shinyCyJS is R/Shiny Package to use cytoscape.js in R environment.
cytoscape.js is a great javascript library for visualize/analysis Graph theory ( network )
if you interested, please refer to this Link.
shinyCyJS is built with cytoscape.js version 3.12.0. (2019/11)
From CRAN (1.0.0)
install.packages('shinyCyJS')From r-universe (1.2.0)
install.packages(
"shinyCyJS",
repos = c("https://jhk0530.r-universe.dev", "https://cloud.r-project.org")
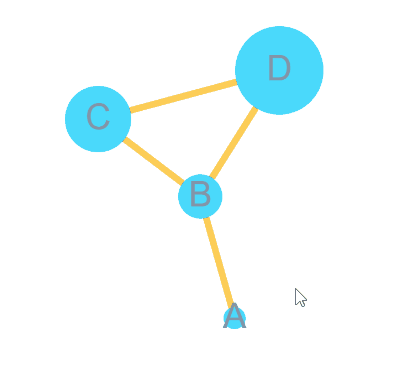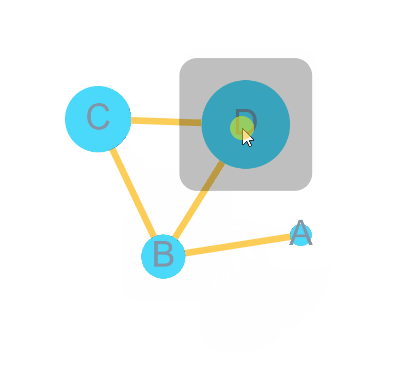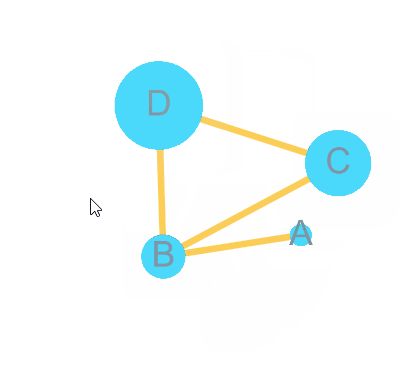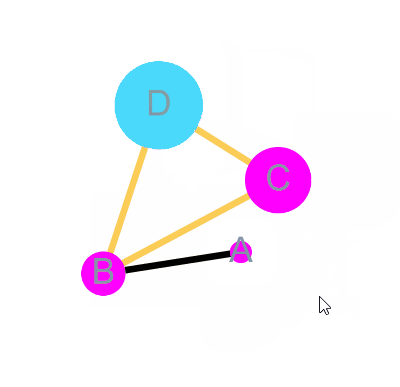
)bulid Graph with node 4 = A, B, C, D and edge = A-B, B-C, C-D, B-D
Code
library(shiny)
library(shinyCyJS)
ui <- function() {
fluidPage(
ShinyCyJSOutput(outputId = "cy")
)
}
server <- function(input, output, session) {
nodes <- data.frame(
id = c("A", "B", "C", "D"),
width = c(10, 20, 30, 40),
height = c(10, 20, 30, 40)
)
edges <- data.frame(
source = c("A", "B", "C", "D"),
target = c("B", "C", "D", "B")
)
nodes <- buildElems(nodes, type = "Node")
edges <- buildElems(edges, type = "Edge")
obj <- shinyCyJS(c(nodes, edges))
output$cy <- renderShinyCyJS(obj)
}
shinyApp(ui, server)see inst/htmlwidgets/shinyCyJS.yaml
Make issue on here