library(admiral)
+library(pharmaversesdtm)
+library(dplyr, warn.conflicts = FALSE)Categories
-Derive a new parameter computed from the value of other parameters +Admiral Hackathon 2023 Revisited
Use admiral::derive_param_computed() like a calculator to derive new parameters/rows based on existing ones
Let’s have a look at the Admiral Hackathon 2023 together.
-Hackathon Feedback Application +Derive a new parameter computed from the value of other parameters
Going through the process of creating a shiny app for the admiral hackathon. The shiny app allows users to check their solutions autonomously, gives feedback, and rates their results.
Use admiral::derive_param_computed() like a calculator to derive new parameters/rows based on existing ones
-Admiral Hackathon 2023 Revisited +Hackathon Feedback Application
admiral
Let’s have a look at the Admiral Hackathon 2023 together.
+Going through the process of creating a shiny app for the admiral hackathon. The shiny app allows users to check their solutions autonomously, gives feedback, and rates their results.
As you can see, the data-frame contains the column GRADE_CRITERIA_CODE which contains comparisons of floating point values. And there was a discrepancy of what Gordon expected to see, and how R actually computed the comparison initially:
Last updated
2023-11-01 15:47:35.340689
+2023-11-27 19:13:46.342818
Details
Last updated
2023-11-01 15:47:31.231853
+2023-11-27 19:13:54.254558
Details
Last updated
2023-11-01 15:47:48.201548
+2023-11-27 19:13:49.940918
Details
Last updated
2023-11-01 15:47:29.564969
+2023-11-27 19:13:33.556149
Details
Last updated
2023-11-01 15:47:45.242711
+2023-11-27 19:13:57.651626
Details
Development workflow
-Every improvement, task, or feature we want to implement on {admiral} starts as an issue on our GitHub repository. It is the centerpiece of our development workflow, along with our developer guides which describe in detail the strategies, conventions, and workflows used in development. The guides help us keep the {admiral} package internally consistent (e.g. naming conventions, function logic) but also ensure that {admiral} adjacent packages follow the same conventions and share the user interface. This is further helped by the implemented CICD pipeline which ensures styling convention and spelling (and much more).
+Every improvement, task, or feature we want to implement on {admiral} starts as an issue on our GitHub repository. It is the centerpiece of our development workflow, along with our developer guides which describe in detail the strategies, conventions, and workflows used in development. The guides help us keep the {admiral} package internally consistent (e.g. naming conventions, function logic) but also ensure that {admiral} adjacent packages follow the same conventions and share the user interface. This is further helped by the implemented CICD pipeline which ensures styling convention and spelling (and much more).
The core package developers team meets once a week (twice a week before a release) to discuss progress and priorities. Here, the role of product lead (currently Edoardo Mancini at Roche) and technical lead (currently Ben Straub at GSK) is to set priorities and track the release schedule. These stand-up meetings are centered around the project-board which gives a complete overview of activities and progress. Issues are mostly self-assigned so developers can really chose what they want to work on.
Last updated
2023-11-01 15:47:53.276819
+2023-11-27 19:13:41.390513
Details
Last updated
2023-11-01 15:47:27.839956
+2023-11-27 19:13:51.677601
Details
Last updated
2023-11-01 15:47:38.469514
+2023-11-27 19:13:40.019302
Details
Last updated
2023-11-01 15:47:37.128939
+2023-11-27 19:13:52.942147
Details
This January and February (2023), the admiral development team and the CDISC Open Source Alliance jointly hosted the admiral hackathon. The idea was to build a community of admiral users, and help participants familiarize themselves with R and admiral. This whole effort was led by Thomas Neitmann and was supported by Zelos Zhu, Sadchla Mascary, and me – Stefan Thoma.
The hackathon event was structured in two parts. First, we offered an Introduction to R for SAS programmers, a three hour workshop for R beginners to get them up to speed. Here we covered practical R basics, talking about how the R-workflow differs from a SAS workflow, and discussed common R functions - mostly from the tidyverse. This ensured that hackathon participants were familiar with core R concepts. The workshop recording and the course materials are available online.
+The main hackathon consisted of several ADAM data generating tasks based on a specs file and synthetic data. Participants were able to solve these tasks in groups at their own pace thanks to a online tool where participants could upload their task specific R scripts and they would get automatic feedback for the data-set produced by their script. Script upload through the feedback application was available all through February, and we offered three additional online meetings throughout the month to discuss challenges and give some tips. If you are interested in learning more about the thoughts that went into the feedback application, you can read about it in this blogpost or check out my public GitHub repository for such an application.
+Introduction to R workshop
+We were really excited to see over 500 people from around 40 countries joining our Introduction to R workshop in January! To get to know prospective users and hackathon participants better, we conducted some polls during the meetings. Below you can see that representatives of many different sorts of organisations joined our Introduction to R workshop:
+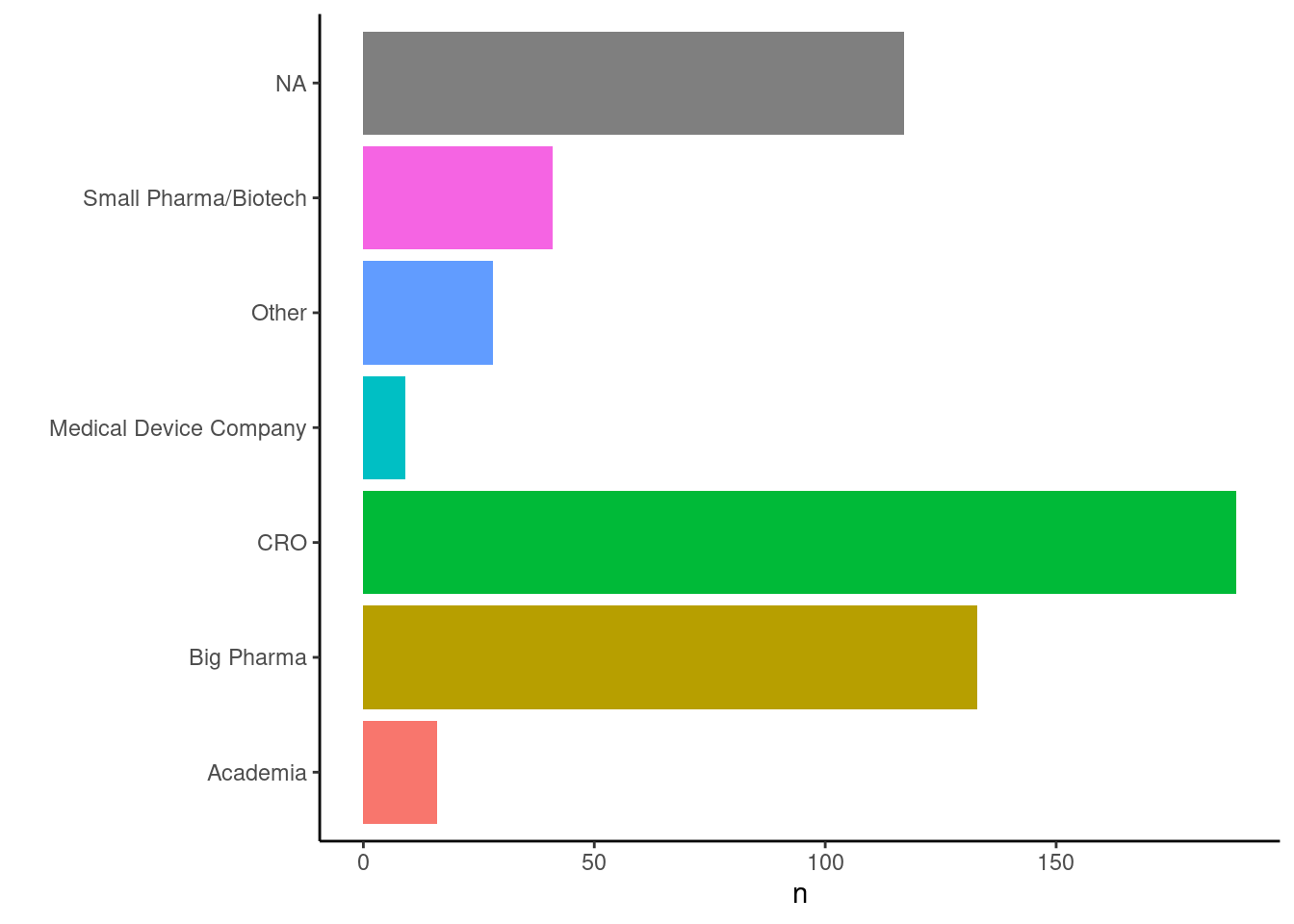
216 out of 402 confirmed that their company is already using R for clinical trial data analysis, the remaining 131 did not answer this question.
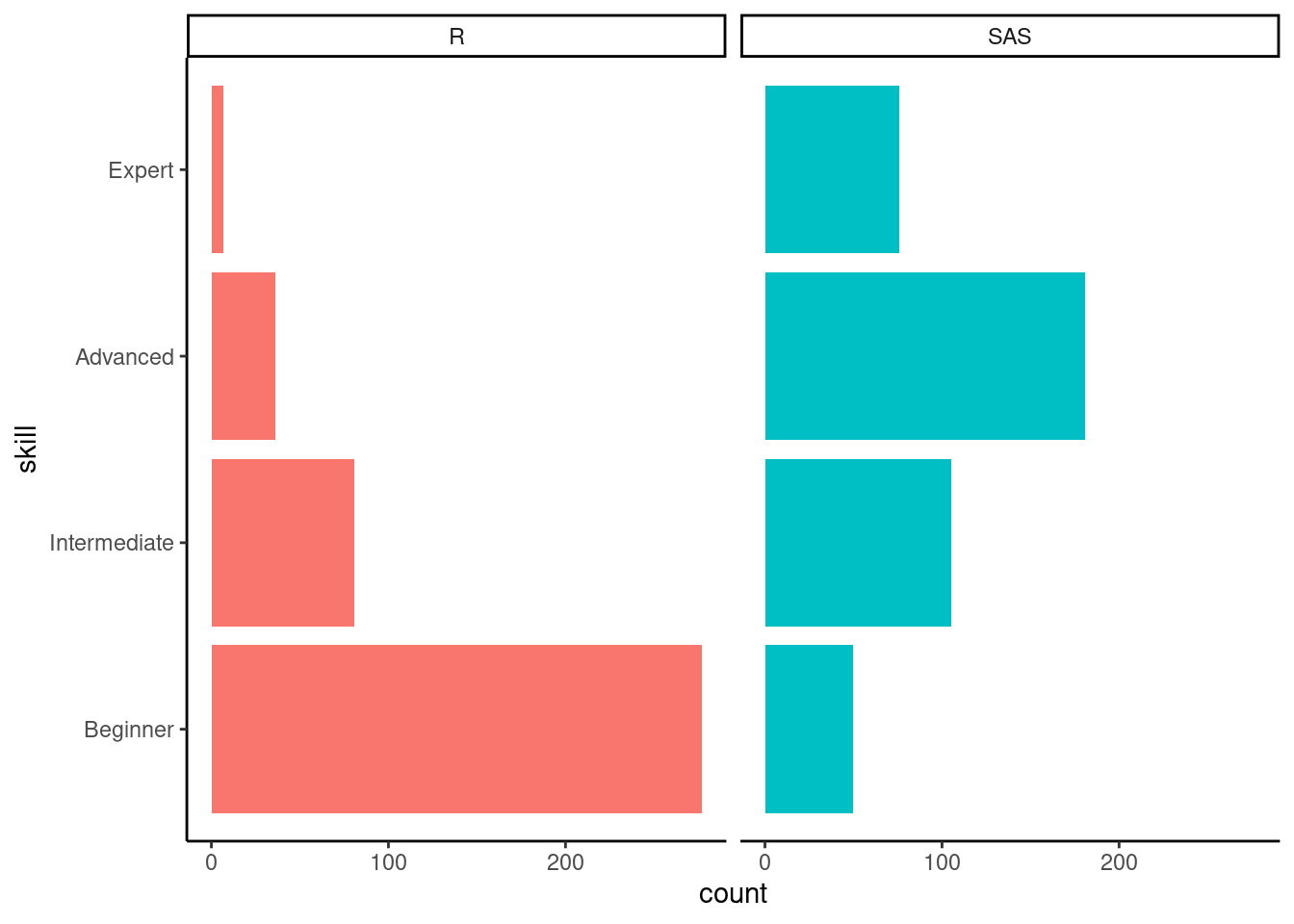
+The target audience for this workshop was programmers who are very familiar with SAS, but not so familiar with R, our polls confirmed this.
+
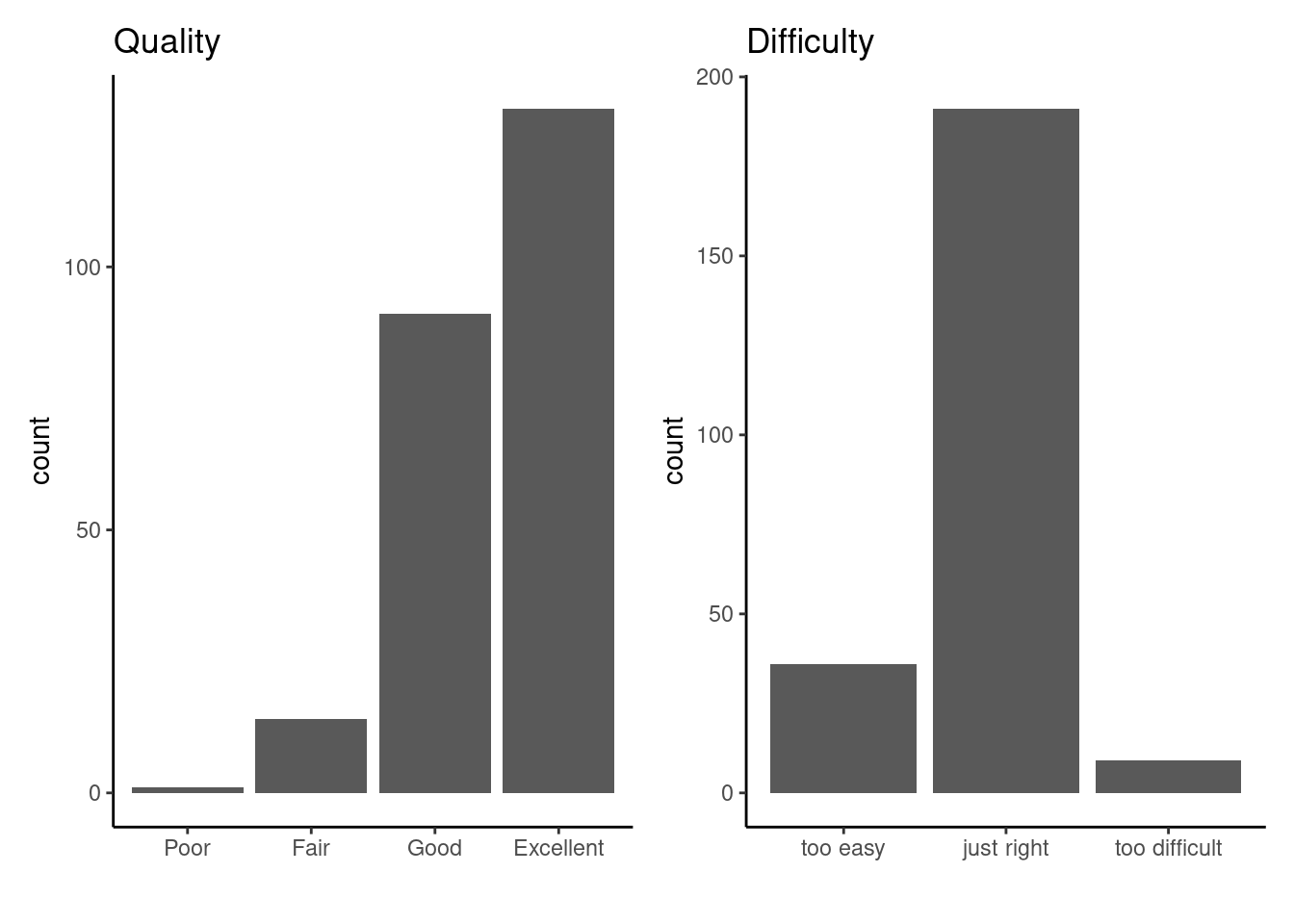
Overall, we were very happy with how the workshop turned out, and participants overall agreed with this sentiment (although there may be a slight survivorship bias…).
+
admiral Hackathon
+Following the kick-off meeting, 371 participants joined the posit (rStudio) workspace that was made available to all participants at no costs by the posit company. About half the participants planned to spend one to two hours per week on the admiral tasks, the other half planned to allocate even more. 15 participants even planned to spend eight hours or more!
+We were really happy to see an overwhelming amount of activity on the slack channel we set up with over 250 members. Not only were people engaging with the materials, but we saw how a community was formed where people were encouraged to ask questions and where community members went out of their way to help each other. Shout-out to our community hero: Jagadish Katam without whom most issues related to the task programming raised by the community would not have been addressed as quickly as they were. Huge thanks from the organizers!
+In the end, a total of 44 teams spanning 87 statistical programmers took part in the admiral hackathon and uploaded solution scripts to the hackathon application solving at least one of the 8 tasks available (ADSL, ADAE, ADLBC, ADVS, ADTTE, ADADAS, ADLBH & ADLBHY). Participants’ scripts were then run on the shiny server and the output data-frame were compared to the solutions we provided. At the read-out there was a live draft of teams to win one-on-one admiral consulting with one of the admiral core developers. Winning probabilities were weighted by the number of points each group received for the quality of their output data-frames and for the number of tasks solved.
Congratulations to the winners:
+-
+
ViiV Team_GSK
+teamspoRt
+TatianaPXL
+Divyasneelam
+AdaMTeamIndia
+Sanofi_BP
+Jagadish(our community hero)
+AZ_WAWA
+
Although this was uncertain during the hackathon we were excited to provide a Certificate of Completion to all participants who uploaded a script to the Web Application.
+A recording of the hackathon readout can be found in the CDISC Open Source Alliance Quarterly Spotlight.
+Conclusion
+Overall, we are very happy with how the hackathon turned out. We were not only positively surprised with the huge audience for the Intro to R workshop (CDISC record breaking) and for the admiral hackathon, but even more so with the engagement of all the participants.
Again, we would like to thank all the organizers, participants, and sponsors for their time and resources and hope to have provided a useful glimpse into our solution for ADAM creation within the end-to-end clinical data analysis open source R framework that the Pharmaverse aims to provide.
+As always, we are very happy to hear more feedback on the hackathon as well as on admiral in general. Simply submit an issue on the admiral GitHub repository. You would like to join the admiral core developers? Please reach out to Edoardo Mancini (product owner) or Ben Straub (technical lead).
Last updated
2023-11-27 19:13:36.963431
+Details
Reuse
Citation
@online{thoma2023,
+ author = {Thoma, Stefan},
+ title = {Admiral {Hackathon} 2023 {Revisited}},
+ date = {2023-06-27},
+ url = {https://pharmaverse.github.io/blog/posts/2023-06-27_hackathon_writeup},
+ langid = {en}
+}
+Last updated
2023-11-01 15:47:33.340574
+2023-11-27 19:13:38.746856
Details
.zip file of the workspace to which hackathon participants had access via posit cloud. For more context you can watch recordings of the hackathon-meetings.
+The hackathon application is still online (although data-upload is switched off) and the GitHub repository is publicly available. The application is embedded into this post right after this paragraph. I have also uploaded to GitHub a .zip file of the workspace to which hackathon participants had access via posit cloud. For more context you can watch recordings of the hackathon-meetings.
Last updated
2023-11-01 15:47:42.927745
+2023-11-27 19:13:48.333469
Details
This January and February (2023), the admiral development team and the CDISC Open Source Alliance jointly hosted the admiral hackathon. The idea was to build a community of admiral users, and help participants familiarize themselves with R and admiral. This whole effort was led by Thomas Neitmann and was supported by Zelos Zhu, Sadchla Mascary, and me – Stefan Thoma.
The hackathon event was structured in two parts. First, we offered an Introduction to R for SAS programmers, a three hour workshop for R beginners to get them up to speed. Here we covered practical R basics, talking about how the R-workflow differs from a SAS workflow, and discussed common R functions - mostly from the tidyverse. This ensured that hackathon participants were familiar with core R concepts. The workshop recording and the course materials are available online.
-The main hackathon consisted of several ADAM data generating tasks based on a specs file and synthetic data. Participants were able to solve these tasks in groups at their own pace thanks to a online tool where participants could upload their task specific R scripts and they would get automatic feedback for the data-set produced by their script. Script upload through the feedback application was available all through February, and we offered three additional online meetings throughout the month to discuss challenges and give some tips. If you are interested in learning more about the thoughts that went into the feedback application, you can read about it in this blogpost or check out my public GitHub repository for such an application.
-Introduction to R workshop
-We were really excited to see over 500 people from around 40 countries joining our Introduction to R workshop in January! To get to know prospective users and hackathon participants better, we conducted some polls during the meetings. Below you can see that representatives of many different sorts of organisations joined our Introduction to R workshop:
-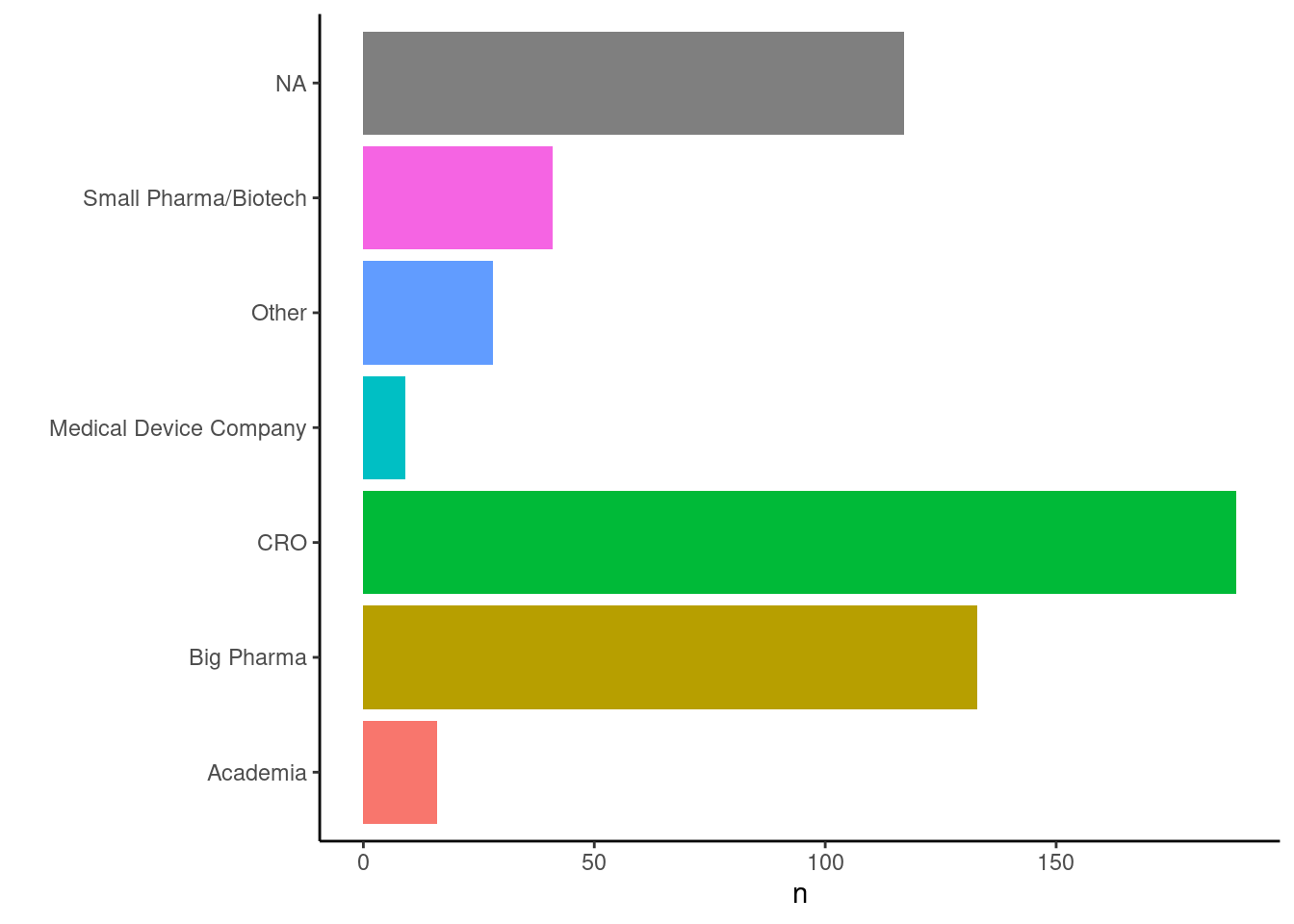
216 out of 402 confirmed that their company is already using R for clinical trial data analysis, the remaining 131 did not answer this question.
-The target audience for this workshop was programmers who are very familiar with SAS, but not so familiar with R, our polls confirmed this.
-
Overall, we were very happy with how the workshop turned out, and participants overall agreed with this sentiment (although there may be a slight survivorship bias…).
-
admiral Hackathon
-Following the kick-off meeting, 371 participants joined the posit (rStudio) workspace that was made available to all participants at no costs by the posit company. About half the participants planned to spend one to two hours per week on the admiral tasks, the other half planned to allocate even more. 15 participants even planned to spend eight hours or more!
-We were really happy to see an overwhelming amount of activity on the slack channel we set up with over 250 members. Not only were people engaging with the materials, but we saw how a community was formed where people were encouraged to ask questions and where community members went out of their way to help each other. Shout-out to our community hero: Jagadish Katam without whom most issues related to the task programming raised by the community would not have been addressed as quickly as they were. Huge thanks from the organizers!
-In the end, a total of 44 teams spanning 87 statistical programmers took part in the admiral hackathon and uploaded solution scripts to the hackathon application solving at least one of the 8 tasks available (ADSL, ADAE, ADLBC, ADVS, ADTTE, ADADAS, ADLBH & ADLBHY). Participants’ scripts were then run on the shiny server and the output data-frame were compared to the solutions we provided. At the read-out there was a live draft of teams to win one-on-one admiral consulting with one of the admiral core developers. Winning probabilities were weighted by the number of points each group received for the quality of their output data-frames and for the number of tasks solved.
Congratulations to the winners:
--
-
ViiV Team_GSK
-teamspoRt
-TatianaPXL
-Divyasneelam
-AdaMTeamIndia
-Sanofi_BP
-Jagadish(our community hero)
-AZ_WAWA
-
Although this was uncertain during the hackathon we were excited to provide a Certificate of Completion to all participants who uploaded a script to the Web Application.
-A recording of the hackathon readout can be found in the CDISC Open Source Alliance Quarterly Spotlight.
-Conclusion
-Overall, we are very happy with how the hackathon turned out. We were not only positively surprised with the huge audience for the Intro to R workshop (CDISC record breaking) and for the admiral hackathon, but even more so with the engagement of all the participants.
Again, we would like to thank all the organizers, participants, and sponsors for their time and resources and hope to have provided a useful glimpse into our solution for ADAM creation within the end-to-end clinical data analysis open source R framework that the Pharmaverse aims to provide.
-As always, we are very happy to hear more feedback on the hackathon as well as on admiral in general. Simply submit an issue on the admiral GitHub repository. You would like to join the admiral core developers? Please reach out to Edoardo Mancini (product owner) or Ben Straub (technical lead).
Last updated
2023-11-01 15:47:51.728905
-Details
Reuse
Citation
@online{thoma2023,
- author = {Thoma, Stefan},
- title = {Admiral {Hackathon} 2023 {Revisited}},
- date = {2023-06-27},
- url = {https://pharmaverse.github.io/blog/posts/2023-06-27_hackathon_writeup},
- langid = {en}
-}
-Table of contents
Run participants R-scripts on the cloud and compare the produced file to a solution file. Let participants autonomously get feedback on their work.
In this blog post I want to highlight some of the thoughts that went into this application. Please keep in mind that this work was done under tight time restraints.
-The hackathon application is still online (although data-upload is switched off) and the GitHub repository is publicly available. The application is embedded into this post right after this paragraph. I have also uploaded to GitHub a .zip file of the workspace to which hackathon participants had access via posit cloud. For more context you can watch recordings of the hackathon-meetings.
The hackathon application is still online (although data-upload is switched off) and the GitHub repository is publicly available. The application is embedded into this post right after this paragraph. I have also uploaded to GitHub a .zip file of the workspace to which hackathon participants had access via posit cloud. For more context you can watch recordings of the hackathon-meetings.
Conclusion
Last updated
2023-11-01 15:47:42.927745
+2023-11-27 19:13:48.333469
Details
Compare
Last updated
2023-11-01 15:47:33.340574
+2023-11-27 19:13:38.746856
Details
Table of contents
This January and February (2023), the admiral development team and the CDISC Open Source Alliance jointly hosted the admiral hackathon. The idea was to build a community of admiral users, and help participants familiarize themselves with R and admiral. This whole effort was led by Thomas Neitmann and was supported by Zelos Zhu, Sadchla Mascary, and me – Stefan Thoma.
The hackathon event was structured in two parts. First, we offered an Introduction to R for SAS programmers, a three hour workshop for R beginners to get them up to speed. Here we covered practical R basics, talking about how the R-workflow differs from a SAS workflow, and discussed common R functions - mostly from the tidyverse. This ensured that hackathon participants were familiar with core R concepts. The workshop recording and the course materials are available online.
+The hackathon event was structured in two parts. First, we offered an Introduction to R for SAS programmers, a three hour workshop for R beginners to get them up to speed. Here we covered practical R basics, talking about how the R-workflow differs from a SAS workflow, and discussed common R functions - mostly from the tidyverse. This ensured that hackathon participants were familiar with core R concepts. The workshop recording and the course materials are available online.
The main hackathon consisted of several ADAM data generating tasks based on a specs file and synthetic data. Participants were able to solve these tasks in groups at their own pace thanks to a online tool where participants could upload their task specific R scripts and they would get automatic feedback for the data-set produced by their script. Script upload through the feedback application was available all through February, and we offered three additional online meetings throughout the month to discuss challenges and give some tips. If you are interested in learning more about the thoughts that went into the feedback application, you can read about it in this blogpost or check out my public GitHub repository for such an application.
Introduction to R workshop
@@ -213,7 +213,7 @@admiral Hac
AZ_WAWAAlthough this was uncertain during the hackathon we were excited to provide a Certificate of Completion to all participants who uploaded a script to the Web Application.
-A recording of the hackathon readout can be found in the CDISC Open Source Alliance Quarterly Spotlight.
+A recording of the hackathon readout can be found in the CDISC Open Source Alliance Quarterly Spotlight.
Conclusion
@@ -233,7 +233,7 @@Conclusion
Last updated
2023-11-01 15:47:51.728905
+2023-11-27 19:13:36.963431
Details
How can I make
Last updated
-2023-11-01 15:47:37.128939
+2023-11-27 19:13:52.942147
Details
diff --git a/posts/2023-07-09_falcon/falcon.html b/posts/2023-07-09_falcon/falcon.html
index 24819813..a1b9e04b 100644
--- a/posts/2023-07-09_falcon/falcon.html
+++ b/posts/2023-07-09_falcon/falcon.html
@@ -196,7 +196,7 @@ Next steps & vision<
Last updated
-2023-11-01 15:47:38.469514
+2023-11-27 19:13:40.019302
Details
diff --git a/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html b/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
index d9ec2ab8..1eea5aae 100644
--- a/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
+++ b/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
@@ -301,7 +301,7 @@ That’s it!
Last updated
-2023-11-01 15:47:27.839956
+2023-11-27 19:13:51.677601
Details
diff --git a/posts/2023-07-14_code_sections/code_sections.html b/posts/2023-07-14_code_sections/code_sections.html
index 73c1bad5..44eaf7d9 100644
--- a/posts/2023-07-14_code_sections/code_sections.html
+++ b/posts/2023-07-14_code_sections/code_sections.html
@@ -294,7 +294,7 @@ Conclusion
Last updated
-2023-11-01 15:47:53.276819
+2023-11-27 19:13:41.390513
Details
diff --git a/posts/2023-07-20_cross_company_dev/cross_industry_dev.html b/posts/2023-07-20_cross_company_dev/cross_industry_dev.html
index c9ed3dcd..62f82d23 100644
--- a/posts/2023-07-20_cross_company_dev/cross_industry_dev.html
+++ b/posts/2023-07-20_cross_company_dev/cross_industry_dev.html
@@ -229,7 +229,7 @@ Insights
Development workflow
-Every improvement, task, or feature we want to implement on {admiral} starts as an issue on our GitHub repository. It is the centerpiece of our development workflow, along with our developer guides which describe in detail the strategies, conventions, and workflows used in development. The guides help us keep the {admiral} package internally consistent (e.g. naming conventions, function logic) but also ensure that {admiral} adjacent packages follow the same conventions and share the user interface. This is further helped by the implemented CICD pipeline which ensures styling convention and spelling (and much more).
+Every improvement, task, or feature we want to implement on {admiral} starts as an issue on our GitHub repository. It is the centerpiece of our development workflow, along with our developer guides which describe in detail the strategies, conventions, and workflows used in development. The guides help us keep the {admiral} package internally consistent (e.g. naming conventions, function logic) but also ensure that {admiral} adjacent packages follow the same conventions and share the user interface. This is further helped by the implemented CICD pipeline which ensures styling convention and spelling (and much more).
The core package developers team meets once a week (twice a week before a release) to discuss progress and priorities. Here, the role of product lead (currently Edoardo Mancini at Roche) and technical lead (currently Ben Straub at GSK) is to set priorities and track the release schedule. These stand-up meetings are centered around the project-board which gives a complete overview of activities and progress. Issues are mostly self-assigned so developers can really chose what they want to work on.
diff --git a/posts/2023-07-24_rounding/rounding.html b/posts/2023-07-24_rounding/rounding.html
index d1b059f0..54a2cb7e 100644
--- a/posts/2023-07-24_rounding/rounding.html
+++ b/posts/2023-07-24_rounding/rounding.html
@@ -467,7 +467,7 @@ Conclusion
Last updated
-2023-11-01 15:47:29.564969
+2023-11-27 19:13:33.556149
Details
diff --git a/posts/2023-08-08_study_day/study_day.html b/posts/2023-08-08_study_day/study_day.html
index 02f167ef..b54e7b65 100644
--- a/posts/2023-08-08_study_day/study_day.html
+++ b/posts/2023-08-08_study_day/study_day.html
@@ -264,7 +264,7 @@ It’s all relative? - Calculating Relative Days using admiral
Last updated
-2023-11-01 15:47:45.242711
+2023-11-27 19:13:57.651626
Details
diff --git a/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html b/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
index fc7f9afa..99f64d55 100644
--- a/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
+++ b/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
@@ -369,7 +369,7 @@ References
Last updated
-2023-11-01 15:47:31.231853
+2023-11-27 19:13:54.254558
Details
diff --git a/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html b/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
index 885d8118..068e0d1c 100644
--- a/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
+++ b/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
@@ -221,7 +221,7 @@ Release, g
Last updated
-2023-11-01 15:47:48.201548
+2023-11-27 19:13:49.940918
Details
diff --git a/posts/2023-10-30_floating_point/floating_point.html b/posts/2023-10-30_floating_point/floating_point.html
index 4b06317b..31c93ef3 100644
--- a/posts/2023-10-30_floating_point/floating_point.html
+++ b/posts/2023-10-30_floating_point/floating_point.html
@@ -107,7 +107,7 @@
-
+
@@ -304,8 +304,8 @@ Issues arising
reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
-
-
+
+
As you can see, the data-frame contains the column GRADE_CRITERIA_CODE which contains comparisons of floating point values. And there was a discrepancy of what Gordon expected to see, and how R actually computed the comparison initially:
@@ -563,8 +563,8 @@ Potential solutions reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
-
-
+
+
Conclusion
Last updated
-2023-11-01 15:47:35.340689
+2023-11-27 19:13:46.342818
Details
diff --git a/posts/2023-11-27_higher_order/admiral.png b/posts/2023-11-27_higher_order/admiral.png
new file mode 100644
index 00000000..cc69ba33
Binary files /dev/null and b/posts/2023-11-27_higher_order/admiral.png differ
diff --git a/posts/2023-11-27_higher_order/higher_order.html b/posts/2023-11-27_higher_order/higher_order.html
new file mode 100644
index 00000000..12fe765b
--- /dev/null
+++ b/posts/2023-11-27_higher_order/higher_order.html
@@ -0,0 +1,1048 @@
+
+
+
+
+
+
+
+
+
+
+
+
+Pharmaverse Blog - Believe in a higher order!
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
\ No newline at end of file
diff --git a/search.json b/search.json
index f352c848..d6425294 100644
--- a/search.json
+++ b/search.json
@@ -1,113 +1,302 @@
[
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html",
- "href": "posts/2023-07-14_code_sections/code_sections.html",
- "title": "How to use Code Sections",
+ "objectID": "posts/2023-08-08_study_day/study_day.html",
+ "href": "posts/2023-08-08_study_day/study_day.html",
+ "title": "It’s all relative? - Calculating Relative Days using admiral",
"section": "",
- "text": "The admiral package embraces a modular style of programming, where blocks of code are pieced together in sequence to create an ADaM dataset. However, with the well-documented advantages of the modular approach comes the recognition that scripts will on average be longer. As such, astute programmers working in RStudio are constantly on the lookout for quick ways to effectively navigate their scripts. Enter code sections!"
+ "text": "Creating --DY variables for your ADaMs is super easy using derive_vars_dy() from the admiral package.\nLet’s build some dummy data with 4 subjects, a start date/time for treatment (TRTSDTM), an analysis start date/time variable (ASTDTM) and an analysis end date variable (AENDT).\nlibrary(admiral)\nlibrary(lubridate)\nlibrary(dplyr)\n\nadam <- tribble(\n ~USUBJID, ~TRTSDTM, ~ASTDTM, ~AENDT,\n \"001\", \"2014-01-17T23:59:59\", \"2014-01-18T13:09:O9\", \"2014-01-20\",\n \"002\", \"2014-02-25T23:59:59\", \"2014-03-18T14:09:O9\", \"2014-03-24\",\n \"003\", \"2014-02-12T23:59:59\", \"2014-02-18T11:03:O9\", \"2014-04-17\",\n \"004\", \"2014-03-17T23:59:59\", \"2014-03-19T13:09:O9\", \"2014-05-04\"\n) %>%\n mutate(\n TRTSDTM = as_datetime(TRTSDTM),\n ASTDTM = as_datetime(ASTDTM),\n AENDT = ymd(AENDT)\n )\nOkay! Next we run our dataset through derive_vars_dy(), specifying:\nderive_vars_dy(\n adam,\n reference_date = TRTSDTM,\n source_vars = exprs(ASTDTM, AENDT)\n)\n\n# A tibble: 4 × 6\n USUBJID TRTSDTM ASTDTM AENDT ASTDY AENDY\n <chr> <dttm> <dttm> <date> <dbl> <dbl>\n1 001 2014-01-17 23:59:59 2014-01-18 13:09:09 2014-01-20 2 4\n2 002 2014-02-25 23:59:59 2014-03-18 14:09:09 2014-03-24 22 28\n3 003 2014-02-12 23:59:59 2014-02-18 11:03:09 2014-04-17 7 65\n4 004 2014-03-17 23:59:59 2014-03-19 13:09:09 2014-05-04 3 49\nThat’s it! We got both our ASTDY and AENDY variables in only a few short lines of code!\nWhat if I want my variables to have a different naming convention?\nEasy! In the source_vars argument if you want your variables to be called DEMOADY and DEMOEDY just do DEMOADY = ASTDTM and DEMOEDY = AENDT and derive_vars_dy() will do the rest!\nderive_vars_dy(\n adam,\n reference_date = TRTSDTM,\n source_vars = exprs(DEMOADY = ASTDTM, DEMOEDY = AENDT)\n)\n\n# A tibble: 4 × 6\n USUBJID TRTSDTM ASTDTM AENDT DEMOADY DEMOEDY\n <chr> <dttm> <dttm> <date> <dbl> <dbl>\n1 001 2014-01-17 23:59:59 2014-01-18 13:09:09 2014-01-20 2 4\n2 002 2014-02-25 23:59:59 2014-03-18 14:09:09 2014-03-24 22 28\n3 003 2014-02-12 23:59:59 2014-02-18 11:03:09 2014-04-17 7 65\n4 004 2014-03-17 23:59:59 2014-03-19 13:09:09 2014-05-04 3 49\nIf you want to get --DT or --DTM variables using admiral then check out derive_vars_dt() and derive_vars_dtm(). If things are messy in your data, e.g. partial dates, both functions have great imputation abilities, which we will cover in an upcoming blog post!"
},
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html#introduction",
- "href": "posts/2023-07-14_code_sections/code_sections.html#introduction",
- "title": "How to use Code Sections",
+ "objectID": "posts/2023-08-08_study_day/study_day.html#last-updated",
+ "href": "posts/2023-08-08_study_day/study_day.html#last-updated",
+ "title": "It’s all relative? - Calculating Relative Days using admiral",
+ "section": "Last updated",
+ "text": "Last updated\n\n2023-11-27 19:13:57.651626"
+ },
+ {
+ "objectID": "posts/2023-08-08_study_day/study_day.html#details",
+ "href": "posts/2023-08-08_study_day/study_day.html#details",
+ "title": "It’s all relative? - Calculating Relative Days using admiral",
+ "section": "Details",
+ "text": "Details\n\nsource code, R environment"
+ },
+ {
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
"section": "",
- "text": "The admiral package embraces a modular style of programming, where blocks of code are pieced together in sequence to create an ADaM dataset. However, with the well-documented advantages of the modular approach comes the recognition that scripts will on average be longer. As such, astute programmers working in RStudio are constantly on the lookout for quick ways to effectively navigate their scripts. Enter code sections!"
+ "text": "There is significant momentum in driving the adoption of R packages in the life sciences industries, in particular, the R Consortium Submissions Working Group is dedicated to promoting the use of R for regulatory submissions to the FDA.\nThe R Consortium Submissions Working Group successfully completed an R-based submission in November 2021 through the eCTD portal (R Submissions Pilot 1). This Pilot was completed on March 10, 2022 after a successful statistical review and evaluation by the FDA staff.\nMoving forward, the Pilot 2 aimed to include a Shiny application that the FDA staff could deploy on their own servers. The R Consortium recently announced that on September 27, 2023, the R Submissions Working Group successfully completed the follow-up to the Pilot 2 R Shiny-based submission and received a response letter from FDA CDER. This marks the first publicly available submission package that includes a Shiny component. The full FDA response letter can be found here.\nThe Shiny application that was sent for the Pilot 2 had the goal to display the 4 Tables, Listings and Figures (TLFs) that were sent for the Pilot 1 with basic filtering functionality.\nThe submission package adhered to the eCTD folder structure and contained 5 deliverables. Among the deliverables was the proprietary R package {pilot2wrappers}, which enables the execution of the Shiny application.\nThe FDA staff were expected to receive the electronic submission packages in the eCTD format, install and load open-source R packages used as dependencies in the included Shiny application, reconstruct and load the submitted Shiny application, and communicate potential improvements in writing.\nIn the following stage, the R Consortium’s R Submission Working Group launched Pilot 4, aiming to investigate innovative technologies like Linux containers and web assembly. These technologies are being explored to package a Shiny application into a self-contained unit, streamlining the transfer and execution processes for the application.\nIn this post, our aim is to outline how we used the Rhino framework to reproduce the Shiny application that was successfully submitted to the FDA for the Pilot 2 project. Additionally, we detail the challenges identified during the process and how we were able to successfully address them by using an open-source package."
},
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html#so-what-are-code-sections-and-why-are-they-useful",
- "href": "posts/2023-07-14_code_sections/code_sections.html#so-what-are-code-sections-and-why-are-they-useful",
- "title": "How to use Code Sections",
- "section": "So, what are code sections and why are they useful?",
- "text": "So, what are code sections and why are they useful?\nCode Sections are separators for long R scripts or functions in RStudio. They can be set up by inserting a comment line followed by four or more dashes in between portions of code, like so:\n\n# First code section ----\n\na <- 1\n\n# Second code section ----\n\nb <- 2\n\n# Third code section ----\n\nc <- 3\n\nRStudio then recognizes the code sections automatically, and enables you to:\n\nCollapse and expand them using the arrow displayed next to the line number, or with the handy shortcuts Alt+L/Shift+Alt+L on Windows or Cmd+Option+L/Cmd+Shift+Option+L on Mac.\nTravel in between them using the navigator at the bottom of the code pane, or by pressing Shift+Alt+J on Windows or Cmd+Shift+Option+J on Mac.\nView an outline of the file using the “Outline” button at the top right of the pane and/or the orange hashtag “Section Navigator” button at the bottom left of the pane.\n\n\n\n\n\n\nCollapsed sections, outline view and the section navigator for the example above.\n\n\n\n\nIt is also possible to create subsections by using two hashtags at the start of a comment line:\n\n# First code section ----\na <- 1\n\n## A code subsection ----\nb <- 2\n\n# Second code section ----\nc <- 3\n\n\n\n\n\n\nCode subsections for the example above.\n\n\n\n\nFor a complete list of Code Sections shortcuts, and for further information, see here."
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#reproducing-the-r-submission-pilot-2-shiny-app-using-rhino",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#reproducing-the-r-submission-pilot-2-shiny-app-using-rhino",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Reproducing the R Submission Pilot 2 Shiny App using Rhino",
+ "text": "Reproducing the R Submission Pilot 2 Shiny App using Rhino\nWhile the original Shiny application submitted to the FDA was wrapped using {Golem}, we replicated the application using our in-house developed framework Rhino. The main motivation was to provide an example of an R Submission that is not an R package and to identify and solve any issues that may arise from this approach.\nOur demo application (FDA-pilot-app) is accessible on our website, alongside other Shiny and Rhinoverse demonstration apps.\n\nThe code for FDA-pilot-app is open-source. You can create your own Rhino-based application by following our tutorial and viewing our workshop, which is available on YouTube."
},
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html#conclusion",
- "href": "posts/2023-07-14_code_sections/code_sections.html#conclusion",
- "title": "How to use Code Sections",
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#brief-introduction-to-rhino",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#brief-introduction-to-rhino",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Brief Introduction to Rhino",
+ "text": "Brief Introduction to Rhino\n\n\n\n\n\nThe Rhino framework was developed by Appsilon to create enterprise-level Shiny applications, consistently and efficiently. This framework allows developers to apply the best software engineering practices, modularize code, test it thoroughly, enhance UI aesthetics, and monitor user adoption effectively.\nRhino provides support in 3 main areas:\n\nClear Code: scalable architecture, modularization based on Box and Shiny modules.\nQuality: comprehensive testing such as unit tests, E2E tests with Cypress and Shinytest2, logging, monitoring and linting.\nAutomation: project startup, CI with GitHub Actions, dependencies management with {renv}, configuration management with config, Sass and JavaScript bundling with ES6 support via Node.js.\n\nRhino is an ideal fit for highly regulated environments such as regulatory submissions or other drug development processes.\n\nFDA-pilot-app structure\nThe structure of this application is available on the github repository. The structure of this Shiny app is the following.\n\nClick here to expand the FDA-pilot-app structure\n\n.\n├── app\n│ ├── view\n│ │ └── demographic_table.R\n| | └── km_plot.R\n| | └── primary_table.R\n| | └── efficacy_table.R\n| | └── completion_table.R\n│ ├── logic\n│ │ └── adam_data.R\n│ │ └── eff_modles.R\n│ │ └── formatters.R\n│ │ └── helpers.R\n│ │ └── kmplot_helpers.R\n│ │ └── Tplyr_helpers.R\n│ ├── data\n│ │ └── adam\n│ │ └── adadas.xpt\n│ │ └── adlbc.xpt\n│ │ └── adsl.xpt\n│ │ └── adtte.xpt\n│ ├── docs\n│ │ └── about.md\n│ ├── js\n│ │ └── index.js\n│ ├── static\n│ │ └── favicon.ico\n│ ├── styles\n│ │ └── main.scss\n│ └── app.R\n├── tests\n│ ├── cypress\n│ │ └── integration\n│ │ └── app.spec.js\n│ ├── testthat\n│ │\n│ └── cypress.json\n├── app.R\n├── rhino_submission.Rproj\n├── dependencies.R\n├── renv.lock\n├── rhino.yml\n└── README.md"
+ },
+ {
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#efficient-submissions-to-the-fda",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#efficient-submissions-to-the-fda",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Efficient Submissions to the FDA",
+ "text": "Efficient Submissions to the FDA\n\n\n\n\n\nTo comply with the Electronic Submission File Formats and Specifications for the eCTD submission, the programming code should carry a “.txt” extension. In the R Submissions Pilot 3 the group did not use {pkglite} as the FDA clarified that “.zip” and “.r” files are acceptable for submission. In this case, we utilized the {pkglite} R package to efficiently pack and unpack the FDA-pilot-app. This approach would facilitate the FDA reviewers in setting up the submission on their systems.\nThis package allows packing R packages to “.txt” files, which are supported for the submission of proprietary packages to the FDA via the eCTD gateway. \n\nPacking the FDA-pilot-app into a .txt file\nThe code below can be used to pack the Shiny application into a .txt file:\n\napp_name <- \"rhinosubmission\"\nrenv_spec <- pkglite::file_spec(\n \"renv\",\n pattern = \"^settings\\\\.dcf$|^activate\\\\.R$\",\n format = \"text\", recursive = FALSE\n)\ntests_spec <- pkglite::file_tests()\napp_spec <- pkglite::file_auto(\"app\")\nroot_spec <- pkglite::file_spec(\n path = \".\",\n pattern = \"^\\\\.Rprofile$|^rhino\\\\.yml$|^renv\\\\.lock$|^dependencies\\\\.R$|^config\\\\.yml$|^app\\\\.R$|^README\\\\.md$|\\\\.Rproj$\",\n all_files = TRUE,\n recursive = FALSE\n)\nwrite(paste0(\"Package: \", app_name), \"DESCRIPTION\", append = TRUE)\npkglite::collate(\n getwd(),\n renv_spec,\n tests_spec,\n app_spec,\n root_spec\n) |> pkglite::pack()\nfile.remove(\"DESCRIPTION\")\n\n\n\nUnpacking the FDA-pilot-app\nThe packed “.txt” file can be unpacked into a Shiny app by using {pkglite} as follows:\n\npkglite::unpack(\"rhinosubmission.txt\")"
+ },
+ {
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#lessons-learned",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#lessons-learned",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Lessons Learned",
+ "text": "Lessons Learned\nOur initial objective was to prove that it would be possible to submit a Shiny application using Rhino through the eCTD gateway. During the rewriting process we identified that this could be done by integrating the open-source {pkglite} package. By following this approach, we concluded that it would be possible to submit a Shiny application through the eCTD gateway. This was also achieved through the successful submission of a package that included a Shiny component in Pilot 2.\nHaving rewritten the R Submissions Pilot 2 Shiny application using Rhino holds major implications for the adoption of our framework within the life sciences. Apart from being a strong, opinionated framework that improves reproducibility and reliability for Shiny development, using Rhino for regulatory submissions could improve the flexibility and speed in the clinical reporting pipeline. This would accelerate the adoption of R/Shiny for submissions to the FDA or other regulatory agencies."
+ },
+ {
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#last-updated",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#last-updated",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Last updated",
+ "text": "Last updated\n\n2023-11-27 19:13:54.254558"
+ },
+ {
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#details",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#details",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Details",
+ "text": "Details\n\nsource code, R environment"
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html",
+ "title": "Blanks and NAs",
+ "section": "",
+ "text": "Reading in SAS-based datasets (.sas7bdat or xpt) into R has users calling the R package haven. A typical call might invoke read_sas() or read_xpt() to bring in your source data to construct your ADaMs or SDTMs.\nUnfortunately, while using haven the character blanks (missing data) found in a typical SAS-based dataset are left as blanks. These blanks will typically prove problematic while using functions like is.na in combination with dplyr::filter() to subset data. Check out Bayer’s SAS2R catalog: handling-of-missing-values for more discussion on missing values and NAs.\nIn the admiral package, we have built a simple function called convert_blanks_to_na() to help us quickly remedy this problem. You can supply an entire dataframe to this function and it will convert any character blanks to NA_character_"
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#loading-packages-and-making-dummy-data",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#loading-packages-and-making-dummy-data",
+ "title": "Blanks and NAs",
+ "section": "Loading Packages and Making Dummy Data",
+ "text": "Loading Packages and Making Dummy Data\n\nlibrary(admiral)\nlibrary(tibble)\nlibrary(dplyr)\n\ndf <- tribble(\n ~USUBJID, ~RFICDTC,\n \"01\", \"2000-01-01\",\n \"02\", \"2001-01-01\",\n \"03\", \"\", # Here we have a character blank\n \"04\", \"2001-01--\",\n \"05\", \"2001---01\",\n \"05\", \"\", # Here we have a character blank\n)\n\ndf\n\n# A tibble: 6 × 2\n USUBJID RFICDTC \n <chr> <chr> \n1 01 \"2000-01-01\"\n2 02 \"2001-01-01\"\n3 03 \"\" \n4 04 \"2001-01--\" \n5 05 \"2001---01\" \n6 05 \"\""
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#a-simple-conversion",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#a-simple-conversion",
+ "title": "Blanks and NAs",
+ "section": "A simple conversion",
+ "text": "A simple conversion\n\ndf_na <- convert_blanks_to_na(df)\n\ndf_na\n\n# A tibble: 6 × 2\n USUBJID RFICDTC \n <chr> <chr> \n1 01 2000-01-01\n2 02 2001-01-01\n3 03 <NA> \n4 04 2001-01-- \n5 05 2001---01 \n6 05 <NA> \n\n\n\ndf_na %>% filter(is.na(RFICDTC))\n\n# A tibble: 2 × 2\n USUBJID RFICDTC\n <chr> <chr> \n1 03 <NA> \n2 05 <NA>"
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#thats-it",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#thats-it",
+ "title": "Blanks and NAs",
+ "section": "That’s it!",
+ "text": "That’s it!\nA simple call to this function can make your derivation life so much easier while working in R if working with SAS-based datasets. In admiral, we make use of this function at the start of all ADaM templates for common ADaM datasets. You can use the function use_ad_template() to get the full R script for the below ADaMs.\n\nlist_all_templates()\n\nExisting ADaM templates in package 'admiral':\n• ADAE\n• ADCM\n• ADEG\n• ADEX\n• ADLB\n• ADLBHY\n• ADMH\n• ADPC\n• ADPP\n• ADPPK\n• ADSL\n• ADVS"
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#last-updated",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#last-updated",
+ "title": "Blanks and NAs",
+ "section": "Last updated",
+ "text": "Last updated\n\n2023-11-27 19:13:51.677601"
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#details",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#details",
+ "title": "Blanks and NAs",
+ "section": "Details",
+ "text": "Details\n\nsource code, R environment"
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html",
+ "href": "posts/2023-06-27__hackathon_app/index.html",
+ "title": "Hackathon Feedback Application",
+ "section": "",
+ "text": "We recently created a shiny application for the admiral hackathon in February 2023. The admiral hackathon was an event designed to make statistical programmers from the pharmaceutical industry more comfortable with the admiral R package which allows users to efficiently transform data from one data standard (SDTM) to another (ADaM).\nHackathon participants formed groups of up to five people and were then tasked to create R-scripts that map the SDTM data to ADaM according to specifics defined in the metadata.\nThe purpose of the shiny app was threefold:\nIn this blog post I want to highlight some of the thoughts that went into this application. Please keep in mind that this work was done under tight time restraints.\nThe hackathon application is still online (although data-upload is switched off) and the GitHub repository is publicly available. The application is embedded into this post right after this paragraph. I have also uploaded to GitHub a .zip file of the workspace to which hackathon participants had access via posit cloud. For more context you can watch recordings of the hackathon-meetings."
