:95399831a206211d6bc40224af1c635cb8790ddd5c7493e0bd03b85711076a53"},
+ {file = "regex-2024.4.16-cp310-cp310-musllinux_1_1_s390x.whl", hash = "sha256:7731728b6568fc286d86745f27f07266de49603a6fdc4d19c87e8c247be452af"},
+ {file = "regex-2024.4.16-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:4facc913e10bdba42ec0aee76d029aedda628161a7ce4116b16680a0413f658a"},
+ {file = "regex-2024.4.16-cp310-cp310-win32.whl", hash = "sha256:911742856ce98d879acbea33fcc03c1d8dc1106234c5e7d068932c945db209c0"},
+ {file = "regex-2024.4.16-cp310-cp310-win_amd64.whl", hash = "sha256:e0a2df336d1135a0b3a67f3bbf78a75f69562c1199ed9935372b82215cddd6e2"},
+ {file = "regex-2024.4.16-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:1210365faba7c2150451eb78ec5687871c796b0f1fa701bfd2a4a25420482d26"},
+ {file = "regex-2024.4.16-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:9ab40412f8cd6f615bfedea40c8bf0407d41bf83b96f6fc9ff34976d6b7037fd"},
+ {file = "regex-2024.4.16-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:fd80d1280d473500d8086d104962a82d77bfbf2b118053824b7be28cd5a79ea5"},
+ {file = "regex-2024.4.16-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:7bb966fdd9217e53abf824f437a5a2d643a38d4fd5fd0ca711b9da683d452969"},
+ {file = "regex-2024.4.16-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:20b7a68444f536365af42a75ccecb7ab41a896a04acf58432db9e206f4e525d6"},
+ {file = "regex-2024.4.16-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:b74586dd0b039c62416034f811d7ee62810174bb70dffcca6439f5236249eb09"},
+ {file = "regex-2024.4.16-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:0c8290b44d8b0af4e77048646c10c6e3aa583c1ca67f3b5ffb6e06cf0c6f0f89"},
+ {file = "regex-2024.4.16-cp311-cp311-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:f2d80a6749724b37853ece57988b39c4e79d2b5fe2869a86e8aeae3bbeef9eb0"},
+ {file = "regex-2024.4.16-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:3a1018e97aeb24e4f939afcd88211ace472ba566efc5bdf53fd8fd7f41fa7170"},
+ {file = "regex-2024.4.16-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:8d015604ee6204e76569d2f44e5a210728fa917115bef0d102f4107e622b08d5"},
+ {file = "regex-2024.4.16-cp311-cp311-musllinux_1_1_ppc64le.whl", hash = "sha256:3d5ac5234fb5053850d79dd8eb1015cb0d7d9ed951fa37aa9e6249a19aa4f336"},
+ {file = "regex-2024.4.16-cp311-cp311-musllinux_1_1_s390x.whl", hash = "sha256:0a38d151e2cdd66d16dab550c22f9521ba79761423b87c01dae0a6e9add79c0d"},
+ {file = "regex-2024.4.16-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:159dc4e59a159cb8e4e8f8961eb1fa5d58f93cb1acd1701d8aff38d45e1a84a6"},
+ {file = "regex-2024.4.16-cp311-cp311-win32.whl", hash = "sha256:ba2336d6548dee3117520545cfe44dc28a250aa091f8281d28804aa8d707d93d"},
+ {file = "regex-2024.4.16-cp311-cp311-win_amd64.whl", hash = "sha256:8f83b6fd3dc3ba94d2b22717f9c8b8512354fd95221ac661784df2769ea9bba9"},
+ {file = "regex-2024.4.16-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:80b696e8972b81edf0af2a259e1b2a4a661f818fae22e5fa4fa1a995fb4a40fd"},
+ {file = "regex-2024.4.16-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:d61ae114d2a2311f61d90c2ef1358518e8f05eafda76eaf9c772a077e0b465ec"},
+ {file = "regex-2024.4.16-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:8ba6745440b9a27336443b0c285d705ce73adb9ec90e2f2004c64d95ab5a7598"},
+ {file = "regex-2024.4.16-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:6295004b2dd37b0835ea5c14a33e00e8cfa3c4add4d587b77287825f3418d310"},
+ {file = "regex-2024.4.16-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:4aba818dcc7263852aabb172ec27b71d2abca02a593b95fa79351b2774eb1d2b"},
+ {file = "regex-2024.4.16-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:d0800631e565c47520aaa04ae38b96abc5196fe8b4aa9bd864445bd2b5848a7a"},
+ {file = "regex-2024.4.16-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:08dea89f859c3df48a440dbdcd7b7155bc675f2fa2ec8c521d02dc69e877db70"},
+ {file = "regex-2024.4.16-cp312-cp312-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:eeaa0b5328b785abc344acc6241cffde50dc394a0644a968add75fcefe15b9d4"},
+ {file = "regex-2024.4.16-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:4e819a806420bc010489f4e741b3036071aba209f2e0989d4750b08b12a9343f"},
+ {file = "regex-2024.4.16-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:c2d0e7cbb6341e830adcbfa2479fdeebbfbb328f11edd6b5675674e7a1e37730"},
+ {file = "regex-2024.4.16-cp312-cp312-musllinux_1_1_ppc64le.whl", hash = "sha256:91797b98f5e34b6a49f54be33f72e2fb658018ae532be2f79f7c63b4ae225145"},
+ {file = "regex-2024.4.16-cp312-cp312-musllinux_1_1_s390x.whl", hash = "sha256:d2da13568eff02b30fd54fccd1e042a70fe920d816616fda4bf54ec705668d81"},
+ {file = "regex-2024.4.16-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:370c68dc5570b394cbaadff50e64d705f64debed30573e5c313c360689b6aadc"},
+ {file = "regex-2024.4.16-cp312-cp312-win32.whl", hash = "sha256:904c883cf10a975b02ab3478bce652f0f5346a2c28d0a8521d97bb23c323cc8b"},
+ {file = "regex-2024.4.16-cp312-cp312-win_amd64.whl", hash = "sha256:785c071c982dce54d44ea0b79cd6dfafddeccdd98cfa5f7b86ef69b381b457d9"},
+ {file = "regex-2024.4.16-cp37-cp37m-macosx_10_9_x86_64.whl", hash = "sha256:e2f142b45c6fed48166faeb4303b4b58c9fcd827da63f4cf0a123c3480ae11fb"},
+ {file = "regex-2024.4.16-cp37-cp37m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:e87ab229332ceb127a165612d839ab87795972102cb9830e5f12b8c9a5c1b508"},
+ {file = "regex-2024.4.16-cp37-cp37m-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:81500ed5af2090b4a9157a59dbc89873a25c33db1bb9a8cf123837dcc9765047"},
+ {file = "regex-2024.4.16-cp37-cp37m-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:b340cccad138ecb363324aa26893963dcabb02bb25e440ebdf42e30963f1a4e0"},
+ {file = "regex-2024.4.16-cp37-cp37m-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:2c72608e70f053643437bd2be0608f7f1c46d4022e4104d76826f0839199347a"},
+ {file = "regex-2024.4.16-cp37-cp37m-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:a01fe2305e6232ef3e8f40bfc0f0f3a04def9aab514910fa4203bafbc0bb4682"},
+ {file = "regex-2024.4.16-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:03576e3a423d19dda13e55598f0fd507b5d660d42c51b02df4e0d97824fdcae3"},
+ {file = "regex-2024.4.16-cp37-cp37m-musllinux_1_1_aarch64.whl", hash = "sha256:549c3584993772e25f02d0656ac48abdda73169fe347263948cf2b1cead622f3"},
+ {file = "regex-2024.4.16-cp37-cp37m-musllinux_1_1_i686.whl", hash = "sha256:34422d5a69a60b7e9a07a690094e824b66f5ddc662a5fc600d65b7c174a05f04"},
+ {file = "regex-2024.4.16-cp37-cp37m-musllinux_1_1_ppc64le.whl", hash = "sha256:5f580c651a72b75c39e311343fe6875d6f58cf51c471a97f15a938d9fe4e0d37"},
+ {file = "regex-2024.4.16-cp37-cp37m-musllinux_1_1_s390x.whl", hash = "sha256:3399dd8a7495bbb2bacd59b84840eef9057826c664472e86c91d675d007137f5"},
+ {file = "regex-2024.4.16-cp37-cp37m-musllinux_1_1_x86_64.whl", hash = "sha256:8d1f86f3f4e2388aa3310b50694ac44daefbd1681def26b4519bd050a398dc5a"},
+ {file = "regex-2024.4.16-cp37-cp37m-win32.whl", hash = "sha256:dd5acc0a7d38fdc7a3a6fd3ad14c880819008ecb3379626e56b163165162cc46"},
+ {file = "regex-2024.4.16-cp37-cp37m-win_amd64.whl", hash = "sha256:ba8122e3bb94ecda29a8de4cf889f600171424ea586847aa92c334772d200331"},
+ {file = "regex-2024.4.16-cp38-cp38-macosx_10_9_universal2.whl", hash = "sha256:743deffdf3b3481da32e8a96887e2aa945ec6685af1cfe2bcc292638c9ba2f48"},
+ {file = "regex-2024.4.16-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:7571f19f4a3fd00af9341c7801d1ad1967fc9c3f5e62402683047e7166b9f2b4"},
+ {file = "regex-2024.4.16-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:df79012ebf6f4efb8d307b1328226aef24ca446b3ff8d0e30202d7ebcb977a8c"},
+ {file = "regex-2024.4.16-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:e757d475953269fbf4b441207bb7dbdd1c43180711b6208e129b637792ac0b93"},
+ {file = "regex-2024.4.16-cp38-cp38-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:4313ab9bf6a81206c8ac28fdfcddc0435299dc88cad12cc6305fd0e78b81f9e4"},
+ {file = "regex-2024.4.16-cp38-cp38-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:d83c2bc678453646f1a18f8db1e927a2d3f4935031b9ad8a76e56760461105dd"},
+ {file = "regex-2024.4.16-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:9df1bfef97db938469ef0a7354b2d591a2d438bc497b2c489471bec0e6baf7c4"},
+ {file = "regex-2024.4.16-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:62120ed0de69b3649cc68e2965376048793f466c5a6c4370fb27c16c1beac22d"},
+ {file = "regex-2024.4.16-cp38-cp38-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:c2ef6f7990b6e8758fe48ad08f7e2f66c8f11dc66e24093304b87cae9037bb4a"},
+ {file = "regex-2024.4.16-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:8fc6976a3395fe4d1fbeb984adaa8ec652a1e12f36b56ec8c236e5117b585427"},
+ {file = "regex-2024.4.16-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:03e68f44340528111067cecf12721c3df4811c67268b897fbe695c95f860ac42"},
+ {file = "regex-2024.4.16-cp38-cp38-musllinux_1_1_ppc64le.whl", hash = "sha256:ec7e0043b91115f427998febaa2beb82c82df708168b35ece3accb610b91fac1"},
+ {file = "regex-2024.4.16-cp38-cp38-musllinux_1_1_s390x.whl", hash = "sha256:c21fc21a4c7480479d12fd8e679b699f744f76bb05f53a1d14182b31f55aac76"},
+ {file = "regex-2024.4.16-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:12f6a3f2f58bb7344751919a1876ee1b976fe08b9ffccb4bbea66f26af6017b9"},
+ {file = "regex-2024.4.16-cp38-cp38-win32.whl", hash = "sha256:479595a4fbe9ed8f8f72c59717e8cf222da2e4c07b6ae5b65411e6302af9708e"},
+ {file = "regex-2024.4.16-cp38-cp38-win_amd64.whl", hash = "sha256:0534b034fba6101611968fae8e856c1698da97ce2efb5c2b895fc8b9e23a5834"},
+ {file = "regex-2024.4.16-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:a7ccdd1c4a3472a7533b0a7aa9ee34c9a2bef859ba86deec07aff2ad7e0c3b94"},
+ {file = "regex-2024.4.16-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:6f2f017c5be19984fbbf55f8af6caba25e62c71293213f044da3ada7091a4455"},
+ {file = "regex-2024.4.16-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:803b8905b52de78b173d3c1e83df0efb929621e7b7c5766c0843704d5332682f"},
+ {file = "regex-2024.4.16-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:684008ec44ad275832a5a152f6e764bbe1914bea10968017b6feaecdad5736e0"},
+ {file = "regex-2024.4.16-cp39-cp39-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:65436dce9fdc0aeeb0a0effe0839cb3d6a05f45aa45a4d9f9c60989beca78b9c"},
+ {file = "regex-2024.4.16-cp39-cp39-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:ea355eb43b11764cf799dda62c658c4d2fdb16af41f59bb1ccfec517b60bcb07"},
+ {file = "regex-2024.4.16-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:98c1165f3809ce7774f05cb74e5408cd3aa93ee8573ae959a97a53db3ca3180d"},
+ {file = "regex-2024.4.16-cp39-cp39-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:cccc79a9be9b64c881f18305a7c715ba199e471a3973faeb7ba84172abb3f317"},
+ {file = "regex-2024.4.16-cp39-cp39-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:00169caa125f35d1bca6045d65a662af0202704489fada95346cfa092ec23f39"},
+ {file = "regex-2024.4.16-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:6cc38067209354e16c5609b66285af17a2863a47585bcf75285cab33d4c3b8df"},
+ {file = "regex-2024.4.16-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:23cff1b267038501b179ccbbd74a821ac4a7192a1852d1d558e562b507d46013"},
+ {file = "regex-2024.4.16-cp39-cp39-musllinux_1_1_ppc64le.whl", hash = "sha256:b9d320b3bf82a39f248769fc7f188e00f93526cc0fe739cfa197868633d44701"},
+ {file = "regex-2024.4.16-cp39-cp39-musllinux_1_1_s390x.whl", hash = "sha256:89ec7f2c08937421bbbb8b48c54096fa4f88347946d4747021ad85f1b3021b3c"},
+ {file = "regex-2024.4.16-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:4918fd5f8b43aa7ec031e0fef1ee02deb80b6afd49c85f0790be1dc4ce34cb50"},
+ {file = "regex-2024.4.16-cp39-cp39-win32.whl", hash = "sha256:684e52023aec43bdf0250e843e1fdd6febbe831bd9d52da72333fa201aaa2335"},
+ {file = "regex-2024.4.16-cp39-cp39-win_amd64.whl", hash = "sha256:e697e1c0238133589e00c244a8b676bc2cfc3ab4961318d902040d099fec7483"},
+ {file = "regex-2024.4.16.tar.gz", hash = "sha256:fa454d26f2e87ad661c4f0c5a5fe4cf6aab1e307d1b94f16ffdfcb089ba685c0"},
+]
+
[[package]]
name = "requests"
version = "2.31.0"
@@ -2912,28 +3235,28 @@ files = [
[[package]]
name = "ruff"
-version = "0.3.4"
+version = "0.3.7"
description = "An extremely fast Python linter and code formatter, written in Rust."
optional = false
python-versions = ">=3.7"
files = [
- {file = "ruff-0.3.4-py3-none-macosx_10_12_x86_64.macosx_11_0_arm64.macosx_10_12_universal2.whl", hash = "sha256:60c870a7d46efcbc8385d27ec07fe534ac32f3b251e4fc44b3cbfd9e09609ef4"},
- {file = "ruff-0.3.4-py3-none-macosx_10_12_x86_64.whl", hash = "sha256:6fc14fa742e1d8f24910e1fff0bd5e26d395b0e0e04cc1b15c7c5e5fe5b4af91"},
- {file = "ruff-0.3.4-py3-none-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d3ee7880f653cc03749a3bfea720cf2a192e4f884925b0cf7eecce82f0ce5854"},
- {file = "ruff-0.3.4-py3-none-manylinux_2_17_armv7l.manylinux2014_armv7l.whl", hash = "sha256:cf133dd744f2470b347f602452a88e70dadfbe0fcfb5fd46e093d55da65f82f7"},
- {file = "ruff-0.3.4-py3-none-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:3f3860057590e810c7ffea75669bdc6927bfd91e29b4baa9258fd48b540a4365"},
- {file = "ruff-0.3.4-py3-none-manylinux_2_17_ppc64.manylinux2014_ppc64.whl", hash = "sha256:986f2377f7cf12efac1f515fc1a5b753c000ed1e0a6de96747cdf2da20a1b369"},
- {file = "ruff-0.3.4-py3-none-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:c4fd98e85869603e65f554fdc5cddf0712e352fe6e61d29d5a6fe087ec82b76c"},
- {file = "ruff-0.3.4-py3-none-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:64abeed785dad51801b423fa51840b1764b35d6c461ea8caef9cf9e5e5ab34d9"},
- {file = "ruff-0.3.4-py3-none-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:df52972138318bc7546d92348a1ee58449bc3f9eaf0db278906eb511889c4b50"},
- {file = "ruff-0.3.4-py3-none-musllinux_1_2_aarch64.whl", hash = "sha256:98e98300056445ba2cc27d0b325fd044dc17fcc38e4e4d2c7711585bd0a958ed"},
- {file = "ruff-0.3.4-py3-none-musllinux_1_2_armv7l.whl", hash = "sha256:519cf6a0ebed244dce1dc8aecd3dc99add7a2ee15bb68cf19588bb5bf58e0488"},
- {file = "ruff-0.3.4-py3-none-musllinux_1_2_i686.whl", hash = "sha256:bb0acfb921030d00070539c038cd24bb1df73a2981e9f55942514af8b17be94e"},
- {file = "ruff-0.3.4-py3-none-musllinux_1_2_x86_64.whl", hash = "sha256:cf187a7e7098233d0d0c71175375c5162f880126c4c716fa28a8ac418dcf3378"},
- {file = "ruff-0.3.4-py3-none-win32.whl", hash = "sha256:af27ac187c0a331e8ef91d84bf1c3c6a5dea97e912a7560ac0cef25c526a4102"},
- {file = "ruff-0.3.4-py3-none-win_amd64.whl", hash = "sha256:de0d5069b165e5a32b3c6ffbb81c350b1e3d3483347196ffdf86dc0ef9e37dd6"},
- {file = "ruff-0.3.4-py3-none-win_arm64.whl", hash = "sha256:6810563cc08ad0096b57c717bd78aeac888a1bfd38654d9113cb3dc4d3f74232"},
- {file = "ruff-0.3.4.tar.gz", hash = "sha256:f0f4484c6541a99862b693e13a151435a279b271cff20e37101116a21e2a1ad1"},
+ {file = "ruff-0.3.7-py3-none-macosx_10_12_x86_64.macosx_11_0_arm64.macosx_10_12_universal2.whl", hash = "sha256:0e8377cccb2f07abd25e84fc5b2cbe48eeb0fea9f1719cad7caedb061d70e5ce"},
+ {file = "ruff-0.3.7-py3-none-macosx_10_12_x86_64.whl", hash = "sha256:15a4d1cc1e64e556fa0d67bfd388fed416b7f3b26d5d1c3e7d192c897e39ba4b"},
+ {file = "ruff-0.3.7-py3-none-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d28bdf3d7dc71dd46929fafeec98ba89b7c3550c3f0978e36389b5631b793663"},
+ {file = "ruff-0.3.7-py3-none-manylinux_2_17_armv7l.manylinux2014_armv7l.whl", hash = "sha256:379b67d4f49774ba679593b232dcd90d9e10f04d96e3c8ce4a28037ae473f7bb"},
+ {file = "ruff-0.3.7-py3-none-manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:c060aea8ad5ef21cdfbbe05475ab5104ce7827b639a78dd55383a6e9895b7c51"},
+ {file = "ruff-0.3.7-py3-none-manylinux_2_17_ppc64.manylinux2014_ppc64.whl", hash = "sha256:ebf8f615dde968272d70502c083ebf963b6781aacd3079081e03b32adfe4d58a"},
+ {file = "ruff-0.3.7-py3-none-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:d48098bd8f5c38897b03604f5428901b65e3c97d40b3952e38637b5404b739a2"},
+ {file = "ruff-0.3.7-py3-none-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:da8a4fda219bf9024692b1bc68c9cff4b80507879ada8769dc7e985755d662ea"},
+ {file = "ruff-0.3.7-py3-none-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:6c44e0149f1d8b48c4d5c33d88c677a4aa22fd09b1683d6a7ff55b816b5d074f"},
+ {file = "ruff-0.3.7-py3-none-musllinux_1_2_aarch64.whl", hash = "sha256:3050ec0af72b709a62ecc2aca941b9cd479a7bf2b36cc4562f0033d688e44fa1"},
+ {file = "ruff-0.3.7-py3-none-musllinux_1_2_armv7l.whl", hash = "sha256:a29cc38e4c1ab00da18a3f6777f8b50099d73326981bb7d182e54a9a21bb4ff7"},
+ {file = "ruff-0.3.7-py3-none-musllinux_1_2_i686.whl", hash = "sha256:5b15cc59c19edca917f51b1956637db47e200b0fc5e6e1878233d3a938384b0b"},
+ {file = "ruff-0.3.7-py3-none-musllinux_1_2_x86_64.whl", hash = "sha256:e491045781b1e38b72c91247cf4634f040f8d0cb3e6d3d64d38dcf43616650b4"},
+ {file = "ruff-0.3.7-py3-none-win32.whl", hash = "sha256:bc931de87593d64fad3a22e201e55ad76271f1d5bfc44e1a1887edd0903c7d9f"},
+ {file = "ruff-0.3.7-py3-none-win_amd64.whl", hash = "sha256:5ef0e501e1e39f35e03c2acb1d1238c595b8bb36cf7a170e7c1df1b73da00e74"},
+ {file = "ruff-0.3.7-py3-none-win_arm64.whl", hash = "sha256:789e144f6dc7019d1f92a812891c645274ed08af6037d11fc65fcbc183b7d59f"},
+ {file = "ruff-0.3.7.tar.gz", hash = "sha256:d5c1aebee5162c2226784800ae031f660c350e7a3402c4d1f8ea4e97e232e3ba"},
]
[[package]]
@@ -3060,18 +3383,18 @@ test = ["asv", "gmpy2", "mpmath", "pytest", "pytest-cov", "pytest-xdist", "sciki
[[package]]
name = "setuptools"
-version = "69.2.0"
+version = "69.5.1"
description = "Easily download, build, install, upgrade, and uninstall Python packages"
optional = false
python-versions = ">=3.8"
files = [
- {file = "setuptools-69.2.0-py3-none-any.whl", hash = "sha256:c21c49fb1042386df081cb5d86759792ab89efca84cf114889191cd09aacc80c"},
- {file = "setuptools-69.2.0.tar.gz", hash = "sha256:0ff4183f8f42cd8fa3acea16c45205521a4ef28f73c6391d8a25e92893134f2e"},
+ {file = "setuptools-69.5.1-py3-none-any.whl", hash = "sha256:c636ac361bc47580504644275c9ad802c50415c7522212252c033bd15f301f32"},
+ {file = "setuptools-69.5.1.tar.gz", hash = "sha256:6c1fccdac05a97e598fb0ae3bbed5904ccb317337a51139dcd51453611bbb987"},
]
[package.extras]
-docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "pygments-github-lexers (==0.0.5)", "rst.linker (>=1.9)", "sphinx (<7.2.5)", "sphinx (>=3.5)", "sphinx-favicon", "sphinx-inline-tabs", "sphinx-lint", "sphinx-notfound-page (>=1,<2)", "sphinx-reredirects", "sphinxcontrib-towncrier"]
-testing = ["build[virtualenv]", "filelock (>=3.4.0)", "importlib-metadata", "ini2toml[lite] (>=0.9)", "jaraco.develop (>=7.21)", "jaraco.envs (>=2.2)", "jaraco.path (>=3.2.0)", "mypy (==1.9)", "packaging (>=23.2)", "pip (>=19.1)", "pytest (>=6)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-home (>=0.5)", "pytest-mypy (>=0.9.1)", "pytest-perf", "pytest-ruff (>=0.2.1)", "pytest-timeout", "pytest-xdist (>=3)", "tomli", "tomli-w (>=1.0.0)", "virtualenv (>=13.0.0)", "wheel"]
+docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "pygments-github-lexers (==0.0.5)", "rst.linker (>=1.9)", "sphinx (>=3.5)", "sphinx-favicon", "sphinx-inline-tabs", "sphinx-lint", "sphinx-notfound-page (>=1,<2)", "sphinx-reredirects", "sphinxcontrib-towncrier"]
+testing = ["build[virtualenv]", "filelock (>=3.4.0)", "importlib-metadata", "ini2toml[lite] (>=0.9)", "jaraco.develop (>=7.21)", "jaraco.envs (>=2.2)", "jaraco.path (>=3.2.0)", "mypy (==1.9)", "packaging (>=23.2)", "pip (>=19.1)", "pytest (>=6,!=8.1.1)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-home (>=0.5)", "pytest-mypy", "pytest-perf", "pytest-ruff (>=0.2.1)", "pytest-timeout", "pytest-xdist (>=3)", "tomli", "tomli-w (>=1.0.0)", "virtualenv (>=13.0.0)", "wheel"]
testing-integration = ["build[virtualenv] (>=1.0.3)", "filelock (>=3.4.0)", "jaraco.envs (>=2.2)", "jaraco.path (>=3.2.0)", "packaging (>=23.2)", "pytest", "pytest-enabler", "pytest-xdist", "tomli", "virtualenv (>=13.0.0)", "wheel"]
[[package]]
@@ -3142,84 +3465,6 @@ docs = ["sphinxcontrib-websupport"]
lint = ["docutils-stubs", "flake8 (>=3.5.0)", "flake8-simplify", "isort", "mypy (>=0.990)", "ruff", "sphinx-lint", "types-requests"]
test = ["cython", "filelock", "html5lib", "pytest (>=4.6)"]
-[[package]]
-name = "sphinx-autoapi"
-version = "3.0.0"
-description = "Sphinx API documentation generator"
-optional = false
-python-versions = ">=3.8"
-files = [
- {file = "sphinx-autoapi-3.0.0.tar.gz", hash = "sha256:09ebd674a32b44467222b0fb8a917b97c89523f20dbf05b52cb8a3f0e15714de"},
- {file = "sphinx_autoapi-3.0.0-py2.py3-none-any.whl", hash = "sha256:ea207793cba1feff7b2ded0e29364f2995a4d157303a98603cee0ce94cea2688"},
-]
-
-[package.dependencies]
-anyascii = "*"
-astroid = [
- {version = ">=2.7", markers = "python_version < \"3.12\""},
- {version = ">=3.0.0a1", markers = "python_version >= \"3.12\""},
-]
-Jinja2 = "*"
-PyYAML = "*"
-sphinx = ">=6.1.0"
-
-[package.extras]
-docs = ["furo", "sphinx", "sphinx-design"]
-
-[[package]]
-name = "sphinx-autobuild"
-version = "2021.3.14"
-description = "Rebuild Sphinx documentation on changes, with live-reload in the browser."
