diff --git a/.github/workflows/main.yml b/.github/workflows/main.yml
index 1dfcdd8..e340fb3 100644
--- a/.github/workflows/main.yml
+++ b/.github/workflows/main.yml
@@ -55,11 +55,6 @@ jobs:
- uses: actions/setup-python@v4
with:
python-version: ${{ matrix.python-version }}
- - name: Debugging
- run: |
- ls -la
- cat Makefile
- make virtualenv
- name: Install project
run: |
make virtualenv
diff --git a/.github/workflows/pylint.yml b/.github/workflows/pylint.yml
index 867fd32..cd625cd 100644
--- a/.github/workflows/pylint.yml
+++ b/.github/workflows/pylint.yml
@@ -21,5 +21,5 @@ jobs:
pip install -r requirements.txt
- name: Analysing the code with pylint
run: |
- pylint --fail-under=4 $(git ls-files '*.py')
+ pylint --fail-under=6 $(git ls-files '*.py')
# pylint $(git ls-files '*.py')
diff --git a/README.md b/README.md
index 6fae346..639414c 100644
--- a/README.md
+++ b/README.md
@@ -55,9 +55,9 @@ These could include visualizing the results for a binary classifier, for which p
|:--------------------------------------------------:|:----------------------------------------------------------:|:-------------------------------------------------:|
| Calibration Curve | Classification Report | Confusion Matrix |
-|  |
|  |
|  |
+|
|
+|  |
|  |
|  |
|:--------------------------------------------------:|:----------------------------------------------------------:|:-------------------------------------------------:|
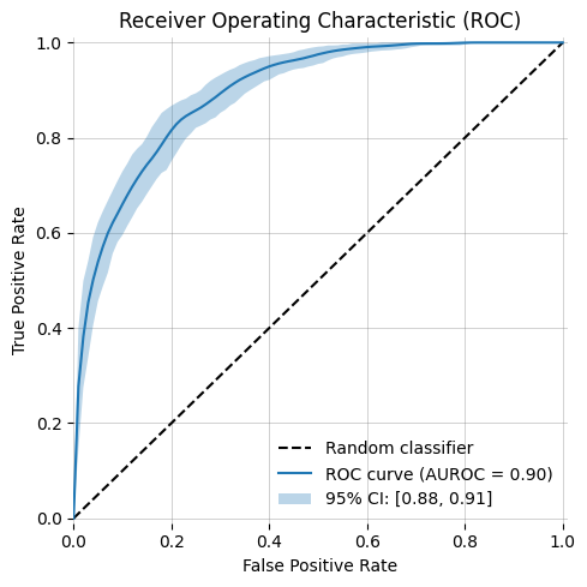
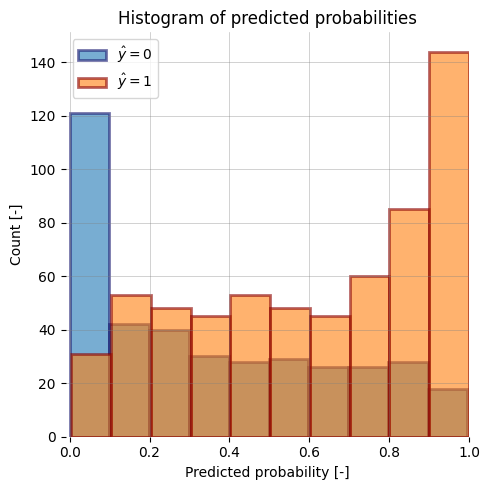
-| ROC Curve (AUROC) | ROC Curve (AUROC) with bootstrapping | y_prob histogram |
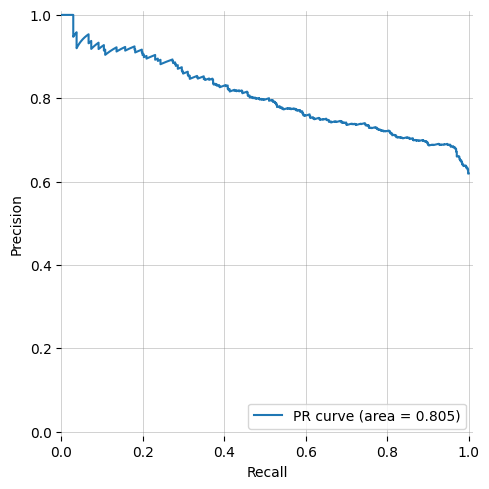
+| ROC Curve (AUROC) with bootstrapping | Precision-Recall Curve | y_prob histogram |
|
|
|:--------------------------------------------------:|:----------------------------------------------------------:|:-------------------------------------------------:|
-| ROC Curve (AUROC) | ROC Curve (AUROC) with bootstrapping | y_prob histogram |
+| ROC Curve (AUROC) with bootstrapping | Precision-Recall Curve | y_prob histogram |
|  |
|  |
|  |
@@ -82,7 +82,7 @@ Install the package via pip.
pip install plotsandgraphs
```
-Alternativelynstall the package from git.
+Alternatively install the package from git.
