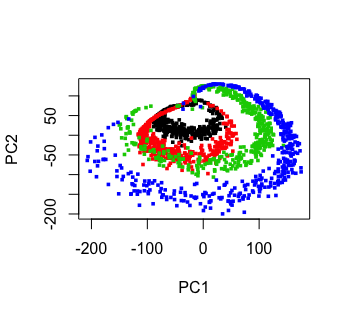
Norm Matloff, Prof. of Computer Science, UC Davis; my bio
(See notice at the end of this document regarding copyright.)
This site is for those who know nothing of R, and maybe even nothing of programming, and seek QUICK, PAINLESS! entree to the world of R.
-
FAST: You'll already be doing good stuff in R -- useful data analysis -- in your very first lesson.
-
For nonprogrammers: If you're comfortable with navigating the Web and viewing charts, you're fine. This tutorial is aimed at you, not experienced C or Python coders.
-
Motivating: Every lesson centers around a real problem to be solved, on real data. The lessons do not consist of a few toy examples, unrelated to the real world. The material is presented in a conversational, story-telling manner.
-
Just the basics, no frills or polemics:
- Notably, in the first few lessons, we do NOT use Integrated Development Environments (IDEs). RStudio, ESS etc. are great, but you shouldn't be burdened with learning R and learning an IDE at the same time, a distraction from the goal of becoming productive in R as fast as possible.
Note that even the excellent course by R-Ladies Sydney, which does start with RStudio, laments that RStudio can be "way too overwhelming."
So, in the initial lessons, we stick to the R command line, and focus on data analysis, not tools such as IDEs, which we will cover as an intermediate-level topic. (Some readers of this tutorial may already be using RStudio or an external editor, and the treatment here will include special instructions for them when needed.)
-
Coverage is mainly limited to base R. So for instance the popular but self-described "opinionated" Tidyverse is not treated at first, partly due to its controversial nature (I am a skeptic), but again mainly because it would be an obstacle to your becoming productive in R quickly. A later lesson to be added will introduce Tidy (defined as dplyr and pipes) and compare it to base-R.
While you can learn a few simple things in Tidy quickly, thinking you are learning a lot, those things are quite limited in scope, and Tidy learners often find difficulty in applying R to real world data. Our tutorial here is aimed at learners whose goal is to USE the R system productively in their own data analysis.
-
Nonpassive approach: Passive learning, just watching the screen, is NO learning. There will be occasional Your Turn sections, in which you the learner must devise and try your own variants on what has been presented. Sometimes the tutorial will be give you some suggestions, but even then, you should cook up your own variants to try. Remember: You get out what you put in! The more actively you work the Your Turn sections, the more powerful you will be as an R coder.
PART I
- Lesson 1: Getting Started
- Lesson 2: First R Steps
- Lesson 3: Vectors and Indices
- Lesson 4: More on Vectors
- Lesson 5: On to Data Frames!
- Lesson 6: The R Factor Class
- Lesson 7: Extracting Rows/Columns from Data Frames
- Lesson 8: More Examples of Extracting Rows, Columns
- Lesson 9: The tapply Function
- Lesson 10: Data Cleaning
- Lesson 11: The R List Class
- Lesson 12: Another Look at the Nile Data
- Lesson 13: Pause to Reflect
- Lesson 14: Introduction to Base R Graphics
- Lesson 15: More on Base Graphics
- Lesson 16: Writing Your Own Functions
- Lesson 17: 'For' Loops
- Lesson 18: Functions with Blocks
- Lesson 19: Text Editing and IDes
- Lesson 20: If, Else, Ifelse
- Lesson 21: Do Pro Athletes Keep Fit?
- Lesson 22: Linear Regression Analysis, I
- Lesson 23: S3 Classes
- Lesson 24: Baseball Player Analysis (cont'd.)
- Lesson 25: R Packages, CRAN, Etc.
PART II
- Lesson 26: A Pause, Before Going on to Advanced Topics
- Lesson 27: A First Look at ggplot2
- Lesson 28: Should You Use Functional Programming?
- Lesson 29 Simple Text Processing, I
- Lesson 30: Simple Text Processing, II
- Lesson 31: Linear Regression Analysis, II
- Lesson 32: Working with the R Date Class
- Lesson 33: Tips on R Coding Style and Strategy
- Lesson 34: The Logistic Model
- Lesson 35: Files and Directories
- Lesson 36: R 'while' Loops
- To Learn More
- Thanks
- Appendix: Installing and Using RStudio
For the time being, the main part of this online course will be this README.md file. It is set up as a potential R package, though, and I may implement that later.
The color figure at the top of this file was generated by our prVis package, run on a famous dataset called Swiss Roll.
-
Nonpassive learning is absolutely key! So even if the output of an R command is shown here, run the command yourself in your R console, by copy-and-pasting from this document into the R console. You will get out of this tutorial what you put in.
-
Similarly, the Your Turn sections are absolutely crucial. Devise your own little examples, and try them! "When in doubt, Try it out!" is a motto I devised for teaching. If you are unclear or curious about something, try it out! Just devise a little experiment, and type in the code. Don't worry -- you won't "break" things.
-
Tip: I cannot teach you how to program. I can merely give you the tools, e.g. R vectors, and some examples. For a given desired programming task, then, you must creatively put these tools together to attain the goal. Treat it like a puzzle! I think you'll find that if you stick with it, you'll find you're pretty good at it. After all, we can all work puzzles.
You'll need to install R, from the R Project site. Start up R, either by clicking an icon or typing 'R' in a terminal window. We are not requiring RStudio here, but if you already have it, start it; you'll be typing into the R console, the Console pane.
As noted, this tutorial will be "bare bones." In particular, there is no script to type your command for you. Instead, you will either copy-and-paste from the text here into the R console, or type them there by hand. (Note that the code lines here will all begin with the R interactive prompt, '>'; that should not be typed.)
This is a Markdown file. You can read it right there on GitHub, which has its own Markdown renderer. Or you can download it to your own machine in Chrome and use the Markdown Reader extension to view it (be sure to enable Allow Access to File URLs).
When you end your R session, exit by typing 'quit()'.
Good luck! And if you have any questions, feel free to e-mail me, at [email protected]
The R command prompt is '>'. Again, it will be shown here, but you don't type it. It is just there in your R window to let you know R is inviting you to submit a command. (If you are using RStudio, you'll see it in the Console pane.)
So, just type '1+1' then hit Enter. Sure enough, it prints out 2 (you were expecting maybe 12108?):
> 1 + 1
[1] 2But what is that '[1]' here? It's just a row label. We'll go into that later, not needed quite yet.
R includes a number of built-in datasets, mainly for illustration purposes. One of them is Nile, 100 years of annual flow data on the Nile River.
Let's find the mean flow:
> mean(Nile)
[1] 919.35Here mean is an example of an R function, and in this case Nile is an argument -- fancy way of saying "input" -- to that function. That output, 919.35, is called the return value or simply value. The act of running the function is termed calling the function.
Another point to note is that we didn't need to call R's print function. We could have typed,
> print(mean(Nile))Function calls in R (and other languages) work "from the inside out." Here we are asking R to find the mean of the Nile data, then print the result.
But whenever we are at the R '>' prompt, any expression we type will be printed out anyway, so there is no need to call print.
Since there are only 100 data points here, it's not unwieldy to print them out. Again, all we have to do is type ``Nile,'' no need to call print:
> Nile
Time Series:
Start = 1871
End = 1970
Frequency = 1
[1] 1120 1160 963 1210 1160 1160 813 1230 1370 1140 995 935 1110 994 1020
[16] 960 1180 799 958 1140 1100 1210 1150 1250 1260 1220 1030 1100 774 840
[31] 874 694 940 833 701 916 692 1020 1050 969 831 726 456 824 702
[46] 1120 1100 832 764 821 768 845 864 862 698 845 744 796 1040 759
[61] 781 865 845 944 984 897 822 1010 771 676 649 846 812 742 801
[76] 1040 860 874 848 890 744 749 838 1050 918 986 797 923 975 815
[91] 1020 906 901 1170 912 746 919 718 714 740Now you can see how the row labels work. There are 15 numbers per row here, so the second row starts with the 16th, indicated by '[16]'. The third row starts with the 31st output number, hence the '[31]' and so on.
Note that a set of numbers such as Nile is called a vector.
R has great graphics, not only in base R but also in wonderful user-contributed packages, such as ggplot2 and lattice. But we'll stick with base-R graphics for now, and save the more powerful yet more complex ggplot2 for a later lesson.
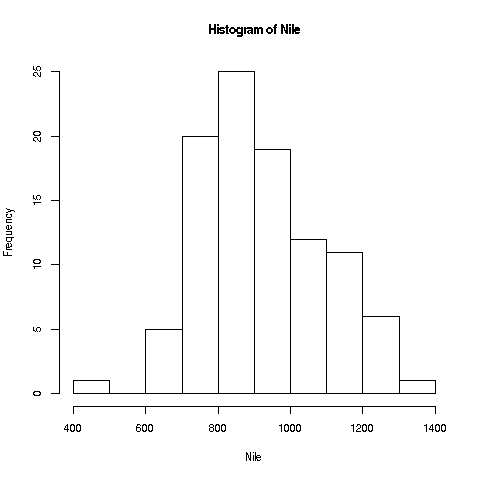
We'll start with a very simple, non-dazzling one, a no-frills histogram:
> hist(Nile)No return value for the hist function (there is one, but it is seldom used, and we won't go into it here), but it does create the graph.
Your Turn: The hist function draws 10 bins for this dataset in the histogram by default, but you can choose other values, by specifying an optional second argument to the function, named breaks. E.g.
> hist(Nile,breaks=20)would draw the histogram with 20 bins. Try plotting using several different large and small values of the number of bins.
Note: The hist function, as with many R functions, has many different options, specifiable via various arguments. For now, we'll just keep things simple, and resist the temptation to explore them all. R has lots of online help, which you can access via '?'. E.g. typing
> ?histwill tell you to full story on all the options available for the hist function. Again, there are far too many for you to digest for now (most users don't ever find a need for the more esoteric ones), but it's a vital resource to know.
Your Turn: Look at the online help for mean and Nile.
Say we want to find the mean river flow after year 1950.
The above output said that the Nile series starts in 1871. That means 1951 will be the 81st year, and the 100th will be 1970. How do we designate the 81st through 100th elements in this data?
Individual elements can be accessed using subscripts or indices (singular is index), which are specified using brackets, e.g.
> Nile[2]
[1] 1160for the second element (the output we saw earlier shows that the second element is indeed 1160). The value 2 here is the index.
The c ("concatenate") function builds a vector, stringing several numbers together. E.g. we can get the 2nd, 5th and 6th elements of Nile:
> Nile[c(2,5,6)]
[1] 1160 1160 1160If we wish to build a vector of consecutive numbers, we can use the "colon" notation:
> Nile[c(2,3,4)]
[1] 1160 963 1210
> Nile[2:4]
[1] 1160 963 1210As you can see, 2:4 is shorter way to specify the vector c(2,3,4).
So, 81:100 means all the numbers from 81 to 100. Thus Nile[81:100] specifies the 81st through 100th elements in the Nile vector.
Then to answer the above question on the mean flow during 1951-1971, we can do
> mean(Nile[81:100])
[1] 877.05NOTE: Observe how the above reasoning process worked. We had a goal, to find the mean river flow after 1950. We knew we had some tools available to us, namely the mean function and R vector indices. We then had to figure out a way to combine these tools in a manner that achieves our goal, which we did.
This is how use of R works in general. As you go through this tutorial, you'll add more and more to your R "toolbox." Then for any given goal, you'll rummage around in that toolbox, and eventually figure out the right set of tools for the goal at hand. Sometimes this will require some patience, but you'll find that the more you do, the more adept you become.
If we plan to do more with that time period, we should make a copy of it:
> n81100 <- Nile[81:100]
> mean(n81100)
[1] 877.05
> sd(n81100)
[1] 125.5583The function sd finds the standard deviation.
Note that we used R's assignment operator here to copy ("assign") those particular Nile elements to n81100. (In most situations, you can use "=" instead of "<-", but why worry about what the exceptions might be? They are arcane, so it's easier just to always use "<-". And though "keyboard shortcuts" for this are possible, again let's just stick to the basics for now.)
Note too that though we will speak of the above operation as having "extracted" the 81st through 100th elements of Nile, we have merely made a copy of those elements. The original vector Nile remains intact.
Tip: We can pretty much choose any name we want; "n81100" just was chosen to easily remember this new vector's provenance. (But names can't include spaces, and must start with a letter.)
Note that n81100 now is a 21-element vector. Its first element is now element 81 of Nile:
> n81100[1]
[1] 744
> Nile[81]
[1] 744Keep in mind that although Nile and n81100 now have identical contents, they are separate vectors; if one changes, the other will not.
Your Turn: Devise and try variants of the above, say finding the mean over the years 1945-1960.
Another oft-used function is length, which gives the number of elements in the vector, e.g.
> length(Nile)
[1] 100Can you guess the value of length(n81100)? Type this expression in at the '>' prompt to check your answer.
Leave R by typing 'q()' or ctrl-d. (Answer no to saving the workspace.)
-
Starting and existing R.
-
Some R functions: mean, hist, length.
-
R vectors, and vector indices.
-
Extracting vector subsets.
-
Forming vectors, using c() and ":".
Not bad for Lesson 1! And needless to say, you'll be using all of these frequently in the subsequent lessons and in your own usage of R.
Continuing along the Nile, say we would like to know in how many years the level exceeded 1200. Let's first introduce R's sum function:
> sum(c(5,12,13))
[1] 30Here the c function built a vector consisting of 5, 12 and 13. That vector was then fed into the sum function, returning 5+12+13 = 30.
By the way, the above is our first example of function composition, where the output of one function, c here, is fed as input into another, sum in this case.
We can now use this to answer our question on the Nile data:
> sum(Nile > 1200)
[1] 7The river level exceeded 1200 in 7 years.
But how in the world did that work? Bear with me a bit here. Let's look at a small example first:
> x <- c(5,12,13)
> x
[1] 5 12 13
> x > 8
[1] FALSE TRUE TRUE
> sum(x > 8)
[1] 2First, notice something odd here, in the expression x > 8. Here x is a vector, 3 elements in length, but 8 is just a number. It would seem that it's nnonsense to ask whether a vector is greater than a number; they're different animals.
But R makes them "the same kind" of animal, by extending that number 8 to a 3-element vector 8,8,8. This is called recycling. This sets up an element-by-element comparison: Then, the 5 in x is compared to the first 8, yielding FALSE i.e. 5 is NOT greater than 8. Then 12 is compared to the second 8, yielding TRUE, and then the comparison of 13 to the third 8 yields another TRUE. So, we get the vector FALSE,TRUE,TRUE.
Fine, but how will sum add up some TRUEs and FALSEs? The answer is that R, like most computer languages, treats TRUE and FALSE as 1 and 0, respectively. So we summed the vector (0,1,1), yielding 2.
Getting back to the question of the number of years in which the Nile flow exceeded 1200, let's look at that expression again:
> sum(Nile > 1200)Since the vector Nile has length 100, that number 1200 will be recycled into a vector of one hundred copies of 1200. The '>' comparison will then yield 100 TRUEs and FALSEs, so summing gives us the number of TRUEs, exactly what we want.
Your Turn: Try a few other experiments of your choice using sum. I'd suggest starting with finding the sum of the first 25 elements in Nile. You may wish to start with experiments on a small vector, say (2,1,1,6,8,5), so you will know that your answers are correct. Remember, you'll learn best nonpassively. Code away!
A question related to how many years had a flow above 1200 is which years had that property. Well, R actually has a which function:
> which(Nile > 1200)
[1] 4 8 9 22 24 25 26So the 4th, 8th, 9th etc. elements in Nile had the queried property. (Note that those were years 1875, 1879 and so on.)
In fact, that gives us another way to get the count of the years with that trait:
> which1200 <- which(Nile > 1200)
> which1200
[1] 4 8 9 22 24 25 26
> length(which1200)
[1] 7Of course, as usual, my choice of the variable name "which1200" was arbirary, just something to help me remember what is stored in that variable.
R's length function does what it says, i.e. finding the length of a vector. In our context, that gives us the count of years with flow above 1200.
And, what were the river flows in those 7 years?
> which1200 <- which(Nile > 1200)
> Nile[which1200]
[1] 1210 1230 1370 1210 1250 1260 1220Finally, something a little fancier. We can combine steps above:
> Nile[Nile > 1200]
[1] 1210 1230 1370 1210 1250 1260 1220We just "eliminated the middle man," which1200. The R interpreter saw our "Nile > 1200", and thus generated the corresponding TRUEs and FALSEs. The R interpreter then treated those TRUEs and FALSEs as subscripts in Nile, thus extracting the desired data.
Now, we might add here, "Don't try this at home, kids." For beginners, it's really easier and more comfortable to break things into steps. Once, you become experienced at R, you may wish to start skipping steps.
Less bold is the notion of negative indices, e.g.
> x <- c(5,12,13,8)
> x[-1]
[1] 12 13 8Here we are asking for all of x except for x[1]. Can you guess what x[c(-1,-4)] evaluates to? Guess first, then try it out.
