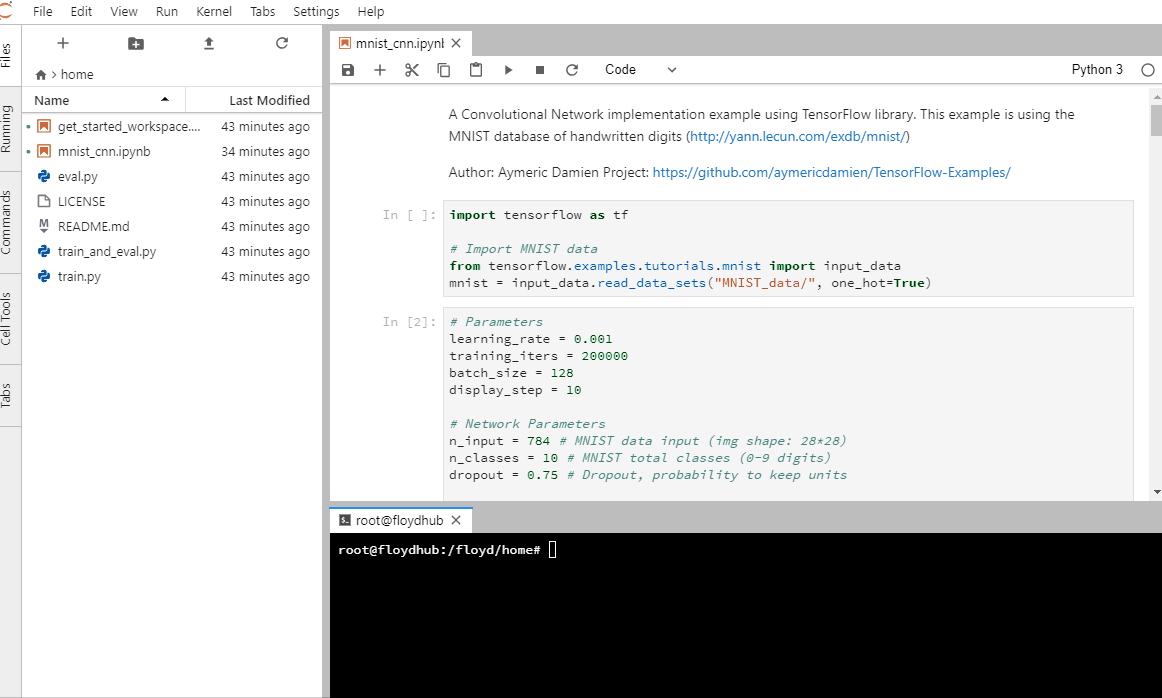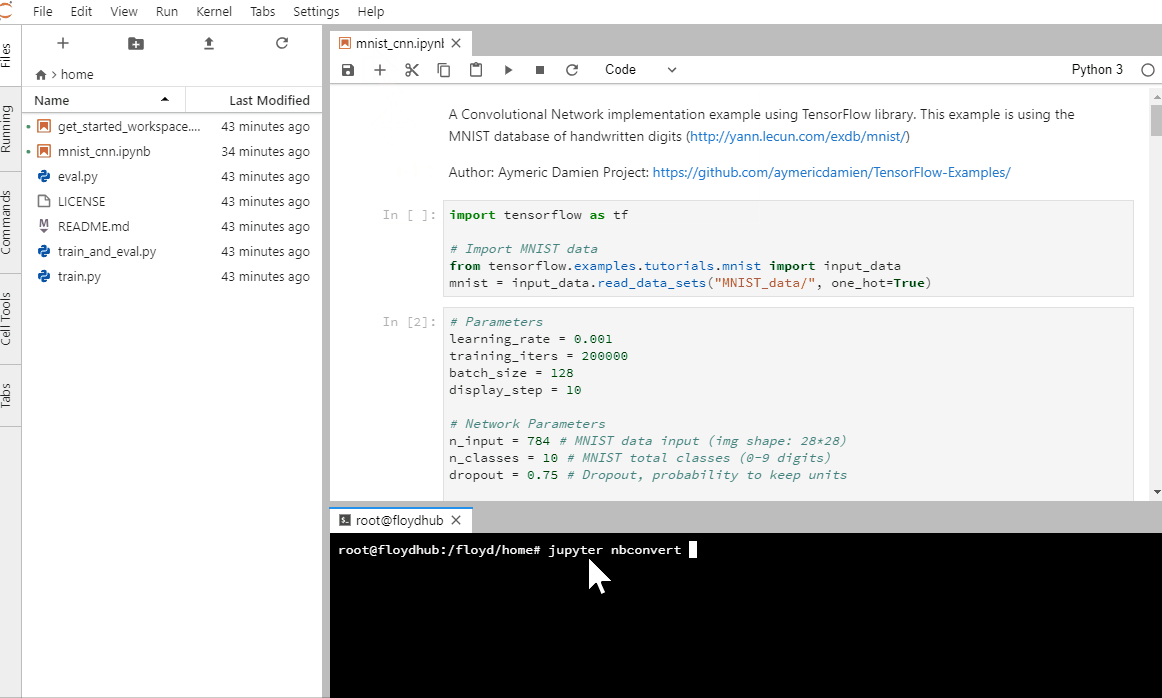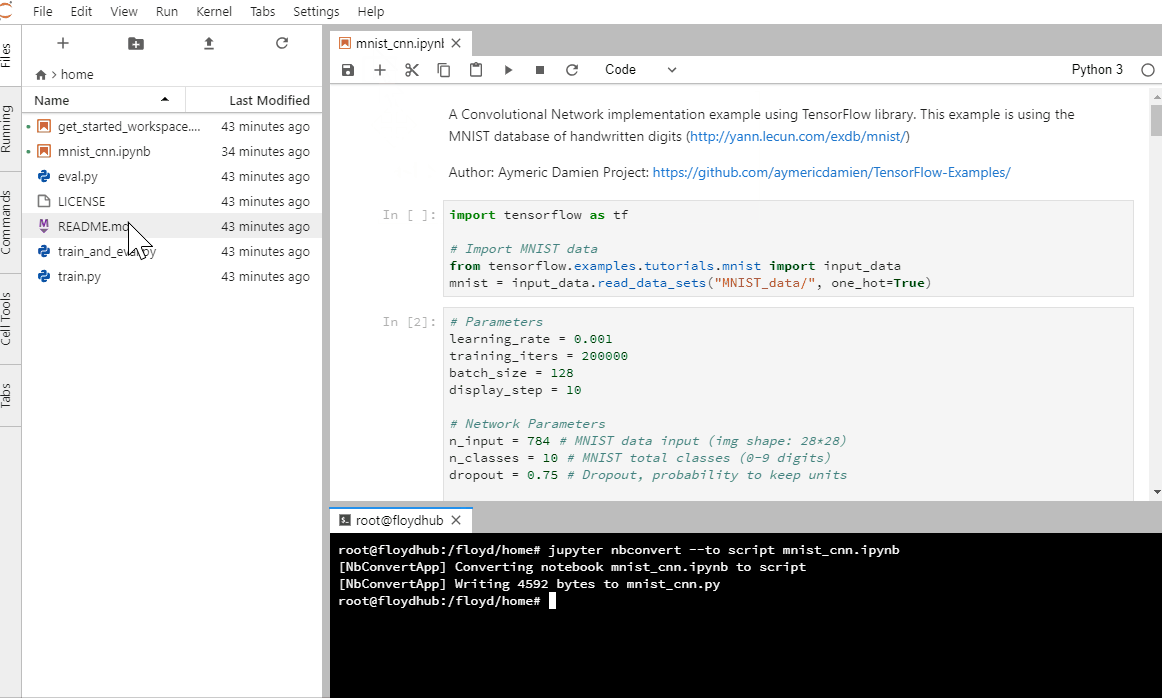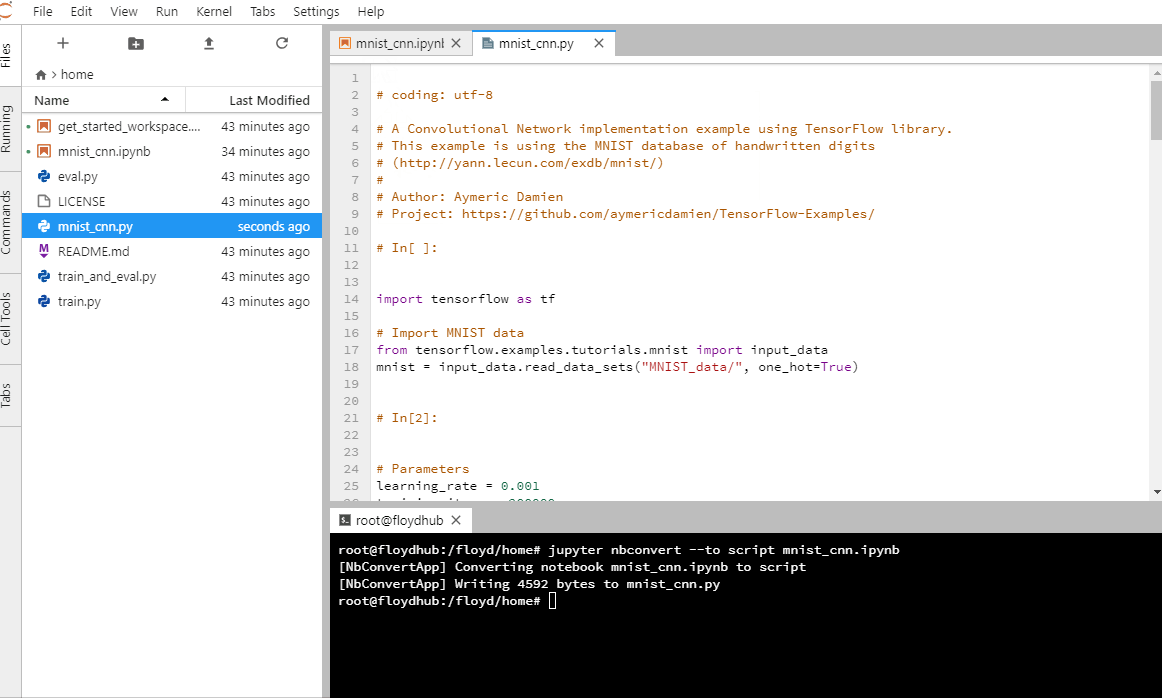
Run this command in your terminal.
jupyter nbconvert --to script <filename>.ipynb
You can paste text in your clipboard into a terminal using the Ctrl + Shift + V shortcut key.
The following don't work:
- Right click -> Paste
Ctrl + V
If you have multiple versions of Notebooks, you can go an intelligent diff on the content (rather than the raw JSON) using the (nbdime plugin)[https://nbdime.readthedocs.io/en/stable/].
Instructions: Run the following commands inside your Workspace console
pip install nbdime
nbdiff <file>.ipynb <file2>.ipynb
You can collapse cell outputs by clicking on the blue bar on the left
Use the argparse library.
Example:
import os
from argparse import ArgumentParser
def build_parser():
parser = ArgumentParser()
# Attributes: Required argument, type=string
parser.add_argument('--checkpoint-dir', type=str,