```bash
git clone https://github.com/joshuawe/plots_and_graphs
cd plots_and_graphs
diff --git a/images/pr_curve.png b/images/pr_curve.png
new file mode 100644
index 0000000..6904ad5
Binary files /dev/null and b/images/pr_curve.png differ
diff --git a/images/y_prob_histogram.png b/images/y_prob_histogram.png
index e094db6..c4a664c 100644
Binary files a/images/y_prob_histogram.png and b/images/y_prob_histogram.png differ
diff --git a/plotsandgraphs/binary_classifier.py b/plotsandgraphs/binary_classifier.py
index 3f81c01..4eb06ef 100644
--- a/plotsandgraphs/binary_classifier.py
+++ b/plotsandgraphs/binary_classifier.py
@@ -1,7 +1,8 @@
+from pathlib import Path
+from typing import Optional
import matplotlib.pyplot as plt
from matplotlib.colors import to_rgba
from matplotlib.figure import Figure
-import seaborn as sns
import numpy as np
import pandas as pd
from sklearn.metrics import (
@@ -15,9 +16,7 @@
)
from sklearn.calibration import calibration_curve
from sklearn.utils import resample
-from pathlib import Path
from tqdm import tqdm
-from typing import Optional
def plot_accuracy(y_true, y_pred, name="", save_fig_path=None) -> Figure:
@@ -39,16 +38,14 @@ def plot_accuracy(y_true, y_pred, name="", save_fig_path=None) -> Figure:
plt.title(title)
plt.tight_layout()
- if save_fig_path != None:
+ if save_fig_path is not None:
path = Path(save_fig_path)
path.parent.mkdir(parents=True, exist_ok=True)
fig.savefig(save_fig_path, bbox_inches="tight")
- return fig, accuracy
+ return fig
-def plot_confusion_matrix(
- y_true: np.ndarray, y_pred: np.ndarray, save_fig_path=None
-) -> Figure:
+def plot_confusion_matrix(y_true: np.ndarray, y_pred: np.ndarray, save_fig_path=None) -> Figure:
import matplotlib.colors as colors
# Compute the confusion matrix
@@ -57,16 +54,14 @@ def plot_confusion_matrix(
cm = cm.astype("float") / cm.sum(axis=1)[:, np.newaxis]
# Create the ConfusionMatrixDisplay instance and plot it
- cmd = ConfusionMatrixDisplay(
- cm, display_labels=["class 0\nnegative", "class 1\npositive"]
- )
+ cmd = ConfusionMatrixDisplay(cm, display_labels=["class 0\nnegative", "class 1\npositive"])
fig, ax = plt.subplots(figsize=(4, 4))
cmd.plot(
cmap="YlOrRd",
values_format="",
colorbar=False,
ax=ax,
- text_kw={"visible": False},
+ # text_kw={"visible": False},
)
cmd.texts_ = []
cmd.text_ = []
@@ -106,7 +101,7 @@ def plot_confusion_matrix(
cbar.outline.set_visible(False)
plt.tight_layout()
- if save_fig_path != None:
+ if save_fig_path is not None:
path = Path(save_fig_path)
path.parent.mkdir(parents=True, exist_ok=True)
fig.savefig(save_fig_path, bbox_inches="tight")
@@ -115,7 +110,7 @@ def plot_confusion_matrix(
def plot_classification_report(
- y_test: np.ndarray,
+ y_true: np.ndarray,
y_pred: np.ndarray,
title="Classification Report",
figsize=(8, 4),
@@ -152,18 +147,13 @@ def plot_classification_report(
import matplotlib as mpl
import matplotlib.colors as colors
import seaborn as sns
- import pathlib
fig, ax = plt.subplots(figsize=figsize)
cmap = "YlOrRd"
- clf_report = classification_report(y_test, y_pred, output_dict=True, **kwargs)
- keys_to_plot = [
- key
- for key in clf_report.keys()
- if key not in ("accuracy", "macro avg", "weighted avg")
- ]
+ clf_report = classification_report(y_true, y_pred, output_dict=True, **kwargs)
+ keys_to_plot = [key for key in clf_report.keys() if key not in ("accuracy", "macro avg", "weighted avg")]
df = pd.DataFrame(clf_report, columns=keys_to_plot).T
# the following line ensures that dataframe are sorted from the majority classes to the minority classes
df.sort_values(by=["support"], inplace=True)
@@ -174,8 +164,8 @@ def plot_classification_report(
mask[:, cols - 1] = True
bounds = np.linspace(0, 1, 11)
- cmap = plt.cm.get_cmap("YlOrRd", len(bounds) + 1)
- norm = colors.BoundaryNorm(bounds, cmap.N) # type: ignore[attr-defined]
+ cmap = plt.cm.get_cmap("YlOrRd", len(bounds) + 1) # type: ignore[assignment]
+ norm = colors.BoundaryNorm(bounds, cmap.N) # type: ignore[attr-defined]
ax = sns.heatmap(
df,
@@ -247,7 +237,7 @@ def plot_classification_report(
plt.yticks(rotation=360)
plt.tight_layout()
- if save_fig_path != None:
+ if save_fig_path is not None:
path = Path(save_fig_path)
path.parent.mkdir(parents=True, exist_ok=True)
fig.savefig(save_fig_path, bbox_inches="tight")
@@ -332,9 +322,7 @@ def plot_roc_curve(
auc_upper = np.quantile(bootstrap_aucs, CI_upper)
auc_lower = np.quantile(bootstrap_aucs, CI_lower)
label = f"{confidence_interval:.0%} CI: [{auc_lower:.2f}, {auc_upper:.2f}]"
- plt.fill_between(
- base_fpr, tprs_lower, tprs_upper, alpha=0.3, label=label, zorder=2
- )
+ plt.fill_between(base_fpr, tprs_lower, tprs_upper, alpha=0.3, label=label, zorder=2)
if highlight_roc_area is True:
print(
@@ -366,9 +354,7 @@ def plot_roc_curve(
return fig
-def plot_calibration_curve(
- y_prob: np.ndarray, y_true: np.ndarray, n_bins=10, save_fig_path=None
-) -> Figure:
+def plot_calibration_curve(y_prob: np.ndarray, y_true: np.ndarray, n_bins=10, save_fig_path=None) -> Figure:
"""
Creates calibration plot for a binary classifier and calculates the ECE.