-optional = false
-python-versions = ">=3.6"
-files = [
- {file = "sphinx-autobuild-2021.3.14.tar.gz", hash = "sha256:de1ca3b66e271d2b5b5140c35034c89e47f263f2cd5db302c9217065f7443f05"},
- {file = "sphinx_autobuild-2021.3.14-py3-none-any.whl", hash = "sha256:8fe8cbfdb75db04475232f05187c776f46f6e9e04cacf1e49ce81bdac649ccac"},
-]
-
-[package.dependencies]
-colorama = "*"
-livereload = "*"
-sphinx = "*"
-
-[package.extras]
-test = ["pytest", "pytest-cov"]
-
-[[package]]
-name = "sphinx-basic-ng"
-version = "1.0.0b2"
-description = "A modern skeleton for Sphinx themes."
-optional = false
-python-versions = ">=3.7"
-files = [
- {file = "sphinx_basic_ng-1.0.0b2-py3-none-any.whl", hash = "sha256:eb09aedbabfb650607e9b4b68c9d240b90b1e1be221d6ad71d61c52e29f7932b"},
- {file = "sphinx_basic_ng-1.0.0b2.tar.gz", hash = "sha256:9ec55a47c90c8c002b5960c57492ec3021f5193cb26cebc2dc4ea226848651c9"},
-]
-
-[package.dependencies]
-sphinx = ">=4.0"
-
-[package.extras]
-docs = ["furo", "ipython", "myst-parser", "sphinx-copybutton", "sphinx-inline-tabs"]
-
-[[package]]
-name = "sphinx-copybutton"
-version = "0.5.2"
-description = "Add a copy button to each of your code cells."
-optional = false
-python-versions = ">=3.7"
-files = [
- {file = "sphinx-copybutton-0.5.2.tar.gz", hash = "sha256:4cf17c82fb9646d1bc9ca92ac280813a3b605d8c421225fd9913154103ee1fbd"},
- {file = "sphinx_copybutton-0.5.2-py3-none-any.whl", hash = "sha256:fb543fd386d917746c9a2c50360c7905b605726b9355cd26e9974857afeae06e"},
-]
-
-[package.dependencies]
-sphinx = ">=1.8"
-
-[package.extras]
-code-style = ["pre-commit (==2.12.1)"]
-rtd = ["ipython", "myst-nb", "sphinx", "sphinx-book-theme", "sphinx-examples"]
-
[[package]]
name = "sphinxcontrib-applehelp"
version = "1.0.4"
@@ -3445,13 +3690,13 @@ files = [
[[package]]
name = "tinycss2"
-version = "1.2.1"
+version = "1.3.0"
description = "A tiny CSS parser"
optional = false
-python-versions = ">=3.7"
+python-versions = ">=3.8"
files = [
- {file = "tinycss2-1.2.1-py3-none-any.whl", hash = "sha256:2b80a96d41e7c3914b8cda8bc7f705a4d9c49275616e886103dd839dfc847847"},
- {file = "tinycss2-1.2.1.tar.gz", hash = "sha256:8cff3a8f066c2ec677c06dbc7b45619804a6938478d9d73c284b29d14ecb0627"},
+ {file = "tinycss2-1.3.0-py3-none-any.whl", hash = "sha256:54a8dbdffb334d536851be0226030e9505965bb2f30f21a4a82c55fb2a80fae7"},
+ {file = "tinycss2-1.3.0.tar.gz", hash = "sha256:152f9acabd296a8375fbca5b84c961ff95971fcfc32e79550c8df8e29118c54d"},
]
[package.dependencies]
@@ -3459,7 +3704,18 @@ webencodings = ">=0.4"
[package.extras]
doc = ["sphinx", "sphinx_rtd_theme"]
-test = ["flake8", "isort", "pytest"]
+test = ["pytest", "ruff"]
+
+[[package]]
+name = "toml"
+version = "0.10.2"
+description = "Python Library for Tom's Obvious, Minimal Language"
+optional = false
+python-versions = ">=2.6, !=3.0.*, !=3.1.*, !=3.2.*"
+files = [
+ {file = "toml-0.10.2-py2.py3-none-any.whl", hash = "sha256:806143ae5bfb6a3c6e736a764057db0e6a0e05e338b5630894a5f779cabb4f9b"},
+ {file = "toml-0.10.2.tar.gz", hash = "sha256:b3bda1d108d5dd99f4a20d24d9c348e91c4db7ab1b749200bded2f839ccbe68f"},
+]
[[package]]
name = "tomli"
@@ -3505,28 +3761,28 @@ files = [
[[package]]
name = "traitlets"
-version = "5.14.2"
+version = "5.14.3"
description = "Traitlets Python configuration system"
optional = false
python-versions = ">=3.8"
files = [
- {file = "traitlets-5.14.2-py3-none-any.whl", hash = "sha256:fcdf85684a772ddeba87db2f398ce00b40ff550d1528c03c14dbf6a02003cd80"},
- {file = "traitlets-5.14.2.tar.gz", hash = "sha256:8cdd83c040dab7d1dee822678e5f5d100b514f7b72b01615b26fc5718916fdf9"},
+ {file = "traitlets-5.14.3-py3-none-any.whl", hash = "sha256:b74e89e397b1ed28cc831db7aea759ba6640cb3de13090ca145426688ff1ac4f"},
+ {file = "traitlets-5.14.3.tar.gz", hash = "sha256:9ed0579d3502c94b4b3732ac120375cda96f923114522847de4b3bb98b96b6b7"},
]
[package.extras]
docs = ["myst-parser", "pydata-sphinx-theme", "sphinx"]
-test = ["argcomplete (>=3.0.3)", "mypy (>=1.7.0)", "pre-commit", "pytest (>=7.0,<8.1)", "pytest-mock", "pytest-mypy-testing"]
+test = ["argcomplete (>=3.0.3)", "mypy (>=1.7.0)", "pre-commit", "pytest (>=7.0,<8.2)", "pytest-mock", "pytest-mypy-testing"]
[[package]]
name = "typing-extensions"
-version = "4.10.0"
+version = "4.11.0"
description = "Backported and Experimental Type Hints for Python 3.8+"
optional = false
python-versions = ">=3.8"
files = [
- {file = "typing_extensions-4.10.0-py3-none-any.whl", hash = "sha256:69b1a937c3a517342112fb4c6df7e72fc39a38e7891a5730ed4985b5214b5475"},
- {file = "typing_extensions-4.10.0.tar.gz", hash = "sha256:b0abd7c89e8fb96f98db18d86106ff1d90ab692004eb746cf6eda2682f91b3cb"},
+ {file = "typing_extensions-4.11.0-py3-none-any.whl", hash = "sha256:c1f94d72897edaf4ce775bb7558d5b79d8126906a14ea5ed1635921406c0387a"},
+ {file = "typing_extensions-4.11.0.tar.gz", hash = "sha256:83f085bd5ca59c80295fc2a82ab5dac679cbe02b9f33f7d83af68e241bea51b0"},
]
[[package]]
@@ -3558,30 +3814,30 @@ socks = ["PySocks (>=1.5.6,!=1.5.7,<2.0)"]
[[package]]
name = "urllib3"
-version = "2.0.7"
+version = "2.2.1"
description = "HTTP library with thread-safe connection pooling, file post, and more."
optional = false
-python-versions = ">=3.7"
+python-versions = ">=3.8"
files = [
- {file = "urllib3-2.0.7-py3-none-any.whl", hash = "sha256:fdb6d215c776278489906c2f8916e6e7d4f5a9b602ccbcfdf7f016fc8da0596e"},
- {file = "urllib3-2.0.7.tar.gz", hash = "sha256:c97dfde1f7bd43a71c8d2a58e369e9b2bf692d1334ea9f9cae55add7d0dd0f84"},
+ {file = "urllib3-2.2.1-py3-none-any.whl", hash = "sha256:450b20ec296a467077128bff42b73080516e71b56ff59a60a02bef2232c4fa9d"},
+ {file = "urllib3-2.2.1.tar.gz", hash = "sha256:d0570876c61ab9e520d776c38acbbb5b05a776d3f9ff98a5c8fd5162a444cf19"},
]
[package.extras]
brotli = ["brotli (>=1.0.9)", "brotlicffi (>=0.8.0)"]
-secure = ["certifi", "cryptography (>=1.9)", "idna (>=2.0.0)", "pyopenssl (>=17.1.0)", "urllib3-secure-extra"]
+h2 = ["h2 (>=4,<5)"]
socks = ["pysocks (>=1.5.6,!=1.5.7,<2.0)"]
zstd = ["zstandard (>=0.18.0)"]
[[package]]
name = "virtualenv"
-version = "20.25.1"
+version = "20.26.0"
description = "Virtual Python Environment builder"
optional = false
python-versions = ">=3.7"
files = [
- {file = "virtualenv-20.25.1-py3-none-any.whl", hash = "sha256:961c026ac520bac5f69acb8ea063e8a4f071bcc9457b9c1f28f6b085c511583a"},
- {file = "virtualenv-20.25.1.tar.gz", hash = "sha256:e08e13ecdca7a0bd53798f356d5831434afa5b07b93f0abdf0797b7a06ffe197"},
+ {file = "virtualenv-20.26.0-py3-none-any.whl", hash = "sha256:0846377ea76e818daaa3e00a4365c018bc3ac9760cbb3544de542885aad61fb3"},
+ {file = "virtualenv-20.26.0.tar.gz", hash = "sha256:ec25a9671a5102c8d2657f62792a27b48f016664c6873f6beed3800008577210"},
]
[package.dependencies]
@@ -3590,24 +3846,9 @@ filelock = ">=3.12.2,<4"
platformdirs = ">=3.9.1,<5"
[package.extras]
-docs = ["furo (>=2023.7.26)", "proselint (>=0.13)", "sphinx (>=7.1.2)", "sphinx-argparse (>=0.4)", "sphinxcontrib-towncrier (>=0.2.1a0)", "towncrier (>=23.6)"]
+docs = ["furo (>=2023.7.26)", "proselint (>=0.13)", "sphinx (>=7.1.2,!=7.3)", "sphinx-argparse (>=0.4)", "sphinxcontrib-towncrier (>=0.2.1a0)", "towncrier (>=23.6)"]
test = ["covdefaults (>=2.3)", "coverage (>=7.2.7)", "coverage-enable-subprocess (>=1)", "flaky (>=3.7)", "packaging (>=23.1)", "pytest (>=7.4)", "pytest-env (>=0.8.2)", "pytest-freezer (>=0.4.8)", "pytest-mock (>=3.11.1)", "pytest-randomly (>=3.12)", "pytest-timeout (>=2.1)", "setuptools (>=68)", "time-machine (>=2.10)"]
-[[package]]
-name = "waitress"
-version = "3.0.0"
-description = "Waitress WSGI server"
-optional = false
-python-versions = ">=3.8.0"
-files = [
- {file = "waitress-3.0.0-py3-none-any.whl", hash = "sha256:2a06f242f4ba0cc563444ca3d1998959447477363a2d7e9b8b4d75d35cfd1669"},
- {file = "waitress-3.0.0.tar.gz", hash = "sha256:005da479b04134cdd9dd602d1ee7c49d79de0537610d653674cc6cbde222b8a1"},
-]
-
-[package.extras]
-docs = ["Sphinx (>=1.8.1)", "docutils", "pylons-sphinx-themes (>=1.0.9)"]
-testing = ["coverage (>=5.0)", "pytest", "pytest-cov"]
-
[[package]]
name = "watchdog"
version = "4.0.0"
@@ -3649,6 +3890,20 @@ files = [
[package.extras]
watchmedo = ["PyYAML (>=3.10)"]
+[[package]]
+name = "wcmatch"
+version = "8.5.1"
+description = "Wildcard/glob file name matcher."
+optional = false
+python-versions = ">=3.8"
+files = [
+ {file = "wcmatch-8.5.1-py3-none-any.whl", hash = "sha256:24c19cedc92bc9c9e27f39db4e1824d72f95bd2cea32b254a47a45b1a1b227ed"},
+ {file = "wcmatch-8.5.1.tar.gz", hash = "sha256:c0088c7f6426cf6bf27e530e2b7b734031905f7e490475fd83c7c5008ab581b3"},
+]
+
+[package.dependencies]
+bracex = ">=2.1.1"
+
[[package]]
name = "wcwidth"
version = "0.2.13"
@@ -3672,39 +3927,18 @@ files = [
]
[[package]]
-name = "webob"
-version = "1.8.7"
-description = "WSGI request and response object"
-optional = false
-python-versions = ">=2.7,!=3.0.*,!=3.1.*,!=3.2.*"
-files = [
- {file = "WebOb-1.8.7-py2.py3-none-any.whl", hash = "sha256:73aae30359291c14fa3b956f8b5ca31960e420c28c1bec002547fb04928cf89b"},
- {file = "WebOb-1.8.7.tar.gz", hash = "sha256:b64ef5141be559cfade448f044fa45c2260351edcb6a8ef6b7e00c7dcef0c323"},
-]
-
-[package.extras]
-docs = ["Sphinx (>=1.7.5)", "pylons-sphinx-themes"]
-testing = ["coverage", "pytest (>=3.1.0)", "pytest-cov", "pytest-xdist"]
-
-[[package]]
-name = "webtest"
-version = "3.0.0"
-description = "Helper to test WSGI applications"
+name = "wheel"
+version = "0.43.0"
+description = "A built-package format for Python"
optional = false
-python-versions = ">=3.6, <4"
+python-versions = ">=3.8"
files = [
- {file = "WebTest-3.0.0-py3-none-any.whl", hash = "sha256:2a001a9efa40d2a7e5d9cd8d1527c75f41814eb6afce2c3d207402547b1e5ead"},
- {file = "WebTest-3.0.0.tar.gz", hash = "sha256:54bd969725838d9861a9fa27f8d971f79d275d94ae255f5c501f53bb6d9929eb"},
+ {file = "wheel-0.43.0-py3-none-any.whl", hash = "sha256:55c570405f142630c6b9f72fe09d9b67cf1477fcf543ae5b8dcb1f5b7377da81"},
+ {file = "wheel-0.43.0.tar.gz", hash = "sha256:465ef92c69fa5c5da2d1cf8ac40559a8c940886afcef87dcf14b9470862f1d85"},
]
-[package.dependencies]
-beautifulsoup4 = "*"
-waitress = ">=0.8.5"
-WebOb = ">=1.2"
-
[package.extras]
-docs = ["Sphinx (>=1.8.1)", "docutils", "pylons-sphinx-themes (>=1.0.8)"]
-tests = ["PasteDeploy", "WSGIProxy2", "coverage", "pyquery", "pytest", "pytest-cov"]
+test = ["pytest (>=6.0.0)", "setuptools (>=65)"]
[[package]]
name = "wrapt"
@@ -3805,7 +4039,7 @@ test = ["pytest", "pytest-cov"]
name = "yarl"
version = "1.9.4"
description = "Yet another URL library"
-optional = false
+optional = true
python-versions = ">=3.7"
files = [
{file = "yarl-1.9.4-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:a8c1df72eb746f4136fe9a2e72b0c9dc1da1cbd23b5372f94b5820ff8ae30e0e"},
@@ -3926,4 +4160,4 @@ collate = ["cytominer-database"]
[metadata]
lock-version = "2.0"
python-versions = ">=3.8,<4.0"
-content-hash = "8cd2e790b6c426bf02ddd240f8cd3d34bc72aa79d23cb012ab51bd48bff15a38"
+content-hash = "70a56ee15e4d18ef704d2dde4ac560fd4089a00a75df0ef9650126aec0e75f27"
diff --git a/pycytominer/__about__.py b/pycytominer/__about__.py
index c76a4b74..cbc13087 100644
--- a/pycytominer/__about__.py
+++ b/pycytominer/__about__.py
@@ -1,6 +1,8 @@
+"""Pycytominer package metadata."""
+
__project__ = "pycytominer"
# These version placeholders are updated during build by poetry-dynamic-versioning
-__version__ = "0.0.0"
-__version_tuple__ = (0, 0, 0)
+__version__ = "1.1.0.post29.dev0+9615c0d"
+__version_tuple__ = (1, 1, 0, "post29", "dev0", "9615c0d")
__license__ = "BSD 3-Clause License"
__author__ = "Pycytominer Contributors"
diff --git a/pycytominer/__config__.py b/pycytominer/__config__.py
index 4ccc6df3..dd6cb0f4 100644
--- a/pycytominer/__config__.py
+++ b/pycytominer/__config__.py
@@ -1,6 +1,4 @@
-"""
-Module used for pycytominer configuration details.
