Path: https://github.com/Image-Py/itk-plgs
Version: 0.1
Author: YXDragon
Email: [email protected]
Keyword: itk, segment
Description: SimpleITK plugin set for ImagePy
you must fill the information upon, and you can not remove or insert line, you can write free below.
Introduction: itk need not much introductions, It is a 2D/3D image segment library. ImagePy is a interactive image processing framework which can wrap any numpy based library esaily. And supporting multi-channels, imagestack, lookuptable, roi, macros recorder...It is a Plugin system(just like ImageJ but more convenient). This project is a wrapper of itk for ImagePy plugins!
Now It is just a start, I wrap little of itk's algrism, aimed to introduct how to wrote ImagePy plugin, The Demo in this document is representative.
I know many numpy based project has a BSD license, but, sorry, I use SimpleITK, so must be under LGPL.
It is ImagePy's MainFrame, like ImageJ. And ImagePy has contains many common function, such as open image, save image, do some filter, do a roi, draw with pencil... It requires wxpython as ui, Numpy as base structure, shapely to treat the roi, and scipy.ndimage to so dome common filter. But this project devotes to do a wrapper for itk
Itk supports many medical format, such as dicom, nii... Now let's add reader and writer plugins for ImagePy.
as itk read any format as a image sequence, but sometimes we need read one slice. so we write a readall, then write a read.
import SimpleITK as sitk
import numpy as np
def readall(path):
image = sitk.ReadImage(path)
arr = sitk.GetArrayFromImage(image)
if arr.dtype == np.int16:
arr = arr.astype(np.int32)
return arr
def read(path):return readall(path)[0]
def write(path, img):
sitk.WriteImage(sitk.GetImageFromArray(img), path)from imagepy.core.util import fileio
# add dicom reader and writer
fileio.add_reader(['dcm'], read)
fileio.add_writer(['dcm'], write)
class OpenDCM(fileio.Reader):
title = 'DICOM Open'
filt = ['DCM']
class SaveDCM(fileio.Writer):
title = 'DICOM Save'
filt = ['DCM']
# add nii reader and writer, because nii is a sequence, so ruse read all, and give as a tuple.
fileio.add_reader(['nii'], (readall,))
fileio.add_writer(['nii'], (write,))
class OpenNII(fileio.Reader):
title = 'NII Open'
filt = ['NII']
class SaveNII(fileio.Reader):
title = 'NII Save'
filt = ['NII']
plgs = [OpenDCM, SaveDCM, '-', OpenNII, SaveNII]import SimpleITK as sitk
from imagepy.core.engine import Filter
class Plugin(Filter):
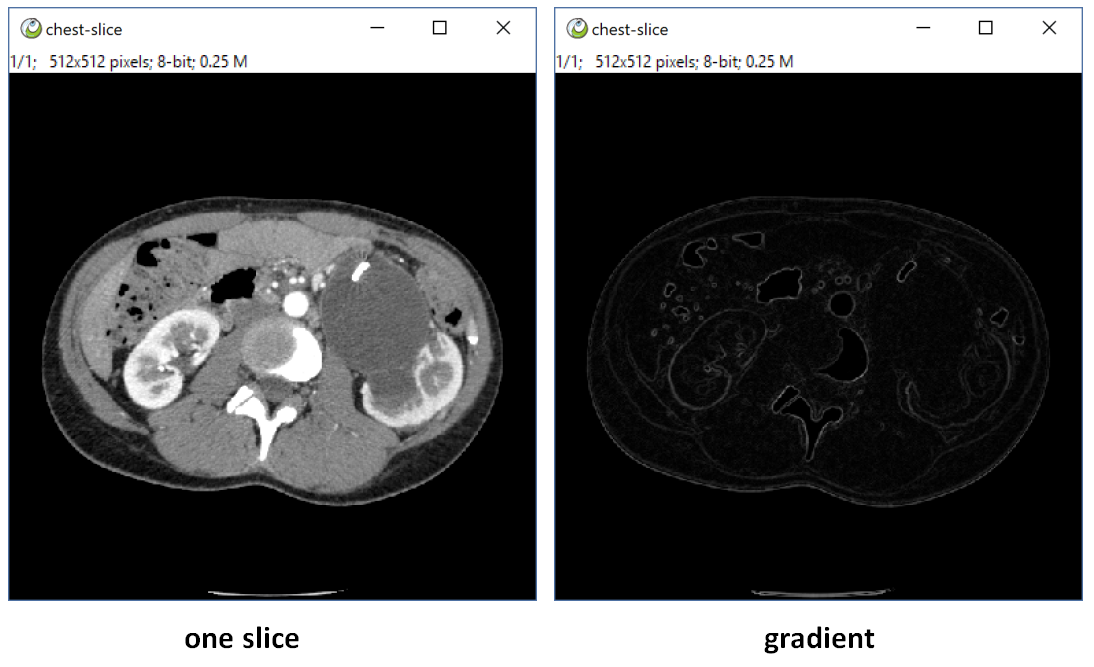
title = 'ITK Gradient Magnitude'
note = ['all', 'auto_msk', 'auto_snap']
def run(self, ips, snap, img, para = None):
img = sitk.GetImageFromArray(img)
img = sitk.GradientMagnitude(img)
return sitk.GetArrayFromImage(img)- class name must be Plugin
- title is necessary, be the plugin's id, show in menus.