@@ -390,9 +376,7 @@ def plot_calibration_curve(
ece : float
The expected calibration error.
"""
- prob_true, prob_pred = calibration_curve(
- y_true, y_prob, n_bins=n_bins, strategy="uniform"
- )
+ prob_true, prob_pred = calibration_curve(y_true, y_prob, n_bins=n_bins, strategy="uniform")
# Find the number of samples in each bin
bin_counts = np.histogram(y_prob, bins=n_bins, range=(0, 1))[0]
@@ -465,7 +449,7 @@ def plot_calibration_curve(
return fig
-def plot_y_prob_histogram(y_prob: np.ndarray, y_true: Optional[np.ndarray]=None, save_fig_path=None) -> Figure:
+def plot_y_prob_histogram(y_prob: np.ndarray, y_true: Optional[np.ndarray] = None, save_fig_path=None) -> Figure:
"""
Provides a histogram for the predicted probabilities of a binary classifier. If ```y_true``` is provided, it divides the ```y_prob``` values into the two classes and plots them jointly into the same plot with different colors.
@@ -485,16 +469,32 @@ def plot_y_prob_histogram(y_prob: np.ndarray, y_true: Optional[np.ndarray]=None,
"""
fig = plt.figure(figsize=(5, 5))
ax = fig.add_subplot(111)
-
+
if y_true is None:
ax.hist(y_prob, bins=10, alpha=0.9, edgecolor="midnightblue", linewidth=2, rwidth=1)
# same histogram as above, but with border lines
# ax.hist(y_prob, bins=10, alpha=0.5, edgecolor='black', linewidth=1.2)
else:
alpha = 0.6
- ax.hist(y_prob[y_true==0], bins=10, alpha=alpha, edgecolor="midnightblue", linewidth=2, rwidth=1, label="$\\hat{y} = 0$")
- ax.hist(y_prob[y_true==1], bins=10, alpha=alpha, edgecolor="darkred", linewidth=2, rwidth=1, label="$\\hat{y} = 1$")
-
+ ax.hist(
+ y_prob[y_true == 0],
+ bins=10,
+ alpha=alpha,
+ edgecolor="midnightblue",
+ linewidth=2,
+ rwidth=1,
+ label="$\\hat{y} = 0$",
+ )
+ ax.hist(
+ y_prob[y_true == 1],
+ bins=10,
+ alpha=alpha,
+ edgecolor="darkred",
+ linewidth=2,
+ rwidth=1,
+ label="$\\hat{y} = 1$",
+ )
+
plt.legend()
ax.set(xlabel="Predicted probability [-]", ylabel="Count [-]", xlim=(-0.01, 1.0))
ax.set_title("Histogram of predicted probabilities")
@@ -505,7 +505,7 @@ def plot_y_prob_histogram(y_prob: np.ndarray, y_true: Optional[np.ndarray]=None,
plt.tight_layout()
# save plot
- if save_fig_path != None:
+ if save_fig_path is not None:
path = Path(save_fig_path)
path.parent.mkdir(parents=True, exist_ok=True)
fig.savefig(save_fig_path, bbox_inches="tight")
diff --git a/plotsandgraphs/compare_distributions.py b/plotsandgraphs/compare_distributions.py
index e9cc9fe..4689245 100644
--- a/plotsandgraphs/compare_distributions.py
+++ b/plotsandgraphs/compare_distributions.py
@@ -1,8 +1,8 @@
+from typing import List, Tuple, Optional
import numpy as np
import matplotlib.pyplot as plt
import matplotlib as mpl
import pandas as pd
-from typing import List, Tuple, Optional
def plot_raincloud(
diff --git a/pyproject.toml b/pyproject.toml
index 7717d35..576db67 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -4,5 +4,8 @@ max-line-length = 120
[tool.pylint."BASIC"]
variable-rgx = "[a-z_][a-z0-9_]{0,30}$|[a-z0-9_]+([A-Z][a-z0-9_]+)*$" # Allow snake case and camel case for variable names