-"""
+"""Module used for pycytominer configuration details."""
import pandas as pd
diff --git a/pycytominer/__init__.py b/pycytominer/__init__.py
index 88937371..843b014d 100644
--- a/pycytominer/__init__.py
+++ b/pycytominer/__init__.py
@@ -1,3 +1,5 @@
+"""Pycytominer is a suite of common functions used to process high dimensional readouts from high-throughput cell experiments."""
+
from pycytominer import __about__, __config__
from .aggregate import aggregate
diff --git a/pycytominer/aggregate.py b/pycytominer/aggregate.py
index bf8403ba..b8e06a78 100644
--- a/pycytominer/aggregate.py
+++ b/pycytominer/aggregate.py
@@ -1,6 +1,4 @@
-"""
-Aggregate profiles based on given grouping variables.
-"""
+"""Aggregate profiles based on given grouping variables."""
from typing import Any, Dict, List, Optional, Union
@@ -67,7 +65,6 @@ def aggregate(
data.
"""
-
# Check that the operation is supported
operation = check_aggregate_operation(operation)
diff --git a/pycytominer/annotate.py b/pycytominer/annotate.py
index d5eb2bcb..e7eca779 100644
--- a/pycytominer/annotate.py
+++ b/pycytominer/annotate.py
@@ -1,6 +1,4 @@
-"""
-Annotates profiles with metadata information
-"""
+"""Annotates profiles with metadata information."""
import os
@@ -77,7 +75,6 @@ def annotate(
DataFrame. If you specify output_file, then write to file and do not return
data.
"""
-
# Load Data
profiles = load_profiles(profiles)
platemap = load_platemap(platemap, add_metadata_id_to_platemap)
diff --git a/pycytominer/consensus.py b/pycytominer/consensus.py
index 7ea19714..933df97d 100644
--- a/pycytominer/consensus.py
+++ b/pycytominer/consensus.py
@@ -1,6 +1,4 @@
-"""
-Acquire consensus signatures for input samples
-"""
+"""Acquire consensus signatures for input samples."""
from pycytominer import aggregate
from pycytominer.cyto_utils import (
diff --git a/pycytominer/cyto_utils/DeepProfiler_processing.py b/pycytominer/cyto_utils/DeepProfiler_processing.py
index e33f1036..254f4291 100644
--- a/pycytominer/cyto_utils/DeepProfiler_processing.py
+++ b/pycytominer/cyto_utils/DeepProfiler_processing.py
@@ -1,6 +1,4 @@
-"""
-Utility function to load and process the output files of a DeepProfiler run.
-"""
+"""Utility function to load and process the output files of a DeepProfiler run."""
import os
import pathlib
@@ -17,7 +15,7 @@
class DeepProfilerData:
- """This class holds all functions needed to load and annotate the DeepProfiler (DP) run.
+ """Class that holds all functions needed to load and annotate the DeepProfiler (DP) run.
Attributes
----------
@@ -58,7 +56,6 @@ def __init__(
See above for all other parameters.
"""
-
self.index_df = pd.read_csv(index_file, dtype=str)
self.profile_dir = profile_dir
self.filename_delimiter = filename_delimiter
@@ -67,9 +64,7 @@ def __init__(
self.file_extension = f".{self.file_extension}"
def build_filenames(self):
- """
- Create file names indicated by plate, well, and site information
- """
+ """Create file names indicated by plate, well, and site information."""
self.filenames = self.index_df.apply(
self.build_filename_from_index, axis="columns"
)
@@ -78,9 +73,7 @@ def build_filenames(self):
]
def build_filename_from_index(self, row):
- """
- Builds the name of the profile files
- """
+ """Build the name of the profile files."""
plate = row["Metadata_Plate"]
well = row["Metadata_Well"]
site = row["Metadata_Site"]
@@ -89,9 +82,10 @@ def build_filename_from_index(self, row):
return filename
def extract_filename_metadata(self, npz_file, delimiter="_"):
- """
- Extract metadata (site, well and plate) from the filename.
- The input format of the file: path/plate/well{delimiter}site.npz
+ """Extract metadata (site, well and plate) from the filename.
+
+ This function is used to extract the metadata from the filename of the npz files.
+ It expects a naming convetion of path/plate/well{delimiter}site.npz.
Arguments
---------
@@ -120,7 +114,7 @@ def extract_filename_metadata(self, npz_file, delimiter="_"):
class AggregateDeepProfiler:
- """This class holds all functions needed to aggregate the DeepProfiler (DP) run.
+ """Class that holds all functions needed to aggregate the DeepProfiler (DP) run.
Attributes
----------
@@ -183,10 +177,9 @@ def __init__(
self.output_file = output_file
def setup_aggregate(self):
- """
- Sets up the file_aggregate attribute. This is a helper function to aggregate_deep().
+ """Set up the file_aggregate attribute.
- the file_aggregate dictionary contains the file locations and metadata for each grouping.
+ A helper function to `aggregate_deep` that aggregate the file_aggregate dictionary contains the file locations and metadata for each grouping.
If for example we are grouping by well then the keys of self.file_aggregate would be:
plate1/well1, plate1/well2, plate2/well1, etc.
"""
@@ -218,7 +211,7 @@ def setup_aggregate(self):
def aggregate_deep(self):
"""
- Main function of this class. Aggregates the profiles into a pandas dataframe.
+ Aggregate the DeepProfiler profiles into a pandas dataframe.
For each key in file_aggregate, the profiles are loaded, concatenated and then aggregated.
If files are missing, we throw a warning but continue the code.
@@ -310,7 +303,9 @@ def aggregate_deep(self):
class SingleCellDeepProfiler:
- """This class holds functions needed to analyze single cells from the DeepProfiler (DP) run. Only pycytominer.normalization() is implemented.
+ """Class that holds functions needed to analyze single cells from the DeepProfiler (DP) run.
+
+ Only pycytominer.normalization() is implemented.
Attributes
----------
@@ -354,15 +349,14 @@ def __init__(
---------
See above for all parameters.
"""
-
self.deep_data = deep_data
def get_single_cells(
self, output=False, location_x_col_index=0, location_y_col_index=1
):
- """
- Sets up the single_cells attribute or output as a variable. This is a helper function to normalize_deep_single_cells().
- single_cells is a pandas dataframe in the format expected by pycytominer.normalize().
+ """Set up a single_cells dataframe in the format expected by pycytominer.normalize().
+
+ Helper function to normalize_deep_single_cells() that sets up the single_cells attribute or outputs it as a dataframe.
Arguments
-----------
@@ -417,7 +411,7 @@ def normalize_deep_single_cells(
spherize_epsilon=1e-6,
):
"""
- Normalizes all cells into a pandas dataframe.
+ Normalize all cells into a pandas dataframe.
For each file in the DP project features folder, the features from each cell are loaded.
These features are put into a profiles dataframe for use in pycytominer.normalize.
diff --git a/pycytominer/cyto_utils/__init__.py b/pycytominer/cyto_utils/__init__.py
index 25456868..f9a0407e 100644
--- a/pycytominer/cyto_utils/__init__.py
+++ b/pycytominer/cyto_utils/__init__.py
@@ -1,3 +1,5 @@
+"""A variety of utility functions for working with cytominer data."""
+
from .output import output
from .util import (
check_compartments,
diff --git a/pycytominer/cyto_utils/annotate_custom.py b/pycytominer/cyto_utils/annotate_custom.py
index a9c9f056..d06edb01 100644
--- a/pycytominer/cyto_utils/annotate_custom.py
+++ b/pycytominer/cyto_utils/annotate_custom.py
@@ -1,10 +1,12 @@
+"""Functions to annotate data frames with custom options according to CMAP specifications."""
+
import numpy as np
def annotate_cmap(
annotated, annotate_join_on, cell_id="unknown", perturbation_mode="none"
):
- """Annotates data frame with custom options according to CMAP specifications
+ """Annotates data frame with custom options according to CMAP specifications.
Parameters
----------
@@ -103,7 +105,7 @@ def annotate_cmap(
def cp_clean(profiles):
- """Specifically clean certain column names derived from different CellProfiler versions
+ """Specifically clean certain column names derived from different CellProfiler versions.
Parameters
----------
@@ -115,7 +117,6 @@ def cp_clean(profiles):
profiles
Renamed to standard metadata
"""
-
profiles = profiles.rename(
{
"Image_Metadata_Plate": "Metadata_Plate",
diff --git a/pycytominer/cyto_utils/cell_locations.py b/pycytominer/cyto_utils/cell_locations.py
index 612ae873..04ee9077 100644
--- a/pycytominer/cyto_utils/cell_locations.py
+++ b/pycytominer/cyto_utils/cell_locations.py
@@ -1,6 +1,4 @@
-"""
-Utility function to augment a metadata file with X,Y locations of cells in each image
-"""
+"""Utility function to augment a metadata file with X,Y locations of cells in each image."""
import pathlib
import pandas as pd
@@ -14,8 +12,7 @@
class CellLocation:
- """This class holds all the functions augment a metadata file with X,Y
- locations of cells in each image.
+ """Class holding all the functions augment a metadata file with X,Y locations of cells in each image.
In the metadata file, which is either a CSV or a Parquet file,
- Each row is single multi-channel image
@@ -107,13 +104,13 @@ def __init__(
)
def _expanduser(self, obj: Union[str, None]):
- """Expand the user home directory in a path"""
+ """Expand the user home directory in a path."""
if obj is not None and isinstance(obj, str) and not obj.startswith("s3://"):
return pathlib.Path(obj).expanduser().as_posix()
return obj
def _parse_s3_path(self, s3_path: str):
- """Parse an S3 path into a bucket and key
+ """Parse an S3 path into a bucket and key.
Parameters
----------
@@ -127,7 +124,6 @@ def _parse_s3_path(self, s3_path: str):
str
The key
"""
-
s3_path = s3_path.replace("s3://", "")
bucket = s3_path.split("/")[0]
@@ -137,7 +133,7 @@ def _parse_s3_path(self, s3_path: str):
return bucket, key
def _s3_file_exists(self, s3_path: str):
- """Check if a file exists on S3
+ """Check if a file exists on S3.
Parameters
----------
@@ -149,7 +145,6 @@ def _s3_file_exists(self, s3_path: str):
bool
True if the file exists on S3, False otherwise
"""
-
bucket, key = self._parse_s3_path(s3_path)
try:
@@ -163,10 +158,7 @@ def _s3_file_exists(self, s3_path: str):
return True
def _download_s3(self, uri: str):
- """
- Download a file from S3 to a temporary file and return the temporary path
- """
-
+ """Download a file from S3 to a temporary file and return the temporary path."""
bucket, key = self._parse_s3_path(uri)
tmp_file = tempfile.NamedTemporaryFile(
@@ -178,14 +170,13 @@ def _download_s3(self, uri: str):
return tmp_file.name
def _load_metadata(self):
- """Load the metadata into a Pandas DataFrame
+ """Load the metadata into a Pandas DataFrame.
Returns
-------
Pandas DataFrame
The metadata loaded into a Pandas DataFrame
"""
-
if not isinstance(self.metadata_input, pd.DataFrame):
# verify that the metadata file is a CSV or a Parquet file
@@ -224,7 +215,7 @@ def _load_metadata(self):
return df
def _create_nested_df(self, df: pd.DataFrame):
- """Create a new column `CellCenters` by nesting the X and Y locations of cell from an image into the row of the image
+ """Create a new column `CellCenters` by nesting the X and Y locations of cell from an image into the row of the image.
Parameters
----------
@@ -235,7 +226,6 @@ def _create_nested_df(self, df: pd.DataFrame):
-------
Pandas DataFrame
"""
-
# define a dictionary to store the output
output_df_list = collections.defaultdict(list)
@@ -271,10 +261,7 @@ def _create_nested_df(self, df: pd.DataFrame):
return pd.DataFrame(output_df_list)
def _get_single_cell_engine(self):
- """
- Get the sqlalchemy.engine.Engine object for the single_cell file
- """
-
+ """Get the sqlalchemy.engine.Engine object for the single_cell file."""
if isinstance(self.single_cell_input, str):
# check if the single_cell file is a SQLite file
if not self.single_cell_input.endswith(".sqlite"):
@@ -298,10 +285,7 @@ def _get_single_cell_engine(self):
return temp_single_cell_input, engine
def _check_single_cell_correctness(self, engine: sqlalchemy.engine.Engine):
- """
- Check that the single_cell file has the required tables and columns
- """
-
+ """Check that the single_cell file has the required tables and columns."""
inspector = sqlalchemy.inspect(engine)
if not all(
@@ -342,9 +326,7 @@ def _check_single_cell_correctness(self, engine: sqlalchemy.engine.Engine):
)
def _get_joined_image_nuclei_tables(self):
- """
- Merge the Image and Nuclei tables in SQL
- """
+ """Merge the Image and Nuclei tables in SQL."""
# get the sqlalchemy.engine.Engine object for the single_cell file
temp_single_cell_input, engine = self._get_single_cell_engine()
@@ -382,18 +364,18 @@ def _get_joined_image_nuclei_tables(self):
return joined_df
def _load_single_cell(self):
- """Load the required columns from the `Image` and `Nuclei` tables in the single_cell file or sqlalchemy.engine.Engine object into a Pandas DataFrame
+ """Load the required columns from the `Image` and `Nuclei` tables in the single_cell file or sqlalchemy.engine.Engine object into a Pandas DataFrame.
Returns
-------
Pandas DataFrame
The required columns from the `Image` and `Nuclei` tables loaded into a Pandas DataFrame
"""
-
return self._create_nested_df(self._get_joined_image_nuclei_tables())
def add_cell_location(self):
"""Add the X,Y locations of all cells to the metadata file in the corresponding row, packed into a single column.
+
Optionally, save the augmented metadata file as a Parquet file.
Returns
@@ -401,7 +383,6 @@ def add_cell_location(self):
Pandas DataFrame
Either a data frame or the path to a Parquet file with the X,Y locations of all cells packed into a single column
"""
-
# If self.augmented_metadata_output is not None and it is a str and the file already exists, there is nothing to do
if (
self.augmented_metadata_output is not None
diff --git a/pycytominer/cyto_utils/cell_locations_cmd.py b/pycytominer/cyto_utils/cell_locations_cmd.py
index f0e08891..3b96965f 100644
--- a/pycytominer/cyto_utils/cell_locations_cmd.py
+++ b/pycytominer/cyto_utils/cell_locations_cmd.py
@@ -1,3 +1,5 @@
+"""CLI for cell location calculations."""
+
from pycytominer.cyto_utils.cell_locations import CellLocation
import fire
diff --git a/pycytominer/cyto_utils/cells.py b/pycytominer/cyto_utils/cells.py
index 4e0a80f9..0e07ae61 100644
--- a/pycytominer/cyto_utils/cells.py
+++ b/pycytominer/cyto_utils/cells.py
@@ -1,3 +1,5 @@
+"""Module containing the SingleCells class, which is used to interact with single cell morphological profiles."""
+
from typing import Dict, Union, Optional
import numpy as np
@@ -25,8 +27,7 @@
class SingleCells:
- """This is a class to interact with single cell morphological profiles. Interaction
- includes aggregation, normalization, and output.
+ """Class to interact with single cell morphological profiles including aggregation, normalization, and output.
Attributes
----------
@@ -115,7 +116,7 @@ def __init__(
object_feature="Metadata_ObjectNumber",
default_datatype_float=np.float64,
):
- """Constructor method"""
+ """Construct a SingleCells object."""
# Check compartments specified
check_compartments(compartments)
@@ -179,21 +180,20 @@ def __init__(
self.load_image(image_table_name=self.image_table_name)
def _check_subsampling(self):
- """Internal method checking if subsampling options were specified correctly.
+ """Check if subsampling options were specified correctly.
Returns
-------
None
Nothing is returned.
"""
-
# Check that the user didn't specify both subset frac and subsample all
assert ( # noqa: S101
self.subsample_frac == 1 or self.subsample_n == "all"
), "Do not set both subsample_frac and subsample_n"
def set_output_file(self, output_file):
- """Setting operation to conveniently rename output file.
+ """Set or modify output file.
Parameters
----------
@@ -205,11 +205,10 @@ def set_output_file(self, output_file):
None
Nothing is returned.
"""
-
self.output_file = output_file
def set_subsample_frac(self, subsample_frac):
- """Setting operation to conveniently update the subsample fraction.
+ """Set or update the subsample fraction.
Parameters
----------
@@ -221,12 +220,11 @@ def set_subsample_frac(self, subsample_frac):
None
Nothing is returned.
"""
-
self.subsample_frac = subsample_frac
self._check_subsampling()
def set_subsample_n(self, subsample_n):
- """Setting operation to conveniently update the subsample n.
+ """Set or update the subsample n.
Parameters
----------
@@ -238,7 +236,6 @@ def set_subsample_n(self, subsample_n):
None
Nothing is returned.
"""
-
try:
self.subsample_n = int(subsample_n)
except ValueError:
@@ -246,7 +243,7 @@ def set_subsample_n(self, subsample_n):
self._check_subsampling()
def set_subsample_random_state(self, random_state):
- """Setting operation to conveniently update the subsample random state.
+ """Set or update the subsample random state.
Parameters
----------
@@ -258,11 +255,10 @@ def set_subsample_random_state(self, random_state):
None
Nothing is returned.
"""
-
self.subsampling_random_state = random_state
def load_image(self, image_table_name=None):
- """Load image table from sqlite file
+ """Load image table from sqlite file.
Returns
-------
@@ -317,7 +313,6 @@ def count_cells(self, compartment="cells", count_subset=False):
pandas.core.frame.DataFrame
DataFrame of cell counts in the experiment.
"""
-
check_compartments(compartment)
if count_subset:
@@ -359,7 +354,6 @@ def subsample_profiles(self, df, rename_col=True):
pandas.core.frame.DataFrame
A subsampled pandas dataframe of single cell profiles.
"""
-
if self.subsampling_random_state is None:
random_state = np.random.randint(0, 10000, size=1)[0]
self.set_subsample_random_state(random_state)
@@ -398,7 +392,6 @@ def get_subsample(self, df=None, compartment="cells", rename_col=True):
None
Nothing is returned.
"""
-
check_compartments(compartment)
query_cols = "TableNumber, ImageNumber, ObjectNumber"
@@ -443,7 +436,7 @@ def split_column_categories(self, col_names):
return meta_cols, feat_cols
def load_compartment(self, compartment):
- """Creates the compartment dataframe.
+ """Create the compartment dataframe.
Note: makes use of default_datatype_float attribute
for setting a default floating point datatype.
@@ -458,7 +451,6 @@ def load_compartment(self, compartment):
pandas.core.frame.DataFrame
Compartment dataframe.
"""
-
# Get data useful to pre-alloc memory
num_cells = self.count_sql_table_rows(compartment)
col_names = self.get_sql_table_col_names(compartment)
@@ -495,7 +487,7 @@ def aggregate_compartment(
add_image_features=False,
n_aggregation_memory_strata=1,
):
- """Aggregate morphological profiles. Uses pycytominer.aggregate()
+ """Aggregate morphological profiles. Uses pycytominer.aggregate().
Parameters
----------
@@ -520,7 +512,6 @@ def aggregate_compartment(
pandas.core.frame.DataFrame
DataFrame of aggregated profiles.
"""
-
check_compartments(compartment)
if (self.subsample_frac < 1 or self.subsample_n != "all") and compute_subsample:
@@ -600,8 +591,7 @@ def _compartment_df_generator(
compartment,
n_aggregation_memory_strata=1,
):
- """A generator function that returns chunks of the entire compartment
- table from disk.
+ """Yield chunks of the entire compartment table from disk.
We want to return dataframes with all compartment entries within unique
combinations of self.merge_cols when aggregated by self.strata
@@ -622,7 +612,6 @@ def _compartment_df_generator(
between chunks, and thus groupby aggregations are valid
"""
-
assert ( # noqa: S101
n_aggregation_memory_strata > 0
), "Number of strata to pull into memory at once (n_aggregation_memory_strata) must be > 0"
@@ -712,7 +701,6 @@ def merge_single_cells(
if output_file=None returns a Pandas dataframe
else will write to file and return the filepath of the file
"""
-
# Load the single cell dataframe by merging on the specific linking columns
sc_df = ""
linking_check_cols = []
@@ -856,7 +844,6 @@ def aggregate_profiles(
if output_file=None) returns a Pandas dataframe
else will write to file and return the filepath of the file
"""
-
if output_file is not None:
self.set_output_file(output_file)
@@ -894,9 +881,7 @@ def aggregate_profiles(
def _sqlite_strata_conditions(df, dtypes, n=1):
- """Given a dataframe where columns are merge_cols and rows are unique
- value combinations that appear as aggregation strata, return a list
- of strings which constitute valid SQLite conditional statements.
+ """Construct a list of strings which constitute valid SQLite conditional statements.
Parameters
----------
@@ -927,13 +912,17 @@ def _sqlite_strata_conditions(df, dtypes, n=1):
[3] | [1, 2]
[4] | [1]
- >>> _sqlite_strata_conditions(df, dtypes={'TableNumber': 'integer', 'ImageNumber': 'integer'}, n=1)
+ >>> _sqlite_strata_conditions(
+ ... df, dtypes={"TableNumber": "integer", "ImageNumber": "integer"}, n=1
+ ... )
["(TableNumber in (1) and ImageNumber in (1))",
"(TableNumber in (2) and ImageNumber in (1, 2, 3))",
"(TableNumber in (3) and ImageNumber in (1, 2))",
"(TableNumber in (4) and ImageNumber in (1))"]
- >>> _sqlite_strata_conditions(df, dtypes={'TableNumber': 'text', 'ImageNumber': 'integer'}, n=2)
+ >>> _sqlite_strata_conditions(
+ ... df, dtypes={"TableNumber": "text", "ImageNumber": "integer"}, n=2
+ ... )
["(TableNumber in ('1') and ImageNumber in (1))
or (TableNumber in ('2') and ImageNumber in (1, 2, 3))",
"(TableNumber in ('3') and ImageNumber in (1, 2))
diff --git a/pycytominer/cyto_utils/collate.py b/pycytominer/cyto_utils/collate.py
index a21752fd..39b3e12b 100644
--- a/pycytominer/cyto_utils/collate.py
+++ b/pycytominer/cyto_utils/collate.py
@@ -1,3 +1,5 @@
+"""Module that provides functions for collating CellProfiler-created CSVs into a single SQLite file."""
