-
Notifications
You must be signed in to change notification settings - Fork 21
Contributing to eddy4R
Contents:
The NEON DevOps approach was fully documented in Metzger et al. (2017), so I will quote the paper below to give a brief description:
NEON’s DevOps framework consists of a periodic sequence (Figure 2) that incorporates these workflow steps. For this purpose we define NEON Science as personnel working directly on the NEON project, and the Science Community, regardless of whether they also work on the NEON project, as anyone producing or using data, algorithms, or research products related to the NEON data themes (Atmosphere; Biogeochemistry; Ecohydrology; Land Cover and Processes; Organisms, Populations, and Communities): The science community contributes algorithms and best practices (1). Implicitly or explicitly, this embodies the DevOps: Plan stage – the algorithms most valued by the community are being incorporated. Together with NEON Science (2), these algorithms are coded in the open-source R computational environment (DevOps: Create stage). DevOps: Verify (testing) and Package (packaging) are performed as the code is compiled into eddy4R packages via the GitHub distributed version control system (3). NEON Science releases an eddy4R version from GitHub, which automatically builds an eddy4R-Docker image on DockerHub as specified in a “Dockerfile” (4; DevOps: Release stage). The eddy4R-Docker image is immediately available for deployment by NEON CI (5; DevOps: Configure & Monitor stages), the Science Community (1) and NEON Science (2) alike. Here the DevOps: Configure (computational resource allocation) & Monitor stages occur. Monitoring of end-user experience is also performed in GitHub (3) via issue-tracking.

How this DevOps cycle is realized in Github is described in the figure below.

The eddy4R repo is read access only; thus, to contribute to eddy4R you must use a "Forking Workflow" as described in the figure below.
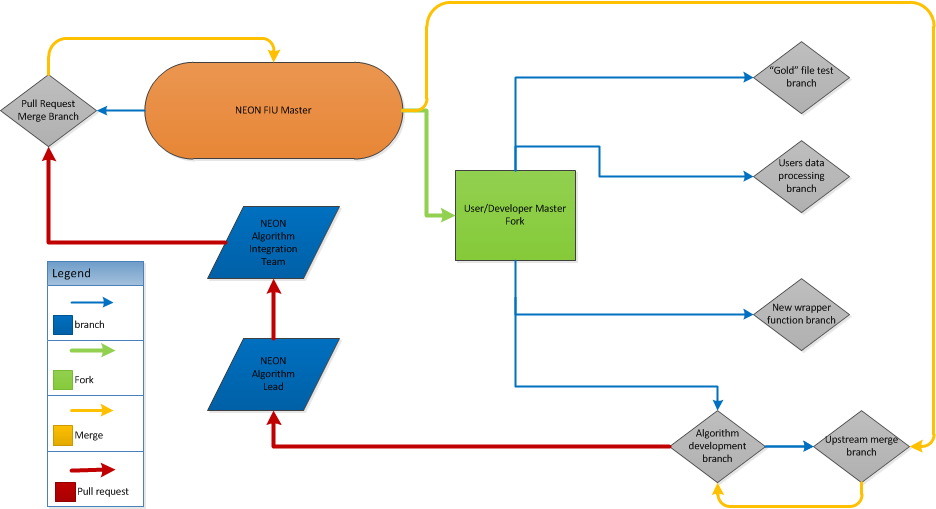

- The eddy4R branching model follows the principles outlined in ["A successful Git branching model"] (https://nvie.com/posts/a-successful-git-branching-model/?) described by Vincent Driessen
- New feature release process
- Open the NEONScience/NEON-FIU-algorithm git project
- Create new release branch from /deve (e.g.
git checkout -b 0.3.0) then push to NEONScience/eddy4r-deve repo (e.g.git push -u origin 0.3.0) - copy dock/eddy4R-base/Dockerfile to root directory, commit, push
- release from release branch (e.g.,
0.3.0) with release tag (e.g.,0.3.0-rc1)
- bug fixes from release branch (e.g.,
0.3.0)- check out branch "/fixDesc" (e.g., fixEcseNa) in personal fork (e.g. ddurden/NEON-FIU-algorithm-ddurden) from target release branch (e.g.,
git checkout -b upstream/0.3.0 fixDesc) - Push bug fix branch "/fixDesc" to github fork (
git push -u origin fixDesc) - resolve bug and test in branch "/fixDesc"
- pull request from branch "/fixDesc" into the release branch (e.g.