+[tool.pylint."MESSAGES CONTROL"]
+disable = "W0621" # Allow redefining names in outer scope
+
[flake8]
max-line-length = 120
diff --git a/src/binary_classifier.py b/src/binary_classifier.py
index de5d636..fdd44ad 100644
--- a/src/binary_classifier.py
+++ b/src/binary_classifier.py
@@ -1,15 +1,14 @@
+from typing import Optional
+from pathlib import Path
import matplotlib.pyplot as plt
from matplotlib.colors import to_rgba
from matplotlib.figure import Figure
-import seaborn as sns
import numpy as np
import pandas as pd
from sklearn.metrics import confusion_matrix, classification_report, ConfusionMatrixDisplay, roc_curve, auc, accuracy_score, precision_recall_curve
from sklearn.calibration import calibration_curve
from sklearn.utils import resample
-from pathlib import Path
from tqdm import tqdm
-from typing import Optional
def plot_accuracy(y_true, y_pred, name='', save_fig_path=None) -> Figure:
@@ -381,7 +380,7 @@ def plot_y_prob_histogram(y_prob: np.ndarray, save_fig_path=None) -> Figure:
plt.tight_layout()
# save plot
- if (save_fig_path != None):
+ if (save_fig_path is not None):
path = Path(save_fig_path)
path.parent.mkdir(parents=True, exist_ok=True)
fig.savefig(save_fig_path, bbox_inches='tight')
diff --git a/src/compare_distributions.py b/src/compare_distributions.py
index 943c3e0..5a9aeb4 100644
--- a/src/compare_distributions.py
+++ b/src/compare_distributions.py
@@ -7,14 +7,14 @@
def plot_raincloud(df: pd.DataFrame,
x_col: str,
- y_col: str,
- colors: List[str] = None,
- order: List[str] = None,
- title: str = None,
- x_label: str = None,
- x_range: Tuple[float, float] = None,
- show_violin = True,
- show_scatter = True,
+ y_col: str,
+ colors: List[str] = None,
+ order: List[str] = None,
+ title: str = None,
+ x_label: str = None,
+ x_range: Tuple[float, float] = None,
+ show_violin = True,
+ show_scatter = True,
show_boxplot = True):
"""
@@ -49,7 +49,6 @@ def plot_raincloud(df: pd.DataFrame,
colors = [mpl.colors.to_hex(cmap(i)) for i in np.linspace(0, 1, len(order))]
else:
assert len(colors) == len(order), 'colors and order must be the same length'
- colors = colors
# Boxplot
if show_boxplot:
diff --git a/tests/__init__.py b/tests/__init__.py
index e69de29..72848bb 100644
--- a/tests/__init__.py
+++ b/tests/__init__.py
@@ -0,0 +1,7 @@
+import os
+
+TEST_RESULTS_PATH = os.path.join(os.path.dirname(__file__), "test_results")
+
+# print cwd in console
+
+# print os.path.dirname(__file__)
diff --git a/tests/test_binary_classifier.py b/tests/test_binary_classifier.py
new file mode 100644
index 0000000..bc878d6
--- /dev/null
+++ b/tests/test_binary_classifier.py
@@ -0,0 +1,143 @@
+from pathlib import Path
+from typing import Tuple
+import numpy as np
+import pytest
+import plotsandgraphs.binary_classifier as binary
+
+TEST_RESULTS_PATH = Path(r"tests\test_results")
+
+
+@pytest.fixture(scope="module")
+def random_data_binary_classifier() -> Tuple[np.ndarray, np.ndarray]:
+ """
+ Create random data for binary classifier tests.