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#permanent-data",
+ "href": "posts/2023-06-27__hackathon_app/index.html#permanent-data",
+ "title": "Hackathon Feedback Application",
+ "section": "Permanent Data",
+ "text": "Permanent Data\nThe biggest challenge you have to consider for this app is the permanent data storage. Shiny apps run on a server. Although we can write files on this server, whenever the app restarts, the files are lost. Therefore, a persistent data storage solution is required.\n\nGoogle drive\nI decided to leverage Google drive using the googledrive package package. This allowed me to save structured data (the team registry and the submission scores) as well as unstructured data (their R-script files).\n\n\n\n\n\n\nAuthentication\n\n\n\nTo access Google drive using the googledrive package we need to authenticate. This can be done interactively using the command googledrive::drive_auth() which takes you to the Google login page. After login you receive an authentication token requested by R.\nFor non-interactive authentication this token must be stored locally. In our case where the shiny app must access the token once deployed, the token must be stored on the project level.\nI have included the authentication procedure I followed in the R folder in google_init.R. You can find more extensive documentation of the non-interactive authentication.\n\n\nThe initial concept was: Each team gets their own folder including the most recent submission for each task, and a .csv file containing team information. To keep track of the submissions and the respective scores we wrote a .csv file in the mock-hackathon folder, so one folder above the team folders.\nSaving the team info as a .csv file worked fine as each team received their own file which – once created – was not touched anymore. As each upload for every team should simply add a row to the submissions.csv file, appending the file would be ideal. This was not possible using the googledrive package package. Instead, for each submission, the submissions file was downloaded, appended, and uploaded again. Unfortunately, this lead to a data loss, as the file was continuously overwritten, especially when two teams would submit simultaneously.\n\n\n\n\n\n\nRecover the Lost Data\n\n\n\nWhenever the submissions.csv file was uploaded, the previous version was sent to the Google drive bin. We ended up with over 3000 submissions.csv files containing a lot of redundant information. I had to write the following chunk to first get the unique file IDs of the 3000 submissions.csv files, create an empty submissions data-frame, and then download each file and add its information to the submisisons data-frame. To keep the data-frame as light as possible, after each append I deleted all duplicate submissions.\n\n# get all task_info.csv ID's\n# each row identifies one file in the trash\ntask_info_master <- drive_find(\n pattern = \"task_info.csv\",\n trashed = TRUE\n)\n\n\n# set up empty df to store all submissions\norigin <- tibble(\n score = numeric(),\n task = character(),\n team = character(),\n email = character(),\n time = character()\n)\n\n# downloads, reads, and returns one csv file given a file id\nget_file <- function(row) {\n tf <- tempfile()\n row %>%\n as_id() %>%\n drive_download(path = tf)\n new <- read_csv(tf) %>%\n select(score, task, team) %>%\n distinct()\n}\n\n\n# quick and dirty for loop to subsequently download each file, extract information\n# merge with previous information and squash it (using distinct()).\nfor (i in 1:nrow(task_info_master)) {\n origin <- rbind(origin, get_file(row = task_info_master[i, ])) %>%\n distinct()\n\n # save progress in a separate file after every 100 downloaded and merged sheets\n if (i %% 100 == 0) {\n print(i)\n write_csv(origin, paste(\"prog_data/task_info_prog_\", i, \".csv\", sep = \"\"))\n # update on progress\n message(i / nrow(task_info_master) * 100)\n }\n}\n\nWhen doing such a time-intensive task, make sure to try it first with only a couple of files to see whether any errors are produced. I am not quite sure how long this took but when I returned from my lunch break everything had finished.\n\n\nIf you want to stay in the Google framework, I recommend using the googlesheets4 package for structured data. googlesheets4 allows appending new information to an already existing sheet without the need to download the file first. As both packages follow the same style, going from one to the other is really simple. googlesheets4 requires authentication as well. However, you can reuse the cached token from the googledrive package authentication by setting gs4_auth(token = drive_token()).\n\n\nSecurity Concerns\nConnecting a public shiny app to your Google account introduces a security vulnerability in general. Especially so because we implemented the upload of files to Google drive. And even more problematic: We run a user generated script and display some of its output. A malicious party might be able to extract the authentication token of our Google account or could upload malware to the drive.\nTo reduce the risk, I simply created an un-associated Google account to host the drive. There are certainly better options available, but this seemed a reasonable solution achieved with very little effort."
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#register-team",
+ "href": "posts/2023-06-27__hackathon_app/index.html#register-team",
+ "title": "Hackathon Feedback Application",
+ "section": "Register Team",
+ "text": "Register Team\nWe wanted to allow users to sign up as teams using the shiny app. The app provides a simple interface where users could input a team name and the number of members. This in turn would open two fields for each user to input their name and email address.\nWe do simple checks to make sure at least one valid email address is supplied, and that the group name is acceptable. The group name cannot be empty, already taken, or contain vulgar words.\nThe team registration itself was adding the team information to the Google sheets file event_info into the sheet teams and to create a team folder in which to store the uploaded R files.\nThe checks and registration is implemented in the register_team() function stored in interact_with_google.R.\n\n\n\nScreenshot of the register team interface\n\n\nThe challenge here was to adapt the number of input fields depending on the number of team members. This means that the team name and email interface must be rendered: First, we check how many team members are part of the group, this is stored in the input$n_members input variable. Then we create a tagList with as many elements as team members. Each element contains two columns, one for the email, one for the member name. This tagList is then returned and displayed to the user.\n\n# render email input UI of the register tab\noutput$name_email <- shiny::renderUI({\n # create field names\n N <- input$n_members\n NAME <- sapply(1:N, function(i) {\n paste0(\"name\", i)\n })\n EMAIL <- sapply(1:N, function(i) {\n paste0(\"email\", i)\n })\n\n output <- tagList()\n\n\n firstsecondthird <- c(\"First\", \"Second\", \"Third\", \"Fourth\", \"Fifth\")\n for (i in 1:N) {\n output[[i]] <- tagList()\n output[[i]] <- fluidRow(\n shiny::h4(paste(firstsecondthird[i], \" Member\")),\n column(6,\n textInput(NAME[i], \"Name\"),\n value = \" \" # displayed default value\n ),\n column(6,\n textInput(EMAIL[i], \"Email\"),\n value = \" \"\n )\n )\n }\n output\n})\n\nThe team information is then uploaded to Google drive. Because some teams have more members than others, we have to create the respective data-frame with the number of team members in mind.\nThe following chunk creates the registration data. Noteworthy here the creation of the NAME and EMAIL variables which depend on the number of members in this team. Further, the user input of these fields is extracted via input[[paste0(NAME[i])]] within a for-loop.\nWe also make the data-creation dependent on the press of the Register Group button and cache some variables.\n\n## registration\nregistrationData <-\n reactive({\n N <- input$n_members\n NAME <- sapply(1:N, function(i) {\n paste0(\"name\", i)\n })\n EMAIL <- sapply(1:N, function(i) {\n paste0(\"email\", i)\n })\n names <- character(0)\n emails <- character(0)\n\n for (i in 1:N) {\n names[i] <- input[[paste0(NAME[i])]]\n emails[i] <- input[[paste0(EMAIL[i])]]\n }\n # create df\n dplyr::tibble(\n team_name = input$team_name,\n n_members = N,\n member_name = names,\n member_email = emails\n )\n }) %>%\n bindCache(input$team_name, input$n_members, input$name1, input$email1) %>%\n bindEvent(input$register) # wait for button press"
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#upload-source-script",
+ "href": "posts/2023-06-27__hackathon_app/index.html#upload-source-script",
+ "title": "Hackathon Feedback Application",
+ "section": "Upload & Source Script",
+ "text": "Upload & Source Script\nTo upload a script, participants had to select their team first. The input options were based on the existing folders on the Google-drive in the mock_hackathon folder. To upload a particular script participants had to also select the task to be solved. The uploaded script is then uploaded to the team folder following a standardised script naming convention.\nThere are different aspects to be aware of when sourcing scripts on a shiny server. For example, you have to anticipate the packages users will include in their uploaded scripts, as their scripts will load but not install packages. Further, you should keep the global environment of your shiny app separate from the environment in which the script is sourced. This is possible by supplying an environment to the source() function, e.g: source(path_to_script, local = new.env())\nAnother thing we had to consider was to replicate the exact folder-structure on the shiny server that participants were working with when creating the scripts, as they were required to source some scripts and to save their file into a specific folder. This was relatively straight forward as we provided participants with a folder structure in the posit cloud instance they were using. They had access to the sdtm folder in which the data was stored, and the adam folder into which they saved their solutions. The structure also included a folder with metadata which was also available on the shiny server.\nFor some tasks, participants required some ADaM-datasets stored in the adam folder, essentially the output from previous tasks. This was achieved by first creating a list mapping tasks to the required ADaM datasets:\n\ndepends_list <- list(\n \"ADADAS\" = c(\"ADSL\"),\n \"ADAE\" = c(\"ADSL\"),\n \"ADLBC\" = c(\"ADSL\"),\n \"ADLBH\" = c(\"ADSL\"),\n \"ADLBHY\" = c(\"ADSL\"),\n \"ADSL\" = NULL,\n \"ADTTE\" = c(\"ADSL\", \"ADAE\"),\n \"ADVS\" = c(\"ADSL\")\n)\n\nThis list is sourced from the R/parameters.R file when initiating the application. We then call the get_depends() function sourced from R/get_depends.R which copies the required files from the key folder (where our solutions to the tasks were stored) to the adam folder. After sourcing the uploaded script the content in the adam folder is deleted."
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#compare-to-solution-file",
+ "href": "posts/2023-06-27__hackathon_app/index.html#compare-to-solution-file",
+ "title": "Hackathon Feedback Application",
+ "section": "Compare to Solution File",
+ "text": "Compare to Solution File\nWe want to compare the file created by participants with our solution (key) file stored in the key folder. The diffdf::diffdf() function allows for easy comparison of two data-frames and directly provides extensive feedback for the user:\n\nlibrary(dplyr)\ndf1 <- tibble(\n numbers = 1:10,\n letters = LETTERS[1:10]\n)\ndf2 <- tibble(\n numbers = 1:10,\n letters = letters[1:10]\n)\n\ndiffdf::diffdf(df1, df2)\n\nWarning in diffdf::diffdf(df1, df2): \nNot all Values Compared Equal\n\n\nDifferences found between the objects!\n\nA summary is given below.\n\nNot all Values Compared Equal\nAll rows are shown in table below\n\n =============================\n Variable No of Differences \n -----------------------------\n letters 10 \n -----------------------------\n\n\nAll rows are shown in table below\n\n ========================================\n VARIABLE ..ROWNUMBER.. BASE COMPARE \n ----------------------------------------\n letters 1 A a \n letters 2 B b \n letters 3 C c \n letters 4 D d \n letters 5 E e \n letters 6 F f \n letters 7 G g \n letters 8 H h \n letters 9 I i \n letters 10 J j \n ----------------------------------------"
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#score",
+ "href": "posts/2023-06-27__hackathon_app/index.html#score",
+ "title": "Hackathon Feedback Application",
+ "section": "Score",
+ "text": "Score\nTo compare submissions between participants we implemented a simple scoring function (score_f()) based on the table comparison by diffdf(). The function can be found in the compare_dfs.R file:\n\nscore_f <- function(df_user, df_key, keys) {\n score <- 10\n diff <- diffdf::diffdf(df_user, df_key, keys = keys)\n if (!diffdf::diffdf_has_issues(diff)) {\n return(score)\n }\n\n # check if there are any differences if the comparison is not strict:\n if (!diffdf::diffdf_has_issues(diffdf::diffdf(df_user,\n df_key,\n keys = keys,\n strict_numeric = FALSE,\n strict_factor = FALSE\n ))) {\n # if differences are not strict, return score - 1\n return(score - 1)\n }\n\n return(round(min(max(score - length(diff) / 3, 1), 9), 2))\n}\n\nEvery comparison starts with a score of 10. We then subtract the length of the comparison object divided by a factor of 3. The length of the comparison object is a simplified way to represent the difference between the two data-frames by one value. Finally, the score is bounded by 1 using max(score, 1).\nThe score is not a perfect capture of the quality of the script uploaded but: 1. helped participants get an idea of how close their data-frame is to the solution file 2. allowed us to raffle prizes based on the merit of submitted r-scripts"
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#reactiveness",
+ "href": "posts/2023-06-27__hackathon_app/index.html#reactiveness",
+ "title": "Hackathon Feedback Application",
+ "section": "Reactiveness",
+ "text": "Reactiveness\nSome of the app functions can take quite some time to execute, e.g. running the uploaded script. Other tasks, e.g. registering a team, do not intrinsically generate user facing outputs. This would make the app using really frustrating, as users would not know whether the app is correctly working or whether it froze.\nWe implemented two small features that made the app more responsive. One is simple loading icons that integrate into the user interface and show that output is being computed – that something is working. The other is a pop up window which communicates whether team registration was successful, and if not, why not.\nWe further aimed to forward errors generated by the uploaded scripts to the user interface, but errors generated by the application itself should be concealed."
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#conclusion",
+ "href": "posts/2023-06-27__hackathon_app/index.html#conclusion",
+ "title": "Hackathon Feedback Application",
"section": "Conclusion",
- "text": "Conclusion\nCode sections are an easy way to navigate long scripts and foster good commenting practices. They are used extensively in the admiral package, but there is no reason that you cannot start using them yourself in your day-to-day R programming!"
+ "text": "Conclusion\nAlthough the application was continuously improved during the hackathon it proved to be a useful resource for participants from day one as it allowed groups to set their own pace. It further allowed admiral developers to gain insights on package usage of a relatively large sample of potential end users. From our perspective, the application provided a great added value to the hackathon and eased the workload of guiding the participants through all the tasks."
},
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html#last-updated",
- "href": "posts/2023-07-14_code_sections/code_sections.html#last-updated",
- "title": "How to use Code Sections",
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#last-updated",
+ "href": "posts/2023-06-27__hackathon_app/index.html#last-updated",
+ "title": "Hackathon Feedback Application",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:53.276819"
+ "text": "Last updated\n\n2023-11-27 19:13:48.333469"
},
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html#details",
- "href": "posts/2023-07-14_code_sections/code_sections.html#details",
- "title": "How to use Code Sections",
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#details",
+ "href": "posts/2023-06-27__hackathon_app/index.html#details",
+ "title": "Hackathon Feedback Application",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html",
- "title": "The pharmaverse (hi)story",
+ "objectID": "posts/2023-11-27_higher_order/higher_order.html",
+ "href": "posts/2023-11-27_higher_order/higher_order.html",
+ "title": "Believe in a higher order!",
"section": "",
- "text": "Pharmaverse: from motivation to present"
+ "text": "Picture the following scenario:\nYou, a budding {admiral} programmer, are finding your groove chaining together modular code blocks to derive variables and parameters in a drive to construct your favorite ADaM dataset, ADAE. Suddenly you notice that one of the flags you are deriving should only use records on or after study day 1. In a moment of mild annoyance, you get to work modifying what was originally a simple call to derive_var_extreme_flag() by first subsetting ADAE to records where AESTDY > 1, then deriving the flag only for the subsetted ADAE, and finally binding the two portions of ADAE back together before continuing on with your program. Miffed by this interruption, you think to yourself: “I wish there was a neater, faster way to do this in stride, that didn’t break my code modularity…”\nIf the above could never be you, then you’ll probably be alright never reading this blog post. However, if you want to learn more about the tools that {admiral} provides to make your life easier in cases like this one, then you are in the right place, since this blog post will highlight how higher order functions can solve such issues.\nA higher order function is a function that takes another function as input. By introducing these higher order functions, {admiral} intends to give the user greater power over derivations, whilst trying to negate the need for both adding additional {admiral} functions/arguments, and the user needing many separate steps.\nThe functions covered in this post are:\n\nrestrict_derivation(): Allows the user to execute a single derivation on a subset of the input dataset.\ncall_derivation(): Allows the user to call a single derivation multiple times with some arguments being fixed across iterations and others varying.\nslice_derivation(): Allows the user to split the input dataset into slices (subsets) and for each slice a single derivation is called separately. Some or all arguments of the derivation may vary depending on the slice.\n\n\n\nThe examples in this blog post require the following packages.\n\nlibrary(admiral)\nlibrary(pharmaversesdtm)\nlibrary(dplyr, warn.conflicts = FALSE)\n\nFor example purpose, the ADSL dataset - which is included in {admiral} - and the SDTM datasets from {pharmaversesdtm} are used.\n\ndata(\"admiral_adsl\")\ndata(\"ae\")\ndata(\"vs\")\nadsl <- admiral_adsl\nae <- convert_blanks_to_na(ae)\nvs <- convert_blanks_to_na(vs)\n\nThe following code creates a minimally viable ADAE dataset to be used where needed in the following examples.\n\nadae <- ae %>%\n left_join(adsl, by = c(\"STUDYID\", \"USUBJID\")) %>%\n derive_vars_dt(\n new_vars_prefix = \"AST\",\n dtc = AESTDTC,\n highest_imputation = \"M\"\n ) %>%\n mutate(\n TRTEMFL = if_else(ASTDT >= TRTSDT, \"Y\", NA_character_),\n TEMP_AESEVN = as.integer(factor(AESEV, levels = c(\"SEVERE\", \"MODERATE\", \"MILD\")))\n )"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#human-history-and-pharmaverse-context",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#human-history-and-pharmaverse-context",
- "title": "The pharmaverse (hi)story",
- "section": "Human history and pharmaverse context",
- "text": "Human history and pharmaverse context\nSince the Australian Aboriginal, the earliest peoples recorded to have inhabited the Earth and who have been in Australia for at least 65,000 to 80,000 years (Encyclopædia Britannica), human beings live in group. Whether to protect yourself, increase your life expectancy or simply share tasks.\nFor most aspects of life, it doesn’t make sense to think, act or work alone for two main reasons:\n\nYou will spend more energy and time;\nSomeone else may be facing (or have faced) the same situation.\n\nThe English poet John Donne used to say “No man is an island entire of itself; every man is a piece of the continent, a part of the main;”. I can’t disagree with him. And I dare say that Ari Siggaard Knoph (Novo Nordisk), Michael Rimler (GSK), Michael Stackhouse (Atorus), Ross Farrugia (Roche), and Sumesh Kalappurakal (Janssen) can’t disagree with him either. They are the founders of pharmaverse, members of its Council and kindly shared their memories of how independent companies, in mid-2020, worked together in the creation of a set of packages developed to support the clinical reporting pipeline.\nIf you are not familiar with this pipeline, the important thing to know is that, in a nutshell, pharmaceutical companies must follow a bunch of standardized procedures and formats (from Clinical Data Interchange Standards Consortium, CDISC) when submitting clinical results to Health Authorities. The focus is on this: different companies seeking the same standards for outputs.\nParaphrasing Ross Farrugia (Roche) Breaking boundaries through open-source collaboration presentation in R/Pharma 2022 and thinking of the development of a new drug, we are talking about a “post-competitive” scenario: the drug has already been discovered and the companies should “just” produce and deliver standardized results."
+ "objectID": "posts/2023-11-27_higher_order/higher_order.html#required-packages",
+ "href": "posts/2023-11-27_higher_order/higher_order.html#required-packages",
+ "title": "Believe in a higher order!",
+ "section": "",
+ "text": "The examples in this blog post require the following packages.\n\nlibrary(admiral)\nlibrary(pharmaversesdtm)\nlibrary(dplyr, warn.conflicts = FALSE)\n\nFor example purpose, the ADSL dataset - which is included in {admiral} - and the SDTM datasets from {pharmaversesdtm} are used.\n\ndata(\"admiral_adsl\")\ndata(\"ae\")\ndata(\"vs\")\nadsl <- admiral_adsl\nae <- convert_blanks_to_na(ae)\nvs <- convert_blanks_to_na(vs)\n\nThe following code creates a minimally viable ADAE dataset to be used where needed in the following examples.\n\nadae <- ae %>%\n left_join(adsl, by = c(\"STUDYID\", \"USUBJID\")) %>%\n derive_vars_dt(\n new_vars_prefix = \"AST\",\n dtc = AESTDTC,\n highest_imputation = \"M\"\n ) %>%\n mutate(\n TRTEMFL = if_else(ASTDT >= TRTSDT, \"Y\", NA_character_),\n TEMP_AESEVN = as.integer(factor(AESEV, levels = c(\"SEVERE\", \"MODERATE\", \"MILD\")))\n )"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#clinical-reporting-outputs",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#clinical-reporting-outputs",
- "title": "The pharmaverse (hi)story",
- "section": "Clinical reporting outputs",
- "text": "Clinical reporting outputs\nRationally, we can say that companies face the same challenges in these steps of the process. Not so intuitively, we can also say they were working in silos on that before 2018. Just as Isaac Newton and Gottfried W. Leibniz independently developed the theory of infinitesimal calculus, pharmaceutical companies were independently working on R solutions for this pipeline.\nBut on August 16 and 17 of the mentioned year above, they gathered at the first edition of R/Pharma conference to discuss R and open-source tooling for drug development (the reasons why open-source is an advantageous approach can be found in this post written by Stefan Thoma). And according to Isabela Velásquez’s article, Pharmaverse: Packages for clinical reporting workflows, one of the most popular questions in this conference was “Is the package code available or on CRAN?”.\nWell, many of them were. And not necessarily at that date, but just to mention a few: pharmaRTF and Tplyr from Atorus, r2rtf from Merck, rtables from Roche, etc. The thing is that, overall, there were almost 10000 other packages as well (today, almost 20000). And that took to another two questions:\n\nWith this overwhelming number of packages on CRAN, how to find the ones related to solving “clinical reporting problems”?\nOnce the packages were found, how to choose which one to use among those that have the same functional purpose?\n\nSo, again, companies re-started to working in silos to find those answers. But now, in collaborative silos and with common goals: create extremely useful packages to solve pharmaceutical-specific gaps once and solve them well!"
+ "objectID": "posts/2023-11-27_higher_order/higher_order.html#last-updated",
+ "href": "posts/2023-11-27_higher_order/higher_order.html#last-updated",
+ "title": "Believe in a higher order!",
+ "section": "Last updated",
+ "text": "Last updated\n\n2023-11-27 19:13:43.938122"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#first-partnerships",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#first-partnerships",
- "title": "The pharmaverse (hi)story",
- "section": "First partnerships",
- "text": "First partnerships\nIn 2020, Michael Stackhouse (Atorus) and Michael Rimler (GSK) talked and formed a partnership between their companies to develop a few more packages, including metacore, to read, store and manipulate metadata for ADaMs/SDTMs in a standardized object; xportr, to create submission compliant SAS transport files and perform pharma specific dataset level validation checks; and logrx (ex-timber), to build log to support reproducibility and traceability of an R script.\nAround the same time, Thomas Neitmann (currently at Denali Therapeutics, then at Roche) and Michael Rimler (GSK) discovered that both were working with ADaM in R, so Thomas Neitmann (currently at Denali Therapeutics, then at Roche), Ross Farrugia (Roche) and Michael Rimler (GSK) saw an opportunity there and GSK started their partnership with Roche to build and release admiral package.\nThe idea of working together, the sense of community, and the appetite from organizations built more and more, with incentive and priority established up into the programming heads council.\nJanssen had a huge effort in building R capabilities going on as well, by releasing tidytlg and envsetup), so eventually Michael Rimler (GSK), Michael Stackhouse (Atorus) and Ross Farrugia (Roche) formalized pharmaverse and formed the council, adding in Sumesh Kalappurakal (Janssen) and Ari Siggaard Knoph (Novo Nordisk) later joined as the fifth council member."
+ "objectID": "posts/2023-11-27_higher_order/higher_order.html#details",
+ "href": "posts/2023-11-27_higher_order/higher_order.html#details",
+ "title": "Believe in a higher order!",
+ "section": "Details",
+ "text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#release-growth-and-developments",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#release-growth-and-developments",
- "title": "The pharmaverse (hi)story",
- "section": "Release, growth and developments",
- "text": "Release, growth and developments\nAt the end of their presentation “Closing the Gap: Creating an End to End R Package Toolkit for the Clinical Reporting Pipeline.”, in R/Pharma 2021, Ben Straub (GSK) and Eli Miller (Atorus) welcomed the community to the pharmaverse, a curated collection of packages developed by various pharmaceutical companies to support open-source clinical workflows.\nFrom the outset, the name pharmaverse was chosen so that it could be a neutral home, unrelated to any company. Also, it was established as not being a consortium, which means that founders don’t own, fund, or maintain the packages. Some individuals and companies maintain them but often allowing for community contributions and being licensed permissively so that there is always a feeling of community ownership. The focus of pharmaverse early on, and today, is on inter organization cooperation, to build an environment where, if organizations identify that they have a joint problem that they want to solve, this is the right space to work on and release it.\nPharmaverse has grown a lot, at the time of writing this post we have >25 packages recommended in pharmaverse, and this has led to a partnership with PHUSE to get support from their organization and platform, and because they are eager to advance and support pharmaverse mission.\nDespite all its structure, it is impossible to say that we have a single solution for each clinical reporting analysis when it comes to pharmaverse, a single pathway is impractical. Instead, it is necessary to accept viable tools fitting different pathways into pharmaverse to direct and give people options as to what might work for them. After all, even though we live together as a community, we still have our own unique internal problems.\n\n\n\nSample of pharmaverse packages"
+ "objectID": "posts/2023-07-09_falcon/falcon.html",
+ "href": "posts/2023-07-09_falcon/falcon.html",
+ "title": "falcon",
+ "section": "",
+ "text": "The {falcon} initiative is an industry collaborative effort under {pharmaverse} that unites Boehringer Ingelheim, Moderna, Roche, and Sanofi with the aspiration of building and open-sourcing a comprehensive catalog of harmonized tables, listings, and graphs (TLGs) for clinical study reporting. By leveraging existing open-source R packages, {falcon} aims to simplify the process of output review, comparison, and meta-analyses, while fostering efficient communication among stakeholders in the pharmaceutical industry."
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#last-updated",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#last-updated",
- "title": "The pharmaverse (hi)story",
+ "objectID": "posts/2023-07-09_falcon/falcon.html#what-is-falcon",
+ "href": "posts/2023-07-09_falcon/falcon.html#what-is-falcon",
+ "title": "falcon",
+ "section": "",
+ "text": "The {falcon} initiative is an industry collaborative effort under {pharmaverse} that unites Boehringer Ingelheim, Moderna, Roche, and Sanofi with the aspiration of building and open-sourcing a comprehensive catalog of harmonized tables, listings, and graphs (TLGs) for clinical study reporting. By leveraging existing open-source R packages, {falcon} aims to simplify the process of output review, comparison, and meta-analyses, while fostering efficient communication among stakeholders in the pharmaceutical industry."
+ },
+ {
+ "objectID": "posts/2023-07-09_falcon/falcon.html#why-do-we-build-it",
+ "href": "posts/2023-07-09_falcon/falcon.html#why-do-we-build-it",
+ "title": "falcon",
+ "section": "Why do we build it?",
+ "text": "Why do we build it?\nThe collaborative effort focuses on improving the clarity, consistency, and accessibility of TLGs by addressing variations and redundancies in their creation and use. This harmonized approach allows for streamlined reporting processes and facilitates effective communication of study results within the industry and to regulatory authorities."
+ },
+ {
+ "objectID": "posts/2023-07-09_falcon/falcon.html#what-has-been-done-so-far",
+ "href": "posts/2023-07-09_falcon/falcon.html#what-has-been-done-so-far",
+ "title": "falcon",
+ "section": "What has been done so far?",
+ "text": "What has been done so far?\nDrawing inspiration from the FDA Standard Safety Tables and Figures Integrated Guide, the {falcon} initiative has successfully developed and open-sourced 11 templates to date. 4 product owners and 11 developers from 4 companies have collaborated to make these templates available and also published them on the official {falcon} website at https://pharmaverse.github.io/falcon/."
+ },
+ {
+ "objectID": "posts/2023-07-09_falcon/falcon.html#next-steps-vision",
+ "href": "posts/2023-07-09_falcon/falcon.html#next-steps-vision",
+ "title": "falcon",
+ "section": "Next steps & vision",
+ "text": "Next steps & vision\nFuture plans for {falcon} involve expanding the catalog through continuous collaboration from participating companies and inviting wider industry engagement. The ultimate goal is to promote harmonization of TLGs for clinical reporting across the pharmaceutical industry, leading to greater efficiency, collaboration, and innovation. Even though templates currently come from a published FDA guide, the collaborating companies are open to share and discuss similarities and differences on analysis concepts and output layouts of their own implementations in clinical reporting, for both safety and efficacy analyses.\nIn addition, while currently all templates were built using {rtables}, {tern}, {rlistings} and drew inspiration from the open-sourced TLG-Catalog, moving forward, we plan to support creating the same templates using alternative table engines such as {gt}.\n{falcon} will be presented at the upcoming PHUSE EU (Standards Implementation stream), where we will share the collaboration journey of {falcon} so far, providing more details on the current progress, long-term vision, and strategies for this initiative. Attendees will gain insights into the challenges and opportunities of harmonizing clinical reporting through open-source collaboration and learn about the potential benefits and future direction of {falcon}."
+ },
+ {
+ "objectID": "posts/2023-07-09_falcon/falcon.html#last-updated",
+ "href": "posts/2023-07-09_falcon/falcon.html#last-updated",
+ "title": "falcon",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:48.201548"
+ "text": "Last updated\n\n2023-11-27 19:13:40.019302"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#details",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#details",
- "title": "The pharmaverse (hi)story",
+ "objectID": "posts/2023-07-09_falcon/falcon.html#details",
+ "href": "posts/2023-07-09_falcon/falcon.html#details",
+ "title": "falcon",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-08-08_study_day/study_day.html",
- "href": "posts/2023-08-08_study_day/study_day.html",
- "title": "It’s all relative? - Calculating Relative Days using admiral",
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html",
+ "title": "Admiral Hackathon 2023 Revisited",
"section": "",
- "text": "Creating --DY variables for your ADaMs is super easy using derive_vars_dy() from the admiral package.\nLet’s build some dummy data with 4 subjects, a start date/time for treatment (TRTSDTM), an analysis start date/time variable (ASTDTM) and an analysis end date variable (AENDT).\nlibrary(admiral)\nlibrary(lubridate)\nlibrary(dplyr)\n\nadam <- tribble(\n ~USUBJID, ~TRTSDTM, ~ASTDTM, ~AENDT,\n \"001\", \"2014-01-17T23:59:59\", \"2014-01-18T13:09:O9\", \"2014-01-20\",\n \"002\", \"2014-02-25T23:59:59\", \"2014-03-18T14:09:O9\", \"2014-03-24\",\n \"003\", \"2014-02-12T23:59:59\", \"2014-02-18T11:03:O9\", \"2014-04-17\",\n \"004\", \"2014-03-17T23:59:59\", \"2014-03-19T13:09:O9\", \"2014-05-04\"\n) %>%\n mutate(\n TRTSDTM = as_datetime(TRTSDTM),\n ASTDTM = as_datetime(ASTDTM),\n AENDT = ymd(AENDT)\n )\nOkay! Next we run our dataset through derive_vars_dy(), specifying:\nderive_vars_dy(\n adam,\n reference_date = TRTSDTM,\n source_vars = exprs(ASTDTM, AENDT)\n)\n\n# A tibble: 4 × 6\n USUBJID TRTSDTM ASTDTM AENDT ASTDY AENDY\n <chr> <dttm> <dttm> <date> <dbl> <dbl>\n1 001 2014-01-17 23:59:59 2014-01-18 13:09:09 2014-01-20 2 4\n2 002 2014-02-25 23:59:59 2014-03-18 14:09:09 2014-03-24 22 28\n3 003 2014-02-12 23:59:59 2014-02-18 11:03:09 2014-04-17 7 65\n4 004 2014-03-17 23:59:59 2014-03-19 13:09:09 2014-05-04 3 49\nThat’s it! We got both our ASTDY and AENDY variables in only a few short lines of code!\nWhat if I want my variables to have a different naming convention?\nEasy! In the source_vars argument if you want your variables to be called DEMOADY and DEMOEDY just do DEMOADY = ASTDTM and DEMOEDY = AENDT and derive_vars_dy() will do the rest!\nderive_vars_dy(\n adam,\n reference_date = TRTSDTM,\n source_vars = exprs(DEMOADY = ASTDTM, DEMOEDY = AENDT)\n)\n\n# A tibble: 4 × 6\n USUBJID TRTSDTM ASTDTM AENDT DEMOADY DEMOEDY\n <chr> <dttm> <dttm> <date> <dbl> <dbl>\n1 001 2014-01-17 23:59:59 2014-01-18 13:09:09 2014-01-20 2 4\n2 002 2014-02-25 23:59:59 2014-03-18 14:09:09 2014-03-24 22 28\n3 003 2014-02-12 23:59:59 2014-02-18 11:03:09 2014-04-17 7 65\n4 004 2014-03-17 23:59:59 2014-03-19 13:09:09 2014-05-04 3 49\nIf you want to get --DT or --DTM variables using admiral then check out derive_vars_dt() and derive_vars_dtm(). If things are messy in your data, e.g. partial dates, both functions have great imputation abilities, which we will cover in an upcoming blog post!"
+ "text": "This January and February (2023), the admiral development team and the CDISC Open Source Alliance jointly hosted the admiral hackathon. The idea was to build a community of admiral users, and help participants familiarize themselves with R and admiral. This whole effort was led by Thomas Neitmann and was supported by Zelos Zhu, Sadchla Mascary, and me – Stefan Thoma.\nThe hackathon event was structured in two parts. First, we offered an Introduction to R for SAS programmers, a three hour workshop for R beginners to get them up to speed. Here we covered practical R basics, talking about how the R-workflow differs from a SAS workflow, and discussed common R functions - mostly from the tidyverse. This ensured that hackathon participants were familiar with core R concepts. The workshop recording and the course materials are available online.\nThe main hackathon consisted of several ADAM data generating tasks based on a specs file and synthetic data. Participants were able to solve these tasks in groups at their own pace thanks to a online tool where participants could upload their task specific R scripts and they would get automatic feedback for the data-set produced by their script. Script upload through the feedback application was available all through February, and we offered three additional online meetings throughout the month to discuss challenges and give some tips. If you are interested in learning more about the thoughts that went into the feedback application, you can read about it in this blogpost or check out my public GitHub repository for such an application."
},
{
- "objectID": "posts/2023-08-08_study_day/study_day.html#last-updated",
- "href": "posts/2023-08-08_study_day/study_day.html#last-updated",
- "title": "It’s all relative? - Calculating Relative Days using admiral",
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html#introduction-to-r-workshop",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html#introduction-to-r-workshop",
+ "title": "Admiral Hackathon 2023 Revisited",
+ "section": "Introduction to R workshop",
+ "text": "Introduction to R workshop\nWe were really excited to see over 500 people from around 40 countries joining our Introduction to R workshop in January! To get to know prospective users and hackathon participants better, we conducted some polls during the meetings. Below you can see that representatives of many different sorts of organisations joined our Introduction to R workshop:\n\n\n\n\n\n216 out of 402 confirmed that their company is already using R for clinical trial data analysis, the remaining 131 did not answer this question.\nThe target audience for this workshop was programmers who are very familiar with SAS, but not so familiar with R, our polls confirmed this.\n\n\n\n\n\nOverall, we were very happy with how the workshop turned out, and participants overall agreed with this sentiment (although there may be a slight survivorship bias…)."
+ },
+ {
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html#admiral-hackathon",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html#admiral-hackathon",
+ "title": "Admiral Hackathon 2023 Revisited",
+ "section": "admiral Hackathon",
+ "text": "admiral Hackathon\nFollowing the kick-off meeting, 371 participants joined the posit (rStudio) workspace that was made available to all participants at no costs by the posit company. About half the participants planned to spend one to two hours per week on the admiral tasks, the other half planned to allocate even more. 15 participants even planned to spend eight hours or more!\nWe were really happy to see an overwhelming amount of activity on the slack channel we set up with over 250 members. Not only were people engaging with the materials, but we saw how a community was formed where people were encouraged to ask questions and where community members went out of their way to help each other. Shout-out to our community hero: Jagadish Katam without whom most issues related to the task programming raised by the community would not have been addressed as quickly as they were. Huge thanks from the organizers!\nIn the end, a total of 44 teams spanning 87 statistical programmers took part in the admiral hackathon and uploaded solution scripts to the hackathon application solving at least one of the 8 tasks available (ADSL, ADAE, ADLBC, ADVS, ADTTE, ADADAS, ADLBH & ADLBHY). Participants’ scripts were then run on the shiny server and the output data-frame were compared to the solutions we provided. At the read-out there was a live draft of teams to win one-on-one admiral consulting with one of the admiral core developers. Winning probabilities were weighted by the number of points each group received for the quality of their output data-frames and for the number of tasks solved.\nCongratulations to the winners:\n\nViiV Team_GSK\nteamspoRt\nTatianaPXL\nDivyasneelam\nAdaMTeamIndia\nSanofi_BP\nJagadish (our community hero)\nAZ_WAWA\n\nAlthough this was uncertain during the hackathon we were excited to provide a Certificate of Completion to all participants who uploaded a script to the Web Application.\nA recording of the hackathon readout can be found in the CDISC Open Source Alliance Quarterly Spotlight."
+ },
+ {
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html#conclusion",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html#conclusion",
+ "title": "Admiral Hackathon 2023 Revisited",
+ "section": "Conclusion",
+ "text": "Conclusion\nOverall, we are very happy with how the hackathon turned out. We were not only positively surprised with the huge audience for the Intro to R workshop (CDISC record breaking) and for the admiral hackathon, but even more so with the engagement of all the participants.\nAgain, we would like to thank all the organizers, participants, and sponsors for their time and resources and hope to have provided a useful glimpse into our solution for ADAM creation within the end-to-end clinical data analysis open source R framework that the Pharmaverse aims to provide.\nAs always, we are very happy to hear more feedback on the hackathon as well as on admiral in general. Simply submit an issue on the admiral GitHub repository. You would like to join the admiral core developers? Please reach out to Edoardo Mancini (product owner) or Ben Straub (technical lead)."
+ },
+ {
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html#last-updated",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html#last-updated",
+ "title": "Admiral Hackathon 2023 Revisited",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:45.242711"
+ "text": "Last updated\n\n2023-11-27 19:13:36.963431"
},
{
- "objectID": "posts/2023-08-08_study_day/study_day.html#details",
- "href": "posts/2023-08-08_study_day/study_day.html#details",
- "title": "It’s all relative? - Calculating Relative Days using admiral",
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html#details",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html#details",
+ "title": "Admiral Hackathon 2023 Revisited",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
@@ -182,88 +371,25 @@
"text": "Using More Than One Imputation Rule for a Variable\nUsing different imputation rules depending on the observation can be done by using the higher-order function slice_derivation(), which applies a derivation function differently (by varying its arguments) in different subsections of a dataset. For example, consider this Vital Signs case where pre-dose records require a different treatment to other records:\n\nvs <- tribble(\n ~VSDTC, ~VSTPT,\n \"2019-08-09T12:34:56\", NA,\n \"2019-10-12\", \"PRE-DOSE\",\n \"2019-11-10\", NA,\n \"2019-12-04\", NA\n) %>%\n slice_derivation(\n derivation = derive_vars_dtm,\n args = params(\n dtc = VSDTC,\n new_vars_prefix = \"A\"\n ),\n derivation_slice(\n filter = VSTPT == \"PRE-DOSE\",\n args = params(time_imputation = \"first\")\n ),\n derivation_slice(\n filter = TRUE,\n args = params(time_imputation = \"last\")\n )\n )\n\n\n\n\n\n\nVSDTC\nVSTPT\nADTM\nATMF\n\n\n\n\n2019-08-09T12:34:56\nNA\n2019-08-09 12:34:56\nNA\n\n\n2019-11-10\nNA\n2019-11-10 23:59:59\nH\n\n\n2019-12-04\nNA\n2019-12-04 23:59:59\nH\n\n\n2019-10-12\nPRE-DOSE\n2019-10-12 00:00:00\nH"
},
{
- "objectID": "posts/2023-06-28_welcome/index.html",
- "href": "posts/2023-06-28_welcome/index.html",
- "title": "Hello pharmaverse",
- "section": "",
- "text": "The communications working group (CWG) seeks to promote and showcase how R can be used in the Clinical Reporting pipeline through short and informative blog posts. These posts will be hosted on this pharmaverse blog and promoted on the pharmaverse slack channels as well as on LinkedIn.\nAs the CWG is a small team, we hope to make the blog development process easy enough that pharmaverse community members will be able to easily write blog posts with guidance from the CWG team."
- },
- {
- "objectID": "posts/2023-06-28_welcome/index.html#purpose",
- "href": "posts/2023-06-28_welcome/index.html#purpose",
- "title": "Hello pharmaverse",
+ "objectID": "index.html",
+ "href": "index.html",
+ "title": "Pharmaverse Blog",
"section": "",
- "text": "The communications working group (CWG) seeks to promote and showcase how R can be used in the Clinical Reporting pipeline through short and informative blog posts. These posts will be hosted on this pharmaverse blog and promoted on the pharmaverse slack channels as well as on LinkedIn.\nAs the CWG is a small team, we hope to make the blog development process easy enough that pharmaverse community members will be able to easily write blog posts with guidance from the CWG team."
- },
- {
- "objectID": "posts/2023-06-28_welcome/index.html#spirit-of-a-blog-post",
- "href": "posts/2023-06-28_welcome/index.html#spirit-of-a-blog-post",
- "title": "Hello pharmaverse",
- "section": "Spirit of a Blog Post",
- "text": "Spirit of a Blog Post\nThe CWG believes that the following 4 points will help guide the creation of Blog Posts.\n\nShort\nPersonalized\nReproducible\nReadable\n\nShort: Posts should aim to be under a 10 minute read. We encourage longer posts to be broken up into multiple posts.\nPersonalized: Posts should have a personality! For example, a person wishing to post on a function in a package needs to differentiate the post from the documentation for function, i.e. we don’t want to just recycle the documentation. How can you add your voice and experience? A bit of cheeky language is also encouraged.\nReproducible: Posts should work with minimal dependencies with data, packages and outside sources. Every dependency introduced in a post adds some risk to the post longevity. As package dependencies change, posts should be built in a way that they can be updated to stay relevant.\nReadable: The CWG sees this site as more of introductory site rather advanced user site. Therefore, the CWG feels that code should be introduced in a way that promotes readability over complexity."
- },
- {
- "objectID": "posts/2023-06-28_welcome/index.html#what-types-of-posts-are-allowed-on-this-site",
- "href": "posts/2023-06-28_welcome/index.html#what-types-of-posts-are-allowed-on-this-site",
- "title": "Hello pharmaverse",
- "section": "What types of posts are allowed on this site?",
- "text": "What types of posts are allowed on this site?\nOverall, we want to stay focus on the Clinical Reporting Pipeline, which we see as the following topics:\n\nPackages in the Clinical Reporting Pipeline\nFunctions from packages in the Clinical Reporting Pipeline\nWider experiences of using R in the Clinical Reporting Pipeline\nConference experiences and the Clinical Reporting Pipeline\n\nHowever, it never hurts to ask if you topic might fit into this medium!\n\nMinimum Post Requirements\n\nA unique image to help showcase the post.\nWorking Code\nSelf-contained data or package data.\nDocumentation of package versions\n\nThat is it! After that you can go wild, but we do ask that it is kept short!"
- },
- {
- "objectID": "posts/2023-06-28_welcome/index.html#how-can-i-make-a-blog-post",
- "href": "posts/2023-06-28_welcome/index.html#how-can-i-make-a-blog-post",
- "title": "Hello pharmaverse",
- "section": "How can I make a Blog Post",
- "text": "How can I make a Blog Post\nStep 1: Reach out to us through pharmaverse/slack or make an issue on our GitHub.\nStep 2: Branch off main\nStep 3: Review the Spirit of the Blog Post in the Pull Request Template\nStep 4: Poke us to do a review!"
+ "text": "Order By\n Default\n \n Title\n \n \n Date - Oldest\n \n \n Date - Newest\n \n \n Author\n \n \n \n \n \n \n \n\n\n\n\n \n\n\n\n\nBelieve in a higher order!\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nA brief foray into the higher order functions in the {admiral} package.\n\n\n\n\n\n\nNov 27, 2023\n\n\nEdoardo Mancini\n\n\n\n\n\n\n \n\n\n\n\nFloating point\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nThe untold story of how admiral deals with floating points.\n\n\n\n\n\n\nOct 30, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nReproducing the R Submissions Pilot 2 Shiny Application using Rhino\n\n\n\n\n\n\n\nsubmission\n\n\ncommunity\n\n\n\n\n\n\n\n\n\n\n\nOct 10, 2023\n\n\nIsmael Rodriguez, Vedha Viyash, André Veríssimo, APPSILON\n\n\n\n\n\n\n \n\n\n\n\nDate/Time Functions and Imputation in {admiral}\n\n\n\n\n\n\n\nadmiral\n\n\nADaM\n\n\nDate/Time\n\n\n\n\nDates, times and imputation can be a frustrating facet of any programming language. Here’s how {admiral} makes all of this easy!\n\n\n\n\n\n\nSep 26, 2023\n\n\nEdoardo Mancini\n\n\n\n\n\n\n \n\n\n\n\nThe pharmaverse (hi)story\n\n\n\n\n\n\n\ncommunity\n\n\n\n\n\n\n\n\n\n\n\nAug 30, 2023\n\n\nNicholas Eugenio\n\n\n\n\n\n\n \n\n\n\n\nRounding\n\n\n\n\n\n\n\nrounding\n\n\n\n\nExploration of some commonly used rounding methods and their corresponding functions in SAS and R, with a focus on ‘round half up’ and reliable solutions for numerical precision challenges\n\n\n\n\n\n\nAug 22, 2023\n\n\nKangjie Zhang\n\n\n\n\n\n\n \n\n\n\n\nIt’s all relative? - Calculating Relative Days using admiral\n\n\n\n\n\n\n\nadmiral\n\n\nADaMs\n\n\n\n\n\n\n\n\n\n\n\nAug 8, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nCross-Industry Open Source Package Development\n\n\n\n\n\n\n\ncommunity\n\n\nadmiral\n\n\n\n\n\n\n\n\n\n\n\nJul 25, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nHow to use Code Sections\n\n\n\n\n\n\n\nADaMs\n\n\nTips and Tricks\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2023\n\n\nEdoardo Mancini\n\n\n\n\n\n\n \n\n\n\n\nBlanks and NAs\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nReading SAS datasets into R. The data is not always as it seems!\n\n\n\n\n\n\nJul 10, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nfalcon\n\n\n\n\n\n\n\nfalcon\n\n\ndisplays\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2023\n\n\nVincent Shen\n\n\n\n\n\n\n \n\n\n\n\nHello pharmaverse\n\n\n\n\n\n\n\ncommunity\n\n\n\n\nShort, fun and user-driven content around the pharmaverse.\n\n\n\n\n\n\nJun 28, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nAdmiral Hackathon 2023 Revisited\n\n\n\n\n\n\n\ncommunity\n\n\nadmiral\n\n\n\n\nLet’s have a look at the Admiral Hackathon 2023 together.\n\n\n\n\n\n\nJun 27, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nDerive a new parameter computed from the value of other parameters\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nUse admiral::derive_param_computed() like a calculator to derive new parameters/rows based on existing ones\n\n\n\n\n\n\nJun 27, 2023\n\n\nKangjie Zhang\n\n\n\n\n\n\n \n\n\n\n\nHackathon Feedback Application\n\n\n\n\n\n\n\nR\n\n\nshiny\n\n\ncommunity\n\n\nadmiral\n\n\n\n\nGoing through the process of creating a shiny app for the admiral hackathon. The shiny app allows users to check their solutions autonomously, gives feedback, and rates their results.\n\n\n\n\n\n\nJun 27, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\nNo matching items"
},
{
- "objectID": "posts/2023-06-28_welcome/index.html#last-updated",
- "href": "posts/2023-06-28_welcome/index.html#last-updated",
- "title": "Hello pharmaverse",
- "section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:37.128939"
- },
- {
- "objectID": "posts/2023-06-28_welcome/index.html#details",
- "href": "posts/2023-06-28_welcome/index.html#details",
- "title": "Hello pharmaverse",
- "section": "Details",
- "text": "Details\n\nsource code, R environment"
- },
- {
- "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html",
- "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html",
- "title": "Derive a new parameter computed from the value of other parameters",
+ "objectID": "R/readme.html",
+ "href": "R/readme.html",
+ "title": "Files in this folder",
"section": "",
- "text": "When creating ADaM Basic Data Structure (BDS) datasets, we often encounter deriving a new parameter based on the analysis values (e.g., AVAL) of other parameters.\nThe admiral function derive_param_computed() adds a parameter computed from the analysis value of other parameters.\nIt works like a calculator to derive new records without worrying about merging and combining datasets, all you need is a derivation formula, which also improves the readability of the code."
+ "text": "Some of these files help in creating/developing blog-posts, others are used by our CICD pipeline.\n\ncreate_blogpost.R: use this script to create a new blogpost based on our blogpost template.\nCICD.R: use this script to spellcheck and stylecheck your blogpost.\n\n\n\n\nhelp_create_blogpost.R: script containing the function(s) used by create_blogpost.R\nswitch.R: Used by CICD spellcheck workflow."
},
{
- "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#introduction",
- "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#introduction",
- "title": "Derive a new parameter computed from the value of other parameters",
+ "objectID": "R/readme.html#development-files",
+ "href": "R/readme.html#development-files",
+ "title": "Files in this folder",
"section": "",
- "text": "When creating ADaM Basic Data Structure (BDS) datasets, we often encounter deriving a new parameter based on the analysis values (e.g., AVAL) of other parameters.\nThe admiral function derive_param_computed() adds a parameter computed from the analysis value of other parameters.\nIt works like a calculator to derive new records without worrying about merging and combining datasets, all you need is a derivation formula, which also improves the readability of the code."
- },
- {
- "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#example",
- "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#example",
- "title": "Derive a new parameter computed from the value of other parameters",
- "section": "Example",
- "text": "Example\nA value level validation use case, where derive_param_computed() is used to validate a derived parameter - PARAMCD = ADPCYMG (Actual Dose per Cycle) in ADEX dataset.\n\nDerivation\nActual Dose per Cycle is derived from the Total Amount of Dose (PARAMCD = TOTDOSE) / Number of Cycles (PARAMCD = NUMCYC)\nIn this example, ADEX.AVAL when ADEX.PARAMCD = ADPCYMG can be derived as:\n\\[\nAVAL (PARAMCD = ADPCYMG) = \\frac{AVAL (PARAMCD = TOTDOSE)}{AVAL (PARAMCD = NUMCYC)}\n\\]\n\n\nLoading Packages and Creating Example Data\n\nlibrary(tibble)\nlibrary(dplyr)\nlibrary(diffdf)\nlibrary(admiral)\n\nadex <- tribble(\n ~USUBJID, ~PARAMCD, ~PARAM, ~AVAL,\n \"101\", \"TOTDOSE\", \"Total Amount of Dose (mg)\", 180,\n \"101\", \"NUMCYC\", \"Number of Cycles\", 3\n)\n\n\n\nDerive New Parameter\n\nadex_admiral <- derive_param_computed(\n adex,\n by_vars = exprs(USUBJID),\n parameters = c(\"TOTDOSE\", \"NUMCYC\"),\n analysis_value = AVAL.TOTDOSE / AVAL.NUMCYC,\n set_values_to = exprs(\n PARAMCD = \"ADPCYMG\",\n PARAM = \"Actual Dose per Cycle (mg)\"\n )\n)\n\nWarning: The `analysis_value` argument of `derive_param_computed()` is deprecated as\nof admiral 0.12.0.\nℹ Please use the `set_values_to` argument instead.\n\n\n\n\n# A tibble: 3 × 4\n USUBJID PARAMCD PARAM AVAL\n <chr> <chr> <chr> <dbl>\n1 101 TOTDOSE Total Amount of Dose (mg) 180\n2 101 NUMCYC Number of Cycles 3\n3 101 ADPCYMG Actual Dose per Cycle (mg) 60\n\n\n\n\nCompare\nFor validation purpose, the diffdf package is used below to mimic SAS proc compare.\n\nadex_expected <- bind_rows(\n adex,\n tribble(\n ~USUBJID, ~PARAMCD, ~PARAM, ~AVAL,\n \"101\", \"ADPCYMG\", \"Actual Dose per Cycle (mg)\", 60\n )\n)\n\ndiffdf(adex_expected, adex_admiral, keys = c(\"USUBJID\", \"PARAMCD\"))\n\nNo issues were found!"
- },
- {
- "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#last-updated",
- "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#last-updated",
- "title": "Derive a new parameter computed from the value of other parameters",
- "section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:33.340574"
- },
- {
- "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#details",
- "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#details",
- "title": "Derive a new parameter computed from the value of other parameters",
- "section": "Details",
- "text": "Details\n\nsource code, R environment"
+ "text": "help_create_blogpost.R: script containing the function(s) used by create_blogpost.R\nswitch.R: Used by CICD spellcheck workflow."
},
{
"objectID": "posts/2023-07-24_rounding/rounding.html",
@@ -298,7 +424,7 @@
"href": "posts/2023-07-24_rounding/rounding.html#last-updated",
"title": "Rounding",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:29.564969"
+ "text": "Last updated\n\n2023-11-27 19:13:33.556149"
},
{
"objectID": "posts/2023-07-24_rounding/rounding.html#details",
@@ -308,114 +434,79 @@
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "R/readme.html",
- "href": "R/readme.html",
- "title": "Files in this folder",
- "section": "",
- "text": "Some of these files help in creating/developing blog-posts, others are used by our CICD pipeline.\n\ncreate_blogpost.R: use this script to create a new blogpost based on our blogpost template.\nCICD.R: use this script to spellcheck and stylecheck your blogpost.\n\n\n\n\nhelp_create_blogpost.R: script containing the function(s) used by create_blogpost.R\nswitch.R: Used by CICD spellcheck workflow."
- },
- {
- "objectID": "R/readme.html#development-files",
- "href": "R/readme.html#development-files",
- "title": "Files in this folder",
- "section": "",
- "text": "help_create_blogpost.R: script containing the function(s) used by create_blogpost.R\nswitch.R: Used by CICD spellcheck workflow."
- },
- {
- "objectID": "index.html",
- "href": "index.html",
- "title": "Pharmaverse Blog",
+ "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html",
+ "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html",
+ "title": "Derive a new parameter computed from the value of other parameters",
"section": "",
- "text": "Order By\n Default\n \n Title\n \n \n Date - Oldest\n \n \n Date - Newest\n \n \n Author\n \n \n \n \n \n \n \n\n\n\n\n \n\n\n\n\nFloating point\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nThe untold story of how admiral deals with floating points.\n\n\n\n\n\n\nOct 30, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nReproducing the R Submissions Pilot 2 Shiny Application using Rhino\n\n\n\n\n\n\n\nsubmission\n\n\ncommunity\n\n\n\n\n\n\n\n\n\n\n\nOct 10, 2023\n\n\nIsmael Rodriguez, Vedha Viyash, André Veríssimo, APPSILON\n\n\n\n\n\n\n \n\n\n\n\nDate/Time Functions and Imputation in {admiral}\n\n\n\n\n\n\n\nadmiral\n\n\nADaM\n\n\nDate/Time\n\n\n\n\nDates, times and imputation can be a frustrating facet of any programming language. Here’s how {admiral} makes all of this easy!\n\n\n\n\n\n\nSep 26, 2023\n\n\nEdoardo Mancini\n\n\n\n\n\n\n \n\n\n\n\nThe pharmaverse (hi)story\n\n\n\n\n\n\n\ncommunity\n\n\n\n\n\n\n\n\n\n\n\nAug 30, 2023\n\n\nNicholas Eugenio\n\n\n\n\n\n\n \n\n\n\n\nRounding\n\n\n\n\n\n\n\nrounding\n\n\n\n\nExploration of some commonly used rounding methods and their corresponding functions in SAS and R, with a focus on ‘round half up’ and reliable solutions for numerical precision challenges\n\n\n\n\n\n\nAug 22, 2023\n\n\nKangjie Zhang\n\n\n\n\n\n\n \n\n\n\n\nIt’s all relative? - Calculating Relative Days using admiral\n\n\n\n\n\n\n\nadmiral\n\n\nADaMs\n\n\n\n\n\n\n\n\n\n\n\nAug 8, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nCross-Industry Open Source Package Development\n\n\n\n\n\n\n\ncommunity\n\n\nadmiral\n\n\n\n\n\n\n\n\n\n\n\nJul 25, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nHow to use Code Sections\n\n\n\n\n\n\n\nADaMs\n\n\nTips and Tricks\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2023\n\n\nEdoardo Mancini\n\n\n\n\n\n\n \n\n\n\n\nBlanks and NAs\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nReading SAS datasets into R. The data is not always as it seems!\n\n\n\n\n\n\nJul 10, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nfalcon\n\n\n\n\n\n\n\nfalcon\n\n\ndisplays\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2023\n\n\nVincent Shen\n\n\n\n\n\n\n \n\n\n\n\nHello pharmaverse\n\n\n\n\n\n\n\ncommunity\n\n\n\n\nShort, fun and user-driven content around the pharmaverse.\n\n\n\n\n\n\nJun 28, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nDerive a new parameter computed from the value of other parameters\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nUse admiral::derive_param_computed() like a calculator to derive new parameters/rows based on existing ones\n\n\n\n\n\n\nJun 27, 2023\n\n\nKangjie Zhang\n\n\n\n\n\n\n \n\n\n\n\nHackathon Feedback Application\n\n\n\n\n\n\n\nR\n\n\nshiny\n\n\ncommunity\n\n\nadmiral\n\n\n\n\nGoing through the process of creating a shiny app for the admiral hackathon. The shiny app allows users to check their solutions autonomously, gives feedback, and rates their results.\n\n\n\n\n\n\nJun 27, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nAdmiral Hackathon 2023 Revisited\n\n\n\n\n\n\n\ncommunity\n\n\nadmiral\n\n\n\n\nLet’s have a look at the Admiral Hackathon 2023 together.\n\n\n\n\n\n\nJun 27, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\nNo matching items"
+ "text": "When creating ADaM Basic Data Structure (BDS) datasets, we often encounter deriving a new parameter based on the analysis values (e.g., AVAL) of other parameters.\nThe admiral function derive_param_computed() adds a parameter computed from the analysis value of other parameters.\nIt works like a calculator to derive new records without worrying about merging and combining datasets, all you need is a derivation formula, which also improves the readability of the code."