Here you've refined your skillset for R vectors, learning R's recycling feature, and two tricks that R users employ for finding counts of things.
Once again, as you progress through this tutorial, you'll see that these things are used a lot in R.
Right after vectors, the next major workhorse of R is the data frame. It's a rectangular table consisting of one row for each data point.
Say we have height, weight and age on each of 100 people. Our data frame would have 100 rows and 3 columns. The entry in, e.g., the second row and third column would be the age of the second person in our data. The second row as a whole would be all the data for that second person, i.e. the height, weight and age of that person.
Note that that row would also be cnsidered a vector. The third column as a whole would be the vector of all ages in our dataset.
As our first example, consider the ToothGrowth dataset built-in to R. Again, you can read about it in the online help by typing
> ?ToothGrowth
``` (The data turn out to be on guinea pigs, with orange juice or
Vitamin C as growth supplements.) Let's take a quick look from the
command line.
``` r
> head(ToothGrowth)
len supp dose
1 4.2 VC 0.5
2 11.5 VC 0.5
3 7.3 VC 0.5
4 5.8 VC 0.5
5 6.4 VC 0.5
6 10.0 VC 0.5R's head function displays (by default) the first 6 rows of the given dataframe. We see there are length, supplement and dosage columns, which the curator of the data decided to name 'len', 'supp' and 'dose'. Each of column is an R vector, or in the case of the second column, a vector-like object called a factor, to be discussed shortly).
Tip: To avoid writing out the long words repeatedly, it's handy to make a copy with a shorter name.
> tg <- ToothGrowthDollar signs are used to denote the individual columns, e.g. ToothGrowth$dose for the dose column. So for instance, we can print out the mean tooth length:
> mean(tg$len)
[1] 18.81333Subscripts/indices in data frames are pairs, specifying row and column numbers. To get the element in row 3, column 1:
> tg[3,1]
[1] 7.3which matches what we saw above in our head example. Or, use the fact that tg$len is a vector:
> tg$len[3]
[1] 7.3The element in row 3, column 1 in the data frame tg is element 3 in the vector tg$letn. This duality between data frames and vectors is often exploited in R.
Your Turn: The above examples are fundamental to R, so you should conduct a few small experiments on your own this time, little variants of the above. The more you do, the better!
For any subset of a data frame d, we can extract whatever rows and columns we want using the format
d[the rows we want, the columns we want]Some data frames don't have column names, but that is no obstacle. We can use column numbers, e.g.
> mean(tg[,1])
[1] 18.81333Note the expression '[,1]'. Since there is a 1 in the second position, we are talking about column 1. And since the first position, before the comma, is empty, no rows are specified -- so all rows are included. That boils down to: all of column 1.
A key feature of R is that one can extract subsets of data frames, just as we extracted subsets of vectors earlier. For instance,
> z <- tg[2:5,c(1,3)]
> z
len dose
2 11.5 0.5
3 7.3 0.5
4 5.8 0.5
5 6.4 0.5Here we extracted rows 2 through 5, and columns 1 and 3, assigning the result to z. To extract those columns but keep all rows, do
> y <- tg[ ,c(1,3)]i.e. leave the row specification field empty.
By the way, note that the three columns are all of the same length, a requirement for data frames. And what is that common length in this case? R's nrow function tells us the number of rows in any data frame:
> nrow(ToothGrowth)
[1] 60Ah, 60 rows (60 guinea pigs, 3 measurements each).
Or, alternatively:
> tg <- ToothGrowth
> length(tg$len)
[1] 60
> length(tg$supp)
[1] 60
> length(tg$dose)
[1] 60So now you know four ways to do the same thing. But isn't one enough? Of course. But in this get-acquainted period, reading all four will help reinforce the knowledge you are now accumulating about R. So, make sure you understand how each of those four approaches produced the number 60.
The head function works on vectors too:
> head(ToothGrowth$len)
[1] 4.2 11.5 7.3 5.8 6.4 10.0Like many R functions, head has an optional second argument, specifying how many elements to print:
> head(ToothGrowth$len,10)
[1] 4.2 11.5 7.3 5.8 6.4 10.0 11.2 11.2 5.2 7.0You can create your own data frames -- good for devising little tests of your understanding -- as follows:
> x <- c(5,12,13)
> y <- c('abc','de','z')
> d <- data.frame(x,y)
> d
x y
1 5 abc
2 12 de
3 13 zLook at that second line! Instead of vectors consisting of numbers, one can form vectors of character strings, complete with indexing capability, e.g.
> y <- c('abc','de','z')
> y[2]
[1] "de"As noted, all the columns in a data frame must be of the same length. Here x consists of 3 numbers, and y consists of 3 character strings. (The string is the unit in the latter. The number of characters in each string is irrelevant.)
One can use negative indices for rows and columns as well, e.g.
> z <- tg[,-2]
> head(z)
len dose
1 4.2 0.5
2 11.5 0.5
3 7.3 0.5
4 5.8 0.5
5 6.4 0.5
6 10.0 0.5Your Turn: Devise your own little examples with the ToothGrowth data. For instance, write code that finds the number of cases in which the tooth length was less than 16. Also, try some examples with another built-in R dataset, faithful. This one involves the Old Faithful geyser in Yellowstone National Park in the US. The first column gives duration of the eruption, and the second has the waiting time since the last eruption. As mentioned, these operations are key features of R, so devise and run as many examples as possible; err on the side of doing too many!
As mentioned, the data frame is the fundamental workhorse of R. It is made up of columns of vectors (of equal lengths), a fact that often comes in handy.
Unlike the single-number indices of vectors, each element in a data frame has 2 indices, a row number and a column number. One can specify sets of rows and columns to extra subframes.
One can use the R nrow function to query the number of rows in a data frame; ncol does the same for the number of columns.
Each object in R has a class. The number 3 is of the 'numeric' class, the character string 'abc' is of the 'character' class, and so on. (In R, class names are quoted; one can use single or double quotation marks.) Note that vectors of numbers are of 'numeric' class too; actually, a single number is considered to be a vector of length 1. So, c('abc','xw'), for instance, is 'character' as well.
Tip: Computers require one to be very, very careful and very, very precise. In that expression c('abc','xw') above, one might wonder why it does not evaluate to 'abcxw'. After all, didn't I say that the 'c' stands for "concatenate"? Yes, but the c function concatenates vectors. Here 'abc' is a vector of length 1 -- we have one character string, and the fact that it consists of 3 characters is irrelevant -- and likewise 'xw' is one character string. So, we are concatenating a 1-element vector with another 1-element vector, resulting in a 2-element vector.
What about tg and tg$supp in the Vitamin C example above? What are their classes?
> class(tg)
[1] "data.frame"
> class(tg$supp)
[1] "factor"R factors are used when we have categorical variables. If in a genetics study, say, we have a variable for hair color, that might comprise four categories: black, brown, red, blond. We can find the list of categories for tg$supp as follows:
> levels(tg$supp)
[1] "OJ" "VC"The categorical variable here is supp, the name the creator of this dataset chose for the supplement column. We see that there are two categories (levels), either orange juice or Vitamin C.
Note carefully that the values of an R factor must be quoted. Either single or double quote marks is fine (though the marks don't show up when we use head).
Factors can sometimes be a bit tricky to work with, but the above is enough for now. Let's see how to apply the notion in the current dataset.
(The reader should cover this lesson especially slowly and carefully. The concepts are simple, but putting them together requires careful inspection.)
First, let's review what we saw in a previous lesson:
> which1200 <- which(Nile > 1200)
> Nile[which1200]
[1] 1210 1230 1370 1210 1250 1260 1220There, we saw how to extract vector elements. We can do something similar to extract data frame rows or columns. Here is how:
Continuing the Vitamin C example, let's compare mean tooth length for the two types of supplements. Here is the code:
> whichOJ <- which(tg$supp == 'OJ')
> whichVC <- which(tg$supp == 'VC')
> mean(tg[whichOJ,1])
[1] 20.66333
> mean(tg[whichVC,1])
[1] 16.96333In the first two lines above, we found which rows in tg (or equivalently, which elements in tg$supp) had the OJ supplement, and recorded those row numbers in whichOJ. Then we did the same for VC.
Now, look at the expression tg[whichOJ,1]. Remember, data frames are accessed with two subscript expressions, one for rows, one for colums, in the format
d[the rows we want, the columns we want]So, tg[whichOJ,1] says to restrict attention to the OJ rows, and only column 1, tooth length. We then find the mean of those restricted numberss. This turned out to be 20.66333. Then do the same for VC.
Again, if we are pretty experienced, we can skip steps:
> tgoj <- tg[tg$supp == 'OJ',]
> tgvc <- tg[tg$supp == 'VC',]
> mean(tgoj$len)
[1] 20.66333
> mean(tgvc$len)
[1] 16.96333Either way, we have the answer to our original question: Orange juice appeared to produce more growth than Vitamin C. (Of course, one might form a confidence interval for the difference etc.)
Just as we learned earlier how to use a sequence of TRUE and FALSE values to extract a parts of a vector, we now see how to do the analogous thing for data frames: We can use a sequence of TRUE and FALSE values to extract a certain rows or columns from a data frame.
It is imperative that the reader fully understand this lesson before continuing, trying some variations of the above example on his/her own. We'll be using this technique often in this tutorial, and it is central to R usage in the real world.
Your turn: Try some of these operations on R's built-in faithful dataset. For instance, find the number of eruptions for which the waiting time was more than 80 minutes.
Often we need to extract rows or columns from a data frame, subject to more than one condition. For instance, say we wish to extract from tg the sub-data frame consisting of OJ and length less than 8.8.
We could do this, using the ampersand symbol '&', which means a logical AND operation:
> tg[tg$supp=='OJ' & tg$len < 8.8,]
len supp dose
37 8.2 OJ 0.5Ah, it turns out that only one case satisfied both conditions.
If we want all rows that satisfy at least one of the conditions, not necessarily both, then we use the OR operator, '|'. Say we want to obtain all rows in which either len is greater than 28 or the treatment dose was 1.0 or both:
> tg[tg$len > 28 | tg$dose == 1.0,]
len supp dose
11 16.5 VC 1
12 16.5 VC 1
13 15.2 VC 1
14 17.3 VC 1
15 22.5 VC 1
16 17.3 VC 1
17 13.6 VC 1
18 14.5 VC 1
19 18.8 VC 1
20 15.5 VC 1
23 33.9 VC 2
26 32.5 VC 2
30 29.5 VC 2
41 19.7 OJ 1
42 23.3 OJ 1
43 23.6 OJ 1
44 26.4 OJ 1
45 20.0 OJ 1
46 25.2 OJ 1
47 25.8 OJ 1
48 21.2 OJ 1
49 14.5 OJ 1
50 27.3 OJ 1
56 30.9 OJ 2
59 29.4 OJ 2By the way, note that the original row numbers are displayed too. For example, the first case satisfying the conditions was row number 11 in the original data frame tg.
> w <- tg[tg$len > 28 | tg$dose == 1.0,]Again, I chose the name 'w' arbitrarily. Names must begin with a letter, and consist only of letters, digits and a few special characters such as '-' or '.'
Note that w is a new data frame, on which we can perform the usual operations, e.g.
> head(w)
len supp dose
11 16.5 VC 1
12 16.5 VC 1
13 15.2 VC 1
14 17.3 VC 1
15 22.5 VC 1
16 17.3 VC 1
> nrow(w)
[1] 25We may only be interested in say, how many cases satisfied the given conditions. As before, we can use nrow for that, as seen here.
As seen early, we can also extract columns. Say our analysis will use only tooth length and dose. We write 'c(1,3)' in the "what columns we want" place, indicating columns 1 and 3:
> lendose <- tg[,c(1,3)]
> head(lendose)
len dose
1 4.2 0.5
2 11.5 0.5
3 7.3 0.5
4 5.8 0.5
5 6.4 0.5
6 10.0 0.5From now on, we would work with lendose instead of tg.
It's a little nicer, though, the specify the columns by name instead of number:
> lendose <- tg[,c('len','dose')]
> head(lendose)
len dose
1 4.2 0.5
2 11.5 0.5
3 7.3 0.5
4 5.8 0.5
5 6.4 0.5
6 10.0 0.5The logical operations work on vectors too. For example, say in the Nile data we wish to know how many years had flows in the extremes, say below 500 or above 1200:
> exts <- Nile[Nile < 800 | Nile > 1300]
> head(exts)
[1] 1370 799 774 694 701 692
> length(exts)
[1] 27By the way, if this count were our only interest, i.e. we have no further use for exts, we can skip assigning to exts, and do things directly:
> length(Nile[Nile < 800 | Nile > 1300])
[1] 27This is fine for advanced, experienced R users, but really, "one step at a time" is better for beginners.
Here we got more practice in manipulating data frames, and were introduced to the logical operators '&' and '|'. We also saw another example of using nrow as a means of counting how many rows satisfy given conditions.
Again, these are all "bread and butter" operations that arise quite freqently in real world R usage.
By the way, note how the essence of R is "combining little things in order to do big things," e.g. combining the subsetting operation, the '&' operator, and nrow to get a count of rows satisfying given conditions. This too is the "bread and butter" of R. It's up to you, the R user, to creatively combine R's little operations (and later, some big ones) to achieve whatever goals you have for your data. Programming is a creative process. It's like a grocery store and cooking: The store has lots of different potential ingredients, and you decide which ones to buy and combine into a meal.
Your turn: Try some of these operations on R's built-in faithful dataset. For instance, find the number of eruptions for which 'eruptions' was greater than 3 and waiting time was more than 80 minutes.
Tip: Often in R there is a shorter, more compact way of doing things. That's the case here; we can use the magical tapply function in the above example. In fact, we can do it in just one line.
> tapply(tg$len,tg$supp,mean)
OJ VC
20.66333 16.96333 In English: "Split the vector tg$len into two groups, according to the value of tg$supp, then apply mean to each group." Note that the result was returned as a vector, which we could save by assigning it to, say z:
> z <- tapply(tg$len,tg$supp,mean)
> z[1]
OJ
20.66333
> z[2]
VC
16.96333 By the way, z is not only a vector, but also a named vector, meaning that its elements have names, in this case 'OJ' and 'VC'.
Saving can be quite handy, because we can use that result in subsequent code.
To make sure it is clear how this works, let's look at a small artificial example:
> x <- c(8,5,12,13)
> g <- c('M',"F",'M','M')Suppose x is the ages of some kids, who are a boy, a girl, then two more boys, as indicated in g. For instance, the 5-year-old is a girl.
Let's call tapply:
> tapply(x,g,mean)
F M
5 11 That call said, "Split x into two piles, according to the corresponding elements of g, and then find the mean in each pile.
Note that it is no accident that x and g had the same number of elements above, 4 each. If on the contrary, g had 5 elements, that fifth element would be useless -- the gender of a nonexistent fifth child's age in x. Similarly, it wouldn't be right if g had had only 3 elements, apparently leaving the fourth child without a specified gender.
Tip: If g had been of the wrong length, we would have gotten an error, "Arguments must be of the same length." This is a common error in R code, so watch out for it, keeping in mind WHY the lengths must be the same.
Instead of mean, we can use any function as that third argument in tapply. Here is another example, using the built-in dataset PlantGrowth:
> tapply(PlantGrowth$weight,PlantGrowth$group,length)
ctrl trt1 trt2
10 10 10 Here tapply split the weight vector into subsets according to the group variable, then called the length function on each subset. We see that each subset had length 10, i.e. the experiment had assigned 10 plants to the control, 10 to treatment 1 and 10 to treatment 2.
Your Turn: One of the most famous built-in R datasets is mtcars, which has various measurements on cars from the 60s and 70s. Lots of opportunties for you to cook up little experiments here! You may wish to start by comparing the mean miles-per-gallon values for 4-, 6- and 8-cylinder cars. Another suggestion would be to find how many cars there are in each cylinder category, using tapply. As usual, the more examples you cook up here, the better!
By the way, the mtcars data frame has a "phantom" column.
> head(mtcars)
mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1That first column seems to give the make (brand) and model of the car. Yes, it does -- but it's not a column. Behold:
> head(mtcars[,1])
[1] 21.0 21.0 22.8 21.4 18.7 18.1Sure enough, column 1 is the mpg data, not the car names. But we see the names there on the far left! The resolution of this seeming contradiction is that those car names are the row names of this data frame:
> row.names(mtcars)
[1] "Mazda RX4" "Mazda RX4 Wag" "Datsun 710"
[4] "Hornet 4 Drive" "Hornet Sportabout" "Valiant"
[7] "Duster 360" "Merc 240D" "Merc 230"
[10] "Merc 280" "Merc 280C" "Merc 450SE"
[13] "Merc 450SL" "Merc 450SLC" "Cadillac Fleetwood"
[16] "Lincoln Continental" "Chrysler Imperial" "Fiat 128"
[19] "Honda Civic" "Toyota Corolla" "Toyota Corona"
[22] "Dodge Challenger" "AMC Javelin" "Camaro Z28"
[25] "Pontiac Firebird" "Fiat X1-9" "Porsche 914-2"
[28] "Lotus Europa" "Ford Pantera L" "Ferrari Dino"
[31] "Maserati Bora" "Volvo 142E" So 'Mazda RX4' was the name of row 1, but not part of the row.