- set the note, which tells ImagePy what to do for you.
- overwrite run method return the result
Filter is one of engines, means need a image, then do some change on it, It has a run method in such type:
- ips is the wrapper of image with some other information (lookup table, roi...)
- snap is a snapshot of the image, if 'auto_snap' in note, ImagePy will copy the image to snap befor run. (for many filter method must be implemented in a buffer)
- img is the current image you are processing.
- para is the parameter you got interactive. (there is no here)
- all means this plugin works for all type image.
- auto_snap means ImagePy do a snapshot befor processing, then you can use Undo.
- auto_msk means when there is a roi on the image, Plugin will only influnce the pixel in.
- more detail information please see ImagePy's README!
# -*- coding: utf-8 -*
import SimpleITK as sitk
from imagepy.core.engine import Filter
class Plugin(Filter):
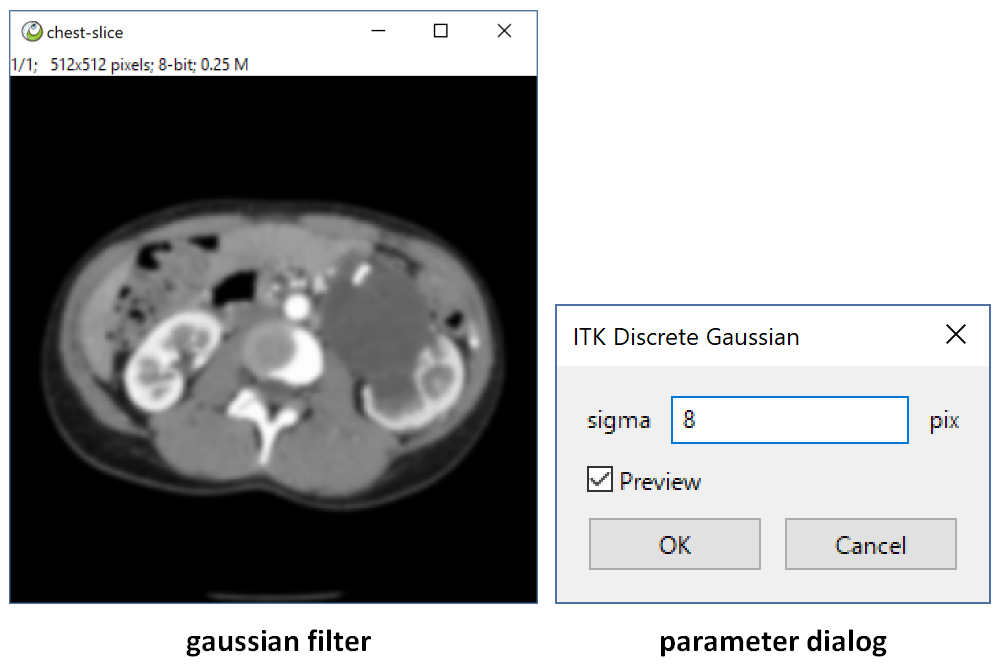
title = 'ITK Discrete Gaussian'
note = ['all', 'auto_msk', 'auto_snap', 'preview']
para = {'sigma':1.0}
view = [(float, (0,10), 1, 'sigma', 'sigma', 'pix')]
def run(self, ips, snap, img, para = None):
itkimg = sitk.GetImageFromArray(snap)
itkimg = sitk.DiscreteGaussian(itkimg, para['sigma'])
return sitk.GetArrayFromImage(itkimg)Many Filter need some parameter. Just like Gaussian. We just need do a little more.