},
{
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html",
- "title": "Blanks and NAs",
+ "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#introduction",
+ "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#introduction",
+ "title": "Derive a new parameter computed from the value of other parameters",
"section": "",
- "text": "Reading in SAS-based datasets (.sas7bdat or xpt) into R has users calling the R package haven. A typical call might invoke read_sas() or read_xpt() to bring in your source data to construct your ADaMs or SDTMs.\nUnfortunately, while using haven the character blanks (missing data) found in a typical SAS-based dataset are left as blanks. These blanks will typically prove problematic while using functions like is.na in combination with dplyr::filter() to subset data. Check out Bayer’s SAS2R catalog: handling-of-missing-values for more discussion on missing values and NAs.\nIn the admiral package, we have built a simple function called convert_blanks_to_na() to help us quickly remedy this problem. You can supply an entire dataframe to this function and it will convert any character blanks to NA_character_"
- },
- {
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#loading-packages-and-making-dummy-data",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#loading-packages-and-making-dummy-data",
- "title": "Blanks and NAs",
- "section": "Loading Packages and Making Dummy Data",
- "text": "Loading Packages and Making Dummy Data\n\nlibrary(admiral)\nlibrary(tibble)\nlibrary(dplyr)\n\ndf <- tribble(\n ~USUBJID, ~RFICDTC,\n \"01\", \"2000-01-01\",\n \"02\", \"2001-01-01\",\n \"03\", \"\", # Here we have a character blank\n \"04\", \"2001-01--\",\n \"05\", \"2001---01\",\n \"05\", \"\", # Here we have a character blank\n)\n\ndf\n\n# A tibble: 6 × 2\n USUBJID RFICDTC \n <chr> <chr> \n1 01 \"2000-01-01\"\n2 02 \"2001-01-01\"\n3 03 \"\" \n4 04 \"2001-01--\" \n5 05 \"2001---01\" \n6 05 \"\""
- },
- {
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#a-simple-conversion",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#a-simple-conversion",
- "title": "Blanks and NAs",
- "section": "A simple conversion",
- "text": "A simple conversion\n\ndf_na <- convert_blanks_to_na(df)\n\ndf_na\n\n# A tibble: 6 × 2\n USUBJID RFICDTC \n <chr> <chr> \n1 01 2000-01-01\n2 02 2001-01-01\n3 03 <NA> \n4 04 2001-01-- \n5 05 2001---01 \n6 05 <NA> \n\n\n\ndf_na %>% filter(is.na(RFICDTC))\n\n# A tibble: 2 × 2\n USUBJID RFICDTC\n <chr> <chr> \n1 03 <NA> \n2 05 <NA>"
+ "text": "When creating ADaM Basic Data Structure (BDS) datasets, we often encounter deriving a new parameter based on the analysis values (e.g., AVAL) of other parameters.\nThe admiral function derive_param_computed() adds a parameter computed from the analysis value of other parameters.\nIt works like a calculator to derive new records without worrying about merging and combining datasets, all you need is a derivation formula, which also improves the readability of the code."
},
{
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#thats-it",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#thats-it",
- "title": "Blanks and NAs",
- "section": "That’s it!",
- "text": "That’s it!\nA simple call to this function can make your derivation life so much easier while working in R if working with SAS-based datasets. In admiral, we make use of this function at the start of all ADaM templates for common ADaM datasets. You can use the function use_ad_template() to get the full R script for the below ADaMs.\n\nlist_all_templates()\n\nExisting ADaM templates in package 'admiral':\n• ADAE\n• ADCM\n• ADEG\n• ADEX\n• ADLB\n• ADLBHY\n• ADMH\n• ADPC\n• ADPP\n• ADPPK\n• ADSL\n• ADVS"
+ "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#example",
+ "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#example",
+ "title": "Derive a new parameter computed from the value of other parameters",
+ "section": "Example",
+ "text": "Example\nA value level validation use case, where derive_param_computed() is used to validate a derived parameter - PARAMCD = ADPCYMG (Actual Dose per Cycle) in ADEX dataset.\n\nDerivation\nActual Dose per Cycle is derived from the Total Amount of Dose (PARAMCD = TOTDOSE) / Number of Cycles (PARAMCD = NUMCYC)\nIn this example, ADEX.AVAL when ADEX.PARAMCD = ADPCYMG can be derived as:\n\\[\nAVAL (PARAMCD = ADPCYMG) = \\frac{AVAL (PARAMCD = TOTDOSE)}{AVAL (PARAMCD = NUMCYC)}\n\\]\n\n\nLoading Packages and Creating Example Data\n\nlibrary(tibble)\nlibrary(dplyr)\nlibrary(diffdf)\nlibrary(admiral)\n\nadex <- tribble(\n ~USUBJID, ~PARAMCD, ~PARAM, ~AVAL,\n \"101\", \"TOTDOSE\", \"Total Amount of Dose (mg)\", 180,\n \"101\", \"NUMCYC\", \"Number of Cycles\", 3\n)\n\n\n\nDerive New Parameter\n\nadex_admiral <- derive_param_computed(\n adex,\n by_vars = exprs(USUBJID),\n parameters = c(\"TOTDOSE\", \"NUMCYC\"),\n analysis_value = AVAL.TOTDOSE / AVAL.NUMCYC,\n set_values_to = exprs(\n PARAMCD = \"ADPCYMG\",\n PARAM = \"Actual Dose per Cycle (mg)\"\n )\n)\n\nWarning: The `analysis_value` argument of `derive_param_computed()` is deprecated as\nof admiral 0.12.0.\nℹ Please use the `set_values_to` argument instead.\n\n\n\n\n# A tibble: 3 × 4\n USUBJID PARAMCD PARAM AVAL\n <chr> <chr> <chr> <dbl>\n1 101 TOTDOSE Total Amount of Dose (mg) 180\n2 101 NUMCYC Number of Cycles 3\n3 101 ADPCYMG Actual Dose per Cycle (mg) 60\n\n\n\n\nCompare\nFor validation purpose, the diffdf package is used below to mimic SAS proc compare.\n\nadex_expected <- bind_rows(\n adex,\n tribble(\n ~USUBJID, ~PARAMCD, ~PARAM, ~AVAL,\n \"101\", \"ADPCYMG\", \"Actual Dose per Cycle (mg)\", 60\n )\n)\n\ndiffdf(adex_expected, adex_admiral, keys = c(\"USUBJID\", \"PARAMCD\"))\n\nNo issues were found!"
},
{
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#last-updated",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#last-updated",
- "title": "Blanks and NAs",
+ "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#last-updated",
+ "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#last-updated",
+ "title": "Derive a new parameter computed from the value of other parameters",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:27.839956"
+ "text": "Last updated\n\n2023-11-27 19:13:38.746856"
},
{
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#details",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#details",
- "title": "Blanks and NAs",
+ "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#details",
+ "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#details",
+ "title": "Derive a new parameter computed from the value of other parameters",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html",
+ "href": "posts/2023-07-14_code_sections/code_sections.html",
+ "title": "How to use Code Sections",
"section": "",
- "text": "There is significant momentum in driving the adoption of R packages in the life sciences industries, in particular, the R Consortium Submissions Working Group is dedicated to promoting the use of R for regulatory submissions to the FDA.\nThe R Consortium Submissions Working Group successfully completed an R-based submission in November 2021 through the eCTD portal (R Submissions Pilot 1). This Pilot was completed on March 10, 2022 after a successful statistical review and evaluation by the FDA staff.\nMoving forward, the Pilot 2 aimed to include a Shiny application that the FDA staff could deploy on their own servers. The R Consortium recently announced that on September 27, 2023, the R Submissions Working Group successfully completed the follow-up to the Pilot 2 R Shiny-based submission and received a response letter from FDA CDER. This marks the first publicly available submission package that includes a Shiny component. The full FDA response letter can be found here.\nThe Shiny application that was sent for the Pilot 2 had the goal to display the 4 Tables, Listings and Figures (TLFs) that were sent for the Pilot 1 with basic filtering functionality.\nThe submission package adhered to the eCTD folder structure and contained 5 deliverables. Among the deliverables was the proprietary R package {pilot2wrappers}, which enables the execution of the Shiny application.\nThe FDA staff were expected to receive the electronic submission packages in the eCTD format, install and load open-source R packages used as dependencies in the included Shiny application, reconstruct and load the submitted Shiny application, and communicate potential improvements in writing.\nIn the following stage, the R Consortium’s R Submission Working Group launched Pilot 4, aiming to investigate innovative technologies like Linux containers and web assembly. These technologies are being explored to package a Shiny application into a self-contained unit, streamlining the transfer and execution processes for the application.\nIn this post, our aim is to outline how we used the Rhino framework to reproduce the Shiny application that was successfully submitted to the FDA for the Pilot 2 project. Additionally, we detail the challenges identified during the process and how we were able to successfully address them by using an open-source package."
- },
- {
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#reproducing-the-r-submission-pilot-2-shiny-app-using-rhino",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#reproducing-the-r-submission-pilot-2-shiny-app-using-rhino",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
- "section": "Reproducing the R Submission Pilot 2 Shiny App using Rhino",
- "text": "Reproducing the R Submission Pilot 2 Shiny App using Rhino\nWhile the original Shiny application submitted to the FDA was wrapped using {Golem}, we replicated the application using our in-house developed framework Rhino. The main motivation was to provide an example of an R Submission that is not an R package and to identify and solve any issues that may arise from this approach.\nOur demo application (FDA-pilot-app) is accessible on our website, alongside other Shiny and Rhinoverse demonstration apps.\n\nThe code for FDA-pilot-app is open-source. You can create your own Rhino-based application by following our tutorial and viewing our workshop, which is available on YouTube."
+ "text": "The admiral package embraces a modular style of programming, where blocks of code are pieced together in sequence to create an ADaM dataset. However, with the well-documented advantages of the modular approach comes the recognition that scripts will on average be longer. As such, astute programmers working in RStudio are constantly on the lookout for quick ways to effectively navigate their scripts. Enter code sections!"
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#brief-introduction-to-rhino",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#brief-introduction-to-rhino",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
- "section": "Brief Introduction to Rhino",
- "text": "Brief Introduction to Rhino\n\n\n\n\n\nThe Rhino framework was developed by Appsilon to create enterprise-level Shiny applications, consistently and efficiently. This framework allows developers to apply the best software engineering practices, modularize code, test it thoroughly, enhance UI aesthetics, and monitor user adoption effectively.\nRhino provides support in 3 main areas:\n\nClear Code: scalable architecture, modularization based on Box and Shiny modules.\nQuality: comprehensive testing such as unit tests, E2E tests with Cypress and Shinytest2, logging, monitoring and linting.\nAutomation: project startup, CI with GitHub Actions, dependencies management with {renv}, configuration management with config, Sass and JavaScript bundling with ES6 support via Node.js.\n\nRhino is an ideal fit for highly regulated environments such as regulatory submissions or other drug development processes.\n\nFDA-pilot-app structure\nThe structure of this application is available on the github repository. The structure of this Shiny app is the following.\n\nClick here to expand the FDA-pilot-app structure\n\n.\n├── app\n│ ├── view\n│ │ └── demographic_table.R\n| | └── km_plot.R\n| | └── primary_table.R\n| | └── efficacy_table.R\n| | └── completion_table.R\n│ ├── logic\n│ │ └── adam_data.R\n│ │ └── eff_modles.R\n│ │ └── formatters.R\n│ │ └── helpers.R\n│ │ └── kmplot_helpers.R\n│ │ └── Tplyr_helpers.R\n│ ├── data\n│ │ └── adam\n│ │ └── adadas.xpt\n│ │ └── adlbc.xpt\n│ │ └── adsl.xpt\n│ │ └── adtte.xpt\n│ ├── docs\n│ │ └── about.md\n│ ├── js\n│ │ └── index.js\n│ ├── static\n│ │ └── favicon.ico\n│ ├── styles\n│ │ └── main.scss\n│ └── app.R\n├── tests\n│ ├── cypress\n│ │ └── integration\n│ │ └── app.spec.js\n│ ├── testthat\n│ │\n│ └── cypress.json\n├── app.R\n├── rhino_submission.Rproj\n├── dependencies.R\n├── renv.lock\n├── rhino.yml\n└── README.md"
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html#introduction",
+ "href": "posts/2023-07-14_code_sections/code_sections.html#introduction",
+ "title": "How to use Code Sections",
+ "section": "",
+ "text": "The admiral package embraces a modular style of programming, where blocks of code are pieced together in sequence to create an ADaM dataset. However, with the well-documented advantages of the modular approach comes the recognition that scripts will on average be longer. As such, astute programmers working in RStudio are constantly on the lookout for quick ways to effectively navigate their scripts. Enter code sections!"
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#efficient-submissions-to-the-fda",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#efficient-submissions-to-the-fda",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
- "section": "Efficient Submissions to the FDA",
- "text": "Efficient Submissions to the FDA\n\n\n\n\n\nTo comply with the Electronic Submission File Formats and Specifications for the eCTD submission, the programming code should carry a “.txt” extension. In the R Submissions Pilot 3 the group did not use {pkglite} as the FDA clarified that “.zip” and “.r” files are acceptable for submission. In this case, we utilized the {pkglite} R package to efficiently pack and unpack the FDA-pilot-app. This approach would facilitate the FDA reviewers in setting up the submission on their systems.\nThis package allows packing R packages to “.txt” files, which are supported for the submission of proprietary packages to the FDA via the eCTD gateway. \n\nPacking the FDA-pilot-app into a .txt file\nThe code below can be used to pack the Shiny application into a .txt file:\n\napp_name <- \"rhinosubmission\"\nrenv_spec <- pkglite::file_spec(\n \"renv\",\n pattern = \"^settings\\\\.dcf$|^activate\\\\.R$\",\n format = \"text\", recursive = FALSE\n)\ntests_spec <- pkglite::file_tests()\napp_spec <- pkglite::file_auto(\"app\")\nroot_spec <- pkglite::file_spec(\n path = \".\",\n pattern = \"^\\\\.Rprofile$|^rhino\\\\.yml$|^renv\\\\.lock$|^dependencies\\\\.R$|^config\\\\.yml$|^app\\\\.R$|^README\\\\.md$|\\\\.Rproj$\",\n all_files = TRUE,\n recursive = FALSE\n)\nwrite(paste0(\"Package: \", app_name), \"DESCRIPTION\", append = TRUE)\npkglite::collate(\n getwd(),\n renv_spec,\n tests_spec,\n app_spec,\n root_spec\n) |> pkglite::pack()\nfile.remove(\"DESCRIPTION\")\n\n\n\nUnpacking the FDA-pilot-app\nThe packed “.txt” file can be unpacked into a Shiny app by using {pkglite} as follows:\n\npkglite::unpack(\"rhinosubmission.txt\")"
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html#so-what-are-code-sections-and-why-are-they-useful",
+ "href": "posts/2023-07-14_code_sections/code_sections.html#so-what-are-code-sections-and-why-are-they-useful",
+ "title": "How to use Code Sections",
+ "section": "So, what are code sections and why are they useful?",
+ "text": "So, what are code sections and why are they useful?\nCode Sections are separators for long R scripts or functions in RStudio. They can be set up by inserting a comment line followed by four or more dashes in between portions of code, like so:\n\n# First code section ----\n\na <- 1\n\n# Second code section ----\n\nb <- 2\n\n# Third code section ----\n\nc <- 3\n\nRStudio then recognizes the code sections automatically, and enables you to:\n\nCollapse and expand them using the arrow displayed next to the line number, or with the handy shortcuts Alt+L/Shift+Alt+L on Windows or Cmd+Option+L/Cmd+Shift+Option+L on Mac.\nTravel in between them using the navigator at the bottom of the code pane, or by pressing Shift+Alt+J on Windows or Cmd+Shift+Option+J on Mac.\nView an outline of the file using the “Outline” button at the top right of the pane and/or the orange hashtag “Section Navigator” button at the bottom left of the pane.\n\n\n\n\n\n\nCollapsed sections, outline view and the section navigator for the example above.\n\n\n\n\nIt is also possible to create subsections by using two hashtags at the start of a comment line:\n\n# First code section ----\na <- 1\n\n## A code subsection ----\nb <- 2\n\n# Second code section ----\nc <- 3\n\n\n\n\n\n\nCode subsections for the example above.\n\n\n\n\nFor a complete list of Code Sections shortcuts, and for further information, see here."
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#lessons-learned",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#lessons-learned",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
- "section": "Lessons Learned",
- "text": "Lessons Learned\nOur initial objective was to prove that it would be possible to submit a Shiny application using Rhino through the eCTD gateway. During the rewriting process we identified that this could be done by integrating the open-source {pkglite} package. By following this approach, we concluded that it would be possible to submit a Shiny application through the eCTD gateway. This was also achieved through the successful submission of a package that included a Shiny component in Pilot 2.\nHaving rewritten the R Submissions Pilot 2 Shiny application using Rhino holds major implications for the adoption of our framework within the life sciences. Apart from being a strong, opinionated framework that improves reproducibility and reliability for Shiny development, using Rhino for regulatory submissions could improve the flexibility and speed in the clinical reporting pipeline. This would accelerate the adoption of R/Shiny for submissions to the FDA or other regulatory agencies."
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html#conclusion",
+ "href": "posts/2023-07-14_code_sections/code_sections.html#conclusion",
+ "title": "How to use Code Sections",
+ "section": "Conclusion",
+ "text": "Conclusion\nCode sections are an easy way to navigate long scripts and foster good commenting practices. They are used extensively in the admiral package, but there is no reason that you cannot start using them yourself in your day-to-day R programming!"
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#last-updated",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#last-updated",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html#last-updated",
+ "href": "posts/2023-07-14_code_sections/code_sections.html#last-updated",
+ "title": "How to use Code Sections",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:31.231853"
+ "text": "Last updated\n\n2023-11-27 19:13:41.390513"
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#details",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#details",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html#details",
+ "href": "posts/2023-07-14_code_sections/code_sections.html#details",
+ "title": "How to use Code Sections",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
@@ -459,7 +550,7 @@
"href": "posts/2023-10-30_floating_point/floating_point.html#last-updated",
"title": "Floating point",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:35.340689"
+ "text": "Last updated\n\n2023-11-27 19:13:46.342818"
},
{
"objectID": "posts/2023-10-30_floating_point/floating_point.html#details",
@@ -469,121 +560,100 @@
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html",
- "href": "posts/2023-07-09_falcon/falcon.html",
- "title": "falcon",
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html",
+ "title": "The pharmaverse (hi)story",
"section": "",
- "text": "The {falcon} initiative is an industry collaborative effort under {pharmaverse} that unites Boehringer Ingelheim, Moderna, Roche, and Sanofi with the aspiration of building and open-sourcing a comprehensive catalog of harmonized tables, listings, and graphs (TLGs) for clinical study reporting. By leveraging existing open-source R packages, {falcon} aims to simplify the process of output review, comparison, and meta-analyses, while fostering efficient communication among stakeholders in the pharmaceutical industry."
+ "text": "Pharmaverse: from motivation to present"
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#what-is-falcon",
- "href": "posts/2023-07-09_falcon/falcon.html#what-is-falcon",
- "title": "falcon",
- "section": "",
- "text": "The {falcon} initiative is an industry collaborative effort under {pharmaverse} that unites Boehringer Ingelheim, Moderna, Roche, and Sanofi with the aspiration of building and open-sourcing a comprehensive catalog of harmonized tables, listings, and graphs (TLGs) for clinical study reporting. By leveraging existing open-source R packages, {falcon} aims to simplify the process of output review, comparison, and meta-analyses, while fostering efficient communication among stakeholders in the pharmaceutical industry."
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#human-history-and-pharmaverse-context",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#human-history-and-pharmaverse-context",
+ "title": "The pharmaverse (hi)story",
+ "section": "Human history and pharmaverse context",
+ "text": "Human history and pharmaverse context\nSince the Australian Aboriginal, the earliest peoples recorded to have inhabited the Earth and who have been in Australia for at least 65,000 to 80,000 years (Encyclopædia Britannica), human beings live in group. Whether to protect yourself, increase your life expectancy or simply share tasks.\nFor most aspects of life, it doesn’t make sense to think, act or work alone for two main reasons:\n\nYou will spend more energy and time;\nSomeone else may be facing (or have faced) the same situation.\n\nThe English poet John Donne used to say “No man is an island entire of itself; every man is a piece of the continent, a part of the main;”. I can’t disagree with him. And I dare say that Ari Siggaard Knoph (Novo Nordisk), Michael Rimler (GSK), Michael Stackhouse (Atorus), Ross Farrugia (Roche), and Sumesh Kalappurakal (Janssen) can’t disagree with him either. They are the founders of pharmaverse, members of its Council and kindly shared their memories of how independent companies, in mid-2020, worked together in the creation of a set of packages developed to support the clinical reporting pipeline.\nIf you are not familiar with this pipeline, the important thing to know is that, in a nutshell, pharmaceutical companies must follow a bunch of standardized procedures and formats (from Clinical Data Interchange Standards Consortium, CDISC) when submitting clinical results to Health Authorities. The focus is on this: different companies seeking the same standards for outputs.\nParaphrasing Ross Farrugia (Roche) Breaking boundaries through open-source collaboration presentation in R/Pharma 2022 and thinking of the development of a new drug, we are talking about a “post-competitive” scenario: the drug has already been discovered and the companies should “just” produce and deliver standardized results."
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#why-do-we-build-it",
- "href": "posts/2023-07-09_falcon/falcon.html#why-do-we-build-it",
- "title": "falcon",
- "section": "Why do we build it?",
- "text": "Why do we build it?\nThe collaborative effort focuses on improving the clarity, consistency, and accessibility of TLGs by addressing variations and redundancies in their creation and use. This harmonized approach allows for streamlined reporting processes and facilitates effective communication of study results within the industry and to regulatory authorities."
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#clinical-reporting-outputs",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#clinical-reporting-outputs",
+ "title": "The pharmaverse (hi)story",
+ "section": "Clinical reporting outputs",
+ "text": "Clinical reporting outputs\nRationally, we can say that companies face the same challenges in these steps of the process. Not so intuitively, we can also say they were working in silos on that before 2018. Just as Isaac Newton and Gottfried W. Leibniz independently developed the theory of infinitesimal calculus, pharmaceutical companies were independently working on R solutions for this pipeline.\nBut on August 16 and 17 of the mentioned year above, they gathered at the first edition of R/Pharma conference to discuss R and open-source tooling for drug development (the reasons why open-source is an advantageous approach can be found in this post written by Stefan Thoma). And according to Isabela Velásquez’s article, Pharmaverse: Packages for clinical reporting workflows, one of the most popular questions in this conference was “Is the package code available or on CRAN?”.\nWell, many of them were. And not necessarily at that date, but just to mention a few: pharmaRTF and Tplyr from Atorus, r2rtf from Merck, rtables from Roche, etc. The thing is that, overall, there were almost 10000 other packages as well (today, almost 20000). And that took to another two questions:\n\nWith this overwhelming number of packages on CRAN, how to find the ones related to solving “clinical reporting problems”?\nOnce the packages were found, how to choose which one to use among those that have the same functional purpose?\n\nSo, again, companies re-started to working in silos to find those answers. But now, in collaborative silos and with common goals: create extremely useful packages to solve pharmaceutical-specific gaps once and solve them well!"
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#what-has-been-done-so-far",
- "href": "posts/2023-07-09_falcon/falcon.html#what-has-been-done-so-far",
- "title": "falcon",
- "section": "What has been done so far?",
- "text": "What has been done so far?\nDrawing inspiration from the FDA Standard Safety Tables and Figures Integrated Guide, the {falcon} initiative has successfully developed and open-sourced 11 templates to date. 4 product owners and 11 developers from 4 companies have collaborated to make these templates available and also published them on the official {falcon} website at https://pharmaverse.github.io/falcon/."
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#first-partnerships",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#first-partnerships",
+ "title": "The pharmaverse (hi)story",
+ "section": "First partnerships",
+ "text": "First partnerships\nIn 2020, Michael Stackhouse (Atorus) and Michael Rimler (GSK) talked and formed a partnership between their companies to develop a few more packages, including metacore, to read, store and manipulate metadata for ADaMs/SDTMs in a standardized object; xportr, to create submission compliant SAS transport files and perform pharma specific dataset level validation checks; and logrx (ex-timber), to build log to support reproducibility and traceability of an R script.\nAround the same time, Thomas Neitmann (currently at Denali Therapeutics, then at Roche) and Michael Rimler (GSK) discovered that both were working with ADaM in R, so Thomas Neitmann (currently at Denali Therapeutics, then at Roche), Ross Farrugia (Roche) and Michael Rimler (GSK) saw an opportunity there and GSK started their partnership with Roche to build and release admiral package.\nThe idea of working together, the sense of community, and the appetite from organizations built more and more, with incentive and priority established up into the programming heads council.\nJanssen had a huge effort in building R capabilities going on as well, by releasing tidytlg and envsetup), so eventually Michael Rimler (GSK), Michael Stackhouse (Atorus) and Ross Farrugia (Roche) formalized pharmaverse and formed the council, adding in Sumesh Kalappurakal (Janssen) and Ari Siggaard Knoph (Novo Nordisk) later joined as the fifth council member."
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#next-steps-vision",
- "href": "posts/2023-07-09_falcon/falcon.html#next-steps-vision",
- "title": "falcon",
- "section": "Next steps & vision",
- "text": "Next steps & vision\nFuture plans for {falcon} involve expanding the catalog through continuous collaboration from participating companies and inviting wider industry engagement. The ultimate goal is to promote harmonization of TLGs for clinical reporting across the pharmaceutical industry, leading to greater efficiency, collaboration, and innovation. Even though templates currently come from a published FDA guide, the collaborating companies are open to share and discuss similarities and differences on analysis concepts and output layouts of their own implementations in clinical reporting, for both safety and efficacy analyses.\nIn addition, while currently all templates were built using {rtables}, {tern}, {rlistings} and drew inspiration from the open-sourced TLG-Catalog, moving forward, we plan to support creating the same templates using alternative table engines such as {gt}.\n{falcon} will be presented at the upcoming PHUSE EU (Standards Implementation stream), where we will share the collaboration journey of {falcon} so far, providing more details on the current progress, long-term vision, and strategies for this initiative. Attendees will gain insights into the challenges and opportunities of harmonizing clinical reporting through open-source collaboration and learn about the potential benefits and future direction of {falcon}."
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#release-growth-and-developments",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#release-growth-and-developments",
+ "title": "The pharmaverse (hi)story",
+ "section": "Release, growth and developments",
+ "text": "Release, growth and developments\nAt the end of their presentation “Closing the Gap: Creating an End to End R Package Toolkit for the Clinical Reporting Pipeline.”, in R/Pharma 2021, Ben Straub (GSK) and Eli Miller (Atorus) welcomed the community to the pharmaverse, a curated collection of packages developed by various pharmaceutical companies to support open-source clinical workflows.\nFrom the outset, the name pharmaverse was chosen so that it could be a neutral home, unrelated to any company. Also, it was established as not being a consortium, which means that founders don’t own, fund, or maintain the packages. Some individuals and companies maintain them but often allowing for community contributions and being licensed permissively so that there is always a feeling of community ownership. The focus of pharmaverse early on, and today, is on inter organization cooperation, to build an environment where, if organizations identify that they have a joint problem that they want to solve, this is the right space to work on and release it.\nPharmaverse has grown a lot, at the time of writing this post we have >25 packages recommended in pharmaverse, and this has led to a partnership with PHUSE to get support from their organization and platform, and because they are eager to advance and support pharmaverse mission.\nDespite all its structure, it is impossible to say that we have a single solution for each clinical reporting analysis when it comes to pharmaverse, a single pathway is impractical. Instead, it is necessary to accept viable tools fitting different pathways into pharmaverse to direct and give people options as to what might work for them. After all, even though we live together as a community, we still have our own unique internal problems.\n\n\n\nSample of pharmaverse packages"
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#last-updated",
- "href": "posts/2023-07-09_falcon/falcon.html#last-updated",
- "title": "falcon",
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#last-updated",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#last-updated",
+ "title": "The pharmaverse (hi)story",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:38.469514"
+ "text": "Last updated\n\n2023-11-27 19:13:49.940918"
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#details",
- "href": "posts/2023-07-09_falcon/falcon.html#details",
- "title": "falcon",
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#details",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#details",
+ "title": "The pharmaverse (hi)story",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html",
- "href": "posts/2023-06-27__hackathon_app/index.html",
- "title": "Hackathon Feedback Application",
+ "objectID": "posts/2023-06-28_welcome/index.html",
+ "href": "posts/2023-06-28_welcome/index.html",
+ "title": "Hello pharmaverse",
"section": "",
- "text": "We recently created a shiny application for the admiral hackathon in February 2023. The admiral hackathon was an event designed to make statistical programmers from the pharmaceutical industry more comfortable with the admiral R package which allows users to efficiently transform data from one data standard (SDTM) to another (ADaM).\nHackathon participants formed groups of up to five people and were then tasked to create R-scripts that map the SDTM data to ADaM according to specifics defined in the metadata.\nThe purpose of the shiny app was threefold:\nIn this blog post I want to highlight some of the thoughts that went into this application. Please keep in mind that this work was done under tight time restraints.\nThe hackathon application is still online (although data-upload is switched off) and the GitHub repository is publicly available. The application is embedded into this post right after this paragraph. I have also uploaded to GitHub a .zip file of the workspace to which hackathon participants had access via posit cloud. For more context you can watch recordings of the hackathon-meetings."
- },
- {
- "objectID": "posts/2023-06-27__hackathon_app/index.html#permanent-data",
- "href": "posts/2023-06-27__hackathon_app/index.html#permanent-data",
- "title": "Hackathon Feedback Application",
- "section": "Permanent Data",
- "text": "Permanent Data\nThe biggest challenge you have to consider for this app is the permanent data storage. Shiny apps run on a server. Although we can write files on this server, whenever the app restarts, the files are lost. Therefore, a persistent data storage solution is required.\n\nGoogle drive\nI decided to leverage Google drive using the googledrive package package. This allowed me to save structured data (the team registry and the submission scores) as well as unstructured data (their R-script files).\n\n\n\n\n\n\nAuthentication\n\n\n\nTo access Google drive using the googledrive package we need to authenticate. This can be done interactively using the command googledrive::drive_auth() which takes you to the Google login page. After login you receive an authentication token requested by R.\nFor non-interactive authentication this token must be stored locally. In our case where the shiny app must access the token once deployed, the token must be stored on the project level.\nI have included the authentication procedure I followed in the R folder in google_init.R. You can find more extensive documentation of the non-interactive authentication.\n\n\nThe initial concept was: Each team gets their own folder including the most recent submission for each task, and a .csv file containing team information. To keep track of the submissions and the respective scores we wrote a .csv file in the mock-hackathon folder, so one folder above the team folders.\nSaving the team info as a .csv file worked fine as each team received their own file which – once created – was not touched anymore. As each upload for every team should simply add a row to the submissions.csv file, appending the file would be ideal. This was not possible using the googledrive package package. Instead, for each submission, the submissions file was downloaded, appended, and uploaded again. Unfortunately, this lead to a data loss, as the file was continuously overwritten, especially when two teams would submit simultaneously.\n\n\n\n\n\n\nRecover the Lost Data\n\n\n\nWhenever the submissions.csv file was uploaded, the previous version was sent to the Google drive bin. We ended up with over 3000 submissions.csv files containing a lot of redundant information. I had to write the following chunk to first get the unique file IDs of the 3000 submissions.csv files, create an empty submissions data-frame, and then download each file and add its information to the submisisons data-frame. To keep the data-frame as light as possible, after each append I deleted all duplicate submissions.\n\n# get all task_info.csv ID's\n# each row identifies one file in the trash\ntask_info_master <- drive_find(\n pattern = \"task_info.csv\",\n trashed = TRUE\n)\n\n\n# set up empty df to store all submissions\norigin <- tibble(\n score = numeric(),\n task = character(),\n team = character(),\n email = character(),\n time = character()\n)\n\n# downloads, reads, and returns one csv file given a file id\nget_file <- function(row) {\n tf <- tempfile()\n row %>%\n as_id() %>%\n drive_download(path = tf)\n new <- read_csv(tf) %>%\n select(score, task, team) %>%\n distinct()\n}\n\n\n# quick and dirty for loop to subsequently download each file, extract information\n# merge with previous information and squash it (using distinct()).\nfor (i in 1:nrow(task_info_master)) {\n origin <- rbind(origin, get_file(row = task_info_master[i, ])) %>%\n distinct()\n\n # save progress in a separate file after every 100 downloaded and merged sheets\n if (i %% 100 == 0) {\n print(i)\n write_csv(origin, paste(\"prog_data/task_info_prog_\", i, \".csv\", sep = \"\"))\n # update on progress\n message(i / nrow(task_info_master) * 100)\n }\n}\n\nWhen doing such a time-intensive task, make sure to try it first with only a couple of files to see whether any errors are produced. I am not quite sure how long this took but when I returned from my lunch break everything had finished.\n\n\nIf you want to stay in the Google framework, I recommend using the googlesheets4 package for structured data. googlesheets4 allows appending new information to an already existing sheet without the need to download the file first. As both packages follow the same style, going from one to the other is really simple. googlesheets4 requires authentication as well. However, you can reuse the cached token from the googledrive package authentication by setting gs4_auth(token = drive_token()).\n\n\nSecurity Concerns\nConnecting a public shiny app to your Google account introduces a security vulnerability in general. Especially so because we implemented the upload of files to Google drive. And even more problematic: We run a user generated script and display some of its output. A malicious party might be able to extract the authentication token of our Google account or could upload malware to the drive.\nTo reduce the risk, I simply created an un-associated Google account to host the drive. There are certainly better options available, but this seemed a reasonable solution achieved with very little effort."
- },
- {
- "objectID": "posts/2023-06-27__hackathon_app/index.html#register-team",
- "href": "posts/2023-06-27__hackathon_app/index.html#register-team",
- "title": "Hackathon Feedback Application",
- "section": "Register Team",
- "text": "Register Team\nWe wanted to allow users to sign up as teams using the shiny app. The app provides a simple interface where users could input a team name and the number of members. This in turn would open two fields for each user to input their name and email address.\nWe do simple checks to make sure at least one valid email address is supplied, and that the group name is acceptable. The group name cannot be empty, already taken, or contain vulgar words.\nThe team registration itself was adding the team information to the Google sheets file event_info into the sheet teams and to create a team folder in which to store the uploaded R files.\nThe checks and registration is implemented in the register_team() function stored in interact_with_google.R.\n\n\n\nScreenshot of the register team interface\n\n\nThe challenge here was to adapt the number of input fields depending on the number of team members. This means that the team name and email interface must be rendered: First, we check how many team members are part of the group, this is stored in the input$n_members input variable. Then we create a tagList with as many elements as team members. Each element contains two columns, one for the email, one for the member name. This tagList is then returned and displayed to the user.\n\n# render email input UI of the register tab\noutput$name_email <- shiny::renderUI({\n # create field names\n N <- input$n_members\n NAME <- sapply(1:N, function(i) {\n paste0(\"name\", i)\n })\n EMAIL <- sapply(1:N, function(i) {\n paste0(\"email\", i)\n })\n\n output <- tagList()\n\n\n firstsecondthird <- c(\"First\", \"Second\", \"Third\", \"Fourth\", \"Fifth\")\n for (i in 1:N) {\n output[[i]] <- tagList()\n output[[i]] <- fluidRow(\n shiny::h4(paste(firstsecondthird[i], \" Member\")),\n column(6,\n textInput(NAME[i], \"Name\"),\n value = \" \" # displayed default value\n ),\n column(6,\n textInput(EMAIL[i], \"Email\"),\n value = \" \"\n )\n )\n }\n output\n})\n\nThe team information is then uploaded to Google drive. Because some teams have more members than others, we have to create the respective data-frame with the number of team members in mind.\nThe following chunk creates the registration data. Noteworthy here the creation of the NAME and EMAIL variables which depend on the number of members in this team. Further, the user input of these fields is extracted via input[[paste0(NAME[i])]] within a for-loop.\nWe also make the data-creation dependent on the press of the Register Group button and cache some variables.\n\n## registration\nregistrationData <-\n reactive({\n N <- input$n_members\n NAME <- sapply(1:N, function(i) {\n paste0(\"name\", i)\n })\n EMAIL <- sapply(1:N, function(i) {\n paste0(\"email\", i)\n })\n names <- character(0)\n emails <- character(0)\n\n for (i in 1:N) {\n names[i] <- input[[paste0(NAME[i])]]\n emails[i] <- input[[paste0(EMAIL[i])]]\n }\n # create df\n dplyr::tibble(\n team_name = input$team_name,\n n_members = N,\n member_name = names,\n member_email = emails\n )\n }) %>%\n bindCache(input$team_name, input$n_members, input$name1, input$email1) %>%\n bindEvent(input$register) # wait for button press"
- },
- {
- "objectID": "posts/2023-06-27__hackathon_app/index.html#upload-source-script",
- "href": "posts/2023-06-27__hackathon_app/index.html#upload-source-script",
- "title": "Hackathon Feedback Application",
- "section": "Upload & Source Script",
- "text": "Upload & Source Script\nTo upload a script, participants had to select their team first. The input options were based on the existing folders on the Google-drive in the mock_hackathon folder. To upload a particular script participants had to also select the task to be solved. The uploaded script is then uploaded to the team folder following a standardised script naming convention.\nThere are different aspects to be aware of when sourcing scripts on a shiny server. For example, you have to anticipate the packages users will include in their uploaded scripts, as their scripts will load but not install packages. Further, you should keep the global environment of your shiny app separate from the environment in which the script is sourced. This is possible by supplying an environment to the source() function, e.g: source(path_to_script, local = new.env())\nAnother thing we had to consider was to replicate the exact folder-structure on the shiny server that participants were working with when creating the scripts, as they were required to source some scripts and to save their file into a specific folder. This was relatively straight forward as we provided participants with a folder structure in the posit cloud instance they were using. They had access to the sdtm folder in which the data was stored, and the adam folder into which they saved their solutions. The structure also included a folder with metadata which was also available on the shiny server.\nFor some tasks, participants required some ADaM-datasets stored in the adam folder, essentially the output from previous tasks. This was achieved by first creating a list mapping tasks to the required ADaM datasets:\n\ndepends_list <- list(\n \"ADADAS\" = c(\"ADSL\"),\n \"ADAE\" = c(\"ADSL\"),\n \"ADLBC\" = c(\"ADSL\"),\n \"ADLBH\" = c(\"ADSL\"),\n \"ADLBHY\" = c(\"ADSL\"),\n \"ADSL\" = NULL,\n \"ADTTE\" = c(\"ADSL\", \"ADAE\"),\n \"ADVS\" = c(\"ADSL\")\n)\n\nThis list is sourced from the R/parameters.R file when initiating the application. We then call the get_depends() function sourced from R/get_depends.R which copies the required files from the key folder (where our solutions to the tasks were stored) to the adam folder. After sourcing the uploaded script the content in the adam folder is deleted."
+ "text": "The communications working group (CWG) seeks to promote and showcase how R can be used in the Clinical Reporting pipeline through short and informative blog posts. These posts will be hosted on this pharmaverse blog and promoted on the pharmaverse slack channels as well as on LinkedIn.\nAs the CWG is a small team, we hope to make the blog development process easy enough that pharmaverse community members will be able to easily write blog posts with guidance from the CWG team."
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#compare-to-solution-file",
- "href": "posts/2023-06-27__hackathon_app/index.html#compare-to-solution-file",
- "title": "Hackathon Feedback Application",
- "section": "Compare to Solution File",
- "text": "Compare to Solution File\nWe want to compare the file created by participants with our solution (key) file stored in the key folder. The diffdf::diffdf() function allows for easy comparison of two data-frames and directly provides extensive feedback for the user:\n\nlibrary(dplyr)\ndf1 <- tibble(\n numbers = 1:10,\n letters = LETTERS[1:10]\n)\ndf2 <- tibble(\n numbers = 1:10,\n letters = letters[1:10]\n)\n\ndiffdf::diffdf(df1, df2)\n\nWarning in diffdf::diffdf(df1, df2): \nNot all Values Compared Equal\n\n\nDifferences found between the objects!\n\nA summary is given below.\n\nNot all Values Compared Equal\nAll rows are shown in table below\n\n =============================\n Variable No of Differences \n -----------------------------\n letters 10 \n -----------------------------\n\n\nAll rows are shown in table below\n\n ========================================\n VARIABLE ..ROWNUMBER.. BASE COMPARE \n ----------------------------------------\n letters 1 A a \n letters 2 B b \n letters 3 C c \n letters 4 D d \n letters 5 E e \n letters 6 F f \n letters 7 G g \n letters 8 H h \n letters 9 I i \n letters 10 J j \n ----------------------------------------"
+ "objectID": "posts/2023-06-28_welcome/index.html#purpose",
+ "href": "posts/2023-06-28_welcome/index.html#purpose",
+ "title": "Hello pharmaverse",
+ "section": "",
+ "text": "The communications working group (CWG) seeks to promote and showcase how R can be used in the Clinical Reporting pipeline through short and informative blog posts. These posts will be hosted on this pharmaverse blog and promoted on the pharmaverse slack channels as well as on LinkedIn.\nAs the CWG is a small team, we hope to make the blog development process easy enough that pharmaverse community members will be able to easily write blog posts with guidance from the CWG team."
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#score",
- "href": "posts/2023-06-27__hackathon_app/index.html#score",
- "title": "Hackathon Feedback Application",
- "section": "Score",
- "text": "Score\nTo compare submissions between participants we implemented a simple scoring function (score_f()) based on the table comparison by diffdf(). The function can be found in the compare_dfs.R file:\n\nscore_f <- function(df_user, df_key, keys) {\n score <- 10\n diff <- diffdf::diffdf(df_user, df_key, keys = keys)\n if (!diffdf::diffdf_has_issues(diff)) {\n return(score)\n }\n\n # check if there are any differences if the comparison is not strict:\n if (!diffdf::diffdf_has_issues(diffdf::diffdf(df_user,\n df_key,\n keys = keys,\n strict_numeric = FALSE,\n strict_factor = FALSE\n ))) {\n # if differences are not strict, return score - 1\n return(score - 1)\n }\n\n return(round(min(max(score - length(diff) / 3, 1), 9), 2))\n}\n\nEvery comparison starts with a score of 10. We then subtract the length of the comparison object divided by a factor of 3. The length of the comparison object is a simplified way to represent the difference between the two data-frames by one value. Finally, the score is bounded by 1 using max(score, 1).\nThe score is not a perfect capture of the quality of the script uploaded but: 1. helped participants get an idea of how close their data-frame is to the solution file 2. allowed us to raffle prizes based on the merit of submitted r-scripts"
+ "objectID": "posts/2023-06-28_welcome/index.html#spirit-of-a-blog-post",
+ "href": "posts/2023-06-28_welcome/index.html#spirit-of-a-blog-post",
+ "title": "Hello pharmaverse",
+ "section": "Spirit of a Blog Post",
+ "text": "Spirit of a Blog Post\nThe CWG believes that the following 4 points will help guide the creation of Blog Posts.\n\nShort\nPersonalized\nReproducible\nReadable\n\nShort: Posts should aim to be under a 10 minute read. We encourage longer posts to be broken up into multiple posts.\nPersonalized: Posts should have a personality! For example, a person wishing to post on a function in a package needs to differentiate the post from the documentation for function, i.e. we don’t want to just recycle the documentation. How can you add your voice and experience? A bit of cheeky language is also encouraged.\nReproducible: Posts should work with minimal dependencies with data, packages and outside sources. Every dependency introduced in a post adds some risk to the post longevity. As package dependencies change, posts should be built in a way that they can be updated to stay relevant.\nReadable: The CWG sees this site as more of introductory site rather advanced user site. Therefore, the CWG feels that code should be introduced in a way that promotes readability over complexity."
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#reactiveness",
- "href": "posts/2023-06-27__hackathon_app/index.html#reactiveness",
- "title": "Hackathon Feedback Application",
- "section": "Reactiveness",
- "text": "Reactiveness\nSome of the app functions can take quite some time to execute, e.g. running the uploaded script. Other tasks, e.g. registering a team, do not intrinsically generate user facing outputs. This would make the app using really frustrating, as users would not know whether the app is correctly working or whether it froze.\nWe implemented two small features that made the app more responsive. One is simple loading icons that integrate into the user interface and show that output is being computed – that something is working. The other is a pop up window which communicates whether team registration was successful, and if not, why not.\nWe further aimed to forward errors generated by the uploaded scripts to the user interface, but errors generated by the application itself should be concealed."
+ "objectID": "posts/2023-06-28_welcome/index.html#what-types-of-posts-are-allowed-on-this-site",
+ "href": "posts/2023-06-28_welcome/index.html#what-types-of-posts-are-allowed-on-this-site",
+ "title": "Hello pharmaverse",
+ "section": "What types of posts are allowed on this site?",
+ "text": "What types of posts are allowed on this site?\nOverall, we want to stay focus on the Clinical Reporting Pipeline, which we see as the following topics:\n\nPackages in the Clinical Reporting Pipeline\nFunctions from packages in the Clinical Reporting Pipeline\nWider experiences of using R in the Clinical Reporting Pipeline\nConference experiences and the Clinical Reporting Pipeline\n\nHowever, it never hurts to ask if you topic might fit into this medium!\n\nMinimum Post Requirements\n\nA unique image to help showcase the post.\nWorking Code\nSelf-contained data or package data.\nDocumentation of package versions\n\nThat is it! After that you can go wild, but we do ask that it is kept short!"
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#conclusion",
- "href": "posts/2023-06-27__hackathon_app/index.html#conclusion",
- "title": "Hackathon Feedback Application",
- "section": "Conclusion",
- "text": "Conclusion\nAlthough the application was continuously improved during the hackathon it proved to be a useful resource for participants from day one as it allowed groups to set their own pace. It further allowed admiral developers to gain insights on package usage of a relatively large sample of potential end users. From our perspective, the application provided a great added value to the hackathon and eased the workload of guiding the participants through all the tasks."
+ "objectID": "posts/2023-06-28_welcome/index.html#how-can-i-make-a-blog-post",
+ "href": "posts/2023-06-28_welcome/index.html#how-can-i-make-a-blog-post",
+ "title": "Hello pharmaverse",
+ "section": "How can I make a Blog Post",
+ "text": "How can I make a Blog Post\nStep 1: Reach out to us through pharmaverse/slack or make an issue on our GitHub.\nStep 2: Branch off main\nStep 3: Review the Spirit of the Blog Post in the Pull Request Template\nStep 4: Poke us to do a review!"
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#last-updated",
- "href": "posts/2023-06-27__hackathon_app/index.html#last-updated",
- "title": "Hackathon Feedback Application",
+ "objectID": "posts/2023-06-28_welcome/index.html#last-updated",
+ "href": "posts/2023-06-28_welcome/index.html#last-updated",
+ "title": "Hello pharmaverse",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:42.927745"
+ "text": "Last updated\n\n2023-11-27 19:13:52.942147"
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#details",
- "href": "posts/2023-06-27__hackathon_app/index.html#details",
- "title": "Hackathon Feedback Application",
+ "objectID": "posts/2023-06-28_welcome/index.html#details",
+ "href": "posts/2023-06-28_welcome/index.html#details",
+ "title": "Hello pharmaverse",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
@@ -621,47 +691,5 @@
"title": "Cross-Industry Open Source Package Development",
"section": "Impact",
"text": "Impact\nFor Roche, cross-industry package development work out in their favor: They get access to software created by specialists and users from across the industry but paying only a fraction of the developmental costs. Of course, they don’t have total developmental control but they do get a seat at the table. Any gaps between the open source {admiral} package and the proprietary Roche workflow were bridged by the internal {admiralroche} package.\nThe switch towards a more language agnostic platform, and open source languages specifically, opens the door to a broad population of university graduates with diverse backgrounds. I personally would not have considered this position five years ago due to a misalignment of skills and job requirements. Working towards an industry standard open source solution will also ensure that skills learned at one company are more easily transferable to external positions, further making the position much more attractive. Access to such a broad pool of potential candidates is clearly beneficial for recruitment at Roche, but also facilitates diversity in teams which makes for a more interesting and effective work place.\nOpen source development comes with much more transparency by definition. Recognition of contributions are built in - anyone can see who did what. This recognition escapes the confines of your company as it is visible to anyone looking at the repository. Anyone can not only see at any time what is being worked on, what discussions are happening and which direction is being taken, but can also participate and contribute. Transparency also applies to errors in the code and how the team is dealing with them. In such an environment it is practically impossible to hide or cover up errors and corrections. Instead, they have to be dealt with publicly and in the open. This openness about errors also helps seeing errors as a natural occurrence that needs to be dealt with. Space for errors encourages learning and is really beneficial for growing both skills and integrity.\nAs you work on a team that spans multiple companies, traditional corporate hierarchies do not apply. Of course, there will always be a sort of hierarchy of experience or skills, but these work in your favor: You will know who to ask for help, and teams are generally very happy for contributors of any skill level. Contributions also need not be in code: Inputs into discussions and domain knowledge contributions are highly valued as well. The flip-side of working in a team without your manager oversight: They may not be directly aware of the work you do. That’s why you have to write blog posts :)\nThe possibility for statistical programmers to pivot towards developing software or writing blog-posts such as this really transforms and broadens their job description. It is this transformation that is reflected by the choice of Roche to re-brand statistical programmers as analytical data scientists. The fact that cross-industry development is being advocated for really lets programmers expand their network outside of their company.\nThe {admiral} project serves as a testament to the power of collaborative open-source development and the potential it holds for the future of work in this industry."
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html",
- "href": "posts/2023-06-27_hackathon_writeup/index.html",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "",
- "text": "This January and February (2023), the admiral development team and the CDISC Open Source Alliance jointly hosted the admiral hackathon. The idea was to build a community of admiral users, and help participants familiarize themselves with R and admiral. This whole effort was led by Thomas Neitmann and was supported by Zelos Zhu, Sadchla Mascary, and me – Stefan Thoma.\nThe hackathon event was structured in two parts. First, we offered an Introduction to R for SAS programmers, a three hour workshop for R beginners to get them up to speed. Here we covered practical R basics, talking about how the R-workflow differs from a SAS workflow, and discussed common R functions - mostly from the tidyverse. This ensured that hackathon participants were familiar with core R concepts. The workshop recording and the course materials are available online.\nThe main hackathon consisted of several ADAM data generating tasks based on a specs file and synthetic data. Participants were able to solve these tasks in groups at their own pace thanks to a online tool where participants could upload their task specific R scripts and they would get automatic feedback for the data-set produced by their script. Script upload through the feedback application was available all through February, and we offered three additional online meetings throughout the month to discuss challenges and give some tips. If you are interested in learning more about the thoughts that went into the feedback application, you can read about it in this blogpost or check out my public GitHub repository for such an application."
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html#introduction-to-r-workshop",
- "href": "posts/2023-06-27_hackathon_writeup/index.html#introduction-to-r-workshop",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "Introduction to R workshop",
- "text": "Introduction to R workshop\nWe were really excited to see over 500 people from around 40 countries joining our Introduction to R workshop in January! To get to know prospective users and hackathon participants better, we conducted some polls during the meetings. Below you can see that representatives of many different sorts of organisations joined our Introduction to R workshop:\n\n\n\n\n\n216 out of 402 confirmed that their company is already using R for clinical trial data analysis, the remaining 131 did not answer this question.\nThe target audience for this workshop was programmers who are very familiar with SAS, but not so familiar with R, our polls confirmed this.\n\n\n\n\n\nOverall, we were very happy with how the workshop turned out, and participants overall agreed with this sentiment (although there may be a slight survivorship bias…)."
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html#admiral-hackathon",
- "href": "posts/2023-06-27_hackathon_writeup/index.html#admiral-hackathon",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "admiral Hackathon",
- "text": "admiral Hackathon\nFollowing the kick-off meeting, 371 participants joined the posit (rStudio) workspace that was made available to all participants at no costs by the posit company. About half the participants planned to spend one to two hours per week on the admiral tasks, the other half planned to allocate even more. 15 participants even planned to spend eight hours or more!\nWe were really happy to see an overwhelming amount of activity on the slack channel we set up with over 250 members. Not only were people engaging with the materials, but we saw how a community was formed where people were encouraged to ask questions and where community members went out of their way to help each other. Shout-out to our community hero: Jagadish Katam without whom most issues related to the task programming raised by the community would not have been addressed as quickly as they were. Huge thanks from the organizers!\nIn the end, a total of 44 teams spanning 87 statistical programmers took part in the admiral hackathon and uploaded solution scripts to the hackathon application solving at least one of the 8 tasks available (ADSL, ADAE, ADLBC, ADVS, ADTTE, ADADAS, ADLBH & ADLBHY). Participants’ scripts were then run on the shiny server and the output data-frame were compared to the solutions we provided. At the read-out there was a live draft of teams to win one-on-one admiral consulting with one of the admiral core developers. Winning probabilities were weighted by the number of points each group received for the quality of their output data-frames and for the number of tasks solved.\nCongratulations to the winners:\n\nViiV Team_GSK\nteamspoRt\nTatianaPXL\nDivyasneelam\nAdaMTeamIndia\nSanofi_BP\nJagadish (our community hero)\nAZ_WAWA\n\nAlthough this was uncertain during the hackathon we were excited to provide a Certificate of Completion to all participants who uploaded a script to the Web Application.\nA recording of the hackathon readout can be found in the CDISC Open Source Alliance Quarterly Spotlight."
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html#conclusion",
- "href": "posts/2023-06-27_hackathon_writeup/index.html#conclusion",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "Conclusion",
- "text": "Conclusion\nOverall, we are very happy with how the hackathon turned out. We were not only positively surprised with the huge audience for the Intro to R workshop (CDISC record breaking) and for the admiral hackathon, but even more so with the engagement of all the participants.\nAgain, we would like to thank all the organizers, participants, and sponsors for their time and resources and hope to have provided a useful glimpse into our solution for ADAM creation within the end-to-end clinical data analysis open source R framework that the Pharmaverse aims to provide.\nAs always, we are very happy to hear more feedback on the hackathon as well as on admiral in general. Simply submit an issue on the admiral GitHub repository. You would like to join the admiral core developers? Please reach out to Edoardo Mancini (product owner) or Ben Straub (technical lead)."
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html#last-updated",
- "href": "posts/2023-06-27_hackathon_writeup/index.html#last-updated",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:51.728905"
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html#details",
- "href": "posts/2023-06-27_hackathon_writeup/index.html#details",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "Details",
- "text": "Details\n\nsource code, R environment"
}
]
\ No newline at end of file
diff --git a/site_libs/htmlwidgets-1.6.2/htmlwidgets.js b/site_libs/htmlwidgets-1.6.3/htmlwidgets.js
similarity index 100%
rename from site_libs/htmlwidgets-1.6.2/htmlwidgets.js
rename to site_libs/htmlwidgets-1.6.3/htmlwidgets.js
diff --git a/sitemap.xml b/sitemap.xml
index a0a76761..e2577287 100644
--- a/sitemap.xml
+++ b/sitemap.xml
@@ -1,67 +1,71 @@
- https://pharmaverse.github.io/blog/posts/2023-07-14_code_sections/code_sections.html
- 2023-11-01T15:47:53.974Z
+ https://pharmaverse.github.io/blog/posts/2023-08-08_study_day/study_day.html
+ 2023-11-27T19:13:58.211Z
- https://pharmaverse.github.io/blog/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
- 2023-11-01T15:47:48.826Z
+ https://pharmaverse.github.io/blog/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
+ 2023-11-27T19:13:54.971Z
- https://pharmaverse.github.io/blog/posts/2023-08-08_study_day/study_day.html
- 2023-11-01T15:47:45.858Z
+ https://pharmaverse.github.io/blog/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
+ 2023-11-27T19:13:52.247Z
- https://pharmaverse.github.io/blog/posts/2023-09-26_date_functions_and_imputation/date_functions_and_imputation.html
- 2023-11-01T15:47:41.734Z
+ https://pharmaverse.github.io/blog/posts/2023-06-27__hackathon_app/index.html
+ 2023-11-27T19:13:49.215Z
- https://pharmaverse.github.io/blog/posts/2023-06-28_welcome/index.html
- 2023-11-01T15:47:37.682Z
+ https://pharmaverse.github.io/blog/posts/2023-11-27_higher_order/higher_order.html
+ 2023-11-27T19:13:44.987Z
- https://pharmaverse.github.io/blog/posts/2023-06-27_admiral/valuelevel/derive_param_computed.html
- 2023-11-01T15:47:33.918Z
+ https://pharmaverse.github.io/blog/posts/2023-07-09_falcon/falcon.html
+ 2023-11-27T19:13:40.491Z
- https://pharmaverse.github.io/blog/posts/2023-07-24_rounding/rounding.html
- 2023-11-01T15:47:30.366Z
+ https://pharmaverse.github.io/blog/posts/2023-06-27_hackathon_writeup/index.html
+ 2023-11-27T19:13:37.491Z
- https://pharmaverse.github.io/blog/R/readme.html
- 2023-11-01T15:47:26.622Z
+ https://pharmaverse.github.io/blog/posts/2023-09-26_date_functions_and_imputation/date_functions_and_imputation.html
+ 2023-11-27T19:13:32.503Z
https://pharmaverse.github.io/blog/index.html
- 2023-11-01T15:47:26.074Z
+ 2023-11-27T19:13:29.559Z
- https://pharmaverse.github.io/blog/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
- 2023-11-01T15:47:28.446Z
+ https://pharmaverse.github.io/blog/R/readme.html
+ 2023-11-27T19:13:30.087Z
- https://pharmaverse.github.io/blog/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
- 2023-11-01T15:47:31.962Z
+ https://pharmaverse.github.io/blog/posts/2023-07-24_rounding/rounding.html
+ 2023-11-27T19:13:34.283Z
- https://pharmaverse.github.io/blog/posts/2023-10-30_floating_point/floating_point.html
- 2023-11-01T15:47:36.342Z
+ https://pharmaverse.github.io/blog/posts/2023-06-27_admiral/valuelevel/derive_param_computed.html
+ 2023-11-27T19:13:39.323Z
- https://pharmaverse.github.io/blog/posts/2023-07-09_falcon/falcon.html
- 2023-11-01T15:47:39.010Z
+ https://pharmaverse.github.io/blog/posts/2023-07-14_code_sections/code_sections.html
+ 2023-11-27T19:13:41.975Z
- https://pharmaverse.github.io/blog/posts/2023-06-27__hackathon_app/index.html
- 2023-11-01T15:47:43.922Z
+ https://pharmaverse.github.io/blog/posts/2023-10-30_floating_point/floating_point.html
+ 2023-11-27T19:13:47.267Z
- https://pharmaverse.github.io/blog/posts/2023-07-20_cross_company_dev/cross_industry_dev.html
- 2023-11-01T15:47:47.398Z
+ https://pharmaverse.github.io/blog/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
+ 2023-11-27T19:13:50.535Z
- https://pharmaverse.github.io/blog/posts/2023-06-27_hackathon_writeup/index.html
- 2023-11-01T15:47:52.282Z
+ https://pharmaverse.github.io/blog/posts/2023-06-28_welcome/index.html
+ 2023-11-27T19:13:53.443Z
+
+
+ https://pharmaverse.github.io/blog/posts/2023-07-20_cross_company_dev/cross_industry_dev.html
+ 2023-11-27T19:13:56.455Z
Last updated
2023-11-01 15:47:37.128939
+2023-11-27 19:13:52.942147
Details
Next steps & vision<
Last updated
-2023-11-01 15:47:38.469514
+2023-11-27 19:13:40.019302
Details
diff --git a/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html b/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
index d9ec2ab8..1eea5aae 100644
--- a/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
+++ b/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
@@ -301,7 +301,7 @@ That’s it!