As with everything else, row.names is a function, and as you can see above, its return value here is a 32-element vector (the data frame had 32 rows, thus 32 row names). The elements of that vector are of class 'character', as is the vector itself.
You can even assign to that vector:
> row.names(mtcars)[7]
[1] "Duster 360"
> row.names(mtcars)[7] <- 'Dustpan'
> row.names(mtcars)[7]
[1] "Dustpan"Inside joke, by the way. Yes, the example is real and significant, but the "Dustpan" thing came from a funny TV commercial at the time.
(If you have some background in programming, it may appear odd to you to have a function call on the left side of an assignment. This is actually common in R. It stems from the fact that '<-' is actually a function! But this is not the place to go into that.)
Your Turn: Try some experiments with the mtcars data, e.g. finding the mean horsepower for 6-cylinder cars.
Tip: As a beginner (and for that matter later on), you should NOT be obsessed with always writing code in the "optimal" way, including in terms of compactness of the code. It's much more important to write something that works and is clear; one can always tweak it later. In this case, though, tapply actually aids clarity, and it is so ubiquitously useful that we have introduced it early in this tutorial. We'll be using it more in later lessons.
Most real-world data is "dirty," i.e. filled with errors. The famous New York taxi trip dataset, for instance, has one trip destination whose lattitude and longitude place it in Antartica! The impact of such erroneous data on one's statistical analysis can be anywhere from mild to disabling. Let's see below how one might ferret out bad data. And along the way, we'll cover several new R concepts.
We'll use the famous Pima Diabetes dataset. Various versions exist, but we'll use the one included in faraway, an R package compiled by Julian Faraway, author of several popular books on statistical regression analysis in R.
I've placed the data file, Pima.csv, on my Web site. Here is how you can read it into R:
> pima <- read.csv('http://heather.cs.ucdavis.edu/FasteR/data/Pima.csv',header=TRUE)The dataset is in a CSV ("comma-separated values") file. Here we read it, and assigned the resulting data frame to a variable we chose to name pima.
Note that second argument, 'header=TRUE'. A header in a file, if one exists, is in the first line in the file. It states what names the columns in the data frame are to have. If the file doesn't have one, set header to FALSE. You can always add names to your data frame later (future lesson).
Tip: It's always good to take a quick look at a new data frame:
> head(pima)
pregnant glucose diastolic triceps insulin bmi diabetes age test
1 6 148 72 35 0 33.6 0.627 50 1
2 1 85 66 29 0 26.6 0.351 31 0
3 8 183 64 0 0 23.3 0.672 32 1
4 1 89 66 23 94 28.1 0.167 21 0
5 0 137 40 35 168 43.1 2.288 33 1
6 5 116 74 0 0 25.6 0.201 30 0
> dim(pima)
[1] 768 9The dim function tells us that there are 768 people in the study, 9 variables measured on each.
Since this is a study of diabetes, let's take a look at the glucose variable. R's table function is quite handy.
> table(pima$glucose)
0 44 56 57 61 62 65 67 68 71 72 73 74 75 76 77 78 79 80 81
5 1 1 2 1 1 1 1 3 4 1 3 4 2 2 2 4 3 6 6
82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101
3 6 10 7 3 7 9 6 11 9 9 7 7 13 8 9 3 17 17 9
102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121
13 9 6 13 14 11 13 12 6 14 13 5 11 10 7 11 6 11 11 6
122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141
12 9 11 14 9 5 11 14 7 5 5 5 6 4 8 8 5 8 5 5
142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161
5 6 7 5 9 7 4 1 3 6 4 2 6 5 3 2 8 2 1 3
162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181
6 3 3 4 3 3 4 1 2 3 1 6 2 2 2 1 1 5 5 5
182 183 184 186 187 188 189 190 191 193 194 195 196 197 198 199
1 3 3 1 4 2 4 1 1 2 3 2 3 4 1 1 Be careful here; the first, third, fifth and so on lines are the glucose values, while the second, fourth, sixth and so on lines are the counts of women having those values. For instance, 3 women had the glucose = 68.
Uh, oh! 5 women in the study had glucose level 0. And 1 had level 44, etc. Presumably 0 is not physiologically possible, and maybe not 44 either.
Let's consider a version of the glucose data that at least excludes these 0s.
> pg <- pima$glucose
> pg1 <- pg[pg > 0]
> length(pg1)
[1] 763As before, the expression "pg > 0" creates a vector of TRUEs and FALSEs. The filtering "pg[pg > 0]" will only pick up the TRUE cases, and sure enough, we see that pg1 has only 763 cases, as opposed to the original 768.
Did removing the 0s make much difference? Turns out it doesn't:
> mean(pg)
[1] 120.8945
> mean(pg1)
[1] 121.6868But still, these things can in fact have major impact in many statistical analyses.
R has a special code for missing values, NA, for situations like this. Rather than removing the 0s, it's better to recode them as NAs. Let's do this, back in the original dataset so we keep all the data in one object:
> pima$glucose[pima$glucose == 0] <- NATip: That's pretty complicated. It's clearer to break things up into smaller steps (I recommend this especially for beginners), as follows:
> glc <- pima$glucose
> z <- glc == 0
> glc[z] <- NA
> pima$glucose <- glcHere is what the code does:
- That first line just makes a copy of the original vector, to avoid clutter in the code.
- The second line determines which elements of glc are 0s, resulting in z being a vector of TRUEs and FALSEs.
- The third line then assigns NA to those elements in glc corresponding to the TRUEs. (Note the recycling of NA.)
- Finally, we need to have the changes in the original data, so we copy glc to it.
Tip: Note again the double-equal sign! If we wish to test whether, say, a and b are equal, the expression must be "a == b", not "a = b"; the latter would do "a <- b". This is a famous beginning programmer's error.
As a check, let's verify that we now have 5 NAs in the glucose variable:
> sum(is.na(pima$glucose))
[1] 5Here the built-in R function is.na will return a vector of TRUEs and FALSEs. Recall that those values can always be treated as 1s and 0s, thus summable. Thus we got our count, 5.
Let's also check that the mean comes out right:
> mean(pima$glucose)
[1] NAWhat went wrong? By default, the mean function will not skip over NA values; thus the mean was reported as NA too. But we can instruct the function to skip the NAs:
> mean(pima$glucose,na.rm=TRUE)
[1] 121.6868Your Turn: Determine which other columns in pima have suspicious 0s, and replace them with NA values.
Now, look again at the plot we made earlier of the Nile flow histogram. There seems to be a gap between the numbers at the low end and the rest. What years did these correspond to? Find the mean of the data, excluding these cases.
We saw earlier how handy the tapply function can be. Let's look at a related one, split.
Earlier we mentioned the built-in dataset mtcars, a data frame. Consider mtcars$mpg, the column containing the miles-per-gallon data. Again, to save typing and avoid clutter in our code, let's make a copy first:
> mtmpg <- mtcars$mpgSuppose we wish to split the original vector into three vectors, one for 4-cylinder cars, one for 6 and one for 8. We could do
> mt4 <- mtmpg[mtcars$cyl == 4]and so on for mt6 and mt8.
Your Turn: In order to keep up, make sure you understand how that line of code works, with the TRUEs and FALSEs etc. First print out the value of mtcars$cyl == 4, and go from there.
But there is a cleaner way:
> mtl <- split(mtmpg,mtcars$cyl)
> mtl
$`4`
[1] 22.8 24.4 22.8 32.4 30.4 33.9 21.5 27.3 26.0 30.4 21.4
$`6`
[1] 21.0 21.0 21.4 18.1 19.2 17.8 19.7
$`8`
[1] 18.7 14.3 16.4 17.3 15.2 10.4 10.4 14.7 15.5 15.2 13.3 19.2 15.8 15.0
> class(mtl)
[1] "list"In English, the call to split said, "Split mtmpg into multiple vectors, with the splitting criterion being the correspond values in mtcars$cyl."
Now mtl, an object of R class "list", contains the 3 vectors. We can access them individually with the dollar sign notation:
> mtl$`4`
[1] 22.8 24.4 22.8 32.4 30.4 33.9 21.5 27.3 26.0 30.4 21.4Or, we can use indices, though now with double brackets:
> mtl[[1]]
[1] 22.8 24.4 22.8 32.4 30.4 33.9 21.5 27.3 26.0 30.4 21.4Looking a little closer:
> head(mtcars$cyl)
[1] 6 6 4 6 8 6 We see that the first car had 6 cylinders, so the
first element of mtmpg, 21.0, was thrown into the 6 pile, i.e.
mtl[[2]] (see above printout of mtl), and so on.
And of course we can make copies for later convenience:
> m4 <- mtl[[1]]
> m6 <- mtl[[2]]
> m8 <- mtl[[3]]Lists are especially good for mixing types together in one package:
> l <- list(a = c(2,5), b = 'sky')
> l
$a
[1] 2 5
$b
[1] "sky"Note that here we can give names to the list elements, 'a' and 'b'. In
forming mtl using split above, the names were assigned
according to the values of the vector beiing split. (In that earlier
case, we also needed backquotes , since the names were numbers.)
If we don't like those default names, we can change them:
> names(mtl) <- c('four','six','eight')
> mtl
$four
[1] 22.8 24.4 22.8 32.4 30.4 33.9 21.5 27.3 26.0 30.4 21.4
$six
[1] 21.0 21.0 21.4 18.1 19.2 17.8 19.7
$eight
[1] 18.7 14.3 16.4 17.3 15.2 10.4 10.4 14.7 15.5 15.2 13.3 19.2 15.8
15.0What if we want, say, the MPG for the third car in the 6-cylinder category?
> mtl[[2]][3]
[1] 21.4The point is that mtl[[2]] is a vector, so if we want element 3 of that vector, we tack on [3].
Or,
> mtl$six[3]
[1] 21.4By the way, it's no coincidence that a dollar sign is used for delineation in both data frames and lists; data frames are lists. Each column is one element of the list. So for instance,
> mtcars[[1]]
[1] 21.0 21.0 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 17.8 16.4 17.3 15.2 10.4
[16] 10.4 14.7 32.4 30.4 33.9 21.5 15.5 15.2 13.3 19.2 27.3 26.0 30.4 15.8 19.7
[31] 15.0 21.4Here we used the double-brackets list notation to get the first element of the list, which is the first column of the data frame.
Your Turn Try using split on the ToothGrowth data, say splitting into groups according to the supplement, and finding various quantities.
Here we'll learn several new concepts, using the Nile data as our starting point.
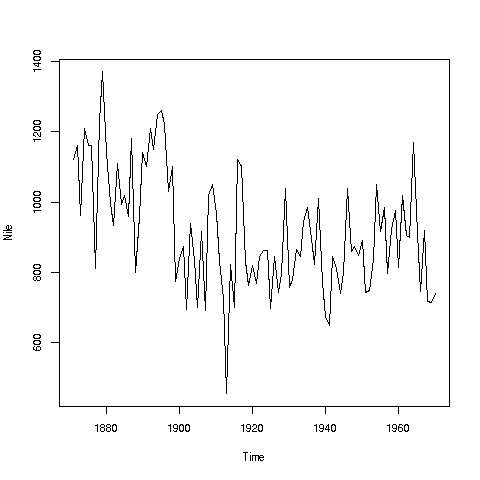
If you look again at the histogram of the Nile we generated, you'll see a gap between the lowest numbers and the rest. In what year(s) did those really low values occur? Let's plot the data against time:
> plot(Nile)Looks like maybe 1912 or so was much lower than the rest. Is this an error? Or was there some big historical event then? This would require more than R to track down, but at least R can tell us which exact year or years correspond to the unusually low flow. Here is how:
We see from the graph that the unusually low value was below 600. We can use R's which function to see when that occurred:
> which(Nile < 600)
[1] 43As before, make sure to understand what happened in this code. The expression "Nile < 600" yields 100 TRUEs and FALSEs. The which then tells us which of those were TRUEs.
So, element 43 is the culprit here, corresponding to year 1871+42=1913. Again, we would have to find supplementary information in order to decide whether this is a genuine value or an error, but at least now we know the exact year.
Of course, since this is a small dataset, we could have just printed out the entire data and visually scanned it for a low number. But what if the length of the data vector had been 100,000 instead of 100? Then the visual approach wouldn't work.
Tip: Remember, a goal of programming is to automate tasks, rather than doing them by hand.
Your Turn: There appear to be some unusually high values as well, e.g. one around 1875. Determine which year this was, using the techniques presented here.
Also, try some similar analysis on the built-in AirPassengers data. Can you guess why those peaks are occurring?
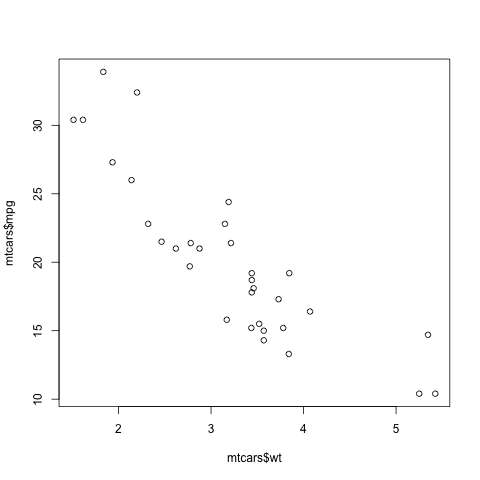
Here is another point: That function plot is not quite so innocuous as it may seem. Let's run the same function, plot, but with two arguments instead of one:
> plot(mtcars$wt,mtcars$mpg)In contrast to the previous plot, in which our data were on the vertical axis and time was on the horizontal, now we are plotting two vectors, against each other. This enables us to explore the relation between car weight and gas mileage.
There are a couple of important points here. First, as we might guess, we see that the heavier cars tended to get poorer gas mileage. But here's more: That plot function is pretty smart!
Why? Well, plot knew to take different actions for different input types. When we fed it a single vector, it plotted those numbers against time (or, against index). When we fed it two vectors, it knew to do a scatter plot.
In fact, plot was even smarter than that. It noticed that Nile is not just of 'numeric' type, but also of another class, 'ts' ("time series"):
> is.numeric(Nile)
[1] TRUE
> class(Nile)
[1] "ts"So, plot put years on the horizontal axis, instead of indices 1,2,3,...
And one more thing: Say we wanted to know the flow in the year 1925. The data start at 1871, so 1925 is 1925 - 1871 = 54 years later. Since the 1871 number is in element 1 of the vector, that means the flow for the year 1925 is in element 1+54 = 55.
> Nile[55]
[1] 698OK, but why did we do this arithmetic ourselves? We should have R do it:
> Nile[1 + 1925 - 1871]
[1] 698R did the computation 1925 - 1871 + 1 itself, yielding 55, then looked up the value of Nile[55]. This is the start of your path to programming -- we try to automate things as much as possible, doing things by hand as little as possible.
Tip: Repeating an earlier point: How does one build a house? There of course is no set formula. One has various tools and materials, and the goal is to put these together in a creative way to produce the end result, the house.
It's the same with R. The tools here are the various functions, e.g. mean and which, and the materials are one's data. One then must creatively put them together to achieve one's goal, say ferreting out patterns in ridership in a public transportation system. Again, it is a creative process; there is no formula for anything. But that is what makes it fun, like solving a puzzle.
And...we can combine various functions in order to build our own functions. This will come in future lessons.
One of the greatest things about R is its graphics capabilities. There are excellent graphics features in base R, and then many contributed packages, with the best known being ggplot2 and lattice. These latter two are quite powerful, and will be the subjects of future lessons, but for now we'll concentrate on the base.
As our example here, we'll use a dataset I compiled on Silicon Valley programmers and engineers, from the US 2000 census. Let's read in the data and take a look at the first records:
> pe <-
read.table('https://raw.githubusercontent.com/matloff/fasteR/master/data/prgeng.txt',header=TRUE)
> head(pe)
age educ occ sex wageinc wkswrkd
1 50.30082 13 102 2 75000 52
2 41.10139 9 101 1 12300 20
3 24.67374 9 102 2 15400 52
4 50.19951 11 100 1 0 52
5 51.18112 11 100 2 160 1
6 57.70413 11 100 1 0 0We used read.table here because the file is not of the CSV type. It uses blank spaces rather than commas as its delineator between fields.
Here educ and occ are codes, for levels of education and different occupations. For now, let's not worry about the specific codes. (You can find them in the Census Bureau document. For instance, search for "Educational Attainment" for the educ variable.)