+
+ Returns
+ -------
+ Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ # create some data
+ n_samples = 1000
+ y_true = np.random.choice(
+ [0, 1], n_samples, p=[0.4, 0.6]
+ ) # the true class labels 0 or 1, with class imbalance 40:60
+
+ y_prob = np.zeros(y_true.shape) # a model's probability of class 1 predictions
+ y_prob[y_true == 1] = np.random.beta(1, 0.6, y_prob[y_true == 1].shape)
+ y_prob[y_true == 0] = np.random.beta(0.5, 1, y_prob[y_true == 0].shape)
+ return y_true, y_prob
+
+
+# Test histogram plot
+def test_hist_plot(random_data_binary_classifier):
+ """
+ Test histogram plot.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ print(TEST_RESULTS_PATH)
+ binary.plot_y_prob_histogram(y_prob, save_fig_path=TEST_RESULTS_PATH / "histogram.png")
+ binary.plot_y_prob_histogram(y_prob, y_true, save_fig_path=TEST_RESULTS_PATH / "histogram_2_classes.png")
+
+
+# test roc curve without bootstrapping
+def test_roc_curve(random_data_binary_classifier):
+ """
+ Test roc curve.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ binary.plot_roc_curve(y_true, y_prob, save_fig_path=TEST_RESULTS_PATH / "roc_curve.png")
+
+
+# test roc curve with bootstrapping
+def test_roc_curve_bootstrap(random_data_binary_classifier):
+ """
+ Test roc curve with bootstrapping.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ binary.plot_roc_curve(
+ y_true, y_prob, n_bootstraps=10000, save_fig_path=TEST_RESULTS_PATH / "roc_curve_bootstrap.png"
+ )
+
+
+# test precision recall curve
+def test_pr_curve(random_data_binary_classifier):
+ """
+ Test precision recall curve.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ binary.plot_pr_curve(y_true, y_prob, save_fig_path=TEST_RESULTS_PATH / "pr_curve.png")
+
+
+# test confusion matrix
+def test_confusion_matrix(random_data_binary_classifier):
+ """
+ Test confusion matrix.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ binary.plot_confusion_matrix(y_true, y_prob, save_fig_path=TEST_RESULTS_PATH / "confusion_matrix.png")
+
+
+# test classification report
+def test_classification_report(random_data_binary_classifier):
+ """
+ Test classification report.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ binary.plot_classification_report(y_true, y_prob, save_fig_path=TEST_RESULTS_PATH / "classification_report.png")
+
+# test calibration curve
+def test_calibration_curve(random_data_binary_classifier):
+ """
+ Test calibration curve.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ binary.plot_calibration_curve(y_prob, y_true, save_fig_path=TEST_RESULTS_PATH / "calibration_curve.png")
+
+# test accuracy
+def test_accuracy(random_data_binary_classifier):
+ """
+ Test accuracy.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ binary.plot_accuracy(y_true, y_prob, save_fig_path=TEST_RESULTS_PATH / "accuracy.png")
diff --git a/tests/test_test.py b/tests/test_test.py
deleted file mode 100644
index b2820fc..0000000
--- a/tests/test_test.py
+++ /dev/null
@@ -1,4 +0,0 @@
-# This is just a test for a test
-
-def test_test():
- assert True
\ No newline at end of file
|
@@ -82,7 +82,7 @@ Install the package via pip.
pip install plotsandgraphs
```
-Alternativelynstall the package from git.
+Alternatively install the package from git.
```bash
git clone https://github.com/joshuawe/plots_and_graphs
cd plots_and_graphs
diff --git a/images/pr_curve.png b/images/pr_curve.png
new file mode 100644
index 0000000..6904ad5
Binary files /dev/null and b/images/pr_curve.png differ
diff --git a/images/y_prob_histogram.png b/images/y_prob_histogram.png
index e094db6..c4a664c 100644
Binary files a/images/y_prob_histogram.png and b/images/y_prob_histogram.png differ
diff --git a/plotsandgraphs/binary_classifier.py b/plotsandgraphs/binary_classifier.py
index 3f81c01..4eb06ef 100644
--- a/plotsandgraphs/binary_classifier.py
+++ b/plotsandgraphs/binary_classifier.py
@@ -1,7 +1,8 @@
+from pathlib import Path
+from typing import Optional
import matplotlib.pyplot as plt
from matplotlib.colors import to_rgba
from matplotlib.figure import Figure
-import seaborn as sns
import numpy as np
import pandas as pd
from sklearn.metrics import (
@@ -15,9 +16,7 @@
)
from sklearn.calibration import calibration_curve
from sklearn.utils import resample
-from pathlib import Path
from tqdm import tqdm
-from typing import Optional
def plot_accuracy(y_true, y_pred, name="", save_fig_path=None) -> Figure:
@@ -39,16 +38,14 @@ def plot_accuracy(y_true, y_pred, name="", save_fig_path=None) -> Figure:
plt.title(title)
plt.tight_layout()
- if save_fig_path != None:
+ if save_fig_path is not None:
path = Path(save_fig_path)