0.3.0)- resolve conflicts (there shouldn't be any, unless another bug-fix was pulled in first)
- perform Travis-CI test; further corrections if test fails
- once Travis-CI successful, merge
- make new release candidate (e.g.,
0.3.0-rc2)
- subsequent pull request from release branch
0.3.0into branchdeve- resolve conflicts
- perform Travis-CI test; further corrections if test fails
- once Travis-CI successful AND new base image build completed on Dockerhub, merge
- in /deve, copy dock/eddy4R-deve/Dockerfile to root directory, commit, push
- delete branch "/fixDesc"
- check out branch "/fixDesc" (e.g., fixEcseNa) in personal fork (e.g. ddurden/NEON-FIU-algorithm-ddurden) from target release branch (e.g.,
- Once the release is stable make the final release from
master- pull request from release branch
0.3.0tomaster - perform Travis-CI test; further corrections if test fails
- once Travis-CI successful, merge
- make official release (e.g.,
0.3.0)
- pull request from release branch
- bug fixes from tagged/final release (e.g.,
0.3.0)- In personal fork (e.g.
ddurden/NEON-FIU-algorithm-ddurden) - check out branch
fixDescfrom target release (e.g.,0.3.0)git fetch --all --tags --prune git tag git checkout -b tags/0.3.0 fixDesc git push -u origin fixDesc - resolve bug and test in branch
fixDesc - pull request from branch
fixDescinto the NEONScience/NEON-FIU-algorithm/master- resolve conflicts (there shouldn't be any, unless another bug-fix was pulled in first)
- perform Travis-CI test; further corrections if test fails
- once Travis-CI successful, merge
- make new official release (e.g.,
0.3.1)
- subsequent pull request from branch
masterinto branchdeve- [merging from branch
masterinstead of from branchfix: in this way branchdevealways has the same commit history as branchmaster, incl. merge commits] - resolve conflicts
- perform Travis-CI test; further corrections if test fails
- once Travis-CI successful AND new base image build completed on Dockerhub, merge
- in
deve, copydock/eddy4R-deve/Dockerfileto root directory, commit, push delete branchfixDesc
- [merging from branch
- In personal fork (e.g.
-
Travis-CI documentation for building and testing an R project: https://docs.travis-ci.com/user/languages/r/
-
eddy4R implementation:
-
Travis-CI tests the eddy4R docker build process, including installation of dependencies and R-package builds, and runs the gold file regression test (flow.test.gold.eddy.R) to ensure changes to the code base do not impact the resulting output (unless expected and verified, which would include an update to the reference gold file).
-
The NEON-FIU-algorithm master repo is currently set to perform testing on all pull requests made to the repo, and the results of the test will be displayed on the pull request github page (see image below). Green checks indicate the pull request passed testing, and red x's indicate a failed test.