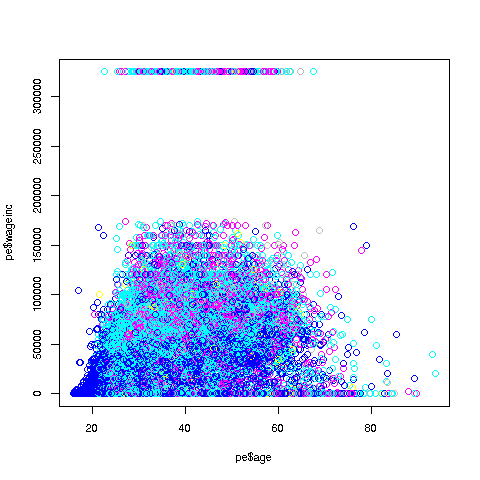
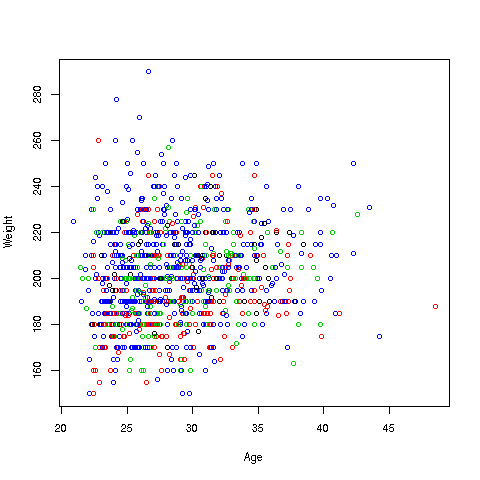
Let's start with a scatter plot of wage vs. age:
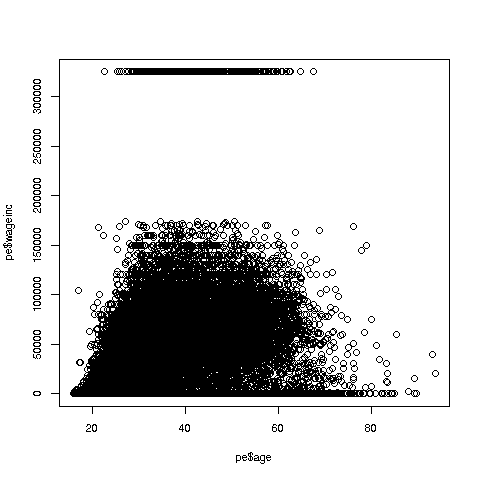
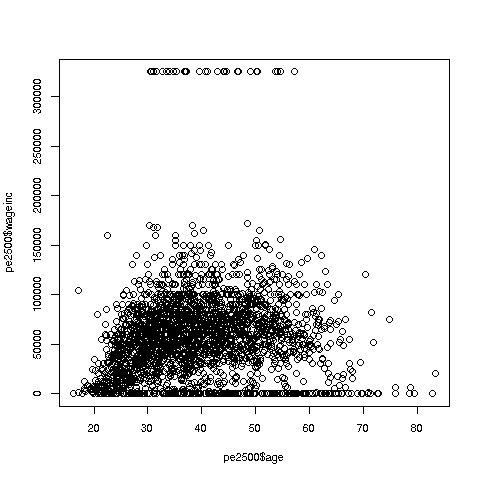
> plot(pe$age,pe$wageinc)Oh no, the dreaded Black Screen Problem! There are about 20,000 data points, thus filling certain parts of the screen. So, let's just plot a random sample, say 2500. (There are other ways of handling the problem, say with smaller dots or alpha blending.)
> indxs <- sample(1:nrow(pe),2500)
> pe2500 <- pe[indxs,]Recall that the nrow() function returns the number of rows in the argument, which in this case is 20090, the number of rows in pe.
R's sample function does what its name implies. Here it randomly samples 2500 of the numbers from 1 to 20090. We then extracted those rows of pe, in a new data frame pe2500.
Tip: Note again that it's clearer to break complex operations into simpler, smaller ones. I could have written the more compact
> pe2500 <- pe[sample(1:nrow(pe),2500),]but it would be hard to read that way. I also use direct function composition sparingly, preferring to break
h(g(f(x),3)into
y <- f(x)
z <- g(y,3)
h(z) So, here is the new plot:
> plot(pe2500$age,pe2500$wageinc)OK, now we are in business. A few things worth noting:
-
The relation between wage and age is not linear, indeed not even monotonic. After the early 40s, one's wage tends to decrease. As with any observational dataset, the underlying factors are complex, but it does seem there is an age discrimination problem in Silicon Valley. (And it is well documented in various studies and litigation.)
-
Note the horizontal streaks at the very top and very bottom of the picture. Some people in the census had 0 income (or close to it), as they were not working. And the census imposed a top wage limit of $350,000 (probably out of privacy concerns).
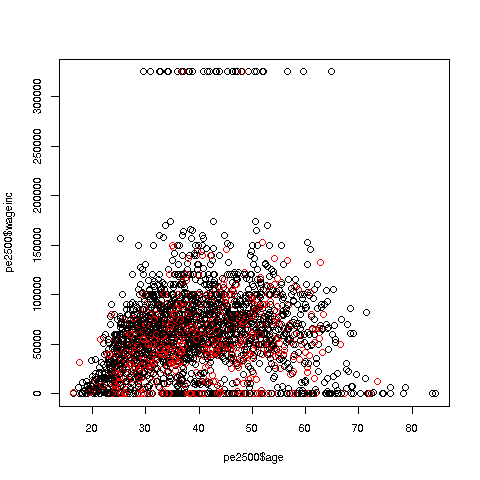
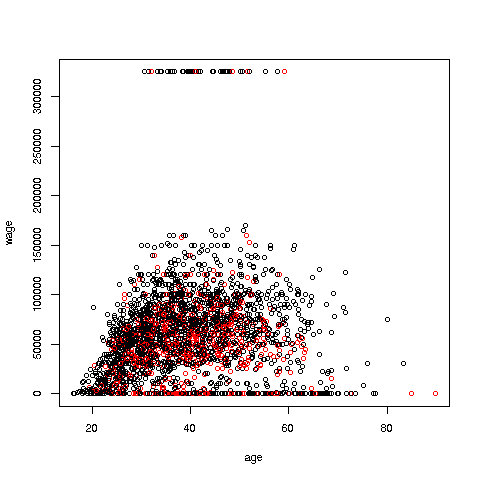
We can break things down by gender, via color coding:
> plot(pe2500$age,pe2500$wageinc,col=as.factor(pe2500$sex))The col argument indicates we wish to color code, in this case by gender. Note that pe2500$sex is a numeric vector, but col requires an R factor; the function as.factor does the conversion.
The red dots are the women. (Details below.) Are they generally paid less than men? There seems to be a hint of that, but detailed statistical analysis is needed (a future lesson).
It would be good to have better labels on the axes, and maybe smaller dots:
> plot(pe2500$age,pe2500$wageinc,col=as.factor(pe2500$sex),xlab='age',ylab='wage',cex=0.6)Here 'xlab' meant "X label" and similarly for 'ylab'. The argument 'cex = 0.6' means "Draw the dots at 60% of default size."
Now, how did the men's dots come out black and the women's red? The men were coded 1, the women 2. So men got color 1 in the default palette, black, and the women color 2, red.
There are many, many other features. More in a future lesson.
Your Turn: Try some scatter plots on various datasets. I suggest first using the above data with wage against age again, but this time color-coding by education level. (By the way, 1-9 codes no college; 10-12 means some college; 13 is a bachelor's degree, 14 a master's, 15 a professional degree and 16 is a doctorate.)
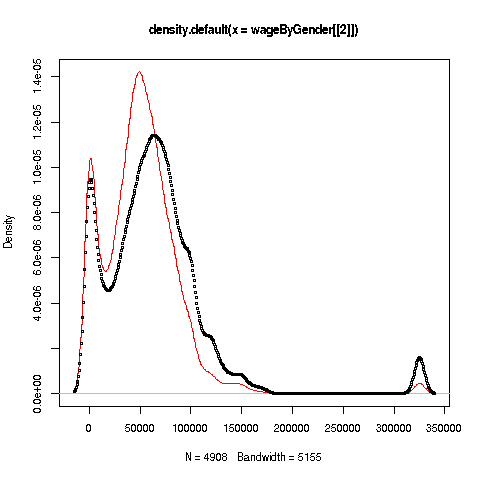
We can also plot multiple histograms on the same graph. But the pictures are more effective using a smoothed version of histograms, available in R's density function. Let's compare men's and women's wages in the census data.
First we use split to separate the data by gender:
> wageByGender <- split(pe$wageinc,pe$sex)
> dm <- density(wageByGender[[1]])
> dw <- density(wageByGender[[2]])So, wageByGender[[1]] will now be the vector of men's wages, and similarly wageByGender[[2]] will have the women's wages.
The density function does not automatically draw a plot; it has the plot information in a return value, which we've assigned to dm and dw here. We can now plot the graph:
> plot(dw,col='red')
> points(dm,cex=0.2)Why did we call the points function instead of plot in that second line? The issue is that calling plot again would destroy the first plot; we merely want to add points to the existing graph.
And why did we plot the women's data first? As you can see, the women's curve is taller, so if we plotted the men first, part of the women's curve would be cut off. Of course, we didn't know that ahead of time, but graphics often is a matter of trial-and-error to get to the picture we really want. (In the case of ggplot2, this is handled automatically by the software.)
Well, then, what does the graph tell us? The peak for women, occurring at a little less than $50,000, seems to be at a lower wage than that for men, at something like $60,000. At salaries around, say, $125,000, there seem to be more men than women. (Black curve higher than red curve. Remember, the curves are just smoothed histograms, so, if a curve is really high at, say 168.0, that means that 168.0 is a very frequently-occurring value.)
Your Turn: Try plotting multiple such curves on the same graph, for other data.
We've seen a number of R's built-in functions so far, but here comes the best part -- you can write your own functions.
Recall a line we had earlier:
> sum(Nile > 1200)This gave us the count of the elements in the Nile data larger than 1200.
Now, say we want the mean of those elements:
> gt1200 <- which(Nile > 1200)
> nileSubsetGT1200 <- Nile[gt1200]
> mean(nileSubsetGT1200)
[1] 1250As before, we could instead write a more compact version,
> mean(Nile[Nile > 1200])
[1] 1250But it's best to do it step by step at first. Let's see how those steps work. Writing the code with line numbers for reference, the code is
1 gt1200Indices <- which(Nile > 1200)
2 nileSubsetGT1200 <- Nile[gt1200Indices]
3 mean(nileSubsetGT1200)Let's review how this works:
-
In line 1, we find the indices in Nile for the elements larger than 1200.
-
In line 2, we extract the subset of Nile consisting of those elements.
-
In line 3, we compute the desired mean.
But we may wish to do this kind thing often, on many datasets etc. Then we have:
Tip: If we have an operation we will use a lot, we should consider writing a function for it.
Say we want to do the above again, but with 1350 instead of 1200. Or, with the tg$len vector from our ToothGrowth example, with 10.2 as our lower bound. We could keep typing the same pattern as above, but if we're going to do this a lot, it's better to write a function for it:
Here is our function:
> mgd <- function(x,d) mean(x[x > d])Here I've used a compact form for convenience. (Otherwise I'd need to use blocks to be covered in a later lesson.) I named it 'mgd' for "mean of elements greater than d," but any name is fine.
Let's try it out, then explain:
> mgd(Nile,1200)
[1] 1250
> mgd(tg$len,10.2)
[1] 21.58125This saved me typing. In the second call, I would have had to type
mean(tg$len[tg$len > 10.2])considerably longer. But even more importantly, I'd have to think about the operation each time I used it; by making a function out of it, I've got it ready to go, all debugged, whenever I need it.
So, how does all this work? Again, look at the code:
> mgd <- function(x,d) mean(x[x > d])
> class(mgd)
[1] "function"There is a lot going on here. Bear with me for a moment, as I bring in a little of the "theory" of R:
Odd to say, but there is a built-in function in R itself named 'function'! We've already seen several built-in R functions, e.g. mean(), sum() and plot(). Well, here is another, function(). We're calling it here. And its job is to build a function. Yes, as I like to say, to my students' amusement,
"The function of the function named function is to build functions! And the class of object returned by function is 'function'!"
So, in the line
> mgd <- function(x,d) mean(x[x > d])we are telling R, "R, I want to write my own function. I'd like to name it 'mgd'; it will have arguments 'x' and 'd', and it will do 'mean(x[x > d])'. Please build the function for me. Thanks in advance, R!"
Here we called function to build a 'function' object, and then assigned to mgd. We can then call the latter, as we saw above, repeated here for convenience:
> mgd(Nile,1200)
[1] 1250In executing
> mgd <- function(x,d) mean(x[x > d])x and d are known as formal arguments, as they are just placeholders. For example, in
> mgd(Nile,1200)we said, "R, please execute mgd with Nile playing the role of x, and 1200 playing the role of d. Here Nile and 1200 are known as the actual arguments.
As with variables, we can pretty much name functions and their arguments as we please.
As you have seen with R's built-in functions, a function will typically have a return value. In our case here, we could arrange that by writing
> mgd <- function(x,d) return(mean(x[x > d]))a bit more complicated than the above version. But the call to return is not needed here, because in any function, R will return the last value computed, in this case the requested mean.
And we can save the function for later use. One way to do this is to call R's save function, which can be used to save any R object:
> save(mgd,file='mean_greater_than_d')The function has now been saved in the indicated file, which will be in whatever folder R is running in right now. We can leave R, and say, come back tomorrow. If we then start R from that same folder, we then run
> load('mean_greater_than_d')and then mgd will be restored, ready for us to use again. (Typically this is not the way people save code, but this is the subject of a later lesson.)
Let's write another function, this one to find the range of a vector, i.e. the difference between the minimal and maximal values:
> rng <- function(y) max(y) - min(y)
> rng(Nile)
[1] 914Here we made use of the built-in R functions max and min.
Tip: Build new functions from old ones (which may in turn depend on other old ones, etc.).
Again, the last item computed is the subtraction, so it will be automatically returned, just what we want. As before, I chose to name the argument y, but it could be anything. However, I did not name the function 'range', as there is already a built-in R function of that name.
Your Turn: Try your hand at writing some simple functions along the lines seen here. You might start by writing a function cgd(), like mgd() above, but returning the count of the number of elements in x that are greater than d. Then may try writing a function n0(x), that returns the number of 0s in the vector x. (Hint: Make use of R's == and sum.) Another suggestion would be a function hld(x,d), which draws a histogram for those elements in the vector x that are less than d. Write at least 4 or 5 functions; the more you write, the easier it will be in later lessons.
Functions are R objects, just as are vectors, lists and so on. Thus, we can print them by just typing their names!
> mgd <- function(x,d) mean(x[x > d])
> mgd
function(x,d) mean(x[x > d])Recall that earlier we found that there were several columns in the Pima dataset that contained values of 0, which were physiologically impossible. These should be coded NA. We saw how to do that recoding for the glucose variable:
> pima$glucose[pima$glucose == 0] <- NABut there are several columns like this, and we'd like to avoid doing this all repeatedly by hand. (What if there were several hundred such columns?) Instead, we'd like to do this programmatically. This can be done with R's for loop construct (which by the way most programming languages have as well).
Let's first check which columns seem appropriate for recoding. Recall that there are 9 columns in this data frame.
> for (i in 1:9) print(sum(pima[,i] == 0))
[1] 111
[1] 5
[1] 35
[1] 227
[1] 374
[1] 11
[1] 0
[1] 0
[1] 500This is known in the programming world as a 'for' loop.
The 'print(etc.)' is called the body of the loop. The 'for (i in 1:9)' part says, "Execute the body of the loop with i = 1, then execute it with i = 2, then i = 3, etc. up through i = 9."
In other words, the above code instructs R to do the following:
i <- 1
print(sum(pima[,i] == 0))
i <- 2
print(sum(pima[,i] == 0))
i <- 3
print(sum(pima[,i] == 0))
i <- 4
print(sum(pima[,i] == 0))
i <- 5
print(sum(pima[,i] == 0))
i <- 6
print(sum(pima[,i] == 0))
i <- 7
print(sum(pima[,i] == 0))
i <- 8
print(sum(pima[,i] == 0))
i <- 9
print(sum(pima[,i] == 0))And this amounts to doing
print(sum(pima[,1] == 0))
print(sum(pima[,2] == 0))
print(sum(pima[,3] == 0))
print(sum(pima[,4] == 0))
print(sum(pima[,5] == 0))
print(sum(pima[,6] == 0))
print(sum(pima[,7] == 0))
print(sum(pima[,8] == 0))
print(sum(pima[,9] == 0))Now, it's worth reviewing what those statements do, say the first. Once again, pima[,1] == 0 yields a vector of TRUEs and FALSEs, each indicating whether the corresponding element of column 1 is 0. When we call sum, TRUEs and FALSEs are treated as 1s and 0s, so we get the total number of TRUEs -- which is a count of the number of elements in that column that are 0, exactly what we wanted.
The variable i in "for (i in 1:9)..." is known as the index of the loop. It's just an ordinary R variable, so name it what you wish. Instead of i, we might name it, say, colNumber.
for (colNumber in 1:9) print(sum(pima[,colNumber] == 0))A technical point: Why did we need the explicit call to print? Didn't we say earlier that just typing an expression at the R '>' prompt will automatically print out the value of the expression? Ah yes -- but we are not at the R prompt here! Yes, in the expanded form we see above,
print(sum(pima[,1] == 0))
print(sum(pima[,2] == 0))
print(sum(pima[,3] == 0))
print(sum(pima[,4] == 0))
print(sum(pima[,5] == 0))
print(sum(pima[,6] == 0))
print(sum(pima[,7] == 0))
print(sum(pima[,8] == 0))
print(sum(pima[,9] == 0))each command would be issued at the prompt. But in the for loop version
for (i in 1:9) print(sum(pima[,i] == 0))we are calling print() from within the loop, not at the prompt. So, the explicit call to print() is needed.