- para is a dict object, which contains the parameter you need.
- view tell ImagePy how to interact when this plugin run, (float, (0,10), 1, 'sigma', 'sigma', 'pix') means it is a float between 0 and 10, title is sigma, corresponding to the sigma parameter with unit pix. More detail information please see ImagePy's README!
Add 'preview' in note, then when you adjust the parameter, ImagePy run this plugin immediately
import SimpleITK as sitk
from imagepy.core.engine import Simple
class Plugin(Simple):
title = 'ITK Gradient Magnitude 3D'
note = ['all', 'stack3d']
def run(self, ips, imgs, para = None):
itkimgs = sitk.GetImageFromArray(imgs)
itkimgs = sitk.GradientMagnitude(itkimgs)
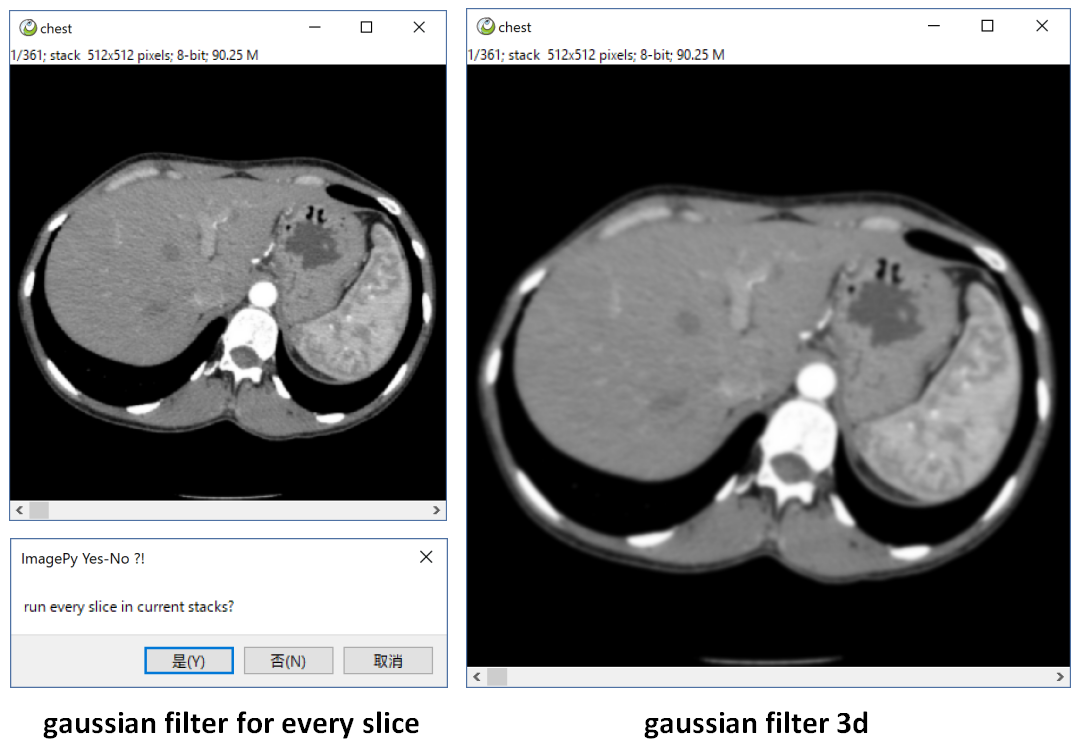
imgs[:] = sitk.GetArrayFromImage(itkimgs)when there is a image sequence, If you run a gaussian filter, it will ask if you want to process every slice. If ok, it will process slice by slice. But a 3d gaussian filter will blur the image sequence by X, Y and Z axis.