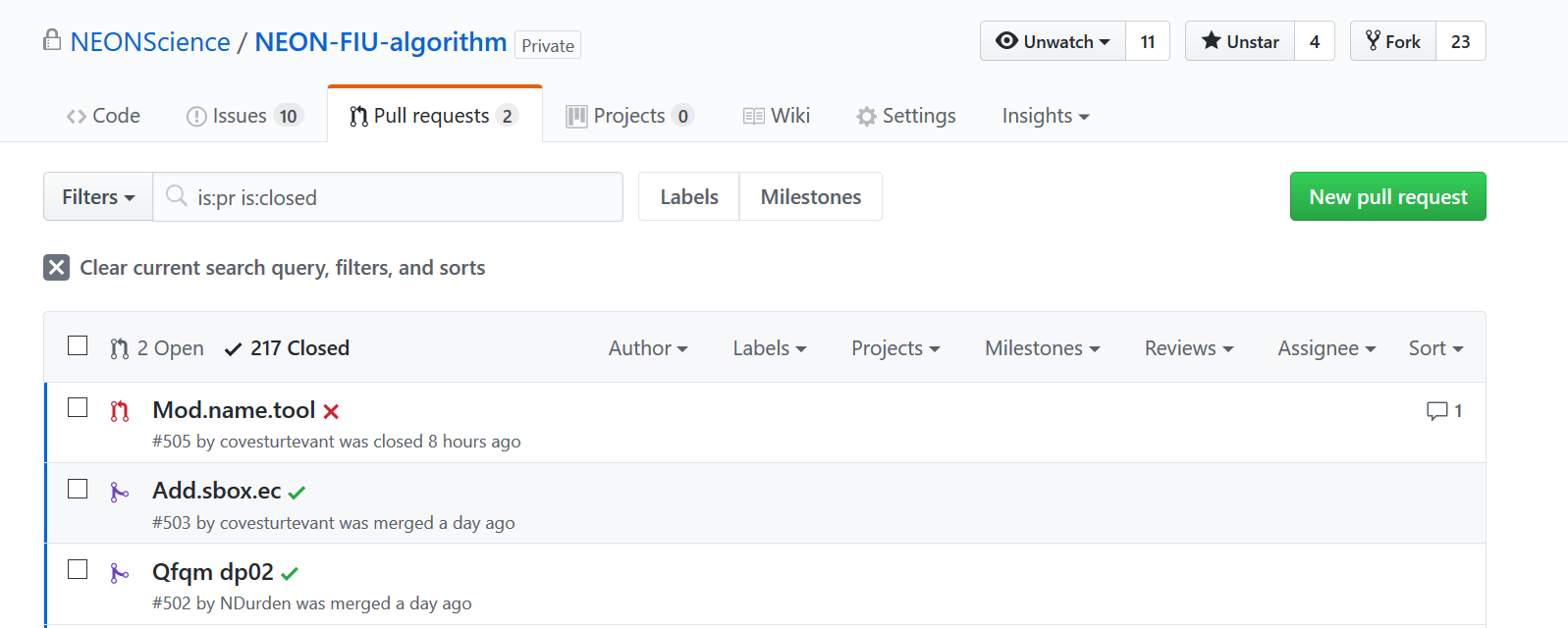
-
Inside a new pull request, the following will be displayed if the automated Travis-CI testing passed.

-
-
Travis-CI users guide: - First, signup for Travis-CI (https://travis-ci.org/) using your github ID.
- Next, you can activate repositories that you would like to perform builds and test on. However, not that the repositories available to you are only public repositories. Travis-CI has a separate webpage (https://travis-ci.com/) for private projects. So, you will need to sign up for a Travis Pro account, which comes with 100 free builds. Once you are signed up, you should be able to sync National Ecological Observatory Network private repos and activate them for building and testing.

-
Once, accounts are activated you should see them in My Repositories tab. The NEONScience/NEON-FIU-algorithm master repo should already be activated. If you want to test your updates prior to sending a pull request, you should activate your fork (e.g. ddurden/NEON-FIU-algorithm-ddurden).

-
Considering you have only 100 free builds, it's suggested that you make some changes to testing settings. These can be found by clicking "More options" ==> "Settings". The most important settings are under "General", I suggest requiring a .yml file be present to build, turn off build branch updates, and turn off build pull request updates (see below for my configuration).