We now see there are a lot of erroneous 0s in this dataset, e.g. 35 of them in column 3. We probably have forgotten which column is which, so let's see, using yet another built-R function:
> colnames(pima)
[1] "pregnant" "glucose" "diastolic" "triceps" "insulin" "bmi"
[7] "diabetes" "age" "test" Ah, so column 3 was 'diastolic'.
Since some women will indeed have had 0 pregnancies, that column should not be recoded. And the last column states whether the test for diabetes came out positive, 1 for yes, 0 for no, so those 0s are legitimate too.
But 0s in columns 2 through 6 ought to be recoded as NAs. And the fact that it's a repetitive action suggests that a for loop can be used there too:
> for (i in 2:6) pima[pima[,i] == 0,i] <- NAYou'll probably find this line quite challenging, but be patient and, as with everything in R, you'll find you can master it.
First, let's write it in more easily digestible (though a bit more involved) form:
> for (i in 2:6) {
+ zeroIndices <- which(pima[,i] == 0)
+ pima[zeroIndices,i] <- NA
+ }You can enter the code for a loop or function etc. line by line at the prompt, as we've done here. R helpfully uses its '+' prompt (which I did not type) to remind me that I am still in the midst of typing the code. (After the '}' I simply hit Enter.)
Here I intended the body of the loop to consist of a block of two statements, not one, so I needed to tell R that, by typing '{' before writing my two statements, then letting R know I was finished with the block, by typing '}'.
For your convenience, below is the code itself, no '+' symbols. You can copy-and-paste into R, with the result as above.
for (i in 2:6) {
zeroIndices <- which(pima[,i] == 0)
pima[zeroIndices,i] <- NA
}
(If you are using RStudio, set up some work space, by selecting File | New File | RScript. Copy-and-paste the above into the empty pane (named SOURCE) that is created, and run it, via Code | Run Region | Run All. If you are using an external text editor, type the code into the editor, save to a file, say x.R, then at the R '>' prompt, type source(x.R).)
So, the block (two lines here) will be executed with i = 2, then 3, 4, 5 and 6. The line
zeroIndices <- which(pima[,i] == 0)determines where the 0s are in column i, and then the line
pima[zeroIndices,i] <- NAreplaces those 0s by NAs.
Tip: Note that I have indented the two lines in the block. This is not required but is considered good for clear code, in order to easily spot the block when you or others read the code.
Sometimes our code needs to leave a loop early, which we can do using the R break construct. Say we are adding cubes of numbers 1,2,3,..., and for some reason want to determine which sum is the first to exceed s:
> f
function(n,s)
{
tot <- 0
for (i in 1:n) {
tot <- tot + i^3
if (tot > s) {
print(i)
break
}
if (i == n) print('failed')
}
}
> f(100,345)
[1] 6
> f(5,345)
[1] "failed"
If our accumulated total meets our goal, we leave the loop.
A better approach is to use 'while' loops, covered later in this tutorial.
Tip: There is a school of thought among some R enthusiasts that one should avoid writing loops, using something called functional programming. We will cover this in Lesson 28, but I do not recommend it for R beginners. As the name implies, functional programming uses functions, and it takes a while for most R beginners to master writing functions. It makes no sense to force beginners to use functional programming before they really can write function code well. I myself, with my several decades as a coder, write some code with loops and some with functional programming. Write in whatever style you feel comfortable with, rather than being a "slave to fashion."
Blocks are usually key in defining functions. Let's generalize the above code in the Loops lesson, writing a function that replaces 0s by NAs in specified columns in general data frames, not just pima as before.
1 zerosToNAs <- function(d,cols)
2 {
3 for (j in cols) {
4 NArows <- which(d[,j] == 0)
5 d[NArows,j] <- NA
6 }
7 d
8 }(We've added line numbers to this display for convenence.)
Here the formal argument d is the data frame to be worked on, and cols specifies the columns in which 0s are to be replaced.
The loop goes through d, one column at a time. Since d[,j] == 0 means all of column j of d, then which(d[,j] == 0) will give us the indices in that column of elements that are 0s. Those indices in turn mean row numbers in d, which we've named NArows. In line 5, then, we replace the 0s we've found in column j by NAs. Before continuing, work through this little example in your mind:
> d <- data.frame(x=c(1,0,3),y=c(0,0,13))
> d
x y
1 1 0
2 0 0
> which(d[,2] == 0)
[1] 1 2 # ah yes; the 0 elements in column 2 are at indices 1 and 2Returning the the above loop code, note that when we reach line 7, we've already finished the loop, and exited from it. So, we are ready to return the new value of d. Recall that we could do this via the expression return(d), but we can save ourselves some typing by simply writing 'd'. That value becomes the last value computed, and R automatically returns that last value.
We could use this in the Pima data:
> pima <- zerosToNAs(pima,2:6)There is an important subtlety here. All of this will produce a new data frame, rather than changing pima itself. That does look odd; isn't d changing, and isn't d the same as pima? Well, no; d is only a separate copy of pima. So, when d changes, pima does not. So, if we want pima to change, we must reassign the output of the function back to pima, as we did above.
Your Turn: Write a function with call form countNAs(dfr), which prints the numbers of NAs in each column of the data frame dfr. You'll need to use the built-in is.na() functon; execute is.na(c(5,NA,13,28,NA)) at the R command prompt to see what it does. Test it on a small artificial dataset that you create.
In trying out our function zeroToNAs above, you probably used your computer's mouse to copy-and-paste from this tutorial into your machine. Your screen would then look like this:
> zerosToNAs <- function(d,cols)
+ {
+ zeroIndices <- which(d[,cols] == 0)
+ d[zeroIndices,cols] <- NA
+ d
+ }
But this is unwieldy. Typing it in line by line is laborious and error-prone. And what if we were to change the code? Must we type in the whole thing again? We really need a text editor for this. Just as we edit, say, reports, we do the same for code.
Here are your choices:
-
If you are already using an IDE, say RStudio, you simply edit in the designated pane.
-
If you are using an external editor, say vim or emacs, just open a new file and use that workspace.
-
For those not using these, we'll just use R's built-in edit function.
Option 3 is fine for now, but eventually you'll want to use either Option 1 or 2. You may wish to start with one of those options now, before going further.
We have details on getting start with RStudio in the Appendix at the end of this document. Warning: As noted earlier, one major R Users Group described RStudio as "overwhelming." But it is quite easy if you resist the temptation (or the exhortations of others) to learn it all at once. As long as you stick to the basics in the Appendix, you'll find it quite easy; you can learn the advanced tricks later.
Consider the following toy example:
f <- function(x,y)
{
s <- x + y
d <- x - y
c(s,d)
}It finds the sum and difference of the inputs, and returns them as a two-element vector.
If you are using RStudio or an external editor, copy-and-paste the above code into the workspace of an empty file.
Or, to create f using edit, we would do the following:
> f <- edit()This would invoke the text editor, which will depend on your machine. It will open your text editor right there in your R window. Type the function code, then save it, using the editor's Save command.
IMPORTANT: Even if you are not using edit, it's important to know what is happening in that command above.
a. edit itself is a function. Its return value is the code you typed in!
b. That code is then assigned to f, which you can now call
If you want to change the function, in the RStudio/external editor case, just edit it there. In the edit case, type
> f <- edit(f)This again opens the text editor, but this time with the current f code showing. You edit the code as desired, then as before, the result is reassigned to f.
How do you then run the code, say for computing f(5,2)?
- If you had created f() using edit(), then execute as usual:
> f(5,2)- If you had used an external text editor, say saving the code into the file a.R, then
> source('a.R')loads file, and then you run as above.
- In RStudio, click on Source, then run as above.
If our Census data example above, it was stated that education codes 0-9 all corresponded to having no college education at all. For instance, 9 means high school graduate, while 6 means schooling through the 10th grade. (Of course, few if any programmers and engineers have educational attainment level below college, but this dataset was extracted from the general data.) 13 means a bachelor's degree.
Suppose we wish to color-code the wage-age graph in an earlier lesson by educational attainment. Let's amalgamate all codes under 13, giving them the code 12.
The straightforward but overly complicated, potentially slower way would be this:
> head(pe$educ,15)
[1] 13 9 9 11 11 11 12 11 14 9 12 13 12 13 6
> for (i in 1:nrow(pe)) {
+ if (pe$educ[i] < 13) pe$educ[i] <- 12
+ }
> head(pe$educ,15)
[1] 13 12 12 12 12 12 12 12 14 12 12 13 12 13 12For pedagogical clarity, I've inserted "before and after" code, using head, to show the educ did indeed change where it should.
The if statement works pretty much like the word "if" in English. First i will be set to 1 in the loop, so R will test whether pe$educ[1] is less than 13. If so, it will reset that element to 12; otherwise, do nothing. Then it will do the same for i equal to 2, and so on. You can see above that, for instance, pe$educ[2] did indeed change from 9 to 12.
But there is a slicker (and actually more standard) way to do this (re-read the data file before running this, so as to be sure the code worked):
> edu <- pe$educ
> pe$educ <- ifelse(edu < 13,12,edu)Tip: Once again, we've broken what could have been one line into two, for clarity.
Now how did that work? As you see above, R's ifelse function has three arguments, and its return value is a new vector, that in this case we've reassigned to pe$educ. Here, edu < 12 produces a vector of TRUEs and FALSEs. For each TRUE, we set the corresponding element of the output to 12; for each FALSE, we set the corresponding element of the output to the corresponding element of edu. That's exactly what we want to happen.
So, we can now produce the desired graph:
> plot(pe$age,pe$wageinc,col=edu)By the way, an ordinary if can be paired with else too. For example, say we need to set y to either -1 or 1, depending on whether x is less than 3. We could write
if (x < 3) y <- -1 else y <- 1One more important point: Using ifelse instead of a loop in the above example is termed vectorization. The name comes from the fact that ifelse operates on vectors, while in the loop we operate on one individual element at a time.
Vectorized code is typically much more compact than loop-based code, as was the case here. In some cases, though certainly not all, the vectorized version will be much faster.
By the way, note the remark above, "ifelse operates on vectors." Let's revisit the above statement with this point in mind.
> pe$educ <- ifelse(edu < 13,12,edu)It would be helpful to keep in mind that both the 13 and the 12 will be recycled, as expained before. The edu vector is 20090 elements long, so in order to be compared on an element-to-element basis, the 13 has to be recycled to a vector consisting of 20090 elements that are each 13. The same holds for the 12.
Here's another example. Say we wish to recode the Nile data to a new vector nile, with values 1, 2 and 3, for the cases in which the value is less than 800, between 800 and 1150 inclusive, or greater than 1150. We could do this:
> nile <- ifelse(Nile > 1150,3,2)
> nile <- ifelse(Nile < 800,1,nile)
# check it
> table(nile)
nile
1 2 3
26 62 12 After the first call to ifelse, the vector nile (not Nile; variable names etc. are case-sensitive) consists of 2s and 3s. The 3s are right, but the 2s need further work, hence the second call.
But let's look closely at the second call, to review some things we've seen before:
-
The expression Nile > 1150 evaluates to a vector of 100 TRUEs and FALSEs.
-
The singleton value 800 is then recycled to one hundred 800s, to set up the '<'. Let's call the result of that '<' operation w.
-
Then ifelse(Nile < 800,1,nile) says, "For each element in the vector w that is TRUE, write down a 1; for each element that is FALSE, write down whatever the corresponding value is in nile."
Well, congratulations! With for and now ifelse, you've really gotten into the programming business. We'll be using them a lot in the coming lessons.
Your Turn: Write a for loop version of the Nile example above.
Many people gain weight as they age. But what about professional athletes? They are supposed to keep fit, after all. Let's explore this using data on professional baseball players. (Dataset courtesy of the UCLA Statistics Dept.)
> mlb <- read.table('https://raw.githubusercontent.com/matloff/fasteR/master/data/mlb.txt',header=TRUE)
> head(mlb)
Name Team Position Height Weight Age PosCategory
1 Adam_Donachie BAL Catcher 74 180 22.99 Catcher
2 Paul_Bako BAL Catcher 74 215 34.69 Catcher
3 Ramon_Hernandez BAL Catcher 72 210 30.78 Catcher
4 Kevin_Millar BAL First_Baseman 72 210 35.43 Infielder
5 Chris_Gomez BAL First_Baseman 73 188 35.71 Infielder
6 Brian_Roberts BAL Second_Baseman 69 176 29.39 Infielder
> class(mlb$Height)
[1] "integer"
> class(mlb$Name)
[1] "factor"Tip: As usual, after reading in the data, we took a look around, glancing at the first few records, and looking at a couple of data types.
Now, as a first try in assessing the question of weight gain over time, let's look at the mean weight for each age group. In order to have groups, we'll round the ages to the nearest integer first, using the R function, round, so that e.g. 21.8 becomes 22 and 35.1 becomes 35.
Let's explore the data using R's table function.
> age <- round(mlb$Age)
> table(age)
age
21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40
2 20 58 80 103 104 106 84 80 74 70 44 44 32 32 22 20 12 6 7
41 42 43 44 49
9 2 2 1 1 Not surprisingly, there are few players of extreme age -- e.g. only two of age 21 and one of age 49. So we don't have a good sampling at those age levels, and may wish to exclude them (which we will do shortly).
Now, how do we find group means? It's a perfect job for the tapply function, in the same way we used it before:
> taout <- tapply(mlb$Weight,age,mean)
> taout
21 22 23 24 25 26 27 28
215.0000 192.8500 196.2241 194.4500 200.2427 200.4327 199.2925 203.9643
29 30 31 32 33 34 35 36
199.4875 204.1757 202.8429 206.7500 203.5909 204.8750 209.6250 205.6364
37 38 39 40 41 42 43 44
203.2000 200.6667 208.3333 207.8571 205.2222 230.5000 229.5000 175.0000
49
188.0000 To review: The call to tapply instructed R to split the mlb$Weight vector according to the corresponding elements in the age vector, and then find the mean in each resulting group. This gives us exactly what we want, the mean weight in each age group.
So, do we see a time trend above? Again, we should dismiss the extreme low and high ages, and we cannot expect a fully consistent upward trend over time, because each mean value is subject to sampling variation. (We view the data as a sample from the population of all professional baseball players, past, present and future.) That said, it does seem there is a slight upward trend; older players tend to be heavier!
By the way, note that taout is vector, but with additional information, in that the elements have names, in this case the ages. In fact, we can extract the names into its own vector if needed:
> names(taout)
[1] "21" "22" "23" "24" "25" "26" "27" "28" "29" "30" "31" "32" "33"
"34" "35"
[16] "36" "37" "38" "39" "40" "41" "42" "43" "44" "49"Let's plot the means against age. We'll just plot the means that are based on larger amounts of data. So we'll restrict it to, say, ages 23 through 35, all of whose means were based on at least 30 players. That age range corresponded to elements 3 through 15 of taout, so here is the code for plotting:
> plot(23:35,taout[3:15])There does indeed seem to be an upward trend in time. Ballplayers should be more careful!
(Though it is far beyond the scope of this tutorial, which is on R rather than statistics, it should be pointed out that interpretation of the regression coefficients must be done with care. It may be, for instance, that heavier players tend to have longer careers. If so, fitting our linear form to data that has many older, heavier players may misleadingly imply that most individual players gain weight as they age. And of course, they would insist the gained weight is all muscle. :-) )
Note again that the plot function noticed that we supplied it with two arguments instead of one, and thus drew a two-dimensional scatter plot. For instance, in taout we see that for age group 25, the mean weight was 200.2427, so there is a dot in the graph for the point (25,200.2427).
Your Turn: There are lots of little experiments you can do on this dataset. For instance, use tapply to find the mean weight for each position; is the stereotype of the "beefy" catcher accurate, i.e. is the mean weight for that position higher than for the others? Another suggestion: Plot the number of players at each age group, to visualize the ages at which the bulk of the players fall.
Looking at the picture in the last lesson, it seems we could draw a straight line through that cloud of points that fits the points pretty well. Here is where linear regression analysis comes in.
We of course cannot go into the details of statistical methodology here, but it will be helpful to at least get a good definition set:
As mentioned, we treat the data as a sample from the (conceptual) population of all players, past, present and future. Accordingly, there is a population mean weight for each age group. It is assumed that those population means, when plotted against age, lie on some straight line.
In other words, our model is
mean weight = β0 + β1 height
where β0 and β1 are the intercept and slope of the population regression line.
So, we need to use the data to estimate the slope and intercept of that straight line, which R's lm ("linear model") function does for us. We'll use the original dataset, since the one with rounded ages was just to guide our intuition.
> lm(Weight ~ Age,data=mlb)
Call:
lm(formula = Weight ~ Age, data = mlb)
Coefficients:
(Intercept) Age
181.4366 0.6936 Here the call instructed R to estimate the regression line of weight against age, based on the mlb data.