Filter aimed at treat a single slice, but if you want to process the whole images, please extends a Simple. It also can process other information. eg. set the look up table, or treat the roi, or save the current image/image sequence.
import SimpleITK as sitk
from imagepy.core.engine import Filter, Simple
import numpy as np
class Plugin(Filter):
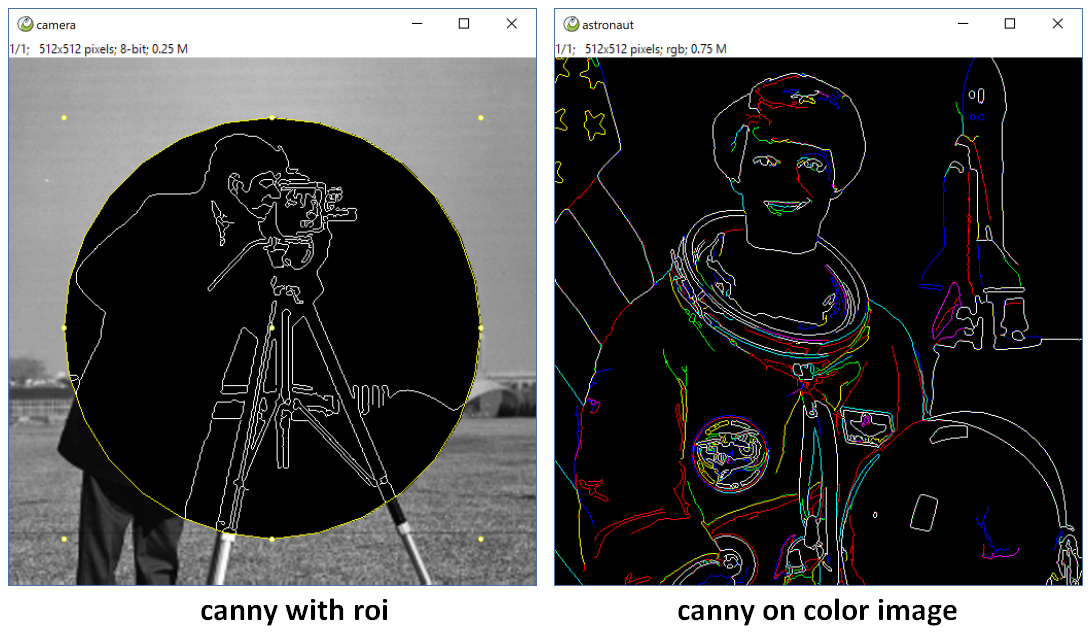
title = 'ITK Canny EdgeDetection'
note = ['all', 'auto_msk', 'auto_snap', '2float', 'preview']
para = {'sigma':1.0, 'low_threshold':10, 'high_threshold':20}
view = [(float, (0,10), 1, 'sigma', 'sigma', 'pix'),
('slide',(0,50), 'low_threshold', 'low_threshold',''),
('slide',(0,50), 'high_threshold', 'high_threshold','')]
def run(self, ips, snap, img, para = None):
img = sitk.GetImageFromArray(snap)
img = sitk.CannyEdgeDetection(img, para['low_threshold'], para['high_threshold'], [para['sigma']]*2)
return sitk.GetArrayFromImage(img)*ips.range[1]You see, we did not write code to treat the color image, but it works, and We can draw a ROI on the image, only the ROI area be changed! And we can undo the lasted operation. Even if it is a imagestack, ImagePy will ask you if run every slice!!
import SimpleITK as sitk
from imagepy.core.engine import Filter, Simple