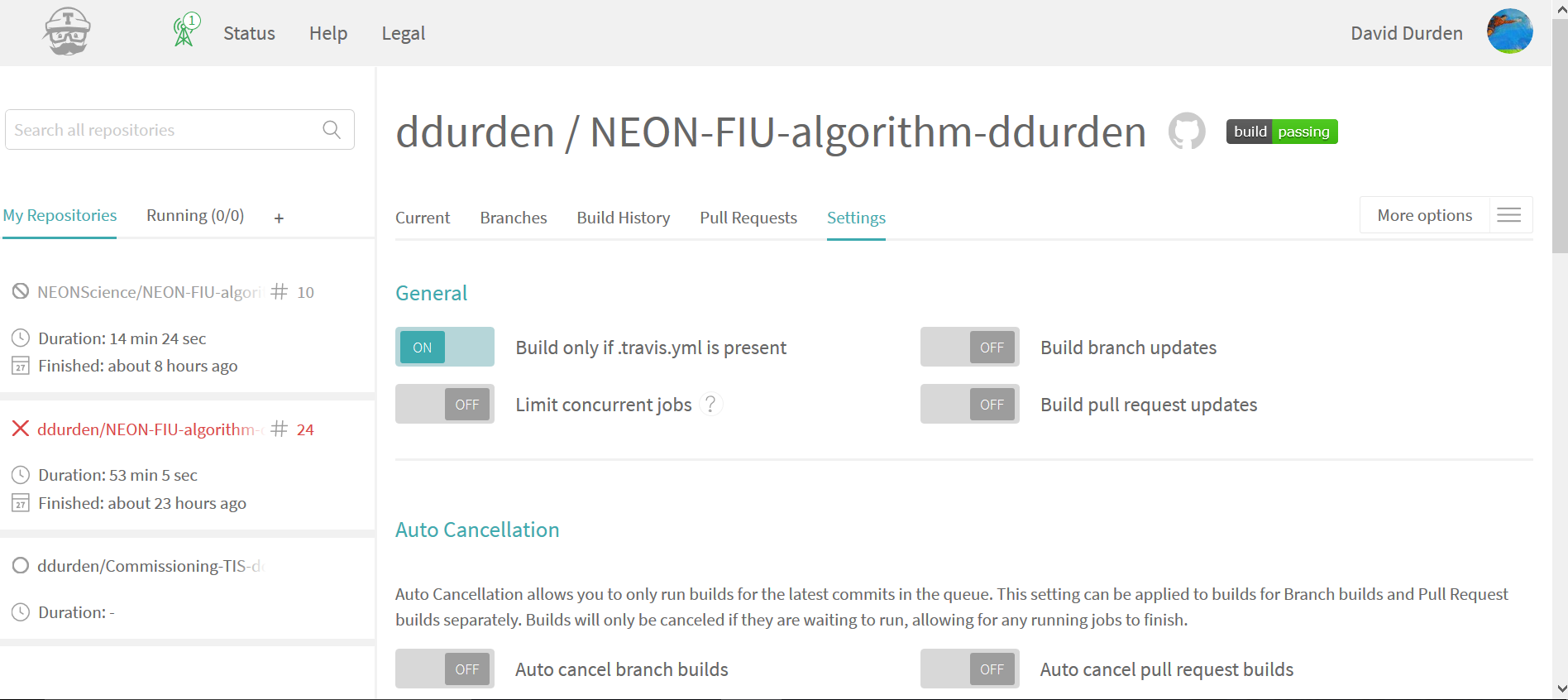
- Then, if you want to test your code changes before submitting the pull request you can trigger a build. This can be found under "More options" ==> "Trigger build", which will give you the option of which branch you would like to build (see below). This will perform a custom build, and will not add any commits to your github code repo.