So the estimated slope and intercept are 0.6936 and 181.4366, respectively. (Remember, these are just sample estimates. We don't know the population values.) R has a provision by which we can draw the line, superimposed on our scatter plot:
> abline(181.4366,0.6936)Your Turn: In the mtcars data, fit a linear model of the regression of MPG against weight; what is the estimated effect of 100 pounds of extra weight?
Tip: Remember, the point of computers is to alleviate us of work. We should avoid doing what the computer could do. For instance, concerning the graph in the last lesson: We had typed
> abline(181.4366,0.6936)but we really shouldn't have to type those numbers in by hand -- and we don't have to. Here's why:
As mentioned earlier, R is an object-oriented language. Everthing is an object, and every object has a class. One of the most common class structures is called 'S3'.
When we call lm, the latter returns an S3 object of 'lm' class:
> lmout <- lm(Weight ~ Age,data=mlb)
> class(lmout)
[1] "lm"A handy way to take a quick glance at the contents of an object is str:
> str(lmout)
List of 12
$ coefficients : Named num [1:2] 181.437 0.694
..- attr(*, "names")= chr [1:2] "(Intercept)" "Age"
...
...
- attr(*, "class")= chr "lm"Our use of ... here is to indicate that we've omitted a lot of the output. But a couple of things stand out even in this excerpt:
-
Our lmout object here is an R list (which is typical of S3 objects). That R list here has 12 elements.
-
But it has an extra attribute, which is the class name, in this case 'lm'. (So the designers of R simply chose to name the class after the function, which is not always the case.)
-
The first of the elements of this R list is named 'coefficients', and it is a vector containing the slope and intercept.
So, we don't have to type the slope and intercept in by hand after all.
> cfs <- lmout$coefficients
> abline(a = cfs[1], b = cfs[2])By the way, abline() is actually a generic function, like print() and plot(). So if we want to be clever, we can add our line to the graph using this approach:
> abline(lmout)Now, what about our original question -- do baseball players gain weight as they age? The answer appears to be yes; for each additional year of age, the estimated mean age increases by about 0.7 pound. That's about 7 pounds in 10 years, rather remarkable.
Again, this is only an estimate -- 181.437 and 0.694 are estimates of the unknown population values β0 and β1. -- generated from sample data. We can get an idea of the accuracy of this estimate by calculating a confidence interval, but we'll leave that for statistics courses.
But we can do more right now. One might ask, Shouldn't we also account for a player's height, not just his age? After all, taller people tend to be heavier. Yes, we should do this:
> lmo <- lm(Weight ~ Height + Age, data=mlb)
> lmo
Call:
lm(formula = Weight ~ Height + Age, data = mlb)
Coefficients:
(Intercept) Height Age
-187.6382 4.9236 0.9115 Here we instruct R to find the estimated regression function of weight, using both height and age as predictors. The '+' doesn't mean addition; it is simply a delimiter between the predictors height and age in our regression specification.
So the new model is
mean weight = β0 + β1 height + β2 age
This says:
estimated mean weight = -187.6382 + 4.9236 height + 0.9115 age
So, under this more refined analysis, things are even more pessimistic; players on average gain about 0.9 pounds per year. And by the way, an extra inch of height corresponds on average to about 4.9 pounds of extra weight; taller players are indeed heavier, as we surmized.
Warning: Though this is not a statistics tutorial per se, an important point should be noted. Regression analysis has two goals, Description and Prediction. Our above analysis was aimed at the former -- we want to describe the nature of fitness issues in pro baseball players. As we saw, a coefficient can change quite a lot when another predictor is added to the model, and in fact can even change sign ("Simpson's Paradox"). Suppose for instance the shorter players tend to have longer careers. If we do not include height in our model, that omission might bias the age coefficient downward. Thus great care must be taken in interpreting coefficients in the Description setting. For Prediction, it is not as much of an issue.
Your Turn: In the mtcars data, fit a linear model of the regression of MPG against weight and horsepower; what is the estimated effect of 100 pounds of extra weight, for fixed horsepower?
This lesson will be a little longer and more detail-oriented. But it will give you more practice on a number of earlier topics, and will also bring in some new R functions for you. Spending extra time on this lesson will pay substantial dividends.
We might wonder whether the regression lines differ much among player positions. (A more statistical approach would be to include interaction terms in the model.) Let's first see what positions are tabulated:
> table(mlb$PosCategory)
Catcher Infielder Outfielder Pitcher
76 210 194 535 Let's fit the regression lines separately for each position type.
There are various ways to do this, involving avoidance of loops to various degrees. But we'll keep it simple, which will be clearer.
First, let's split the data by position. You might at first think this is easily done using the split function, but that doesn't work, since that function is for splitting vectors. Here we wish to split a data frame.
So what can be done instead? We need to think creatively here. One solution is this:
We need to determine the row numbers of the catchers, the row numbers of the infielders and so on. So we can take all the row numbers, 1:nrow(mlb), and apply split to that vector!
> rownums <- split(1:nrow(mlb),mlb$PosCategory)Tip: As usual, following an intricate operation like this, we should glance at the result:
> str(rownums)
List of 4
$ Catcher : int [1:76] 1 2 3 35 36 66 67 68 101 102 ...
$ Infielder : int [1:210] 4 5 6 7 8 9 37 38 39 40 ...
$ Outfielder: int [1:194] 10 11 12 13 14 15 16 43 44 45 ...
$ Pitcher : int [1:535] 17 18 19 20 21 22 23 24 25 26 ...So the output is an R list; no surprise there, as we knew before before that split produces an R list. Also not surprisingly, the elements of the list are named "Catcher" etc. So for example, the third outfielder is in row 12 of the data frame.
Tip: The idea here, using split on 1:nrow(mlb), was a bit of a trick. Actually, it is a common ploy for experienced R coders, but you might ask, "How could a novice come up with this idea?" The answer, as noted several times already here, is that programming is a creative process. Creativity may not come quickly! In some case, one might need to mull over a problem for a long time before coming up with a solution. Don't give up! The more you think about a problem, the more skilled you will get, even if you sometimes come up empty-handed. And of course, there are many forums on the Web at which you can ask questions, e.g. Stack Overflow.
Tip: Now, remember, a nice thing about R lists is that we can reference their elements in various ways. The first element above, for instance, is any of rownums$Catcher, rownums[['Catcher']] and rownums[[1]], This versatility is great, as for example we can use the latter two forms to write loops.
And a loop is exactly what we need here. We want to call lm four times, once for each position. We could do this, say, with a loop beginning with
for (i in 1:4)to iterate through the four position types, but it will be clearer if we use the names:
for (pos in c('Catcher','Infielder','Outfielder','Pitcher'))Just an ordinary 'for' loop. Recall such loops are of the form
for (variable in vector)...Instead of having a numeric vector, e.g. the 1:4 above, we now have a character vector, which each element of the vector being a character string, but the principles are the same.
We could have lm and print calls in the body of the loop. But let's be a little fancier, building up a data frame with the output. We'll start with an empty frame, and keep adding rows to it.
Our code is
posNames <- c('Catcher','Infielder','Outfielder','Pitcher')
m <- data.frame()
for (pos in posNames) {
posRows <- rownums[[pos]]
lmo <- lm(Weight ~ Age, data = mlb[posRows,])
newrow <- lmo$coefficients
m <- rbind(m,newrow)
}Here is the output:
> m
X180.828029016113 X0.794925225995348
1 180.8280 0.7949252
2 170.2466 0.8589593
3 176.2884 0.7883343
4 185.5994 0.6543904Some key things to note here.
-
The overall strategy is to start with an empty data frame, then keep adding rows to it, one row of regression coefficients per playing position.
-
In order to add rows to m, we used R's rbind ("row bind") function. The expression rbind(m,newrow) forms a new data frame, by tacking newrow onto m. Here we reassign the result back to m, also a common operation. (Note carefully: The rbind operation did not change m; it merely created a new data frame. To update m, we needed to assign that new data frame to m.) By the way, there is also a cbind function for columns.
-
In the call to lm, we used mlb[rownums[[pos]],] instead of mlb as previously, since here we wanted to fit a regression line on each position subgroup. So, we restricted attention to only those rows of mlb for which the position was equal to the current value of pos.
So, what happens is: m is initially an empty data frame. Then the loop, for its first iteration, sets pos to 'Catcher'. Then a regression line will be fit to the rows of mlb that are for catchers. That line is returned to us from lm, and we assign it to lmo. (Once again, the name is arbitrary; I chose this one to symbolize "lm output.") We extract the coefficients and tack them on at the end of m.
Tip: This is a very common design pattern in R (and most other languages)
Nice output, with the two columns aligned. But those column names are awful, and the row labels should be nicer than 1,2,3,4. We can fix these things:
> row.names(m) <- posNames
> names(m) <- c('intercept','slope')
> m
intercept slope
Catcher 180.8280 0.7949252
Infielder 170.2466 0.8589593
Outfielder 176.2884 0.7883343
Pitcher 185.5994 0.6543904What happened here? We earlier saw the built-in row.names function, so that setting row names was easy. But what about the column names? Recall that a data frame is actually an R list, consisting of several vectors of the same length, which form the columns. So, names(m) is the names of the columns.
So with a little finessing here, we got some nicely-formatted output. Moreover, we now have our results in a data frame for further use. For instance, we may wish to plot the four lines on the same graph, and we would use rows of the data frame as input.
A little more finessing is possible. Look at the line
posNames <- c('Catcher','Infielder','Outfielder','Pitcher')We're using a computer! We shouldn't have to type out these names by hand, as I did in this line. In fact, we already have them in one of our R objects, rownums; recall our earlier check:
> str(rownums)
List of 4
$ Catcher : int [1:76] 1 2 3 35 36 66 67 68 101 102 ...
$ Infielder : int [1:210] 4 5 6 7 8 9 37 38 39 40 ...
$ Outfielder: int [1:194] 10 11 12 13 14 15 16 43 44 45 ...
$ Pitcher : int [1:535] 17 18 19 20 21 22 23 24 25 26 ...The elements of the R list rownums are the names of the positions! So, the better way to set posNames is
posNames <- names(rownums)Tip: Again, the reader may be thinking, "How in the world would I have been able to realize this?" Again, the answer is that as you acquire more experience in coding, you will be more and more ability to come up with insights like this. Patience!
Finally, what about those numerical results? There is substantial variation in those estimated slopes, but again, they are only estimates. The question of whether there is substantial variation at the population level is one of statistical inference, beyond the scope of this R course.
One of the great things about R is that are tens of thousands of packages that were developed by users and then contributed to the CRAN repository. As of December 2020, there were nearly 17,000 packages there. If you need to do some special operation in R, say spatial data analysis, it may well be in there. You might take the CRAN Task Views as your starting point, or simply use Google, e.g. plugging in the search term "CRAN spatial data." Other good sources of public R packages are Bioconductor and useRs' personal GitHub pages.
Below, we'll introduce one of the most popular user-contributed packages, ggplot2. But first, how does one install and load packages?
First, one needs a place to put the packages. UseRs often designate a special folder/directory for their packages (both those they download and ones they write themselves). I use 'R' in my home directory for that purpose, but if you don't specify a folder, your package installer will choose one for you. It won't matter as long as you are consistent. I'll assume you don't specify a package folder.
To install, say, ggplot2, you can type at the R prompt,
> install.packages('ggplot2')Or in RStudio, choose Tools | Install Packages...
When you want to use one of your installed packages, you need to tell R to load it, e.g. by typing at the R prompt,
> library(ggplot2)In RStudio, click the Packages button and select the one you want; there may be a delay while R makes a list of all your packages.
Later, you'll write your own R packages. We won't cover that here, but there are many good tutorials for this on the Web.
At this point, you have a pretty good grounding in R. You are capable of doing lots of things in R. It may be all you need, but even if not, you know enough to ask a question online if you get stuck on something.
The remaining topics are more advanced, and lessons will be somewhat longer and more detailed that the previous ones. But you are still strongly encouraged to go through them, as they will not only cover new topics but also give you deeper insight into the earlier material.
Now, on to ggplot2.
The ggplot2 package was written by Hadley Wickham, who later became Chief Scientist at RStudio. It's highly complex, with well over 400 functions, and rather abstract, but quite powerful. We will touch on it at various points in this tutorial, while staying with base-R graphics when it is easier to go that route.
Now to build up to using ggplot2, let's do a bit more with base-R graphics first, continuing with our weight/age investigation of the ballplayers. To begin, let's do a scatter plot of weight against age, color-coded by position. We could type
> plot(mlb$Age,mlb$Weight,col=mlb$PosCategory)but to save some typing, let's use R's with function (we'll change the point size while we are at it):
> with(mlb,plot(Age,Weight,col=PosCategory,cex=0.6))By writing with, we tell R to take Age, Weight and PosCategory in the context of mlb.
Here is how we can do it in ggplot2:
First, I make an empty plot, based on the data frame mlb:
> p <- ggplot(mlb)Nothing will appear on the screen. The package displays only when you "print" the plot:
> pThis will just display an empty plot. (Try it.) By the way, recall that any expression you type, even 1 + 1, will be evaluated and printed to the screen. Here the plot (albeit) empty is printed to the screen.
Now let's do something useful:
> p + geom_point(aes(x = Age, y = Weight, col = PosCategory),cex=0.6)What happened here? Quite a bit, actually, so let's take this slowly.
-
We took our existing (blank) plot, p, and by writing the'+' sign, directed ggplot2 to add to the plot p.
-
Now, WHAT do we want added? We are saying, "ggplot2, please add to the plot p whatever geom_point() returns."
-
Note that geom_point() is a ggplot2 function. Its task is to produce scatter plots.
-
Here are the details on the arguments to geom_point:
-
We want to plot weight against height. We do not need to specify what data frame these two variables are from, as we already stated that the plot p is for the data frame mlb.
-
We are also specifying that the color coding will be according to the player position, again from mlb.
-
-
When R evaluates that entire expression, p + geom_point(aes(x = Age, y = Weight, col = PosCategory),cex=0.6), the result will be another ggplot2 graph object. Since we typed that expression at the '>' prompt, it was then printed to the screen as seen above.
-
There is one mystery left, though: What does the function aes ('aesthetic") do? And why is the expression cex=0.6 NOT an argument to aes? Unfortunately, there are no easy answers to these questions, and in a rare exception to our rule of explaining all, we will just have to leave this as something that must be done.
One nice thing is that we automatically got a legend printed to the right of the graph, so we know which color corresponds to which position. We can do this in base-R graphics too, but need to set an argument for it in plot.
Earlier in this tutorial, we've found R's tapply function to be quite handy. There are several others in this family, notably apply, lapply and sapply. In addition, there are other related functions, such as do.call and Reduce(). And there are a number of counterparts in the Tidyverse purrr package. All of these go under the aegis of functional programming (FP).
To many, FP is intended as a higher-level replacement for loops, and some members of the R community view that as desirable, even a must. I personally take a more moderate point of view, but before discussing the controversy, let's see how FP works as a loop-replacement.
As a simple example, say we have a nonnegative integer vector x, and want code that counts doubles each element that is greater than 9. Of course, this is something we should not use a loop with in the first place. We should take advantage of R's vectorization capabilities:
x <- ifelse(x > 9,2*x,x)
But let's ignore vectorization, for the sake of illustrating the issues, and write up a loop version:
for (i in 1:5) if (x[i] > 9) x[i] <- 2 * x[i]Now, how would we replace this loop by a call to R's sapply function? The latter has the call form
sapply(X,FUN)where X is an R factor and FUN is a function. We will assume here that FUN returns a number, not a vector or other R object. The action of the function is to apply FUN on each element of X, producing a new vector. (It of course can be reassigned to the old one.)
The key is defining FUN:
doubleIt <- function(z) if(z > 9) return(2*z) else return(z)
sapply(x,doubleIt)Let's check:
> x <- c(5,12,13,8,88)
> x <- sapply(x,doubleIt)
> x
[1] 5 24 26 8 176Or, we can use what is called an anonymous function:
> x <- c(5,12,13,8,88)
> x <- sapply(x,function(z) if(z > 9) return(2*z) else return(z))
> x
[1] 5 24 26 8 176Instead of defining the function separately, we define it right there in the second argument of sapply.
Now let's consider something more elaborate. Recall our earlier baseball player example, in which we wanted to fit separate regression lines to each of the four player position categories. We used a loop, which for convenience I'll duplicate here:
rownums <- split(1:nrow(mlb),mlb$PosCategory)
posNames <- c('Catcher','Infielder','Outfielder','Pitcher')
m <- data.frame()
for (pos in posNames) {
lmo <- lm(Weight ~ Age, data = mlb[rownums[[pos]],])
newrow <- lmo$coefficients
m <- rbind(m,newrow)
}How might we do this in FP? We've seen the tapply function a couple of times already. Now let's turn to lapply ("list apply"). The call form is
lapply(VectorOrList,FUN)This first argument must be a vector or list, and the second argument must be the name of a one-argument function. This calls FUN on each element of VetorcOrList, placing the return values in a new list.