Last updated
-2023-11-01 15:47:27.839956
+2023-11-27 19:13:51.677601
Details
diff --git a/posts/2023-07-14_code_sections/code_sections.html b/posts/2023-07-14_code_sections/code_sections.html
index 73c1bad5..44eaf7d9 100644
--- a/posts/2023-07-14_code_sections/code_sections.html
+++ b/posts/2023-07-14_code_sections/code_sections.html
@@ -294,7 +294,7 @@ Conclusion
Last updated
-2023-11-01 15:47:53.276819
+2023-11-27 19:13:41.390513
Details
diff --git a/posts/2023-07-20_cross_company_dev/cross_industry_dev.html b/posts/2023-07-20_cross_company_dev/cross_industry_dev.html
index c9ed3dcd..62f82d23 100644
--- a/posts/2023-07-20_cross_company_dev/cross_industry_dev.html
+++ b/posts/2023-07-20_cross_company_dev/cross_industry_dev.html
@@ -229,7 +229,7 @@ Insights
Development workflow
-Every improvement, task, or feature we want to implement on {admiral} starts as an issue on our GitHub repository. It is the centerpiece of our development workflow, along with our developer guides which describe in detail the strategies, conventions, and workflows used in development. The guides help us keep the {admiral} package internally consistent (e.g. naming conventions, function logic) but also ensure that {admiral} adjacent packages follow the same conventions and share the user interface. This is further helped by the implemented CICD pipeline which ensures styling convention and spelling (and much more).
+Every improvement, task, or feature we want to implement on {admiral} starts as an issue on our GitHub repository. It is the centerpiece of our development workflow, along with our developer guides which describe in detail the strategies, conventions, and workflows used in development. The guides help us keep the {admiral} package internally consistent (e.g. naming conventions, function logic) but also ensure that {admiral} adjacent packages follow the same conventions and share the user interface. This is further helped by the implemented CICD pipeline which ensures styling convention and spelling (and much more).
The core package developers team meets once a week (twice a week before a release) to discuss progress and priorities. Here, the role of product lead (currently Edoardo Mancini at Roche) and technical lead (currently Ben Straub at GSK) is to set priorities and track the release schedule. These stand-up meetings are centered around the project-board which gives a complete overview of activities and progress. Issues are mostly self-assigned so developers can really chose what they want to work on.
diff --git a/posts/2023-07-24_rounding/rounding.html b/posts/2023-07-24_rounding/rounding.html
index d1b059f0..54a2cb7e 100644
--- a/posts/2023-07-24_rounding/rounding.html
+++ b/posts/2023-07-24_rounding/rounding.html
@@ -467,7 +467,7 @@ Conclusion
Last updated
-2023-11-01 15:47:29.564969
+2023-11-27 19:13:33.556149
Details
diff --git a/posts/2023-08-08_study_day/study_day.html b/posts/2023-08-08_study_day/study_day.html
index 02f167ef..b54e7b65 100644
--- a/posts/2023-08-08_study_day/study_day.html
+++ b/posts/2023-08-08_study_day/study_day.html
@@ -264,7 +264,7 @@ It’s all relative? - Calculating Relative Days using admiral
Last updated
-2023-11-01 15:47:45.242711
+2023-11-27 19:13:57.651626
Details
diff --git a/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html b/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
index fc7f9afa..99f64d55 100644
--- a/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
+++ b/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
@@ -369,7 +369,7 @@ References
Last updated
-2023-11-01 15:47:31.231853
+2023-11-27 19:13:54.254558
Details
diff --git a/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html b/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
index 885d8118..068e0d1c 100644
--- a/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
+++ b/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
@@ -221,7 +221,7 @@ Release, g
Last updated
-2023-11-01 15:47:48.201548
+2023-11-27 19:13:49.940918
Details
diff --git a/posts/2023-10-30_floating_point/floating_point.html b/posts/2023-10-30_floating_point/floating_point.html
index 4b06317b..31c93ef3 100644
--- a/posts/2023-10-30_floating_point/floating_point.html
+++ b/posts/2023-10-30_floating_point/floating_point.html
@@ -107,7 +107,7 @@
-
+
@@ -304,8 +304,8 @@ Issues arising
reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
-
-
+
+
As you can see, the data-frame contains the column GRADE_CRITERIA_CODE which contains comparisons of floating point values. And there was a discrepancy of what Gordon expected to see, and how R actually computed the comparison initially:
@@ -563,8 +563,8 @@ Potential solutions reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
-
-
+
+
Conclusion
Last updated
-2023-11-01 15:47:35.340689
+2023-11-27 19:13:46.342818
Details
diff --git a/posts/2023-11-27_higher_order/admiral.png b/posts/2023-11-27_higher_order/admiral.png
new file mode 100644
index 00000000..cc69ba33
Binary files /dev/null and b/posts/2023-11-27_higher_order/admiral.png differ
diff --git a/posts/2023-11-27_higher_order/higher_order.html b/posts/2023-11-27_higher_order/higher_order.html
new file mode 100644
index 00000000..12fe765b
--- /dev/null
+++ b/posts/2023-11-27_higher_order/higher_order.html
@@ -0,0 +1,1048 @@
+
+
+
+
+
+
+
+
+
+
+
+
+Pharmaverse Blog - Believe in a higher order!
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
\ No newline at end of file
diff --git a/search.json b/search.json
index f352c848..d6425294 100644
--- a/search.json
+++ b/search.json
@@ -1,113 +1,302 @@
[
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html",
- "href": "posts/2023-07-14_code_sections/code_sections.html",
- "title": "How to use Code Sections",
+ "objectID": "posts/2023-08-08_study_day/study_day.html",
+ "href": "posts/2023-08-08_study_day/study_day.html",
+ "title": "It’s all relative? - Calculating Relative Days using admiral",
"section": "",
- "text": "The admiral package embraces a modular style of programming, where blocks of code are pieced together in sequence to create an ADaM dataset. However, with the well-documented advantages of the modular approach comes the recognition that scripts will on average be longer. As such, astute programmers working in RStudio are constantly on the lookout for quick ways to effectively navigate their scripts. Enter code sections!"
+ "text": "Creating --DY variables for your ADaMs is super easy using derive_vars_dy() from the admiral package.\nLet’s build some dummy data with 4 subjects, a start date/time for treatment (TRTSDTM), an analysis start date/time variable (ASTDTM) and an analysis end date variable (AENDT).\nlibrary(admiral)\nlibrary(lubridate)\nlibrary(dplyr)\n\nadam <- tribble(\n ~USUBJID, ~TRTSDTM, ~ASTDTM, ~AENDT,\n \"001\", \"2014-01-17T23:59:59\", \"2014-01-18T13:09:O9\", \"2014-01-20\",\n \"002\", \"2014-02-25T23:59:59\", \"2014-03-18T14:09:O9\", \"2014-03-24\",\n \"003\", \"2014-02-12T23:59:59\", \"2014-02-18T11:03:O9\", \"2014-04-17\",\n \"004\", \"2014-03-17T23:59:59\", \"2014-03-19T13:09:O9\", \"2014-05-04\"\n) %>%\n mutate(\n TRTSDTM = as_datetime(TRTSDTM),\n ASTDTM = as_datetime(ASTDTM),\n AENDT = ymd(AENDT)\n )\nOkay! Next we run our dataset through derive_vars_dy(), specifying:\nderive_vars_dy(\n adam,\n reference_date = TRTSDTM,\n source_vars = exprs(ASTDTM, AENDT)\n)\n\n# A tibble: 4 × 6\n USUBJID TRTSDTM ASTDTM AENDT ASTDY AENDY\n <chr> <dttm> <dttm> <date> <dbl> <dbl>\n1 001 2014-01-17 23:59:59 2014-01-18 13:09:09 2014-01-20 2 4\n2 002 2014-02-25 23:59:59 2014-03-18 14:09:09 2014-03-24 22 28\n3 003 2014-02-12 23:59:59 2014-02-18 11:03:09 2014-04-17 7 65\n4 004 2014-03-17 23:59:59 2014-03-19 13:09:09 2014-05-04 3 49\nThat’s it! We got both our ASTDY and AENDY variables in only a few short lines of code!\nWhat if I want my variables to have a different naming convention?\nEasy! In the source_vars argument if you want your variables to be called DEMOADY and DEMOEDY just do DEMOADY = ASTDTM and DEMOEDY = AENDT and derive_vars_dy() will do the rest!\nderive_vars_dy(\n adam,\n reference_date = TRTSDTM,\n source_vars = exprs(DEMOADY = ASTDTM, DEMOEDY = AENDT)\n)\n\n# A tibble: 4 × 6\n USUBJID TRTSDTM ASTDTM AENDT DEMOADY DEMOEDY\n <chr> <dttm> <dttm> <date> <dbl> <dbl>\n1 001 2014-01-17 23:59:59 2014-01-18 13:09:09 2014-01-20 2 4\n2 002 2014-02-25 23:59:59 2014-03-18 14:09:09 2014-03-24 22 28\n3 003 2014-02-12 23:59:59 2014-02-18 11:03:09 2014-04-17 7 65\n4 004 2014-03-17 23:59:59 2014-03-19 13:09:09 2014-05-04 3 49\nIf you want to get --DT or --DTM variables using admiral then check out derive_vars_dt() and derive_vars_dtm(). If things are messy in your data, e.g. partial dates, both functions have great imputation abilities, which we will cover in an upcoming blog post!"
},
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html#introduction",
- "href": "posts/2023-07-14_code_sections/code_sections.html#introduction",
- "title": "How to use Code Sections",
+ "objectID": "posts/2023-08-08_study_day/study_day.html#last-updated",
+ "href": "posts/2023-08-08_study_day/study_day.html#last-updated",
+ "title": "It’s all relative? - Calculating Relative Days using admiral",
+ "section": "Last updated",
+ "text": "Last updated\n\n2023-11-27 19:13:57.651626"
+ },
+ {
+ "objectID": "posts/2023-08-08_study_day/study_day.html#details",
+ "href": "posts/2023-08-08_study_day/study_day.html#details",
+ "title": "It’s all relative? - Calculating Relative Days using admiral",
+ "section": "Details",
+ "text": "Details\n\nsource code, R environment"
+ },
+ {
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
"section": "",
- "text": "The admiral package embraces a modular style of programming, where blocks of code are pieced together in sequence to create an ADaM dataset. However, with the well-documented advantages of the modular approach comes the recognition that scripts will on average be longer. As such, astute programmers working in RStudio are constantly on the lookout for quick ways to effectively navigate their scripts. Enter code sections!"
+ "text": "There is significant momentum in driving the adoption of R packages in the life sciences industries, in particular, the R Consortium Submissions Working Group is dedicated to promoting the use of R for regulatory submissions to the FDA.\nThe R Consortium Submissions Working Group successfully completed an R-based submission in November 2021 through the eCTD portal (R Submissions Pilot 1). This Pilot was completed on March 10, 2022 after a successful statistical review and evaluation by the FDA staff.\nMoving forward, the Pilot 2 aimed to include a Shiny application that the FDA staff could deploy on their own servers. The R Consortium recently announced that on September 27, 2023, the R Submissions Working Group successfully completed the follow-up to the Pilot 2 R Shiny-based submission and received a response letter from FDA CDER. This marks the first publicly available submission package that includes a Shiny component. The full FDA response letter can be found here.\nThe Shiny application that was sent for the Pilot 2 had the goal to display the 4 Tables, Listings and Figures (TLFs) that were sent for the Pilot 1 with basic filtering functionality.\nThe submission package adhered to the eCTD folder structure and contained 5 deliverables. Among the deliverables was the proprietary R package {pilot2wrappers}, which enables the execution of the Shiny application.\nThe FDA staff were expected to receive the electronic submission packages in the eCTD format, install and load open-source R packages used as dependencies in the included Shiny application, reconstruct and load the submitted Shiny application, and communicate potential improvements in writing.\nIn the following stage, the R Consortium’s R Submission Working Group launched Pilot 4, aiming to investigate innovative technologies like Linux containers and web assembly. These technologies are being explored to package a Shiny application into a self-contained unit, streamlining the transfer and execution processes for the application.\nIn this post, our aim is to outline how we used the Rhino framework to reproduce the Shiny application that was successfully submitted to the FDA for the Pilot 2 project. Additionally, we detail the challenges identified during the process and how we were able to successfully address them by using an open-source package."
},
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html#so-what-are-code-sections-and-why-are-they-useful",
- "href": "posts/2023-07-14_code_sections/code_sections.html#so-what-are-code-sections-and-why-are-they-useful",
- "title": "How to use Code Sections",
- "section": "So, what are code sections and why are they useful?",
- "text": "So, what are code sections and why are they useful?\nCode Sections are separators for long R scripts or functions in RStudio. They can be set up by inserting a comment line followed by four or more dashes in between portions of code, like so:\n\n# First code section ----\n\na <- 1\n\n# Second code section ----\n\nb <- 2\n\n# Third code section ----\n\nc <- 3\n\nRStudio then recognizes the code sections automatically, and enables you to:\n\nCollapse and expand them using the arrow displayed next to the line number, or with the handy shortcuts Alt+L/Shift+Alt+L on Windows or Cmd+Option+L/Cmd+Shift+Option+L on Mac.\nTravel in between them using the navigator at the bottom of the code pane, or by pressing Shift+Alt+J on Windows or Cmd+Shift+Option+J on Mac.\nView an outline of the file using the “Outline” button at the top right of the pane and/or the orange hashtag “Section Navigator” button at the bottom left of the pane.\n\n\n\n\n\n\nCollapsed sections, outline view and the section navigator for the example above.\n\n\n\n\nIt is also possible to create subsections by using two hashtags at the start of a comment line:\n\n# First code section ----\na <- 1\n\n## A code subsection ----\nb <- 2\n\n# Second code section ----\nc <- 3\n\n\n\n\n\n\nCode subsections for the example above.\n\n\n\n\nFor a complete list of Code Sections shortcuts, and for further information, see here."
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#reproducing-the-r-submission-pilot-2-shiny-app-using-rhino",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#reproducing-the-r-submission-pilot-2-shiny-app-using-rhino",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Reproducing the R Submission Pilot 2 Shiny App using Rhino",
+ "text": "Reproducing the R Submission Pilot 2 Shiny App using Rhino\nWhile the original Shiny application submitted to the FDA was wrapped using {Golem}, we replicated the application using our in-house developed framework Rhino. The main motivation was to provide an example of an R Submission that is not an R package and to identify and solve any issues that may arise from this approach.\nOur demo application (FDA-pilot-app) is accessible on our website, alongside other Shiny and Rhinoverse demonstration apps.\n\nThe code for FDA-pilot-app is open-source. You can create your own Rhino-based application by following our tutorial and viewing our workshop, which is available on YouTube."
},
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html#conclusion",
- "href": "posts/2023-07-14_code_sections/code_sections.html#conclusion",
- "title": "How to use Code Sections",
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#brief-introduction-to-rhino",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#brief-introduction-to-rhino",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Brief Introduction to Rhino",
+ "text": "Brief Introduction to Rhino\n\n\n\n\n\nThe Rhino framework was developed by Appsilon to create enterprise-level Shiny applications, consistently and efficiently. This framework allows developers to apply the best software engineering practices, modularize code, test it thoroughly, enhance UI aesthetics, and monitor user adoption effectively.\nRhino provides support in 3 main areas:\n\nClear Code: scalable architecture, modularization based on Box and Shiny modules.\nQuality: comprehensive testing such as unit tests, E2E tests with Cypress and Shinytest2, logging, monitoring and linting.\nAutomation: project startup, CI with GitHub Actions, dependencies management with {renv}, configuration management with config, Sass and JavaScript bundling with ES6 support via Node.js.\n\nRhino is an ideal fit for highly regulated environments such as regulatory submissions or other drug development processes.\n\nFDA-pilot-app structure\nThe structure of this application is available on the github repository. The structure of this Shiny app is the following.\n\nClick here to expand the FDA-pilot-app structure\n\n.\n├── app\n│ ├── view\n│ │ └── demographic_table.R\n| | └── km_plot.R\n| | └── primary_table.R\n| | └── efficacy_table.R\n| | └── completion_table.R\n│ ├── logic\n│ │ └── adam_data.R\n│ │ └── eff_modles.R\n│ │ └── formatters.R\n│ │ └── helpers.R\n│ │ └── kmplot_helpers.R\n│ │ └── Tplyr_helpers.R\n│ ├── data\n│ │ └── adam\n│ │ └── adadas.xpt\n│ │ └── adlbc.xpt\n│ │ └── adsl.xpt\n│ │ └── adtte.xpt\n│ ├── docs\n│ │ └── about.md\n│ ├── js\n│ │ └── index.js\n│ ├── static\n│ │ └── favicon.ico\n│ ├── styles\n│ │ └── main.scss\n│ └── app.R\n├── tests\n│ ├── cypress\n│ │ └── integration\n│ │ └── app.spec.js\n│ ├── testthat\n│ │\n│ └── cypress.json\n├── app.R\n├── rhino_submission.Rproj\n├── dependencies.R\n├── renv.lock\n├── rhino.yml\n└── README.md"
+ },
+ {
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#efficient-submissions-to-the-fda",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#efficient-submissions-to-the-fda",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Efficient Submissions to the FDA",
+ "text": "Efficient Submissions to the FDA\n\n\n\n\n\nTo comply with the Electronic Submission File Formats and Specifications for the eCTD submission, the programming code should carry a “.txt” extension. In the R Submissions Pilot 3 the group did not use {pkglite} as the FDA clarified that “.zip” and “.r” files are acceptable for submission. In this case, we utilized the {pkglite} R package to efficiently pack and unpack the FDA-pilot-app. This approach would facilitate the FDA reviewers in setting up the submission on their systems.\nThis package allows packing R packages to “.txt” files, which are supported for the submission of proprietary packages to the FDA via the eCTD gateway. \n\nPacking the FDA-pilot-app into a .txt file\nThe code below can be used to pack the Shiny application into a .txt file:\n\napp_name <- \"rhinosubmission\"\nrenv_spec <- pkglite::file_spec(\n \"renv\",\n pattern = \"^settings\\\\.dcf$|^activate\\\\.R$\",\n format = \"text\", recursive = FALSE\n)\ntests_spec <- pkglite::file_tests()\napp_spec <- pkglite::file_auto(\"app\")\nroot_spec <- pkglite::file_spec(\n path = \".\",\n pattern = \"^\\\\.Rprofile$|^rhino\\\\.yml$|^renv\\\\.lock$|^dependencies\\\\.R$|^config\\\\.yml$|^app\\\\.R$|^README\\\\.md$|\\\\.Rproj$\",\n all_files = TRUE,\n recursive = FALSE\n)\nwrite(paste0(\"Package: \", app_name), \"DESCRIPTION\", append = TRUE)\npkglite::collate(\n getwd(),\n renv_spec,\n tests_spec,\n app_spec,\n root_spec\n) |> pkglite::pack()\nfile.remove(\"DESCRIPTION\")\n\n\n\nUnpacking the FDA-pilot-app\nThe packed “.txt” file can be unpacked into a Shiny app by using {pkglite} as follows:\n\npkglite::unpack(\"rhinosubmission.txt\")"
+ },
+ {
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#lessons-learned",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#lessons-learned",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Lessons Learned",
+ "text": "Lessons Learned\nOur initial objective was to prove that it would be possible to submit a Shiny application using Rhino through the eCTD gateway. During the rewriting process we identified that this could be done by integrating the open-source {pkglite} package. By following this approach, we concluded that it would be possible to submit a Shiny application through the eCTD gateway. This was also achieved through the successful submission of a package that included a Shiny component in Pilot 2.\nHaving rewritten the R Submissions Pilot 2 Shiny application using Rhino holds major implications for the adoption of our framework within the life sciences. Apart from being a strong, opinionated framework that improves reproducibility and reliability for Shiny development, using Rhino for regulatory submissions could improve the flexibility and speed in the clinical reporting pipeline. This would accelerate the adoption of R/Shiny for submissions to the FDA or other regulatory agencies."
+ },
+ {
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#last-updated",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#last-updated",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Last updated",
+ "text": "Last updated\n\n2023-11-27 19:13:54.254558"
+ },
+ {
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#details",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#details",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Details",
+ "text": "Details\n\nsource code, R environment"
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html",
+ "title": "Blanks and NAs",
+ "section": "",
+ "text": "Reading in SAS-based datasets (.sas7bdat or xpt) into R has users calling the R package haven. A typical call might invoke read_sas() or read_xpt() to bring in your source data to construct your ADaMs or SDTMs.\nUnfortunately, while using haven the character blanks (missing data) found in a typical SAS-based dataset are left as blanks. These blanks will typically prove problematic while using functions like is.na in combination with dplyr::filter() to subset data. Check out Bayer’s SAS2R catalog: handling-of-missing-values for more discussion on missing values and NAs.\nIn the admiral package, we have built a simple function called convert_blanks_to_na() to help us quickly remedy this problem. You can supply an entire dataframe to this function and it will convert any character blanks to NA_character_"
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#loading-packages-and-making-dummy-data",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#loading-packages-and-making-dummy-data",
+ "title": "Blanks and NAs",
+ "section": "Loading Packages and Making Dummy Data",
+ "text": "Loading Packages and Making Dummy Data\n\nlibrary(admiral)\nlibrary(tibble)\nlibrary(dplyr)\n\ndf <- tribble(\n ~USUBJID, ~RFICDTC,\n \"01\", \"2000-01-01\",\n \"02\", \"2001-01-01\",\n \"03\", \"\", # Here we have a character blank\n \"04\", \"2001-01--\",\n \"05\", \"2001---01\",\n \"05\", \"\", # Here we have a character blank\n)\n\ndf\n\n# A tibble: 6 × 2\n USUBJID RFICDTC \n <chr> <chr> \n1 01 \"2000-01-01\"\n2 02 \"2001-01-01\"\n3 03 \"\" \n4 04 \"2001-01--\" \n5 05 \"2001---01\" \n6 05 \"\""
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#a-simple-conversion",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#a-simple-conversion",
+ "title": "Blanks and NAs",
+ "section": "A simple conversion",
+ "text": "A simple conversion\n\ndf_na <- convert_blanks_to_na(df)\n\ndf_na\n\n# A tibble: 6 × 2\n USUBJID RFICDTC \n <chr> <chr> \n1 01 2000-01-01\n2 02 2001-01-01\n3 03 <NA> \n4 04 2001-01-- \n5 05 2001---01 \n6 05 <NA> \n\n\n\ndf_na %>% filter(is.na(RFICDTC))\n\n# A tibble: 2 × 2\n USUBJID RFICDTC\n <chr> <chr> \n1 03 <NA> \n2 05 <NA>"
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#thats-it",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#thats-it",
+ "title": "Blanks and NAs",
+ "section": "That’s it!",
+ "text": "That’s it!\nA simple call to this function can make your derivation life so much easier while working in R if working with SAS-based datasets. In admiral, we make use of this function at the start of all ADaM templates for common ADaM datasets. You can use the function use_ad_template() to get the full R script for the below ADaMs.\n\nlist_all_templates()\n\nExisting ADaM templates in package 'admiral':\n• ADAE\n• ADCM\n• ADEG\n• ADEX\n• ADLB\n• ADLBHY\n• ADMH\n• ADPC\n• ADPP\n• ADPPK\n• ADSL\n• ADVS"
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#last-updated",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#last-updated",
+ "title": "Blanks and NAs",
+ "section": "Last updated",
+ "text": "Last updated\n\n2023-11-27 19:13:51.677601"
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#details",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#details",
+ "title": "Blanks and NAs",
+ "section": "Details",
+ "text": "Details\n\nsource code, R environment"
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html",
+ "href": "posts/2023-06-27__hackathon_app/index.html",
+ "title": "Hackathon Feedback Application",
+ "section": "",
+ "text": "We recently created a shiny application for the admiral hackathon in February 2023. The admiral hackathon was an event designed to make statistical programmers from the pharmaceutical industry more comfortable with the admiral R package which allows users to efficiently transform data from one data standard (SDTM) to another (ADaM).\nHackathon participants formed groups of up to five people and were then tasked to create R-scripts that map the SDTM data to ADaM according to specifics defined in the metadata.\nThe purpose of the shiny app was threefold:\nIn this blog post I want to highlight some of the thoughts that went into this application. Please keep in mind that this work was done under tight time restraints.\nThe hackathon application is still online (although data-upload is switched off) and the GitHub repository is publicly available. The application is embedded into this post right after this paragraph. I have also uploaded to GitHub a .zip file of the workspace to which hackathon participants had access via posit cloud. For more context you can watch recordings of the hackathon-meetings."
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#permanent-data",
+ "href": "posts/2023-06-27__hackathon_app/index.html#permanent-data",
+ "title": "Hackathon Feedback Application",
+ "section": "Permanent Data",
+ "text": "Permanent Data\nThe biggest challenge you have to consider for this app is the permanent data storage. Shiny apps run on a server. Although we can write files on this server, whenever the app restarts, the files are lost. Therefore, a persistent data storage solution is required.\n\nGoogle drive\nI decided to leverage Google drive using the googledrive package package. This allowed me to save structured data (the team registry and the submission scores) as well as unstructured data (their R-script files).\n\n\n\n\n\n\nAuthentication\n\n\n\nTo access Google drive using the googledrive package we need to authenticate. This can be done interactively using the command googledrive::drive_auth() which takes you to the Google login page. After login you receive an authentication token requested by R.\nFor non-interactive authentication this token must be stored locally. In our case where the shiny app must access the token once deployed, the token must be stored on the project level.\nI have included the authentication procedure I followed in the R folder in google_init.R. You can find more extensive documentation of the non-interactive authentication.\n\n\nThe initial concept was: Each team gets their own folder including the most recent submission for each task, and a .csv file containing team information. To keep track of the submissions and the respective scores we wrote a .csv file in the mock-hackathon folder, so one folder above the team folders.\nSaving the team info as a .csv file worked fine as each team received their own file which – once created – was not touched anymore. As each upload for every team should simply add a row to the submissions.csv file, appending the file would be ideal. This was not possible using the googledrive package package. Instead, for each submission, the submissions file was downloaded, appended, and uploaded again. Unfortunately, this lead to a data loss, as the file was continuously overwritten, especially when two teams would submit simultaneously.\n\n\n\n\n\n\nRecover the Lost Data\n\n\n\nWhenever the submissions.csv file was uploaded, the previous version was sent to the Google drive bin. We ended up with over 3000 submissions.csv files containing a lot of redundant information. I had to write the following chunk to first get the unique file IDs of the 3000 submissions.csv files, create an empty submissions data-frame, and then download each file and add its information to the submisisons data-frame. To keep the data-frame as light as possible, after each append I deleted all duplicate submissions.\n\n# get all task_info.csv ID's\n# each row identifies one file in the trash\ntask_info_master <- drive_find(\n pattern = \"task_info.csv\",\n trashed = TRUE\n)\n\n\n# set up empty df to store all submissions\norigin <- tibble(\n score = numeric(),\n task = character(),\n team = character(),\n email = character(),\n time = character()\n)\n\n# downloads, reads, and returns one csv file given a file id\nget_file <- function(row) {\n tf <- tempfile()\n row %>%\n as_id() %>%\n drive_download(path = tf)\n new <- read_csv(tf) %>%\n select(score, task, team) %>%\n distinct()\n}\n\n\n# quick and dirty for loop to subsequently download each file, extract information\n# merge with previous information and squash it (using distinct()).\nfor (i in 1:nrow(task_info_master)) {\n origin <- rbind(origin, get_file(row = task_info_master[i, ])) %>%\n distinct()\n\n # save progress in a separate file after every 100 downloaded and merged sheets\n if (i %% 100 == 0) {\n print(i)\n write_csv(origin, paste(\"prog_data/task_info_prog_\", i, \".csv\", sep = \"\"))\n # update on progress\n message(i / nrow(task_info_master) * 100)\n }\n}\n\nWhen doing such a time-intensive task, make sure to try it first with only a couple of files to see whether any errors are produced. I am not quite sure how long this took but when I returned from my lunch break everything had finished.\n\n\nIf you want to stay in the Google framework, I recommend using the googlesheets4 package for structured data. googlesheets4 allows appending new information to an already existing sheet without the need to download the file first. As both packages follow the same style, going from one to the other is really simple. googlesheets4 requires authentication as well. However, you can reuse the cached token from the googledrive package authentication by setting gs4_auth(token = drive_token()).\n\n\nSecurity Concerns\nConnecting a public shiny app to your Google account introduces a security vulnerability in general. Especially so because we implemented the upload of files to Google drive. And even more problematic: We run a user generated script and display some of its output. A malicious party might be able to extract the authentication token of our Google account or could upload malware to the drive.\nTo reduce the risk, I simply created an un-associated Google account to host the drive. There are certainly better options available, but this seemed a reasonable solution achieved with very little effort."
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#register-team",
+ "href": "posts/2023-06-27__hackathon_app/index.html#register-team",
+ "title": "Hackathon Feedback Application",
+ "section": "Register Team",
+ "text": "Register Team\nWe wanted to allow users to sign up as teams using the shiny app. The app provides a simple interface where users could input a team name and the number of members. This in turn would open two fields for each user to input their name and email address.\nWe do simple checks to make sure at least one valid email address is supplied, and that the group name is acceptable. The group name cannot be empty, already taken, or contain vulgar words.\nThe team registration itself was adding the team information to the Google sheets file event_info into the sheet teams and to create a team folder in which to store the uploaded R files.\nThe checks and registration is implemented in the register_team() function stored in interact_with_google.R.\n\n\n\nScreenshot of the register team interface\n\n\nThe challenge here was to adapt the number of input fields depending on the number of team members. This means that the team name and email interface must be rendered: First, we check how many team members are part of the group, this is stored in the input$n_members input variable. Then we create a tagList with as many elements as team members. Each element contains two columns, one for the email, one for the member name. This tagList is then returned and displayed to the user.\n\n# render email input UI of the register tab\noutput$name_email <- shiny::renderUI({\n # create field names\n N <- input$n_members\n NAME <- sapply(1:N, function(i) {\n paste0(\"name\", i)\n })\n EMAIL <- sapply(1:N, function(i) {\n paste0(\"email\", i)\n })\n\n output <- tagList()\n\n\n firstsecondthird <- c(\"First\", \"Second\", \"Third\", \"Fourth\", \"Fifth\")\n for (i in 1:N) {\n output[[i]] <- tagList()\n output[[i]] <- fluidRow(\n shiny::h4(paste(firstsecondthird[i], \" Member\")),\n column(6,\n textInput(NAME[i], \"Name\"),\n value = \" \" # displayed default value\n ),\n column(6,\n textInput(EMAIL[i], \"Email\"),\n value = \" \"\n )\n )\n }\n output\n})\n\nThe team information is then uploaded to Google drive. Because some teams have more members than others, we have to create the respective data-frame with the number of team members in mind.\nThe following chunk creates the registration data. Noteworthy here the creation of the NAME and EMAIL variables which depend on the number of members in this team. Further, the user input of these fields is extracted via input[[paste0(NAME[i])]] within a for-loop.\nWe also make the data-creation dependent on the press of the Register Group button and cache some variables.\n\n## registration\nregistrationData <-\n reactive({\n N <- input$n_members\n NAME <- sapply(1:N, function(i) {\n paste0(\"name\", i)\n })\n EMAIL <- sapply(1:N, function(i) {\n paste0(\"email\", i)\n })\n names <- character(0)\n emails <- character(0)\n\n for (i in 1:N) {\n names[i] <- input[[paste0(NAME[i])]]\n emails[i] <- input[[paste0(EMAIL[i])]]\n }\n # create df\n dplyr::tibble(\n team_name = input$team_name,\n n_members = N,\n member_name = names,\n member_email = emails\n )\n }) %>%\n bindCache(input$team_name, input$n_members, input$name1, input$email1) %>%\n bindEvent(input$register) # wait for button press"
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#upload-source-script",
+ "href": "posts/2023-06-27__hackathon_app/index.html#upload-source-script",
+ "title": "Hackathon Feedback Application",
+ "section": "Upload & Source Script",
+ "text": "Upload & Source Script\nTo upload a script, participants had to select their team first. The input options were based on the existing folders on the Google-drive in the mock_hackathon folder. To upload a particular script participants had to also select the task to be solved. The uploaded script is then uploaded to the team folder following a standardised script naming convention.\nThere are different aspects to be aware of when sourcing scripts on a shiny server. For example, you have to anticipate the packages users will include in their uploaded scripts, as their scripts will load but not install packages. Further, you should keep the global environment of your shiny app separate from the environment in which the script is sourced. This is possible by supplying an environment to the source() function, e.g: source(path_to_script, local = new.env())\nAnother thing we had to consider was to replicate the exact folder-structure on the shiny server that participants were working with when creating the scripts, as they were required to source some scripts and to save their file into a specific folder. This was relatively straight forward as we provided participants with a folder structure in the posit cloud instance they were using. They had access to the sdtm folder in which the data was stored, and the adam folder into which they saved their solutions. The structure also included a folder with metadata which was also available on the shiny server.\nFor some tasks, participants required some ADaM-datasets stored in the adam folder, essentially the output from previous tasks. This was achieved by first creating a list mapping tasks to the required ADaM datasets:\n\ndepends_list <- list(\n \"ADADAS\" = c(\"ADSL\"),\n \"ADAE\" = c(\"ADSL\"),\n \"ADLBC\" = c(\"ADSL\"),\n \"ADLBH\" = c(\"ADSL\"),\n \"ADLBHY\" = c(\"ADSL\"),\n \"ADSL\" = NULL,\n \"ADTTE\" = c(\"ADSL\", \"ADAE\"),\n \"ADVS\" = c(\"ADSL\")\n)\n\nThis list is sourced from the R/parameters.R file when initiating the application. We then call the get_depends() function sourced from R/get_depends.R which copies the required files from the key folder (where our solutions to the tasks were stored) to the adam folder. After sourcing the uploaded script the content in the adam folder is deleted."
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#compare-to-solution-file",
+ "href": "posts/2023-06-27__hackathon_app/index.html#compare-to-solution-file",
+ "title": "Hackathon Feedback Application",
+ "section": "Compare to Solution File",
+ "text": "Compare to Solution File\nWe want to compare the file created by participants with our solution (key) file stored in the key folder. The diffdf::diffdf() function allows for easy comparison of two data-frames and directly provides extensive feedback for the user:\n\nlibrary(dplyr)\ndf1 <- tibble(\n numbers = 1:10,\n letters = LETTERS[1:10]\n)\ndf2 <- tibble(\n numbers = 1:10,\n letters = letters[1:10]\n)\n\ndiffdf::diffdf(df1, df2)\n\nWarning in diffdf::diffdf(df1, df2): \nNot all Values Compared Equal\n\n\nDifferences found between the objects!\n\nA summary is given below.\n\nNot all Values Compared Equal\nAll rows are shown in table below\n\n =============================\n Variable No of Differences \n -----------------------------\n letters 10 \n -----------------------------\n\n\nAll rows are shown in table below\n\n ========================================\n VARIABLE ..ROWNUMBER.. BASE COMPARE \n ----------------------------------------\n letters 1 A a \n letters 2 B b \n letters 3 C c \n letters 4 D d \n letters 5 E e \n letters 6 F f \n letters 7 G g \n letters 8 H h \n letters 9 I i \n letters 10 J j \n ----------------------------------------"
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#score",
+ "href": "posts/2023-06-27__hackathon_app/index.html#score",
+ "title": "Hackathon Feedback Application",
+ "section": "Score",
+ "text": "Score\nTo compare submissions between participants we implemented a simple scoring function (score_f()) based on the table comparison by diffdf(). The function can be found in the compare_dfs.R file:\n\nscore_f <- function(df_user, df_key, keys) {\n score <- 10\n diff <- diffdf::diffdf(df_user, df_key, keys = keys)\n if (!diffdf::diffdf_has_issues(diff)) {\n return(score)\n }\n\n # check if there are any differences if the comparison is not strict:\n if (!diffdf::diffdf_has_issues(diffdf::diffdf(df_user,\n df_key,\n keys = keys,\n strict_numeric = FALSE,\n strict_factor = FALSE\n ))) {\n # if differences are not strict, return score - 1\n return(score - 1)\n }\n\n return(round(min(max(score - length(diff) / 3, 1), 9), 2))\n}\n\nEvery comparison starts with a score of 10. We then subtract the length of the comparison object divided by a factor of 3. The length of the comparison object is a simplified way to represent the difference between the two data-frames by one value. Finally, the score is bounded by 1 using max(score, 1).\nThe score is not a perfect capture of the quality of the script uploaded but: 1. helped participants get an idea of how close their data-frame is to the solution file 2. allowed us to raffle prizes based on the merit of submitted r-scripts"
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#reactiveness",
+ "href": "posts/2023-06-27__hackathon_app/index.html#reactiveness",
+ "title": "Hackathon Feedback Application",
+ "section": "Reactiveness",
+ "text": "Reactiveness\nSome of the app functions can take quite some time to execute, e.g. running the uploaded script. Other tasks, e.g. registering a team, do not intrinsically generate user facing outputs. This would make the app using really frustrating, as users would not know whether the app is correctly working or whether it froze.\nWe implemented two small features that made the app more responsive. One is simple loading icons that integrate into the user interface and show that output is being computed – that something is working. The other is a pop up window which communicates whether team registration was successful, and if not, why not.\nWe further aimed to forward errors generated by the uploaded scripts to the user interface, but errors generated by the application itself should be concealed."
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#conclusion",
+ "href": "posts/2023-06-27__hackathon_app/index.html#conclusion",
+ "title": "Hackathon Feedback Application",
"section": "Conclusion",
- "text": "Conclusion\nCode sections are an easy way to navigate long scripts and foster good commenting practices. They are used extensively in the admiral package, but there is no reason that you cannot start using them yourself in your day-to-day R programming!"
+ "text": "Conclusion\nAlthough the application was continuously improved during the hackathon it proved to be a useful resource for participants from day one as it allowed groups to set their own pace. It further allowed admiral developers to gain insights on package usage of a relatively large sample of potential end users. From our perspective, the application provided a great added value to the hackathon and eased the workload of guiding the participants through all the tasks."
},
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html#last-updated",
- "href": "posts/2023-07-14_code_sections/code_sections.html#last-updated",
- "title": "How to use Code Sections",
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#last-updated",
+ "href": "posts/2023-06-27__hackathon_app/index.html#last-updated",
+ "title": "Hackathon Feedback Application",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:53.276819"
+ "text": "Last updated\n\n2023-11-27 19:13:48.333469"
},
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html#details",
- "href": "posts/2023-07-14_code_sections/code_sections.html#details",
- "title": "How to use Code Sections",
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#details",
+ "href": "posts/2023-06-27__hackathon_app/index.html#details",
+ "title": "Hackathon Feedback Application",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html",
- "title": "The pharmaverse (hi)story",
+ "objectID": "posts/2023-11-27_higher_order/higher_order.html",
+ "href": "posts/2023-11-27_higher_order/higher_order.html",
+ "title": "Believe in a higher order!",
"section": "",
- "text": "Pharmaverse: from motivation to present"
+ "text": "Picture the following scenario:\nYou, a budding {admiral} programmer, are finding your groove chaining together modular code blocks to derive variables and parameters in a drive to construct your favorite ADaM dataset, ADAE. Suddenly you notice that one of the flags you are deriving should only use records on or after study day 1. In a moment of mild annoyance, you get to work modifying what was originally a simple call to derive_var_extreme_flag() by first subsetting ADAE to records where AESTDY > 1, then deriving the flag only for the subsetted ADAE, and finally binding the two portions of ADAE back together before continuing on with your program. Miffed by this interruption, you think to yourself: “I wish there was a neater, faster way to do this in stride, that didn’t break my code modularity…”\nIf the above could never be you, then you’ll probably be alright never reading this blog post. However, if you want to learn more about the tools that {admiral} provides to make your life easier in cases like this one, then you are in the right place, since this blog post will highlight how higher order functions can solve such issues.\nA higher order function is a function that takes another function as input. By introducing these higher order functions, {admiral} intends to give the user greater power over derivations, whilst trying to negate the need for both adding additional {admiral} functions/arguments, and the user needing many separate steps.\nThe functions covered in this post are:\n\nrestrict_derivation(): Allows the user to execute a single derivation on a subset of the input dataset.\ncall_derivation(): Allows the user to call a single derivation multiple times with some arguments being fixed across iterations and others varying.\nslice_derivation(): Allows the user to split the input dataset into slices (subsets) and for each slice a single derivation is called separately. Some or all arguments of the derivation may vary depending on the slice.\n\n\n\nThe examples in this blog post require the following packages.\n\nlibrary(admiral)\nlibrary(pharmaversesdtm)\nlibrary(dplyr, warn.conflicts = FALSE)\n\nFor example purpose, the ADSL dataset - which is included in {admiral} - and the SDTM datasets from {pharmaversesdtm} are used.\n\ndata(\"admiral_adsl\")\ndata(\"ae\")\ndata(\"vs\")\nadsl <- admiral_adsl\nae <- convert_blanks_to_na(ae)\nvs <- convert_blanks_to_na(vs)\n\nThe following code creates a minimally viable ADAE dataset to be used where needed in the following examples.\n\nadae <- ae %>%\n left_join(adsl, by = c(\"STUDYID\", \"USUBJID\")) %>%\n derive_vars_dt(\n new_vars_prefix = \"AST\",\n dtc = AESTDTC,\n highest_imputation = \"M\"\n ) %>%\n mutate(\n TRTEMFL = if_else(ASTDT >= TRTSDT, \"Y\", NA_character_),\n TEMP_AESEVN = as.integer(factor(AESEV, levels = c(\"SEVERE\", \"MODERATE\", \"MILD\")))\n )"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#human-history-and-pharmaverse-context",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#human-history-and-pharmaverse-context",
- "title": "The pharmaverse (hi)story",
- "section": "Human history and pharmaverse context",
- "text": "Human history and pharmaverse context\nSince the Australian Aboriginal, the earliest peoples recorded to have inhabited the Earth and who have been in Australia for at least 65,000 to 80,000 years (Encyclopædia Britannica), human beings live in group. Whether to protect yourself, increase your life expectancy or simply share tasks.\nFor most aspects of life, it doesn’t make sense to think, act or work alone for two main reasons:\n\nYou will spend more energy and time;\nSomeone else may be facing (or have faced) the same situation.\n\nThe English poet John Donne used to say “No man is an island entire of itself; every man is a piece of the continent, a part of the main;”. I can’t disagree with him. And I dare say that Ari Siggaard Knoph (Novo Nordisk), Michael Rimler (GSK), Michael Stackhouse (Atorus), Ross Farrugia (Roche), and Sumesh Kalappurakal (Janssen) can’t disagree with him either. They are the founders of pharmaverse, members of its Council and kindly shared their memories of how independent companies, in mid-2020, worked together in the creation of a set of packages developed to support the clinical reporting pipeline.\nIf you are not familiar with this pipeline, the important thing to know is that, in a nutshell, pharmaceutical companies must follow a bunch of standardized procedures and formats (from Clinical Data Interchange Standards Consortium, CDISC) when submitting clinical results to Health Authorities. The focus is on this: different companies seeking the same standards for outputs.\nParaphrasing Ross Farrugia (Roche) Breaking boundaries through open-source collaboration presentation in R/Pharma 2022 and thinking of the development of a new drug, we are talking about a “post-competitive” scenario: the drug has already been discovered and the companies should “just” produce and deliver standardized results."
+ "objectID": "posts/2023-11-27_higher_order/higher_order.html#required-packages",
+ "href": "posts/2023-11-27_higher_order/higher_order.html#required-packages",
+ "title": "Believe in a higher order!",
+ "section": "",
+ "text": "The examples in this blog post require the following packages.\n\nlibrary(admiral)\nlibrary(pharmaversesdtm)\nlibrary(dplyr, warn.conflicts = FALSE)\n\nFor example purpose, the ADSL dataset - which is included in {admiral} - and the SDTM datasets from {pharmaversesdtm} are used.\n\ndata(\"admiral_adsl\")\ndata(\"ae\")\ndata(\"vs\")\nadsl <- admiral_adsl\nae <- convert_blanks_to_na(ae)\nvs <- convert_blanks_to_na(vs)\n\nThe following code creates a minimally viable ADAE dataset to be used where needed in the following examples.\n\nadae <- ae %>%\n left_join(adsl, by = c(\"STUDYID\", \"USUBJID\")) %>%\n derive_vars_dt(\n new_vars_prefix = \"AST\",\n dtc = AESTDTC,\n highest_imputation = \"M\"\n ) %>%\n mutate(\n TRTEMFL = if_else(ASTDT >= TRTSDT, \"Y\", NA_character_),\n TEMP_AESEVN = as.integer(factor(AESEV, levels = c(\"SEVERE\", \"MODERATE\", \"MILD\")))\n )"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#clinical-reporting-outputs",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#clinical-reporting-outputs",
- "title": "The pharmaverse (hi)story",
- "section": "Clinical reporting outputs",
- "text": "Clinical reporting outputs\nRationally, we can say that companies face the same challenges in these steps of the process. Not so intuitively, we can also say they were working in silos on that before 2018. Just as Isaac Newton and Gottfried W. Leibniz independently developed the theory of infinitesimal calculus, pharmaceutical companies were independently working on R solutions for this pipeline.\nBut on August 16 and 17 of the mentioned year above, they gathered at the first edition of R/Pharma conference to discuss R and open-source tooling for drug development (the reasons why open-source is an advantageous approach can be found in this post written by Stefan Thoma). And according to Isabela Velásquez’s article, Pharmaverse: Packages for clinical reporting workflows, one of the most popular questions in this conference was “Is the package code available or on CRAN?”.\nWell, many of them were. And not necessarily at that date, but just to mention a few: pharmaRTF and Tplyr from Atorus, r2rtf from Merck, rtables from Roche, etc. The thing is that, overall, there were almost 10000 other packages as well (today, almost 20000). And that took to another two questions:\n\nWith this overwhelming number of packages on CRAN, how to find the ones related to solving “clinical reporting problems”?\nOnce the packages were found, how to choose which one to use among those that have the same functional purpose?\n\nSo, again, companies re-started to working in silos to find those answers. But now, in collaborative silos and with common goals: create extremely useful packages to solve pharmaceutical-specific gaps once and solve them well!"
+ "objectID": "posts/2023-11-27_higher_order/higher_order.html#last-updated",
+ "href": "posts/2023-11-27_higher_order/higher_order.html#last-updated",
+ "title": "Believe in a higher order!",
+ "section": "Last updated",
+ "text": "Last updated\n\n2023-11-27 19:13:43.938122"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#first-partnerships",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#first-partnerships",
- "title": "The pharmaverse (hi)story",
- "section": "First partnerships",
- "text": "First partnerships\nIn 2020, Michael Stackhouse (Atorus) and Michael Rimler (GSK) talked and formed a partnership between their companies to develop a few more packages, including metacore, to read, store and manipulate metadata for ADaMs/SDTMs in a standardized object; xportr, to create submission compliant SAS transport files and perform pharma specific dataset level validation checks; and logrx (ex-timber), to build log to support reproducibility and traceability of an R script.\nAround the same time, Thomas Neitmann (currently at Denali Therapeutics, then at Roche) and Michael Rimler (GSK) discovered that both were working with ADaM in R, so Thomas Neitmann (currently at Denali Therapeutics, then at Roche), Ross Farrugia (Roche) and Michael Rimler (GSK) saw an opportunity there and GSK started their partnership with Roche to build and release admiral package.\nThe idea of working together, the sense of community, and the appetite from organizations built more and more, with incentive and priority established up into the programming heads council.\nJanssen had a huge effort in building R capabilities going on as well, by releasing tidytlg and envsetup), so eventually Michael Rimler (GSK), Michael Stackhouse (Atorus) and Ross Farrugia (Roche) formalized pharmaverse and formed the council, adding in Sumesh Kalappurakal (Janssen) and Ari Siggaard Knoph (Novo Nordisk) later joined as the fifth council member."
+ "objectID": "posts/2023-11-27_higher_order/higher_order.html#details",
+ "href": "posts/2023-11-27_higher_order/higher_order.html#details",
+ "title": "Believe in a higher order!",
+ "section": "Details",
+ "text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#release-growth-and-developments",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#release-growth-and-developments",
- "title": "The pharmaverse (hi)story",
- "section": "Release, growth and developments",
- "text": "Release, growth and developments\nAt the end of their presentation “Closing the Gap: Creating an End to End R Package Toolkit for the Clinical Reporting Pipeline.”, in R/Pharma 2021, Ben Straub (GSK) and Eli Miller (Atorus) welcomed the community to the pharmaverse, a curated collection of packages developed by various pharmaceutical companies to support open-source clinical workflows.\nFrom the outset, the name pharmaverse was chosen so that it could be a neutral home, unrelated to any company. Also, it was established as not being a consortium, which means that founders don’t own, fund, or maintain the packages. Some individuals and companies maintain them but often allowing for community contributions and being licensed permissively so that there is always a feeling of community ownership. The focus of pharmaverse early on, and today, is on inter organization cooperation, to build an environment where, if organizations identify that they have a joint problem that they want to solve, this is the right space to work on and release it.\nPharmaverse has grown a lot, at the time of writing this post we have >25 packages recommended in pharmaverse, and this has led to a partnership with PHUSE to get support from their organization and platform, and because they are eager to advance and support pharmaverse mission.\nDespite all its structure, it is impossible to say that we have a single solution for each clinical reporting analysis when it comes to pharmaverse, a single pathway is impractical. Instead, it is necessary to accept viable tools fitting different pathways into pharmaverse to direct and give people options as to what might work for them. After all, even though we live together as a community, we still have our own unique internal problems.\n\n\n\nSample of pharmaverse packages"
+ "objectID": "posts/2023-07-09_falcon/falcon.html",
+ "href": "posts/2023-07-09_falcon/falcon.html",
+ "title": "falcon",
+ "section": "",
+ "text": "The {falcon} initiative is an industry collaborative effort under {pharmaverse} that unites Boehringer Ingelheim, Moderna, Roche, and Sanofi with the aspiration of building and open-sourcing a comprehensive catalog of harmonized tables, listings, and graphs (TLGs) for clinical study reporting. By leveraging existing open-source R packages, {falcon} aims to simplify the process of output review, comparison, and meta-analyses, while fostering efficient communication among stakeholders in the pharmaceutical industry."
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#last-updated",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#last-updated",
- "title": "The pharmaverse (hi)story",
+ "objectID": "posts/2023-07-09_falcon/falcon.html#what-is-falcon",
+ "href": "posts/2023-07-09_falcon/falcon.html#what-is-falcon",
+ "title": "falcon",
+ "section": "",
+ "text": "The {falcon} initiative is an industry collaborative effort under {pharmaverse} that unites Boehringer Ingelheim, Moderna, Roche, and Sanofi with the aspiration of building and open-sourcing a comprehensive catalog of harmonized tables, listings, and graphs (TLGs) for clinical study reporting. By leveraging existing open-source R packages, {falcon} aims to simplify the process of output review, comparison, and meta-analyses, while fostering efficient communication among stakeholders in the pharmaceutical industry."
+ },
+ {
+ "objectID": "posts/2023-07-09_falcon/falcon.html#why-do-we-build-it",
+ "href": "posts/2023-07-09_falcon/falcon.html#why-do-we-build-it",
+ "title": "falcon",
+ "section": "Why do we build it?",
+ "text": "Why do we build it?\nThe collaborative effort focuses on improving the clarity, consistency, and accessibility of TLGs by addressing variations and redundancies in their creation and use. This harmonized approach allows for streamlined reporting processes and facilitates effective communication of study results within the industry and to regulatory authorities."
+ },
+ {
+ "objectID": "posts/2023-07-09_falcon/falcon.html#what-has-been-done-so-far",
+ "href": "posts/2023-07-09_falcon/falcon.html#what-has-been-done-so-far",
+ "title": "falcon",
+ "section": "What has been done so far?",
+ "text": "What has been done so far?\nDrawing inspiration from the FDA Standard Safety Tables and Figures Integrated Guide, the {falcon} initiative has successfully developed and open-sourced 11 templates to date. 4 product owners and 11 developers from 4 companies have collaborated to make these templates available and also published them on the official {falcon} website at https://pharmaverse.github.io/falcon/."
+ },
+ {
+ "objectID": "posts/2023-07-09_falcon/falcon.html#next-steps-vision",
+ "href": "posts/2023-07-09_falcon/falcon.html#next-steps-vision",
+ "title": "falcon",
+ "section": "Next steps & vision",
+ "text": "Next steps & vision\nFuture plans for {falcon} involve expanding the catalog through continuous collaboration from participating companies and inviting wider industry engagement. The ultimate goal is to promote harmonization of TLGs for clinical reporting across the pharmaceutical industry, leading to greater efficiency, collaboration, and innovation. Even though templates currently come from a published FDA guide, the collaborating companies are open to share and discuss similarities and differences on analysis concepts and output layouts of their own implementations in clinical reporting, for both safety and efficacy analyses.\nIn addition, while currently all templates were built using {rtables}, {tern}, {rlistings} and drew inspiration from the open-sourced TLG-Catalog, moving forward, we plan to support creating the same templates using alternative table engines such as {gt}.\n{falcon} will be presented at the upcoming PHUSE EU (Standards Implementation stream), where we will share the collaboration journey of {falcon} so far, providing more details on the current progress, long-term vision, and strategies for this initiative. Attendees will gain insights into the challenges and opportunities of harmonizing clinical reporting through open-source collaboration and learn about the potential benefits and future direction of {falcon}."
+ },
+ {
+ "objectID": "posts/2023-07-09_falcon/falcon.html#last-updated",
+ "href": "posts/2023-07-09_falcon/falcon.html#last-updated",
+ "title": "falcon",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:48.201548"
+ "text": "Last updated\n\n2023-11-27 19:13:40.019302"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#details",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#details",
- "title": "The pharmaverse (hi)story",
+ "objectID": "posts/2023-07-09_falcon/falcon.html#details",
+ "href": "posts/2023-07-09_falcon/falcon.html#details",
+ "title": "falcon",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-08-08_study_day/study_day.html",
- "href": "posts/2023-08-08_study_day/study_day.html",
- "title": "It’s all relative? - Calculating Relative Days using admiral",
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html",
+ "title": "Admiral Hackathon 2023 Revisited",
"section": "",
- "text": "Creating --DY variables for your ADaMs is super easy using derive_vars_dy() from the admiral package.\nLet’s build some dummy data with 4 subjects, a start date/time for treatment (TRTSDTM), an analysis start date/time variable (ASTDTM) and an analysis end date variable (AENDT).\nlibrary(admiral)\nlibrary(lubridate)\nlibrary(dplyr)\n\nadam <- tribble(\n ~USUBJID, ~TRTSDTM, ~ASTDTM, ~AENDT,\n \"001\", \"2014-01-17T23:59:59\", \"2014-01-18T13:09:O9\", \"2014-01-20\",\n \"002\", \"2014-02-25T23:59:59\", \"2014-03-18T14:09:O9\", \"2014-03-24\",\n \"003\", \"2014-02-12T23:59:59\", \"2014-02-18T11:03:O9\", \"2014-04-17\",\n \"004\", \"2014-03-17T23:59:59\", \"2014-03-19T13:09:O9\", \"2014-05-04\"\n) %>%\n mutate(\n TRTSDTM = as_datetime(TRTSDTM),\n ASTDTM = as_datetime(ASTDTM),\n AENDT = ymd(AENDT)\n )\nOkay! Next we run our dataset through derive_vars_dy(), specifying:\nderive_vars_dy(\n adam,\n reference_date = TRTSDTM,\n source_vars = exprs(ASTDTM, AENDT)\n)\n\n# A tibble: 4 × 6\n USUBJID TRTSDTM ASTDTM AENDT ASTDY AENDY\n <chr> <dttm> <dttm> <date> <dbl> <dbl>\n1 001 2014-01-17 23:59:59 2014-01-18 13:09:09 2014-01-20 2 4\n2 002 2014-02-25 23:59:59 2014-03-18 14:09:09 2014-03-24 22 28\n3 003 2014-02-12 23:59:59 2014-02-18 11:03:09 2014-04-17 7 65\n4 004 2014-03-17 23:59:59 2014-03-19 13:09:09 2014-05-04 3 49\nThat’s it! We got both our ASTDY and AENDY variables in only a few short lines of code!\nWhat if I want my variables to have a different naming convention?\nEasy! In the source_vars argument if you want your variables to be called DEMOADY and DEMOEDY just do DEMOADY = ASTDTM and DEMOEDY = AENDT and derive_vars_dy() will do the rest!\nderive_vars_dy(\n adam,\n reference_date = TRTSDTM,\n source_vars = exprs(DEMOADY = ASTDTM, DEMOEDY = AENDT)\n)\n\n# A tibble: 4 × 6\n USUBJID TRTSDTM ASTDTM AENDT DEMOADY DEMOEDY\n <chr> <dttm> <dttm> <date> <dbl> <dbl>\n1 001 2014-01-17 23:59:59 2014-01-18 13:09:09 2014-01-20 2 4\n2 002 2014-02-25 23:59:59 2014-03-18 14:09:09 2014-03-24 22 28\n3 003 2014-02-12 23:59:59 2014-02-18 11:03:09 2014-04-17 7 65\n4 004 2014-03-17 23:59:59 2014-03-19 13:09:09 2014-05-04 3 49\nIf you want to get --DT or --DTM variables using admiral then check out derive_vars_dt() and derive_vars_dtm(). If things are messy in your data, e.g. partial dates, both functions have great imputation abilities, which we will cover in an upcoming blog post!"