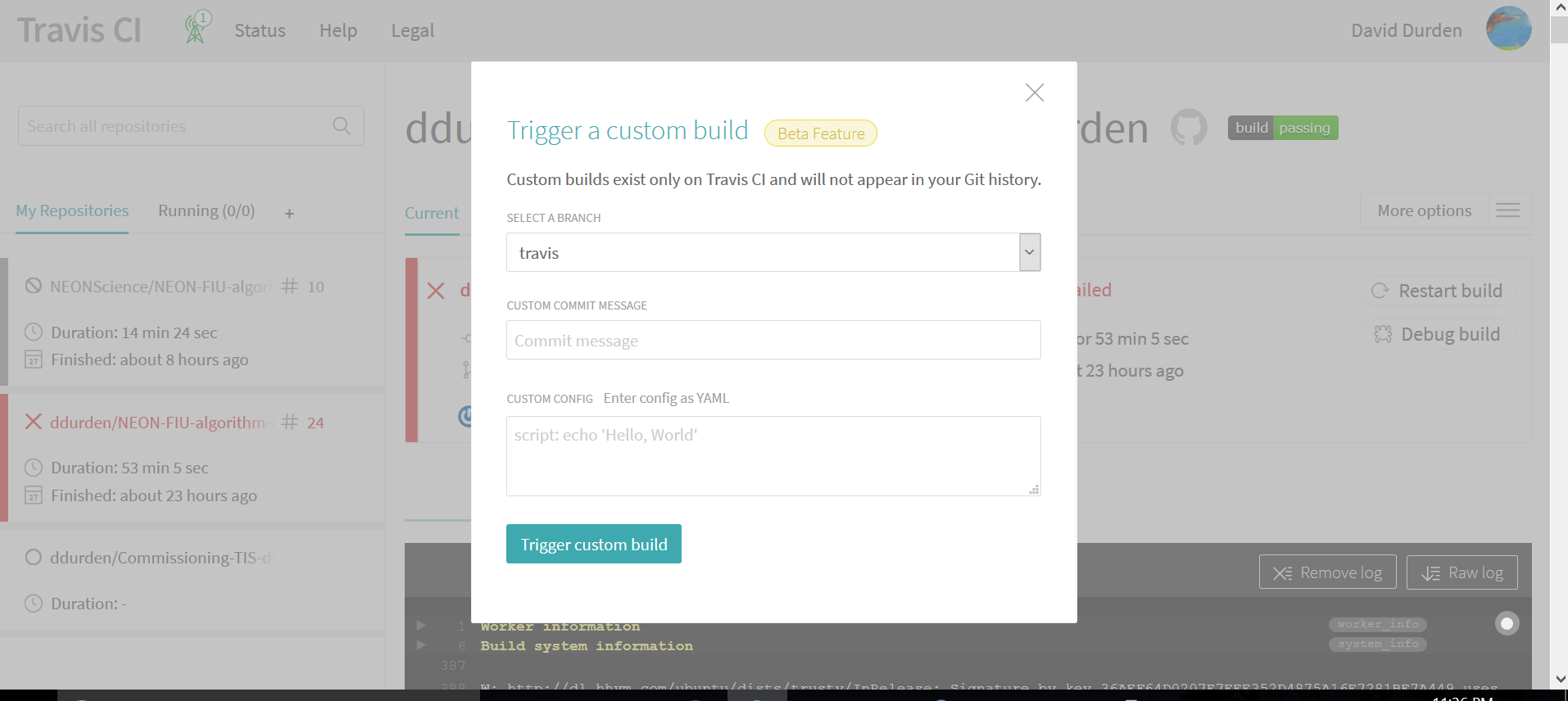
-
Additional Notes: - Travis_CI for R is usually performed on a package basis, not a suite of packages - use devtools::use_travis() to automatically add a minimal .travis.yml to your package, add .travis.yml to .Rbuildignore, and add the code for the badge to your README.