+
import os
import pathlib
import subprocess
@@ -6,7 +8,7 @@
def run_check_errors(cmd):
- """Run a system command, and exit if an error occurred, otherwise continue"""
+ """Run a system command, and exit if an error occurred, otherwise continue."""
if isinstance(cmd, str):
cmd = cmd.split()
output = subprocess.run(cmd, capture_output=True, text=True) # noqa: S603
@@ -34,7 +36,7 @@ def collate(
image_feature_categories=["Granularity", "Texture", "ImageQuality", "Threshold"],
printtoscreen=True,
):
- """Collate the CellProfiler-created CSVs into a single SQLite file by calling cytominer-database
+ """Collate the CellProfiler-created CSVs into a single SQLite file by calling cytominer-database.
Parameters
----------
@@ -67,7 +69,6 @@ def collate(
printtoscreen: bool, optional, default True
Whether or not to print output to the terminal
"""
-
from pycytominer.cyto_utils.cells import SingleCells
# Check if optional dependency cytominer-database is installed
diff --git a/pycytominer/cyto_utils/collate_cmd.py b/pycytominer/cyto_utils/collate_cmd.py
index 7819d4bd..6f012a01 100644
--- a/pycytominer/cyto_utils/collate_cmd.py
+++ b/pycytominer/cyto_utils/collate_cmd.py
@@ -1,3 +1,5 @@
+"""Command line interface for collate function in pycytominer.cyto_utils.collate."""
+
import argparse
from pycytominer.cyto_utils.collate import collate
diff --git a/pycytominer/cyto_utils/cp_image_features.py b/pycytominer/cyto_utils/cp_image_features.py
index 1e9f29b2..b11db2aa 100644
--- a/pycytominer/cyto_utils/cp_image_features.py
+++ b/pycytominer/cyto_utils/cp_image_features.py
@@ -1,13 +1,11 @@
-"""
-Functions for counting the number of fields and aggregating other images features
-"""
+"""Functions for counting the number of fields and aggregating other images features."""
import numpy as np
from pycytominer import aggregate
def aggregate_fields_count(image_df, strata, fields_of_view_feature):
- """Compute the number of fields per well and create a new column called Metadata_Site_Count
+ """Compute the number of fields per well and create a new column called Metadata_Site_Count.
Parameters
----------
@@ -24,7 +22,6 @@ def aggregate_fields_count(image_df, strata, fields_of_view_feature):
DataFrame with the Metadata_Site_Count column.
"""
-
fields_count_df = image_df.loc[:, list(np.union1d(strata, fields_of_view_feature))]
fields_count_df = (
@@ -62,7 +59,6 @@ def aggregate_image_count_features(
remove_cols : list of str
Columns to remove from the image table before aggregating using aggregate_image_features()
"""
-
count_features = list(
image_features_df.columns[
image_features_df.columns.str.startswith("Metadata_" + str(count_prefix))
@@ -112,7 +108,6 @@ def aggregate_image_features(
DataFrame of aggregated image features.
"""
-
# Aggregate image count features
if count_prefix in image_feature_categories:
df, remove_cols = aggregate_image_count_features(
diff --git a/pycytominer/cyto_utils/features.py b/pycytominer/cyto_utils/features.py
index 449873df..304e71f9 100644
--- a/pycytominer/cyto_utils/features.py
+++ b/pycytominer/cyto_utils/features.py
@@ -1,6 +1,4 @@
-"""
-Utility function to manipulate cell profiler features
-"""
+"""Utility function to manipulate cell profiler features."""
import os
import pandas as pd
@@ -25,7 +23,6 @@ def get_blocklist_features(blocklist_file=blocklist_file, population_df=None):
blocklist_features : list of str
Features to exclude from downstream analysis.
"""
-
blocklist = pd.read_csv(blocklist_file)
assert any( # noqa: S101
@@ -48,7 +45,7 @@ def label_compartment(cp_features, compartment, metadata_cols):
cp_features : list of str
All features being used.
compartment : str
- Measured compartment.
+ Measured compartment.
metadata_cols : list
Columns that should be considered metadata.
@@ -57,7 +54,6 @@ def label_compartment(cp_features, compartment, metadata_cols):
cp_features: list of str
Recoded column names with appropriate metadata and compartment labels.
"""
-
compartment = compartment.Title()
avail_compartments = ["Cells", "Cytoplasm", "Nuceli", "Image", "Barcode"]
@@ -97,7 +93,6 @@ def infer_cp_features(
features: list of str
List of Cell Painting features.
"""
-
compartments = convert_compartment_format_to_list(compartments)
compartments = [x.title() for x in compartments]
@@ -135,7 +130,6 @@ def count_na_features(population_df, features):
-------
Dataframe of NA counts per feature
"""
-
return pd.DataFrame(population_df.loc[:, features].isna().sum(), columns=["num_na"])
@@ -156,7 +150,7 @@ def drop_outlier_features(
"Metadata_treatment == 'control'" (include all quotes).
If "all", use all samples to calculate.
outlier_cutoff : int or float, default 500
- see https://github.com/cytomining/pycytominer/issues/237 for details.
+ see https://github.com/cytomining/pycytominer/issues/237 for details.
Threshold to remove features if absolute values is greater
Returns
@@ -164,7 +158,6 @@ def drop_outlier_features(
outlier_features: list of str
Features greater than the threshold.
"""
-
# Subset dataframe
if samples != "all":
population_df.query(samples, inplace=True)
@@ -186,7 +179,7 @@ def drop_outlier_features(
def convert_compartment_format_to_list(compartments):
- """Converts compartment to a list.
+ """Convert cell painting compartments to a list.
Parameters
----------
@@ -198,7 +191,6 @@ def convert_compartment_format_to_list(compartments):
compartments : list of str
List of Cell Painting compartments.
"""
-
if isinstance(compartments, list):
compartments = [x.lower() for x in compartments]
elif isinstance(compartments, str):
diff --git a/pycytominer/cyto_utils/load.py b/pycytominer/cyto_utils/load.py
index 7abdd06d..198fd990 100644
--- a/pycytominer/cyto_utils/load.py
+++ b/pycytominer/cyto_utils/load.py
@@ -1,3 +1,5 @@
+"""Module for loading data from various file formats."""
+
import csv
import gzip
import pathlib
@@ -7,7 +9,7 @@
def is_path_a_parquet_file(file: Union[str, pathlib.PurePath]) -> bool:
- """Checks if the provided file path is a parquet file.
+ """Check if the provided file path is a parquet file.
Identify parquet files by inspecting the file extensions.
If the file does not end with `parquet`, this will return False, else True.
@@ -30,7 +32,6 @@ def is_path_a_parquet_file(file: Union[str, pathlib.PurePath]) -> bool:
FileNotFoundError
Raised if the provided path in the `file` does not exist
"""
-
file = pathlib.PurePath(file)
try:
# strict=true tests if path exists
@@ -47,7 +48,7 @@ def is_path_a_parquet_file(file: Union[str, pathlib.PurePath]) -> bool:
def infer_delim(file: str):
"""
- Sniff the delimiter in the given file
+ Sniff the delimiter in the given file.
Parameters
----------
@@ -72,7 +73,7 @@ def infer_delim(file: str):
def load_profiles(profiles):
"""
- Unless a dataframe is provided, load the given profile dataframe from path or string
+ Unless a dataframe is provided, load the given profile dataframe from path or string.
Parameters
----------
@@ -83,8 +84,8 @@ def load_profiles(profiles):
------
pandas DataFrame of profiles
- Raises:
- -------
+ Raises
+ ------
FileNotFoundError
Raised if the provided profile does not exists
"""
@@ -102,7 +103,7 @@ def load_profiles(profiles):
def load_platemap(platemap, add_metadata_id=True):
"""
- Unless a dataframe is provided, load the given platemap dataframe from path or string
+ Unless a dataframe is provided, load the given platemap dataframe from path or string.
Parameters
----------
diff --git a/pycytominer/cyto_utils/modz.py b/pycytominer/cyto_utils/modz.py
index 6e598ed5..5f719f0b 100644
--- a/pycytominer/cyto_utils/modz.py
+++ b/pycytominer/cyto_utils/modz.py
@@ -1,3 +1,5 @@
+"""Module for performing a modified z score transformation."""
+
import numpy as np
import pandas as pd
from pycytominer.cyto_utils.util import (
@@ -88,7 +90,7 @@ def modz(
min_weight=0.01,
precision=4,
):
- """Collapse replicates into a consensus signature using a weighted transformation
+ """Collapse replicates into a consensus signature using a weighted transformation.
Parameters
----------
diff --git a/pycytominer/cyto_utils/output.py b/pycytominer/cyto_utils/output.py
index aa5290a0..d0cd4f4d 100644
--- a/pycytominer/cyto_utils/output.py
+++ b/pycytominer/cyto_utils/output.py
@@ -1,6 +1,4 @@
-"""
-Utility function to compress output data
-"""
+"""Utility function to compress output data."""
from typing import Dict, Union, Optional
@@ -18,7 +16,7 @@ def output(
compression_options: Union[str, Dict] = {"method": "gzip", "mtime": 1},
**kwargs,
):
- """Given an output file and compression options, write file to disk
+ """Given an output file and compression options, write file to disk.
Parameters
----------
@@ -78,7 +76,6 @@ def output(
float_format=None,
)
"""
-
if output_type == "csv":
compression_options = set_compression_method(compression=compression_options)
@@ -99,7 +96,7 @@ def output(
def set_compression_method(compression: Union[str, Dict]):
- """Set the compression options
+ """Set the compression options.
Parameters
----------
@@ -112,7 +109,6 @@ def set_compression_method(compression: Union[str, Dict]):
compression, dict
A formated dictionary expected by output()
"""
-
if compression is None:
compression = {"method": None}
@@ -124,7 +120,7 @@ def set_compression_method(compression: Union[str, Dict]):
def check_compression_method(compression: str):
- """Ensure compression options are set properly
+ """Ensure compression options are set properly.
Parameters
----------
diff --git a/pycytominer/cyto_utils/single_cell_ingest_utils.py b/pycytominer/cyto_utils/single_cell_ingest_utils.py
index 2a5a5308..d9dcc070 100644
--- a/pycytominer/cyto_utils/single_cell_ingest_utils.py
+++ b/pycytominer/cyto_utils/single_cell_ingest_utils.py
@@ -1,9 +1,11 @@
+"""Utility functions for single cell ingest."""
+
from collections import Counter
from pycytominer.cyto_utils import get_default_compartments
def get_default_linking_cols():
- """Define the standard experiment linking columns between tables
+ """Define the standard experiment linking columns between tables.
Returns
-------
@@ -26,7 +28,7 @@ def get_default_linking_cols():
def assert_linking_cols_complete(linking_cols="default", compartments="default"):
- """Confirm that the linking cols and compartments are compatible
+ """Confirm that the linking cols and compartments are compatible.
Parameters
----------
@@ -75,8 +77,7 @@ def assert_linking_cols_complete(linking_cols="default", compartments="default")
def provide_linking_cols_feature_name_update(linking_cols="default"):
- """Output a dictionary to use to update pandas dataframe column names. The linking
- cols must be Metadata.
+ """Output a dictionary to use to update pandas dataframe column names from linking cols in the Metadata.
Parameters
----------
diff --git a/pycytominer/cyto_utils/util.py b/pycytominer/cyto_utils/util.py
index c550e61f..3a7302ad 100644
--- a/pycytominer/cyto_utils/util.py
+++ b/pycytominer/cyto_utils/util.py
@@ -1,6 +1,4 @@
-"""
-Miscellaneous utility functions
-"""
+"""Miscellaneous utility functions."""
import os
import warnings
@@ -16,7 +14,7 @@
def get_default_compartments():
- """Returns default compartments.
+ """Return default compartments.
Returns
-------
@@ -24,12 +22,11 @@ def get_default_compartments():
Default compartments.
"""
-
return ["cells", "cytoplasm", "nuclei"]
def check_compartments(compartments):
- """Checks if the input compartments are noncanonical compartments.
+ """Check if the input compartments are noncanonical compartments.
Parameters
----------
@@ -42,7 +39,6 @@ def check_compartments(compartments):
Nothing is returned.
"""
-
default_compartments = get_default_compartments()
compartments = convert_compartment_format_to_list(compartments)
@@ -60,13 +56,13 @@ def check_compartments(compartments):
def load_known_metadata_dictionary(metadata_file=default_metadata_file):
- """From a tab separated text file (two columns: ["compartment", "feature"]), load
- previously known metadata columns per compartment.
+ """Load previously known metadata columns per compartment from metadata text file.
Parameters
----------
metadata_file : str, optional
- File location of the metadata text file. Uses a default dictionary if you do not specify.
+ File location of the metadata text file which should be a tab-separated file with two columns: ["compartment", "feature"].
+ If not provided, the default metadata file will be used.
Returns
-------
@@ -74,7 +70,6 @@ def load_known_metadata_dictionary(metadata_file=default_metadata_file):
Compartment (keys) mappings to previously known metadata (values).
"""
-
metadata_dict = {}
with open(metadata_file) as meta_fh:
next(meta_fh)
@@ -103,7 +98,6 @@ def check_correlation_method(method):
Correctly formatted correlation method.
"""
-
method = method.lower()
avail_methods = ["pearson", "spearman", "kendall"]
assert ( # noqa: S101
@@ -127,7 +121,6 @@ def check_aggregate_operation(operation):
Correctly formatted operation method.
"""
-
operation = operation.lower()
avail_ops = ["mean", "median"]
assert ( # noqa: S101
@@ -151,7 +144,6 @@ def check_consensus_operation(operation):
Correctly formatted operation method.
"""
-
operation = operation.lower()
avail_ops = ["modz"] # All aggregation operations are also supported
try:
@@ -178,7 +170,6 @@ def check_fields_of_view_format(fields_of_view):
Correctly formatted fields_of_view variable.
"""
-
if fields_of_view != "all":
if isinstance(fields_of_view, list):
if all(isinstance(x, int) for x in fields_of_view):
@@ -214,7 +205,6 @@ def check_fields_of_view(data_fields_of_view, input_fields_of_view):
Nothing is returned.
"""
-
try:
assert len( # noqa: S101
list(np.intersect1d(data_fields_of_view, input_fields_of_view))
@@ -226,7 +216,7 @@ def check_fields_of_view(data_fields_of_view, input_fields_of_view):
def check_image_features(image_features, image_columns):
- """Confirm that the input list of image features are present in the image table
+ """Confirm that the input list of image features are present in the image table.
Parameters
----------
@@ -240,7 +230,6 @@ def check_image_features(image_features, image_columns):
None
Nothing is returned.
"""
-
if "Image" in list({img_col.split("_")[0] for img_col in image_columns}):
# Image has already been prepended to most, but not all, columns
level = 1
@@ -281,7 +270,6 @@ def extract_image_features(image_feature_categories, image_df, image_cols, strat
Correctly formatted image feature categories.
"""
-
# Check if the input image feature groups are valid.
check_image_features(image_feature_categories, list(image_df.columns))
@@ -320,13 +308,13 @@ def get_pairwise_correlation(population_df, method="pearson"):
Includes metadata and observation features.
method : str, default "pearson"
Which correlation matrix to use to test cutoff.
+
Returns
-------
list of str
Features to exclude from the population_df.
"""
-
# Check that the input method is supported
method = check_correlation_method(method)
diff --git a/pycytominer/cyto_utils/write_gct.py b/pycytominer/cyto_utils/write_gct.py
index 812811bd..02bba65b 100644
--- a/pycytominer/cyto_utils/write_gct.py
+++ b/pycytominer/cyto_utils/write_gct.py
@@ -1,7 +1,8 @@
-"""
+"""Module to write a gct file from a pandas DataFrame.
+
Transform profiles into a gct (Gene Cluster Text) file
A gct is a tab deliminted text file that traditionally stores gene expression data
-File Format Description: https://clue.io/connectopedia/gct_format
+File Format Description: https://clue.io/connectopedia/gct_format.
Modified from cytominer_scripts "write_gcg" written in R
https://github.com/broadinstitute/cytominer_scripts/blob/master/write_gct.R
@@ -21,7 +22,7 @@ def write_gct(
feature_metadata=None,
version="#1.3",
):
- """Convert profiles to a .gct file
+ """Convert profiles to a .gct file.
Parameters
----------
@@ -47,7 +48,6 @@ def write_gct(
None
Writes gct to file
"""
-
# Note, only version 1.3 is currently supported
assert version == "#1.3", "Only version #1.3 is currently supported." # noqa: S101
diff --git a/pycytominer/feature_select.py b/pycytominer/feature_select.py
index c1fd87d1..9d164e7b 100644
--- a/pycytominer/feature_select.py
+++ b/pycytominer/feature_select.py
@@ -1,6 +1,4 @@
-"""
-Select features to use in downstream analysis based on specified selection method
-"""
+"""Select features to use in downstream analysis based on specified selection method."""
from pycytominer.cyto_utils import (
drop_outlier_features,
@@ -37,7 +35,7 @@ def feature_select(
noise_removal_perturb_groups=None,
noise_removal_stdev_cutoff=None,
):
- """Performs feature selection based on the given operation.
+ """Perform feature selection based on the given operation.
Parameters
----------
@@ -100,7 +98,6 @@ def feature_select(
data.
"""
-
all_ops = [
"variance_threshold",
"correlation_threshold",
diff --git a/pycytominer/normalize.py b/pycytominer/normalize.py
index 7a83ca5f..28d4b8bc 100644
--- a/pycytominer/normalize.py
+++ b/pycytominer/normalize.py
@@ -1,6 +1,4 @@
-"""
-Normalize observation features based on specified normalization method
-"""
+"""Normalize observation features based on specified normalization method."""
import pandas as pd
from sklearn.preprocessing import RobustScaler, StandardScaler
@@ -25,7 +23,7 @@ def normalize(
spherize_method="ZCA-cor",
spherize_epsilon=1e-6,
):
- """Normalize profiling features
+ """Normalize profiling features.
Parameters
----------
@@ -58,15 +56,13 @@ def normalize(
If provided, will write normalized profiles as a specified file type (either CSV or parquet).
If not specified and output_file is provided, then the file will be outputed as CSV as default.
compression_options : str or dict, optional
- Contains compression options as input to
- pd.DataFrame.to_csv(compression=compression_options). pandas version >= 1.2.
+ Contains compression options as input to `pd.DataFrame.to_csv(compression=compression_options)`.
float_format : str, optional
Decimal precision to use in writing output file as input to
pd.DataFrame.to_csv(float_format=float_format). For example, use "%.3g" for 3
decimal precision.
mad_robustize_epsilon: float, optional
- The mad_robustize fudge factor parameter. The function only uses
- this variable if method = "mad_robustize". Set this to 0 if
+ The mad_robustize fudge factor parameter. The function only uses this variable if method = "mad_robustize". Set this to 0 if
mad_robustize generates features with large values.
spherize_center : bool
If the function should center data before sphering (aka whitening). The
@@ -88,38 +84,37 @@ def normalize(
Examples
--------
+ ```python
import pandas as pd
from pycytominer import normalize
- data_df = pd.DataFrame(
- {
- "Metadata_plate": ["a", "a", "a", "a", "b", "b", "b", "b"],
- "Metadata_treatment": [
- "drug",
- "drug",
- "control",
- "control",
- "drug",
- "drug",
- "control",
- "control",
- ],
- "x": [1, 2, 8, 2, 5, 5, 5, 1],
- "y": [3, 1, 7, 4, 5, 9, 6, 1],
- "z": [1, 8, 2, 5, 6, 22, 2, 2],
- "zz": [14, 46, 1, 6, 30, 100, 2, 2],
- }
- ).reset_index(drop=True)
+ data_df = pd.DataFrame({
+ "Metadata_plate": ["a", "a", "a", "a", "b", "b", "b", "b"],
+ "Metadata_treatment": [
+ "drug",
+ "drug",
+ "control",
+ "control",
+ "drug",
+ "drug",
+ "control",
+ "control",
+ ],
+ "x": [1, 2, 8, 2, 5, 5, 5, 1],
+ "y": [3, 1, 7, 4, 5, 9, 6, 1],
+ "z": [1, 8, 2, 5, 6, 22, 2, 2],
+ "zz": [14, 46, 1, 6, 30, 100, 2, 2],
+ }).reset_index(drop=True)
normalized_df = normalize(
profiles=data_df,
features=["x", "y", "z", "zz"],
meta_features="infer",
samples="Metadata_treatment == 'control'",
- method="standardize"
+ method="standardize",
)
+ ```
"""
-
# Load Data
profiles = load_profiles(profiles)
diff --git a/pycytominer/operations/__init__.py b/pycytominer/operations/__init__.py
index 39a59b30..07f5bba8 100644
--- a/pycytominer/operations/__init__.py
+++ b/pycytominer/operations/__init__.py
@@ -1,3 +1,5 @@
+"""Module containing statistical operations for data processing."""