+ "text": "This January and February (2023), the admiral development team and the CDISC Open Source Alliance jointly hosted the admiral hackathon. The idea was to build a community of admiral users, and help participants familiarize themselves with R and admiral. This whole effort was led by Thomas Neitmann and was supported by Zelos Zhu, Sadchla Mascary, and me – Stefan Thoma.\nThe hackathon event was structured in two parts. First, we offered an Introduction to R for SAS programmers, a three hour workshop for R beginners to get them up to speed. Here we covered practical R basics, talking about how the R-workflow differs from a SAS workflow, and discussed common R functions - mostly from the tidyverse. This ensured that hackathon participants were familiar with core R concepts. The workshop recording and the course materials are available online.\nThe main hackathon consisted of several ADAM data generating tasks based on a specs file and synthetic data. Participants were able to solve these tasks in groups at their own pace thanks to a online tool where participants could upload their task specific R scripts and they would get automatic feedback for the data-set produced by their script. Script upload through the feedback application was available all through February, and we offered three additional online meetings throughout the month to discuss challenges and give some tips. If you are interested in learning more about the thoughts that went into the feedback application, you can read about it in this blogpost or check out my public GitHub repository for such an application."
},
{
- "objectID": "posts/2023-08-08_study_day/study_day.html#last-updated",
- "href": "posts/2023-08-08_study_day/study_day.html#last-updated",
- "title": "It’s all relative? - Calculating Relative Days using admiral",
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html#introduction-to-r-workshop",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html#introduction-to-r-workshop",
+ "title": "Admiral Hackathon 2023 Revisited",
+ "section": "Introduction to R workshop",
+ "text": "Introduction to R workshop\nWe were really excited to see over 500 people from around 40 countries joining our Introduction to R workshop in January! To get to know prospective users and hackathon participants better, we conducted some polls during the meetings. Below you can see that representatives of many different sorts of organisations joined our Introduction to R workshop:\n\n\n\n\n\n216 out of 402 confirmed that their company is already using R for clinical trial data analysis, the remaining 131 did not answer this question.\nThe target audience for this workshop was programmers who are very familiar with SAS, but not so familiar with R, our polls confirmed this.\n\n\n\n\n\nOverall, we were very happy with how the workshop turned out, and participants overall agreed with this sentiment (although there may be a slight survivorship bias…)."
+ },
+ {
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html#admiral-hackathon",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html#admiral-hackathon",
+ "title": "Admiral Hackathon 2023 Revisited",
+ "section": "admiral Hackathon",
+ "text": "admiral Hackathon\nFollowing the kick-off meeting, 371 participants joined the posit (rStudio) workspace that was made available to all participants at no costs by the posit company. About half the participants planned to spend one to two hours per week on the admiral tasks, the other half planned to allocate even more. 15 participants even planned to spend eight hours or more!\nWe were really happy to see an overwhelming amount of activity on the slack channel we set up with over 250 members. Not only were people engaging with the materials, but we saw how a community was formed where people were encouraged to ask questions and where community members went out of their way to help each other. Shout-out to our community hero: Jagadish Katam without whom most issues related to the task programming raised by the community would not have been addressed as quickly as they were. Huge thanks from the organizers!\nIn the end, a total of 44 teams spanning 87 statistical programmers took part in the admiral hackathon and uploaded solution scripts to the hackathon application solving at least one of the 8 tasks available (ADSL, ADAE, ADLBC, ADVS, ADTTE, ADADAS, ADLBH & ADLBHY). Participants’ scripts were then run on the shiny server and the output data-frame were compared to the solutions we provided. At the read-out there was a live draft of teams to win one-on-one admiral consulting with one of the admiral core developers. Winning probabilities were weighted by the number of points each group received for the quality of their output data-frames and for the number of tasks solved.\nCongratulations to the winners:\n\nViiV Team_GSK\nteamspoRt\nTatianaPXL\nDivyasneelam\nAdaMTeamIndia\nSanofi_BP\nJagadish (our community hero)\nAZ_WAWA\n\nAlthough this was uncertain during the hackathon we were excited to provide a Certificate of Completion to all participants who uploaded a script to the Web Application.\nA recording of the hackathon readout can be found in the CDISC Open Source Alliance Quarterly Spotlight."
+ },
+ {
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html#conclusion",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html#conclusion",
+ "title": "Admiral Hackathon 2023 Revisited",
+ "section": "Conclusion",
+ "text": "Conclusion\nOverall, we are very happy with how the hackathon turned out. We were not only positively surprised with the huge audience for the Intro to R workshop (CDISC record breaking) and for the admiral hackathon, but even more so with the engagement of all the participants.\nAgain, we would like to thank all the organizers, participants, and sponsors for their time and resources and hope to have provided a useful glimpse into our solution for ADAM creation within the end-to-end clinical data analysis open source R framework that the Pharmaverse aims to provide.\nAs always, we are very happy to hear more feedback on the hackathon as well as on admiral in general. Simply submit an issue on the admiral GitHub repository. You would like to join the admiral core developers? Please reach out to Edoardo Mancini (product owner) or Ben Straub (technical lead)."
+ },
+ {
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html#last-updated",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html#last-updated",
+ "title": "Admiral Hackathon 2023 Revisited",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:45.242711"
+ "text": "Last updated\n\n2023-11-27 19:13:36.963431"
},
{
- "objectID": "posts/2023-08-08_study_day/study_day.html#details",
- "href": "posts/2023-08-08_study_day/study_day.html#details",
- "title": "It’s all relative? - Calculating Relative Days using admiral",
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html#details",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html#details",
+ "title": "Admiral Hackathon 2023 Revisited",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
@@ -182,88 +371,25 @@
"text": "Using More Than One Imputation Rule for a Variable\nUsing different imputation rules depending on the observation can be done by using the higher-order function slice_derivation(), which applies a derivation function differently (by varying its arguments) in different subsections of a dataset. For example, consider this Vital Signs case where pre-dose records require a different treatment to other records:\n\nvs <- tribble(\n ~VSDTC, ~VSTPT,\n \"2019-08-09T12:34:56\", NA,\n \"2019-10-12\", \"PRE-DOSE\",\n \"2019-11-10\", NA,\n \"2019-12-04\", NA\n) %>%\n slice_derivation(\n derivation = derive_vars_dtm,\n args = params(\n dtc = VSDTC,\n new_vars_prefix = \"A\"\n ),\n derivation_slice(\n filter = VSTPT == \"PRE-DOSE\",\n args = params(time_imputation = \"first\")\n ),\n derivation_slice(\n filter = TRUE,\n args = params(time_imputation = \"last\")\n )\n )\n\n\n\n\n\n\nVSDTC\nVSTPT\nADTM\nATMF\n\n\n\n\n2019-08-09T12:34:56\nNA\n2019-08-09 12:34:56\nNA\n\n\n2019-11-10\nNA\n2019-11-10 23:59:59\nH\n\n\n2019-12-04\nNA\n2019-12-04 23:59:59\nH\n\n\n2019-10-12\nPRE-DOSE\n2019-10-12 00:00:00\nH"
},
{
- "objectID": "posts/2023-06-28_welcome/index.html",
- "href": "posts/2023-06-28_welcome/index.html",
- "title": "Hello pharmaverse",
- "section": "",
- "text": "The communications working group (CWG) seeks to promote and showcase how R can be used in the Clinical Reporting pipeline through short and informative blog posts. These posts will be hosted on this pharmaverse blog and promoted on the pharmaverse slack channels as well as on LinkedIn.\nAs the CWG is a small team, we hope to make the blog development process easy enough that pharmaverse community members will be able to easily write blog posts with guidance from the CWG team."
- },
- {
- "objectID": "posts/2023-06-28_welcome/index.html#purpose",
- "href": "posts/2023-06-28_welcome/index.html#purpose",
- "title": "Hello pharmaverse",
+ "objectID": "index.html",
+ "href": "index.html",
+ "title": "Pharmaverse Blog",
"section": "",
- "text": "The communications working group (CWG) seeks to promote and showcase how R can be used in the Clinical Reporting pipeline through short and informative blog posts. These posts will be hosted on this pharmaverse blog and promoted on the pharmaverse slack channels as well as on LinkedIn.\nAs the CWG is a small team, we hope to make the blog development process easy enough that pharmaverse community members will be able to easily write blog posts with guidance from the CWG team."
- },
- {
- "objectID": "posts/2023-06-28_welcome/index.html#spirit-of-a-blog-post",
- "href": "posts/2023-06-28_welcome/index.html#spirit-of-a-blog-post",
- "title": "Hello pharmaverse",
- "section": "Spirit of a Blog Post",
- "text": "Spirit of a Blog Post\nThe CWG believes that the following 4 points will help guide the creation of Blog Posts.\n\nShort\nPersonalized\nReproducible\nReadable\n\nShort: Posts should aim to be under a 10 minute read. We encourage longer posts to be broken up into multiple posts.\nPersonalized: Posts should have a personality! For example, a person wishing to post on a function in a package needs to differentiate the post from the documentation for function, i.e. we don’t want to just recycle the documentation. How can you add your voice and experience? A bit of cheeky language is also encouraged.\nReproducible: Posts should work with minimal dependencies with data, packages and outside sources. Every dependency introduced in a post adds some risk to the post longevity. As package dependencies change, posts should be built in a way that they can be updated to stay relevant.\nReadable: The CWG sees this site as more of introductory site rather advanced user site. Therefore, the CWG feels that code should be introduced in a way that promotes readability over complexity."
- },
- {
- "objectID": "posts/2023-06-28_welcome/index.html#what-types-of-posts-are-allowed-on-this-site",
- "href": "posts/2023-06-28_welcome/index.html#what-types-of-posts-are-allowed-on-this-site",
- "title": "Hello pharmaverse",
- "section": "What types of posts are allowed on this site?",
- "text": "What types of posts are allowed on this site?\nOverall, we want to stay focus on the Clinical Reporting Pipeline, which we see as the following topics:\n\nPackages in the Clinical Reporting Pipeline\nFunctions from packages in the Clinical Reporting Pipeline\nWider experiences of using R in the Clinical Reporting Pipeline\nConference experiences and the Clinical Reporting Pipeline\n\nHowever, it never hurts to ask if you topic might fit into this medium!\n\nMinimum Post Requirements\n\nA unique image to help showcase the post.\nWorking Code\nSelf-contained data or package data.\nDocumentation of package versions\n\nThat is it! After that you can go wild, but we do ask that it is kept short!"
- },
- {
- "objectID": "posts/2023-06-28_welcome/index.html#how-can-i-make-a-blog-post",
- "href": "posts/2023-06-28_welcome/index.html#how-can-i-make-a-blog-post",
- "title": "Hello pharmaverse",
- "section": "How can I make a Blog Post",
- "text": "How can I make a Blog Post\nStep 1: Reach out to us through pharmaverse/slack or make an issue on our GitHub.\nStep 2: Branch off main\nStep 3: Review the Spirit of the Blog Post in the Pull Request Template\nStep 4: Poke us to do a review!"
+ "text": "Order By\n Default\n \n Title\n \n \n Date - Oldest\n \n \n Date - Newest\n \n \n Author\n \n \n \n \n \n \n \n\n\n\n\n \n\n\n\n\nBelieve in a higher order!\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nA brief foray into the higher order functions in the {admiral} package.\n\n\n\n\n\n\nNov 27, 2023\n\n\nEdoardo Mancini\n\n\n\n\n\n\n \n\n\n\n\nFloating point\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nThe untold story of how admiral deals with floating points.\n\n\n\n\n\n\nOct 30, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nReproducing the R Submissions Pilot 2 Shiny Application using Rhino\n\n\n\n\n\n\n\nsubmission\n\n\ncommunity\n\n\n\n\n\n\n\n\n\n\n\nOct 10, 2023\n\n\nIsmael Rodriguez, Vedha Viyash, André Veríssimo, APPSILON\n\n\n\n\n\n\n \n\n\n\n\nDate/Time Functions and Imputation in {admiral}\n\n\n\n\n\n\n\nadmiral\n\n\nADaM\n\n\nDate/Time\n\n\n\n\nDates, times and imputation can be a frustrating facet of any programming language. Here’s how {admiral} makes all of this easy!\n\n\n\n\n\n\nSep 26, 2023\n\n\nEdoardo Mancini\n\n\n\n\n\n\n \n\n\n\n\nThe pharmaverse (hi)story\n\n\n\n\n\n\n\ncommunity\n\n\n\n\n\n\n\n\n\n\n\nAug 30, 2023\n\n\nNicholas Eugenio\n\n\n\n\n\n\n \n\n\n\n\nRounding\n\n\n\n\n\n\n\nrounding\n\n\n\n\nExploration of some commonly used rounding methods and their corresponding functions in SAS and R, with a focus on ‘round half up’ and reliable solutions for numerical precision challenges\n\n\n\n\n\n\nAug 22, 2023\n\n\nKangjie Zhang\n\n\n\n\n\n\n \n\n\n\n\nIt’s all relative? - Calculating Relative Days using admiral\n\n\n\n\n\n\n\nadmiral\n\n\nADaMs\n\n\n\n\n\n\n\n\n\n\n\nAug 8, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nCross-Industry Open Source Package Development\n\n\n\n\n\n\n\ncommunity\n\n\nadmiral\n\n\n\n\n\n\n\n\n\n\n\nJul 25, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nHow to use Code Sections\n\n\n\n\n\n\n\nADaMs\n\n\nTips and Tricks\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2023\n\n\nEdoardo Mancini\n\n\n\n\n\n\n \n\n\n\n\nBlanks and NAs\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nReading SAS datasets into R. The data is not always as it seems!\n\n\n\n\n\n\nJul 10, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nfalcon\n\n\n\n\n\n\n\nfalcon\n\n\ndisplays\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2023\n\n\nVincent Shen\n\n\n\n\n\n\n \n\n\n\n\nHello pharmaverse\n\n\n\n\n\n\n\ncommunity\n\n\n\n\nShort, fun and user-driven content around the pharmaverse.\n\n\n\n\n\n\nJun 28, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nAdmiral Hackathon 2023 Revisited\n\n\n\n\n\n\n\ncommunity\n\n\nadmiral\n\n\n\n\nLet’s have a look at the Admiral Hackathon 2023 together.\n\n\n\n\n\n\nJun 27, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nDerive a new parameter computed from the value of other parameters\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nUse admiral::derive_param_computed() like a calculator to derive new parameters/rows based on existing ones\n\n\n\n\n\n\nJun 27, 2023\n\n\nKangjie Zhang\n\n\n\n\n\n\n \n\n\n\n\nHackathon Feedback Application\n\n\n\n\n\n\n\nR\n\n\nshiny\n\n\ncommunity\n\n\nadmiral\n\n\n\n\nGoing through the process of creating a shiny app for the admiral hackathon. The shiny app allows users to check their solutions autonomously, gives feedback, and rates their results.\n\n\n\n\n\n\nJun 27, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\nNo matching items"
},
{
- "objectID": "posts/2023-06-28_welcome/index.html#last-updated",
- "href": "posts/2023-06-28_welcome/index.html#last-updated",
- "title": "Hello pharmaverse",
- "section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:37.128939"
- },
- {
- "objectID": "posts/2023-06-28_welcome/index.html#details",
- "href": "posts/2023-06-28_welcome/index.html#details",
- "title": "Hello pharmaverse",
- "section": "Details",
- "text": "Details\n\nsource code, R environment"
- },
- {
- "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html",
- "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html",
- "title": "Derive a new parameter computed from the value of other parameters",
+ "objectID": "R/readme.html",
+ "href": "R/readme.html",
+ "title": "Files in this folder",
"section": "",
- "text": "When creating ADaM Basic Data Structure (BDS) datasets, we often encounter deriving a new parameter based on the analysis values (e.g., AVAL) of other parameters.\nThe admiral function derive_param_computed() adds a parameter computed from the analysis value of other parameters.\nIt works like a calculator to derive new records without worrying about merging and combining datasets, all you need is a derivation formula, which also improves the readability of the code."
+ "text": "Some of these files help in creating/developing blog-posts, others are used by our CICD pipeline.\n\ncreate_blogpost.R: use this script to create a new blogpost based on our blogpost template.\nCICD.R: use this script to spellcheck and stylecheck your blogpost.\n\n\n\n\nhelp_create_blogpost.R: script containing the function(s) used by create_blogpost.R\nswitch.R: Used by CICD spellcheck workflow."
},
{
- "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#introduction",
- "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#introduction",
- "title": "Derive a new parameter computed from the value of other parameters",
+ "objectID": "R/readme.html#development-files",
+ "href": "R/readme.html#development-files",
+ "title": "Files in this folder",
"section": "",
- "text": "When creating ADaM Basic Data Structure (BDS) datasets, we often encounter deriving a new parameter based on the analysis values (e.g., AVAL) of other parameters.\nThe admiral function derive_param_computed() adds a parameter computed from the analysis value of other parameters.\nIt works like a calculator to derive new records without worrying about merging and combining datasets, all you need is a derivation formula, which also improves the readability of the code."
- },
- {
- "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#example",
- "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#example",
- "title": "Derive a new parameter computed from the value of other parameters",
- "section": "Example",
- "text": "Example\nA value level validation use case, where derive_param_computed() is used to validate a derived parameter - PARAMCD = ADPCYMG (Actual Dose per Cycle) in ADEX dataset.\n\nDerivation\nActual Dose per Cycle is derived from the Total Amount of Dose (PARAMCD = TOTDOSE) / Number of Cycles (PARAMCD = NUMCYC)\nIn this example, ADEX.AVAL when ADEX.PARAMCD = ADPCYMG can be derived as:\n\\[\nAVAL (PARAMCD = ADPCYMG) = \\frac{AVAL (PARAMCD = TOTDOSE)}{AVAL (PARAMCD = NUMCYC)}\n\\]\n\n\nLoading Packages and Creating Example Data\n\nlibrary(tibble)\nlibrary(dplyr)\nlibrary(diffdf)\nlibrary(admiral)\n\nadex <- tribble(\n ~USUBJID, ~PARAMCD, ~PARAM, ~AVAL,\n \"101\", \"TOTDOSE\", \"Total Amount of Dose (mg)\", 180,\n \"101\", \"NUMCYC\", \"Number of Cycles\", 3\n)\n\n\n\nDerive New Parameter\n\nadex_admiral <- derive_param_computed(\n adex,\n by_vars = exprs(USUBJID),\n parameters = c(\"TOTDOSE\", \"NUMCYC\"),\n analysis_value = AVAL.TOTDOSE / AVAL.NUMCYC,\n set_values_to = exprs(\n PARAMCD = \"ADPCYMG\",\n PARAM = \"Actual Dose per Cycle (mg)\"\n )\n)\n\nWarning: The `analysis_value` argument of `derive_param_computed()` is deprecated as\nof admiral 0.12.0.\nℹ Please use the `set_values_to` argument instead.\n\n\n\n\n# A tibble: 3 × 4\n USUBJID PARAMCD PARAM AVAL\n <chr> <chr> <chr> <dbl>\n1 101 TOTDOSE Total Amount of Dose (mg) 180\n2 101 NUMCYC Number of Cycles 3\n3 101 ADPCYMG Actual Dose per Cycle (mg) 60\n\n\n\n\nCompare\nFor validation purpose, the diffdf package is used below to mimic SAS proc compare.\n\nadex_expected <- bind_rows(\n adex,\n tribble(\n ~USUBJID, ~PARAMCD, ~PARAM, ~AVAL,\n \"101\", \"ADPCYMG\", \"Actual Dose per Cycle (mg)\", 60\n )\n)\n\ndiffdf(adex_expected, adex_admiral, keys = c(\"USUBJID\", \"PARAMCD\"))\n\nNo issues were found!"
- },
- {
- "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#last-updated",
- "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#last-updated",
- "title": "Derive a new parameter computed from the value of other parameters",
- "section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:33.340574"
- },
- {
- "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#details",
- "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#details",
- "title": "Derive a new parameter computed from the value of other parameters",
- "section": "Details",
- "text": "Details\n\nsource code, R environment"
+ "text": "help_create_blogpost.R: script containing the function(s) used by create_blogpost.R\nswitch.R: Used by CICD spellcheck workflow."
},
{
"objectID": "posts/2023-07-24_rounding/rounding.html",
@@ -298,7 +424,7 @@
"href": "posts/2023-07-24_rounding/rounding.html#last-updated",
"title": "Rounding",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:29.564969"
+ "text": "Last updated\n\n2023-11-27 19:13:33.556149"
},
{
"objectID": "posts/2023-07-24_rounding/rounding.html#details",
@@ -308,114 +434,79 @@
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "R/readme.html",
- "href": "R/readme.html",
- "title": "Files in this folder",
- "section": "",
- "text": "Some of these files help in creating/developing blog-posts, others are used by our CICD pipeline.\n\ncreate_blogpost.R: use this script to create a new blogpost based on our blogpost template.\nCICD.R: use this script to spellcheck and stylecheck your blogpost.\n\n\n\n\nhelp_create_blogpost.R: script containing the function(s) used by create_blogpost.R\nswitch.R: Used by CICD spellcheck workflow."
- },
- {
- "objectID": "R/readme.html#development-files",
- "href": "R/readme.html#development-files",
- "title": "Files in this folder",
- "section": "",
- "text": "help_create_blogpost.R: script containing the function(s) used by create_blogpost.R\nswitch.R: Used by CICD spellcheck workflow."
- },
- {
- "objectID": "index.html",
- "href": "index.html",
- "title": "Pharmaverse Blog",
+ "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html",
+ "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html",
+ "title": "Derive a new parameter computed from the value of other parameters",
"section": "",
- "text": "Order By\n Default\n \n Title\n \n \n Date - Oldest\n \n \n Date - Newest\n \n \n Author\n \n \n \n \n \n \n \n\n\n\n\n \n\n\n\n\nFloating point\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nThe untold story of how admiral deals with floating points.\n\n\n\n\n\n\nOct 30, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nReproducing the R Submissions Pilot 2 Shiny Application using Rhino\n\n\n\n\n\n\n\nsubmission\n\n\ncommunity\n\n\n\n\n\n\n\n\n\n\n\nOct 10, 2023\n\n\nIsmael Rodriguez, Vedha Viyash, André Veríssimo, APPSILON\n\n\n\n\n\n\n \n\n\n\n\nDate/Time Functions and Imputation in {admiral}\n\n\n\n\n\n\n\nadmiral\n\n\nADaM\n\n\nDate/Time\n\n\n\n\nDates, times and imputation can be a frustrating facet of any programming language. Here’s how {admiral} makes all of this easy!\n\n\n\n\n\n\nSep 26, 2023\n\n\nEdoardo Mancini\n\n\n\n\n\n\n \n\n\n\n\nThe pharmaverse (hi)story\n\n\n\n\n\n\n\ncommunity\n\n\n\n\n\n\n\n\n\n\n\nAug 30, 2023\n\n\nNicholas Eugenio\n\n\n\n\n\n\n \n\n\n\n\nRounding\n\n\n\n\n\n\n\nrounding\n\n\n\n\nExploration of some commonly used rounding methods and their corresponding functions in SAS and R, with a focus on ‘round half up’ and reliable solutions for numerical precision challenges\n\n\n\n\n\n\nAug 22, 2023\n\n\nKangjie Zhang\n\n\n\n\n\n\n \n\n\n\n\nIt’s all relative? - Calculating Relative Days using admiral\n\n\n\n\n\n\n\nadmiral\n\n\nADaMs\n\n\n\n\n\n\n\n\n\n\n\nAug 8, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nCross-Industry Open Source Package Development\n\n\n\n\n\n\n\ncommunity\n\n\nadmiral\n\n\n\n\n\n\n\n\n\n\n\nJul 25, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nHow to use Code Sections\n\n\n\n\n\n\n\nADaMs\n\n\nTips and Tricks\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2023\n\n\nEdoardo Mancini\n\n\n\n\n\n\n \n\n\n\n\nBlanks and NAs\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nReading SAS datasets into R. The data is not always as it seems!\n\n\n\n\n\n\nJul 10, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nfalcon\n\n\n\n\n\n\n\nfalcon\n\n\ndisplays\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2023\n\n\nVincent Shen\n\n\n\n\n\n\n \n\n\n\n\nHello pharmaverse\n\n\n\n\n\n\n\ncommunity\n\n\n\n\nShort, fun and user-driven content around the pharmaverse.\n\n\n\n\n\n\nJun 28, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nDerive a new parameter computed from the value of other parameters\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nUse admiral::derive_param_computed() like a calculator to derive new parameters/rows based on existing ones\n\n\n\n\n\n\nJun 27, 2023\n\n\nKangjie Zhang\n\n\n\n\n\n\n \n\n\n\n\nHackathon Feedback Application\n\n\n\n\n\n\n\nR\n\n\nshiny\n\n\ncommunity\n\n\nadmiral\n\n\n\n\nGoing through the process of creating a shiny app for the admiral hackathon. The shiny app allows users to check their solutions autonomously, gives feedback, and rates their results.\n\n\n\n\n\n\nJun 27, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nAdmiral Hackathon 2023 Revisited\n\n\n\n\n\n\n\ncommunity\n\n\nadmiral\n\n\n\n\nLet’s have a look at the Admiral Hackathon 2023 together.\n\n\n\n\n\n\nJun 27, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\nNo matching items"
+ "text": "When creating ADaM Basic Data Structure (BDS) datasets, we often encounter deriving a new parameter based on the analysis values (e.g., AVAL) of other parameters.\nThe admiral function derive_param_computed() adds a parameter computed from the analysis value of other parameters.\nIt works like a calculator to derive new records without worrying about merging and combining datasets, all you need is a derivation formula, which also improves the readability of the code."
},
{
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html",
- "title": "Blanks and NAs",
+ "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#introduction",
+ "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#introduction",
+ "title": "Derive a new parameter computed from the value of other parameters",
"section": "",
- "text": "Reading in SAS-based datasets (.sas7bdat or xpt) into R has users calling the R package haven. A typical call might invoke read_sas() or read_xpt() to bring in your source data to construct your ADaMs or SDTMs.\nUnfortunately, while using haven the character blanks (missing data) found in a typical SAS-based dataset are left as blanks. These blanks will typically prove problematic while using functions like is.na in combination with dplyr::filter() to subset data. Check out Bayer’s SAS2R catalog: handling-of-missing-values for more discussion on missing values and NAs.\nIn the admiral package, we have built a simple function called convert_blanks_to_na() to help us quickly remedy this problem. You can supply an entire dataframe to this function and it will convert any character blanks to NA_character_"
- },
- {
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#loading-packages-and-making-dummy-data",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#loading-packages-and-making-dummy-data",
- "title": "Blanks and NAs",
- "section": "Loading Packages and Making Dummy Data",
- "text": "Loading Packages and Making Dummy Data\n\nlibrary(admiral)\nlibrary(tibble)\nlibrary(dplyr)\n\ndf <- tribble(\n ~USUBJID, ~RFICDTC,\n \"01\", \"2000-01-01\",\n \"02\", \"2001-01-01\",\n \"03\", \"\", # Here we have a character blank\n \"04\", \"2001-01--\",\n \"05\", \"2001---01\",\n \"05\", \"\", # Here we have a character blank\n)\n\ndf\n\n# A tibble: 6 × 2\n USUBJID RFICDTC \n <chr> <chr> \n1 01 \"2000-01-01\"\n2 02 \"2001-01-01\"\n3 03 \"\" \n4 04 \"2001-01--\" \n5 05 \"2001---01\" \n6 05 \"\""
- },
- {
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#a-simple-conversion",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#a-simple-conversion",
- "title": "Blanks and NAs",
- "section": "A simple conversion",
- "text": "A simple conversion\n\ndf_na <- convert_blanks_to_na(df)\n\ndf_na\n\n# A tibble: 6 × 2\n USUBJID RFICDTC \n <chr> <chr> \n1 01 2000-01-01\n2 02 2001-01-01\n3 03 <NA> \n4 04 2001-01-- \n5 05 2001---01 \n6 05 <NA> \n\n\n\ndf_na %>% filter(is.na(RFICDTC))\n\n# A tibble: 2 × 2\n USUBJID RFICDTC\n <chr> <chr> \n1 03 <NA> \n2 05 <NA>"
+ "text": "When creating ADaM Basic Data Structure (BDS) datasets, we often encounter deriving a new parameter based on the analysis values (e.g., AVAL) of other parameters.\nThe admiral function derive_param_computed() adds a parameter computed from the analysis value of other parameters.\nIt works like a calculator to derive new records without worrying about merging and combining datasets, all you need is a derivation formula, which also improves the readability of the code."
},
{
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#thats-it",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#thats-it",
- "title": "Blanks and NAs",
- "section": "That’s it!",
- "text": "That’s it!\nA simple call to this function can make your derivation life so much easier while working in R if working with SAS-based datasets. In admiral, we make use of this function at the start of all ADaM templates for common ADaM datasets. You can use the function use_ad_template() to get the full R script for the below ADaMs.\n\nlist_all_templates()\n\nExisting ADaM templates in package 'admiral':\n• ADAE\n• ADCM\n• ADEG\n• ADEX\n• ADLB\n• ADLBHY\n• ADMH\n• ADPC\n• ADPP\n• ADPPK\n• ADSL\n• ADVS"
+ "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#example",
+ "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#example",
+ "title": "Derive a new parameter computed from the value of other parameters",
+ "section": "Example",
+ "text": "Example\nA value level validation use case, where derive_param_computed() is used to validate a derived parameter - PARAMCD = ADPCYMG (Actual Dose per Cycle) in ADEX dataset.\n\nDerivation\nActual Dose per Cycle is derived from the Total Amount of Dose (PARAMCD = TOTDOSE) / Number of Cycles (PARAMCD = NUMCYC)\nIn this example, ADEX.AVAL when ADEX.PARAMCD = ADPCYMG can be derived as:\n\\[\nAVAL (PARAMCD = ADPCYMG) = \\frac{AVAL (PARAMCD = TOTDOSE)}{AVAL (PARAMCD = NUMCYC)}\n\\]\n\n\nLoading Packages and Creating Example Data\n\nlibrary(tibble)\nlibrary(dplyr)\nlibrary(diffdf)\nlibrary(admiral)\n\nadex <- tribble(\n ~USUBJID, ~PARAMCD, ~PARAM, ~AVAL,\n \"101\", \"TOTDOSE\", \"Total Amount of Dose (mg)\", 180,\n \"101\", \"NUMCYC\", \"Number of Cycles\", 3\n)\n\n\n\nDerive New Parameter\n\nadex_admiral <- derive_param_computed(\n adex,\n by_vars = exprs(USUBJID),\n parameters = c(\"TOTDOSE\", \"NUMCYC\"),\n analysis_value = AVAL.TOTDOSE / AVAL.NUMCYC,\n set_values_to = exprs(\n PARAMCD = \"ADPCYMG\",\n PARAM = \"Actual Dose per Cycle (mg)\"\n )\n)\n\nWarning: The `analysis_value` argument of `derive_param_computed()` is deprecated as\nof admiral 0.12.0.\nℹ Please use the `set_values_to` argument instead.\n\n\n\n\n# A tibble: 3 × 4\n USUBJID PARAMCD PARAM AVAL\n <chr> <chr> <chr> <dbl>\n1 101 TOTDOSE Total Amount of Dose (mg) 180\n2 101 NUMCYC Number of Cycles 3\n3 101 ADPCYMG Actual Dose per Cycle (mg) 60\n\n\n\n\nCompare\nFor validation purpose, the diffdf package is used below to mimic SAS proc compare.\n\nadex_expected <- bind_rows(\n adex,\n tribble(\n ~USUBJID, ~PARAMCD, ~PARAM, ~AVAL,\n \"101\", \"ADPCYMG\", \"Actual Dose per Cycle (mg)\", 60\n )\n)\n\ndiffdf(adex_expected, adex_admiral, keys = c(\"USUBJID\", \"PARAMCD\"))\n\nNo issues were found!"
},
{
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#last-updated",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#last-updated",
- "title": "Blanks and NAs",
+ "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#last-updated",
+ "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#last-updated",
+ "title": "Derive a new parameter computed from the value of other parameters",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:27.839956"
+ "text": "Last updated\n\n2023-11-27 19:13:38.746856"
},
{
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#details",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#details",
- "title": "Blanks and NAs",
+ "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#details",
+ "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#details",
+ "title": "Derive a new parameter computed from the value of other parameters",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html",
+ "href": "posts/2023-07-14_code_sections/code_sections.html",
+ "title": "How to use Code Sections",
"section": "",
- "text": "There is significant momentum in driving the adoption of R packages in the life sciences industries, in particular, the R Consortium Submissions Working Group is dedicated to promoting the use of R for regulatory submissions to the FDA.\nThe R Consortium Submissions Working Group successfully completed an R-based submission in November 2021 through the eCTD portal (R Submissions Pilot 1). This Pilot was completed on March 10, 2022 after a successful statistical review and evaluation by the FDA staff.\nMoving forward, the Pilot 2 aimed to include a Shiny application that the FDA staff could deploy on their own servers. The R Consortium recently announced that on September 27, 2023, the R Submissions Working Group successfully completed the follow-up to the Pilot 2 R Shiny-based submission and received a response letter from FDA CDER. This marks the first publicly available submission package that includes a Shiny component. The full FDA response letter can be found here.\nThe Shiny application that was sent for the Pilot 2 had the goal to display the 4 Tables, Listings and Figures (TLFs) that were sent for the Pilot 1 with basic filtering functionality.\nThe submission package adhered to the eCTD folder structure and contained 5 deliverables. Among the deliverables was the proprietary R package {pilot2wrappers}, which enables the execution of the Shiny application.\nThe FDA staff were expected to receive the electronic submission packages in the eCTD format, install and load open-source R packages used as dependencies in the included Shiny application, reconstruct and load the submitted Shiny application, and communicate potential improvements in writing.\nIn the following stage, the R Consortium’s R Submission Working Group launched Pilot 4, aiming to investigate innovative technologies like Linux containers and web assembly. These technologies are being explored to package a Shiny application into a self-contained unit, streamlining the transfer and execution processes for the application.\nIn this post, our aim is to outline how we used the Rhino framework to reproduce the Shiny application that was successfully submitted to the FDA for the Pilot 2 project. Additionally, we detail the challenges identified during the process and how we were able to successfully address them by using an open-source package."
- },
- {
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#reproducing-the-r-submission-pilot-2-shiny-app-using-rhino",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#reproducing-the-r-submission-pilot-2-shiny-app-using-rhino",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
- "section": "Reproducing the R Submission Pilot 2 Shiny App using Rhino",
- "text": "Reproducing the R Submission Pilot 2 Shiny App using Rhino\nWhile the original Shiny application submitted to the FDA was wrapped using {Golem}, we replicated the application using our in-house developed framework Rhino. The main motivation was to provide an example of an R Submission that is not an R package and to identify and solve any issues that may arise from this approach.\nOur demo application (FDA-pilot-app) is accessible on our website, alongside other Shiny and Rhinoverse demonstration apps.\n\nThe code for FDA-pilot-app is open-source. You can create your own Rhino-based application by following our tutorial and viewing our workshop, which is available on YouTube."
+ "text": "The admiral package embraces a modular style of programming, where blocks of code are pieced together in sequence to create an ADaM dataset. However, with the well-documented advantages of the modular approach comes the recognition that scripts will on average be longer. As such, astute programmers working in RStudio are constantly on the lookout for quick ways to effectively navigate their scripts. Enter code sections!"
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#brief-introduction-to-rhino",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#brief-introduction-to-rhino",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
- "section": "Brief Introduction to Rhino",
- "text": "Brief Introduction to Rhino\n\n\n\n\n\nThe Rhino framework was developed by Appsilon to create enterprise-level Shiny applications, consistently and efficiently. This framework allows developers to apply the best software engineering practices, modularize code, test it thoroughly, enhance UI aesthetics, and monitor user adoption effectively.\nRhino provides support in 3 main areas:\n\nClear Code: scalable architecture, modularization based on Box and Shiny modules.\nQuality: comprehensive testing such as unit tests, E2E tests with Cypress and Shinytest2, logging, monitoring and linting.\nAutomation: project startup, CI with GitHub Actions, dependencies management with {renv}, configuration management with config, Sass and JavaScript bundling with ES6 support via Node.js.\n\nRhino is an ideal fit for highly regulated environments such as regulatory submissions or other drug development processes.\n\nFDA-pilot-app structure\nThe structure of this application is available on the github repository. The structure of this Shiny app is the following.\n\nClick here to expand the FDA-pilot-app structure\n\n.\n├── app\n│ ├── view\n│ │ └── demographic_table.R\n| | └── km_plot.R\n| | └── primary_table.R\n| | └── efficacy_table.R\n| | └── completion_table.R\n│ ├── logic\n│ │ └── adam_data.R\n│ │ └── eff_modles.R\n│ │ └── formatters.R\n│ │ └── helpers.R\n│ │ └── kmplot_helpers.R\n│ │ └── Tplyr_helpers.R\n│ ├── data\n│ │ └── adam\n│ │ └── adadas.xpt\n│ │ └── adlbc.xpt\n│ │ └── adsl.xpt\n│ │ └── adtte.xpt\n│ ├── docs\n│ │ └── about.md\n│ ├── js\n│ │ └── index.js\n│ ├── static\n│ │ └── favicon.ico\n│ ├── styles\n│ │ └── main.scss\n│ └── app.R\n├── tests\n│ ├── cypress\n│ │ └── integration\n│ │ └── app.spec.js\n│ ├── testthat\n│ │\n│ └── cypress.json\n├── app.R\n├── rhino_submission.Rproj\n├── dependencies.R\n├── renv.lock\n├── rhino.yml\n└── README.md"
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html#introduction",
+ "href": "posts/2023-07-14_code_sections/code_sections.html#introduction",
+ "title": "How to use Code Sections",
+ "section": "",
+ "text": "The admiral package embraces a modular style of programming, where blocks of code are pieced together in sequence to create an ADaM dataset. However, with the well-documented advantages of the modular approach comes the recognition that scripts will on average be longer. As such, astute programmers working in RStudio are constantly on the lookout for quick ways to effectively navigate their scripts. Enter code sections!"
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#efficient-submissions-to-the-fda",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#efficient-submissions-to-the-fda",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
- "section": "Efficient Submissions to the FDA",
- "text": "Efficient Submissions to the FDA\n\n\n\n\n\nTo comply with the Electronic Submission File Formats and Specifications for the eCTD submission, the programming code should carry a “.txt” extension. In the R Submissions Pilot 3 the group did not use {pkglite} as the FDA clarified that “.zip” and “.r” files are acceptable for submission. In this case, we utilized the {pkglite} R package to efficiently pack and unpack the FDA-pilot-app. This approach would facilitate the FDA reviewers in setting up the submission on their systems.\nThis package allows packing R packages to “.txt” files, which are supported for the submission of proprietary packages to the FDA via the eCTD gateway. \n\nPacking the FDA-pilot-app into a .txt file\nThe code below can be used to pack the Shiny application into a .txt file:\n\napp_name <- \"rhinosubmission\"\nrenv_spec <- pkglite::file_spec(\n \"renv\",\n pattern = \"^settings\\\\.dcf$|^activate\\\\.R$\",\n format = \"text\", recursive = FALSE\n)\ntests_spec <- pkglite::file_tests()\napp_spec <- pkglite::file_auto(\"app\")\nroot_spec <- pkglite::file_spec(\n path = \".\",\n pattern = \"^\\\\.Rprofile$|^rhino\\\\.yml$|^renv\\\\.lock$|^dependencies\\\\.R$|^config\\\\.yml$|^app\\\\.R$|^README\\\\.md$|\\\\.Rproj$\",\n all_files = TRUE,\n recursive = FALSE\n)\nwrite(paste0(\"Package: \", app_name), \"DESCRIPTION\", append = TRUE)\npkglite::collate(\n getwd(),\n renv_spec,\n tests_spec,\n app_spec,\n root_spec\n) |> pkglite::pack()\nfile.remove(\"DESCRIPTION\")\n\n\n\nUnpacking the FDA-pilot-app\nThe packed “.txt” file can be unpacked into a Shiny app by using {pkglite} as follows:\n\npkglite::unpack(\"rhinosubmission.txt\")"
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html#so-what-are-code-sections-and-why-are-they-useful",
+ "href": "posts/2023-07-14_code_sections/code_sections.html#so-what-are-code-sections-and-why-are-they-useful",
+ "title": "How to use Code Sections",
+ "section": "So, what are code sections and why are they useful?",
+ "text": "So, what are code sections and why are they useful?\nCode Sections are separators for long R scripts or functions in RStudio. They can be set up by inserting a comment line followed by four or more dashes in between portions of code, like so:\n\n# First code section ----\n\na <- 1\n\n# Second code section ----\n\nb <- 2\n\n# Third code section ----\n\nc <- 3\n\nRStudio then recognizes the code sections automatically, and enables you to:\n\nCollapse and expand them using the arrow displayed next to the line number, or with the handy shortcuts Alt+L/Shift+Alt+L on Windows or Cmd+Option+L/Cmd+Shift+Option+L on Mac.\nTravel in between them using the navigator at the bottom of the code pane, or by pressing Shift+Alt+J on Windows or Cmd+Shift+Option+J on Mac.\nView an outline of the file using the “Outline” button at the top right of the pane and/or the orange hashtag “Section Navigator” button at the bottom left of the pane.\n\n\n\n\n\n\nCollapsed sections, outline view and the section navigator for the example above.\n\n\n\n\nIt is also possible to create subsections by using two hashtags at the start of a comment line:\n\n# First code section ----\na <- 1\n\n## A code subsection ----\nb <- 2\n\n# Second code section ----\nc <- 3\n\n\n\n\n\n\nCode subsections for the example above.\n\n\n\n\nFor a complete list of Code Sections shortcuts, and for further information, see here."
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#lessons-learned",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#lessons-learned",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
- "section": "Lessons Learned",
- "text": "Lessons Learned\nOur initial objective was to prove that it would be possible to submit a Shiny application using Rhino through the eCTD gateway. During the rewriting process we identified that this could be done by integrating the open-source {pkglite} package. By following this approach, we concluded that it would be possible to submit a Shiny application through the eCTD gateway. This was also achieved through the successful submission of a package that included a Shiny component in Pilot 2.\nHaving rewritten the R Submissions Pilot 2 Shiny application using Rhino holds major implications for the adoption of our framework within the life sciences. Apart from being a strong, opinionated framework that improves reproducibility and reliability for Shiny development, using Rhino for regulatory submissions could improve the flexibility and speed in the clinical reporting pipeline. This would accelerate the adoption of R/Shiny for submissions to the FDA or other regulatory agencies."
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html#conclusion",
+ "href": "posts/2023-07-14_code_sections/code_sections.html#conclusion",
+ "title": "How to use Code Sections",
+ "section": "Conclusion",
+ "text": "Conclusion\nCode sections are an easy way to navigate long scripts and foster good commenting practices. They are used extensively in the admiral package, but there is no reason that you cannot start using them yourself in your day-to-day R programming!"
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#last-updated",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#last-updated",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html#last-updated",
+ "href": "posts/2023-07-14_code_sections/code_sections.html#last-updated",
+ "title": "How to use Code Sections",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:31.231853"
+ "text": "Last updated\n\n2023-11-27 19:13:41.390513"
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#details",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#details",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html#details",
+ "href": "posts/2023-07-14_code_sections/code_sections.html#details",
+ "title": "How to use Code Sections",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
@@ -459,7 +550,7 @@
"href": "posts/2023-10-30_floating_point/floating_point.html#last-updated",
"title": "Floating point",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:35.340689"
+ "text": "Last updated\n\n2023-11-27 19:13:46.342818"
},
{
"objectID": "posts/2023-10-30_floating_point/floating_point.html#details",
@@ -469,121 +560,100 @@
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html",
- "href": "posts/2023-07-09_falcon/falcon.html",
- "title": "falcon",
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html",
+ "title": "The pharmaverse (hi)story",
"section": "",
- "text": "The {falcon} initiative is an industry collaborative effort under {pharmaverse} that unites Boehringer Ingelheim, Moderna, Roche, and Sanofi with the aspiration of building and open-sourcing a comprehensive catalog of harmonized tables, listings, and graphs (TLGs) for clinical study reporting. By leveraging existing open-source R packages, {falcon} aims to simplify the process of output review, comparison, and meta-analyses, while fostering efficient communication among stakeholders in the pharmaceutical industry."
+ "text": "Pharmaverse: from motivation to present"
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#what-is-falcon",
- "href": "posts/2023-07-09_falcon/falcon.html#what-is-falcon",
- "title": "falcon",
- "section": "",
- "text": "The {falcon} initiative is an industry collaborative effort under {pharmaverse} that unites Boehringer Ingelheim, Moderna, Roche, and Sanofi with the aspiration of building and open-sourcing a comprehensive catalog of harmonized tables, listings, and graphs (TLGs) for clinical study reporting. By leveraging existing open-source R packages, {falcon} aims to simplify the process of output review, comparison, and meta-analyses, while fostering efficient communication among stakeholders in the pharmaceutical industry."
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#human-history-and-pharmaverse-context",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#human-history-and-pharmaverse-context",
+ "title": "The pharmaverse (hi)story",
+ "section": "Human history and pharmaverse context",
+ "text": "Human history and pharmaverse context\nSince the Australian Aboriginal, the earliest peoples recorded to have inhabited the Earth and who have been in Australia for at least 65,000 to 80,000 years (Encyclopædia Britannica), human beings live in group. Whether to protect yourself, increase your life expectancy or simply share tasks.\nFor most aspects of life, it doesn’t make sense to think, act or work alone for two main reasons:\n\nYou will spend more energy and time;\nSomeone else may be facing (or have faced) the same situation.\n\nThe English poet John Donne used to say “No man is an island entire of itself; every man is a piece of the continent, a part of the main;”. I can’t disagree with him. And I dare say that Ari Siggaard Knoph (Novo Nordisk), Michael Rimler (GSK), Michael Stackhouse (Atorus), Ross Farrugia (Roche), and Sumesh Kalappurakal (Janssen) can’t disagree with him either. They are the founders of pharmaverse, members of its Council and kindly shared their memories of how independent companies, in mid-2020, worked together in the creation of a set of packages developed to support the clinical reporting pipeline.\nIf you are not familiar with this pipeline, the important thing to know is that, in a nutshell, pharmaceutical companies must follow a bunch of standardized procedures and formats (from Clinical Data Interchange Standards Consortium, CDISC) when submitting clinical results to Health Authorities. The focus is on this: different companies seeking the same standards for outputs.\nParaphrasing Ross Farrugia (Roche) Breaking boundaries through open-source collaboration presentation in R/Pharma 2022 and thinking of the development of a new drug, we are talking about a “post-competitive” scenario: the drug has already been discovered and the companies should “just” produce and deliver standardized results."
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#why-do-we-build-it",
- "href": "posts/2023-07-09_falcon/falcon.html#why-do-we-build-it",
- "title": "falcon",
- "section": "Why do we build it?",
- "text": "Why do we build it?\nThe collaborative effort focuses on improving the clarity, consistency, and accessibility of TLGs by addressing variations and redundancies in their creation and use. This harmonized approach allows for streamlined reporting processes and facilitates effective communication of study results within the industry and to regulatory authorities."
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#clinical-reporting-outputs",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#clinical-reporting-outputs",
+ "title": "The pharmaverse (hi)story",
+ "section": "Clinical reporting outputs",
+ "text": "Clinical reporting outputs\nRationally, we can say that companies face the same challenges in these steps of the process. Not so intuitively, we can also say they were working in silos on that before 2018. Just as Isaac Newton and Gottfried W. Leibniz independently developed the theory of infinitesimal calculus, pharmaceutical companies were independently working on R solutions for this pipeline.\nBut on August 16 and 17 of the mentioned year above, they gathered at the first edition of R/Pharma conference to discuss R and open-source tooling for drug development (the reasons why open-source is an advantageous approach can be found in this post written by Stefan Thoma). And according to Isabela Velásquez’s article, Pharmaverse: Packages for clinical reporting workflows, one of the most popular questions in this conference was “Is the package code available or on CRAN?”.\nWell, many of them were. And not necessarily at that date, but just to mention a few: pharmaRTF and Tplyr from Atorus, r2rtf from Merck, rtables from Roche, etc. The thing is that, overall, there were almost 10000 other packages as well (today, almost 20000). And that took to another two questions:\n\nWith this overwhelming number of packages on CRAN, how to find the ones related to solving “clinical reporting problems”?\nOnce the packages were found, how to choose which one to use among those that have the same functional purpose?\n\nSo, again, companies re-started to working in silos to find those answers. But now, in collaborative silos and with common goals: create extremely useful packages to solve pharmaceutical-specific gaps once and solve them well!"
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#what-has-been-done-so-far",
- "href": "posts/2023-07-09_falcon/falcon.html#what-has-been-done-so-far",
- "title": "falcon",
- "section": "What has been done so far?",
- "text": "What has been done so far?\nDrawing inspiration from the FDA Standard Safety Tables and Figures Integrated Guide, the {falcon} initiative has successfully developed and open-sourced 11 templates to date. 4 product owners and 11 developers from 4 companies have collaborated to make these templates available and also published them on the official {falcon} website at https://pharmaverse.github.io/falcon/."
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#first-partnerships",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#first-partnerships",
+ "title": "The pharmaverse (hi)story",
+ "section": "First partnerships",
+ "text": "First partnerships\nIn 2020, Michael Stackhouse (Atorus) and Michael Rimler (GSK) talked and formed a partnership between their companies to develop a few more packages, including metacore, to read, store and manipulate metadata for ADaMs/SDTMs in a standardized object; xportr, to create submission compliant SAS transport files and perform pharma specific dataset level validation checks; and logrx (ex-timber), to build log to support reproducibility and traceability of an R script.\nAround the same time, Thomas Neitmann (currently at Denali Therapeutics, then at Roche) and Michael Rimler (GSK) discovered that both were working with ADaM in R, so Thomas Neitmann (currently at Denali Therapeutics, then at Roche), Ross Farrugia (Roche) and Michael Rimler (GSK) saw an opportunity there and GSK started their partnership with Roche to build and release admiral package.\nThe idea of working together, the sense of community, and the appetite from organizations built more and more, with incentive and priority established up into the programming heads council.\nJanssen had a huge effort in building R capabilities going on as well, by releasing tidytlg and envsetup), so eventually Michael Rimler (GSK), Michael Stackhouse (Atorus) and Ross Farrugia (Roche) formalized pharmaverse and formed the council, adding in Sumesh Kalappurakal (Janssen) and Ari Siggaard Knoph (Novo Nordisk) later joined as the fifth council member."
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#next-steps-vision",
- "href": "posts/2023-07-09_falcon/falcon.html#next-steps-vision",
- "title": "falcon",
- "section": "Next steps & vision",
- "text": "Next steps & vision\nFuture plans for {falcon} involve expanding the catalog through continuous collaboration from participating companies and inviting wider industry engagement. The ultimate goal is to promote harmonization of TLGs for clinical reporting across the pharmaceutical industry, leading to greater efficiency, collaboration, and innovation. Even though templates currently come from a published FDA guide, the collaborating companies are open to share and discuss similarities and differences on analysis concepts and output layouts of their own implementations in clinical reporting, for both safety and efficacy analyses.\nIn addition, while currently all templates were built using {rtables}, {tern}, {rlistings} and drew inspiration from the open-sourced TLG-Catalog, moving forward, we plan to support creating the same templates using alternative table engines such as {gt}.\n{falcon} will be presented at the upcoming PHUSE EU (Standards Implementation stream), where we will share the collaboration journey of {falcon} so far, providing more details on the current progress, long-term vision, and strategies for this initiative. Attendees will gain insights into the challenges and opportunities of harmonizing clinical reporting through open-source collaboration and learn about the potential benefits and future direction of {falcon}."
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#release-growth-and-developments",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#release-growth-and-developments",
+ "title": "The pharmaverse (hi)story",
+ "section": "Release, growth and developments",
+ "text": "Release, growth and developments\nAt the end of their presentation “Closing the Gap: Creating an End to End R Package Toolkit for the Clinical Reporting Pipeline.”, in R/Pharma 2021, Ben Straub (GSK) and Eli Miller (Atorus) welcomed the community to the pharmaverse, a curated collection of packages developed by various pharmaceutical companies to support open-source clinical workflows.\nFrom the outset, the name pharmaverse was chosen so that it could be a neutral home, unrelated to any company. Also, it was established as not being a consortium, which means that founders don’t own, fund, or maintain the packages. Some individuals and companies maintain them but often allowing for community contributions and being licensed permissively so that there is always a feeling of community ownership. The focus of pharmaverse early on, and today, is on inter organization cooperation, to build an environment where, if organizations identify that they have a joint problem that they want to solve, this is the right space to work on and release it.\nPharmaverse has grown a lot, at the time of writing this post we have >25 packages recommended in pharmaverse, and this has led to a partnership with PHUSE to get support from their organization and platform, and because they are eager to advance and support pharmaverse mission.\nDespite all its structure, it is impossible to say that we have a single solution for each clinical reporting analysis when it comes to pharmaverse, a single pathway is impractical. Instead, it is necessary to accept viable tools fitting different pathways into pharmaverse to direct and give people options as to what might work for them. After all, even though we live together as a community, we still have our own unique internal problems.\n\n\n\nSample of pharmaverse packages"
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#last-updated",
- "href": "posts/2023-07-09_falcon/falcon.html#last-updated",
- "title": "falcon",
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#last-updated",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#last-updated",
+ "title": "The pharmaverse (hi)story",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:38.469514"
+ "text": "Last updated\n\n2023-11-27 19:13:49.940918"
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#details",
- "href": "posts/2023-07-09_falcon/falcon.html#details",
- "title": "falcon",
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#details",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#details",
+ "title": "The pharmaverse (hi)story",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html",
- "href": "posts/2023-06-27__hackathon_app/index.html",
- "title": "Hackathon Feedback Application",
+ "objectID": "posts/2023-06-28_welcome/index.html",
+ "href": "posts/2023-06-28_welcome/index.html",
+ "title": "Hello pharmaverse",
"section": "",
- "text": "We recently created a shiny application for the admiral hackathon in February 2023. The admiral hackathon was an event designed to make statistical programmers from the pharmaceutical industry more comfortable with the admiral R package which allows users to efficiently transform data from one data standard (SDTM) to another (ADaM).\nHackathon participants formed groups of up to five people and were then tasked to create R-scripts that map the SDTM data to ADaM according to specifics defined in the metadata.\nThe purpose of the shiny app was threefold:\nIn this blog post I want to highlight some of the thoughts that went into this application. Please keep in mind that this work was done under tight time restraints.\nThe hackathon application is still online (although data-upload is switched off) and the GitHub repository is publicly available. The application is embedded into this post right after this paragraph. I have also uploaded to GitHub a .zip file of the workspace to which hackathon participants had access via posit cloud. For more context you can watch recordings of the hackathon-meetings."