import numpy as np
class Plugin(Filter):
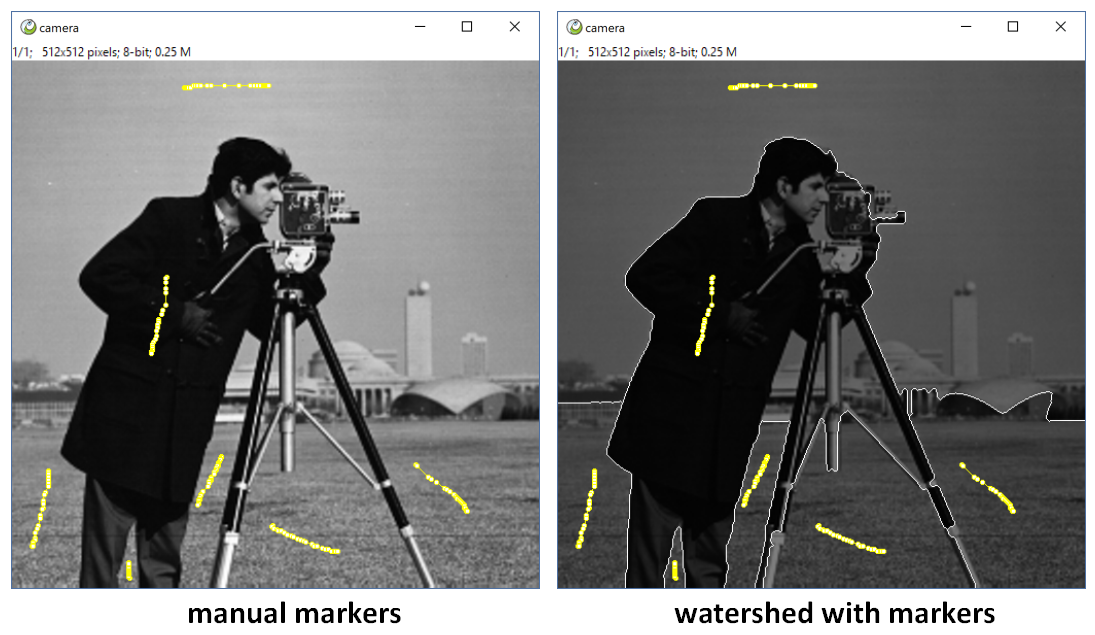
title = 'ITK Watershed Manual Marker'
note = ['8-bit', 'not_slice', 'auto_snap', 'req_roi']
para = {'sigma':2}
view = [(int, (0,10), 0, 'sigma', 'sigma', 'pix')]
def run(self, ips, snap, img, para = None):
itkimg = sitk.GetImageFromArray(img)
itkimg = sitk.DiscreteGaussian(itkimg, para['sigma'])
itkimg = sitk.GradientMagnitude(itkimg)
itkmarker = sitk.GetImageFromArray(ips.get_msk().astype(np.uint16))
itkmarker = sitk.ConnectedComponent(itkmarker, fullyConnected=True)
lineimg = sitk.MorphologicalWatershedFromMarkers(itkimg, itkmarker, markWatershedLine=True)
labels = sitk.GetArrayFromImage(lineimg)
return np.where(labels==0, ips.range[1], 0)ImagePy support ROI, you can use tool to draw a roi(point, line, polygon...), And We can use ips.roi access it, and ips.get_msk(mode='in') to get the roi mask image. mode can be 'in','out',or int means a sketch with specific width. Then use the mask as marker to do a watershed,
- req_roi means this plugin need a roi, ImagePy will check for you, if ther is not, interrupt the plugin.
- not_slice tells Imagepy need not to iterate slices if it is a stack, because this interactive is just ok for specific image, there is no need to go through.