+
from .correlation_threshold import correlation_threshold
from .get_na_columns import get_na_columns
from .noise_removal import noise_removal
diff --git a/pycytominer/operations/correlation_threshold.py b/pycytominer/operations/correlation_threshold.py
index 7c4522ba..d6526949 100644
--- a/pycytominer/operations/correlation_threshold.py
+++ b/pycytominer/operations/correlation_threshold.py
@@ -1,6 +1,7 @@
-"""
-Returns list of features such that no two features have a correlation greater than a
-specified threshold
+"""Module for correlation threshold operation.
+
+The correlation threshold operation list of features such that no two features have a correlation greater than a
+specified threshold.
"""
from pycytominer.cyto_utils import (
@@ -13,7 +14,7 @@
def correlation_threshold(
population_df, features="infer", samples="all", threshold=0.9, method="pearson"
):
- """Exclude features that have correlations above a certain threshold
+ """Exclude features that have correlations above a certain threshold.
Parameters
----------
@@ -38,7 +39,6 @@ def correlation_threshold(
excluded_features : list of str
List of features to exclude from the population_df.
"""
-
# Check that the input method is supported
method = check_correlation_method(method)
@@ -80,8 +80,9 @@ def correlation_threshold(
def determine_high_cor_pair(correlation_row, sorted_correlation_pairs):
- """Select highest correlated variable given a correlation row with columns:
- ["pair_a", "pair_b", "correlation"]. For use in a pandas.apply().
+ """Select highest correlated variable given a correlation row.
+
+ From a row with columns: ["pair_a", "pair_b", "correlation"]. For use in a pandas.apply().
Parameters
----------
@@ -94,7 +95,6 @@ def determine_high_cor_pair(correlation_row, sorted_correlation_pairs):
-------
The feature that has a lower total correlation sum with all other features
"""
-
pair_a = correlation_row["pair_a"]
pair_b = correlation_row["pair_b"]
diff --git a/pycytominer/operations/get_na_columns.py b/pycytominer/operations/get_na_columns.py
index ad36c377..69393318 100644
--- a/pycytominer/operations/get_na_columns.py
+++ b/pycytominer/operations/get_na_columns.py
@@ -1,13 +1,14 @@
-"""
+"""Function to get columns with NA values above a certain threshold.
+
Remove variables with specified threshold of NA values
-Note: This was called `drop_na_columns` in cytominer for R
+Note: This was called `drop_na_columns` in cytominer for R.
"""
from pycytominer.cyto_utils.features import infer_cp_features
def get_na_columns(population_df, features="infer", samples="all", cutoff=0.05):
- """Get features that have more NA values than cutoff defined
+ """Get features that have more NA values than cutoff defined.
Parameters
----------
@@ -30,7 +31,6 @@ def get_na_columns(population_df, features="infer", samples="all", cutoff=0.05):
excluded_features : list of str
List of features to exclude from the population_df.
"""
-
if samples != "all":
population_df.query(samples, inplace=True)
diff --git a/pycytominer/operations/noise_removal.py b/pycytominer/operations/noise_removal.py
index aba1a29e..f52c03ff 100644
--- a/pycytominer/operations/noise_removal.py
+++ b/pycytominer/operations/noise_removal.py
@@ -1,6 +1,4 @@
-"""
-Remove noisy features, as defined by features with excessive standard deviation within the same perturbation group.
-"""
+"""Remove noisy features, as defined by features with excessive standard deviation within the same perturbation group."""
from pycytominer.cyto_utils import infer_cp_features
@@ -12,7 +10,7 @@ def noise_removal(
samples="all",
noise_removal_stdev_cutoff=0.8,
):
- """
+ """Remove features with excessive standard deviation within the same perturbation group.
Parameters
----------
diff --git a/pycytominer/operations/transform.py b/pycytominer/operations/transform.py
index 83a889a4..5077d977 100644
--- a/pycytominer/operations/transform.py
+++ b/pycytominer/operations/transform.py
@@ -13,14 +13,14 @@
class Spherize(BaseEstimator, TransformerMixin):
- """Class to apply a sphering transform (aka whitening) data in the base sklearn
- transform API. Note, this implementation is modified/inspired from the following
- sources:
+ """Class to apply a sphering transform (aka whitening) data in the base sklearn transform API.
+
+ This implementation is modified/inspired from the following sources:
1) A custom function written by Juan C. Caicedo
2) A custom ZCA function at https://github.com/mwv/zca
3) Notes from Niranj Chandrasekaran (https://github.com/cytomining/pycytominer/issues/90)
4) The R package "whitening" written by Strimmer et al (http://strimmerlab.org/software/whitening/)
- 5) Kessy et al. 2016 "Optimal Whitening and Decorrelation" [1]_
+ 5) Kessy et al. 2016 "Optimal Whitening and Decorrelation" [1]_.
Attributes
----------
@@ -33,7 +33,8 @@ class Spherize(BaseEstimator, TransformerMixin):
"""
def __init__(self, epsilon=1e-6, center=True, method="ZCA", return_numpy=False):
- """
+ """Construct a Spherize object.
+
Parameters
----------
epsilon : float, default 1e-6
@@ -65,7 +66,7 @@ def __init__(self, epsilon=1e-6, center=True, method="ZCA", return_numpy=False):
raise ValueError("PCA-cor and ZCA-cor require center=True")
def fit(self, X, y=None):
- """Identify the sphering transform given self.X
+ """Identify the sphering transform given self.X.
Parameters
----------
@@ -221,7 +222,7 @@ def fit(self, X, y=None):
return self
def transform(self, X, y=None):
- """Perform the sphering transform
+ """Perform the sphering transform.
Parameters
----------
@@ -235,7 +236,6 @@ def transform(self, X, y=None):
pandas.core.frame.DataFrame
Spherized dataframe
"""
-
columns = X.columns
# Get Numpy representation of the DataFrame
@@ -261,7 +261,7 @@ def transform(self, X, y=None):
class RobustMAD(BaseEstimator, TransformerMixin):
- """Class to perform a "Robust" normalization with respect to median and mad
+ """Class to perform a "Robust" normalization with respect to median and mad.
scaled = (x - median) / mad
@@ -298,7 +298,7 @@ def fit(self, X, y=None):
return self
def transform(self, X, copy=None):
- """Apply the RobustMAD calculation
+ """Apply the RobustMAD calculation.
Parameters
----------
diff --git a/pycytominer/operations/variance_threshold.py b/pycytominer/operations/variance_threshold.py
index 67d3b767..d4b999fc 100644
--- a/pycytominer/operations/variance_threshold.py
+++ b/pycytominer/operations/variance_threshold.py
@@ -1,6 +1,6 @@
-"""
-Remove variables with near-zero variance.
-Modified from caret::nearZeroVar()
+"""Remove variables with near-zero variance.
+
+Modified from caret::nearZeroVar().
"""
import numpy as np
@@ -11,7 +11,7 @@
def variance_threshold(
population_df, features="infer", samples="all", freq_cut=0.05, unique_cut=0.01
):
- """Exclude features that have low variance (low information content)
+ """Exclude features that have low variance (low information content).
Parameters
----------
@@ -42,7 +42,6 @@ def variance_threshold(
List of features to exclude from the population_df.
"""
-
if not 0 <= freq_cut <= 1:
raise ValueError("freq_cut variable must be between (0 and 1)")
if not 0 <= unique_cut <= 1:
@@ -80,7 +79,6 @@ def variance_threshold(
def calculate_frequency(feature_column, freq_cut):
"""Calculate frequency of second most common to most common feature.
- Used in pandas.apply()
Parameters
----------
@@ -97,7 +95,6 @@ def calculate_frequency(feature_column, freq_cut):
Feature name if it passes threshold, "NA" otherwise
"""
-
val_count = feature_column.value_counts()
try:
max_count = val_count.iloc[0]
diff --git a/pyproject.toml b/pyproject.toml
index a8ea5428..7452a30b 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -1,7 +1,7 @@
[tool.poetry]
name = "pycytominer"
# This version is a placeholder updated during build by poetry-dynamic-versioning
-version = "0.0.0"
+version = "1.1.0.post29.dev0+9615c0d"
description = "Python package for processing image-based profiling data"
authors = [
"Erik Serrano",
@@ -39,7 +39,9 @@ license = "BSD-3-Clause"
readme = "README.md"
homepage = "https://pycytominer.readthedocs.io/"
repository = "https://github.com/cytomining/pycytominer"
-include = [{ path = "poetry.lock", format = "sdist" }]
+include = [
+ { path = "poetry.lock", format = "sdist" },
+]
[tool.poetry.dependencies]
python = ">=3.8,<4.0"
@@ -78,22 +80,18 @@ ruff = "^0.3.4"
optional = true
[tool.poetry.group.docs.dependencies]
-mkdocs = "*"
-sphinx = "~7.1"
-sphinx-autobuild = "^2021.3.14"
-sphinx-autoapi = "^3.0.0"
+mkdocs = "^1.6.0"
ipython = "^8.12"
-groundwork-sphinx-theme = "^1.1.1"
-sphinx-copybutton = "^0.5.2"
nbsphinx = "^0.9.3"
-m2r2 = "^0.3.3.post2"
-furo = "^2023.9.10"
mock = "^5.1.0"
-autodoc = "^0.5.0"
dunamai = "^1.19.0"
+mkdocs-material = "^9.5.19"
+mkdocstrings = {extras = ["python"], version = "^0.23.0"}
+mkdocs-include-markdown-plugin = "^6.0.5"
+mkdocs-jupyter = "^0.24.7"
[tool.poetry-dynamic-versioning]
-enable = true
+enable = false
style = "pep440"
vcs = "git"
@@ -117,6 +115,10 @@ version_files = [
target-version = "py38"
line-length = 88
fix = true
+extend-include = ["*.ipynb"]
+exclude = [
+ "walkthroughs/nbconverted/*",
+]
[tool.ruff.lint]
select = [
@@ -144,6 +146,8 @@ select = [
"S",
# isort
"I",
+ # pydocstyle
+ "D",
# The following checks are disabled, but we are working towards enabling them in the future.
# flake8-bugbear
# "B",
@@ -158,12 +162,21 @@ ignore = [
# DoNotAssignLambda
"E731",
]
+exclude = ["*.ipynb"]
+
+[tool.ruff.lint.pydocstyle]
+# Use Google-style docstrings.
+convention = "numpy"
[tool.ruff.lint.per-file-ignores]
# Ignore `E402` and `F401` (unusued imports) in all `__init__.py` files
"__init__.py" = ["E402", "F401"]
-# Ignore assert statements in tests
-"tests/*" = ["S101"]
+"tests/*" = [
+ # Allow assert statements in tests
+ "S101",
+ # Disable docstring checks in tests
+ "D",
+]
"pycytominer/cyto_utils/*" = [
# I (isort) is ignored due to circular dependencies in the cyto_utils module
"I",
@@ -173,6 +186,8 @@ ignore = [
[tool.ruff.format]
preview = true
+# Enable reformatting of code snippets in docstrings.
+docstring-code-format = true
[tool.pytest.ini_options]
testpaths = "tests"
diff --git a/tests/test_aggregate.py b/tests/test_aggregate.py
index d1a08889..694b7f16 100644
--- a/tests/test_aggregate.py
+++ b/tests/test_aggregate.py
@@ -49,9 +49,7 @@
def test_aggregate_median_allvar():
- """
- Testing aggregate pycytominer function
- """
+ """Testing aggregate pycytominer function."""
aggregate_result = aggregate(
population_df=data_df, strata=["g"], features="infer", operation="median"
)
@@ -78,9 +76,7 @@ def test_aggregate_median_allvar():
def test_aggregate_mean_allvar():
- """
- Testing aggregate pycytominer function
- """
+ """Testing aggregate pycytominer function."""
aggregate_result = aggregate(
population_df=data_df, strata=["g"], features="infer", operation="mean"
)
@@ -95,9 +91,7 @@ def test_aggregate_mean_allvar():
def test_aggregate_median_subsetvar():
- """
- Testing aggregate pycytominer function
- """
+ """Testing aggregate pycytominer function."""
aggregate_result = aggregate(
population_df=data_df, strata=["g"], features=["Cells_x"], operation="median"
)
@@ -109,9 +103,7 @@ def test_aggregate_median_subsetvar():
def test_aggregate_mean_subsetvar():
- """
- Testing aggregate pycytominer function
- """
+ """Testing aggregate pycytominer function."""
aggregate_result = aggregate(
population_df=data_df, strata=["g"], features=["Cells_x"], operation="mean"
)
@@ -123,10 +115,7 @@ def test_aggregate_mean_subsetvar():
def test_aggregate_median_dtype_confirm():
- """
- Testing aggregate pycytominer function
- """
-
+ """Testing aggregate pycytominer function."""
# Convert dtype of one variable to object
data_dtype_df = data_df.copy()
data_dtype_df.Cells_x = data_dtype_df.Cells_x.astype(str)
@@ -145,10 +134,7 @@ def test_aggregate_median_dtype_confirm():
def test_aggregate_median_with_missing_values():
- """
- Testing aggregate pycytominer function
- """
-
+ """Testing aggregate pycytominer function."""
# Convert dtype of one variable to object
data_dtype_df = data_missing_df.copy()
data_dtype_df.Cells_x = data_dtype_df.Cells_x.astype(str)
@@ -167,10 +153,7 @@ def test_aggregate_median_with_missing_values():
def test_aggregate_compute_object_count():
- """
- Testing aggregate pycytominer function
- """
-
+ """Testing aggregate pycytominer function."""
aggregate_result = aggregate(
population_df=data_df,
strata=["g"],
@@ -212,10 +195,7 @@ def test_aggregate_compute_object_count():
def test_aggregate_incorrect_object_feature():
- """
- Testing aggregate pycytominer function
- """
-
+ """Testing aggregate pycytominer function."""
incorrect_object_feature = "DOES NOT EXIST"
with pytest.raises(KeyError) as err:
@@ -250,10 +230,7 @@ def test_aggregate_incorrect_object_feature():
def test_custom_objectnumber_feature():
- """
- Testing aggregate pycytominer function
- """
-
+ """Testing aggregate pycytominer function."""
data_df_copy = data_df.copy().rename(
columns={"Metadata_ObjectNumber": "Custom_ObjectNumber_Feature"}
)
@@ -287,9 +264,7 @@ def test_custom_objectnumber_feature():
def test_output_type():
- """
- Testing aggregate pycytominer function
- """
+ """Testing aggregate pycytominer function."""
# dictionary with the output name associated with the file type
output_dict = {"csv": test_output_file_csv, "parquet": test_output_file_parquet}
diff --git a/tests/test_cyto_utils/conftest.py b/tests/test_cyto_utils/conftest.py
index 20ac6cc9..78422f70 100644
--- a/tests/test_cyto_utils/conftest.py
+++ b/tests/test_cyto_utils/conftest.py
@@ -1,6 +1,4 @@
-"""
-conftest.py for pytest
-"""
+"""conftest.py for pytest."""
import os
import pathlib
@@ -14,10 +12,7 @@
@pytest.fixture(name="data_dir_cell_locations")
def fixture_data_dir_cell_locations() -> str:
- """
- Provide a data directory for cell_locations test data
- """
-
+ """Provide a data directory for cell_locations test data."""
return (
f"{pathlib.Path(__file__).parent.parent}/test_data/cell_locations_example_data"
)
@@ -25,9 +20,7 @@ def fixture_data_dir_cell_locations() -> str:
@pytest.fixture(name="metadata_input_file")
def fixture_metadata_input_file(data_dir_cell_locations: str) -> str:
- """
- Provide a metadata input file for cell_locations test data
- """
+ """Provide a metadata input file for cell_locations test data."""
return os.path.join(
data_dir_cell_locations, "test_BR00126114_load_data_with_illum.parquet"
)
@@ -35,33 +28,25 @@ def fixture_metadata_input_file(data_dir_cell_locations: str) -> str:
@pytest.fixture(name="single_cell_input_file")
def fixture_single_cell_input_file(data_dir_cell_locations: str) -> str:
- """
- Provide a single cell input file for cell_locations test data
- """
+ """Provide a single cell input file for cell_locations test data."""
return os.path.join(data_dir_cell_locations, "test_BR00126114.sqlite")
@pytest.fixture(name="metadata_input_file_s3")
def fixture_metadata_input_file_s3() -> str:
- """
- Provide a metadata input file for cell_locations test data
- """
+ """Provide a metadata input file for cell_locations test data."""
return "s3://cellpainting-gallery/cpg0016-jump/source_4/workspace/load_data_csv/2021_08_23_Batch12/BR00126114/load_data_with_illum.parquet"
@pytest.fixture(name="single_cell_input_file_s3")
def fixture_single_cell_input_file_s3() -> str:
- """
- Provide a single cell input file for cell_locations test data
- """
+ """Provide a single cell input file for cell_locations test data."""
return "s3://cellpainting-gallery/cpg0016-jump/source_4/workspace/backend/2021_08_23_Batch12/BR00126114/BR00126114.sqlite"
@pytest.fixture(name="metadata_input_dataframe")
def fixture_metadata_input_dataframe(metadata_input_file: str) -> pd.DataFrame:
- """
- Provide a metadata input file for cell_locations test data
- """
+ """Provide a metadata input file for cell_locations test data."""
return pd.read_parquet(metadata_input_file)
@@ -69,9 +54,7 @@ def fixture_metadata_input_dataframe(metadata_input_file: str) -> pd.DataFrame:
def fixture_single_cell_input_engine(
single_cell_input_file: str,
) -> sqlalchemy.engine.Engine:
- """
- Provide a single cell input file for cell_locations test data
- """
+ """Provide a single cell input file for cell_locations test data."""
return sqlalchemy.create_engine(f"sqlite:///{single_cell_input_file}")
@@ -80,9 +63,7 @@ def fixture_cell_loc_obj1(
metadata_input_file: str,
single_cell_input_file: str,
) -> CellLocation:
- """
- Provide a CellLocation object with file inputs
- """
+ """Provide a CellLocation object with file inputs."""
return CellLocation(
metadata_input=metadata_input_file,
single_cell_input=single_cell_input_file,
@@ -94,9 +75,7 @@ def fixture_cell_loc_obj2(
metadata_input_dataframe: pd.DataFrame,
single_cell_input_engine: sqlalchemy.engine.Engine,
) -> CellLocation:
- """
- Provide a CellLocation object with in-memory inputs
- """
+ """Provide a CellLocation object with in-memory inputs."""
return CellLocation(
metadata_input=metadata_input_dataframe,
single_cell_input=single_cell_input_engine,
@@ -108,9 +87,7 @@ def fixture_cell_loc_obj3(
metadata_input_file_s3: str,
single_cell_input_file_s3: str,
) -> CellLocation:
- """
- Provide a CellLocation object with s3 inputs
- """
+ """Provide a CellLocation object with s3 inputs."""
return CellLocation(
metadata_input=metadata_input_file_s3,
single_cell_input=single_cell_input_file_s3,
diff --git a/tests/test_cyto_utils/test_DeepProfiler_processing.py b/tests/test_cyto_utils/test_DeepProfiler_processing.py
index 600006d3..cbf51c3b 100644
--- a/tests/test_cyto_utils/test_DeepProfiler_processing.py
+++ b/tests/test_cyto_utils/test_DeepProfiler_processing.py
@@ -1,6 +1,4 @@
-"""
-This tests the output from a DeepProfiler run (May 2021)
-"""
+"""This tests the output from a DeepProfiler run (May 2021)."""
import os
import pathlib
@@ -24,7 +22,7 @@
@pytest.fixture(scope="session")
def deep_profiler_data(tmp_path_factory):
- """This fixture returns the DeepProfilerData object and the output folder"""
+ """This fixture returns the DeepProfilerData object and the output folder."""
example_project_dir = ROOT_DIR / "tests" / "test_data" / "DeepProfiler_example_data"
profile_dir = example_project_dir / "outputs" / "results" / "features"
index_file = example_project_dir / "inputs" / "metadata" / "test_index.csv"
@@ -41,7 +39,7 @@ def deep_profiler_data(tmp_path_factory):
@pytest.fixture(scope="session")
def single_cell_deep_profiler(deep_profiler_data):
- """This fixture returns the single cell data and the SingleCellDeepProfiler object"""
+ """This fixture returns the single cell data and the SingleCellDeepProfiler object."""
deep_data, output_folder = deep_profiler_data
# compile single cell data from DP run
@@ -52,7 +50,7 @@ def single_cell_deep_profiler(deep_profiler_data):
def test_single_cell(single_cell_deep_profiler):
- """Test output from SingleCellDeepProfiler.get_single_cells()"""
+ """Test output from SingleCellDeepProfiler.get_single_cells()."""
single_cells, single_cells_DP, output_folder = single_cell_deep_profiler
meta_cols = [x for x in single_cells.columns if x.startswith("Location_")]
@@ -66,8 +64,7 @@ def test_single_cell(single_cell_deep_profiler):
def test_single_cell_normalize(single_cell_deep_profiler):
- """Test output from SingleCellDeepProfiler.normalize_deep_single_cells()"""
-
+ """Test output from SingleCellDeepProfiler.normalize_deep_single_cells()."""
single_cells, single_cells_DP, output_folder = single_cell_deep_profiler
# normalize single cell data with DP processing
diff --git a/tests/test_cyto_utils/test_cell_locations.py b/tests/test_cyto_utils/test_cell_locations.py
index e8100598..622c4121 100644
--- a/tests/test_cyto_utils/test_cell_locations.py
+++ b/tests/test_cyto_utils/test_cell_locations.py
@@ -1,4 +1,4 @@
-"""This tests the output from CellLocation class"""
+"""This tests the output from CellLocation class."""
from typing import List, Type
@@ -14,7 +14,6 @@ def get_metadata_input_dataframe(cell_loc: CellLocation) -> pd.DataFrame:
Gathers the metadata input dataframe given various conditions
from a CellLocation object.