- },
- {
- "objectID": "posts/2023-06-27__hackathon_app/index.html#permanent-data",
- "href": "posts/2023-06-27__hackathon_app/index.html#permanent-data",
- "title": "Hackathon Feedback Application",
- "section": "Permanent Data",
- "text": "Permanent Data\nThe biggest challenge you have to consider for this app is the permanent data storage. Shiny apps run on a server. Although we can write files on this server, whenever the app restarts, the files are lost. Therefore, a persistent data storage solution is required.\n\nGoogle drive\nI decided to leverage Google drive using the googledrive package package. This allowed me to save structured data (the team registry and the submission scores) as well as unstructured data (their R-script files).\n\n\n\n\n\n\nAuthentication\n\n\n\nTo access Google drive using the googledrive package we need to authenticate. This can be done interactively using the command googledrive::drive_auth() which takes you to the Google login page. After login you receive an authentication token requested by R.\nFor non-interactive authentication this token must be stored locally. In our case where the shiny app must access the token once deployed, the token must be stored on the project level.\nI have included the authentication procedure I followed in the R folder in google_init.R. You can find more extensive documentation of the non-interactive authentication.\n\n\nThe initial concept was: Each team gets their own folder including the most recent submission for each task, and a .csv file containing team information. To keep track of the submissions and the respective scores we wrote a .csv file in the mock-hackathon folder, so one folder above the team folders.\nSaving the team info as a .csv file worked fine as each team received their own file which – once created – was not touched anymore. As each upload for every team should simply add a row to the submissions.csv file, appending the file would be ideal. This was not possible using the googledrive package package. Instead, for each submission, the submissions file was downloaded, appended, and uploaded again. Unfortunately, this lead to a data loss, as the file was continuously overwritten, especially when two teams would submit simultaneously.\n\n\n\n\n\n\nRecover the Lost Data\n\n\n\nWhenever the submissions.csv file was uploaded, the previous version was sent to the Google drive bin. We ended up with over 3000 submissions.csv files containing a lot of redundant information. I had to write the following chunk to first get the unique file IDs of the 3000 submissions.csv files, create an empty submissions data-frame, and then download each file and add its information to the submisisons data-frame. To keep the data-frame as light as possible, after each append I deleted all duplicate submissions.\n\n# get all task_info.csv ID's\n# each row identifies one file in the trash\ntask_info_master <- drive_find(\n pattern = \"task_info.csv\",\n trashed = TRUE\n)\n\n\n# set up empty df to store all submissions\norigin <- tibble(\n score = numeric(),\n task = character(),\n team = character(),\n email = character(),\n time = character()\n)\n\n# downloads, reads, and returns one csv file given a file id\nget_file <- function(row) {\n tf <- tempfile()\n row %>%\n as_id() %>%\n drive_download(path = tf)\n new <- read_csv(tf) %>%\n select(score, task, team) %>%\n distinct()\n}\n\n\n# quick and dirty for loop to subsequently download each file, extract information\n# merge with previous information and squash it (using distinct()).\nfor (i in 1:nrow(task_info_master)) {\n origin <- rbind(origin, get_file(row = task_info_master[i, ])) %>%\n distinct()\n\n # save progress in a separate file after every 100 downloaded and merged sheets\n if (i %% 100 == 0) {\n print(i)\n write_csv(origin, paste(\"prog_data/task_info_prog_\", i, \".csv\", sep = \"\"))\n # update on progress\n message(i / nrow(task_info_master) * 100)\n }\n}\n\nWhen doing such a time-intensive task, make sure to try it first with only a couple of files to see whether any errors are produced. I am not quite sure how long this took but when I returned from my lunch break everything had finished.\n\n\nIf you want to stay in the Google framework, I recommend using the googlesheets4 package for structured data. googlesheets4 allows appending new information to an already existing sheet without the need to download the file first. As both packages follow the same style, going from one to the other is really simple. googlesheets4 requires authentication as well. However, you can reuse the cached token from the googledrive package authentication by setting gs4_auth(token = drive_token()).\n\n\nSecurity Concerns\nConnecting a public shiny app to your Google account introduces a security vulnerability in general. Especially so because we implemented the upload of files to Google drive. And even more problematic: We run a user generated script and display some of its output. A malicious party might be able to extract the authentication token of our Google account or could upload malware to the drive.\nTo reduce the risk, I simply created an un-associated Google account to host the drive. There are certainly better options available, but this seemed a reasonable solution achieved with very little effort."
- },
- {
- "objectID": "posts/2023-06-27__hackathon_app/index.html#register-team",
- "href": "posts/2023-06-27__hackathon_app/index.html#register-team",
- "title": "Hackathon Feedback Application",
- "section": "Register Team",
- "text": "Register Team\nWe wanted to allow users to sign up as teams using the shiny app. The app provides a simple interface where users could input a team name and the number of members. This in turn would open two fields for each user to input their name and email address.\nWe do simple checks to make sure at least one valid email address is supplied, and that the group name is acceptable. The group name cannot be empty, already taken, or contain vulgar words.\nThe team registration itself was adding the team information to the Google sheets file event_info into the sheet teams and to create a team folder in which to store the uploaded R files.\nThe checks and registration is implemented in the register_team() function stored in interact_with_google.R.\n\n\n\nScreenshot of the register team interface\n\n\nThe challenge here was to adapt the number of input fields depending on the number of team members. This means that the team name and email interface must be rendered: First, we check how many team members are part of the group, this is stored in the input$n_members input variable. Then we create a tagList with as many elements as team members. Each element contains two columns, one for the email, one for the member name. This tagList is then returned and displayed to the user.\n\n# render email input UI of the register tab\noutput$name_email <- shiny::renderUI({\n # create field names\n N <- input$n_members\n NAME <- sapply(1:N, function(i) {\n paste0(\"name\", i)\n })\n EMAIL <- sapply(1:N, function(i) {\n paste0(\"email\", i)\n })\n\n output <- tagList()\n\n\n firstsecondthird <- c(\"First\", \"Second\", \"Third\", \"Fourth\", \"Fifth\")\n for (i in 1:N) {\n output[[i]] <- tagList()\n output[[i]] <- fluidRow(\n shiny::h4(paste(firstsecondthird[i], \" Member\")),\n column(6,\n textInput(NAME[i], \"Name\"),\n value = \" \" # displayed default value\n ),\n column(6,\n textInput(EMAIL[i], \"Email\"),\n value = \" \"\n )\n )\n }\n output\n})\n\nThe team information is then uploaded to Google drive. Because some teams have more members than others, we have to create the respective data-frame with the number of team members in mind.\nThe following chunk creates the registration data. Noteworthy here the creation of the NAME and EMAIL variables which depend on the number of members in this team. Further, the user input of these fields is extracted via input[[paste0(NAME[i])]] within a for-loop.\nWe also make the data-creation dependent on the press of the Register Group button and cache some variables.\n\n## registration\nregistrationData <-\n reactive({\n N <- input$n_members\n NAME <- sapply(1:N, function(i) {\n paste0(\"name\", i)\n })\n EMAIL <- sapply(1:N, function(i) {\n paste0(\"email\", i)\n })\n names <- character(0)\n emails <- character(0)\n\n for (i in 1:N) {\n names[i] <- input[[paste0(NAME[i])]]\n emails[i] <- input[[paste0(EMAIL[i])]]\n }\n # create df\n dplyr::tibble(\n team_name = input$team_name,\n n_members = N,\n member_name = names,\n member_email = emails\n )\n }) %>%\n bindCache(input$team_name, input$n_members, input$name1, input$email1) %>%\n bindEvent(input$register) # wait for button press"
- },
- {
- "objectID": "posts/2023-06-27__hackathon_app/index.html#upload-source-script",
- "href": "posts/2023-06-27__hackathon_app/index.html#upload-source-script",
- "title": "Hackathon Feedback Application",
- "section": "Upload & Source Script",
- "text": "Upload & Source Script\nTo upload a script, participants had to select their team first. The input options were based on the existing folders on the Google-drive in the mock_hackathon folder. To upload a particular script participants had to also select the task to be solved. The uploaded script is then uploaded to the team folder following a standardised script naming convention.\nThere are different aspects to be aware of when sourcing scripts on a shiny server. For example, you have to anticipate the packages users will include in their uploaded scripts, as their scripts will load but not install packages. Further, you should keep the global environment of your shiny app separate from the environment in which the script is sourced. This is possible by supplying an environment to the source() function, e.g: source(path_to_script, local = new.env())\nAnother thing we had to consider was to replicate the exact folder-structure on the shiny server that participants were working with when creating the scripts, as they were required to source some scripts and to save their file into a specific folder. This was relatively straight forward as we provided participants with a folder structure in the posit cloud instance they were using. They had access to the sdtm folder in which the data was stored, and the adam folder into which they saved their solutions. The structure also included a folder with metadata which was also available on the shiny server.\nFor some tasks, participants required some ADaM-datasets stored in the adam folder, essentially the output from previous tasks. This was achieved by first creating a list mapping tasks to the required ADaM datasets:\n\ndepends_list <- list(\n \"ADADAS\" = c(\"ADSL\"),\n \"ADAE\" = c(\"ADSL\"),\n \"ADLBC\" = c(\"ADSL\"),\n \"ADLBH\" = c(\"ADSL\"),\n \"ADLBHY\" = c(\"ADSL\"),\n \"ADSL\" = NULL,\n \"ADTTE\" = c(\"ADSL\", \"ADAE\"),\n \"ADVS\" = c(\"ADSL\")\n)\n\nThis list is sourced from the R/parameters.R file when initiating the application. We then call the get_depends() function sourced from R/get_depends.R which copies the required files from the key folder (where our solutions to the tasks were stored) to the adam folder. After sourcing the uploaded script the content in the adam folder is deleted."
+ "text": "The communications working group (CWG) seeks to promote and showcase how R can be used in the Clinical Reporting pipeline through short and informative blog posts. These posts will be hosted on this pharmaverse blog and promoted on the pharmaverse slack channels as well as on LinkedIn.\nAs the CWG is a small team, we hope to make the blog development process easy enough that pharmaverse community members will be able to easily write blog posts with guidance from the CWG team."
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#compare-to-solution-file",
- "href": "posts/2023-06-27__hackathon_app/index.html#compare-to-solution-file",
- "title": "Hackathon Feedback Application",
- "section": "Compare to Solution File",
- "text": "Compare to Solution File\nWe want to compare the file created by participants with our solution (key) file stored in the key folder. The diffdf::diffdf() function allows for easy comparison of two data-frames and directly provides extensive feedback for the user:\n\nlibrary(dplyr)\ndf1 <- tibble(\n numbers = 1:10,\n letters = LETTERS[1:10]\n)\ndf2 <- tibble(\n numbers = 1:10,\n letters = letters[1:10]\n)\n\ndiffdf::diffdf(df1, df2)\n\nWarning in diffdf::diffdf(df1, df2): \nNot all Values Compared Equal\n\n\nDifferences found between the objects!\n\nA summary is given below.\n\nNot all Values Compared Equal\nAll rows are shown in table below\n\n =============================\n Variable No of Differences \n -----------------------------\n letters 10 \n -----------------------------\n\n\nAll rows are shown in table below\n\n ========================================\n VARIABLE ..ROWNUMBER.. BASE COMPARE \n ----------------------------------------\n letters 1 A a \n letters 2 B b \n letters 3 C c \n letters 4 D d \n letters 5 E e \n letters 6 F f \n letters 7 G g \n letters 8 H h \n letters 9 I i \n letters 10 J j \n ----------------------------------------"
+ "objectID": "posts/2023-06-28_welcome/index.html#purpose",
+ "href": "posts/2023-06-28_welcome/index.html#purpose",
+ "title": "Hello pharmaverse",
+ "section": "",
+ "text": "The communications working group (CWG) seeks to promote and showcase how R can be used in the Clinical Reporting pipeline through short and informative blog posts. These posts will be hosted on this pharmaverse blog and promoted on the pharmaverse slack channels as well as on LinkedIn.\nAs the CWG is a small team, we hope to make the blog development process easy enough that pharmaverse community members will be able to easily write blog posts with guidance from the CWG team."
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#score",
- "href": "posts/2023-06-27__hackathon_app/index.html#score",
- "title": "Hackathon Feedback Application",
- "section": "Score",
- "text": "Score\nTo compare submissions between participants we implemented a simple scoring function (score_f()) based on the table comparison by diffdf(). The function can be found in the compare_dfs.R file:\n\nscore_f <- function(df_user, df_key, keys) {\n score <- 10\n diff <- diffdf::diffdf(df_user, df_key, keys = keys)\n if (!diffdf::diffdf_has_issues(diff)) {\n return(score)\n }\n\n # check if there are any differences if the comparison is not strict:\n if (!diffdf::diffdf_has_issues(diffdf::diffdf(df_user,\n df_key,\n keys = keys,\n strict_numeric = FALSE,\n strict_factor = FALSE\n ))) {\n # if differences are not strict, return score - 1\n return(score - 1)\n }\n\n return(round(min(max(score - length(diff) / 3, 1), 9), 2))\n}\n\nEvery comparison starts with a score of 10. We then subtract the length of the comparison object divided by a factor of 3. The length of the comparison object is a simplified way to represent the difference between the two data-frames by one value. Finally, the score is bounded by 1 using max(score, 1).\nThe score is not a perfect capture of the quality of the script uploaded but: 1. helped participants get an idea of how close their data-frame is to the solution file 2. allowed us to raffle prizes based on the merit of submitted r-scripts"
+ "objectID": "posts/2023-06-28_welcome/index.html#spirit-of-a-blog-post",
+ "href": "posts/2023-06-28_welcome/index.html#spirit-of-a-blog-post",
+ "title": "Hello pharmaverse",
+ "section": "Spirit of a Blog Post",
+ "text": "Spirit of a Blog Post\nThe CWG believes that the following 4 points will help guide the creation of Blog Posts.\n\nShort\nPersonalized\nReproducible\nReadable\n\nShort: Posts should aim to be under a 10 minute read. We encourage longer posts to be broken up into multiple posts.\nPersonalized: Posts should have a personality! For example, a person wishing to post on a function in a package needs to differentiate the post from the documentation for function, i.e. we don’t want to just recycle the documentation. How can you add your voice and experience? A bit of cheeky language is also encouraged.\nReproducible: Posts should work with minimal dependencies with data, packages and outside sources. Every dependency introduced in a post adds some risk to the post longevity. As package dependencies change, posts should be built in a way that they can be updated to stay relevant.\nReadable: The CWG sees this site as more of introductory site rather advanced user site. Therefore, the CWG feels that code should be introduced in a way that promotes readability over complexity."
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#reactiveness",
- "href": "posts/2023-06-27__hackathon_app/index.html#reactiveness",
- "title": "Hackathon Feedback Application",
- "section": "Reactiveness",
- "text": "Reactiveness\nSome of the app functions can take quite some time to execute, e.g. running the uploaded script. Other tasks, e.g. registering a team, do not intrinsically generate user facing outputs. This would make the app using really frustrating, as users would not know whether the app is correctly working or whether it froze.\nWe implemented two small features that made the app more responsive. One is simple loading icons that integrate into the user interface and show that output is being computed – that something is working. The other is a pop up window which communicates whether team registration was successful, and if not, why not.\nWe further aimed to forward errors generated by the uploaded scripts to the user interface, but errors generated by the application itself should be concealed."
+ "objectID": "posts/2023-06-28_welcome/index.html#what-types-of-posts-are-allowed-on-this-site",
+ "href": "posts/2023-06-28_welcome/index.html#what-types-of-posts-are-allowed-on-this-site",
+ "title": "Hello pharmaverse",
+ "section": "What types of posts are allowed on this site?",
+ "text": "What types of posts are allowed on this site?\nOverall, we want to stay focus on the Clinical Reporting Pipeline, which we see as the following topics:\n\nPackages in the Clinical Reporting Pipeline\nFunctions from packages in the Clinical Reporting Pipeline\nWider experiences of using R in the Clinical Reporting Pipeline\nConference experiences and the Clinical Reporting Pipeline\n\nHowever, it never hurts to ask if you topic might fit into this medium!\n\nMinimum Post Requirements\n\nA unique image to help showcase the post.\nWorking Code\nSelf-contained data or package data.\nDocumentation of package versions\n\nThat is it! After that you can go wild, but we do ask that it is kept short!"
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#conclusion",
- "href": "posts/2023-06-27__hackathon_app/index.html#conclusion",
- "title": "Hackathon Feedback Application",
- "section": "Conclusion",
- "text": "Conclusion\nAlthough the application was continuously improved during the hackathon it proved to be a useful resource for participants from day one as it allowed groups to set their own pace. It further allowed admiral developers to gain insights on package usage of a relatively large sample of potential end users. From our perspective, the application provided a great added value to the hackathon and eased the workload of guiding the participants through all the tasks."
+ "objectID": "posts/2023-06-28_welcome/index.html#how-can-i-make-a-blog-post",
+ "href": "posts/2023-06-28_welcome/index.html#how-can-i-make-a-blog-post",
+ "title": "Hello pharmaverse",
+ "section": "How can I make a Blog Post",
+ "text": "How can I make a Blog Post\nStep 1: Reach out to us through pharmaverse/slack or make an issue on our GitHub.\nStep 2: Branch off main\nStep 3: Review the Spirit of the Blog Post in the Pull Request Template\nStep 4: Poke us to do a review!"
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#last-updated",
- "href": "posts/2023-06-27__hackathon_app/index.html#last-updated",
- "title": "Hackathon Feedback Application",
+ "objectID": "posts/2023-06-28_welcome/index.html#last-updated",
+ "href": "posts/2023-06-28_welcome/index.html#last-updated",
+ "title": "Hello pharmaverse",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:42.927745"
+ "text": "Last updated\n\n2023-11-27 19:13:52.942147"
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#details",
- "href": "posts/2023-06-27__hackathon_app/index.html#details",
- "title": "Hackathon Feedback Application",
+ "objectID": "posts/2023-06-28_welcome/index.html#details",
+ "href": "posts/2023-06-28_welcome/index.html#details",
+ "title": "Hello pharmaverse",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
@@ -621,47 +691,5 @@
"title": "Cross-Industry Open Source Package Development",
"section": "Impact",
"text": "Impact\nFor Roche, cross-industry package development work out in their favor: They get access to software created by specialists and users from across the industry but paying only a fraction of the developmental costs. Of course, they don’t have total developmental control but they do get a seat at the table. Any gaps between the open source {admiral} package and the proprietary Roche workflow were bridged by the internal {admiralroche} package.\nThe switch towards a more language agnostic platform, and open source languages specifically, opens the door to a broad population of university graduates with diverse backgrounds. I personally would not have considered this position five years ago due to a misalignment of skills and job requirements. Working towards an industry standard open source solution will also ensure that skills learned at one company are more easily transferable to external positions, further making the position much more attractive. Access to such a broad pool of potential candidates is clearly beneficial for recruitment at Roche, but also facilitates diversity in teams which makes for a more interesting and effective work place.\nOpen source development comes with much more transparency by definition. Recognition of contributions are built in - anyone can see who did what. This recognition escapes the confines of your company as it is visible to anyone looking at the repository. Anyone can not only see at any time what is being worked on, what discussions are happening and which direction is being taken, but can also participate and contribute. Transparency also applies to errors in the code and how the team is dealing with them. In such an environment it is practically impossible to hide or cover up errors and corrections. Instead, they have to be dealt with publicly and in the open. This openness about errors also helps seeing errors as a natural occurrence that needs to be dealt with. Space for errors encourages learning and is really beneficial for growing both skills and integrity.\nAs you work on a team that spans multiple companies, traditional corporate hierarchies do not apply. Of course, there will always be a sort of hierarchy of experience or skills, but these work in your favor: You will know who to ask for help, and teams are generally very happy for contributors of any skill level. Contributions also need not be in code: Inputs into discussions and domain knowledge contributions are highly valued as well. The flip-side of working in a team without your manager oversight: They may not be directly aware of the work you do. That’s why you have to write blog posts :)\nThe possibility for statistical programmers to pivot towards developing software or writing blog-posts such as this really transforms and broadens their job description. It is this transformation that is reflected by the choice of Roche to re-brand statistical programmers as analytical data scientists. The fact that cross-industry development is being advocated for really lets programmers expand their network outside of their company.\nThe {admiral} project serves as a testament to the power of collaborative open-source development and the potential it holds for the future of work in this industry."
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html",
- "href": "posts/2023-06-27_hackathon_writeup/index.html",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "",
- "text": "This January and February (2023), the admiral development team and the CDISC Open Source Alliance jointly hosted the admiral hackathon. The idea was to build a community of admiral users, and help participants familiarize themselves with R and admiral. This whole effort was led by Thomas Neitmann and was supported by Zelos Zhu, Sadchla Mascary, and me – Stefan Thoma.\nThe hackathon event was structured in two parts. First, we offered an Introduction to R for SAS programmers, a three hour workshop for R beginners to get them up to speed. Here we covered practical R basics, talking about how the R-workflow differs from a SAS workflow, and discussed common R functions - mostly from the tidyverse. This ensured that hackathon participants were familiar with core R concepts. The workshop recording and the course materials are available online.\nThe main hackathon consisted of several ADAM data generating tasks based on a specs file and synthetic data. Participants were able to solve these tasks in groups at their own pace thanks to a online tool where participants could upload their task specific R scripts and they would get automatic feedback for the data-set produced by their script. Script upload through the feedback application was available all through February, and we offered three additional online meetings throughout the month to discuss challenges and give some tips. If you are interested in learning more about the thoughts that went into the feedback application, you can read about it in this blogpost or check out my public GitHub repository for such an application."
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html#introduction-to-r-workshop",
- "href": "posts/2023-06-27_hackathon_writeup/index.html#introduction-to-r-workshop",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "Introduction to R workshop",
- "text": "Introduction to R workshop\nWe were really excited to see over 500 people from around 40 countries joining our Introduction to R workshop in January! To get to know prospective users and hackathon participants better, we conducted some polls during the meetings. Below you can see that representatives of many different sorts of organisations joined our Introduction to R workshop:\n\n\n\n\n\n216 out of 402 confirmed that their company is already using R for clinical trial data analysis, the remaining 131 did not answer this question.\nThe target audience for this workshop was programmers who are very familiar with SAS, but not so familiar with R, our polls confirmed this.\n\n\n\n\n\nOverall, we were very happy with how the workshop turned out, and participants overall agreed with this sentiment (although there may be a slight survivorship bias…)."
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html#admiral-hackathon",
- "href": "posts/2023-06-27_hackathon_writeup/index.html#admiral-hackathon",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "admiral Hackathon",
- "text": "admiral Hackathon\nFollowing the kick-off meeting, 371 participants joined the posit (rStudio) workspace that was made available to all participants at no costs by the posit company. About half the participants planned to spend one to two hours per week on the admiral tasks, the other half planned to allocate even more. 15 participants even planned to spend eight hours or more!\nWe were really happy to see an overwhelming amount of activity on the slack channel we set up with over 250 members. Not only were people engaging with the materials, but we saw how a community was formed where people were encouraged to ask questions and where community members went out of their way to help each other. Shout-out to our community hero: Jagadish Katam without whom most issues related to the task programming raised by the community would not have been addressed as quickly as they were. Huge thanks from the organizers!\nIn the end, a total of 44 teams spanning 87 statistical programmers took part in the admiral hackathon and uploaded solution scripts to the hackathon application solving at least one of the 8 tasks available (ADSL, ADAE, ADLBC, ADVS, ADTTE, ADADAS, ADLBH & ADLBHY). Participants’ scripts were then run on the shiny server and the output data-frame were compared to the solutions we provided. At the read-out there was a live draft of teams to win one-on-one admiral consulting with one of the admiral core developers. Winning probabilities were weighted by the number of points each group received for the quality of their output data-frames and for the number of tasks solved.\nCongratulations to the winners:\n\nViiV Team_GSK\nteamspoRt\nTatianaPXL\nDivyasneelam\nAdaMTeamIndia\nSanofi_BP\nJagadish (our community hero)\nAZ_WAWA\n\nAlthough this was uncertain during the hackathon we were excited to provide a Certificate of Completion to all participants who uploaded a script to the Web Application.\nA recording of the hackathon readout can be found in the CDISC Open Source Alliance Quarterly Spotlight."
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html#conclusion",
- "href": "posts/2023-06-27_hackathon_writeup/index.html#conclusion",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "Conclusion",
- "text": "Conclusion\nOverall, we are very happy with how the hackathon turned out. We were not only positively surprised with the huge audience for the Intro to R workshop (CDISC record breaking) and for the admiral hackathon, but even more so with the engagement of all the participants.\nAgain, we would like to thank all the organizers, participants, and sponsors for their time and resources and hope to have provided a useful glimpse into our solution for ADAM creation within the end-to-end clinical data analysis open source R framework that the Pharmaverse aims to provide.\nAs always, we are very happy to hear more feedback on the hackathon as well as on admiral in general. Simply submit an issue on the admiral GitHub repository. You would like to join the admiral core developers? Please reach out to Edoardo Mancini (product owner) or Ben Straub (technical lead)."
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html#last-updated",
- "href": "posts/2023-06-27_hackathon_writeup/index.html#last-updated",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:51.728905"
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html#details",
- "href": "posts/2023-06-27_hackathon_writeup/index.html#details",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "Details",
- "text": "Details\n\nsource code, R environment"
}
]
\ No newline at end of file
diff --git a/site_libs/htmlwidgets-1.6.2/htmlwidgets.js b/site_libs/htmlwidgets-1.6.3/htmlwidgets.js
similarity index 100%
rename from site_libs/htmlwidgets-1.6.2/htmlwidgets.js
rename to site_libs/htmlwidgets-1.6.3/htmlwidgets.js
diff --git a/sitemap.xml b/sitemap.xml
index a0a76761..e2577287 100644
--- a/sitemap.xml
+++ b/sitemap.xml
@@ -1,67 +1,71 @@
- https://pharmaverse.github.io/blog/posts/2023-07-14_code_sections/code_sections.html
- 2023-11-01T15:47:53.974Z
+ https://pharmaverse.github.io/blog/posts/2023-08-08_study_day/study_day.html
+ 2023-11-27T19:13:58.211Z
- https://pharmaverse.github.io/blog/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
- 2023-11-01T15:47:48.826Z
+ https://pharmaverse.github.io/blog/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
+ 2023-11-27T19:13:54.971Z
- https://pharmaverse.github.io/blog/posts/2023-08-08_study_day/study_day.html
- 2023-11-01T15:47:45.858Z
+ https://pharmaverse.github.io/blog/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
+ 2023-11-27T19:13:52.247Z
- https://pharmaverse.github.io/blog/posts/2023-09-26_date_functions_and_imputation/date_functions_and_imputation.html
- 2023-11-01T15:47:41.734Z
+ https://pharmaverse.github.io/blog/posts/2023-06-27__hackathon_app/index.html
+ 2023-11-27T19:13:49.215Z
- https://pharmaverse.github.io/blog/posts/2023-06-28_welcome/index.html
- 2023-11-01T15:47:37.682Z
+ https://pharmaverse.github.io/blog/posts/2023-11-27_higher_order/higher_order.html
+ 2023-11-27T19:13:44.987Z
- https://pharmaverse.github.io/blog/posts/2023-06-27_admiral/valuelevel/derive_param_computed.html
- 2023-11-01T15:47:33.918Z
+ https://pharmaverse.github.io/blog/posts/2023-07-09_falcon/falcon.html
+ 2023-11-27T19:13:40.491Z
- https://pharmaverse.github.io/blog/posts/2023-07-24_rounding/rounding.html
- 2023-11-01T15:47:30.366Z
+ https://pharmaverse.github.io/blog/posts/2023-06-27_hackathon_writeup/index.html
+ 2023-11-27T19:13:37.491Z
- https://pharmaverse.github.io/blog/R/readme.html
- 2023-11-01T15:47:26.622Z
+ https://pharmaverse.github.io/blog/posts/2023-09-26_date_functions_and_imputation/date_functions_and_imputation.html
+ 2023-11-27T19:13:32.503Z
https://pharmaverse.github.io/blog/index.html
- 2023-11-01T15:47:26.074Z
+ 2023-11-27T19:13:29.559Z
- https://pharmaverse.github.io/blog/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
- 2023-11-01T15:47:28.446Z
+ https://pharmaverse.github.io/blog/R/readme.html
+ 2023-11-27T19:13:30.087Z
- https://pharmaverse.github.io/blog/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
- 2023-11-01T15:47:31.962Z
+ https://pharmaverse.github.io/blog/posts/2023-07-24_rounding/rounding.html
+ 2023-11-27T19:13:34.283Z
- https://pharmaverse.github.io/blog/posts/2023-10-30_floating_point/floating_point.html
- 2023-11-01T15:47:36.342Z
+ https://pharmaverse.github.io/blog/posts/2023-06-27_admiral/valuelevel/derive_param_computed.html
+ 2023-11-27T19:13:39.323Z
- https://pharmaverse.github.io/blog/posts/2023-07-09_falcon/falcon.html
- 2023-11-01T15:47:39.010Z
+ https://pharmaverse.github.io/blog/posts/2023-07-14_code_sections/code_sections.html
+ 2023-11-27T19:13:41.975Z
- https://pharmaverse.github.io/blog/posts/2023-06-27__hackathon_app/index.html
- 2023-11-01T15:47:43.922Z
+ https://pharmaverse.github.io/blog/posts/2023-10-30_floating_point/floating_point.html
+ 2023-11-27T19:13:47.267Z
- https://pharmaverse.github.io/blog/posts/2023-07-20_cross_company_dev/cross_industry_dev.html
- 2023-11-01T15:47:47.398Z
+ https://pharmaverse.github.io/blog/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
+ 2023-11-27T19:13:50.535Z
- https://pharmaverse.github.io/blog/posts/2023-06-27_hackathon_writeup/index.html
- 2023-11-01T15:47:52.282Z
+ https://pharmaverse.github.io/blog/posts/2023-06-28_welcome/index.html
+ 2023-11-27T19:13:53.443Z
+
+
+ https://pharmaverse.github.io/blog/posts/2023-07-20_cross_company_dev/cross_industry_dev.html
+ 2023-11-27T19:13:56.455Z
Last updated
2023-11-01 15:47:38.469514
+2023-11-27 19:13:40.019302
Details
That’s it!
Last updated
2023-11-01 15:47:27.839956
+2023-11-27 19:13:51.677601
Details
Conclusion
Last updated
2023-11-01 15:47:53.276819
+2023-11-27 19:13:41.390513
Details
Insights
Development workflow
-Every improvement, task, or feature we want to implement on {admiral} starts as an issue on our GitHub repository. It is the centerpiece of our development workflow, along with our developer guides which describe in detail the strategies, conventions, and workflows used in development. The guides help us keep the {admiral} package internally consistent (e.g. naming conventions, function logic) but also ensure that {admiral} adjacent packages follow the same conventions and share the user interface. This is further helped by the implemented CICD pipeline which ensures styling convention and spelling (and much more).
+Every improvement, task, or feature we want to implement on {admiral} starts as an issue on our GitHub repository. It is the centerpiece of our development workflow, along with our developer guides which describe in detail the strategies, conventions, and workflows used in development. The guides help us keep the {admiral} package internally consistent (e.g. naming conventions, function logic) but also ensure that {admiral} adjacent packages follow the same conventions and share the user interface. This is further helped by the implemented CICD pipeline which ensures styling convention and spelling (and much more).
The core package developers team meets once a week (twice a week before a release) to discuss progress and priorities. Here, the role of product lead (currently Edoardo Mancini at Roche) and technical lead (currently Ben Straub at GSK) is to set priorities and track the release schedule. These stand-up meetings are centered around the project-board which gives a complete overview of activities and progress. Issues are mostly self-assigned so developers can really chose what they want to work on.
Conclusion
Last updated
2023-11-01 15:47:29.564969
+2023-11-27 19:13:33.556149
Details
It’s all relative? - Calculating Relative Days using admiral
Last updated
-2023-11-01 15:47:45.242711
+2023-11-27 19:13:57.651626
Details
diff --git a/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html b/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
index fc7f9afa..99f64d55 100644
--- a/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
+++ b/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
@@ -369,7 +369,7 @@ References
Last updated
-2023-11-01 15:47:31.231853
+2023-11-27 19:13:54.254558
Details
diff --git a/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html b/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
index 885d8118..068e0d1c 100644
--- a/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
+++ b/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
@@ -221,7 +221,7 @@ Release, g
Last updated
-2023-11-01 15:47:48.201548
+2023-11-27 19:13:49.940918
Details
diff --git a/posts/2023-10-30_floating_point/floating_point.html b/posts/2023-10-30_floating_point/floating_point.html
index 4b06317b..31c93ef3 100644
--- a/posts/2023-10-30_floating_point/floating_point.html
+++ b/posts/2023-10-30_floating_point/floating_point.html
@@ -107,7 +107,7 @@
-
+
@@ -304,8 +304,8 @@ Issues arising
reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
-
-
+
+
As you can see, the data-frame contains the column GRADE_CRITERIA_CODE which contains comparisons of floating point values. And there was a discrepancy of what Gordon expected to see, and how R actually computed the comparison initially:
@@ -563,8 +563,8 @@ Potential solutions reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
-
-
+
+
Conclusion
Last updated
-2023-11-01 15:47:35.340689
+2023-11-27 19:13:46.342818
Details
diff --git a/posts/2023-11-27_higher_order/admiral.png b/posts/2023-11-27_higher_order/admiral.png
new file mode 100644
index 00000000..cc69ba33
Binary files /dev/null and b/posts/2023-11-27_higher_order/admiral.png differ
diff --git a/posts/2023-11-27_higher_order/higher_order.html b/posts/2023-11-27_higher_order/higher_order.html
new file mode 100644
index 00000000..12fe765b
--- /dev/null
+++ b/posts/2023-11-27_higher_order/higher_order.html
@@ -0,0 +1,1048 @@
+
+
+
+
+
+
+
+
+
+
+
+
+Pharmaverse Blog - Believe in a higher order!
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
\ No newline at end of file
diff --git a/search.json b/search.json
index f352c848..d6425294 100644
--- a/search.json
+++ b/search.json
@@ -1,113 +1,302 @@
[
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html",
- "href": "posts/2023-07-14_code_sections/code_sections.html",
- "title": "How to use Code Sections",
+ "objectID": "posts/2023-08-08_study_day/study_day.html",
+ "href": "posts/2023-08-08_study_day/study_day.html",
+ "title": "It’s all relative? - Calculating Relative Days using admiral",
"section": "",
- "text": "The admiral package embraces a modular style of programming, where blocks of code are pieced together in sequence to create an ADaM dataset. However, with the well-documented advantages of the modular approach comes the recognition that scripts will on average be longer. As such, astute programmers working in RStudio are constantly on the lookout for quick ways to effectively navigate their scripts. Enter code sections!"
+ "text": "Creating --DY variables for your ADaMs is super easy using derive_vars_dy() from the admiral package.\nLet’s build some dummy data with 4 subjects, a start date/time for treatment (TRTSDTM), an analysis start date/time variable (ASTDTM) and an analysis end date variable (AENDT).\nlibrary(admiral)\nlibrary(lubridate)\nlibrary(dplyr)\n\nadam <- tribble(\n ~USUBJID, ~TRTSDTM, ~ASTDTM, ~AENDT,\n \"001\", \"2014-01-17T23:59:59\", \"2014-01-18T13:09:O9\", \"2014-01-20\",\n \"002\", \"2014-02-25T23:59:59\", \"2014-03-18T14:09:O9\", \"2014-03-24\",\n \"003\", \"2014-02-12T23:59:59\", \"2014-02-18T11:03:O9\", \"2014-04-17\",\n \"004\", \"2014-03-17T23:59:59\", \"2014-03-19T13:09:O9\", \"2014-05-04\"\n) %>%\n mutate(\n TRTSDTM = as_datetime(TRTSDTM),\n ASTDTM = as_datetime(ASTDTM),\n AENDT = ymd(AENDT)\n )\nOkay! Next we run our dataset through derive_vars_dy(), specifying:\nderive_vars_dy(\n adam,\n reference_date = TRTSDTM,\n source_vars = exprs(ASTDTM, AENDT)\n)\n\n# A tibble: 4 × 6\n USUBJID TRTSDTM ASTDTM AENDT ASTDY AENDY\n <chr> <dttm> <dttm> <date> <dbl> <dbl>\n1 001 2014-01-17 23:59:59 2014-01-18 13:09:09 2014-01-20 2 4\n2 002 2014-02-25 23:59:59 2014-03-18 14:09:09 2014-03-24 22 28\n3 003 2014-02-12 23:59:59 2014-02-18 11:03:09 2014-04-17 7 65\n4 004 2014-03-17 23:59:59 2014-03-19 13:09:09 2014-05-04 3 49\nThat’s it! We got both our ASTDY and AENDY variables in only a few short lines of code!\nWhat if I want my variables to have a different naming convention?\nEasy! In the source_vars argument if you want your variables to be called DEMOADY and DEMOEDY just do DEMOADY = ASTDTM and DEMOEDY = AENDT and derive_vars_dy() will do the rest!\nderive_vars_dy(\n adam,\n reference_date = TRTSDTM,\n source_vars = exprs(DEMOADY = ASTDTM, DEMOEDY = AENDT)\n)\n\n# A tibble: 4 × 6\n USUBJID TRTSDTM ASTDTM AENDT DEMOADY DEMOEDY\n <chr> <dttm> <dttm> <date> <dbl> <dbl>\n1 001 2014-01-17 23:59:59 2014-01-18 13:09:09 2014-01-20 2 4\n2 002 2014-02-25 23:59:59 2014-03-18 14:09:09 2014-03-24 22 28\n3 003 2014-02-12 23:59:59 2014-02-18 11:03:09 2014-04-17 7 65\n4 004 2014-03-17 23:59:59 2014-03-19 13:09:09 2014-05-04 3 49\nIf you want to get --DT or --DTM variables using admiral then check out derive_vars_dt() and derive_vars_dtm(). If things are messy in your data, e.g. partial dates, both functions have great imputation abilities, which we will cover in an upcoming blog post!"
},
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html#introduction",
- "href": "posts/2023-07-14_code_sections/code_sections.html#introduction",
- "title": "How to use Code Sections",
+ "objectID": "posts/2023-08-08_study_day/study_day.html#last-updated",
+ "href": "posts/2023-08-08_study_day/study_day.html#last-updated",
+ "title": "It’s all relative? - Calculating Relative Days using admiral",
+ "section": "Last updated",
+ "text": "Last updated\n\n2023-11-27 19:13:57.651626"
+ },
+ {
+ "objectID": "posts/2023-08-08_study_day/study_day.html#details",
+ "href": "posts/2023-08-08_study_day/study_day.html#details",
+ "title": "It’s all relative? - Calculating Relative Days using admiral",
+ "section": "Details",
+ "text": "Details\n\nsource code, R environment"
+ },
+ {
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
"section": "",
- "text": "The admiral package embraces a modular style of programming, where blocks of code are pieced together in sequence to create an ADaM dataset. However, with the well-documented advantages of the modular approach comes the recognition that scripts will on average be longer. As such, astute programmers working in RStudio are constantly on the lookout for quick ways to effectively navigate their scripts. Enter code sections!"
+ "text": "There is significant momentum in driving the adoption of R packages in the life sciences industries, in particular, the R Consortium Submissions Working Group is dedicated to promoting the use of R for regulatory submissions to the FDA.\nThe R Consortium Submissions Working Group successfully completed an R-based submission in November 2021 through the eCTD portal (R Submissions Pilot 1). This Pilot was completed on March 10, 2022 after a successful statistical review and evaluation by the FDA staff.\nMoving forward, the Pilot 2 aimed to include a Shiny application that the FDA staff could deploy on their own servers. The R Consortium recently announced that on September 27, 2023, the R Submissions Working Group successfully completed the follow-up to the Pilot 2 R Shiny-based submission and received a response letter from FDA CDER. This marks the first publicly available submission package that includes a Shiny component. The full FDA response letter can be found here.\nThe Shiny application that was sent for the Pilot 2 had the goal to display the 4 Tables, Listings and Figures (TLFs) that were sent for the Pilot 1 with basic filtering functionality.\nThe submission package adhered to the eCTD folder structure and contained 5 deliverables. Among the deliverables was the proprietary R package {pilot2wrappers}, which enables the execution of the Shiny application.\nThe FDA staff were expected to receive the electronic submission packages in the eCTD format, install and load open-source R packages used as dependencies in the included Shiny application, reconstruct and load the submitted Shiny application, and communicate potential improvements in writing.\nIn the following stage, the R Consortium’s R Submission Working Group launched Pilot 4, aiming to investigate innovative technologies like Linux containers and web assembly. These technologies are being explored to package a Shiny application into a self-contained unit, streamlining the transfer and execution processes for the application.\nIn this post, our aim is to outline how we used the Rhino framework to reproduce the Shiny application that was successfully submitted to the FDA for the Pilot 2 project. Additionally, we detail the challenges identified during the process and how we were able to successfully address them by using an open-source package."
},
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html#so-what-are-code-sections-and-why-are-they-useful",
- "href": "posts/2023-07-14_code_sections/code_sections.html#so-what-are-code-sections-and-why-are-they-useful",
- "title": "How to use Code Sections",
- "section": "So, what are code sections and why are they useful?",
- "text": "So, what are code sections and why are they useful?\nCode Sections are separators for long R scripts or functions in RStudio. They can be set up by inserting a comment line followed by four or more dashes in between portions of code, like so:\n\n# First code section ----\n\na <- 1\n\n# Second code section ----\n\nb <- 2\n\n# Third code section ----\n\nc <- 3\n\nRStudio then recognizes the code sections automatically, and enables you to:\n\nCollapse and expand them using the arrow displayed next to the line number, or with the handy shortcuts Alt+L/Shift+Alt+L on Windows or Cmd+Option+L/Cmd+Shift+Option+L on Mac.\nTravel in between them using the navigator at the bottom of the code pane, or by pressing Shift+Alt+J on Windows or Cmd+Shift+Option+J on Mac.\nView an outline of the file using the “Outline” button at the top right of the pane and/or the orange hashtag “Section Navigator” button at the bottom left of the pane.\n\n\n\n\n\n\nCollapsed sections, outline view and the section navigator for the example above.\n\n\n\n\nIt is also possible to create subsections by using two hashtags at the start of a comment line:\n\n# First code section ----\na <- 1\n\n## A code subsection ----\nb <- 2\n\n# Second code section ----\nc <- 3\n\n\n\n\n\n\nCode subsections for the example above.\n\n\n\n\nFor a complete list of Code Sections shortcuts, and for further information, see here."
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#reproducing-the-r-submission-pilot-2-shiny-app-using-rhino",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#reproducing-the-r-submission-pilot-2-shiny-app-using-rhino",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Reproducing the R Submission Pilot 2 Shiny App using Rhino",
+ "text": "Reproducing the R Submission Pilot 2 Shiny App using Rhino\nWhile the original Shiny application submitted to the FDA was wrapped using {Golem}, we replicated the application using our in-house developed framework Rhino. The main motivation was to provide an example of an R Submission that is not an R package and to identify and solve any issues that may arise from this approach.\nOur demo application (FDA-pilot-app) is accessible on our website, alongside other Shiny and Rhinoverse demonstration apps.\n\nThe code for FDA-pilot-app is open-source. You can create your own Rhino-based application by following our tutorial and viewing our workshop, which is available on YouTube."
},
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html#conclusion",
- "href": "posts/2023-07-14_code_sections/code_sections.html#conclusion",
- "title": "How to use Code Sections",
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#brief-introduction-to-rhino",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#brief-introduction-to-rhino",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Brief Introduction to Rhino",
+ "text": "Brief Introduction to Rhino\n\n\n\n\n\nThe Rhino framework was developed by Appsilon to create enterprise-level Shiny applications, consistently and efficiently. This framework allows developers to apply the best software engineering practices, modularize code, test it thoroughly, enhance UI aesthetics, and monitor user adoption effectively.\nRhino provides support in 3 main areas:\n\nClear Code: scalable architecture, modularization based on Box and Shiny modules.\nQuality: comprehensive testing such as unit tests, E2E tests with Cypress and Shinytest2, logging, monitoring and linting.\nAutomation: project startup, CI with GitHub Actions, dependencies management with {renv}, configuration management with config, Sass and JavaScript bundling with ES6 support via Node.js.\n\nRhino is an ideal fit for highly regulated environments such as regulatory submissions or other drug development processes.\n\nFDA-pilot-app structure\nThe structure of this application is available on the github repository. The structure of this Shiny app is the following.\n\nClick here to expand the FDA-pilot-app structure\n\n.\n├── app\n│ ├── view\n│ │ └── demographic_table.R\n| | └── km_plot.R\n| | └── primary_table.R\n| | └── efficacy_table.R\n| | └── completion_table.R\n│ ├── logic\n│ │ └── adam_data.R\n│ │ └── eff_modles.R\n│ │ └── formatters.R\n│ │ └── helpers.R\n│ │ └── kmplot_helpers.R\n│ │ └── Tplyr_helpers.R\n│ ├── data\n│ │ └── adam\n│ │ └── adadas.xpt\n│ │ └── adlbc.xpt\n│ │ └── adsl.xpt\n│ │ └── adtte.xpt\n│ ├── docs\n│ │ └── about.md\n│ ├── js\n│ │ └── index.js\n│ ├── static\n│ │ └── favicon.ico\n│ ├── styles\n│ │ └── main.scss\n│ └── app.R\n├── tests\n│ ├── cypress\n│ │ └── integration\n│ │ └── app.spec.js\n│ ├── testthat\n│ │\n│ └── cypress.json\n├── app.R\n├── rhino_submission.Rproj\n├── dependencies.R\n├── renv.lock\n├── rhino.yml\n└── README.md"
+ },
+ {
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#efficient-submissions-to-the-fda",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#efficient-submissions-to-the-fda",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Efficient Submissions to the FDA",
+ "text": "Efficient Submissions to the FDA\n\n\n\n\n\nTo comply with the Electronic Submission File Formats and Specifications for the eCTD submission, the programming code should carry a “.txt” extension. In the R Submissions Pilot 3 the group did not use {pkglite} as the FDA clarified that “.zip” and “.r” files are acceptable for submission. In this case, we utilized the {pkglite} R package to efficiently pack and unpack the FDA-pilot-app. This approach would facilitate the FDA reviewers in setting up the submission on their systems.\nThis package allows packing R packages to “.txt” files, which are supported for the submission of proprietary packages to the FDA via the eCTD gateway. \n\nPacking the FDA-pilot-app into a .txt file\nThe code below can be used to pack the Shiny application into a .txt file:\n\napp_name <- \"rhinosubmission\"\nrenv_spec <- pkglite::file_spec(\n \"renv\",\n pattern = \"^settings\\\\.dcf$|^activate\\\\.R$\",\n format = \"text\", recursive = FALSE\n)\ntests_spec <- pkglite::file_tests()\napp_spec <- pkglite::file_auto(\"app\")\nroot_spec <- pkglite::file_spec(\n path = \".\",\n pattern = \"^\\\\.Rprofile$|^rhino\\\\.yml$|^renv\\\\.lock$|^dependencies\\\\.R$|^config\\\\.yml$|^app\\\\.R$|^README\\\\.md$|\\\\.Rproj$\",\n all_files = TRUE,\n recursive = FALSE\n)\nwrite(paste0(\"Package: \", app_name), \"DESCRIPTION\", append = TRUE)\npkglite::collate(\n getwd(),\n renv_spec,\n tests_spec,\n app_spec,\n root_spec\n) |> pkglite::pack()\nfile.remove(\"DESCRIPTION\")\n\n\n\nUnpacking the FDA-pilot-app\nThe packed “.txt” file can be unpacked into a Shiny app by using {pkglite} as follows:\n\npkglite::unpack(\"rhinosubmission.txt\")"
+ },
+ {
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#lessons-learned",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#lessons-learned",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Lessons Learned",
+ "text": "Lessons Learned\nOur initial objective was to prove that it would be possible to submit a Shiny application using Rhino through the eCTD gateway. During the rewriting process we identified that this could be done by integrating the open-source {pkglite} package. By following this approach, we concluded that it would be possible to submit a Shiny application through the eCTD gateway. This was also achieved through the successful submission of a package that included a Shiny component in Pilot 2.\nHaving rewritten the R Submissions Pilot 2 Shiny application using Rhino holds major implications for the adoption of our framework within the life sciences. Apart from being a strong, opinionated framework that improves reproducibility and reliability for Shiny development, using Rhino for regulatory submissions could improve the flexibility and speed in the clinical reporting pipeline. This would accelerate the adoption of R/Shiny for submissions to the FDA or other regulatory agencies."
+ },
+ {
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#last-updated",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#last-updated",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Last updated",
+ "text": "Last updated\n\n2023-11-27 19:13:54.254558"
+ },
+ {
+ "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#details",
+ "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#details",
+ "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "section": "Details",
+ "text": "Details\n\nsource code, R environment"
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html",
+ "title": "Blanks and NAs",
+ "section": "",
+ "text": "Reading in SAS-based datasets (.sas7bdat or xpt) into R has users calling the R package haven. A typical call might invoke read_sas() or read_xpt() to bring in your source data to construct your ADaMs or SDTMs.\nUnfortunately, while using haven the character blanks (missing data) found in a typical SAS-based dataset are left as blanks. These blanks will typically prove problematic while using functions like is.na in combination with dplyr::filter() to subset data. Check out Bayer’s SAS2R catalog: handling-of-missing-values for more discussion on missing values and NAs.\nIn the admiral package, we have built a simple function called convert_blanks_to_na() to help us quickly remedy this problem. You can supply an entire dataframe to this function and it will convert any character blanks to NA_character_"
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#loading-packages-and-making-dummy-data",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#loading-packages-and-making-dummy-data",
+ "title": "Blanks and NAs",
+ "section": "Loading Packages and Making Dummy Data",
+ "text": "Loading Packages and Making Dummy Data\n\nlibrary(admiral)\nlibrary(tibble)\nlibrary(dplyr)\n\ndf <- tribble(\n ~USUBJID, ~RFICDTC,\n \"01\", \"2000-01-01\",\n \"02\", \"2001-01-01\",\n \"03\", \"\", # Here we have a character blank\n \"04\", \"2001-01--\",\n \"05\", \"2001---01\",\n \"05\", \"\", # Here we have a character blank\n)\n\ndf\n\n# A tibble: 6 × 2\n USUBJID RFICDTC \n <chr> <chr> \n1 01 \"2000-01-01\"\n2 02 \"2001-01-01\"\n3 03 \"\" \n4 04 \"2001-01--\" \n5 05 \"2001---01\" \n6 05 \"\""
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#a-simple-conversion",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#a-simple-conversion",
+ "title": "Blanks and NAs",
+ "section": "A simple conversion",
+ "text": "A simple conversion\n\ndf_na <- convert_blanks_to_na(df)\n\ndf_na\n\n# A tibble: 6 × 2\n USUBJID RFICDTC \n <chr> <chr> \n1 01 2000-01-01\n2 02 2001-01-01\n3 03 <NA> \n4 04 2001-01-- \n5 05 2001---01 \n6 05 <NA> \n\n\n\ndf_na %>% filter(is.na(RFICDTC))\n\n# A tibble: 2 × 2\n USUBJID RFICDTC\n <chr> <chr> \n1 03 <NA> \n2 05 <NA>"
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#thats-it",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#thats-it",
+ "title": "Blanks and NAs",
+ "section": "That’s it!",
+ "text": "That’s it!\nA simple call to this function can make your derivation life so much easier while working in R if working with SAS-based datasets. In admiral, we make use of this function at the start of all ADaM templates for common ADaM datasets. You can use the function use_ad_template() to get the full R script for the below ADaMs.\n\nlist_all_templates()\n\nExisting ADaM templates in package 'admiral':\n• ADAE\n• ADCM\n• ADEG\n• ADEX\n• ADLB\n• ADLBHY\n• ADMH\n• ADPC\n• ADPP\n• ADPPK\n• ADSL\n• ADVS"
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#last-updated",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#last-updated",
+ "title": "Blanks and NAs",
+ "section": "Last updated",
+ "text": "Last updated\n\n2023-11-27 19:13:51.677601"
+ },
+ {
+ "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#details",
+ "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#details",
+ "title": "Blanks and NAs",
+ "section": "Details",
+ "text": "Details\n\nsource code, R environment"
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html",
+ "href": "posts/2023-06-27__hackathon_app/index.html",
+ "title": "Hackathon Feedback Application",
+ "section": "",
+ "text": "We recently created a shiny application for the admiral hackathon in February 2023. The admiral hackathon was an event designed to make statistical programmers from the pharmaceutical industry more comfortable with the admiral R package which allows users to efficiently transform data from one data standard (SDTM) to another (ADaM).\nHackathon participants formed groups of up to five people and were then tasked to create R-scripts that map the SDTM data to ADaM according to specifics defined in the metadata.\nThe purpose of the shiny app was threefold:\nIn this blog post I want to highlight some of the thoughts that went into this application. Please keep in mind that this work was done under tight time restraints.\nThe hackathon application is still online (although data-upload is switched off) and the GitHub repository is publicly available. The application is embedded into this post right after this paragraph. I have also uploaded to GitHub a .zip file of the workspace to which hackathon participants had access via posit cloud. For more context you can watch recordings of the hackathon-meetings."
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#permanent-data",
+ "href": "posts/2023-06-27__hackathon_app/index.html#permanent-data",
+ "title": "Hackathon Feedback Application",
+ "section": "Permanent Data",
+ "text": "Permanent Data\nThe biggest challenge you have to consider for this app is the permanent data storage. Shiny apps run on a server. Although we can write files on this server, whenever the app restarts, the files are lost. Therefore, a persistent data storage solution is required.\n\nGoogle drive\nI decided to leverage Google drive using the googledrive package package. This allowed me to save structured data (the team registry and the submission scores) as well as unstructured data (their R-script files).\n\n\n\n\n\n\nAuthentication\n\n\n\nTo access Google drive using the googledrive package we need to authenticate. This can be done interactively using the command googledrive::drive_auth() which takes you to the Google login page. After login you receive an authentication token requested by R.\nFor non-interactive authentication this token must be stored locally. In our case where the shiny app must access the token once deployed, the token must be stored on the project level.\nI have included the authentication procedure I followed in the R folder in google_init.R. You can find more extensive documentation of the non-interactive authentication.\n\n\nThe initial concept was: Each team gets their own folder including the most recent submission for each task, and a .csv file containing team information. To keep track of the submissions and the respective scores we wrote a .csv file in the mock-hackathon folder, so one folder above the team folders.\nSaving the team info as a .csv file worked fine as each team received their own file which – once created – was not touched anymore. As each upload for every team should simply add a row to the submissions.csv file, appending the file would be ideal. This was not possible using the googledrive package package. Instead, for each submission, the submissions file was downloaded, appended, and uploaded again. Unfortunately, this lead to a data loss, as the file was continuously overwritten, especially when two teams would submit simultaneously.\n\n\n\n\n\n\nRecover the Lost Data\n\n\n\nWhenever the submissions.csv file was uploaded, the previous version was sent to the Google drive bin. We ended up with over 3000 submissions.csv files containing a lot of redundant information. I had to write the following chunk to first get the unique file IDs of the 3000 submissions.csv files, create an empty submissions data-frame, and then download each file and add its information to the submisisons data-frame. To keep the data-frame as light as possible, after each append I deleted all duplicate submissions.\n\n# get all task_info.csv ID's\n# each row identifies one file in the trash\ntask_info_master <- drive_find(\n pattern = \"task_info.csv\",\n trashed = TRUE\n)\n\n\n# set up empty df to store all submissions\norigin <- tibble(\n score = numeric(),\n task = character(),\n team = character(),\n email = character(),\n time = character()\n)\n\n# downloads, reads, and returns one csv file given a file id\nget_file <- function(row) {\n tf <- tempfile()\n row %>%\n as_id() %>%\n drive_download(path = tf)\n new <- read_csv(tf) %>%\n select(score, task, team) %>%\n distinct()\n}\n\n\n# quick and dirty for loop to subsequently download each file, extract information\n# merge with previous information and squash it (using distinct()).\nfor (i in 1:nrow(task_info_master)) {\n origin <- rbind(origin, get_file(row = task_info_master[i, ])) %>%\n distinct()\n\n # save progress in a separate file after every 100 downloaded and merged sheets\n if (i %% 100 == 0) {\n print(i)\n write_csv(origin, paste(\"prog_data/task_info_prog_\", i, \".csv\", sep = \"\"))\n # update on progress\n message(i / nrow(task_info_master) * 100)\n }\n}\n\nWhen doing such a time-intensive task, make sure to try it first with only a couple of files to see whether any errors are produced. I am not quite sure how long this took but when I returned from my lunch break everything had finished.\n\n\nIf you want to stay in the Google framework, I recommend using the googlesheets4 package for structured data. googlesheets4 allows appending new information to an already existing sheet without the need to download the file first. As both packages follow the same style, going from one to the other is really simple. googlesheets4 requires authentication as well. However, you can reuse the cached token from the googledrive package authentication by setting gs4_auth(token = drive_token()).\n\n\nSecurity Concerns\nConnecting a public shiny app to your Google account introduces a security vulnerability in general. Especially so because we implemented the upload of files to Google drive. And even more problematic: We run a user generated script and display some of its output. A malicious party might be able to extract the authentication token of our Google account or could upload malware to the drive.\nTo reduce the risk, I simply created an un-associated Google account to host the drive. There are certainly better options available, but this seemed a reasonable solution achieved with very little effort."