How might we use that here? Well, lapply, as the name implies, is aimed at working on lists. Do we have any? Why yes, rownums is a list!
And indeed, we do want to take some action on each element of that list: We want to fit a linear regression model to the rows in that element. It is natural, then, to take for FUN the following function:
zlm <- function(rws) lm(Weight ~ Age, data=mlb[rws,])$coefficientsHere rws is a set of row numbers, e.g. those for the pitchers. This function calls lm on those rows, i.e. on the data mlb[rws,], then extracts the regression coefficients.
The code then is
> zlm <- function(rws) lm(Weight ~ Age, data=mlb[rws,])$coefficients
> w <- lapply(rownums,zlm)The call to lapply then says, run zlm on each set of rows we see in rownums, placing the coefficient vectors in an output list. Specifically: Recall that the first element of rownums was rownums[['catcher']]. So, first lapply will make the call
zlm(rownums[['Catcher']])which will fit the desired regression model on the catcher data. Then lapply will do
zlm(rownums[['Infielder']])and so on. The outputs of the four lm calls will be returned in an R list, which we have assigned to w above. Let's check the first one:
> w[[1]]
(Intercept) Age
180.8280290 0.7949252 jibing with m[1,] in our data-frame/loop appraoch above.
Well, then, what did we accomplish -- if anything -- by using lapply here rather than our earlier approach using a loop? Certainly the lapply version did make for more compact code, just 2 lines. But we had to think a harder to come up with the idea. Also, printing it out is less compact:
> w
$Catcher
(Intercept) Age
180.8280290 0.7949252
$Infielder
(Intercept) Age
170.2465739 0.8589593
$Outfielder
(Intercept) Age
176.2884016 0.7883343
$Pitcher
(Intercept) Age
185.5993689 0.6543904 (Actually, we can use sapply here instead of lapply, with a nicer printing.)
So, should beginning R coders use FP? Actually, even I, with decades of coding experience, take a moderate approach. The criterion for loop-based (LB) code vs. FP should be to ask these questions:
-
Would FP code be easier to write than LB in this case?
-
Would FP code be easier to debug than LB in this case?
-
Would FP code be easier to read -- either by others, or by myself 6 months from now -- than LB in this case?
For the code in this tutorial in which we've used tapply, I believe the answers to the above questions are definitely Yes. But for the lapply example above, I would say the answer is No -- especially for beginning coders, but even for myself.
Beginners are in the process of learning functions. FP by definition is based on writing functions, thus making FP a more abstract and difficult process. And I certainly disagree with the doctrinnaire view of some that one should never write loops.
My recommendation is to take things on a case-by-case basis.
Now, let's turn to another central function in the apply family. Not surprisingly, it's named apply! It is usually used on matrix objects (like data frames, but with the contents being all of the same type, e.g. all numerical), on either rows or columnṡ, but can be used on data frames too.
The call form is
apply(d,rc,g)Here R will apply the function g to each row (rc = 1) or column (rc = 2) of the data d. If the function g returns a number, then apply will return a vector.
Let's find the max values for the variables in the pima data:
> apply(pima,2,max)
pregnant glucose diastolic triceps insulin bmi diabetes
age
17.00 199.00 122.00 99.00 846.00 67.10 2.42
81.00
test
1.00 Note that to use the apply family, you can either use a built-in R function, e.g. max here, or one you write yourself, such as zlm above.
The R apply family includes other functions as well, They are quite useful, but don't use them solely for the sake of avoiding writing a loop. More compact code may not be easier.
These days, text processing is big in the Data Science field, e.g. in Natural Language Processing applications. In this lesson, we'll do a simple yet practical example, in order to illustrate some key functions in base-R. (R has many packages for advanced text work, such as tm.)
Our example will cover reading in a file of text, and compiling a word count, i.e. calculating the number of times each word appears. This kind of task is at the core of many text classification algorithms.
The file is here. It's basically the About section of the R Project home page. Here are the first few lines:
What is R?
Introduction to R
R is a language and environment for statistical computing and graphics.
Now, how can we read the file? For instance, read.table won't work, as it expects the same number of nonblank fields on each line. As you can see above, our file has a variable number of such fields per line.
Instead, we read the lines of the file via a function named, not surprisingly, readLines:
> abt <- readLines('https://raw.githubusercontent.com/matloff/fasteR/master/data/aboutR.txt')So, what exactly is in abt now? Let's turn to our usual inspection tools, str and head.
> str(abt)
chr [1:70] "" "What is R?" "" "Introduction to R" "" ...So, abt is a vector of 70 elements, of type character. Each element of this vector is one line from the file:
> head(abt)
[1] ""
[2] "What is R?"
[3] ""
[4] "Introduction to R"
[5] ""
[6] " R is a language and environment for statistical computing and graphics."
The first line in the file was empty, so abt[1] is "", and so on.
Now, to count the number of words in the file, we'll need a way to count the number in each line, which we will then sum. R's strsplit function will serve us well here, e.g. for line 4:
> y <- strsplit(abt[4],' ')
> y
[[1]]
[1] "Introduction" "to" "R" (That second argument, ' ', means we want the blank character to be our splitting delimiter.
Good, it split the line into the three words on that line, "Introduction", "to", and "R".
But be careful! What is that [[1]] doing there? Remember, the double bracket notation is for R lists. So, strsplit has split abt[4] a list with one element, and that element is in turn the three-element character vector c("Introduction","to","R"). So for instance,
> y[[1]][2]
[1] "to"
> y[[1]][3]
[1] "R"Why the R list form? Well, strsplit can be applied to the entire character vector abt, yielding a list of 70 elements; the i-th such element will contain the split form of the i-th line in the file, e.g. line 6:
> w <- strsplit(abt,' ')
> w[[6]] # example
[1] "" "" "" "R" "is"
[6] "a" "language" "and" "environment" "for"
[11] "statistical" "computing" "and" "graphics." Yep, that's the split form of line 6.
The material beginning with the # sign is what is called a comment in the programming world. It does not get executed by R, but it is a memo to us, the programmer, a note to help us remember what we did.
Comments are extremely important. When we read our code six months from now, we will have forgotten most of it, so comments help us reorient. The same holds if someone else reads our code. Comments -- meaningful comments -- are key to good coding. More on this in a future lesson.
But we also see another snag. The above output tells us that R took line 6, which has 11 words, and split into 14 words -- the first 3 of which are empty words "". This is because the first three characters in line 6 are blanks. When there is more than one consecutive blank, the strsplit function treats the excess blanks as "words." (This comes as quite a surprise to Python programmers.)
So, how to fix it? Answering that question will give us a chance to learn more about R in general.
For that particular line, we could do, say, is remove those empty "words" as follows:
> z <- w[[6]]
> z <- z[z != ""]
> z
[1] "R" "is" "a" "language" "and"
[6] "environment" "for" "statistical" "computing" "and"
[11] "graphics." R's '!=' means "not equal to." By the way, '!' means "not" in R, e.g.
> 3 < 8
[1] TRUE
> !(3 < 8)
[1] FALSESo what we did to z above followed our usual pattern:
-
The expression z != "" yields a bunch of TRUEs and FALSEs.
-
The operation z[bunch of TRUEs and FALSEs] extracts those elements of z at which there are TRUEs, which are exactly the ones we want to keep here.
- Tip: When you write some code that looks like it will be generally useful, make a function out of it, and save it for future use. The above code to delete the empty "words" sounds like something worth keeping. So, let's write it in function form:
extractNonemptyWords <- function(s)
{
z <- strsplit(s,' ')[[1]]
z[z != ""]
}Recall that in R functions, the last computed value is automatically returned. The expression z[z != ""] evaluates to the set of nonempty words, and it is returned.
Tip: As mentioned, we should probably save that function for future use. We could save it using the save function as discussed earlier, but it would be better to save our "home grown" functions in one or more packages, maybe even submitting them to CRAN. More on this in a later lesson.
We'll continue with this example in the next lesson, but first, time for a Your Turn session.
Your Turn: That [[1]] expression in the body of extractNonemptyWords was crucial! Try the code without it, and see if you can explain the result, which is not what we desire. Tip: This illustrates a common error for beginners and veterans alike. The error message probably won't be helpful! So keep this frequent error in mind, both when you're writing code and viewing cryptic error messages.
We can then call our extractNonemptyWords function on each line of the file, say in a loop. We'll do this in the next section.
Your Turn: Write a function with call form delNAs(x), that returns x with NAs deleted.
So, let's use our extractNonemptyWords function on our abt vector. Here's a loop way to do it:
allWords <- NULL # start with empty vector
for (i in 1:70) {
thisLine <- extractNonemptyWords(abt[i])
allWords <- c(allWords,thisLine)
}Note that the result, i.e. the final value of allWords, will be one long vector, consisting of all the words in the file.
As usual, it is a must to inspect the result, say the first 25 elements:
> head(allWords,25)
[1] "What" "is" "R?"
[4] "Introduction" "to" "R"
[7] "R" "is" "a"
[10] "language" "and" "environment"
[13] "for" "statistical" "computing"
[16] "and" "graphics." "It"
[19] "is" "a" "GNU"
[22] "project" "which" "is"
[25] "similar" Good, all the words seem to be there, and the "" are NOT there, just as desired. But how to get the word counts? Why, it's our old friend, tapply!
> q <- tapply(allWords,allWords,length)
> head(q,25)
; …) “environment” (easily) (formerly
1 1 1 1 1
(including (linear * © a
1 1 5 1 13
about accretion activity. add additional
3 1 1 1 1
Advanced algorithmic allows an analysis
1 1 1 5 2
analysis, and are around arrays,
2 27 4 1 1 Actually, this really the same pattern we saw before, with the length function as our third argument. It may look a little odd that the first two arguments are identical, but it makes sense:
-
We split up the allWords vector into piles, according to the second argument, which happens to be the same vector.
-
We apply the length function to each pile, giving us the count in each pile, exactly what we needed.
Tip: In coding, certain patterns do arise often, one did here. In fact, there are coding books with "design patterns" in their titles. Take note when you see the same pattern a lot.
We're not fully done yet. For instance, we have a punctuation problem, where periods, commas and so on are considered parts of words, such as the period in allWords[17] seen above, 'graphics.' We also probably should change capital letters to lower
For major usage, we should consider using one of the advanced R packages in text processing. For instance, the tm package has a removePunctuation function. But let's see how we can do this with the basics.
We'll use R's gsub function. It's call form, as we'll use it, is
gsub(string_to_change,replacement,input_vector,fixed=TRUE)E.g.
> a <- c('abc','de.')
> gsub('.','',a,fixed=TRUE) # replace '.' by empty stringW
[1] "abc" "de" (The fixed argument is complex, and pops up in all the R string manipulation packages. This again is something you should use for now, and look into when you become more skilled at R.)
So, to remove all periods in allWords, we can do:
> awNoPers <- gsub('.','',allWords,fixed=TRUE)
> awNoPers[17] # check that it worked
[1] "graphics"Continuing our look at linear regression analysis using R, let's look at the famous bike sharing data. (See the latter site for the documentation; e.g. temperature has been scaled, rather than measured in degrees.) It's in a .zip file, so it will need a little extra preprocessing:
# fetch from Web, and store the downloaded data to the file 'bike.sip'
> download.file('https://archive.ics.uci.edu/ml/machine-learning-databases/00275/Bike-Sharing-Dataset.zip','bike.zip')
> unzip('bike.zip') # out come the files 'day.csv' and 'hour.csv'
> day <- read.csv('day.csv',header=TRUE)
> head(day) # always take a look around!
instant dteday season yr mnth holiday weekday workingday weathersit
1 1 2011-01-01 1 0 1 0 6 0 2
2 2 2011-01-02 1 0 1 0 0 0 2
3 3 2011-01-03 1 0 1 0 1 1 1
4 4 2011-01-04 1 0 1 0 2 1 1
5 5 2011-01-05 1 0 1 0 3 1 1
6 6 2011-01-06 1 0 1 0 4 1 1
temp atemp hum windspeed casual registered cnt
1 0.344167 0.363625 0.805833 0.1604460 331 654 985
2 0.363478 0.353739 0.696087 0.2485390 131 670 801
3 0.196364 0.189405 0.437273 0.2483090 120 1229 1349
4 0.200000 0.212122 0.590435 0.1602960 108 1454 1562
5 0.226957 0.229270 0.436957 0.1869000 82 1518 1600
6 0.204348 0.233209 0.518261 0.0895652 88 1518 1606By the way, the weather variables have been rescaled to the interval [0,1]. A value of 0.28, for instance, means 28% of the way from the minimum to the maximum value of this variable.
One new concept here is the presence of indicator variables, more informally known as dummy variables. These are variables taking only the values 0 and 1, with a 1 "indicating" that some trait is present. For instance, the holiday variable is either 1 or 0, depending on whether that day was a holiday (which might affect the demand for bikes that day).
Those who manage this bike sharing service may wish to predict future demand for bikes, say the next day, to aid in their planning. As an example, let's try to predict the number of casual riders from some weather variables and the dummy variable workingday.
> day1 <- day[,c(8,10,12:14)]
> head(day1)
workingday temp hum windspeed casual
1 0 0.344167 0.805833 0.1604460 331
2 0 0.363478 0.696087 0.2485390 131
3 1 0.196364 0.437273 0.2483090 120
4 1 0.200000 0.590435 0.1602960 108
5 1 0.226957 0.436957 0.1869000 82
6 1 0.204348 0.518261 0.0895652 88
> lmout <- lm(casual ~ .,data=day1)
> lmout
...
Coefficients:
(Intercept) workingday temp hum windspeed
1063.6 -806.6 2149.5 -812.7 -1145.3
The expression "casual ~ ." means, "regress casual against all the other variables in this dataset.
These numbers make sense. The negative coefficient for workingday says that, all else equal, there tend to be fewer casual riders on a work day.
By the way, we probably should expect fewer riders on very cold or very hot days, so we may wish to add a quadratic term to the model, say by doing
day1$temp2 <- temp^2 # the caret symbol means exponentiation,
# i.e. 2nd power hereThis would add the indicated column to day1. But we will not pursue this for now.
One of the very important features of R is generic functions. These are functions that take on different roles for objects of different classes. One such example is the plot function we saw earlier.
Try typing "plot(lmout)" at the R prompt. You will be shown several plots desribing the fitted regression model. What happened was that the function plot is just a placeholder. When we type "plot(lmout)" R says, "Hmm, what kind of object is lmout? Oh, it's of class 'lm'. So I'm going to transfer (dispatch) this call to one involving a special plot function for that class, plot.lm." This is in contrast to our previous calls to plot, which were invoked on vectors; those calls were dispatched to plot.default.
Another generic function is summary:
> summary(Nile)
Min. 1st Qu. Median Mean 3rd Qu. Max.
456.0 798.5 893.5 919.4 1032.5 1370.0
> summary(lmout)
...
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1063.55 101.37 10.492 < 2e-16 ***
workingday -806.63 33.41 -24.143 < 2e-16 ***
temp 2149.52 86.32 24.901 < 2e-16 ***
hum -812.74 112.98 -7.194 1.57e-12 ***
windspeed -1145.31 208.55 -5.492 5.51e-08 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
...In the first case, the call to summary, invoked on a vector, was dispatched to summary.default, while in the second the transfer was to summary.lm. In both cases, whoever it was in the R development team who wrote these functions decided what summary information should be printed out automatically.
Again, the purpose of this tutorial is to present R, not statistics. The interested reader should consult a statistics book regarding p-values and confidence intervals. The former are show in the last column of the above summary. An approximate 95% confidence interval for, say, the population coefficient of humidity is -812.74 plus or minus 1.96 times the standard error, 112.98. Note carefully that p-values have long been considered to be poor methodology; see the ASA statement.
Another important generic function is predict. Say we want to predict casual for a work day in which temp, hum and windspeed are 0.26, 0.55, 0.18, respectively.
> newCase <- data.frame(workingday=1, temp=0.26, hum=0.55, windspeed=0.18)
> predict(lmout,newCase)
1
162.6296 The predict function, which here is predict.lm, assumes that the new cases to be predicted are supplied as a data frame, with the same column names as with the original data.
In the bike sharing data, dates were included, in day$dteday. As noted, some of those were holidays, indicated in the holiday column. Let's see how many holidays there were:
> hds <- day$dteday[day$holiday == 1]
> hds
[1] 2011-01-17 2011-02-21 2011-04-15 2011-05-30 2011-07-04 2011-09-05
[7] 2011-10-10 2011-11-11 2011-11-24 2011-12-26 2012-01-02 2012-01-16
[13] 2012-02-20 2012-04-16 2012-05-28 2012-07-04 2012-09-03 2012-10-08
[19] 2012-11-12 2012-11-22 2012-12-25Once again, let's review how the above code works. The expression "[day$holiday == 1]" yields a bunch of TRUEs and FALSEs. Using them as indices in the vecotr day$dteday gives us exactly the dates that are holidays.
We see above that there were 21 holidays during the time period of the data. But we can do more. First, what kind of object is hds above?