path.parent.mkdir(parents=True, exist_ok=True)
fig.savefig(save_fig_path, bbox_inches="tight")
- return fig, accuracy
+ return fig
-def plot_confusion_matrix(
- y_true: np.ndarray, y_pred: np.ndarray, save_fig_path=None
-) -> Figure:
+def plot_confusion_matrix(y_true: np.ndarray, y_pred: np.ndarray, save_fig_path=None) -> Figure:
import matplotlib.colors as colors
# Compute the confusion matrix
@@ -57,16 +54,14 @@ def plot_confusion_matrix(
cm = cm.astype("float") / cm.sum(axis=1)[:, np.newaxis]
# Create the ConfusionMatrixDisplay instance and plot it
- cmd = ConfusionMatrixDisplay(
- cm, display_labels=["class 0\nnegative", "class 1\npositive"]
- )
+ cmd = ConfusionMatrixDisplay(cm, display_labels=["class 0\nnegative", "class 1\npositive"])
fig, ax = plt.subplots(figsize=(4, 4))
cmd.plot(
cmap="YlOrRd",
values_format="",
colorbar=False,
ax=ax,
- text_kw={"visible": False},
+ # text_kw={"visible": False},
)
cmd.texts_ = []
cmd.text_ = []
@@ -106,7 +101,7 @@ def plot_confusion_matrix(
cbar.outline.set_visible(False)
plt.tight_layout()
- if save_fig_path != None:
+ if save_fig_path is not None:
path = Path(save_fig_path)
path.parent.mkdir(parents=True, exist_ok=True)
fig.savefig(save_fig_path, bbox_inches="tight")
@@ -115,7 +110,7 @@ def plot_confusion_matrix(
def plot_classification_report(
- y_test: np.ndarray,
+ y_true: np.ndarray,
y_pred: np.ndarray,
title="Classification Report",
figsize=(8, 4),
@@ -152,18 +147,13 @@ def plot_classification_report(
import matplotlib as mpl
import matplotlib.colors as colors
import seaborn as sns
- import pathlib
fig, ax = plt.subplots(figsize=figsize)
cmap = "YlOrRd"
- clf_report = classification_report(y_test, y_pred, output_dict=True, **kwargs)
- keys_to_plot = [
- key
- for key in clf_report.keys()
- if key not in ("accuracy", "macro avg", "weighted avg")
- ]
+ clf_report = classification_report(y_true, y_pred, output_dict=True, **kwargs)
+ keys_to_plot = [key for key in clf_report.keys() if key not in ("accuracy", "macro avg", "weighted avg")]
df = pd.DataFrame(clf_report, columns=keys_to_plot).T
# the following line ensures that dataframe are sorted from the majority classes to the minority classes
df.sort_values(by=["support"], inplace=True)
@@ -174,8 +164,8 @@ def plot_classification_report(
mask[:, cols - 1] = True
bounds = np.linspace(0, 1, 11)
- cmap = plt.cm.get_cmap("YlOrRd", len(bounds) + 1)
- norm = colors.BoundaryNorm(bounds, cmap.N) # type: ignore[attr-defined]
+ cmap = plt.cm.get_cmap("YlOrRd", len(bounds) + 1) # type: ignore[assignment]
+ norm = colors.BoundaryNorm(bounds, cmap.N) # type: ignore[attr-defined]
ax = sns.heatmap(
df,
@@ -247,7 +237,7 @@ def plot_classification_report(
plt.yticks(rotation=360)
plt.tight_layout()
- if save_fig_path != None:
+ if save_fig_path is not None:
path = Path(save_fig_path)
path.parent.mkdir(parents=True, exist_ok=True)
fig.savefig(save_fig_path, bbox_inches="tight")
@@ -332,9 +322,7 @@ def plot_roc_curve(
auc_upper = np.quantile(bootstrap_aucs, CI_upper)
auc_lower = np.quantile(bootstrap_aucs, CI_lower)
label = f"{confidence_interval:.0%} CI: [{auc_lower:.2f}, {auc_upper:.2f}]"
- plt.fill_between(
- base_fpr, tprs_lower, tprs_upper, alpha=0.3, label=label, zorder=2
- )
+ plt.fill_between(base_fpr, tprs_lower, tprs_upper, alpha=0.3, label=label, zorder=2)
if highlight_roc_area is True:
print(
@@ -366,9 +354,7 @@ def plot_roc_curve(
return fig
-def plot_calibration_curve(
- y_prob: np.ndarray, y_true: np.ndarray, n_bins=10, save_fig_path=None
-) -> Figure:
+def plot_calibration_curve(y_prob: np.ndarray, y_true: np.ndarray, n_bins=10, save_fig_path=None) -> Figure:
"""
Creates calibration plot for a binary classifier and calculates the ECE.
@@ -390,9 +376,7 @@ def plot_calibration_curve(
ece : float
The expected calibration error.
"""
- prob_true, prob_pred = calibration_curve(
- y_true, y_prob, n_bins=n_bins, strategy="uniform"
- )
+ prob_true, prob_pred = calibration_curve(y_true, y_prob, n_bins=n_bins, strategy="uniform")
# Find the number of samples in each bin
bin_counts = np.histogram(y_prob, bins=n_bins, range=(0, 1))[0]
@@ -465,7 +449,7 @@ def plot_calibration_curve(
return fig
-def plot_y_prob_histogram(y_prob: np.ndarray, y_true: Optional[np.ndarray]=None, save_fig_path=None) -> Figure:
+def plot_y_prob_histogram(y_prob: np.ndarray, y_true: Optional[np.ndarray] = None, save_fig_path=None) -> Figure:
"""
Provides a histogram for the predicted probabilities of a binary classifier. If ```y_true``` is provided, it divides the ```y_prob``` values into the two classes and plots them jointly into the same plot with different colors.