"""
-
return (
pd.read_parquet(
cell_loc.metadata_input,
@@ -41,10 +40,7 @@ def test_output_shape_and_required_columns(
cell_loc_param: List[str],
request: Type[FixtureRequest],
):
- """
- This tests the shape of the output from CellLocation class and verifies that the required columns are present
- """
-
+ """This tests the shape of the output from CellLocation class and verifies that the required columns are present."""
cls_cell_loc = request.getfixturevalue(cell_loc_param)
cell_loc = cls_cell_loc.add_cell_location()
metadata_input_dataframe = get_metadata_input_dataframe(cell_loc=cls_cell_loc)
@@ -74,10 +70,7 @@ def test_output_value_correctness(
cell_loc_param: List[str],
request: Type[FixtureRequest],
):
- """
- This tests the correctness of the values in the output from CellLocation class by comparing the values in the output to the values in the input
- """
-
+ """This tests the correctness of the values in the output from CellLocation class by comparing the values in the output to the values in the input."""
cls_cell_loc = request.getfixturevalue(cell_loc_param)
cell_loc = cls_cell_loc.add_cell_location()
metadata_input_dataframe = get_metadata_input_dataframe(cell_loc=cls_cell_loc)
diff --git a/tests/test_cyto_utils/test_cells.py b/tests/test_cyto_utils/test_cells.py
index 395be7df..ac63fe01 100644
--- a/tests/test_cyto_utils/test_cells.py
+++ b/tests/test_cyto_utils/test_cells.py
@@ -205,9 +205,7 @@ def build_random_data(
def test_SingleCells_init():
- """
- Testing initialization of SingleCells
- """
+ """Testing initialization of SingleCells."""
assert AP.sql_file == TMP_SQLITE_FILE
assert AP.strata == ["Metadata_Plate", "Metadata_Well"]
assert AP.merge_cols == ["TableNumber", "ImageNumber"]
@@ -234,9 +232,7 @@ def test_SingleCells_init():
def test_SingleCells_reset_variables():
- """
- Testing initialization of SingleCells
- """
+ """Testing initialization of SingleCells."""
ap_switch = SingleCells(sql_file=TMP_SQLITE_FILE)
assert ap_switch.subsample_frac == 1
assert ap_switch.subsample_n == "all"
@@ -529,10 +525,7 @@ def test_merge_single_cells_subsample():
def test_merge_single_cells_annotate():
- """
- Tests SingleCells.merge_single_cells using optional annotate functionality
- """
-
+ """Tests SingleCells.merge_single_cells using optional annotate functionality."""
expected_sc_merged_df = annotate(
profiles=AP.merge_single_cells(),
platemap=PLATEMAP_DF,
@@ -546,10 +539,7 @@ def test_merge_single_cells_annotate():
def test_merge_single_cells_cytominer_database_test_file():
- """
- Tests SingleCells.merge_single_cells using cytominer-database test file
- """
-
+ """Tests SingleCells.merge_single_cells using cytominer-database test file."""
# read test file based on cytominer-database exports
sql_path = pathlib.Path(
f"{os.path.dirname(__file__)}/../test_data/cytominer_database_example_data/test_SQ00014613.sqlite",
@@ -1007,9 +997,7 @@ def test_add_image_features():
def test_load_non_canonical_image_table():
- """
- Loading an image table with non-canonical image table name
- """
+ """Loading an image table with non-canonical image table name."""
# test for exception loading image table with default table name "image"
with pytest.raises(OperationalError):
AP_IMAGE_DIFF_NAME.load_image()
diff --git a/tests/test_cyto_utils/test_features_count_na.py b/tests/test_cyto_utils/test_features_count_na.py
index 2232ece6..4de9be20 100644
--- a/tests/test_cyto_utils/test_features_count_na.py
+++ b/tests/test_cyto_utils/test_features_count_na.py
@@ -12,9 +12,7 @@
def test_count_na_features():
- """
- Testing count_na_features pycytominer function
- """
+ """Testing count_na_features pycytominer function."""
count_na_features_result = count_na_features(
population_df=data_df, features=["x", "zz"]
)
diff --git a/tests/test_cyto_utils/test_output.py b/tests/test_cyto_utils/test_output.py
index 72d5a9cf..bf8aec40 100644
--- a/tests/test_cyto_utils/test_output.py
+++ b/tests/test_cyto_utils/test_output.py
@@ -75,10 +75,7 @@ def test_output_tsv():
def test_output_parquet():
- """
- Tests using output function with parquet type
- """
-
+ """Tests using output function with parquet type."""
output_filename = pathlib.Path(f"{TMPDIR}/test_output.parquet")
# test with base output arguments and
diff --git a/tests/test_feature_select.py b/tests/test_feature_select.py
index 8cc6a205..b4f8e7c1 100644
--- a/tests/test_feature_select.py
+++ b/tests/test_feature_select.py
@@ -74,9 +74,7 @@
def test_feature_select_noise_removal():
- """
- Testing noise_removal feature selection operation
- """
+ """Testing noise_removal feature selection operation."""
# Set perturbation groups for the test dataframes
data_df_groups = ["a", "a", "a", "b", "b", "b"]
@@ -223,9 +221,7 @@ def test_feature_select_noise_removal():
def test_feature_select_get_na_columns():
- """
- Testing feature_select and get_na_columns pycytominer function
- """
+ """Testing feature_select and get_na_columns pycytominer function."""
result = feature_select(
data_na_df, features=data_na_df.columns.tolist(), operation="drop_na_columns"
)
@@ -256,9 +252,7 @@ def test_feature_select_get_na_columns():
def test_feature_select_get_na_columns_feature_infer():
- """
- Testing feature_select and get_na_columns pycytominer function
- """
+ """Testing feature_select and get_na_columns pycytominer function."""
result = feature_select(
data_feature_infer_df,
features="infer",
@@ -288,9 +282,7 @@ def test_feature_select_get_na_columns_feature_infer():
def test_feature_select_variance_threshold():
- """
- Testing feature_select and variance_threshold pycytominer function
- """
+ """Testing feature_select and variance_threshold pycytominer function."""
result = feature_select(
data_unique_test_df,
features=data_unique_test_df.columns.tolist(),
@@ -331,10 +323,7 @@ def test_feature_select_variance_threshold():
def test_feature_select_correlation_threshold():
- """
- Testing feature_select and correlation_threshold pycytominer function
- """
-
+ """Testing feature_select and correlation_threshold pycytominer function."""
result = feature_select(
data_df, features=data_df.columns.tolist(), operation="correlation_threshold"
)
@@ -417,10 +406,7 @@ def test_feature_select_compress():
def test_feature_select_blocklist():
- """
- Testing feature_select and get_na_columns pycytominer function
- """
-
+ """Testing feature_select and get_na_columns pycytominer function."""
data_blocklist_df = pd.DataFrame({
"Nuclei_Correlation_Manders_AGP_DNA": [1, 3, 8, 5, 2, 2],
"y": [1, 2, 8, 5, 2, 1],
@@ -442,9 +428,7 @@ def test_feature_select_blocklist():
def test_feature_select_drop_outlier():
- """
- Testing feature_select and get_na_columns pycytominer function
- """
+ """Testing feature_select and get_na_columns pycytominer function."""
result = feature_select(
data_outlier_df, features="infer", operation="drop_outliers"
)
@@ -467,9 +451,7 @@ def test_feature_select_drop_outlier():
def test_output_type():
- """
- Testing feature_select pycytominer function
- """
+ """Testing feature_select pycytominer function."""
# dictionary with the output name associated with the file type
output_dict = {"csv": output_test_file_csv, "parquet": output_test_file_parquet}
diff --git a/tests/test_normalize.py b/tests/test_normalize.py
index 223a0992..b7e8082f 100644
--- a/tests/test_normalize.py
+++ b/tests/test_normalize.py
@@ -84,7 +84,7 @@ def test_normalize_standardize_allsamples():
Testing normalize pycytominer function
method = "standardize"
meta_features = None
- samples="all"
+ samples="all".
"""
normalize_result = normalize(
profiles=data_df.copy(),
@@ -120,7 +120,7 @@ def test_normalize_standardize_ctrlsamples():
Testing normalize pycytominer function
method = "standardize"
meta_features = None
- samples="Metadata_treatment == 'control'"
+ samples="Metadata_treatment == 'control'".
"""
normalize_result = normalize(
profiles=data_df.copy(),
@@ -156,7 +156,7 @@ def test_normalize_robustize_allsamples():
Testing normalize pycytominer function
method = "robustize"
meta_features = None
- samples="all"
+ samples="all".
"""
normalize_result = normalize(
profiles=data_df.copy(),
@@ -192,7 +192,7 @@ def test_normalize_robustize_ctrlsamples():
Testing normalize pycytominer function
method = "robustize"
meta_features = None
- samples="Metadata_treatment == 'control'"
+ samples="Metadata_treatment == 'control'".
"""
normalize_result = normalize(
profiles=data_df.copy(),
@@ -228,7 +228,7 @@ def test_normalize_robustize_mad_allsamples():
Testing normalize pycytominer function
method = "mad_robustize"
meta_features = None
- samples="all"
+ samples="all".
"""
normalize_result = normalize(
profiles=data_df.copy(),
@@ -264,7 +264,7 @@ def test_normalize_robustize_mad_allsamples_novar():
Testing normalize pycytominer function
method = "mad_robustize"
meta_features = None
- samples="all"
+ samples="all".
"""
features = ["x", "y", "z", "zz", "yy"]
@@ -308,7 +308,7 @@ def test_normalize_standardize_allsamples_fromfile():
data_file provided
method = "standardize"
meta_features = None
- samples="all"
+ samples="all".
"""
normalize_result = normalize(
profiles=data_file,
@@ -356,7 +356,7 @@ def test_normalize_standardize_allsamples_output():
data_file provided
method = "standardize"
meta_features = None
- samples="all"
+ samples="all".
"""
out_normalize_file = os.path.join(tmpdir, "test_normalize_output.csv")
@@ -501,9 +501,7 @@ def test_normalize_spherize():
def test_spherize_epsilon():
- """
- Test that epsilon is successfully passed to the spherize transform method
- """
+ """Test that epsilon is successfully passed to the spherize transform method."""
sphere_norm_df = normalize(
data_spherize_df, features=["a", "b", "c", "d"], meta_features=["id"]
)
@@ -532,9 +530,7 @@ def test_spherize_epsilon():
def test_output_type():
- """
- Testing normalize pycytominer function with output
- """
+ """Testing normalize pycytominer function with output."""
# dictionary with the output name associated with the file type
output_dict = {"csv": output_test_file_csv, "parquet": output_test_file_parquet}
diff --git a/tests/test_operations/test_get_na_columns.py b/tests/test_operations/test_get_na_columns.py
index 9c8bd557..555d9c01 100644
--- a/tests/test_operations/test_get_na_columns.py
+++ b/tests/test_operations/test_get_na_columns.py
@@ -13,9 +13,7 @@
def test_get_na_columns():
- """
- Testing get_na_columns pycytominer function
- """
+ """Testing get_na_columns pycytominer function."""
get_na_columns_result = get_na_columns(
population_df=data_df, features=["x", "y", "zz"], cutoff=0.4
)
@@ -41,9 +39,7 @@ def test_get_na_columns():
def test_get_na_columns_sample():
- """
- Testing get_na_columns pycyominer function with samples option
- """
+ """Testing get_na_columns pycyominer function with samples option."""
data_sample_id_df = data_df.assign(
Metadata_sample=[f"sample_{x}" for x in range(0, data_df.shape[0])]
)
diff --git a/tests/test_operations/test_transform.py b/tests/test_operations/test_transform.py
index d66e1615..85a9f6e0 100644
--- a/tests/test_operations/test_transform.py
+++ b/tests/test_operations/test_transform.py
@@ -79,9 +79,7 @@ def test_spherize_precenter():
def test_robust_mad():
- """
- Testing the RobustMAD class
- """
+ """Testing the RobustMAD class."""
scaler = RobustMAD()
scaler = scaler.fit(data_df)
transform_df = scaler.transform(data_df)
diff --git a/tests/test_operations/test_variance_threshold.py b/tests/test_operations/test_variance_threshold.py
index a1a19764..ceb4e97f 100644
--- a/tests/test_operations/test_variance_threshold.py
+++ b/tests/test_operations/test_variance_threshold.py
@@ -28,9 +28,7 @@
def test_calculate_frequency():
- """
- Testing calculate_frequency pycytominer function for variance threshold calculation
- """
+ """Testing calculate_frequency pycytominer function for variance threshold calculation."""
freq_cut = 0.05
excluded_features_freq = data_df.apply(
diff --git a/walkthroughs/nbconverted/single_cell_usage.py b/walkthroughs/nbconverted/single_cell_usage.py
deleted file mode 100644
index c21c8691..00000000
--- a/walkthroughs/nbconverted/single_cell_usage.py
+++ /dev/null
@@ -1,242 +0,0 @@
-#!/usr/bin/env python
-
-# # Single-cell Profiling Walkthrough
-#
-# Welcome to this walkthrough where we will guide you through the process of extracting single cell morphology features using the [pycytominer](https://github.com/cytomining/pycytominer) API.
-#
-# For this walkthrough, we will be working with the NF1-Schwann cell morphology dataset.
-# If you would like the more information about this dataset, you can refer to this [repository](https://github.com/WayScience/nf1_cellpainting_data)
-#
-# From the mentioned repo, we specifically used this [dataset](https://github.com/WayScience/nf1_cellpainting_data/tree/main/2.cellprofiler_analysis/analysis_output) and the associated [metadata](https://github.com/WayScience/nf1_cellpainting_data/tree/main/0.download_data/metadata) to generate the walkthrough.
-#
-#
-# Let's get started with the walkthrough below!
-
-# In[1]:
-
-
-import pathlib
-
-# ignore mix type warnings from pandas
-import warnings
-
-import pandas as pd
-
-from pycytominer import annotate, feature_select, normalize
-
-# pycytominer imports
-from pycytominer.cyto_utils.cells import SingleCells
-
-warnings.filterwarnings("ignore")
-
-
-# ## About the inputs
-#
-#
-# In this section, we will set up the expected input and output paths that will be generated throughout this walkthrough. Let's take a look at the explanation of these inputs and outputs.
-#
-# For this workflow, we have two main inputs:
-#
-# - **plate_data** (SQLite file): This contains the quantified single-cell morphology features that we'll be working with.
-# - **plate_map** (CSV file): This contains additional information related to the cells, providing valuable context of our single-cell morphology dataset.
-#
-# Now, let's explore the outputs generated in this workflow. In this walkthrough, we will be generating four profiles:
-#
-# - **sc_profiles_path**: This refers to the single-cell morphology profile that will be generated.
-# - **anno_profiles_path**: This corresponds to the annotated single-cell morphology profile, meaning the information from the **plate_map** is added to each single-cell
-# - **norm_profiles_path**: This represents the normalized single-cell morphology profile.
-# - **feat_profiles_path**: Lastly, this refers to the selected features from the single-cell morphology profile.
-#
-# **Note**: All profiles are outputted as `.csv` files, however, users can output these files as `parquet` or other file file formats.
-# For more information about output formats, please refer the documentation [here](https://pycytominer.readthedocs.io/en/latest/pycytominer.cyto_utils.html#pycytominer.cyto_utils.output.output)
-#
-# These profiles will serve as important outputs that will help us analyze and interpret the single-cell morphology data effectively. Now that we have a clear understanding of the inputs and outputs, let's proceed further in our walkthrough.
-
-# In[2]:
-
-
-# Setting file paths
-data_dir = pathlib.Path("./data/").resolve(strict=True)
-metadata_dir = (data_dir / "metadata").resolve(strict=True)
-out_dir = pathlib.Path("results")
-out_dir.mkdir(exist_ok=True)
-
-# input file paths
-plate_data = pathlib.Path("./data/nf1_data.sqlite").resolve(strict=True)
-plate_map = (metadata_dir / "platemap_NF1_CP.csv").resolve(strict=True)
-
-# setting output paths
-sc_profiles_path = out_dir / "nf1_single_cell_profile.csv.gz"
-anno_profiles_path = out_dir / "nf1_annotated_profile.csv.gz"
-norm_profiles_path = out_dir / "nf1_normalized_profile.csv.gz"
-feat_profiles_path = out_dir / "nf1_features_profile.csv.gz"
-
-
-# ## Generating Merged Single-cell Morphology Dataset
-#
-# In this section of the walkthrough, our goal is to load the NF1 dataset and create a merged single-cell morphology dataset.
-#
-# Currently, the NF1 morphology `SQLite` dataset was generated by using [CellProfiler's](https://github.com/CellProfiler/CellProfiler) [ExportToDatabase](https://cellprofiler-manual.s3.amazonaws.com/CellProfiler-4.2.5/modules/fileprocessing.html?highlight=exporttodatabase#module-cellprofiler.modules.exporttodatabase) function, where each table represents a different compartment, such as Image, Cell, Nucleus, and Cytoplasm.
-#
-# To achieve this, we will utilize the `SingleCells` class, which offers a range of functionalities specifically designed for single-cell analysis. You can find detailed documentation on these functionalities [here](https://pycytominer.readthedocs.io/en/latest/pycytominer.cyto_utils.html#pycytominer.cyto_utils.cells.SingleCells).
-#
-# However, for our purpose in this walkthrough, we will focus on using the `SingleCells` class to merge all the tables within the NF1 sqlite file into a merged single-cell morphology dataset.
-#
-# ### Updating defaults
-# Before we proceed further, it is important to update the default parameters in the `SingleCells`class to accommodate the table name changes in our NF1 dataset.
-#
-# Since the table names in our NF1 dataset differ from the default table names recognized by the `SingleCells` class, we need to make adjustments to ensure proper recognition of these table name changes.
-
-# In[3]:
-
-
-# update compartment names and strata
-strata = ["Image_Metadata_Well", "Image_Metadata_Plate"]
-compartments = ["Per_Cells", "Per_Cytoplasm", "Per_Nuclei"]
-
-# Updating linking columns for merging all compartments
-linking_cols = {
- "Per_Cytoplasm": {
- "Per_Cells": "Cytoplasm_Parent_Cells",
- "Per_Nuclei": "Cytoplasm_Parent_Nuclei",
- },
- "Per_Cells": {"Per_Cytoplasm": "Cells_Number_Object_Number"},
- "Per_Nuclei": {"Per_Cytoplasm": "Nuclei_Number_Object_Number"},
-}
-
-
-# Now that we have stored the updated parameters, we can use them as inputs for `SingleCells` class to merge all the NF1 sqlite tables into a single consolidated dataset.
-#
-# This is done through the `merge_single_cells` method. For more infromation about `merge_single_cells` please refer to the documentation [here](https://pycytominer.readthedocs.io/en/latest/pycytominer.cyto_utils.html#pycytominer.cyto_utils.cells.SingleCells.merge_single_cells)
-
-# In[4]:
-
-
-# setting up sqlite address
-sqlite_address = f"sqlite:///{plate_data!s}"
-
-# loading single cell morphology data into pycyotminer's SingleCells Object
-single_cell_profile = SingleCells(
- sql_file=sqlite_address,
- compartments=compartments,
- compartment_linking_cols=linking_cols,
- image_table_name="Per_Image",
- strata=strata,
- merge_cols=["ImageNumber"],
- image_cols="ImageNumber",
- load_image_data=True,
-)
-
-# merging all sqlite table into a single tabular dataset (csv) and save as
-# compressed csv file
-single_cell_profile.merge_single_cells(
- sc_output_file=sc_profiles_path, compression_options="gzip"
-)
-
-
-# Now that we have created our merged single-cell profile, let's move on to the next step: loading our `platemaps`.
-#
-# `Platemaps` provide us with additional information that is crucial for our analysis. They contain details such as well positions, genotypes, gene names, perturbation types, and more. In other words, platemaps serve as a valuable source of metadata for our single-cell morphology profile.
-#
-
-# In[5]:
-
-
-# loading plate map and display it
-platemap_df = pd.read_csv(plate_map)
-
-# displaying platemap
-print(platemap_df.columns.tolist())
-
-
-# ## Annotation
-#
-# In this step of the walkthrough, we will combine the metadata with the merged single-cell morphology dataset. To accomplish this, we will utilize the `annotation` function provided by `pycytominer`.
-#
-# The `annotation` function takes two inputs: the merged single-cell morphology dataset and its associated plate map. By combining these two datasets, we will generate an annotated_profile that contains enriched information.
-#
-# More information about the `annotation` function can be found [here](https://pycytominer.readthedocs.io/en/latest/pycytominer.html#module-pycytominer.annotate)
-#
-
-# In[6]:
-
-
-# annotating merged single-cell profile with metadata
-annotate(
- profiles=sc_profiles_path,
- platemap=platemap_df,
- join_on=["Metadata_well_position", "Image_Metadata_Well"],
- output_file=anno_profiles_path,
- compression_options="gzip",
-)
-
-# save message display
-print(f"Annotated profile saved in: {anno_profiles_path}")
-
-
-# ## Normalization Step
-#
-# The next step is to normalize our dataset using the `normalize` function provided by `pycytominer`.
-# More information regards `pycytominer`'s `normalize` function can be found [here](https://pycytominer.readthedocs.io/en/latest/pycytominer.html#module-pycytominer.normalize)
-#
-# Normalization is a critical preprocessing step that improves the quality of our dataset. It addresses two key challenges: mitigating the impact of outliers and handling variations in value scales. By normalizing the data, we ensure that our downstream analysis is not heavily influenced by these factors.