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#register-team",
+ "href": "posts/2023-06-27__hackathon_app/index.html#register-team",
+ "title": "Hackathon Feedback Application",
+ "section": "Register Team",
+ "text": "Register Team\nWe wanted to allow users to sign up as teams using the shiny app. The app provides a simple interface where users could input a team name and the number of members. This in turn would open two fields for each user to input their name and email address.\nWe do simple checks to make sure at least one valid email address is supplied, and that the group name is acceptable. The group name cannot be empty, already taken, or contain vulgar words.\nThe team registration itself was adding the team information to the Google sheets file event_info into the sheet teams and to create a team folder in which to store the uploaded R files.\nThe checks and registration is implemented in the register_team() function stored in interact_with_google.R.\n\n\n\nScreenshot of the register team interface\n\n\nThe challenge here was to adapt the number of input fields depending on the number of team members. This means that the team name and email interface must be rendered: First, we check how many team members are part of the group, this is stored in the input$n_members input variable. Then we create a tagList with as many elements as team members. Each element contains two columns, one for the email, one for the member name. This tagList is then returned and displayed to the user.\n\n# render email input UI of the register tab\noutput$name_email <- shiny::renderUI({\n # create field names\n N <- input$n_members\n NAME <- sapply(1:N, function(i) {\n paste0(\"name\", i)\n })\n EMAIL <- sapply(1:N, function(i) {\n paste0(\"email\", i)\n })\n\n output <- tagList()\n\n\n firstsecondthird <- c(\"First\", \"Second\", \"Third\", \"Fourth\", \"Fifth\")\n for (i in 1:N) {\n output[[i]] <- tagList()\n output[[i]] <- fluidRow(\n shiny::h4(paste(firstsecondthird[i], \" Member\")),\n column(6,\n textInput(NAME[i], \"Name\"),\n value = \" \" # displayed default value\n ),\n column(6,\n textInput(EMAIL[i], \"Email\"),\n value = \" \"\n )\n )\n }\n output\n})\n\nThe team information is then uploaded to Google drive. Because some teams have more members than others, we have to create the respective data-frame with the number of team members in mind.\nThe following chunk creates the registration data. Noteworthy here the creation of the NAME and EMAIL variables which depend on the number of members in this team. Further, the user input of these fields is extracted via input[[paste0(NAME[i])]] within a for-loop.\nWe also make the data-creation dependent on the press of the Register Group button and cache some variables.\n\n## registration\nregistrationData <-\n reactive({\n N <- input$n_members\n NAME <- sapply(1:N, function(i) {\n paste0(\"name\", i)\n })\n EMAIL <- sapply(1:N, function(i) {\n paste0(\"email\", i)\n })\n names <- character(0)\n emails <- character(0)\n\n for (i in 1:N) {\n names[i] <- input[[paste0(NAME[i])]]\n emails[i] <- input[[paste0(EMAIL[i])]]\n }\n # create df\n dplyr::tibble(\n team_name = input$team_name,\n n_members = N,\n member_name = names,\n member_email = emails\n )\n }) %>%\n bindCache(input$team_name, input$n_members, input$name1, input$email1) %>%\n bindEvent(input$register) # wait for button press"
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#upload-source-script",
+ "href": "posts/2023-06-27__hackathon_app/index.html#upload-source-script",
+ "title": "Hackathon Feedback Application",
+ "section": "Upload & Source Script",
+ "text": "Upload & Source Script\nTo upload a script, participants had to select their team first. The input options were based on the existing folders on the Google-drive in the mock_hackathon folder. To upload a particular script participants had to also select the task to be solved. The uploaded script is then uploaded to the team folder following a standardised script naming convention.\nThere are different aspects to be aware of when sourcing scripts on a shiny server. For example, you have to anticipate the packages users will include in their uploaded scripts, as their scripts will load but not install packages. Further, you should keep the global environment of your shiny app separate from the environment in which the script is sourced. This is possible by supplying an environment to the source() function, e.g: source(path_to_script, local = new.env())\nAnother thing we had to consider was to replicate the exact folder-structure on the shiny server that participants were working with when creating the scripts, as they were required to source some scripts and to save their file into a specific folder. This was relatively straight forward as we provided participants with a folder structure in the posit cloud instance they were using. They had access to the sdtm folder in which the data was stored, and the adam folder into which they saved their solutions. The structure also included a folder with metadata which was also available on the shiny server.\nFor some tasks, participants required some ADaM-datasets stored in the adam folder, essentially the output from previous tasks. This was achieved by first creating a list mapping tasks to the required ADaM datasets:\n\ndepends_list <- list(\n \"ADADAS\" = c(\"ADSL\"),\n \"ADAE\" = c(\"ADSL\"),\n \"ADLBC\" = c(\"ADSL\"),\n \"ADLBH\" = c(\"ADSL\"),\n \"ADLBHY\" = c(\"ADSL\"),\n \"ADSL\" = NULL,\n \"ADTTE\" = c(\"ADSL\", \"ADAE\"),\n \"ADVS\" = c(\"ADSL\")\n)\n\nThis list is sourced from the R/parameters.R file when initiating the application. We then call the get_depends() function sourced from R/get_depends.R which copies the required files from the key folder (where our solutions to the tasks were stored) to the adam folder. After sourcing the uploaded script the content in the adam folder is deleted."
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#compare-to-solution-file",
+ "href": "posts/2023-06-27__hackathon_app/index.html#compare-to-solution-file",
+ "title": "Hackathon Feedback Application",
+ "section": "Compare to Solution File",
+ "text": "Compare to Solution File\nWe want to compare the file created by participants with our solution (key) file stored in the key folder. The diffdf::diffdf() function allows for easy comparison of two data-frames and directly provides extensive feedback for the user:\n\nlibrary(dplyr)\ndf1 <- tibble(\n numbers = 1:10,\n letters = LETTERS[1:10]\n)\ndf2 <- tibble(\n numbers = 1:10,\n letters = letters[1:10]\n)\n\ndiffdf::diffdf(df1, df2)\n\nWarning in diffdf::diffdf(df1, df2): \nNot all Values Compared Equal\n\n\nDifferences found between the objects!\n\nA summary is given below.\n\nNot all Values Compared Equal\nAll rows are shown in table below\n\n =============================\n Variable No of Differences \n -----------------------------\n letters 10 \n -----------------------------\n\n\nAll rows are shown in table below\n\n ========================================\n VARIABLE ..ROWNUMBER.. BASE COMPARE \n ----------------------------------------\n letters 1 A a \n letters 2 B b \n letters 3 C c \n letters 4 D d \n letters 5 E e \n letters 6 F f \n letters 7 G g \n letters 8 H h \n letters 9 I i \n letters 10 J j \n ----------------------------------------"
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#score",
+ "href": "posts/2023-06-27__hackathon_app/index.html#score",
+ "title": "Hackathon Feedback Application",
+ "section": "Score",
+ "text": "Score\nTo compare submissions between participants we implemented a simple scoring function (score_f()) based on the table comparison by diffdf(). The function can be found in the compare_dfs.R file:\n\nscore_f <- function(df_user, df_key, keys) {\n score <- 10\n diff <- diffdf::diffdf(df_user, df_key, keys = keys)\n if (!diffdf::diffdf_has_issues(diff)) {\n return(score)\n }\n\n # check if there are any differences if the comparison is not strict:\n if (!diffdf::diffdf_has_issues(diffdf::diffdf(df_user,\n df_key,\n keys = keys,\n strict_numeric = FALSE,\n strict_factor = FALSE\n ))) {\n # if differences are not strict, return score - 1\n return(score - 1)\n }\n\n return(round(min(max(score - length(diff) / 3, 1), 9), 2))\n}\n\nEvery comparison starts with a score of 10. We then subtract the length of the comparison object divided by a factor of 3. The length of the comparison object is a simplified way to represent the difference between the two data-frames by one value. Finally, the score is bounded by 1 using max(score, 1).\nThe score is not a perfect capture of the quality of the script uploaded but: 1. helped participants get an idea of how close their data-frame is to the solution file 2. allowed us to raffle prizes based on the merit of submitted r-scripts"
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#reactiveness",
+ "href": "posts/2023-06-27__hackathon_app/index.html#reactiveness",
+ "title": "Hackathon Feedback Application",
+ "section": "Reactiveness",
+ "text": "Reactiveness\nSome of the app functions can take quite some time to execute, e.g. running the uploaded script. Other tasks, e.g. registering a team, do not intrinsically generate user facing outputs. This would make the app using really frustrating, as users would not know whether the app is correctly working or whether it froze.\nWe implemented two small features that made the app more responsive. One is simple loading icons that integrate into the user interface and show that output is being computed – that something is working. The other is a pop up window which communicates whether team registration was successful, and if not, why not.\nWe further aimed to forward errors generated by the uploaded scripts to the user interface, but errors generated by the application itself should be concealed."
+ },
+ {
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#conclusion",
+ "href": "posts/2023-06-27__hackathon_app/index.html#conclusion",
+ "title": "Hackathon Feedback Application",
"section": "Conclusion",
- "text": "Conclusion\nCode sections are an easy way to navigate long scripts and foster good commenting practices. They are used extensively in the admiral package, but there is no reason that you cannot start using them yourself in your day-to-day R programming!"
+ "text": "Conclusion\nAlthough the application was continuously improved during the hackathon it proved to be a useful resource for participants from day one as it allowed groups to set their own pace. It further allowed admiral developers to gain insights on package usage of a relatively large sample of potential end users. From our perspective, the application provided a great added value to the hackathon and eased the workload of guiding the participants through all the tasks."
},
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html#last-updated",
- "href": "posts/2023-07-14_code_sections/code_sections.html#last-updated",
- "title": "How to use Code Sections",
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#last-updated",
+ "href": "posts/2023-06-27__hackathon_app/index.html#last-updated",
+ "title": "Hackathon Feedback Application",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:53.276819"
+ "text": "Last updated\n\n2023-11-27 19:13:48.333469"
},
{
- "objectID": "posts/2023-07-14_code_sections/code_sections.html#details",
- "href": "posts/2023-07-14_code_sections/code_sections.html#details",
- "title": "How to use Code Sections",
+ "objectID": "posts/2023-06-27__hackathon_app/index.html#details",
+ "href": "posts/2023-06-27__hackathon_app/index.html#details",
+ "title": "Hackathon Feedback Application",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html",
- "title": "The pharmaverse (hi)story",
+ "objectID": "posts/2023-11-27_higher_order/higher_order.html",
+ "href": "posts/2023-11-27_higher_order/higher_order.html",
+ "title": "Believe in a higher order!",
"section": "",
- "text": "Pharmaverse: from motivation to present"
+ "text": "Picture the following scenario:\nYou, a budding {admiral} programmer, are finding your groove chaining together modular code blocks to derive variables and parameters in a drive to construct your favorite ADaM dataset, ADAE. Suddenly you notice that one of the flags you are deriving should only use records on or after study day 1. In a moment of mild annoyance, you get to work modifying what was originally a simple call to derive_var_extreme_flag() by first subsetting ADAE to records where AESTDY > 1, then deriving the flag only for the subsetted ADAE, and finally binding the two portions of ADAE back together before continuing on with your program. Miffed by this interruption, you think to yourself: “I wish there was a neater, faster way to do this in stride, that didn’t break my code modularity…”\nIf the above could never be you, then you’ll probably be alright never reading this blog post. However, if you want to learn more about the tools that {admiral} provides to make your life easier in cases like this one, then you are in the right place, since this blog post will highlight how higher order functions can solve such issues.\nA higher order function is a function that takes another function as input. By introducing these higher order functions, {admiral} intends to give the user greater power over derivations, whilst trying to negate the need for both adding additional {admiral} functions/arguments, and the user needing many separate steps.\nThe functions covered in this post are:\n\nrestrict_derivation(): Allows the user to execute a single derivation on a subset of the input dataset.\ncall_derivation(): Allows the user to call a single derivation multiple times with some arguments being fixed across iterations and others varying.\nslice_derivation(): Allows the user to split the input dataset into slices (subsets) and for each slice a single derivation is called separately. Some or all arguments of the derivation may vary depending on the slice.\n\n\n\nThe examples in this blog post require the following packages.\n\nlibrary(admiral)\nlibrary(pharmaversesdtm)\nlibrary(dplyr, warn.conflicts = FALSE)\n\nFor example purpose, the ADSL dataset - which is included in {admiral} - and the SDTM datasets from {pharmaversesdtm} are used.\n\ndata(\"admiral_adsl\")\ndata(\"ae\")\ndata(\"vs\")\nadsl <- admiral_adsl\nae <- convert_blanks_to_na(ae)\nvs <- convert_blanks_to_na(vs)\n\nThe following code creates a minimally viable ADAE dataset to be used where needed in the following examples.\n\nadae <- ae %>%\n left_join(adsl, by = c(\"STUDYID\", \"USUBJID\")) %>%\n derive_vars_dt(\n new_vars_prefix = \"AST\",\n dtc = AESTDTC,\n highest_imputation = \"M\"\n ) %>%\n mutate(\n TRTEMFL = if_else(ASTDT >= TRTSDT, \"Y\", NA_character_),\n TEMP_AESEVN = as.integer(factor(AESEV, levels = c(\"SEVERE\", \"MODERATE\", \"MILD\")))\n )"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#human-history-and-pharmaverse-context",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#human-history-and-pharmaverse-context",
- "title": "The pharmaverse (hi)story",
- "section": "Human history and pharmaverse context",
- "text": "Human history and pharmaverse context\nSince the Australian Aboriginal, the earliest peoples recorded to have inhabited the Earth and who have been in Australia for at least 65,000 to 80,000 years (Encyclopædia Britannica), human beings live in group. Whether to protect yourself, increase your life expectancy or simply share tasks.\nFor most aspects of life, it doesn’t make sense to think, act or work alone for two main reasons:\n\nYou will spend more energy and time;\nSomeone else may be facing (or have faced) the same situation.\n\nThe English poet John Donne used to say “No man is an island entire of itself; every man is a piece of the continent, a part of the main;”. I can’t disagree with him. And I dare say that Ari Siggaard Knoph (Novo Nordisk), Michael Rimler (GSK), Michael Stackhouse (Atorus), Ross Farrugia (Roche), and Sumesh Kalappurakal (Janssen) can’t disagree with him either. They are the founders of pharmaverse, members of its Council and kindly shared their memories of how independent companies, in mid-2020, worked together in the creation of a set of packages developed to support the clinical reporting pipeline.\nIf you are not familiar with this pipeline, the important thing to know is that, in a nutshell, pharmaceutical companies must follow a bunch of standardized procedures and formats (from Clinical Data Interchange Standards Consortium, CDISC) when submitting clinical results to Health Authorities. The focus is on this: different companies seeking the same standards for outputs.\nParaphrasing Ross Farrugia (Roche) Breaking boundaries through open-source collaboration presentation in R/Pharma 2022 and thinking of the development of a new drug, we are talking about a “post-competitive” scenario: the drug has already been discovered and the companies should “just” produce and deliver standardized results."
+ "objectID": "posts/2023-11-27_higher_order/higher_order.html#required-packages",
+ "href": "posts/2023-11-27_higher_order/higher_order.html#required-packages",
+ "title": "Believe in a higher order!",
+ "section": "",
+ "text": "The examples in this blog post require the following packages.\n\nlibrary(admiral)\nlibrary(pharmaversesdtm)\nlibrary(dplyr, warn.conflicts = FALSE)\n\nFor example purpose, the ADSL dataset - which is included in {admiral} - and the SDTM datasets from {pharmaversesdtm} are used.\n\ndata(\"admiral_adsl\")\ndata(\"ae\")\ndata(\"vs\")\nadsl <- admiral_adsl\nae <- convert_blanks_to_na(ae)\nvs <- convert_blanks_to_na(vs)\n\nThe following code creates a minimally viable ADAE dataset to be used where needed in the following examples.\n\nadae <- ae %>%\n left_join(adsl, by = c(\"STUDYID\", \"USUBJID\")) %>%\n derive_vars_dt(\n new_vars_prefix = \"AST\",\n dtc = AESTDTC,\n highest_imputation = \"M\"\n ) %>%\n mutate(\n TRTEMFL = if_else(ASTDT >= TRTSDT, \"Y\", NA_character_),\n TEMP_AESEVN = as.integer(factor(AESEV, levels = c(\"SEVERE\", \"MODERATE\", \"MILD\")))\n )"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#clinical-reporting-outputs",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#clinical-reporting-outputs",
- "title": "The pharmaverse (hi)story",
- "section": "Clinical reporting outputs",
- "text": "Clinical reporting outputs\nRationally, we can say that companies face the same challenges in these steps of the process. Not so intuitively, we can also say they were working in silos on that before 2018. Just as Isaac Newton and Gottfried W. Leibniz independently developed the theory of infinitesimal calculus, pharmaceutical companies were independently working on R solutions for this pipeline.\nBut on August 16 and 17 of the mentioned year above, they gathered at the first edition of R/Pharma conference to discuss R and open-source tooling for drug development (the reasons why open-source is an advantageous approach can be found in this post written by Stefan Thoma). And according to Isabela Velásquez’s article, Pharmaverse: Packages for clinical reporting workflows, one of the most popular questions in this conference was “Is the package code available or on CRAN?”.\nWell, many of them were. And not necessarily at that date, but just to mention a few: pharmaRTF and Tplyr from Atorus, r2rtf from Merck, rtables from Roche, etc. The thing is that, overall, there were almost 10000 other packages as well (today, almost 20000). And that took to another two questions:\n\nWith this overwhelming number of packages on CRAN, how to find the ones related to solving “clinical reporting problems”?\nOnce the packages were found, how to choose which one to use among those that have the same functional purpose?\n\nSo, again, companies re-started to working in silos to find those answers. But now, in collaborative silos and with common goals: create extremely useful packages to solve pharmaceutical-specific gaps once and solve them well!"
+ "objectID": "posts/2023-11-27_higher_order/higher_order.html#last-updated",
+ "href": "posts/2023-11-27_higher_order/higher_order.html#last-updated",
+ "title": "Believe in a higher order!",
+ "section": "Last updated",
+ "text": "Last updated\n\n2023-11-27 19:13:43.938122"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#first-partnerships",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#first-partnerships",
- "title": "The pharmaverse (hi)story",
- "section": "First partnerships",
- "text": "First partnerships\nIn 2020, Michael Stackhouse (Atorus) and Michael Rimler (GSK) talked and formed a partnership between their companies to develop a few more packages, including metacore, to read, store and manipulate metadata for ADaMs/SDTMs in a standardized object; xportr, to create submission compliant SAS transport files and perform pharma specific dataset level validation checks; and logrx (ex-timber), to build log to support reproducibility and traceability of an R script.\nAround the same time, Thomas Neitmann (currently at Denali Therapeutics, then at Roche) and Michael Rimler (GSK) discovered that both were working with ADaM in R, so Thomas Neitmann (currently at Denali Therapeutics, then at Roche), Ross Farrugia (Roche) and Michael Rimler (GSK) saw an opportunity there and GSK started their partnership with Roche to build and release admiral package.\nThe idea of working together, the sense of community, and the appetite from organizations built more and more, with incentive and priority established up into the programming heads council.\nJanssen had a huge effort in building R capabilities going on as well, by releasing tidytlg and envsetup), so eventually Michael Rimler (GSK), Michael Stackhouse (Atorus) and Ross Farrugia (Roche) formalized pharmaverse and formed the council, adding in Sumesh Kalappurakal (Janssen) and Ari Siggaard Knoph (Novo Nordisk) later joined as the fifth council member."
+ "objectID": "posts/2023-11-27_higher_order/higher_order.html#details",
+ "href": "posts/2023-11-27_higher_order/higher_order.html#details",
+ "title": "Believe in a higher order!",
+ "section": "Details",
+ "text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#release-growth-and-developments",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#release-growth-and-developments",
- "title": "The pharmaverse (hi)story",
- "section": "Release, growth and developments",
- "text": "Release, growth and developments\nAt the end of their presentation “Closing the Gap: Creating an End to End R Package Toolkit for the Clinical Reporting Pipeline.”, in R/Pharma 2021, Ben Straub (GSK) and Eli Miller (Atorus) welcomed the community to the pharmaverse, a curated collection of packages developed by various pharmaceutical companies to support open-source clinical workflows.\nFrom the outset, the name pharmaverse was chosen so that it could be a neutral home, unrelated to any company. Also, it was established as not being a consortium, which means that founders don’t own, fund, or maintain the packages. Some individuals and companies maintain them but often allowing for community contributions and being licensed permissively so that there is always a feeling of community ownership. The focus of pharmaverse early on, and today, is on inter organization cooperation, to build an environment where, if organizations identify that they have a joint problem that they want to solve, this is the right space to work on and release it.\nPharmaverse has grown a lot, at the time of writing this post we have >25 packages recommended in pharmaverse, and this has led to a partnership with PHUSE to get support from their organization and platform, and because they are eager to advance and support pharmaverse mission.\nDespite all its structure, it is impossible to say that we have a single solution for each clinical reporting analysis when it comes to pharmaverse, a single pathway is impractical. Instead, it is necessary to accept viable tools fitting different pathways into pharmaverse to direct and give people options as to what might work for them. After all, even though we live together as a community, we still have our own unique internal problems.\n\n\n\nSample of pharmaverse packages"
+ "objectID": "posts/2023-07-09_falcon/falcon.html",
+ "href": "posts/2023-07-09_falcon/falcon.html",
+ "title": "falcon",
+ "section": "",
+ "text": "The {falcon} initiative is an industry collaborative effort under {pharmaverse} that unites Boehringer Ingelheim, Moderna, Roche, and Sanofi with the aspiration of building and open-sourcing a comprehensive catalog of harmonized tables, listings, and graphs (TLGs) for clinical study reporting. By leveraging existing open-source R packages, {falcon} aims to simplify the process of output review, comparison, and meta-analyses, while fostering efficient communication among stakeholders in the pharmaceutical industry."
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#last-updated",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#last-updated",
- "title": "The pharmaverse (hi)story",
+ "objectID": "posts/2023-07-09_falcon/falcon.html#what-is-falcon",
+ "href": "posts/2023-07-09_falcon/falcon.html#what-is-falcon",
+ "title": "falcon",
+ "section": "",
+ "text": "The {falcon} initiative is an industry collaborative effort under {pharmaverse} that unites Boehringer Ingelheim, Moderna, Roche, and Sanofi with the aspiration of building and open-sourcing a comprehensive catalog of harmonized tables, listings, and graphs (TLGs) for clinical study reporting. By leveraging existing open-source R packages, {falcon} aims to simplify the process of output review, comparison, and meta-analyses, while fostering efficient communication among stakeholders in the pharmaceutical industry."
+ },
+ {
+ "objectID": "posts/2023-07-09_falcon/falcon.html#why-do-we-build-it",
+ "href": "posts/2023-07-09_falcon/falcon.html#why-do-we-build-it",
+ "title": "falcon",
+ "section": "Why do we build it?",
+ "text": "Why do we build it?\nThe collaborative effort focuses on improving the clarity, consistency, and accessibility of TLGs by addressing variations and redundancies in their creation and use. This harmonized approach allows for streamlined reporting processes and facilitates effective communication of study results within the industry and to regulatory authorities."
+ },
+ {
+ "objectID": "posts/2023-07-09_falcon/falcon.html#what-has-been-done-so-far",
+ "href": "posts/2023-07-09_falcon/falcon.html#what-has-been-done-so-far",
+ "title": "falcon",
+ "section": "What has been done so far?",
+ "text": "What has been done so far?\nDrawing inspiration from the FDA Standard Safety Tables and Figures Integrated Guide, the {falcon} initiative has successfully developed and open-sourced 11 templates to date. 4 product owners and 11 developers from 4 companies have collaborated to make these templates available and also published them on the official {falcon} website at https://pharmaverse.github.io/falcon/."
+ },
+ {
+ "objectID": "posts/2023-07-09_falcon/falcon.html#next-steps-vision",
+ "href": "posts/2023-07-09_falcon/falcon.html#next-steps-vision",
+ "title": "falcon",
+ "section": "Next steps & vision",
+ "text": "Next steps & vision\nFuture plans for {falcon} involve expanding the catalog through continuous collaboration from participating companies and inviting wider industry engagement. The ultimate goal is to promote harmonization of TLGs for clinical reporting across the pharmaceutical industry, leading to greater efficiency, collaboration, and innovation. Even though templates currently come from a published FDA guide, the collaborating companies are open to share and discuss similarities and differences on analysis concepts and output layouts of their own implementations in clinical reporting, for both safety and efficacy analyses.\nIn addition, while currently all templates were built using {rtables}, {tern}, {rlistings} and drew inspiration from the open-sourced TLG-Catalog, moving forward, we plan to support creating the same templates using alternative table engines such as {gt}.\n{falcon} will be presented at the upcoming PHUSE EU (Standards Implementation stream), where we will share the collaboration journey of {falcon} so far, providing more details on the current progress, long-term vision, and strategies for this initiative. Attendees will gain insights into the challenges and opportunities of harmonizing clinical reporting through open-source collaboration and learn about the potential benefits and future direction of {falcon}."
+ },
+ {
+ "objectID": "posts/2023-07-09_falcon/falcon.html#last-updated",
+ "href": "posts/2023-07-09_falcon/falcon.html#last-updated",
+ "title": "falcon",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:48.201548"
+ "text": "Last updated\n\n2023-11-27 19:13:40.019302"
},
{
- "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#details",
- "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#details",
- "title": "The pharmaverse (hi)story",
+ "objectID": "posts/2023-07-09_falcon/falcon.html#details",
+ "href": "posts/2023-07-09_falcon/falcon.html#details",
+ "title": "falcon",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-08-08_study_day/study_day.html",
- "href": "posts/2023-08-08_study_day/study_day.html",
- "title": "It’s all relative? - Calculating Relative Days using admiral",
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html",
+ "title": "Admiral Hackathon 2023 Revisited",
"section": "",
- "text": "Creating --DY variables for your ADaMs is super easy using derive_vars_dy() from the admiral package.\nLet’s build some dummy data with 4 subjects, a start date/time for treatment (TRTSDTM), an analysis start date/time variable (ASTDTM) and an analysis end date variable (AENDT).\nlibrary(admiral)\nlibrary(lubridate)\nlibrary(dplyr)\n\nadam <- tribble(\n ~USUBJID, ~TRTSDTM, ~ASTDTM, ~AENDT,\n \"001\", \"2014-01-17T23:59:59\", \"2014-01-18T13:09:O9\", \"2014-01-20\",\n \"002\", \"2014-02-25T23:59:59\", \"2014-03-18T14:09:O9\", \"2014-03-24\",\n \"003\", \"2014-02-12T23:59:59\", \"2014-02-18T11:03:O9\", \"2014-04-17\",\n \"004\", \"2014-03-17T23:59:59\", \"2014-03-19T13:09:O9\", \"2014-05-04\"\n) %>%\n mutate(\n TRTSDTM = as_datetime(TRTSDTM),\n ASTDTM = as_datetime(ASTDTM),\n AENDT = ymd(AENDT)\n )\nOkay! Next we run our dataset through derive_vars_dy(), specifying:\nderive_vars_dy(\n adam,\n reference_date = TRTSDTM,\n source_vars = exprs(ASTDTM, AENDT)\n)\n\n# A tibble: 4 × 6\n USUBJID TRTSDTM ASTDTM AENDT ASTDY AENDY\n <chr> <dttm> <dttm> <date> <dbl> <dbl>\n1 001 2014-01-17 23:59:59 2014-01-18 13:09:09 2014-01-20 2 4\n2 002 2014-02-25 23:59:59 2014-03-18 14:09:09 2014-03-24 22 28\n3 003 2014-02-12 23:59:59 2014-02-18 11:03:09 2014-04-17 7 65\n4 004 2014-03-17 23:59:59 2014-03-19 13:09:09 2014-05-04 3 49\nThat’s it! We got both our ASTDY and AENDY variables in only a few short lines of code!\nWhat if I want my variables to have a different naming convention?\nEasy! In the source_vars argument if you want your variables to be called DEMOADY and DEMOEDY just do DEMOADY = ASTDTM and DEMOEDY = AENDT and derive_vars_dy() will do the rest!\nderive_vars_dy(\n adam,\n reference_date = TRTSDTM,\n source_vars = exprs(DEMOADY = ASTDTM, DEMOEDY = AENDT)\n)\n\n# A tibble: 4 × 6\n USUBJID TRTSDTM ASTDTM AENDT DEMOADY DEMOEDY\n <chr> <dttm> <dttm> <date> <dbl> <dbl>\n1 001 2014-01-17 23:59:59 2014-01-18 13:09:09 2014-01-20 2 4\n2 002 2014-02-25 23:59:59 2014-03-18 14:09:09 2014-03-24 22 28\n3 003 2014-02-12 23:59:59 2014-02-18 11:03:09 2014-04-17 7 65\n4 004 2014-03-17 23:59:59 2014-03-19 13:09:09 2014-05-04 3 49\nIf you want to get --DT or --DTM variables using admiral then check out derive_vars_dt() and derive_vars_dtm(). If things are messy in your data, e.g. partial dates, both functions have great imputation abilities, which we will cover in an upcoming blog post!"
+ "text": "This January and February (2023), the admiral development team and the CDISC Open Source Alliance jointly hosted the admiral hackathon. The idea was to build a community of admiral users, and help participants familiarize themselves with R and admiral. This whole effort was led by Thomas Neitmann and was supported by Zelos Zhu, Sadchla Mascary, and me – Stefan Thoma.\nThe hackathon event was structured in two parts. First, we offered an Introduction to R for SAS programmers, a three hour workshop for R beginners to get them up to speed. Here we covered practical R basics, talking about how the R-workflow differs from a SAS workflow, and discussed common R functions - mostly from the tidyverse. This ensured that hackathon participants were familiar with core R concepts. The workshop recording and the course materials are available online.\nThe main hackathon consisted of several ADAM data generating tasks based on a specs file and synthetic data. Participants were able to solve these tasks in groups at their own pace thanks to a online tool where participants could upload their task specific R scripts and they would get automatic feedback for the data-set produced by their script. Script upload through the feedback application was available all through February, and we offered three additional online meetings throughout the month to discuss challenges and give some tips. If you are interested in learning more about the thoughts that went into the feedback application, you can read about it in this blogpost or check out my public GitHub repository for such an application."
},
{
- "objectID": "posts/2023-08-08_study_day/study_day.html#last-updated",
- "href": "posts/2023-08-08_study_day/study_day.html#last-updated",
- "title": "It’s all relative? - Calculating Relative Days using admiral",
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html#introduction-to-r-workshop",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html#introduction-to-r-workshop",
+ "title": "Admiral Hackathon 2023 Revisited",
+ "section": "Introduction to R workshop",
+ "text": "Introduction to R workshop\nWe were really excited to see over 500 people from around 40 countries joining our Introduction to R workshop in January! To get to know prospective users and hackathon participants better, we conducted some polls during the meetings. Below you can see that representatives of many different sorts of organisations joined our Introduction to R workshop:\n\n\n\n\n\n216 out of 402 confirmed that their company is already using R for clinical trial data analysis, the remaining 131 did not answer this question.\nThe target audience for this workshop was programmers who are very familiar with SAS, but not so familiar with R, our polls confirmed this.\n\n\n\n\n\nOverall, we were very happy with how the workshop turned out, and participants overall agreed with this sentiment (although there may be a slight survivorship bias…)."
+ },
+ {
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html#admiral-hackathon",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html#admiral-hackathon",
+ "title": "Admiral Hackathon 2023 Revisited",
+ "section": "admiral Hackathon",
+ "text": "admiral Hackathon\nFollowing the kick-off meeting, 371 participants joined the posit (rStudio) workspace that was made available to all participants at no costs by the posit company. About half the participants planned to spend one to two hours per week on the admiral tasks, the other half planned to allocate even more. 15 participants even planned to spend eight hours or more!\nWe were really happy to see an overwhelming amount of activity on the slack channel we set up with over 250 members. Not only were people engaging with the materials, but we saw how a community was formed where people were encouraged to ask questions and where community members went out of their way to help each other. Shout-out to our community hero: Jagadish Katam without whom most issues related to the task programming raised by the community would not have been addressed as quickly as they were. Huge thanks from the organizers!\nIn the end, a total of 44 teams spanning 87 statistical programmers took part in the admiral hackathon and uploaded solution scripts to the hackathon application solving at least one of the 8 tasks available (ADSL, ADAE, ADLBC, ADVS, ADTTE, ADADAS, ADLBH & ADLBHY). Participants’ scripts were then run on the shiny server and the output data-frame were compared to the solutions we provided. At the read-out there was a live draft of teams to win one-on-one admiral consulting with one of the admiral core developers. Winning probabilities were weighted by the number of points each group received for the quality of their output data-frames and for the number of tasks solved.\nCongratulations to the winners:\n\nViiV Team_GSK\nteamspoRt\nTatianaPXL\nDivyasneelam\nAdaMTeamIndia\nSanofi_BP\nJagadish (our community hero)\nAZ_WAWA\n\nAlthough this was uncertain during the hackathon we were excited to provide a Certificate of Completion to all participants who uploaded a script to the Web Application.\nA recording of the hackathon readout can be found in the CDISC Open Source Alliance Quarterly Spotlight."
+ },
+ {
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html#conclusion",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html#conclusion",
+ "title": "Admiral Hackathon 2023 Revisited",
+ "section": "Conclusion",
+ "text": "Conclusion\nOverall, we are very happy with how the hackathon turned out. We were not only positively surprised with the huge audience for the Intro to R workshop (CDISC record breaking) and for the admiral hackathon, but even more so with the engagement of all the participants.\nAgain, we would like to thank all the organizers, participants, and sponsors for their time and resources and hope to have provided a useful glimpse into our solution for ADAM creation within the end-to-end clinical data analysis open source R framework that the Pharmaverse aims to provide.\nAs always, we are very happy to hear more feedback on the hackathon as well as on admiral in general. Simply submit an issue on the admiral GitHub repository. You would like to join the admiral core developers? Please reach out to Edoardo Mancini (product owner) or Ben Straub (technical lead)."
+ },
+ {
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html#last-updated",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html#last-updated",
+ "title": "Admiral Hackathon 2023 Revisited",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:45.242711"
+ "text": "Last updated\n\n2023-11-27 19:13:36.963431"
},
{
- "objectID": "posts/2023-08-08_study_day/study_day.html#details",
- "href": "posts/2023-08-08_study_day/study_day.html#details",
- "title": "It’s all relative? - Calculating Relative Days using admiral",
+ "objectID": "posts/2023-06-27_hackathon_writeup/index.html#details",
+ "href": "posts/2023-06-27_hackathon_writeup/index.html#details",
+ "title": "Admiral Hackathon 2023 Revisited",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
@@ -182,88 +371,25 @@
"text": "Using More Than One Imputation Rule for a Variable\nUsing different imputation rules depending on the observation can be done by using the higher-order function slice_derivation(), which applies a derivation function differently (by varying its arguments) in different subsections of a dataset. For example, consider this Vital Signs case where pre-dose records require a different treatment to other records:\n\nvs <- tribble(\n ~VSDTC, ~VSTPT,\n \"2019-08-09T12:34:56\", NA,\n \"2019-10-12\", \"PRE-DOSE\",\n \"2019-11-10\", NA,\n \"2019-12-04\", NA\n) %>%\n slice_derivation(\n derivation = derive_vars_dtm,\n args = params(\n dtc = VSDTC,\n new_vars_prefix = \"A\"\n ),\n derivation_slice(\n filter = VSTPT == \"PRE-DOSE\",\n args = params(time_imputation = \"first\")\n ),\n derivation_slice(\n filter = TRUE,\n args = params(time_imputation = \"last\")\n )\n )\n\n\n\n\n\n\nVSDTC\nVSTPT\nADTM\nATMF\n\n\n\n\n2019-08-09T12:34:56\nNA\n2019-08-09 12:34:56\nNA\n\n\n2019-11-10\nNA\n2019-11-10 23:59:59\nH\n\n\n2019-12-04\nNA\n2019-12-04 23:59:59\nH\n\n\n2019-10-12\nPRE-DOSE\n2019-10-12 00:00:00\nH"
},
{
- "objectID": "posts/2023-06-28_welcome/index.html",
- "href": "posts/2023-06-28_welcome/index.html",
- "title": "Hello pharmaverse",
- "section": "",
- "text": "The communications working group (CWG) seeks to promote and showcase how R can be used in the Clinical Reporting pipeline through short and informative blog posts. These posts will be hosted on this pharmaverse blog and promoted on the pharmaverse slack channels as well as on LinkedIn.\nAs the CWG is a small team, we hope to make the blog development process easy enough that pharmaverse community members will be able to easily write blog posts with guidance from the CWG team."
- },
- {
- "objectID": "posts/2023-06-28_welcome/index.html#purpose",
- "href": "posts/2023-06-28_welcome/index.html#purpose",
- "title": "Hello pharmaverse",
+ "objectID": "index.html",
+ "href": "index.html",
+ "title": "Pharmaverse Blog",
"section": "",
- "text": "The communications working group (CWG) seeks to promote and showcase how R can be used in the Clinical Reporting pipeline through short and informative blog posts. These posts will be hosted on this pharmaverse blog and promoted on the pharmaverse slack channels as well as on LinkedIn.\nAs the CWG is a small team, we hope to make the blog development process easy enough that pharmaverse community members will be able to easily write blog posts with guidance from the CWG team."
- },
- {
- "objectID": "posts/2023-06-28_welcome/index.html#spirit-of-a-blog-post",
- "href": "posts/2023-06-28_welcome/index.html#spirit-of-a-blog-post",
- "title": "Hello pharmaverse",
- "section": "Spirit of a Blog Post",
- "text": "Spirit of a Blog Post\nThe CWG believes that the following 4 points will help guide the creation of Blog Posts.\n\nShort\nPersonalized\nReproducible\nReadable\n\nShort: Posts should aim to be under a 10 minute read. We encourage longer posts to be broken up into multiple posts.\nPersonalized: Posts should have a personality! For example, a person wishing to post on a function in a package needs to differentiate the post from the documentation for function, i.e. we don’t want to just recycle the documentation. How can you add your voice and experience? A bit of cheeky language is also encouraged.\nReproducible: Posts should work with minimal dependencies with data, packages and outside sources. Every dependency introduced in a post adds some risk to the post longevity. As package dependencies change, posts should be built in a way that they can be updated to stay relevant.\nReadable: The CWG sees this site as more of introductory site rather advanced user site. Therefore, the CWG feels that code should be introduced in a way that promotes readability over complexity."
- },
- {
- "objectID": "posts/2023-06-28_welcome/index.html#what-types-of-posts-are-allowed-on-this-site",
- "href": "posts/2023-06-28_welcome/index.html#what-types-of-posts-are-allowed-on-this-site",
- "title": "Hello pharmaverse",
- "section": "What types of posts are allowed on this site?",
- "text": "What types of posts are allowed on this site?\nOverall, we want to stay focus on the Clinical Reporting Pipeline, which we see as the following topics:\n\nPackages in the Clinical Reporting Pipeline\nFunctions from packages in the Clinical Reporting Pipeline\nWider experiences of using R in the Clinical Reporting Pipeline\nConference experiences and the Clinical Reporting Pipeline\n\nHowever, it never hurts to ask if you topic might fit into this medium!\n\nMinimum Post Requirements\n\nA unique image to help showcase the post.\nWorking Code\nSelf-contained data or package data.\nDocumentation of package versions\n\nThat is it! After that you can go wild, but we do ask that it is kept short!"
- },
- {
- "objectID": "posts/2023-06-28_welcome/index.html#how-can-i-make-a-blog-post",
- "href": "posts/2023-06-28_welcome/index.html#how-can-i-make-a-blog-post",
- "title": "Hello pharmaverse",
- "section": "How can I make a Blog Post",
- "text": "How can I make a Blog Post\nStep 1: Reach out to us through pharmaverse/slack or make an issue on our GitHub.\nStep 2: Branch off main\nStep 3: Review the Spirit of the Blog Post in the Pull Request Template\nStep 4: Poke us to do a review!"
+ "text": "Order By\n Default\n \n Title\n \n \n Date - Oldest\n \n \n Date - Newest\n \n \n Author\n \n \n \n \n \n \n \n\n\n\n\n \n\n\n\n\nBelieve in a higher order!\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nA brief foray into the higher order functions in the {admiral} package.\n\n\n\n\n\n\nNov 27, 2023\n\n\nEdoardo Mancini\n\n\n\n\n\n\n \n\n\n\n\nFloating point\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nThe untold story of how admiral deals with floating points.\n\n\n\n\n\n\nOct 30, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nReproducing the R Submissions Pilot 2 Shiny Application using Rhino\n\n\n\n\n\n\n\nsubmission\n\n\ncommunity\n\n\n\n\n\n\n\n\n\n\n\nOct 10, 2023\n\n\nIsmael Rodriguez, Vedha Viyash, André Veríssimo, APPSILON\n\n\n\n\n\n\n \n\n\n\n\nDate/Time Functions and Imputation in {admiral}\n\n\n\n\n\n\n\nadmiral\n\n\nADaM\n\n\nDate/Time\n\n\n\n\nDates, times and imputation can be a frustrating facet of any programming language. Here’s how {admiral} makes all of this easy!\n\n\n\n\n\n\nSep 26, 2023\n\n\nEdoardo Mancini\n\n\n\n\n\n\n \n\n\n\n\nThe pharmaverse (hi)story\n\n\n\n\n\n\n\ncommunity\n\n\n\n\n\n\n\n\n\n\n\nAug 30, 2023\n\n\nNicholas Eugenio\n\n\n\n\n\n\n \n\n\n\n\nRounding\n\n\n\n\n\n\n\nrounding\n\n\n\n\nExploration of some commonly used rounding methods and their corresponding functions in SAS and R, with a focus on ‘round half up’ and reliable solutions for numerical precision challenges\n\n\n\n\n\n\nAug 22, 2023\n\n\nKangjie Zhang\n\n\n\n\n\n\n \n\n\n\n\nIt’s all relative? - Calculating Relative Days using admiral\n\n\n\n\n\n\n\nadmiral\n\n\nADaMs\n\n\n\n\n\n\n\n\n\n\n\nAug 8, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nCross-Industry Open Source Package Development\n\n\n\n\n\n\n\ncommunity\n\n\nadmiral\n\n\n\n\n\n\n\n\n\n\n\nJul 25, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nHow to use Code Sections\n\n\n\n\n\n\n\nADaMs\n\n\nTips and Tricks\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2023\n\n\nEdoardo Mancini\n\n\n\n\n\n\n \n\n\n\n\nBlanks and NAs\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nReading SAS datasets into R. The data is not always as it seems!\n\n\n\n\n\n\nJul 10, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nfalcon\n\n\n\n\n\n\n\nfalcon\n\n\ndisplays\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2023\n\n\nVincent Shen\n\n\n\n\n\n\n \n\n\n\n\nHello pharmaverse\n\n\n\n\n\n\n\ncommunity\n\n\n\n\nShort, fun and user-driven content around the pharmaverse.\n\n\n\n\n\n\nJun 28, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nAdmiral Hackathon 2023 Revisited\n\n\n\n\n\n\n\ncommunity\n\n\nadmiral\n\n\n\n\nLet’s have a look at the Admiral Hackathon 2023 together.\n\n\n\n\n\n\nJun 27, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nDerive a new parameter computed from the value of other parameters\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nUse admiral::derive_param_computed() like a calculator to derive new parameters/rows based on existing ones\n\n\n\n\n\n\nJun 27, 2023\n\n\nKangjie Zhang\n\n\n\n\n\n\n \n\n\n\n\nHackathon Feedback Application\n\n\n\n\n\n\n\nR\n\n\nshiny\n\n\ncommunity\n\n\nadmiral\n\n\n\n\nGoing through the process of creating a shiny app for the admiral hackathon. The shiny app allows users to check their solutions autonomously, gives feedback, and rates their results.\n\n\n\n\n\n\nJun 27, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\nNo matching items"
},
{
- "objectID": "posts/2023-06-28_welcome/index.html#last-updated",
- "href": "posts/2023-06-28_welcome/index.html#last-updated",
- "title": "Hello pharmaverse",
- "section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:37.128939"
- },
- {
- "objectID": "posts/2023-06-28_welcome/index.html#details",
- "href": "posts/2023-06-28_welcome/index.html#details",
- "title": "Hello pharmaverse",
- "section": "Details",
- "text": "Details\n\nsource code, R environment"
- },
- {
- "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html",
- "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html",
- "title": "Derive a new parameter computed from the value of other parameters",
+ "objectID": "R/readme.html",
+ "href": "R/readme.html",
+ "title": "Files in this folder",
"section": "",
- "text": "When creating ADaM Basic Data Structure (BDS) datasets, we often encounter deriving a new parameter based on the analysis values (e.g., AVAL) of other parameters.\nThe admiral function derive_param_computed() adds a parameter computed from the analysis value of other parameters.\nIt works like a calculator to derive new records without worrying about merging and combining datasets, all you need is a derivation formula, which also improves the readability of the code."
+ "text": "Some of these files help in creating/developing blog-posts, others are used by our CICD pipeline.\n\ncreate_blogpost.R: use this script to create a new blogpost based on our blogpost template.\nCICD.R: use this script to spellcheck and stylecheck your blogpost.\n\n\n\n\nhelp_create_blogpost.R: script containing the function(s) used by create_blogpost.R\nswitch.R: Used by CICD spellcheck workflow."
},
{
- "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#introduction",
- "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#introduction",
- "title": "Derive a new parameter computed from the value of other parameters",
+ "objectID": "R/readme.html#development-files",
+ "href": "R/readme.html#development-files",
+ "title": "Files in this folder",
"section": "",
- "text": "When creating ADaM Basic Data Structure (BDS) datasets, we often encounter deriving a new parameter based on the analysis values (e.g., AVAL) of other parameters.\nThe admiral function derive_param_computed() adds a parameter computed from the analysis value of other parameters.\nIt works like a calculator to derive new records without worrying about merging and combining datasets, all you need is a derivation formula, which also improves the readability of the code."
- },
- {
- "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#example",
- "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#example",
- "title": "Derive a new parameter computed from the value of other parameters",
- "section": "Example",
- "text": "Example\nA value level validation use case, where derive_param_computed() is used to validate a derived parameter - PARAMCD = ADPCYMG (Actual Dose per Cycle) in ADEX dataset.\n\nDerivation\nActual Dose per Cycle is derived from the Total Amount of Dose (PARAMCD = TOTDOSE) / Number of Cycles (PARAMCD = NUMCYC)\nIn this example, ADEX.AVAL when ADEX.PARAMCD = ADPCYMG can be derived as:\n\\[\nAVAL (PARAMCD = ADPCYMG) = \\frac{AVAL (PARAMCD = TOTDOSE)}{AVAL (PARAMCD = NUMCYC)}\n\\]\n\n\nLoading Packages and Creating Example Data\n\nlibrary(tibble)\nlibrary(dplyr)\nlibrary(diffdf)\nlibrary(admiral)\n\nadex <- tribble(\n ~USUBJID, ~PARAMCD, ~PARAM, ~AVAL,\n \"101\", \"TOTDOSE\", \"Total Amount of Dose (mg)\", 180,\n \"101\", \"NUMCYC\", \"Number of Cycles\", 3\n)\n\n\n\nDerive New Parameter\n\nadex_admiral <- derive_param_computed(\n adex,\n by_vars = exprs(USUBJID),\n parameters = c(\"TOTDOSE\", \"NUMCYC\"),\n analysis_value = AVAL.TOTDOSE / AVAL.NUMCYC,\n set_values_to = exprs(\n PARAMCD = \"ADPCYMG\",\n PARAM = \"Actual Dose per Cycle (mg)\"\n )\n)\n\nWarning: The `analysis_value` argument of `derive_param_computed()` is deprecated as\nof admiral 0.12.0.\nℹ Please use the `set_values_to` argument instead.\n\n\n\n\n# A tibble: 3 × 4\n USUBJID PARAMCD PARAM AVAL\n <chr> <chr> <chr> <dbl>\n1 101 TOTDOSE Total Amount of Dose (mg) 180\n2 101 NUMCYC Number of Cycles 3\n3 101 ADPCYMG Actual Dose per Cycle (mg) 60\n\n\n\n\nCompare\nFor validation purpose, the diffdf package is used below to mimic SAS proc compare.\n\nadex_expected <- bind_rows(\n adex,\n tribble(\n ~USUBJID, ~PARAMCD, ~PARAM, ~AVAL,\n \"101\", \"ADPCYMG\", \"Actual Dose per Cycle (mg)\", 60\n )\n)\n\ndiffdf(adex_expected, adex_admiral, keys = c(\"USUBJID\", \"PARAMCD\"))\n\nNo issues were found!"
- },
- {
- "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#last-updated",
- "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#last-updated",
- "title": "Derive a new parameter computed from the value of other parameters",
- "section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:33.340574"
- },
- {
- "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#details",
- "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#details",
- "title": "Derive a new parameter computed from the value of other parameters",
- "section": "Details",
- "text": "Details\n\nsource code, R environment"
+ "text": "help_create_blogpost.R: script containing the function(s) used by create_blogpost.R\nswitch.R: Used by CICD spellcheck workflow."
},
{
"objectID": "posts/2023-07-24_rounding/rounding.html",
@@ -298,7 +424,7 @@
"href": "posts/2023-07-24_rounding/rounding.html#last-updated",
"title": "Rounding",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:29.564969"
+ "text": "Last updated\n\n2023-11-27 19:13:33.556149"
},
{
"objectID": "posts/2023-07-24_rounding/rounding.html#details",
@@ -308,114 +434,79 @@
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "R/readme.html",
- "href": "R/readme.html",
- "title": "Files in this folder",
- "section": "",
- "text": "Some of these files help in creating/developing blog-posts, others are used by our CICD pipeline.\n\ncreate_blogpost.R: use this script to create a new blogpost based on our blogpost template.\nCICD.R: use this script to spellcheck and stylecheck your blogpost.\n\n\n\n\nhelp_create_blogpost.R: script containing the function(s) used by create_blogpost.R\nswitch.R: Used by CICD spellcheck workflow."
- },
- {
- "objectID": "R/readme.html#development-files",
- "href": "R/readme.html#development-files",
- "title": "Files in this folder",
- "section": "",
- "text": "help_create_blogpost.R: script containing the function(s) used by create_blogpost.R\nswitch.R: Used by CICD spellcheck workflow."
- },
- {
- "objectID": "index.html",
- "href": "index.html",
- "title": "Pharmaverse Blog",
+ "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html",
+ "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html",
+ "title": "Derive a new parameter computed from the value of other parameters",
"section": "",
- "text": "Order By\n Default\n \n Title\n \n \n Date - Oldest\n \n \n Date - Newest\n \n \n Author\n \n \n \n \n \n \n \n\n\n\n\n \n\n\n\n\nFloating point\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nThe untold story of how admiral deals with floating points.\n\n\n\n\n\n\nOct 30, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nReproducing the R Submissions Pilot 2 Shiny Application using Rhino\n\n\n\n\n\n\n\nsubmission\n\n\ncommunity\n\n\n\n\n\n\n\n\n\n\n\nOct 10, 2023\n\n\nIsmael Rodriguez, Vedha Viyash, André Veríssimo, APPSILON\n\n\n\n\n\n\n \n\n\n\n\nDate/Time Functions and Imputation in {admiral}\n\n\n\n\n\n\n\nadmiral\n\n\nADaM\n\n\nDate/Time\n\n\n\n\nDates, times and imputation can be a frustrating facet of any programming language. Here’s how {admiral} makes all of this easy!\n\n\n\n\n\n\nSep 26, 2023\n\n\nEdoardo Mancini\n\n\n\n\n\n\n \n\n\n\n\nThe pharmaverse (hi)story\n\n\n\n\n\n\n\ncommunity\n\n\n\n\n\n\n\n\n\n\n\nAug 30, 2023\n\n\nNicholas Eugenio\n\n\n\n\n\n\n \n\n\n\n\nRounding\n\n\n\n\n\n\n\nrounding\n\n\n\n\nExploration of some commonly used rounding methods and their corresponding functions in SAS and R, with a focus on ‘round half up’ and reliable solutions for numerical precision challenges\n\n\n\n\n\n\nAug 22, 2023\n\n\nKangjie Zhang\n\n\n\n\n\n\n \n\n\n\n\nIt’s all relative? - Calculating Relative Days using admiral\n\n\n\n\n\n\n\nadmiral\n\n\nADaMs\n\n\n\n\n\n\n\n\n\n\n\nAug 8, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nCross-Industry Open Source Package Development\n\n\n\n\n\n\n\ncommunity\n\n\nadmiral\n\n\n\n\n\n\n\n\n\n\n\nJul 25, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nHow to use Code Sections\n\n\n\n\n\n\n\nADaMs\n\n\nTips and Tricks\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2023\n\n\nEdoardo Mancini\n\n\n\n\n\n\n \n\n\n\n\nBlanks and NAs\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nReading SAS datasets into R. The data is not always as it seems!\n\n\n\n\n\n\nJul 10, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nfalcon\n\n\n\n\n\n\n\nfalcon\n\n\ndisplays\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2023\n\n\nVincent Shen\n\n\n\n\n\n\n \n\n\n\n\nHello pharmaverse\n\n\n\n\n\n\n\ncommunity\n\n\n\n\nShort, fun and user-driven content around the pharmaverse.\n\n\n\n\n\n\nJun 28, 2023\n\n\nBen Straub\n\n\n\n\n\n\n \n\n\n\n\nDerive a new parameter computed from the value of other parameters\n\n\n\n\n\n\n\nadmiral\n\n\n\n\nUse admiral::derive_param_computed() like a calculator to derive new parameters/rows based on existing ones\n\n\n\n\n\n\nJun 27, 2023\n\n\nKangjie Zhang\n\n\n\n\n\n\n \n\n\n\n\nHackathon Feedback Application\n\n\n\n\n\n\n\nR\n\n\nshiny\n\n\ncommunity\n\n\nadmiral\n\n\n\n\nGoing through the process of creating a shiny app for the admiral hackathon. The shiny app allows users to check their solutions autonomously, gives feedback, and rates their results.\n\n\n\n\n\n\nJun 27, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\n \n\n\n\n\nAdmiral Hackathon 2023 Revisited\n\n\n\n\n\n\n\ncommunity\n\n\nadmiral\n\n\n\n\nLet’s have a look at the Admiral Hackathon 2023 together.\n\n\n\n\n\n\nJun 27, 2023\n\n\nStefan Thoma\n\n\n\n\n\n\nNo matching items"
+ "text": "When creating ADaM Basic Data Structure (BDS) datasets, we often encounter deriving a new parameter based on the analysis values (e.g., AVAL) of other parameters.\nThe admiral function derive_param_computed() adds a parameter computed from the analysis value of other parameters.\nIt works like a calculator to derive new records without worrying about merging and combining datasets, all you need is a derivation formula, which also improves the readability of the code."
},
{
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html",
- "title": "Blanks and NAs",
+ "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#introduction",
+ "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#introduction",
+ "title": "Derive a new parameter computed from the value of other parameters",
"section": "",
- "text": "Reading in SAS-based datasets (.sas7bdat or xpt) into R has users calling the R package haven. A typical call might invoke read_sas() or read_xpt() to bring in your source data to construct your ADaMs or SDTMs.\nUnfortunately, while using haven the character blanks (missing data) found in a typical SAS-based dataset are left as blanks. These blanks will typically prove problematic while using functions like is.na in combination with dplyr::filter() to subset data. Check out Bayer’s SAS2R catalog: handling-of-missing-values for more discussion on missing values and NAs.\nIn the admiral package, we have built a simple function called convert_blanks_to_na() to help us quickly remedy this problem. You can supply an entire dataframe to this function and it will convert any character blanks to NA_character_"
- },
- {
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#loading-packages-and-making-dummy-data",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#loading-packages-and-making-dummy-data",
- "title": "Blanks and NAs",
- "section": "Loading Packages and Making Dummy Data",
- "text": "Loading Packages and Making Dummy Data\n\nlibrary(admiral)\nlibrary(tibble)\nlibrary(dplyr)\n\ndf <- tribble(\n ~USUBJID, ~RFICDTC,\n \"01\", \"2000-01-01\",\n \"02\", \"2001-01-01\",\n \"03\", \"\", # Here we have a character blank\n \"04\", \"2001-01--\",\n \"05\", \"2001---01\",\n \"05\", \"\", # Here we have a character blank\n)\n\ndf\n\n# A tibble: 6 × 2\n USUBJID RFICDTC \n <chr> <chr> \n1 01 \"2000-01-01\"\n2 02 \"2001-01-01\"\n3 03 \"\" \n4 04 \"2001-01--\" \n5 05 \"2001---01\" \n6 05 \"\""
- },
- {
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#a-simple-conversion",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#a-simple-conversion",
- "title": "Blanks and NAs",
- "section": "A simple conversion",
- "text": "A simple conversion\n\ndf_na <- convert_blanks_to_na(df)\n\ndf_na\n\n# A tibble: 6 × 2\n USUBJID RFICDTC \n <chr> <chr> \n1 01 2000-01-01\n2 02 2001-01-01\n3 03 <NA> \n4 04 2001-01-- \n5 05 2001---01 \n6 05 <NA> \n\n\n\ndf_na %>% filter(is.na(RFICDTC))\n\n# A tibble: 2 × 2\n USUBJID RFICDTC\n <chr> <chr> \n1 03 <NA> \n2 05 <NA>"
+ "text": "When creating ADaM Basic Data Structure (BDS) datasets, we often encounter deriving a new parameter based on the analysis values (e.g., AVAL) of other parameters.\nThe admiral function derive_param_computed() adds a parameter computed from the analysis value of other parameters.\nIt works like a calculator to derive new records without worrying about merging and combining datasets, all you need is a derivation formula, which also improves the readability of the code."