@@ -485,16 +469,32 @@ def plot_y_prob_histogram(y_prob: np.ndarray, y_true: Optional[np.ndarray]=None,
"""
fig = plt.figure(figsize=(5, 5))
ax = fig.add_subplot(111)
-
+
if y_true is None:
ax.hist(y_prob, bins=10, alpha=0.9, edgecolor="midnightblue", linewidth=2, rwidth=1)
# same histogram as above, but with border lines
# ax.hist(y_prob, bins=10, alpha=0.5, edgecolor='black', linewidth=1.2)
else:
alpha = 0.6
- ax.hist(y_prob[y_true==0], bins=10, alpha=alpha, edgecolor="midnightblue", linewidth=2, rwidth=1, label="$\\hat{y} = 0$")
- ax.hist(y_prob[y_true==1], bins=10, alpha=alpha, edgecolor="darkred", linewidth=2, rwidth=1, label="$\\hat{y} = 1$")
-
+ ax.hist(
+ y_prob[y_true == 0],
+ bins=10,
+ alpha=alpha,
+ edgecolor="midnightblue",
+ linewidth=2,
+ rwidth=1,
+ label="$\\hat{y} = 0$",
+ )
+ ax.hist(
+ y_prob[y_true == 1],
+ bins=10,
+ alpha=alpha,
+ edgecolor="darkred",
+ linewidth=2,
+ rwidth=1,
+ label="$\\hat{y} = 1$",
+ )
+
plt.legend()
ax.set(xlabel="Predicted probability [-]", ylabel="Count [-]", xlim=(-0.01, 1.0))
ax.set_title("Histogram of predicted probabilities")
@@ -505,7 +505,7 @@ def plot_y_prob_histogram(y_prob: np.ndarray, y_true: Optional[np.ndarray]=None,
plt.tight_layout()
# save plot
- if save_fig_path != None:
+ if save_fig_path is not None:
path = Path(save_fig_path)
path.parent.mkdir(parents=True, exist_ok=True)
fig.savefig(save_fig_path, bbox_inches="tight")
diff --git a/plotsandgraphs/compare_distributions.py b/plotsandgraphs/compare_distributions.py
index e9cc9fe..4689245 100644
--- a/plotsandgraphs/compare_distributions.py
+++ b/plotsandgraphs/compare_distributions.py
@@ -1,8 +1,8 @@
+from typing import List, Tuple, Optional
import numpy as np
import matplotlib.pyplot as plt
import matplotlib as mpl
import pandas as pd
-from typing import List, Tuple, Optional
def plot_raincloud(
diff --git a/pyproject.toml b/pyproject.toml
index 7717d35..576db67 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -4,5 +4,8 @@ max-line-length = 120
[tool.pylint."BASIC"]
variable-rgx = "[a-z_][a-z0-9_]{0,30}$|[a-z0-9_]+([A-Z][a-z0-9_]+)*$" # Allow snake case and camel case for variable names
+[tool.pylint."MESSAGES CONTROL"]
+disable = "W0621" # Allow redefining names in outer scope
+
[flake8]
max-line-length = 120
diff --git a/src/binary_classifier.py b/src/binary_classifier.py
index de5d636..fdd44ad 100644
--- a/src/binary_classifier.py
+++ b/src/binary_classifier.py
@@ -1,15 +1,14 @@
+from typing import Optional
+from pathlib import Path
import matplotlib.pyplot as plt
from matplotlib.colors import to_rgba
from matplotlib.figure import Figure
-import seaborn as sns
import numpy as np
import pandas as pd
from sklearn.metrics import confusion_matrix, classification_report, ConfusionMatrixDisplay, roc_curve, auc, accuracy_score, precision_recall_curve
from sklearn.calibration import calibration_curve
from sklearn.utils import resample
-from pathlib import Path
from tqdm import tqdm
-from typing import Optional
def plot_accuracy(y_true, y_pred, name='', save_fig_path=None) -> Figure:
@@ -381,7 +380,7 @@ def plot_y_prob_histogram(y_prob: np.ndarray, save_fig_path=None) -> Figure:
plt.tight_layout()
# save plot
- if (save_fig_path != None):
+ if (save_fig_path is not None):
path = Path(save_fig_path)
path.parent.mkdir(parents=True, exist_ok=True)
fig.savefig(save_fig_path, bbox_inches='tight')
diff --git a/src/compare_distributions.py b/src/compare_distributions.py
index 943c3e0..5a9aeb4 100644
--- a/src/compare_distributions.py
+++ b/src/compare_distributions.py
@@ -7,14 +7,14 @@
def plot_raincloud(df: pd.DataFrame,
x_col: str,
- y_col: str,
- colors: List[str] = None,
- order: List[str] = None,
- title: str = None,
- x_label: str = None,
- x_range: Tuple[float, float] = None,
- show_violin = True,
- show_scatter = True,
+ y_col: str,
+ colors: List[str] = None,
+ order: List[str] = None,
+ title: str = None,
+ x_label: str = None,
+ x_range: Tuple[float, float] = None,
+ show_violin = True,
+ show_scatter = True,
show_boxplot = True):
"""
@@ -49,7 +49,6 @@ def plot_raincloud(df: pd.DataFrame,
colors = [mpl.colors.to_hex(cmap(i)) for i in np.linspace(0, 1, len(order))]
else:
assert len(colors) == len(order), 'colors and order must be the same length'
- colors = colors
# Boxplot
if show_boxplot:
diff --git a/tests/__init__.py b/tests/__init__.py
index e69de29..72848bb 100644
--- a/tests/__init__.py
+++ b/tests/__init__.py
@@ -0,0 +1,7 @@
+import os
+
+TEST_RESULTS_PATH = os.path.join(os.path.dirname(__file__), "test_results")
+
+# print cwd in console
+
+# print os.path.dirname(__file__)
diff --git a/tests/test_binary_classifier.py b/tests/test_binary_classifier.py
new file mode 100644
index 0000000..bc878d6
--- /dev/null
+++ b/tests/test_binary_classifier.py
@@ -0,0 +1,143 @@
+from pathlib import Path
+from typing import Tuple
+import numpy as np
+import pytest
+import plotsandgraphs.binary_classifier as binary
+
+TEST_RESULTS_PATH = Path(r"tests\test_results")
+
+
+@pytest.fixture(scope="module")
+def random_data_binary_classifier() -> Tuple[np.ndarray, np.ndarray]:
+ """
+ Create random data for binary classifier tests.