-#
-# Additionally, normalization plays a crucial role in determining feature importance (which is crucial for our last step). By bringing all features to a comparable scale, it enables the identification of important features without biases caused by outliers or widely-scaled values.
-#
-# To normalize our annotated single-cell morphology profile, we will utilize the `normalize` function from `pycytominer`. This function is specifically designed to handle the normalization process for cytometry data.
-
-# In[7]:
-
-
-# normalize dataset
-normalize(
- profiles=anno_profiles_path,
- features="infer",
- image_features=False,
- meta_features="infer",
- samples="all",
- method="standardize",
- output_file=norm_profiles_path,
- compression_options="gzip",
-)
-
-# save message display
-print(f"Normalized profile saved in: {norm_profiles_path}")
-
-
-# ## Feature Selection
-#
-#
-# In the final section of our walkthrough, we will utilize the normalized dataset to extract important morphological features and generate a selected features profile.
-#
-# To accomplish this, we will make use of the `feature_select` function provided by `pycytominer`.
-# Using `pycytominer`'s `feature_select` function to our dataset, we can identify the most informative morphological features that contribute significantly to the variations observed in our data. These selected features will be utilized to create our feature profile.
-#
-# For more detailed information about the `feature_select` function, its parameters, and its capabilities, please refer to the documentation available [here](https://pycytominer.readthedocs.io/en/latest/pycytominer.html#module-pycytominer.feature_select).
-#
-
-# In[8]:
-
-
-# creating selected features profile
-feature_select(
- profiles=norm_profiles_path,
- features="infer",
- image_features=False,
- samples="all",
- operation=["variance_threshold", "correlation_threshold", "blocklist"],
- output_file=feat_profiles_path,
- compression_options="gzip",
-)
-
-# save message display
-print(f"Selected features profile saved in: {feat_profiles_path}")
-
-
-# Congratulations! You have successfully completed our walkthrough. We hope that it has provided you with a basic understanding of how to analyze cell morphology features using `pycytominer`.
-#
-# By following the steps outlined in this walkthrough, you have gained valuable insights into processing high-dimensional single-cell morphology data with ease using `pycytominer`. However, please keep in mind that `pycytominer` offers a wide range of functionalities beyond what we covered here. We encourage you to explore the documentation to discover more advanced features and techniques.
-#
-# If you have any questions or need further assistance, don't hesitate to visit the `pycytominer` repository and post your question in the [issues](https://github.com/cytomining/pycytominer/issues) section. The community is there to support you and provide guidance.
-#
-# Now that you have the knowledge and tools to analyze cell morphology features, have fun exploring and mining your data!
diff --git a/walkthroughs/single_cell_usage.ipynb b/walkthroughs/single_cell_usage.ipynb
index 440f4259..0f1c943e 100644
--- a/walkthroughs/single_cell_usage.ipynb
+++ b/walkthroughs/single_cell_usage.ipynb
@@ -35,7 +35,7 @@
"# ignore mix type warnings from pandas\n",
"import warnings\n",
"\n",
- "warnings.filterwarnings(\"ignore\")\n"
+ "warnings.filterwarnings(\"ignore\")"
]
},
{
@@ -86,7 +86,7 @@
"sc_profiles_path = out_dir / \"nf1_single_cell_profile.csv.gz\"\n",
"anno_profiles_path = out_dir / \"nf1_annotated_profile.csv.gz\"\n",
"norm_profiles_path = out_dir / \"nf1_normalized_profile.csv.gz\"\n",
- "feat_profiles_path = out_dir / \"nf1_features_profile.csv.gz\"\n"
+ "feat_profiles_path = out_dir / \"nf1_features_profile.csv.gz\""
]
},
{
@@ -128,7 +128,7 @@
" },\n",
" \"Per_Cells\": {\"Per_Cytoplasm\": \"Cells_Number_Object_Number\"},\n",
" \"Per_Nuclei\": {\"Per_Cytoplasm\": \"Nuclei_Number_Object_Number\"},\n",
- "}\n"
+ "}"
]
},
{
@@ -177,7 +177,7 @@
"# compressed csv file\n",
"single_cell_profile.merge_single_cells(\n",
" sc_output_file=sc_profiles_path, compression_options=\"gzip\"\n",
- ")\n"
+ ")"
]
},
{
@@ -208,7 +208,7 @@
"platemap_df = pd.read_csv(plate_map)\n",
"\n",
"# displaying platemap\n",
- "print(platemap_df.columns.tolist())\n"
+ "print(platemap_df.columns.tolist())"
]
},
{
@@ -249,7 +249,7 @@
")\n",
"\n",
"# save message display\n",
- "print(f\"Annotated profile saved in: {anno_profiles_path}\")\n"
+ "print(f\"Annotated profile saved in: {anno_profiles_path}\")"
]
},
{
@@ -296,7 +296,7 @@
")\n",
"\n",
"# save message display\n",
- "print(f\"Normalized profile saved in: {norm_profiles_path}\")\n"
+ "print(f\"Normalized profile saved in: {norm_profiles_path}\")"
]
},
{
@@ -341,7 +341,7 @@
")\n",
"\n",
"# save message display\n",
- "print(f\"Selected features profile saved in: {feat_profiles_path}\")\n"
+ "print(f\"Selected features profile saved in: {feat_profiles_path}\")"
]
},
{
 +- [Data processing for image-based profiling](#data-processing-for-image-based-profiling)
+ - [Installation](##installation)
+ - [Frameworks](#frameworks)
+ - [API](#api)
+ - [Usage](#usage)
+ - [Pipeline orchestration](#pipeline-orchestration)
+ - [Other functionality](#other-functionality)
+ - [CellProfiler CSV collation](#cellprofiler-csv-collation)
+ - [Creating a cell locations lookup table](#creating-a-cell-locations-lookup-table)
+ - [Generating a GCT file for morpheus](#generating-a-gct-file-for-morpheus)
+ - [Citing pycytominer](#citing-pycytominer)
+
# Data processing for image-based profiling
[](https://github.com/cytomining/pycytominer/actions/workflows/integration-test.yml?query=branch%3Amain)
@@ -111,19 +123,7 @@ And, more specifically than that, image-based profiling readouts from [CellProfi
Therefore, we have included some custom tools in `pycytominer/cyto_utils` that provides other functionality:
-- [Data processing for image-based profiling](#data-processing-for-image-based-profiling)
- - [Installation](#installation)
- - [Frameworks](#frameworks)
- - [API](#api)
- - [Usage](#usage)
- - [Pipeline orchestration](#pipeline-orchestration)
- - [Other functionality](#other-functionality)
- - [CellProfiler CSV collation](#cellprofiler-csv-collation)
- - [Creating a cell locations lookup table](#creating-a-cell-locations-lookup-table)
- - [Generating a GCT file for morpheus](#generating-a-gct-file-for-morpheus)
- - [Citing pycytominer](#citing-pycytominer)
-
-Note, [`pycytominer.cyto_utils.cells.SingleCells()`](pycytominer/cyto_utils/cells.py) contains code to interact with single-cell SQLite files, which are output from CellProfiler.
+Note, [`pycytominer.cyto_utils.cells.SingleCells()`](./pycytominer/cyto_utils/cells.py) contains code to interact with single-cell SQLite files, which are output from CellProfiler.
Processing capabilities for SQLite files depends on SQLite file size and your available computational resources (for ex. memory and cores).
### CellProfiler CSV collation
diff --git a/docs/README.md b/docs/README.md
new file mode 100644
index 00000000..7f5067ed
--- /dev/null
+++ b/docs/README.md
@@ -0,0 +1,204 @@
+
+- [Data processing for image-based profiling](#data-processing-for-image-based-profiling)
+ - [Installation](##installation)
+ - [Frameworks](#frameworks)
+ - [API](#api)
+ - [Usage](#usage)
+ - [Pipeline orchestration](#pipeline-orchestration)
+ - [Other functionality](#other-functionality)
+ - [CellProfiler CSV collation](#cellprofiler-csv-collation)
+ - [Creating a cell locations lookup table](#creating-a-cell-locations-lookup-table)
+ - [Generating a GCT file for morpheus](#generating-a-gct-file-for-morpheus)
+ - [Citing pycytominer](#citing-pycytominer)
+
# Data processing for image-based profiling
[](https://github.com/cytomining/pycytominer/actions/workflows/integration-test.yml?query=branch%3Amain)
@@ -111,19 +123,7 @@ And, more specifically than that, image-based profiling readouts from [CellProfi
Therefore, we have included some custom tools in `pycytominer/cyto_utils` that provides other functionality:
-- [Data processing for image-based profiling](#data-processing-for-image-based-profiling)
- - [Installation](#installation)
- - [Frameworks](#frameworks)
- - [API](#api)
- - [Usage](#usage)
- - [Pipeline orchestration](#pipeline-orchestration)
- - [Other functionality](#other-functionality)
- - [CellProfiler CSV collation](#cellprofiler-csv-collation)
- - [Creating a cell locations lookup table](#creating-a-cell-locations-lookup-table)
- - [Generating a GCT file for morpheus](#generating-a-gct-file-for-morpheus)
- - [Citing pycytominer](#citing-pycytominer)
-
-Note, [`pycytominer.cyto_utils.cells.SingleCells()`](pycytominer/cyto_utils/cells.py) contains code to interact with single-cell SQLite files, which are output from CellProfiler.
+Note, [`pycytominer.cyto_utils.cells.SingleCells()`](./pycytominer/cyto_utils/cells.py) contains code to interact with single-cell SQLite files, which are output from CellProfiler.
Processing capabilities for SQLite files depends on SQLite file size and your available computational resources (for ex. memory and cores).
### CellProfiler CSV collation
diff --git a/docs/README.md b/docs/README.md
new file mode 100644
index 00000000..7f5067ed
--- /dev/null
+++ b/docs/README.md
@@ -0,0 +1,204 @@
+ +
+# Data processing for image-based profiling
+
+[](https://github.com/cytomining/pycytominer/actions/workflows/integration-test.yml?query=branch%3Amain)
+[](https://codecov.io/github/cytomining/pycytominer?branch=main)
+[](https://github.com/astral-sh/ruff)
+[](https://pycytominer.readthedocs.io/)
+[](https://doi.org/10.48550/arXiv.2311.13417)
+
+Pycytominer is a suite of common functions used to process high dimensional readouts from high-throughput cell experiments.
+The tool is most often used for processing data through the following pipeline:
+
+
+
+# Data processing for image-based profiling
+
+[](https://github.com/cytomining/pycytominer/actions/workflows/integration-test.yml?query=branch%3Amain)
+[](https://codecov.io/github/cytomining/pycytominer?branch=main)
+[](https://github.com/astral-sh/ruff)
+[](https://pycytominer.readthedocs.io/)
+[](https://doi.org/10.48550/arXiv.2311.13417)
+
+Pycytominer is a suite of common functions used to process high dimensional readouts from high-throughput cell experiments.
+The tool is most often used for processing data through the following pipeline:
+
+ +
+[Click here for high resolution pipeline image](https://github.com/cytomining/pycytominer/blob/main/media/pipeline.png)
+
+Image data flow from a microscope to cell segmentation and feature extraction tools (e.g. CellProfiler or DeepProfiler).
+From here, additional single cell processing tools curate the single cell readouts into a form manageable for pycytominer input.
+For CellProfiler, we use [cytominer-database](https://github.com/cytomining/cytominer-database) or [CytoTable](https://github.com/cytomining/CytoTable).
+For DeepProfiler, we include single cell processing tools in [pycytominer.cyto_utils](cyto_utils.md).
+
+From the single cell output, pycytominer performs five steps using a simple API (described below), before passing along data to [cytominer-eval](https://github.com/cytomining/cytominer-eval) for quality and perturbation strength evaluation.
+
+## Installation
+
+You can install pycytominer via pip:
+
+```bash
+pip install pycytominer
+```
+
+or conda:
+
+```bash
+conda install -c conda-forge pycytominer
+```
+
+## Frameworks
+
+Pycytominer is primarily built on top of [pandas](https://pandas.pydata.org/docs/index.html), also using aspects of SQLAlchemy, sklearn, and pyarrow.
+
+Pycytominer currently supports [parquet](https://parquet.apache.org/) and compressed text file (e.g. `.csv.gz`) i/o.
+
+## API
+
+Pycytominer has five major processing functions:
+
+1. Aggregate - Average single-cell profiles based on metadata information (most often "well").
+2. Annotate - Append metadata (most often from the platemap file) to the feature profile
+3. Normalize - Transform input feature data into consistent distributions
+4. Feature select - Exclude non-informative or redundant features
+5. Consensus - Average aggregated profiles by replicates to form a "consensus signature"
+
+The API is consistent for each of these functions:
+
+```python
+# Each function takes as input a pandas DataFrame or file path
+# and transforms the input data based on the provided options and methods
+df = function(
+ profiles_or_path,
+ features,
+ samples,
+ method,
+ output_file,
+ additional_options...
+)
+```
+
+Each processing function has unique arguments, see our [documentation](https://pycytominer.readthedocs.io/) for more details.
+
+## Usage
+
+The default way to use pycytominer is within python scripts, and using pycytominer is simple and fun.
+
+```python
+# Real world example
+import pandas as pd
+import pycytominer
+
+commit = "da8ae6a3bc103346095d61b4ee02f08fc85a5d98"
+url = f"https://media.githubusercontent.com/media/broadinstitute/lincs-cell-painting/{commit}/profiles/2016_04_01_a549_48hr_batch1/SQ00014812/SQ00014812_augmented.csv.gz"
+
+df = pd.read_csv(url)
+
+normalized_df = pycytominer.normalize(
+ profiles=df,
+ method="standardize",
+ samples="Metadata_broad_sample == 'DMSO'"
+)
+```
+
+### Pipeline orchestration
+
+Pycytominer is a collection of different functions with no explicit link between steps.
+However, some options exist to use pycytominer within a pipeline framework.
+
+| Project | Format | Environment | pycytominer usage |
+| :------------------------------------------------------------------------------- | :-------- | :------------------- | :---------------------- |
+| [Profiling-recipe](https://github.com/cytomining/profiling-recipe) | yaml | agnostic | full pipeline support |
+| [CellProfiler-on-Terra](https://github.com/broadinstitute/cellprofiler-on-Terra) | WDL | google cloud / Terra | single-cell aggregation |
+| [CytoSnake](https://github.com/WayScience/CytoSnake) | snakemake | agnostic | full pipeline support |
+
+A separate project called [AuSPICES](https://github.com/broadinstitute/AuSPICEs) offers pipeline support up to image feature extraction.
+
+## Other functionality
+
+Pycytominer was written with a goal of processing any high-throughput image-based profiling data.
+However, the initial use case was developed for processing image-based profiling experiments specifically.
+And, more specifically than that, image-based profiling readouts from [CellProfiler](https://github.com/CellProfiler) measurements from [Cell Painting](https://www.nature.com/articles/nprot.2016.105) data.
+
+Therefore, we have included some custom tools in `pycytominer/cyto_utils` that provides other functionality:
+
+Note, [`pycytominer.cyto_utils.cells.SingleCells()`](cyto_utils.md##pycytominer.cyto_utils.cells) contains code to interact with single-cell SQLite files, which are output from CellProfiler.
+Processing capabilities for SQLite files depends on SQLite file size and your available computational resources (for ex. memory and cores).
+
+### CellProfiler CSV collation
+
+If running your images on a cluster, unless you have a MySQL or similar large database set up then you will likely end up with lots of different folders from the different cluster runs (often one per well or one per site), each one containing an `Image.csv`, `Nuclei.csv`, etc.
+In order to look at full plates, therefore, we first need to collate all of these CSVs into a single file (currently SQLite) per plate.
+We currently do this with a library called [cytominer-database](https://github.com/cytomining/cytominer-database).
+
+If you want to perform this data collation inside pycytominer using the `cyto_utils` function `collate` (and/or you want to be able to run the tests and have them all pass!), you will need `cytominer-database==0.3.4`; this will change your installation commands slightly:
+
+```bash
+# Example for general case commit:
+pip install "pycytominer[collate]"
+
+# Example for specific commit:
+pip install "pycytominer[collate] @ git+https://github.com/cytomining/pycytominer@77d93a3a551a438799a97ba57d49b19de0a293ab"
+```
+
+If using `pycytominer` in a conda environment, in order to run `collate.py`, you will also want to make sure to add `cytominer-database=0.3.4` to your list of dependencies.
+
+### Creating a cell locations lookup table
+
+The `CellLocation` class offers a convenient way to augment a [LoadData](https://cellprofiler-manual.s3.amazonaws.com/CPmanual/LoadData.html) file with X,Y locations of cells in each image.
+The locations information is obtained from a single cell SQLite file.
+
+To use this functionality, you will need to modify your installation command, similar to above:
+
+```bash
+# Example for general case commit:
+pip install "pycytominer[cell_locations]"
+```
+
+Example using this functionality:
+
+```bash
+metadata_input="s3://cellpainting-gallery/test-cpg0016-jump/source_4/workspace/load_data_csv/2021_08_23_Batch12/BR00126114/test_BR00126114_load_data_with_illum.parquet"
+single_single_cell_input="s3://cellpainting-gallery/test-cpg0016-jump/source_4/workspace/backend/2021_08_23_Batch12/BR00126114/test_BR00126114.sqlite"
+augmented_metadata_output="~/Desktop/load_data_with_illum_and_cell_location_subset.parquet"
+
+python \
+ -m pycytominer.cyto_utils.cell_locations_cmd \
+ --metadata_input ${metadata_input} \
+ --single_cell_input ${single_single_cell_input} \
+ --augmented_metadata_output ${augmented_metadata_output} \
+ add_cell_location
+
+# Check the output
+
+python -c "import pandas as pd; print(pd.read_parquet('${augmented_metadata_output}').head())"
+
+# It should look something like this (depends on the width of your terminal):
+
+# Metadata_Plate Metadata_Well Metadata_Site ... PathName_OrigRNA ImageNumber CellCenters
+# 0 BR00126114 A01 1 ... s3://cellpainting-gallery/cpg0016-jump/source_... 1 [{'Nuclei_Location_Center_X': 943.512129380054...
+# 1 BR00126114 A01 2 ... s3://cellpainting-gallery/cpg0016-jump/source_... 2 [{'Nuclei_Location_Center_X': 29.9516027655562...
+```
+
+### Generating a GCT file for morpheus
+
+The software [morpheus](https://software.broadinstitute.org/morpheus/) enables profile visualization in the form of interactive heatmaps.
+Pycytominer can convert profiles into a `.gct` file for drag-and-drop input into morpheus.
+
+```python
+# Real world example
+import pandas as pd
+import pycytominer
+
+commit = "da8ae6a3bc103346095d61b4ee02f08fc85a5d98"
+plate = "SQ00014812"
+url = f"https://media.githubusercontent.com/media/broadinstitute/lincs-cell-painting/{commit}/profiles/2016_04_01_a549_48hr_batch1/{plate}/{plate}_normalized_feature_select.csv.gz"
+
+df = pd.read_csv(url)
+output_file = f"{plate}.gct"
+
+pycytominer.cyto_utils.write_gct(
+ profiles=df,
+ output_file=output_file
+)
+```
+
+## Citing pycytominer
+
+If you have used `pycytominer` in your project, please use the citation below.
+You can also find the citation in the 'cite this repository' link at the top right under `about` section.
+
+APA:
+
+```text
+Serrano, E., Chandrasekaran, N., Bunten, D., Brewer, K., Tomkinson, J., Kern, R., Bornholdt, M., Fleming, S., Pei, R., Arevalo, J., Tsang, H., Rubinetti, V., Tromans-Coia, C., Becker, T., Weisbart, E., Bunne, C., Kalinin, A. A., Senft, R., Taylor, S. J., Jamali, N., Adeboye, A., Abbasi, H. S., Goodman, A., Caicedo, J., Carpenter, A. E., Cimini, B. A., Singh, S., & Way, G. P. Reproducible image-based profiling with Pycytominer. https://doi.org/10.48550/arXiv.2311.13417
+```
diff --git a/docs/conf.py b/docs/conf.py
index be8581a2..af5e0eec 100644
--- a/docs/conf.py
+++ b/docs/conf.py
@@ -1,4 +1,4 @@
-# Configuration file for the Sphinx documentation builder.
+"""Configuration file for the Sphinx documentation builder."""
#
# This file only contains a selection of the most common options. For a full
# list see the documentation:
diff --git a/docs/cyto_utils.md b/docs/cyto_utils.md
new file mode 100644
index 00000000..0251bb86
--- /dev/null
+++ b/docs/cyto_utils.md
@@ -0,0 +1,5 @@
+# Cyto utilities
+
+Functions enabling smooth interaction with CellProfiler and DeepProfiler output formats.
+
+::: pycytominer.cyto_utils
diff --git a/docs/functions.md b/docs/functions.md
new file mode 100644
index 00000000..ef4a65ab
--- /dev/null
+++ b/docs/functions.md
@@ -0,0 +1,16 @@
+# Main Functions
+
+
+
+
+::: pycytominer
+ options:
+ members:
+ - aggregate
+ - annotate
+ - consensus
+ - feature_select
+ - normalize
+
+
+
diff --git a/docs/index.rst b/docs/index.rst
deleted file mode 100644
index 460d88b8..00000000
--- a/docs/index.rst
+++ /dev/null
@@ -1,22 +0,0 @@
-.. pycytominer documentation main file, created by
- sphinx-quickstart on Fri Feb 19 14:44:29 2021.
- You can adapt this file completely to your liking, but it should at least
- contain the root `toctree` directive.