- Adding system requirements can be done by
- Adding docker to services in the travis.yml:
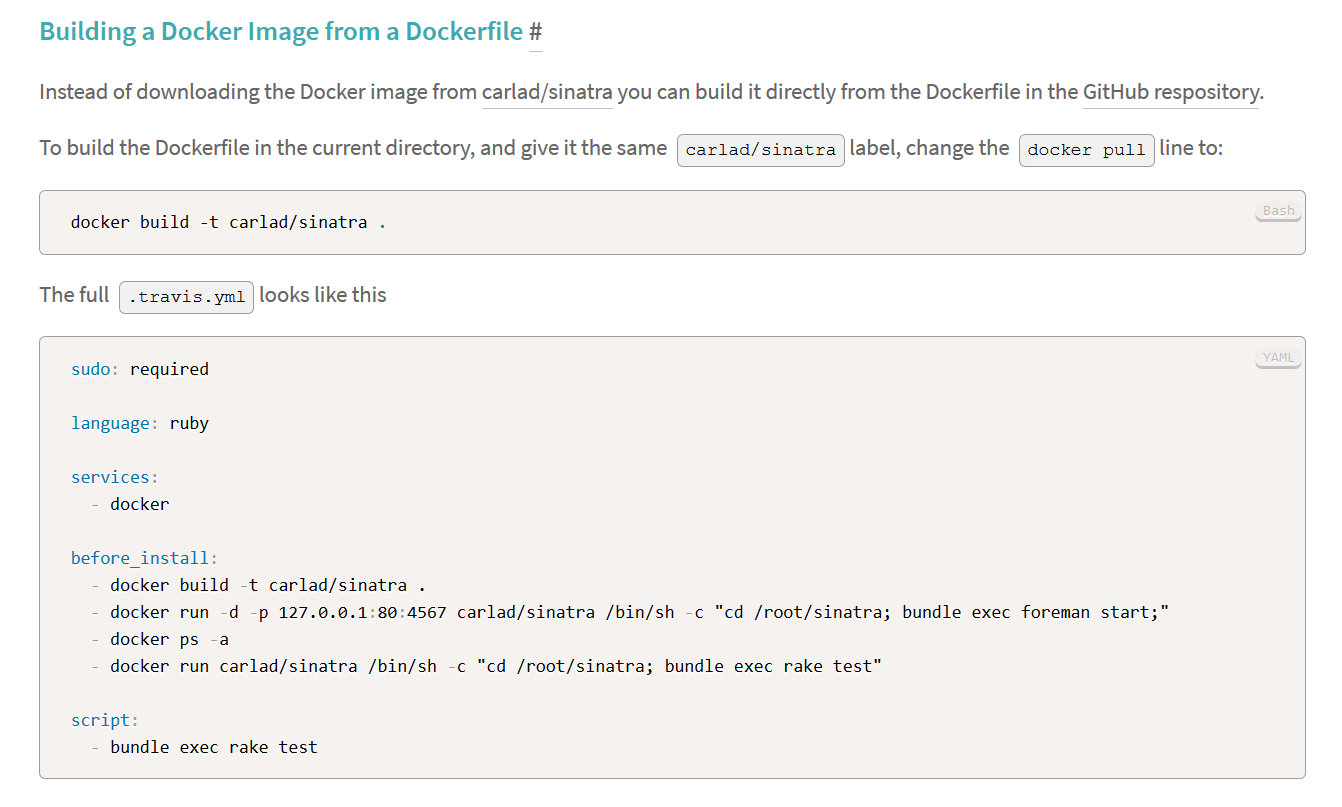
-
Future improvements:
- Travis-CI is integrated with testthat package - True unit testing functionality. Relatively simple to automate for packages, but would require some work up front. - Can be automated for directories or packages - R CMD 'check' will perform the package test
-
Resources:
- Hadley Wickham’s R Packages book has a section on Travis that is short but helpful; it is within the section on automated checking that also has relevant helpful information. The R docs at Travis itself are also quite helpful, although when I am having a real headscratcher of a problem I always wish there was more detail.
To inquire with repository maintainers on questions or ideas that fall outside the Github Issue Tracker workflow, please contact us @ [email protected]

The National Ecological Observatory Network is a project solely funded by the National Science Foundation and managed under cooperative agreement by Battelle. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
GNU AFFERO GENERAL PUBLIC LICENSE Version 3, 19 November 2007
Information and documents contained within this repository are available as-is. Codes or documents, or their use, may not be supported or maintained under any program or service and may not be compatible with data currently available from the NEON Data Portal.