+
+ Returns
+ -------
+ Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ # create some data
+ n_samples = 1000
+ y_true = np.random.choice(
+ [0, 1], n_samples, p=[0.4, 0.6]
+ ) # the true class labels 0 or 1, with class imbalance 40:60
+
+ y_prob = np.zeros(y_true.shape) # a model's probability of class 1 predictions
+ y_prob[y_true == 1] = np.random.beta(1, 0.6, y_prob[y_true == 1].shape)
+ y_prob[y_true == 0] = np.random.beta(0.5, 1, y_prob[y_true == 0].shape)
+ return y_true, y_prob
+
+
+# Test histogram plot
+def test_hist_plot(random_data_binary_classifier):
+ """
+ Test histogram plot.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ print(TEST_RESULTS_PATH)
+ binary.plot_y_prob_histogram(y_prob, save_fig_path=TEST_RESULTS_PATH / "histogram.png")
+ binary.plot_y_prob_histogram(y_prob, y_true, save_fig_path=TEST_RESULTS_PATH / "histogram_2_classes.png")
+
+
+# test roc curve without bootstrapping
+def test_roc_curve(random_data_binary_classifier):
+ """
+ Test roc curve.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ binary.plot_roc_curve(y_true, y_prob, save_fig_path=TEST_RESULTS_PATH / "roc_curve.png")
+
+
+# test roc curve with bootstrapping
+def test_roc_curve_bootstrap(random_data_binary_classifier):
+ """
+ Test roc curve with bootstrapping.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ binary.plot_roc_curve(
+ y_true, y_prob, n_bootstraps=10000, save_fig_path=TEST_RESULTS_PATH / "roc_curve_bootstrap.png"
+ )
+
+
+# test precision recall curve
+def test_pr_curve(random_data_binary_classifier):
+ """
+ Test precision recall curve.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ binary.plot_pr_curve(y_true, y_prob, save_fig_path=TEST_RESULTS_PATH / "pr_curve.png")
+
+
+# test confusion matrix
+def test_confusion_matrix(random_data_binary_classifier):
+ """
+ Test confusion matrix.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ binary.plot_confusion_matrix(y_true, y_prob, save_fig_path=TEST_RESULTS_PATH / "confusion_matrix.png")
+
+
+# test classification report
+def test_classification_report(random_data_binary_classifier):
+ """
+ Test classification report.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ binary.plot_classification_report(y_true, y_prob, save_fig_path=TEST_RESULTS_PATH / "classification_report.png")
+
+# test calibration curve
+def test_calibration_curve(random_data_binary_classifier):
+ """
+ Test calibration curve.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ binary.plot_calibration_curve(y_prob, y_true, save_fig_path=TEST_RESULTS_PATH / "calibration_curve.png")
+
+# test accuracy
+def test_accuracy(random_data_binary_classifier):
+ """
+ Test accuracy.
+
+ Parameters
+ ----------
+ random_data_binary_classifier : Tuple[np.ndarray, np.ndarray]
+ The simulated data.
+ """
+ y_true, y_prob = random_data_binary_classifier
+ binary.plot_accuracy(y_true, y_prob, save_fig_path=TEST_RESULTS_PATH / "accuracy.png")
diff --git a/tests/test_test.py b/tests/test_test.py
deleted file mode 100644
index b2820fc..0000000
--- a/tests/test_test.py
+++ /dev/null
@@ -1,4 +0,0 @@
-# This is just a test for a test
-
-def test_test():
- assert True
\ No newline at end of file
 |
|  |
|  |
+|
|
+|  |
|  |
|  |
|:--------------------------------------------------:|:----------------------------------------------------------:|:-------------------------------------------------:|
-| ROC Curve (AUROC) | ROC Curve (AUROC) with bootstrapping | y_prob histogram |
+| ROC Curve (AUROC) with bootstrapping | Precision-Recall Curve | y_prob histogram |
|
|
|:--------------------------------------------------:|:----------------------------------------------------------:|:-------------------------------------------------:|
-| ROC Curve (AUROC) | ROC Curve (AUROC) with bootstrapping | y_prob histogram |
+| ROC Curve (AUROC) with bootstrapping | Precision-Recall Curve | y_prob histogram |
|  |
|