dest='checkpoint_dir', help='dir to save checkpoint in',
required=True)
# Attributes: Optional argument, type=string
parser.add_argument('--train-path', type=str,
dest='train_path', help='path to training images folder',
default='/floyd/input/train')
# Attributes: boolean variable
parser.add_argument('--slow', dest='slow', action='store_true',
help='gatys\' approach (for debugging, not supported)',
default=False)
# Attributes: int variable, with default value
parser.add_argument('--epochs', type=int,
dest='epochs', help='num epochs',
default=100)
# Attributes: float variable, with default value
parser.add_argument('--content-weight', type=float,
dest='content_weight',
help='content weight (default %(default)s)',
default=0.75)
return parser
def check_opts(opts):
assert os.path.exists(opts.checkpoint_dir), "checkpoint dir not found!"
assert opts.epochs > 0
def main():
parser = build_parser()
options = parser.parse_args()
check_opts(options)
print("Num epochs: ", options.epochs)
View the complete API of the add_argument method
git init
git config user.name "ENTER_USERNAME"
git config user.email "ENTER_EMAIL"
git add .
git commit -m "First commit"
# Create a new repo on Github: https://help.github.com/articles/creating-a-new-repository/
git remote add origin <<URL>> # Replace URL with your git URL (e.g. https://github.com/floydhub/mnist-demo.git)
git push origin master
# Enter your username and password. NOTE: If you have 2-factor auth (2FA) enabled on GitHub, you need to use a personal access token (https://github.com/settings/tokens) instead of a password.
FloydHub automatically creates a .gitignore file for you. If you have files that you don't want to check in (e.g. data, models, Jupyter Notebook checkpoints, etc.), you can add them to it.
Note: .gitignore is a hidden file. You can open it from your Workspace Terminal using the nano terminal editor command.
root@floydhub:/floyd/home# nano .gitignore