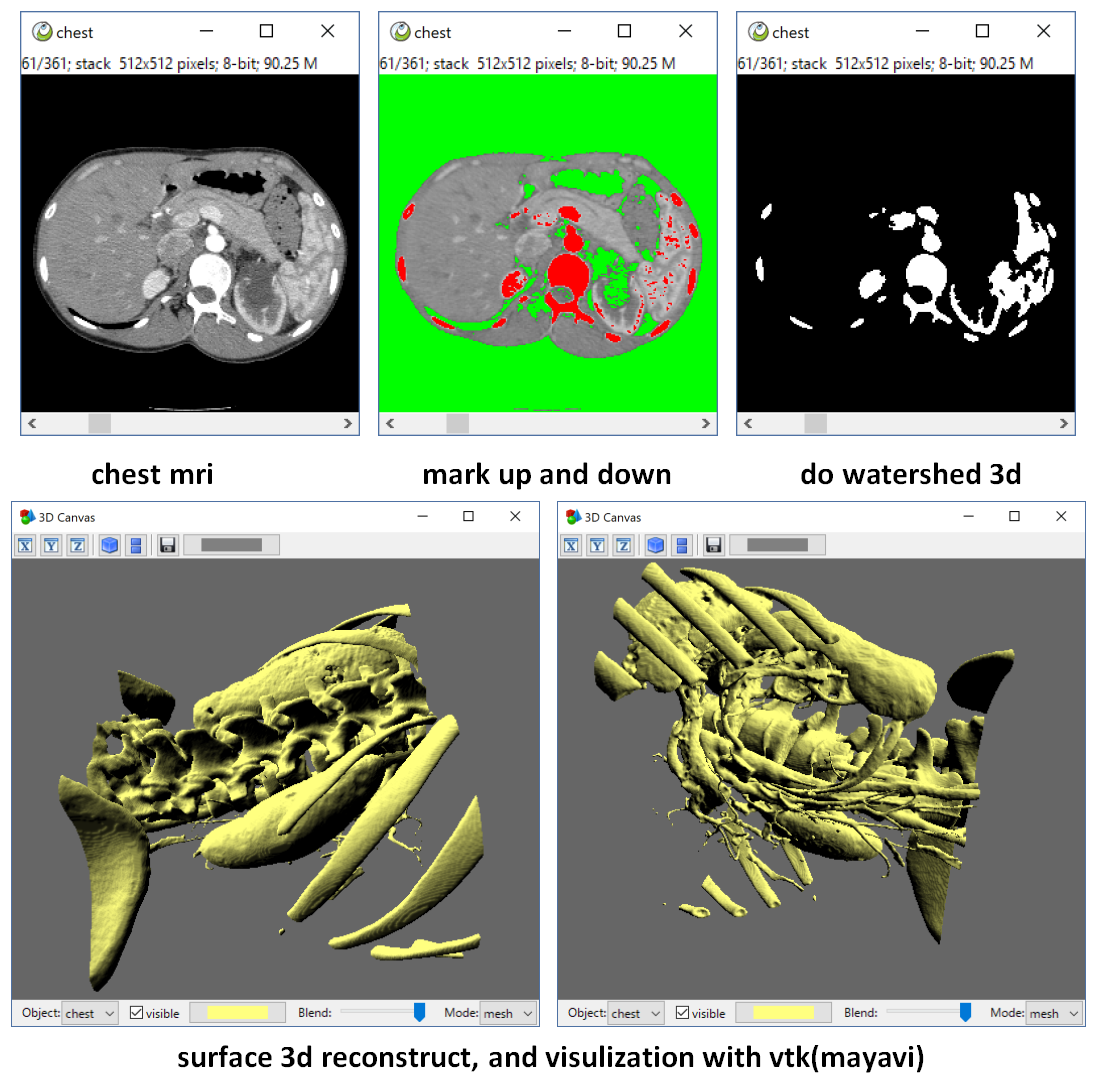
we can also do a 3d watershed, mark the up and down as two markers, then watershed on the 3d gradient image. we can get a perfect mask. Then do a 3d surface reconstruction with vtk(mayavi).
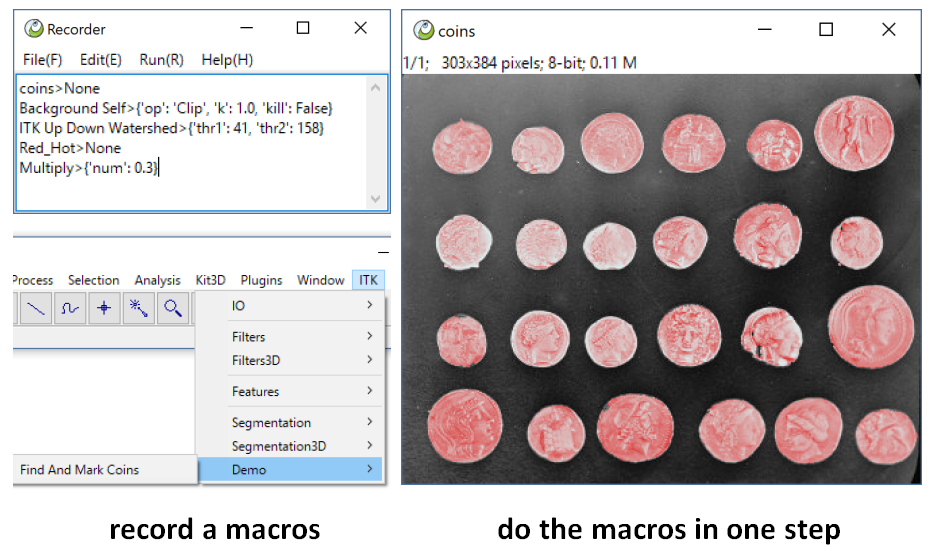
Macros is one of engines, It is a text file with every line as: "PluginID > {parameter}", If the parameter is None and the Plugin need parameter, IPy will show dialog to interact, if the parameter is given, ImagePy just run use the given parameter.
We can Open the Plugin > Macros > Macros Recorder to record the operate. Then save as a file with .mc extent under the menus folder. It will be parsed as a menu when started next. This is Macros, We never need to implements ourself.
Then we Try the Find And Mark Coins macros, Wow!, It run the command sequence automatically!
We can use Macros to do some bat processing, what more? It can be used as a good tutorial, We just implement the basic method, and use macros to show this method can solve such problem!!!
you can also write a markdown file, and lay it under any sub folder of menus, when ImagePy setup, It will be loaded as a menus too, when click it, the markdown page will show.