-
-.. mdinclude:: ../README.md
-
-.. toctree::
- :maxdepth: 2
- :caption: Contents:
-
- install
- walkthrough
- tutorial
- modules
-
-Indices and tables
-==================
-
-* :ref:`genindex`
-* :ref:`modindex`
-* :ref:`search`
diff --git a/docs/install.rst b/docs/install.md
similarity index 71%
rename from docs/install.rst
rename to docs/install.md
index 99213db9..9f2963ab 100644
--- a/docs/install.rst
+++ b/docs/install.md
@@ -1,14 +1,13 @@
-Install
-=======
+# Installation
To install pycytominer, use pip:
-.. code-block:: bash
-
+```bash
pip install pycytominer
+```
You can also install pycytominer with conda:
-.. code-block:: bash
-
+```bash
conda install -c conda-forge pycytominer
+```
diff --git a/docs/modules.rst b/docs/modules.rst
deleted file mode 100644
index bec89387..00000000
--- a/docs/modules.rst
+++ /dev/null
@@ -1,7 +0,0 @@
-pycytominer
-===========
-
-.. toctree::
- :maxdepth: 4
-
- pycytominer
diff --git a/docs/operations.md b/docs/operations.md
new file mode 100644
index 00000000..ba31c6ed
--- /dev/null
+++ b/docs/operations.md
@@ -0,0 +1,6 @@
+# Operations
+
+We do not recommend interacting with these functions directly.
+The core pycytominer API uses these operations internally.
+
+::: pycytominer.operations
diff --git a/docs/pycytominer.cyto_utils.rst b/docs/pycytominer.cyto_utils.rst
deleted file mode 100644
index 8618d60d..00000000
--- a/docs/pycytominer.cyto_utils.rst
+++ /dev/null
@@ -1,76 +0,0 @@
-Cyto utilities
-==============
-
-Functions enabling smooth interaction with CellProfiler and DeepProfiler output formats.
-
-pycytominer.cyto\_utils.cells module
-------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.cells
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.collate module
---------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.collate
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.features module
----------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.features
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.load module
------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.load
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.modz module
------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.modz
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.output module
--------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.output
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.util module
------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.util
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.write\_gct module
------------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.write_gct
- :members:
- :undoc-members:
- :show-inheritance:
-
-Module contents
----------------
-
-.. automodule:: pycytominer.cyto_utils
- :members:
- :undoc-members:
- :show-inheritance:
diff --git a/docs/pycytominer.operations.rst b/docs/pycytominer.operations.rst
deleted file mode 100644
index 062919d6..00000000
--- a/docs/pycytominer.operations.rst
+++ /dev/null
@@ -1,45 +0,0 @@
-Operations
-==========
-
-We do not recommend interacting with these functions directly.
-The core pycytominer API uses these operations internally.
-
-pycytominer.operations.correlation\_threshold module
-----------------------------------------------------
-
-.. automodule:: pycytominer.operations.correlation_threshold
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.operations.get\_na\_columns module
-----------------------------------------------
-
-.. automodule:: pycytominer.operations.get_na_columns
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.operations.transform module
----------------------------------------
-
-.. automodule:: pycytominer.operations.transform
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.operations.variance\_threshold module
--------------------------------------------------
-
-.. automodule:: pycytominer.operations.variance_threshold
- :members:
- :undoc-members:
- :show-inheritance:
-
-Module contents
----------------
-
-.. automodule:: pycytominer.operations
- :members:
- :undoc-members:
- :show-inheritance:
diff --git a/docs/pycytominer.rst b/docs/pycytominer.rst
deleted file mode 100644
index 01b61229..00000000
--- a/docs/pycytominer.rst
+++ /dev/null
@@ -1,59 +0,0 @@
-Core functions
-==============
-
-pycytominer.aggregate module
-----------------------------
-
-.. automodule:: pycytominer.aggregate
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.annotate module
----------------------------
-
-.. automodule:: pycytominer.annotate
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.normalize module
-----------------------------
-
-.. automodule:: pycytominer.normalize
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.feature\_select module
-----------------------------------
-
-.. automodule:: pycytominer.feature_select
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.consensus module
-----------------------------
-
-.. automodule:: pycytominer.consensus
- :members:
- :undoc-members:
- :show-inheritance:
-
-Helper functions
-----------------
-
-.. toctree::
- :maxdepth: 4
-
- pycytominer.cyto_utils
- pycytominer.operations
-
-Module contents
----------------
-
-.. automodule:: pycytominer
- :members:
- :undoc-members:
- :show-inheritance:
diff --git a/docs/tutorial.md b/docs/tutorial.md
new file mode 100644
index 00000000..492e0008
--- /dev/null
+++ b/docs/tutorial.md
@@ -0,0 +1,13 @@
+# Tutorials
+
+This [tutorial](https://github.com/cytomining/pipeline-examples#readme) shows how to run a image-based profiling pipeline using pycytominer. Using IPython notebooks, it walks through the following steps:
+
+- Downloading a dataset of single cell [CellProfiler](https://cellprofiler.org/) profiles.
+- Processing the profiles using PyCytominer. This includes the following steps:
+- Data initialization
+- Single cell aggregation to create well-level profiles
+- Addition of experiment metadata to the well-level profiles
+- Profile normalization
+- Feature selection
+- Forming consensus signatures
+- Evaluating the profile quality using [cytominer-eval](https://github.com/cytomining/cytominer-eval>).
diff --git a/docs/tutorial.rst b/docs/tutorial.rst
deleted file mode 100644
index da5d5f6f..00000000
--- a/docs/tutorial.rst
+++ /dev/null
@@ -1,14 +0,0 @@
-Tutorials
-=========
-
-`This
+
+[Click here for high resolution pipeline image](https://github.com/cytomining/pycytominer/blob/main/media/pipeline.png)
+
+Image data flow from a microscope to cell segmentation and feature extraction tools (e.g. CellProfiler or DeepProfiler).
+From here, additional single cell processing tools curate the single cell readouts into a form manageable for pycytominer input.
+For CellProfiler, we use [cytominer-database](https://github.com/cytomining/cytominer-database) or [CytoTable](https://github.com/cytomining/CytoTable).
+For DeepProfiler, we include single cell processing tools in [pycytominer.cyto_utils](cyto_utils.md).
+
+From the single cell output, pycytominer performs five steps using a simple API (described below), before passing along data to [cytominer-eval](https://github.com/cytomining/cytominer-eval) for quality and perturbation strength evaluation.
+
+## Installation
+
+You can install pycytominer via pip:
+
+```bash
+pip install pycytominer
+```
+
+or conda:
+
+```bash
+conda install -c conda-forge pycytominer
+```
+
+## Frameworks
+
+Pycytominer is primarily built on top of [pandas](https://pandas.pydata.org/docs/index.html), also using aspects of SQLAlchemy, sklearn, and pyarrow.
+
+Pycytominer currently supports [parquet](https://parquet.apache.org/) and compressed text file (e.g. `.csv.gz`) i/o.
+
+## API
+
+Pycytominer has five major processing functions:
+
+1. Aggregate - Average single-cell profiles based on metadata information (most often "well").
+2. Annotate - Append metadata (most often from the platemap file) to the feature profile
+3. Normalize - Transform input feature data into consistent distributions
+4. Feature select - Exclude non-informative or redundant features
+5. Consensus - Average aggregated profiles by replicates to form a "consensus signature"
+
+The API is consistent for each of these functions:
+
+```python
+# Each function takes as input a pandas DataFrame or file path
+# and transforms the input data based on the provided options and methods
+df = function(
+ profiles_or_path,
+ features,
+ samples,
+ method,
+ output_file,
+ additional_options...
+)
+```
+
+Each processing function has unique arguments, see our [documentation](https://pycytominer.readthedocs.io/) for more details.
+
+## Usage
+
+The default way to use pycytominer is within python scripts, and using pycytominer is simple and fun.
+
+```python
+# Real world example
+import pandas as pd
+import pycytominer
+
+commit = "da8ae6a3bc103346095d61b4ee02f08fc85a5d98"
+url = f"https://media.githubusercontent.com/media/broadinstitute/lincs-cell-painting/{commit}/profiles/2016_04_01_a549_48hr_batch1/SQ00014812/SQ00014812_augmented.csv.gz"
+
+df = pd.read_csv(url)
+
+normalized_df = pycytominer.normalize(
+ profiles=df,
+ method="standardize",
+ samples="Metadata_broad_sample == 'DMSO'"
+)
+```
+
+### Pipeline orchestration
+
+Pycytominer is a collection of different functions with no explicit link between steps.
+However, some options exist to use pycytominer within a pipeline framework.
+
+| Project | Format | Environment | pycytominer usage |
+| :------------------------------------------------------------------------------- | :-------- | :------------------- | :---------------------- |
+| [Profiling-recipe](https://github.com/cytomining/profiling-recipe) | yaml | agnostic | full pipeline support |
+| [CellProfiler-on-Terra](https://github.com/broadinstitute/cellprofiler-on-Terra) | WDL | google cloud / Terra | single-cell aggregation |
+| [CytoSnake](https://github.com/WayScience/CytoSnake) | snakemake | agnostic | full pipeline support |
+
+A separate project called [AuSPICES](https://github.com/broadinstitute/AuSPICEs) offers pipeline support up to image feature extraction.
+
+## Other functionality
+
+Pycytominer was written with a goal of processing any high-throughput image-based profiling data.
+However, the initial use case was developed for processing image-based profiling experiments specifically.
+And, more specifically than that, image-based profiling readouts from [CellProfiler](https://github.com/CellProfiler) measurements from [Cell Painting](https://www.nature.com/articles/nprot.2016.105) data.
+
+Therefore, we have included some custom tools in `pycytominer/cyto_utils` that provides other functionality:
+
+Note, [`pycytominer.cyto_utils.cells.SingleCells()`](cyto_utils.md##pycytominer.cyto_utils.cells) contains code to interact with single-cell SQLite files, which are output from CellProfiler.
+Processing capabilities for SQLite files depends on SQLite file size and your available computational resources (for ex. memory and cores).
+
+### CellProfiler CSV collation
+
+If running your images on a cluster, unless you have a MySQL or similar large database set up then you will likely end up with lots of different folders from the different cluster runs (often one per well or one per site), each one containing an `Image.csv`, `Nuclei.csv`, etc.
+In order to look at full plates, therefore, we first need to collate all of these CSVs into a single file (currently SQLite) per plate.
+We currently do this with a library called [cytominer-database](https://github.com/cytomining/cytominer-database).
+
+If you want to perform this data collation inside pycytominer using the `cyto_utils` function `collate` (and/or you want to be able to run the tests and have them all pass!), you will need `cytominer-database==0.3.4`; this will change your installation commands slightly:
+
+```bash
+# Example for general case commit:
+pip install "pycytominer[collate]"
+
+# Example for specific commit:
+pip install "pycytominer[collate] @ git+https://github.com/cytomining/pycytominer@77d93a3a551a438799a97ba57d49b19de0a293ab"
+```
+
+If using `pycytominer` in a conda environment, in order to run `collate.py`, you will also want to make sure to add `cytominer-database=0.3.4` to your list of dependencies.
+
+### Creating a cell locations lookup table
+
+The `CellLocation` class offers a convenient way to augment a [LoadData](https://cellprofiler-manual.s3.amazonaws.com/CPmanual/LoadData.html) file with X,Y locations of cells in each image.
+The locations information is obtained from a single cell SQLite file.
+
+To use this functionality, you will need to modify your installation command, similar to above:
+
+```bash
+# Example for general case commit:
+pip install "pycytominer[cell_locations]"
+```
+
+Example using this functionality:
+
+```bash
+metadata_input="s3://cellpainting-gallery/test-cpg0016-jump/source_4/workspace/load_data_csv/2021_08_23_Batch12/BR00126114/test_BR00126114_load_data_with_illum.parquet"
+single_single_cell_input="s3://cellpainting-gallery/test-cpg0016-jump/source_4/workspace/backend/2021_08_23_Batch12/BR00126114/test_BR00126114.sqlite"
+augmented_metadata_output="~/Desktop/load_data_with_illum_and_cell_location_subset.parquet"
+
+python \
+ -m pycytominer.cyto_utils.cell_locations_cmd \
+ --metadata_input ${metadata_input} \
+ --single_cell_input ${single_single_cell_input} \
+ --augmented_metadata_output ${augmented_metadata_output} \
+ add_cell_location
+
+# Check the output
+
+python -c "import pandas as pd; print(pd.read_parquet('${augmented_metadata_output}').head())"
+
+# It should look something like this (depends on the width of your terminal):
+
+# Metadata_Plate Metadata_Well Metadata_Site ... PathName_OrigRNA ImageNumber CellCenters
+# 0 BR00126114 A01 1 ... s3://cellpainting-gallery/cpg0016-jump/source_... 1 [{'Nuclei_Location_Center_X': 943.512129380054...
+# 1 BR00126114 A01 2 ... s3://cellpainting-gallery/cpg0016-jump/source_... 2 [{'Nuclei_Location_Center_X': 29.9516027655562...
+```
+
+### Generating a GCT file for morpheus
+
+The software [morpheus](https://software.broadinstitute.org/morpheus/) enables profile visualization in the form of interactive heatmaps.
+Pycytominer can convert profiles into a `.gct` file for drag-and-drop input into morpheus.
+
+```python
+# Real world example
+import pandas as pd
+import pycytominer
+
+commit = "da8ae6a3bc103346095d61b4ee02f08fc85a5d98"
+plate = "SQ00014812"
+url = f"https://media.githubusercontent.com/media/broadinstitute/lincs-cell-painting/{commit}/profiles/2016_04_01_a549_48hr_batch1/{plate}/{plate}_normalized_feature_select.csv.gz"
+
+df = pd.read_csv(url)
+output_file = f"{plate}.gct"
+
+pycytominer.cyto_utils.write_gct(
+ profiles=df,
+ output_file=output_file
+)
+```
+
+## Citing pycytominer
+
+If you have used `pycytominer` in your project, please use the citation below.
+You can also find the citation in the 'cite this repository' link at the top right under `about` section.
+
+APA:
+
+```text
+Serrano, E., Chandrasekaran, N., Bunten, D., Brewer, K., Tomkinson, J., Kern, R., Bornholdt, M., Fleming, S., Pei, R., Arevalo, J., Tsang, H., Rubinetti, V., Tromans-Coia, C., Becker, T., Weisbart, E., Bunne, C., Kalinin, A. A., Senft, R., Taylor, S. J., Jamali, N., Adeboye, A., Abbasi, H. S., Goodman, A., Caicedo, J., Carpenter, A. E., Cimini, B. A., Singh, S., & Way, G. P. Reproducible image-based profiling with Pycytominer. https://doi.org/10.48550/arXiv.2311.13417
+```
diff --git a/docs/conf.py b/docs/conf.py
index be8581a2..af5e0eec 100644
--- a/docs/conf.py
+++ b/docs/conf.py
@@ -1,4 +1,4 @@
-# Configuration file for the Sphinx documentation builder.
+"""Configuration file for the Sphinx documentation builder."""
#
# This file only contains a selection of the most common options. For a full
# list see the documentation:
diff --git a/docs/cyto_utils.md b/docs/cyto_utils.md
new file mode 100644
index 00000000..0251bb86
--- /dev/null
+++ b/docs/cyto_utils.md
@@ -0,0 +1,5 @@
+# Cyto utilities
+
+Functions enabling smooth interaction with CellProfiler and DeepProfiler output formats.
+
+::: pycytominer.cyto_utils
diff --git a/docs/functions.md b/docs/functions.md
new file mode 100644
index 00000000..ef4a65ab
--- /dev/null
+++ b/docs/functions.md
@@ -0,0 +1,16 @@
+# Main Functions
+
+
+
+
+::: pycytominer
+ options:
+ members:
+ - aggregate
+ - annotate
+ - consensus
+ - feature_select
+ - normalize
+
+
+
diff --git a/docs/index.rst b/docs/index.rst
deleted file mode 100644
index 460d88b8..00000000
--- a/docs/index.rst
+++ /dev/null
@@ -1,22 +0,0 @@
-.. pycytominer documentation main file, created by
- sphinx-quickstart on Fri Feb 19 14:44:29 2021.
- You can adapt this file completely to your liking, but it should at least
- contain the root `toctree` directive.
-
-.. mdinclude:: ../README.md
-
-.. toctree::
- :maxdepth: 2
- :caption: Contents:
-
- install
- walkthrough
- tutorial
- modules
-
-Indices and tables
-==================
-
-* :ref:`genindex`
-* :ref:`modindex`
-* :ref:`search`
diff --git a/docs/install.rst b/docs/install.md
similarity index 71%
rename from docs/install.rst
rename to docs/install.md
index 99213db9..9f2963ab 100644
--- a/docs/install.rst
+++ b/docs/install.md
@@ -1,14 +1,13 @@
-Install
-=======
+# Installation
To install pycytominer, use pip:
-.. code-block:: bash
-
+```bash
pip install pycytominer
+```
You can also install pycytominer with conda:
-.. code-block:: bash
-
+```bash
conda install -c conda-forge pycytominer
+```
diff --git a/docs/modules.rst b/docs/modules.rst
deleted file mode 100644
index bec89387..00000000
--- a/docs/modules.rst
+++ /dev/null
@@ -1,7 +0,0 @@
-pycytominer
-===========
-
-.. toctree::
- :maxdepth: 4
-
- pycytominer
diff --git a/docs/operations.md b/docs/operations.md
new file mode 100644
index 00000000..ba31c6ed
--- /dev/null
+++ b/docs/operations.md
@@ -0,0 +1,6 @@
+# Operations
+
+We do not recommend interacting with these functions directly.
+The core pycytominer API uses these operations internally.
+
+::: pycytominer.operations
diff --git a/docs/pycytominer.cyto_utils.rst b/docs/pycytominer.cyto_utils.rst
deleted file mode 100644
index 8618d60d..00000000
--- a/docs/pycytominer.cyto_utils.rst
+++ /dev/null
@@ -1,76 +0,0 @@
-Cyto utilities
-==============
-
-Functions enabling smooth interaction with CellProfiler and DeepProfiler output formats.
-
-pycytominer.cyto\_utils.cells module
-------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.cells
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.collate module
---------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.collate
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.features module
----------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.features
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.load module
------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.load
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.modz module
------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.modz
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.output module
--------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.output
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.util module
------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.util
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.cyto\_utils.write\_gct module
------------------------------------------
-
-.. automodule:: pycytominer.cyto_utils.write_gct
- :members:
- :undoc-members:
- :show-inheritance:
-
-Module contents
----------------
-
-.. automodule:: pycytominer.cyto_utils
- :members:
- :undoc-members:
- :show-inheritance:
diff --git a/docs/pycytominer.operations.rst b/docs/pycytominer.operations.rst
deleted file mode 100644
index 062919d6..00000000
--- a/docs/pycytominer.operations.rst
+++ /dev/null
@@ -1,45 +0,0 @@
-Operations
-==========
-
-We do not recommend interacting with these functions directly.
-The core pycytominer API uses these operations internally.
-
-pycytominer.operations.correlation\_threshold module
-----------------------------------------------------
-
-.. automodule:: pycytominer.operations.correlation_threshold
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.operations.get\_na\_columns module
-----------------------------------------------
-
-.. automodule:: pycytominer.operations.get_na_columns
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.operations.transform module
----------------------------------------
-
-.. automodule:: pycytominer.operations.transform
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.operations.variance\_threshold module
--------------------------------------------------
-
-.. automodule:: pycytominer.operations.variance_threshold
- :members:
- :undoc-members:
- :show-inheritance:
-
-Module contents
----------------
-
-.. automodule:: pycytominer.operations
- :members:
- :undoc-members:
- :show-inheritance:
diff --git a/docs/pycytominer.rst b/docs/pycytominer.rst
deleted file mode 100644
index 01b61229..00000000
--- a/docs/pycytominer.rst
+++ /dev/null
@@ -1,59 +0,0 @@
-Core functions
-==============
-
-pycytominer.aggregate module
-----------------------------
-
-.. automodule:: pycytominer.aggregate
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.annotate module
----------------------------
-
-.. automodule:: pycytominer.annotate
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.normalize module
-----------------------------
-
-.. automodule:: pycytominer.normalize
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.feature\_select module
-----------------------------------
-
-.. automodule:: pycytominer.feature_select
- :members:
- :undoc-members:
- :show-inheritance:
-
-pycytominer.consensus module
-----------------------------
-
-.. automodule:: pycytominer.consensus
- :members:
- :undoc-members:
- :show-inheritance:
-
-Helper functions
-----------------
-
-.. toctree::
- :maxdepth: 4
-
- pycytominer.cyto_utils
- pycytominer.operations
-
-Module contents
----------------
-
-.. automodule:: pycytominer
- :members:
- :undoc-members:
- :show-inheritance:
diff --git a/docs/tutorial.md b/docs/tutorial.md
new file mode 100644
index 00000000..492e0008
--- /dev/null
+++ b/docs/tutorial.md
@@ -0,0 +1,13 @@
+# Tutorials
+
+This [tutorial](https://github.com/cytomining/pipeline-examples#readme) shows how to run a image-based profiling pipeline using pycytominer. Using IPython notebooks, it walks through the following steps:
+
+- Downloading a dataset of single cell [CellProfiler](https://cellprofiler.org/) profiles.
+- Processing the profiles using PyCytominer. This includes the following steps:
+- Data initialization
+- Single cell aggregation to create well-level profiles
+- Addition of experiment metadata to the well-level profiles
+- Profile normalization
+- Feature selection
+- Forming consensus signatures
+- Evaluating the profile quality using [cytominer-eval](https://github.com/cytomining/cytominer-eval>).
diff --git a/docs/tutorial.rst b/docs/tutorial.rst
deleted file mode 100644
index da5d5f6f..00000000
--- a/docs/tutorial.rst
+++ /dev/null
@@ -1,14 +0,0 @@
-Tutorials
-=========
-
-`This