> class(hds)
[1] "factor"Fine, but R has a special class for date data, not surprisingly called 'Date'. Let's convert to that class:
> hd <- as.Date(hds)
> class(hd)
[1] "Date"
> hd
[1] "2011-01-17" "2011-02-21" "2011-04-15" "2011-05-30" "2011-07-04"
[6] "2011-09-05" "2011-10-10" "2011-11-11" "2011-11-24" "2011-12-26"
[11] "2012-01-02" "2012-01-16" "2012-02-20" "2012-04-16" "2012-05-28"
[16] "2012-07-04" "2012-09-03" "2012-10-08" "2012-11-12" "2012-11-22"
[21] "2012-12-25"Though it prints out just as before, there are extra properties now, and even bettery, in POSIX form:
> hp <- as.POSIXlt(hd)
> hp
[1] "2011-01-17 UTC" "2011-02-21 UTC" "2011-04-15 UTC" "2011-05-30 UTC"
[5] "2011-07-04 UTC" "2011-09-05 UTC" "2011-10-10 UTC" "2011-11-11 UTC"
[9] "2011-11-24 UTC" "2011-12-26 UTC" "2012-01-02 UTC" "2012-01-16 UTC"
[13] "2012-02-20 UTC" "2012-04-16 UTC" "2012-05-28 UTC" "2012-07-04 UTC"
[17] "2012-09-03 UTC" "2012-10-08 UTC" "2012-11-12 UTC" "2012-11-22 UTC"
[21] "2012-12-25 UTC"
> hp[16]$wday # what day of the week was July 4, 2012?
[1] 3
# ah, Wednesday (code 3)(The UTC parts are the times of day, which we had not supplied.)
There are many operations that can be done on R dates. The above is just a little sample.
Programming is a creative activity, and thus different programmers will have different coding styles. Some people feel so strongly that they will publish there own particular style guides, such as this one by the R community at Google. Mine is here`.
Needless to say, style is a matter of personal taste. But:
Style IS important for any code you intend to use again, for two reasons:
-
You will quickly forget how your code works.
-
If you share your code with others, you need to make its workings clear to them.
Equally important is strategy, the way you approach a coding project.
There is no magic formula on how to write code. As noted earlier, I cannot teach yow how to code. I can only show you how the ingredients work -- loops, variables, functions, if/else etc. -- and you must creatively put them together into code that achieves goals. It's like solving a big puzzle, and like many big puzzles, you may need to ponder the problem for quite a while, gaining insights here and there until it's finally done. Yet, as with coding style, there are strategies that we all agree on.
So in spite of great individual variation, there are common aspects that everyone agrees with, which we'll discuss in this lesson.
Comment your code:
In any programming course for Computer Science students, this is absolutely central. If a student turns in a programming assignment with few or no comments, it will get a failing grade. If comments are needed for clarity and readability for CS students, who are presumably strong programmers, then R users who are not expert programmers need comments even more.
A style guide at a top university computer science department puts it well:
Commenting involves placing Human Readable Descriptions inside of computer programs detailing what the Code is doing. Proper use of commenting can make code maintenance much easier, as well as helping make finding bugs faster. Further, commenting is very important when writing functions that other people will use. Remember, well documented code is as important as correctly working code.
(Also see specific tips on commenting, later in that document.)
Don't be under the illusion that your code is self-documenting; it isn't! A typical comment might look like this:
w <- f(w)
# at this point, the data frame w will consist of the original rows for
# people over age 65 and who are homeownersAt the top of each source file, insert comments giving the reader an overview of the contents.
This will typically an overview of the roles of each major function, how the functions interact with each other, what the main data structures are, and so on.
I strongly recommend that you write these comments at the top of a file BEFORE you start coding (and of course modifying it as you do write code). This will really help you focus during the coding process.
Indent your code:
if (x < y) {
x <- y^2
z <- x + y
}is much easier to read than
if (x < y) {
x <- y^2
z <- x + y
}Write your code in top-down fashion:
If you have a function f that is more than, say, a dozen lines long, break its code into calls to smaller functions, say g and h. Then f will consist of those calls, plus some "glue" lines to deal with the return values and so on. Of course, it's a matter of taste as to break things up that way, but the point is that it makes your code both easier to read (by others, or by yourself later), and even more important, easier to write. Breaking up the code like this makes it read like an outline.
Don't skimp on attending to the "corner cases":
Computer Science people talk about "corner cases," meaning special situations in which code may fail in spite of being generally sound.
For instance, consider this code:
> i <- 5
> 1:i
[1] 1 2 3 4 5But what about the special case in which i = 0?
> i <- 0
> 1:i
[1] 1 0This may not be what you wanted. You probably should insert a check, say
if (i >= 1) i:5and maybe also code to handle the erroneous case. This will depend on the situation, but the main point is to be aware of possible corner cases.
Use a debugging tool:
More on this in a later lesson!
In our earlier examples of regression analysis, we were predicting a continuous variable such as human weight. But what if we wish to predict a dichotomous varible, i.e. one recording which of two outcomes occurs?
Consider the Pima dataset from earlier examples. Say we are predicting whether someone has -- or will later develop -- diabetes. This is coded in the test column of the dataset, 1 for having the disease, 0 for not.
As a simple example, say we try to predict test from the variables bim and age. A linear model would be
mean test = β0 + β1 bmi + β2 age
Remember, test takes on the values 1 and 0. What happens when we take the average of a bunch of 1s and 0s? The answer is that we get the proportion of 1s. For instance, the mean of the numbers 1,0,1,1 is 3/4, which is exactly the proportion of 1s in that data.
In statististical terms, what the above equation is doing is expressing the probability of a 1 -- i.e. the probability of having diabetes -- in terms of Body Mass Index and age.
Not a bad model, but one troubling point is that the right-hand side could evaluate to a number less than 0 or greater than 1, which would be impossible for a probability. In order to deal with that problem, we might use a logistic model, as follows.
Define the logistic function to be
l(t) = 1 / (1 + e-t)
We then modify the above equation to
probability of diabetes = l(β0 + β1 bmi + β2 age)
As before, the statistical details are beyond the scope of this R tutorial, but here is how you estimate the coefficients βi using R:
> glout <- glm(test ~ bmi + age, data=pima, family=binomial)
> summary(glout)
...
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -5.40378 0.51530 -10.487 < 2e-16 ***
bmi 0.09825 0.01248 7.874 3.45e-15 ***
age 0.04561 0.00694 6.571 4.98e-11 ***
...Let's explore those estimated βi a bit. Consider women with about average BMI, say 32, and compare 30-year-olds to those of age 40.
> l <- function(t) 1 / (1 + exp(-t))
> l(-5.40378 + 32*0.09825 + 30*0.04561)
[1] 0.2908045
> l(-5.40378 + 32*0.09825 + 40*0.04561)
[1] 0.3928424So, the risk of diabetes increases substantial over that 10-year period, but this population and BMI level.
Note: On Unix-family systems such as Linux, the Windows term folder is said to be a directory. You will frequently see this in Mac discussions as well. (The Mac OS is a Unix-family system.) We will typically use the term directory here, as that is what R uses.
In assmebling a dataset for my regtools package, I needed to collect the records of several of my course offerings. I started in a directory that had one subdirectory for each offering. In turn, there was a file named Results. As an intermediate step, wanted to find all such files, placing the text for each one in an R list gFiles. Only some specific columns of each file will be retained. (The discussion here is a slightly adapted version.)
The chief R functions I used were:
-
list.dirs(): Returns a character vector with the names of all the directories (i.e. subdirectories) within the current directory.
-
dir(): Returns a character vector with the names of all files within the current directory.
-
%in%: Determines whether a specified object is an element in a specified vector.
-
setwd(): Changes to the specified directory.
Here is the code:
getData <- function() {
currDir <- getwd() # leave a trail of bread crumbs
dirs <- list.dirs(recursive=FALSE)
numCourseOfferings <- 0
# create empty R list, into which we'll store our course records
resultsFiles <- list()
for (d in dirs) {
setwd(d) # descend into d directory
# check if there is a Results file there
fls <- dir()
if (!('Results' %in% fls)) { # not there, skip this dir
setwd(currDir)
next
}
# ah, there is such a file; increment our count
numCourseOfferings <- numCourseOfferings + 1
# open it
resultsLines <- readLines('Results')
# delete the comment lines; look at 1st character in each line
resultsLines <- delComments(resultsLines)
resultsFiles[[numCourseOfferings]] <- extractCols(resultsLines)
setwd(currDir)
}
resultsFiles # return all the grades records
}Before we go into the details, note the following:
-
The code is written in a top-down manner. Much of the work of getData() is offloaded to other functions (code not shown), delComments() and extractCols().
-
There are lots of comments!
Now, consider the line
dirs <- list.dirs(recursive=FALSE)As mentioned, list.dirs() will determine all the subdirectories within the current directory. But what about subdirectories of subdirectories, and subdirectories of subdirectories of subdirectories, and so on? Setting recursive to FALSE means we want only first-level subdirectories.
So, the line
for (d in dirs) {will then have us process each (first-level) directory, one by one.
When we enter one of those subdirectories, the line
fls <- dir()will determine all the files there, storing the result as a character vector fls.
Then, as the comment notes, the lines
if (!('Results' %in% fls)) { # not there, skip this dir
setwd(currDir)
next
}will, in the event that there is no Results file in this subdirectory, skip this subdirectory. The R keyword next says, "Go to the next iteration of this loop," which here means to process the next subdirectory. Note that to prepare for that, we need to move back to the original directory:
setwd(currDir)On the other hand, if this subdirectory does contain a file named Results, the remaining code increments our count of such files, reads in the found file, and assigns its contents as a new element of our resultsFiles list.
We've seen R for loops in previous lessons, but there's another kind of loop, while. It keeps iterating until some specified condition is met. We don't know how many iterations will be needed, unlike the for case, with a fixed number of iterations.
As our example, consider AirPassengers, which consists of number of air travelers in thousands, in monthly data from January 1949. As usual, let's glance at it first:
> str(airpass)
Time-Series [1:144] from 1949 to 1961: 112 118 132 129 121 135 148 148 136 119 ...Suppose we wish to know when the cumulative number of passengers first exceeded 10 million. A crude way would be to use R's cumsum ("cumulative sums") function:
> cumsum(airpass)
[1] 112 230 362 491 612 747 895 1043 1179 1298 1402 1520
[13] 1635 1761 1902 2037 2162 2311 2481 2651 2809 2942 3056 3196
[25] 3341 3491 3669 3832 4004 4182 4381 4580 4764 4926 5072 5238
[37] 5409 5589 5782 5963 6146 6364 6594 6836 7045 7236 7408 7602
[49] 7798 7994 8230 8465 8694 8937 9201 9473 9710 9921 10101 10302
[61] 10506 10694 10929 11156 11390 11654 11956 12249 12508 12737 12940 13169
[73] 13411 13644 13911 14180 14450 14765 15129 15476 15788 16062 16299 16577
[85] 16861 17138 17455 17768 18086 18460 18873 19278 19633 19939 20210 20516
[97] 20831 21132 21488 21836 22191 22613 23078 23545 23949 24296 24601 24937
[109] 25277 25595 25957 26305 26668 27103 27594 28099 28503 28862 29172 29509
[121] 29869 30211 30617 31013 31433 31905 32453 33012 33475 33882 34244 34649
[133] 35066 35457 35876 36337 36809 37344 37966 38572 39080 39541 39931 40363We see that that occurred in the 59th month. But though this approach would be convenient, it also would be wasteful: We are calculating all the cumulative sums, even though we don't need them all. In a really long vector, this could be slow. Here is a less wasteful way:
tot <- 0
> i <- 0
> while (i <= length(airpass) && tot < 10000) {
+ i <- i + 1
+ tot <- tot + airpass[i]
+ }
> i
[1] 59So, the while loop keeps iterating until we get the desired cumulative total.
Key points here:
-
The '&&' operator stands for "and".
-
The condition within the 'while' says that (a) we are not yet at the end of the airpass vector, AND (b) our total is still less than
- Note the need for the condition i <= length(airpass). It's possible that tot will never exceed 10000 (not true here, but we wouldn't know that a priori), so we need that condition so that the loop doesn't iterate forever!
There's more, though. The cumsum function is vectorized, so using it, though seemingly wasteful, may actually be faster than the loop
These are books and other resources that I myself consult a lot (yes, I do consult my own books; can't keep it all in my head :-) ), plus others I recommend.
Nonprogramming Coverage of R
-
Andrie de Vries and Joris Meys, R For Dummies (second edition), For Dummies
-
Jaren Lander, R for Everyone: Advanced Analytics and Graphics (second ed.), Addison-Wesley
R Programming and Language
-
John Chambers, Software for Data Analysis: Programming with R, Springer
-
Dirk Eddelbuettel, Seamless R and C++ Integration with Rcpp, Springer
-
Colin Gillespie and Robin Lovelace, Efficient R Programming: A Practical Guide to Smarter Programming
-
Norm Matloff, The Art of R Programming, NSP
-
Norm Matloff, Parallel Computation for Data Science, CRC
-
Hadley Wickham, Advanced R (second edition), CRC
Data Science with R
-
Nina Zumel and John Mount, Practical Data Science with R, Manning (2nd ed.)
-
Hadley Wickham and Garrett Grolemund, R for Data Science: Import, Tidy, Transform, Visualize, and Model Data, O'Reilly
Graphics in R
-
Winston Chang, R Graphics Cookbook: Practical Recipes for Visualizing Data, O'Reilly
-
Paul Murrell, R Graphics (third edition), CRC
-
Deepayan Sarkar, Lattice: Multivariate Data Visualization with R, Springer
-
Hadley Wickham, ggplot2: Elegant Graphics for Data Analysis, Springer
Regression Analysis and Machine Learning, Using R
-
Francis Chollet and JJ Allaire, Deep Learning in R, Manning
-
Julian Faraway, Linear Models with R, CRC
-
Julian Faraway, Extending the Linear Model with R, CRC
-
John Fox and Sanford Weisberg, An R Companion to Applied Regression, SAGE
-
Frank Harrell, Regression Modeling Strategies: With Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis, Springer
-
Gareth James, Daniela Witten, Trevor Hastie, Robert Tibshirani, Introduction to Statistical Learning: with Applications in R, Springer, 2nd ed.
-
Max Kuhn, Applied Predictive Modeling, Springer
-
Max Kuhn and Kjell Johnson, Feature Engineering and Selection: A Practical Approach for Predictive Models, CRC
-
Norm Matloff, Statistical Regression and Classification: from Linear Models to Machine Learning, CRC
-
Norm Matloff, The Art of Machine Learning: Algorithms+Data+R, NSP, coming soon
Other
-
Rob Hyndman and George Athanasopoulos, Forecasting: Principles and Practice, OTexts
-
Ted Kwartler, Text Mining in Practice with R
-
Norm Matloff, Probability and Statistics for Data Science: Math + R + Data, CRC
-
Julia Silge and David Robinson, Text Mining with R: A Tidy Approach, O'Reilly
-
Yihui Xie et al, R Markdown: The Definitive Guide, CRC
Web
I also would recommend various Web tutorials:
-
Szilard Palka, CEU Business Analytics program: Use Case Seminar 2 with Szilard Pafka (2019- 05-08)
-
Hadley Wickham, the Tidyverse
This tutorial has benefited from feedback from (in alphabetical order) Reese Goding, Ira Sharenow, and Aaron Wichman, as well as various anonymous suggestions.
An interactive development environment (IDE) is a software tool that enables editing, saving and running your code, as well as related actions such as installing packages.
The real "power users" tend to use either Emacs Speaks Statistics (ESS), a plugin for the Emacs editor, or Nvim-r,, a plugin for the vim editor. However, since this tutorial is aimed at those with little or no prior coding background, we will not cover them. Instead, we introduce RStudio. Here are some pros and cons:
-
RStudio is very highly popular, especially in the US and Australia/New Zealand. Indeed, for many users, RStudio is R.
-
Lots of help available on the Web, and in R User Groups that have been established in many major cities. Has numerous features, keyboard shortcuts etc.
-
That however also has a downside, since as noted earlier, the compexity of RStudio can be "overwhelming."
In light of that last point, we recommend that you NOT try to learn RStudio to any degree of complexity at the outset. Just learn how to create, load, run, and save files of R code, the simple stuff, which should be easy. Leave the advanced features for later.
There are many tutorials on the Web for installing RStudio.
This one is pretty good,
for all major platforms.
If your screen has an RStudio icon, click it. Otherwise type 'rstudio' into a terminal window.
Again, there is a lot more one can do than the following, but we'll stick to the absolute basics.
Note the pane in the lower-left portion of the RStudio screen. By default, that is the Console pane, containing the usual R '>' prompt. You can use it just as we have throughout this tutorial. Note too that this is where your R output will appear.
Everything here involves files, where we store our R code (scripts).
creating a new code file: File | New File | R Script will create an empty window pane, ready to be filled with code. Start typing!
saving a code file: File | Save will save the contents of the pane. If it's a new file, you'll be asked to give the file a name. Make sure to note what folder the file will be in, so you know where to read it from later.
running code: To run the code in your current window, choose Code | Run Region | Run All.
exiting RStudio:
File | Quit Session...
The document is covered by a
Creative Commons license,
Creative Commons Attribution-No Derivative Works 3.0 United States