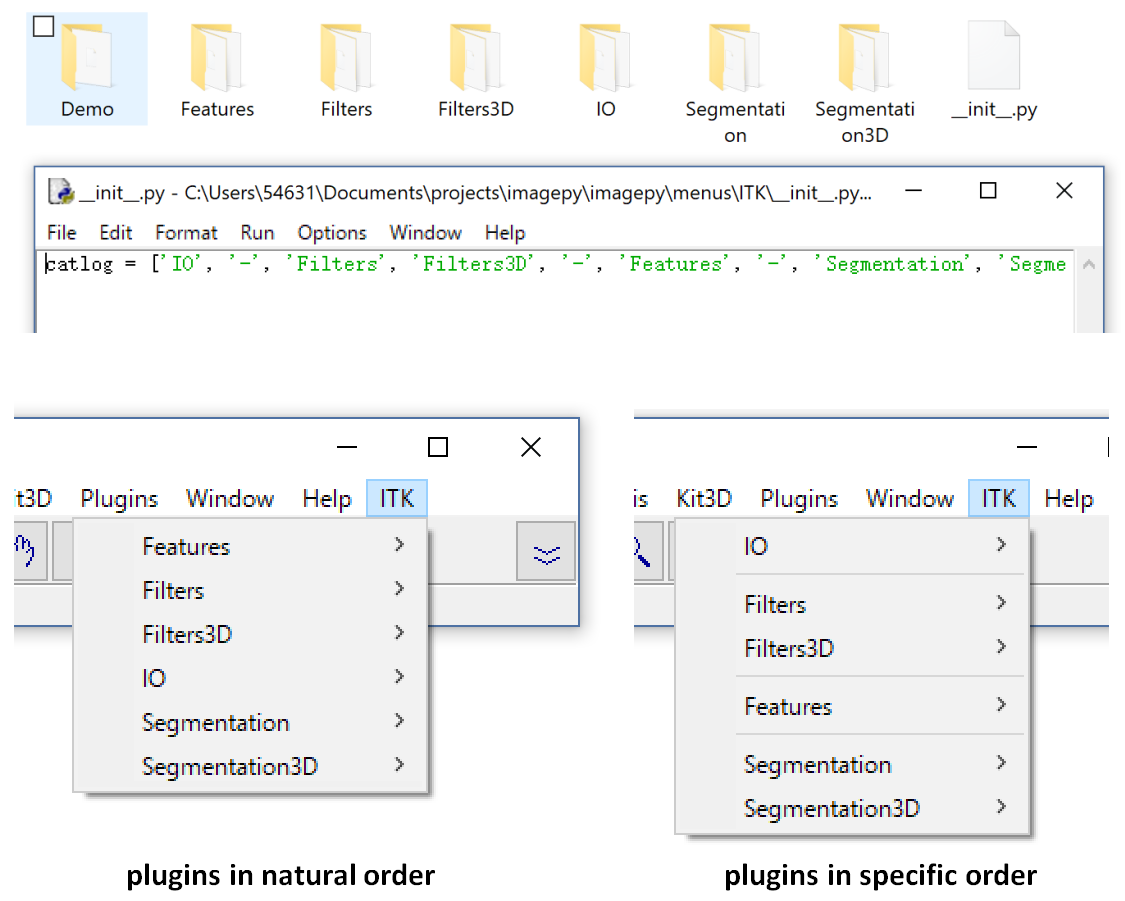
ImagePy is a plugin framework. The Catlog will be parsed as the corresponding menus. You just copy package under the menus folder or it's sub folder. But the question is, Our function will be in a disordered order. So we can add a list called catlog under every init file.
Now ITK menu is before the Help, and the IO is the first Item, then Filters, Features, Segmentation. and if we put '-' in catlog, It will be parsed as a spliter line.
ImagePy can wrap any numpy based libraries, can generate table esaily. In the basic version, It contains scikit-image, and I want to build a opencv-plgs and a itk-plgs. then integrate them, It will be powerful than Fiji, and be more esaily to extend.
OK! That is a start, I want more developer can join. I think it is significative to let ITK be esaier to approach, Benifit more scientists who does not master programming. But I cannot do it by myself, My English is not so good, and have little spare time, But I will do my best!
as simple itk cannot treat numpy array directly, so we must use GetImageFromArray and GetArrayFromImage, Did you have any better method?