},
{
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#thats-it",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#thats-it",
- "title": "Blanks and NAs",
- "section": "That’s it!",
- "text": "That’s it!\nA simple call to this function can make your derivation life so much easier while working in R if working with SAS-based datasets. In admiral, we make use of this function at the start of all ADaM templates for common ADaM datasets. You can use the function use_ad_template() to get the full R script for the below ADaMs.\n\nlist_all_templates()\n\nExisting ADaM templates in package 'admiral':\n• ADAE\n• ADCM\n• ADEG\n• ADEX\n• ADLB\n• ADLBHY\n• ADMH\n• ADPC\n• ADPP\n• ADPPK\n• ADSL\n• ADVS"
+ "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#example",
+ "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#example",
+ "title": "Derive a new parameter computed from the value of other parameters",
+ "section": "Example",
+ "text": "Example\nA value level validation use case, where derive_param_computed() is used to validate a derived parameter - PARAMCD = ADPCYMG (Actual Dose per Cycle) in ADEX dataset.\n\nDerivation\nActual Dose per Cycle is derived from the Total Amount of Dose (PARAMCD = TOTDOSE) / Number of Cycles (PARAMCD = NUMCYC)\nIn this example, ADEX.AVAL when ADEX.PARAMCD = ADPCYMG can be derived as:\n\\[\nAVAL (PARAMCD = ADPCYMG) = \\frac{AVAL (PARAMCD = TOTDOSE)}{AVAL (PARAMCD = NUMCYC)}\n\\]\n\n\nLoading Packages and Creating Example Data\n\nlibrary(tibble)\nlibrary(dplyr)\nlibrary(diffdf)\nlibrary(admiral)\n\nadex <- tribble(\n ~USUBJID, ~PARAMCD, ~PARAM, ~AVAL,\n \"101\", \"TOTDOSE\", \"Total Amount of Dose (mg)\", 180,\n \"101\", \"NUMCYC\", \"Number of Cycles\", 3\n)\n\n\n\nDerive New Parameter\n\nadex_admiral <- derive_param_computed(\n adex,\n by_vars = exprs(USUBJID),\n parameters = c(\"TOTDOSE\", \"NUMCYC\"),\n analysis_value = AVAL.TOTDOSE / AVAL.NUMCYC,\n set_values_to = exprs(\n PARAMCD = \"ADPCYMG\",\n PARAM = \"Actual Dose per Cycle (mg)\"\n )\n)\n\nWarning: The `analysis_value` argument of `derive_param_computed()` is deprecated as\nof admiral 0.12.0.\nℹ Please use the `set_values_to` argument instead.\n\n\n\n\n# A tibble: 3 × 4\n USUBJID PARAMCD PARAM AVAL\n <chr> <chr> <chr> <dbl>\n1 101 TOTDOSE Total Amount of Dose (mg) 180\n2 101 NUMCYC Number of Cycles 3\n3 101 ADPCYMG Actual Dose per Cycle (mg) 60\n\n\n\n\nCompare\nFor validation purpose, the diffdf package is used below to mimic SAS proc compare.\n\nadex_expected <- bind_rows(\n adex,\n tribble(\n ~USUBJID, ~PARAMCD, ~PARAM, ~AVAL,\n \"101\", \"ADPCYMG\", \"Actual Dose per Cycle (mg)\", 60\n )\n)\n\ndiffdf(adex_expected, adex_admiral, keys = c(\"USUBJID\", \"PARAMCD\"))\n\nNo issues were found!"
},
{
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#last-updated",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#last-updated",
- "title": "Blanks and NAs",
+ "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#last-updated",
+ "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#last-updated",
+ "title": "Derive a new parameter computed from the value of other parameters",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:27.839956"
+ "text": "Last updated\n\n2023-11-27 19:13:38.746856"
},
{
- "objectID": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#details",
- "href": "posts/2023-07-10_blanks_and_nas/blanks_and_nas.html#details",
- "title": "Blanks and NAs",
+ "objectID": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#details",
+ "href": "posts/2023-06-27_admiral/valuelevel/derive_param_computed.html#details",
+ "title": "Derive a new parameter computed from the value of other parameters",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html",
+ "href": "posts/2023-07-14_code_sections/code_sections.html",
+ "title": "How to use Code Sections",
"section": "",
- "text": "There is significant momentum in driving the adoption of R packages in the life sciences industries, in particular, the R Consortium Submissions Working Group is dedicated to promoting the use of R for regulatory submissions to the FDA.\nThe R Consortium Submissions Working Group successfully completed an R-based submission in November 2021 through the eCTD portal (R Submissions Pilot 1). This Pilot was completed on March 10, 2022 after a successful statistical review and evaluation by the FDA staff.\nMoving forward, the Pilot 2 aimed to include a Shiny application that the FDA staff could deploy on their own servers. The R Consortium recently announced that on September 27, 2023, the R Submissions Working Group successfully completed the follow-up to the Pilot 2 R Shiny-based submission and received a response letter from FDA CDER. This marks the first publicly available submission package that includes a Shiny component. The full FDA response letter can be found here.\nThe Shiny application that was sent for the Pilot 2 had the goal to display the 4 Tables, Listings and Figures (TLFs) that were sent for the Pilot 1 with basic filtering functionality.\nThe submission package adhered to the eCTD folder structure and contained 5 deliverables. Among the deliverables was the proprietary R package {pilot2wrappers}, which enables the execution of the Shiny application.\nThe FDA staff were expected to receive the electronic submission packages in the eCTD format, install and load open-source R packages used as dependencies in the included Shiny application, reconstruct and load the submitted Shiny application, and communicate potential improvements in writing.\nIn the following stage, the R Consortium’s R Submission Working Group launched Pilot 4, aiming to investigate innovative technologies like Linux containers and web assembly. These technologies are being explored to package a Shiny application into a self-contained unit, streamlining the transfer and execution processes for the application.\nIn this post, our aim is to outline how we used the Rhino framework to reproduce the Shiny application that was successfully submitted to the FDA for the Pilot 2 project. Additionally, we detail the challenges identified during the process and how we were able to successfully address them by using an open-source package."
- },
- {
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#reproducing-the-r-submission-pilot-2-shiny-app-using-rhino",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#reproducing-the-r-submission-pilot-2-shiny-app-using-rhino",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
- "section": "Reproducing the R Submission Pilot 2 Shiny App using Rhino",
- "text": "Reproducing the R Submission Pilot 2 Shiny App using Rhino\nWhile the original Shiny application submitted to the FDA was wrapped using {Golem}, we replicated the application using our in-house developed framework Rhino. The main motivation was to provide an example of an R Submission that is not an R package and to identify and solve any issues that may arise from this approach.\nOur demo application (FDA-pilot-app) is accessible on our website, alongside other Shiny and Rhinoverse demonstration apps.\n\nThe code for FDA-pilot-app is open-source. You can create your own Rhino-based application by following our tutorial and viewing our workshop, which is available on YouTube."
+ "text": "The admiral package embraces a modular style of programming, where blocks of code are pieced together in sequence to create an ADaM dataset. However, with the well-documented advantages of the modular approach comes the recognition that scripts will on average be longer. As such, astute programmers working in RStudio are constantly on the lookout for quick ways to effectively navigate their scripts. Enter code sections!"
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#brief-introduction-to-rhino",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#brief-introduction-to-rhino",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
- "section": "Brief Introduction to Rhino",
- "text": "Brief Introduction to Rhino\n\n\n\n\n\nThe Rhino framework was developed by Appsilon to create enterprise-level Shiny applications, consistently and efficiently. This framework allows developers to apply the best software engineering practices, modularize code, test it thoroughly, enhance UI aesthetics, and monitor user adoption effectively.\nRhino provides support in 3 main areas:\n\nClear Code: scalable architecture, modularization based on Box and Shiny modules.\nQuality: comprehensive testing such as unit tests, E2E tests with Cypress and Shinytest2, logging, monitoring and linting.\nAutomation: project startup, CI with GitHub Actions, dependencies management with {renv}, configuration management with config, Sass and JavaScript bundling with ES6 support via Node.js.\n\nRhino is an ideal fit for highly regulated environments such as regulatory submissions or other drug development processes.\n\nFDA-pilot-app structure\nThe structure of this application is available on the github repository. The structure of this Shiny app is the following.\n\nClick here to expand the FDA-pilot-app structure\n\n.\n├── app\n│ ├── view\n│ │ └── demographic_table.R\n| | └── km_plot.R\n| | └── primary_table.R\n| | └── efficacy_table.R\n| | └── completion_table.R\n│ ├── logic\n│ │ └── adam_data.R\n│ │ └── eff_modles.R\n│ │ └── formatters.R\n│ │ └── helpers.R\n│ │ └── kmplot_helpers.R\n│ │ └── Tplyr_helpers.R\n│ ├── data\n│ │ └── adam\n│ │ └── adadas.xpt\n│ │ └── adlbc.xpt\n│ │ └── adsl.xpt\n│ │ └── adtte.xpt\n│ ├── docs\n│ │ └── about.md\n│ ├── js\n│ │ └── index.js\n│ ├── static\n│ │ └── favicon.ico\n│ ├── styles\n│ │ └── main.scss\n│ └── app.R\n├── tests\n│ ├── cypress\n│ │ └── integration\n│ │ └── app.spec.js\n│ ├── testthat\n│ │\n│ └── cypress.json\n├── app.R\n├── rhino_submission.Rproj\n├── dependencies.R\n├── renv.lock\n├── rhino.yml\n└── README.md"
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html#introduction",
+ "href": "posts/2023-07-14_code_sections/code_sections.html#introduction",
+ "title": "How to use Code Sections",
+ "section": "",
+ "text": "The admiral package embraces a modular style of programming, where blocks of code are pieced together in sequence to create an ADaM dataset. However, with the well-documented advantages of the modular approach comes the recognition that scripts will on average be longer. As such, astute programmers working in RStudio are constantly on the lookout for quick ways to effectively navigate their scripts. Enter code sections!"
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#efficient-submissions-to-the-fda",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#efficient-submissions-to-the-fda",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
- "section": "Efficient Submissions to the FDA",
- "text": "Efficient Submissions to the FDA\n\n\n\n\n\nTo comply with the Electronic Submission File Formats and Specifications for the eCTD submission, the programming code should carry a “.txt” extension. In the R Submissions Pilot 3 the group did not use {pkglite} as the FDA clarified that “.zip” and “.r” files are acceptable for submission. In this case, we utilized the {pkglite} R package to efficiently pack and unpack the FDA-pilot-app. This approach would facilitate the FDA reviewers in setting up the submission on their systems.\nThis package allows packing R packages to “.txt” files, which are supported for the submission of proprietary packages to the FDA via the eCTD gateway. \n\nPacking the FDA-pilot-app into a .txt file\nThe code below can be used to pack the Shiny application into a .txt file:\n\napp_name <- \"rhinosubmission\"\nrenv_spec <- pkglite::file_spec(\n \"renv\",\n pattern = \"^settings\\\\.dcf$|^activate\\\\.R$\",\n format = \"text\", recursive = FALSE\n)\ntests_spec <- pkglite::file_tests()\napp_spec <- pkglite::file_auto(\"app\")\nroot_spec <- pkglite::file_spec(\n path = \".\",\n pattern = \"^\\\\.Rprofile$|^rhino\\\\.yml$|^renv\\\\.lock$|^dependencies\\\\.R$|^config\\\\.yml$|^app\\\\.R$|^README\\\\.md$|\\\\.Rproj$\",\n all_files = TRUE,\n recursive = FALSE\n)\nwrite(paste0(\"Package: \", app_name), \"DESCRIPTION\", append = TRUE)\npkglite::collate(\n getwd(),\n renv_spec,\n tests_spec,\n app_spec,\n root_spec\n) |> pkglite::pack()\nfile.remove(\"DESCRIPTION\")\n\n\n\nUnpacking the FDA-pilot-app\nThe packed “.txt” file can be unpacked into a Shiny app by using {pkglite} as follows:\n\npkglite::unpack(\"rhinosubmission.txt\")"
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html#so-what-are-code-sections-and-why-are-they-useful",
+ "href": "posts/2023-07-14_code_sections/code_sections.html#so-what-are-code-sections-and-why-are-they-useful",
+ "title": "How to use Code Sections",
+ "section": "So, what are code sections and why are they useful?",
+ "text": "So, what are code sections and why are they useful?\nCode Sections are separators for long R scripts or functions in RStudio. They can be set up by inserting a comment line followed by four or more dashes in between portions of code, like so:\n\n# First code section ----\n\na <- 1\n\n# Second code section ----\n\nb <- 2\n\n# Third code section ----\n\nc <- 3\n\nRStudio then recognizes the code sections automatically, and enables you to:\n\nCollapse and expand them using the arrow displayed next to the line number, or with the handy shortcuts Alt+L/Shift+Alt+L on Windows or Cmd+Option+L/Cmd+Shift+Option+L on Mac.\nTravel in between them using the navigator at the bottom of the code pane, or by pressing Shift+Alt+J on Windows or Cmd+Shift+Option+J on Mac.\nView an outline of the file using the “Outline” button at the top right of the pane and/or the orange hashtag “Section Navigator” button at the bottom left of the pane.\n\n\n\n\n\n\nCollapsed sections, outline view and the section navigator for the example above.\n\n\n\n\nIt is also possible to create subsections by using two hashtags at the start of a comment line:\n\n# First code section ----\na <- 1\n\n## A code subsection ----\nb <- 2\n\n# Second code section ----\nc <- 3\n\n\n\n\n\n\nCode subsections for the example above.\n\n\n\n\nFor a complete list of Code Sections shortcuts, and for further information, see here."
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#lessons-learned",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#lessons-learned",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
- "section": "Lessons Learned",
- "text": "Lessons Learned\nOur initial objective was to prove that it would be possible to submit a Shiny application using Rhino through the eCTD gateway. During the rewriting process we identified that this could be done by integrating the open-source {pkglite} package. By following this approach, we concluded that it would be possible to submit a Shiny application through the eCTD gateway. This was also achieved through the successful submission of a package that included a Shiny component in Pilot 2.\nHaving rewritten the R Submissions Pilot 2 Shiny application using Rhino holds major implications for the adoption of our framework within the life sciences. Apart from being a strong, opinionated framework that improves reproducibility and reliability for Shiny development, using Rhino for regulatory submissions could improve the flexibility and speed in the clinical reporting pipeline. This would accelerate the adoption of R/Shiny for submissions to the FDA or other regulatory agencies."
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html#conclusion",
+ "href": "posts/2023-07-14_code_sections/code_sections.html#conclusion",
+ "title": "How to use Code Sections",
+ "section": "Conclusion",
+ "text": "Conclusion\nCode sections are an easy way to navigate long scripts and foster good commenting practices. They are used extensively in the admiral package, but there is no reason that you cannot start using them yourself in your day-to-day R programming!"
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#last-updated",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#last-updated",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html#last-updated",
+ "href": "posts/2023-07-14_code_sections/code_sections.html#last-updated",
+ "title": "How to use Code Sections",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:31.231853"
+ "text": "Last updated\n\n2023-11-27 19:13:41.390513"
},
{
- "objectID": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#details",
- "href": "posts/2023-08-14_rhino_submission_2/rhino_submission_2.html#details",
- "title": "Reproducing the R Submissions Pilot 2 Shiny Application using Rhino",
+ "objectID": "posts/2023-07-14_code_sections/code_sections.html#details",
+ "href": "posts/2023-07-14_code_sections/code_sections.html#details",
+ "title": "How to use Code Sections",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
@@ -459,7 +550,7 @@
"href": "posts/2023-10-30_floating_point/floating_point.html#last-updated",
"title": "Floating point",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:35.340689"
+ "text": "Last updated\n\n2023-11-27 19:13:46.342818"
},
{
"objectID": "posts/2023-10-30_floating_point/floating_point.html#details",
@@ -469,121 +560,100 @@
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html",
- "href": "posts/2023-07-09_falcon/falcon.html",
- "title": "falcon",
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html",
+ "title": "The pharmaverse (hi)story",
"section": "",
- "text": "The {falcon} initiative is an industry collaborative effort under {pharmaverse} that unites Boehringer Ingelheim, Moderna, Roche, and Sanofi with the aspiration of building and open-sourcing a comprehensive catalog of harmonized tables, listings, and graphs (TLGs) for clinical study reporting. By leveraging existing open-source R packages, {falcon} aims to simplify the process of output review, comparison, and meta-analyses, while fostering efficient communication among stakeholders in the pharmaceutical industry."
+ "text": "Pharmaverse: from motivation to present"
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#what-is-falcon",
- "href": "posts/2023-07-09_falcon/falcon.html#what-is-falcon",
- "title": "falcon",
- "section": "",
- "text": "The {falcon} initiative is an industry collaborative effort under {pharmaverse} that unites Boehringer Ingelheim, Moderna, Roche, and Sanofi with the aspiration of building and open-sourcing a comprehensive catalog of harmonized tables, listings, and graphs (TLGs) for clinical study reporting. By leveraging existing open-source R packages, {falcon} aims to simplify the process of output review, comparison, and meta-analyses, while fostering efficient communication among stakeholders in the pharmaceutical industry."
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#human-history-and-pharmaverse-context",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#human-history-and-pharmaverse-context",
+ "title": "The pharmaverse (hi)story",
+ "section": "Human history and pharmaverse context",
+ "text": "Human history and pharmaverse context\nSince the Australian Aboriginal, the earliest peoples recorded to have inhabited the Earth and who have been in Australia for at least 65,000 to 80,000 years (Encyclopædia Britannica), human beings live in group. Whether to protect yourself, increase your life expectancy or simply share tasks.\nFor most aspects of life, it doesn’t make sense to think, act or work alone for two main reasons:\n\nYou will spend more energy and time;\nSomeone else may be facing (or have faced) the same situation.\n\nThe English poet John Donne used to say “No man is an island entire of itself; every man is a piece of the continent, a part of the main;”. I can’t disagree with him. And I dare say that Ari Siggaard Knoph (Novo Nordisk), Michael Rimler (GSK), Michael Stackhouse (Atorus), Ross Farrugia (Roche), and Sumesh Kalappurakal (Janssen) can’t disagree with him either. They are the founders of pharmaverse, members of its Council and kindly shared their memories of how independent companies, in mid-2020, worked together in the creation of a set of packages developed to support the clinical reporting pipeline.\nIf you are not familiar with this pipeline, the important thing to know is that, in a nutshell, pharmaceutical companies must follow a bunch of standardized procedures and formats (from Clinical Data Interchange Standards Consortium, CDISC) when submitting clinical results to Health Authorities. The focus is on this: different companies seeking the same standards for outputs.\nParaphrasing Ross Farrugia (Roche) Breaking boundaries through open-source collaboration presentation in R/Pharma 2022 and thinking of the development of a new drug, we are talking about a “post-competitive” scenario: the drug has already been discovered and the companies should “just” produce and deliver standardized results."
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#why-do-we-build-it",
- "href": "posts/2023-07-09_falcon/falcon.html#why-do-we-build-it",
- "title": "falcon",
- "section": "Why do we build it?",
- "text": "Why do we build it?\nThe collaborative effort focuses on improving the clarity, consistency, and accessibility of TLGs by addressing variations and redundancies in their creation and use. This harmonized approach allows for streamlined reporting processes and facilitates effective communication of study results within the industry and to regulatory authorities."
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#clinical-reporting-outputs",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#clinical-reporting-outputs",
+ "title": "The pharmaverse (hi)story",
+ "section": "Clinical reporting outputs",
+ "text": "Clinical reporting outputs\nRationally, we can say that companies face the same challenges in these steps of the process. Not so intuitively, we can also say they were working in silos on that before 2018. Just as Isaac Newton and Gottfried W. Leibniz independently developed the theory of infinitesimal calculus, pharmaceutical companies were independently working on R solutions for this pipeline.\nBut on August 16 and 17 of the mentioned year above, they gathered at the first edition of R/Pharma conference to discuss R and open-source tooling for drug development (the reasons why open-source is an advantageous approach can be found in this post written by Stefan Thoma). And according to Isabela Velásquez’s article, Pharmaverse: Packages for clinical reporting workflows, one of the most popular questions in this conference was “Is the package code available or on CRAN?”.\nWell, many of them were. And not necessarily at that date, but just to mention a few: pharmaRTF and Tplyr from Atorus, r2rtf from Merck, rtables from Roche, etc. The thing is that, overall, there were almost 10000 other packages as well (today, almost 20000). And that took to another two questions:\n\nWith this overwhelming number of packages on CRAN, how to find the ones related to solving “clinical reporting problems”?\nOnce the packages were found, how to choose which one to use among those that have the same functional purpose?\n\nSo, again, companies re-started to working in silos to find those answers. But now, in collaborative silos and with common goals: create extremely useful packages to solve pharmaceutical-specific gaps once and solve them well!"
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#what-has-been-done-so-far",
- "href": "posts/2023-07-09_falcon/falcon.html#what-has-been-done-so-far",
- "title": "falcon",
- "section": "What has been done so far?",
- "text": "What has been done so far?\nDrawing inspiration from the FDA Standard Safety Tables and Figures Integrated Guide, the {falcon} initiative has successfully developed and open-sourced 11 templates to date. 4 product owners and 11 developers from 4 companies have collaborated to make these templates available and also published them on the official {falcon} website at https://pharmaverse.github.io/falcon/."
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#first-partnerships",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#first-partnerships",
+ "title": "The pharmaverse (hi)story",
+ "section": "First partnerships",
+ "text": "First partnerships\nIn 2020, Michael Stackhouse (Atorus) and Michael Rimler (GSK) talked and formed a partnership between their companies to develop a few more packages, including metacore, to read, store and manipulate metadata for ADaMs/SDTMs in a standardized object; xportr, to create submission compliant SAS transport files and perform pharma specific dataset level validation checks; and logrx (ex-timber), to build log to support reproducibility and traceability of an R script.\nAround the same time, Thomas Neitmann (currently at Denali Therapeutics, then at Roche) and Michael Rimler (GSK) discovered that both were working with ADaM in R, so Thomas Neitmann (currently at Denali Therapeutics, then at Roche), Ross Farrugia (Roche) and Michael Rimler (GSK) saw an opportunity there and GSK started their partnership with Roche to build and release admiral package.\nThe idea of working together, the sense of community, and the appetite from organizations built more and more, with incentive and priority established up into the programming heads council.\nJanssen had a huge effort in building R capabilities going on as well, by releasing tidytlg and envsetup), so eventually Michael Rimler (GSK), Michael Stackhouse (Atorus) and Ross Farrugia (Roche) formalized pharmaverse and formed the council, adding in Sumesh Kalappurakal (Janssen) and Ari Siggaard Knoph (Novo Nordisk) later joined as the fifth council member."
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#next-steps-vision",
- "href": "posts/2023-07-09_falcon/falcon.html#next-steps-vision",
- "title": "falcon",
- "section": "Next steps & vision",
- "text": "Next steps & vision\nFuture plans for {falcon} involve expanding the catalog through continuous collaboration from participating companies and inviting wider industry engagement. The ultimate goal is to promote harmonization of TLGs for clinical reporting across the pharmaceutical industry, leading to greater efficiency, collaboration, and innovation. Even though templates currently come from a published FDA guide, the collaborating companies are open to share and discuss similarities and differences on analysis concepts and output layouts of their own implementations in clinical reporting, for both safety and efficacy analyses.\nIn addition, while currently all templates were built using {rtables}, {tern}, {rlistings} and drew inspiration from the open-sourced TLG-Catalog, moving forward, we plan to support creating the same templates using alternative table engines such as {gt}.\n{falcon} will be presented at the upcoming PHUSE EU (Standards Implementation stream), where we will share the collaboration journey of {falcon} so far, providing more details on the current progress, long-term vision, and strategies for this initiative. Attendees will gain insights into the challenges and opportunities of harmonizing clinical reporting through open-source collaboration and learn about the potential benefits and future direction of {falcon}."
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#release-growth-and-developments",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#release-growth-and-developments",
+ "title": "The pharmaverse (hi)story",
+ "section": "Release, growth and developments",
+ "text": "Release, growth and developments\nAt the end of their presentation “Closing the Gap: Creating an End to End R Package Toolkit for the Clinical Reporting Pipeline.”, in R/Pharma 2021, Ben Straub (GSK) and Eli Miller (Atorus) welcomed the community to the pharmaverse, a curated collection of packages developed by various pharmaceutical companies to support open-source clinical workflows.\nFrom the outset, the name pharmaverse was chosen so that it could be a neutral home, unrelated to any company. Also, it was established as not being a consortium, which means that founders don’t own, fund, or maintain the packages. Some individuals and companies maintain them but often allowing for community contributions and being licensed permissively so that there is always a feeling of community ownership. The focus of pharmaverse early on, and today, is on inter organization cooperation, to build an environment where, if organizations identify that they have a joint problem that they want to solve, this is the right space to work on and release it.\nPharmaverse has grown a lot, at the time of writing this post we have >25 packages recommended in pharmaverse, and this has led to a partnership with PHUSE to get support from their organization and platform, and because they are eager to advance and support pharmaverse mission.\nDespite all its structure, it is impossible to say that we have a single solution for each clinical reporting analysis when it comes to pharmaverse, a single pathway is impractical. Instead, it is necessary to accept viable tools fitting different pathways into pharmaverse to direct and give people options as to what might work for them. After all, even though we live together as a community, we still have our own unique internal problems.\n\n\n\nSample of pharmaverse packages"
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#last-updated",
- "href": "posts/2023-07-09_falcon/falcon.html#last-updated",
- "title": "falcon",
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#last-updated",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#last-updated",
+ "title": "The pharmaverse (hi)story",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:38.469514"
+ "text": "Last updated\n\n2023-11-27 19:13:49.940918"
},
{
- "objectID": "posts/2023-07-09_falcon/falcon.html#details",
- "href": "posts/2023-07-09_falcon/falcon.html#details",
- "title": "falcon",
+ "objectID": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#details",
+ "href": "posts/2023-10-10_pharmaverse_story/pharmaverse_story.html#details",
+ "title": "The pharmaverse (hi)story",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html",
- "href": "posts/2023-06-27__hackathon_app/index.html",
- "title": "Hackathon Feedback Application",
+ "objectID": "posts/2023-06-28_welcome/index.html",
+ "href": "posts/2023-06-28_welcome/index.html",
+ "title": "Hello pharmaverse",
"section": "",
- "text": "We recently created a shiny application for the admiral hackathon in February 2023. The admiral hackathon was an event designed to make statistical programmers from the pharmaceutical industry more comfortable with the admiral R package which allows users to efficiently transform data from one data standard (SDTM) to another (ADaM).\nHackathon participants formed groups of up to five people and were then tasked to create R-scripts that map the SDTM data to ADaM according to specifics defined in the metadata.\nThe purpose of the shiny app was threefold:\nIn this blog post I want to highlight some of the thoughts that went into this application. Please keep in mind that this work was done under tight time restraints.\nThe hackathon application is still online (although data-upload is switched off) and the GitHub repository is publicly available. The application is embedded into this post right after this paragraph. I have also uploaded to GitHub a .zip file of the workspace to which hackathon participants had access via posit cloud. For more context you can watch recordings of the hackathon-meetings."
- },
- {
- "objectID": "posts/2023-06-27__hackathon_app/index.html#permanent-data",
- "href": "posts/2023-06-27__hackathon_app/index.html#permanent-data",
- "title": "Hackathon Feedback Application",
- "section": "Permanent Data",
- "text": "Permanent Data\nThe biggest challenge you have to consider for this app is the permanent data storage. Shiny apps run on a server. Although we can write files on this server, whenever the app restarts, the files are lost. Therefore, a persistent data storage solution is required.\n\nGoogle drive\nI decided to leverage Google drive using the googledrive package package. This allowed me to save structured data (the team registry and the submission scores) as well as unstructured data (their R-script files).\n\n\n\n\n\n\nAuthentication\n\n\n\nTo access Google drive using the googledrive package we need to authenticate. This can be done interactively using the command googledrive::drive_auth() which takes you to the Google login page. After login you receive an authentication token requested by R.\nFor non-interactive authentication this token must be stored locally. In our case where the shiny app must access the token once deployed, the token must be stored on the project level.\nI have included the authentication procedure I followed in the R folder in google_init.R. You can find more extensive documentation of the non-interactive authentication.\n\n\nThe initial concept was: Each team gets their own folder including the most recent submission for each task, and a .csv file containing team information. To keep track of the submissions and the respective scores we wrote a .csv file in the mock-hackathon folder, so one folder above the team folders.\nSaving the team info as a .csv file worked fine as each team received their own file which – once created – was not touched anymore. As each upload for every team should simply add a row to the submissions.csv file, appending the file would be ideal. This was not possible using the googledrive package package. Instead, for each submission, the submissions file was downloaded, appended, and uploaded again. Unfortunately, this lead to a data loss, as the file was continuously overwritten, especially when two teams would submit simultaneously.\n\n\n\n\n\n\nRecover the Lost Data\n\n\n\nWhenever the submissions.csv file was uploaded, the previous version was sent to the Google drive bin. We ended up with over 3000 submissions.csv files containing a lot of redundant information. I had to write the following chunk to first get the unique file IDs of the 3000 submissions.csv files, create an empty submissions data-frame, and then download each file and add its information to the submisisons data-frame. To keep the data-frame as light as possible, after each append I deleted all duplicate submissions.\n\n# get all task_info.csv ID's\n# each row identifies one file in the trash\ntask_info_master <- drive_find(\n pattern = \"task_info.csv\",\n trashed = TRUE\n)\n\n\n# set up empty df to store all submissions\norigin <- tibble(\n score = numeric(),\n task = character(),\n team = character(),\n email = character(),\n time = character()\n)\n\n# downloads, reads, and returns one csv file given a file id\nget_file <- function(row) {\n tf <- tempfile()\n row %>%\n as_id() %>%\n drive_download(path = tf)\n new <- read_csv(tf) %>%\n select(score, task, team) %>%\n distinct()\n}\n\n\n# quick and dirty for loop to subsequently download each file, extract information\n# merge with previous information and squash it (using distinct()).\nfor (i in 1:nrow(task_info_master)) {\n origin <- rbind(origin, get_file(row = task_info_master[i, ])) %>%\n distinct()\n\n # save progress in a separate file after every 100 downloaded and merged sheets\n if (i %% 100 == 0) {\n print(i)\n write_csv(origin, paste(\"prog_data/task_info_prog_\", i, \".csv\", sep = \"\"))\n # update on progress\n message(i / nrow(task_info_master) * 100)\n }\n}\n\nWhen doing such a time-intensive task, make sure to try it first with only a couple of files to see whether any errors are produced. I am not quite sure how long this took but when I returned from my lunch break everything had finished.\n\n\nIf you want to stay in the Google framework, I recommend using the googlesheets4 package for structured data. googlesheets4 allows appending new information to an already existing sheet without the need to download the file first. As both packages follow the same style, going from one to the other is really simple. googlesheets4 requires authentication as well. However, you can reuse the cached token from the googledrive package authentication by setting gs4_auth(token = drive_token()).\n\n\nSecurity Concerns\nConnecting a public shiny app to your Google account introduces a security vulnerability in general. Especially so because we implemented the upload of files to Google drive. And even more problematic: We run a user generated script and display some of its output. A malicious party might be able to extract the authentication token of our Google account or could upload malware to the drive.\nTo reduce the risk, I simply created an un-associated Google account to host the drive. There are certainly better options available, but this seemed a reasonable solution achieved with very little effort."
- },
- {
- "objectID": "posts/2023-06-27__hackathon_app/index.html#register-team",
- "href": "posts/2023-06-27__hackathon_app/index.html#register-team",
- "title": "Hackathon Feedback Application",
- "section": "Register Team",
- "text": "Register Team\nWe wanted to allow users to sign up as teams using the shiny app. The app provides a simple interface where users could input a team name and the number of members. This in turn would open two fields for each user to input their name and email address.\nWe do simple checks to make sure at least one valid email address is supplied, and that the group name is acceptable. The group name cannot be empty, already taken, or contain vulgar words.\nThe team registration itself was adding the team information to the Google sheets file event_info into the sheet teams and to create a team folder in which to store the uploaded R files.\nThe checks and registration is implemented in the register_team() function stored in interact_with_google.R.\n\n\n\nScreenshot of the register team interface\n\n\nThe challenge here was to adapt the number of input fields depending on the number of team members. This means that the team name and email interface must be rendered: First, we check how many team members are part of the group, this is stored in the input$n_members input variable. Then we create a tagList with as many elements as team members. Each element contains two columns, one for the email, one for the member name. This tagList is then returned and displayed to the user.\n\n# render email input UI of the register tab\noutput$name_email <- shiny::renderUI({\n # create field names\n N <- input$n_members\n NAME <- sapply(1:N, function(i) {\n paste0(\"name\", i)\n })\n EMAIL <- sapply(1:N, function(i) {\n paste0(\"email\", i)\n })\n\n output <- tagList()\n\n\n firstsecondthird <- c(\"First\", \"Second\", \"Third\", \"Fourth\", \"Fifth\")\n for (i in 1:N) {\n output[[i]] <- tagList()\n output[[i]] <- fluidRow(\n shiny::h4(paste(firstsecondthird[i], \" Member\")),\n column(6,\n textInput(NAME[i], \"Name\"),\n value = \" \" # displayed default value\n ),\n column(6,\n textInput(EMAIL[i], \"Email\"),\n value = \" \"\n )\n )\n }\n output\n})\n\nThe team information is then uploaded to Google drive. Because some teams have more members than others, we have to create the respective data-frame with the number of team members in mind.\nThe following chunk creates the registration data. Noteworthy here the creation of the NAME and EMAIL variables which depend on the number of members in this team. Further, the user input of these fields is extracted via input[[paste0(NAME[i])]] within a for-loop.\nWe also make the data-creation dependent on the press of the Register Group button and cache some variables.\n\n## registration\nregistrationData <-\n reactive({\n N <- input$n_members\n NAME <- sapply(1:N, function(i) {\n paste0(\"name\", i)\n })\n EMAIL <- sapply(1:N, function(i) {\n paste0(\"email\", i)\n })\n names <- character(0)\n emails <- character(0)\n\n for (i in 1:N) {\n names[i] <- input[[paste0(NAME[i])]]\n emails[i] <- input[[paste0(EMAIL[i])]]\n }\n # create df\n dplyr::tibble(\n team_name = input$team_name,\n n_members = N,\n member_name = names,\n member_email = emails\n )\n }) %>%\n bindCache(input$team_name, input$n_members, input$name1, input$email1) %>%\n bindEvent(input$register) # wait for button press"
- },
- {
- "objectID": "posts/2023-06-27__hackathon_app/index.html#upload-source-script",
- "href": "posts/2023-06-27__hackathon_app/index.html#upload-source-script",
- "title": "Hackathon Feedback Application",
- "section": "Upload & Source Script",
- "text": "Upload & Source Script\nTo upload a script, participants had to select their team first. The input options were based on the existing folders on the Google-drive in the mock_hackathon folder. To upload a particular script participants had to also select the task to be solved. The uploaded script is then uploaded to the team folder following a standardised script naming convention.\nThere are different aspects to be aware of when sourcing scripts on a shiny server. For example, you have to anticipate the packages users will include in their uploaded scripts, as their scripts will load but not install packages. Further, you should keep the global environment of your shiny app separate from the environment in which the script is sourced. This is possible by supplying an environment to the source() function, e.g: source(path_to_script, local = new.env())\nAnother thing we had to consider was to replicate the exact folder-structure on the shiny server that participants were working with when creating the scripts, as they were required to source some scripts and to save their file into a specific folder. This was relatively straight forward as we provided participants with a folder structure in the posit cloud instance they were using. They had access to the sdtm folder in which the data was stored, and the adam folder into which they saved their solutions. The structure also included a folder with metadata which was also available on the shiny server.\nFor some tasks, participants required some ADaM-datasets stored in the adam folder, essentially the output from previous tasks. This was achieved by first creating a list mapping tasks to the required ADaM datasets:\n\ndepends_list <- list(\n \"ADADAS\" = c(\"ADSL\"),\n \"ADAE\" = c(\"ADSL\"),\n \"ADLBC\" = c(\"ADSL\"),\n \"ADLBH\" = c(\"ADSL\"),\n \"ADLBHY\" = c(\"ADSL\"),\n \"ADSL\" = NULL,\n \"ADTTE\" = c(\"ADSL\", \"ADAE\"),\n \"ADVS\" = c(\"ADSL\")\n)\n\nThis list is sourced from the R/parameters.R file when initiating the application. We then call the get_depends() function sourced from R/get_depends.R which copies the required files from the key folder (where our solutions to the tasks were stored) to the adam folder. After sourcing the uploaded script the content in the adam folder is deleted."
+ "text": "The communications working group (CWG) seeks to promote and showcase how R can be used in the Clinical Reporting pipeline through short and informative blog posts. These posts will be hosted on this pharmaverse blog and promoted on the pharmaverse slack channels as well as on LinkedIn.\nAs the CWG is a small team, we hope to make the blog development process easy enough that pharmaverse community members will be able to easily write blog posts with guidance from the CWG team."
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#compare-to-solution-file",
- "href": "posts/2023-06-27__hackathon_app/index.html#compare-to-solution-file",
- "title": "Hackathon Feedback Application",
- "section": "Compare to Solution File",
- "text": "Compare to Solution File\nWe want to compare the file created by participants with our solution (key) file stored in the key folder. The diffdf::diffdf() function allows for easy comparison of two data-frames and directly provides extensive feedback for the user:\n\nlibrary(dplyr)\ndf1 <- tibble(\n numbers = 1:10,\n letters = LETTERS[1:10]\n)\ndf2 <- tibble(\n numbers = 1:10,\n letters = letters[1:10]\n)\n\ndiffdf::diffdf(df1, df2)\n\nWarning in diffdf::diffdf(df1, df2): \nNot all Values Compared Equal\n\n\nDifferences found between the objects!\n\nA summary is given below.\n\nNot all Values Compared Equal\nAll rows are shown in table below\n\n =============================\n Variable No of Differences \n -----------------------------\n letters 10 \n -----------------------------\n\n\nAll rows are shown in table below\n\n ========================================\n VARIABLE ..ROWNUMBER.. BASE COMPARE \n ----------------------------------------\n letters 1 A a \n letters 2 B b \n letters 3 C c \n letters 4 D d \n letters 5 E e \n letters 6 F f \n letters 7 G g \n letters 8 H h \n letters 9 I i \n letters 10 J j \n ----------------------------------------"
+ "objectID": "posts/2023-06-28_welcome/index.html#purpose",
+ "href": "posts/2023-06-28_welcome/index.html#purpose",
+ "title": "Hello pharmaverse",
+ "section": "",
+ "text": "The communications working group (CWG) seeks to promote and showcase how R can be used in the Clinical Reporting pipeline through short and informative blog posts. These posts will be hosted on this pharmaverse blog and promoted on the pharmaverse slack channels as well as on LinkedIn.\nAs the CWG is a small team, we hope to make the blog development process easy enough that pharmaverse community members will be able to easily write blog posts with guidance from the CWG team."
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#score",
- "href": "posts/2023-06-27__hackathon_app/index.html#score",
- "title": "Hackathon Feedback Application",
- "section": "Score",
- "text": "Score\nTo compare submissions between participants we implemented a simple scoring function (score_f()) based on the table comparison by diffdf(). The function can be found in the compare_dfs.R file:\n\nscore_f <- function(df_user, df_key, keys) {\n score <- 10\n diff <- diffdf::diffdf(df_user, df_key, keys = keys)\n if (!diffdf::diffdf_has_issues(diff)) {\n return(score)\n }\n\n # check if there are any differences if the comparison is not strict:\n if (!diffdf::diffdf_has_issues(diffdf::diffdf(df_user,\n df_key,\n keys = keys,\n strict_numeric = FALSE,\n strict_factor = FALSE\n ))) {\n # if differences are not strict, return score - 1\n return(score - 1)\n }\n\n return(round(min(max(score - length(diff) / 3, 1), 9), 2))\n}\n\nEvery comparison starts with a score of 10. We then subtract the length of the comparison object divided by a factor of 3. The length of the comparison object is a simplified way to represent the difference between the two data-frames by one value. Finally, the score is bounded by 1 using max(score, 1).\nThe score is not a perfect capture of the quality of the script uploaded but: 1. helped participants get an idea of how close their data-frame is to the solution file 2. allowed us to raffle prizes based on the merit of submitted r-scripts"
+ "objectID": "posts/2023-06-28_welcome/index.html#spirit-of-a-blog-post",
+ "href": "posts/2023-06-28_welcome/index.html#spirit-of-a-blog-post",
+ "title": "Hello pharmaverse",
+ "section": "Spirit of a Blog Post",
+ "text": "Spirit of a Blog Post\nThe CWG believes that the following 4 points will help guide the creation of Blog Posts.\n\nShort\nPersonalized\nReproducible\nReadable\n\nShort: Posts should aim to be under a 10 minute read. We encourage longer posts to be broken up into multiple posts.\nPersonalized: Posts should have a personality! For example, a person wishing to post on a function in a package needs to differentiate the post from the documentation for function, i.e. we don’t want to just recycle the documentation. How can you add your voice and experience? A bit of cheeky language is also encouraged.\nReproducible: Posts should work with minimal dependencies with data, packages and outside sources. Every dependency introduced in a post adds some risk to the post longevity. As package dependencies change, posts should be built in a way that they can be updated to stay relevant.\nReadable: The CWG sees this site as more of introductory site rather advanced user site. Therefore, the CWG feels that code should be introduced in a way that promotes readability over complexity."
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#reactiveness",
- "href": "posts/2023-06-27__hackathon_app/index.html#reactiveness",
- "title": "Hackathon Feedback Application",
- "section": "Reactiveness",
- "text": "Reactiveness\nSome of the app functions can take quite some time to execute, e.g. running the uploaded script. Other tasks, e.g. registering a team, do not intrinsically generate user facing outputs. This would make the app using really frustrating, as users would not know whether the app is correctly working or whether it froze.\nWe implemented two small features that made the app more responsive. One is simple loading icons that integrate into the user interface and show that output is being computed – that something is working. The other is a pop up window which communicates whether team registration was successful, and if not, why not.\nWe further aimed to forward errors generated by the uploaded scripts to the user interface, but errors generated by the application itself should be concealed."
+ "objectID": "posts/2023-06-28_welcome/index.html#what-types-of-posts-are-allowed-on-this-site",
+ "href": "posts/2023-06-28_welcome/index.html#what-types-of-posts-are-allowed-on-this-site",
+ "title": "Hello pharmaverse",
+ "section": "What types of posts are allowed on this site?",
+ "text": "What types of posts are allowed on this site?\nOverall, we want to stay focus on the Clinical Reporting Pipeline, which we see as the following topics:\n\nPackages in the Clinical Reporting Pipeline\nFunctions from packages in the Clinical Reporting Pipeline\nWider experiences of using R in the Clinical Reporting Pipeline\nConference experiences and the Clinical Reporting Pipeline\n\nHowever, it never hurts to ask if you topic might fit into this medium!\n\nMinimum Post Requirements\n\nA unique image to help showcase the post.\nWorking Code\nSelf-contained data or package data.\nDocumentation of package versions\n\nThat is it! After that you can go wild, but we do ask that it is kept short!"
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#conclusion",
- "href": "posts/2023-06-27__hackathon_app/index.html#conclusion",
- "title": "Hackathon Feedback Application",
- "section": "Conclusion",
- "text": "Conclusion\nAlthough the application was continuously improved during the hackathon it proved to be a useful resource for participants from day one as it allowed groups to set their own pace. It further allowed admiral developers to gain insights on package usage of a relatively large sample of potential end users. From our perspective, the application provided a great added value to the hackathon and eased the workload of guiding the participants through all the tasks."
+ "objectID": "posts/2023-06-28_welcome/index.html#how-can-i-make-a-blog-post",
+ "href": "posts/2023-06-28_welcome/index.html#how-can-i-make-a-blog-post",
+ "title": "Hello pharmaverse",
+ "section": "How can I make a Blog Post",
+ "text": "How can I make a Blog Post\nStep 1: Reach out to us through pharmaverse/slack or make an issue on our GitHub.\nStep 2: Branch off main\nStep 3: Review the Spirit of the Blog Post in the Pull Request Template\nStep 4: Poke us to do a review!"
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#last-updated",
- "href": "posts/2023-06-27__hackathon_app/index.html#last-updated",
- "title": "Hackathon Feedback Application",
+ "objectID": "posts/2023-06-28_welcome/index.html#last-updated",
+ "href": "posts/2023-06-28_welcome/index.html#last-updated",
+ "title": "Hello pharmaverse",
"section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:42.927745"
+ "text": "Last updated\n\n2023-11-27 19:13:52.942147"
},
{
- "objectID": "posts/2023-06-27__hackathon_app/index.html#details",
- "href": "posts/2023-06-27__hackathon_app/index.html#details",
- "title": "Hackathon Feedback Application",
+ "objectID": "posts/2023-06-28_welcome/index.html#details",
+ "href": "posts/2023-06-28_welcome/index.html#details",
+ "title": "Hello pharmaverse",
"section": "Details",
"text": "Details\n\nsource code, R environment"
},
@@ -621,47 +691,5 @@
"title": "Cross-Industry Open Source Package Development",
"section": "Impact",
"text": "Impact\nFor Roche, cross-industry package development work out in their favor: They get access to software created by specialists and users from across the industry but paying only a fraction of the developmental costs. Of course, they don’t have total developmental control but they do get a seat at the table. Any gaps between the open source {admiral} package and the proprietary Roche workflow were bridged by the internal {admiralroche} package.\nThe switch towards a more language agnostic platform, and open source languages specifically, opens the door to a broad population of university graduates with diverse backgrounds. I personally would not have considered this position five years ago due to a misalignment of skills and job requirements. Working towards an industry standard open source solution will also ensure that skills learned at one company are more easily transferable to external positions, further making the position much more attractive. Access to such a broad pool of potential candidates is clearly beneficial for recruitment at Roche, but also facilitates diversity in teams which makes for a more interesting and effective work place.\nOpen source development comes with much more transparency by definition. Recognition of contributions are built in - anyone can see who did what. This recognition escapes the confines of your company as it is visible to anyone looking at the repository. Anyone can not only see at any time what is being worked on, what discussions are happening and which direction is being taken, but can also participate and contribute. Transparency also applies to errors in the code and how the team is dealing with them. In such an environment it is practically impossible to hide or cover up errors and corrections. Instead, they have to be dealt with publicly and in the open. This openness about errors also helps seeing errors as a natural occurrence that needs to be dealt with. Space for errors encourages learning and is really beneficial for growing both skills and integrity.\nAs you work on a team that spans multiple companies, traditional corporate hierarchies do not apply. Of course, there will always be a sort of hierarchy of experience or skills, but these work in your favor: You will know who to ask for help, and teams are generally very happy for contributors of any skill level. Contributions also need not be in code: Inputs into discussions and domain knowledge contributions are highly valued as well. The flip-side of working in a team without your manager oversight: They may not be directly aware of the work you do. That’s why you have to write blog posts :)\nThe possibility for statistical programmers to pivot towards developing software or writing blog-posts such as this really transforms and broadens their job description. It is this transformation that is reflected by the choice of Roche to re-brand statistical programmers as analytical data scientists. The fact that cross-industry development is being advocated for really lets programmers expand their network outside of their company.\nThe {admiral} project serves as a testament to the power of collaborative open-source development and the potential it holds for the future of work in this industry."
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html",
- "href": "posts/2023-06-27_hackathon_writeup/index.html",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "",
- "text": "This January and February (2023), the admiral development team and the CDISC Open Source Alliance jointly hosted the admiral hackathon. The idea was to build a community of admiral users, and help participants familiarize themselves with R and admiral. This whole effort was led by Thomas Neitmann and was supported by Zelos Zhu, Sadchla Mascary, and me – Stefan Thoma.\nThe hackathon event was structured in two parts. First, we offered an Introduction to R for SAS programmers, a three hour workshop for R beginners to get them up to speed. Here we covered practical R basics, talking about how the R-workflow differs from a SAS workflow, and discussed common R functions - mostly from the tidyverse. This ensured that hackathon participants were familiar with core R concepts. The workshop recording and the course materials are available online.\nThe main hackathon consisted of several ADAM data generating tasks based on a specs file and synthetic data. Participants were able to solve these tasks in groups at their own pace thanks to a online tool where participants could upload their task specific R scripts and they would get automatic feedback for the data-set produced by their script. Script upload through the feedback application was available all through February, and we offered three additional online meetings throughout the month to discuss challenges and give some tips. If you are interested in learning more about the thoughts that went into the feedback application, you can read about it in this blogpost or check out my public GitHub repository for such an application."
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html#introduction-to-r-workshop",
- "href": "posts/2023-06-27_hackathon_writeup/index.html#introduction-to-r-workshop",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "Introduction to R workshop",
- "text": "Introduction to R workshop\nWe were really excited to see over 500 people from around 40 countries joining our Introduction to R workshop in January! To get to know prospective users and hackathon participants better, we conducted some polls during the meetings. Below you can see that representatives of many different sorts of organisations joined our Introduction to R workshop:\n\n\n\n\n\n216 out of 402 confirmed that their company is already using R for clinical trial data analysis, the remaining 131 did not answer this question.\nThe target audience for this workshop was programmers who are very familiar with SAS, but not so familiar with R, our polls confirmed this.\n\n\n\n\n\nOverall, we were very happy with how the workshop turned out, and participants overall agreed with this sentiment (although there may be a slight survivorship bias…)."
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html#admiral-hackathon",
- "href": "posts/2023-06-27_hackathon_writeup/index.html#admiral-hackathon",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "admiral Hackathon",
- "text": "admiral Hackathon\nFollowing the kick-off meeting, 371 participants joined the posit (rStudio) workspace that was made available to all participants at no costs by the posit company. About half the participants planned to spend one to two hours per week on the admiral tasks, the other half planned to allocate even more. 15 participants even planned to spend eight hours or more!\nWe were really happy to see an overwhelming amount of activity on the slack channel we set up with over 250 members. Not only were people engaging with the materials, but we saw how a community was formed where people were encouraged to ask questions and where community members went out of their way to help each other. Shout-out to our community hero: Jagadish Katam without whom most issues related to the task programming raised by the community would not have been addressed as quickly as they were. Huge thanks from the organizers!\nIn the end, a total of 44 teams spanning 87 statistical programmers took part in the admiral hackathon and uploaded solution scripts to the hackathon application solving at least one of the 8 tasks available (ADSL, ADAE, ADLBC, ADVS, ADTTE, ADADAS, ADLBH & ADLBHY). Participants’ scripts were then run on the shiny server and the output data-frame were compared to the solutions we provided. At the read-out there was a live draft of teams to win one-on-one admiral consulting with one of the admiral core developers. Winning probabilities were weighted by the number of points each group received for the quality of their output data-frames and for the number of tasks solved.\nCongratulations to the winners:\n\nViiV Team_GSK\nteamspoRt\nTatianaPXL\nDivyasneelam\nAdaMTeamIndia\nSanofi_BP\nJagadish (our community hero)\nAZ_WAWA\n\nAlthough this was uncertain during the hackathon we were excited to provide a Certificate of Completion to all participants who uploaded a script to the Web Application.\nA recording of the hackathon readout can be found in the CDISC Open Source Alliance Quarterly Spotlight."
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html#conclusion",
- "href": "posts/2023-06-27_hackathon_writeup/index.html#conclusion",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "Conclusion",
- "text": "Conclusion\nOverall, we are very happy with how the hackathon turned out. We were not only positively surprised with the huge audience for the Intro to R workshop (CDISC record breaking) and for the admiral hackathon, but even more so with the engagement of all the participants.\nAgain, we would like to thank all the organizers, participants, and sponsors for their time and resources and hope to have provided a useful glimpse into our solution for ADAM creation within the end-to-end clinical data analysis open source R framework that the Pharmaverse aims to provide.\nAs always, we are very happy to hear more feedback on the hackathon as well as on admiral in general. Simply submit an issue on the admiral GitHub repository. You would like to join the admiral core developers? Please reach out to Edoardo Mancini (product owner) or Ben Straub (technical lead)."
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html#last-updated",
- "href": "posts/2023-06-27_hackathon_writeup/index.html#last-updated",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "Last updated",
- "text": "Last updated\n\n2023-11-01 15:47:51.728905"
- },
- {
- "objectID": "posts/2023-06-27_hackathon_writeup/index.html#details",
- "href": "posts/2023-06-27_hackathon_writeup/index.html#details",
- "title": "Admiral Hackathon 2023 Revisited",
- "section": "Details",
- "text": "Details\n\nsource code, R environment"
}
]
\ No newline at end of file
diff --git a/site_libs/htmlwidgets-1.6.2/htmlwidgets.js b/site_libs/htmlwidgets-1.6.3/htmlwidgets.js
similarity index 100%
rename from site_libs/htmlwidgets-1.6.2/htmlwidgets.js
rename to site_libs/htmlwidgets-1.6.3/htmlwidgets.js
diff --git a/sitemap.xml b/sitemap.xml
index a0a76761..e2577287 100644
--- a/sitemap.xml
+++ b/sitemap.xml
@@ -1,67 +1,71 @@
- https://pharmaverse.github.io/blog/posts/2023-07-14_code_sections/code_sections.html
- 2023-11-01T15:47:53.974Z
+ https://pharmaverse.github.io/blog/posts/2023-08-08_study_day/study_day.html
+ 2023-11-27T19:13:58.211Z
- https://pharmaverse.github.io/blog/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
- 2023-11-01T15:47:48.826Z
+ https://pharmaverse.github.io/blog/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
+ 2023-11-27T19:13:54.971Z
- https://pharmaverse.github.io/blog/posts/2023-08-08_study_day/study_day.html
- 2023-11-01T15:47:45.858Z
+ https://pharmaverse.github.io/blog/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
+ 2023-11-27T19:13:52.247Z
- https://pharmaverse.github.io/blog/posts/2023-09-26_date_functions_and_imputation/date_functions_and_imputation.html
- 2023-11-01T15:47:41.734Z
+ https://pharmaverse.github.io/blog/posts/2023-06-27__hackathon_app/index.html
+ 2023-11-27T19:13:49.215Z
- https://pharmaverse.github.io/blog/posts/2023-06-28_welcome/index.html
- 2023-11-01T15:47:37.682Z
+ https://pharmaverse.github.io/blog/posts/2023-11-27_higher_order/higher_order.html
+ 2023-11-27T19:13:44.987Z
- https://pharmaverse.github.io/blog/posts/2023-06-27_admiral/valuelevel/derive_param_computed.html
- 2023-11-01T15:47:33.918Z
+ https://pharmaverse.github.io/blog/posts/2023-07-09_falcon/falcon.html
+ 2023-11-27T19:13:40.491Z
- https://pharmaverse.github.io/blog/posts/2023-07-24_rounding/rounding.html
- 2023-11-01T15:47:30.366Z
+ https://pharmaverse.github.io/blog/posts/2023-06-27_hackathon_writeup/index.html
+ 2023-11-27T19:13:37.491Z
- https://pharmaverse.github.io/blog/R/readme.html
- 2023-11-01T15:47:26.622Z
+ https://pharmaverse.github.io/blog/posts/2023-09-26_date_functions_and_imputation/date_functions_and_imputation.html
+ 2023-11-27T19:13:32.503Z
https://pharmaverse.github.io/blog/index.html
- 2023-11-01T15:47:26.074Z
+ 2023-11-27T19:13:29.559Z
- https://pharmaverse.github.io/blog/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
- 2023-11-01T15:47:28.446Z
+ https://pharmaverse.github.io/blog/R/readme.html
+ 2023-11-27T19:13:30.087Z
- https://pharmaverse.github.io/blog/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
- 2023-11-01T15:47:31.962Z
+ https://pharmaverse.github.io/blog/posts/2023-07-24_rounding/rounding.html
+ 2023-11-27T19:13:34.283Z
- https://pharmaverse.github.io/blog/posts/2023-10-30_floating_point/floating_point.html
- 2023-11-01T15:47:36.342Z
+ https://pharmaverse.github.io/blog/posts/2023-06-27_admiral/valuelevel/derive_param_computed.html
+ 2023-11-27T19:13:39.323Z
- https://pharmaverse.github.io/blog/posts/2023-07-09_falcon/falcon.html
- 2023-11-01T15:47:39.010Z
+ https://pharmaverse.github.io/blog/posts/2023-07-14_code_sections/code_sections.html
+ 2023-11-27T19:13:41.975Z
- https://pharmaverse.github.io/blog/posts/2023-06-27__hackathon_app/index.html
- 2023-11-01T15:47:43.922Z
+ https://pharmaverse.github.io/blog/posts/2023-10-30_floating_point/floating_point.html
+ 2023-11-27T19:13:47.267Z
- https://pharmaverse.github.io/blog/posts/2023-07-20_cross_company_dev/cross_industry_dev.html
- 2023-11-01T15:47:47.398Z
+ https://pharmaverse.github.io/blog/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
+ 2023-11-27T19:13:50.535Z
- https://pharmaverse.github.io/blog/posts/2023-06-27_hackathon_writeup/index.html
- 2023-11-01T15:47:52.282Z
+ https://pharmaverse.github.io/blog/posts/2023-06-28_welcome/index.html
+ 2023-11-27T19:13:53.443Z
+
+
+ https://pharmaverse.github.io/blog/posts/2023-07-20_cross_company_dev/cross_industry_dev.html
+ 2023-11-27T19:13:56.455Z
Last updated
2023-11-01 15:47:45.242711
+2023-11-27 19:13:57.651626
Details
References
Last updated
2023-11-01 15:47:31.231853
+2023-11-27 19:13:54.254558
Details
Release, g
Last updated
-2023-11-01 15:47:48.201548
+2023-11-27 19:13:49.940918
Details
diff --git a/posts/2023-10-30_floating_point/floating_point.html b/posts/2023-10-30_floating_point/floating_point.html
index 4b06317b..31c93ef3 100644
--- a/posts/2023-10-30_floating_point/floating_point.html
+++ b/posts/2023-10-30_floating_point/floating_point.html
@@ -107,7 +107,7 @@
-
+
@@ -304,8 +304,8 @@ Issues arising
reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
-
-
+
+
Last updated
2023-11-01 15:47:48.201548
+2023-11-27 19:13:49.940918
Details
Issues arising
reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)As you can see, the data-frame contains the column GRADE_CRITERIA_CODE which contains comparisons of floating point values. And there was a discrepancy of what Gordon expected to see, and how R actually computed the comparison initially:
Potential solutions reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
Conclusion
Last updated
2023-11-01 15:47:35.340689
+2023-11-27 19:13:46.342818












