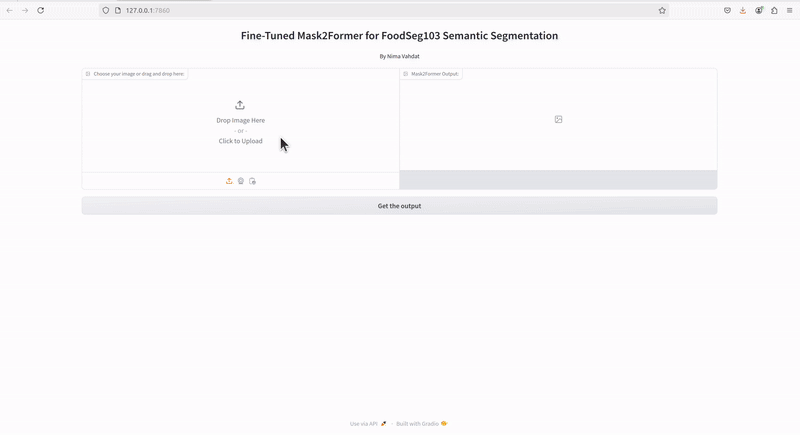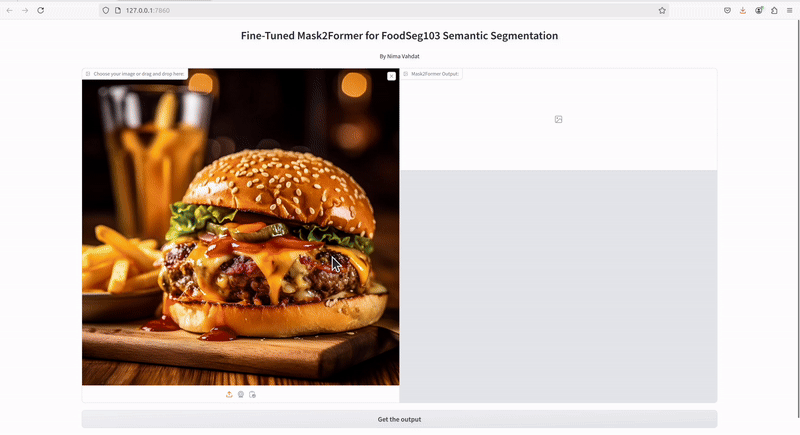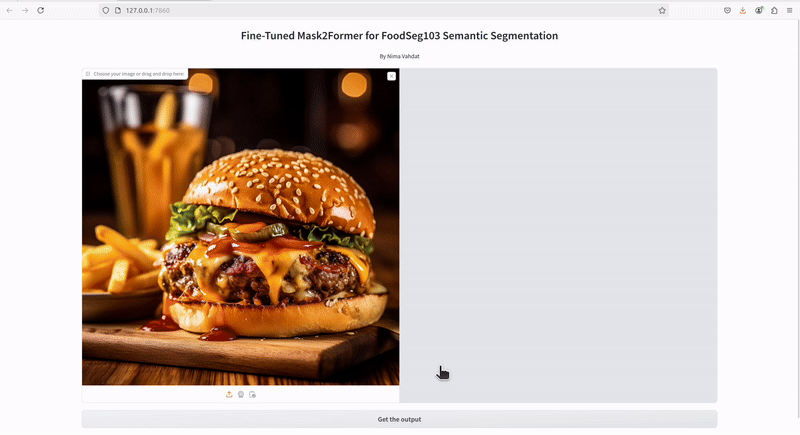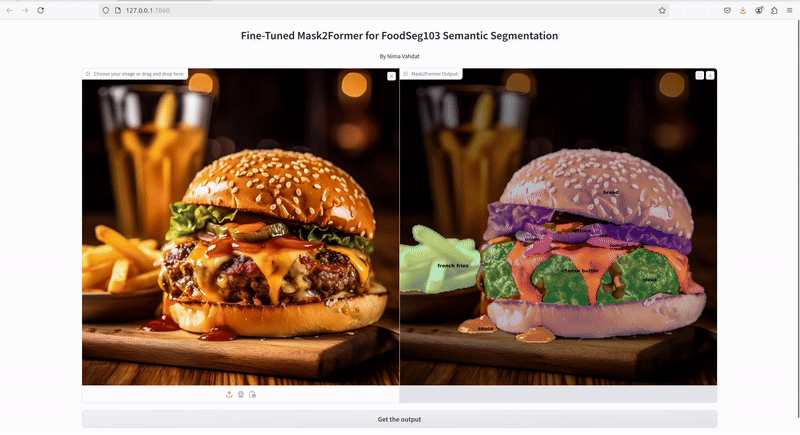
This project focuses on fine-tuning the Mask2Former model for semantic segmentation specifically on the FoodSeg103 dataset. The goal was to enhance the model's performance in identifying and segmenting various food items from images. The project also includes deploying the fine-tuned model and creating a user-friendly GUI with Gradio for interactive inference.
See the Gradio interface in action with the GIF below. 🍴✨
-
Clone the Repository:
git clone https://github.com/NimaVahdat/FoodSeg_mask2former.git cd FoodSeg_mask2former -
Install Dependencies:
pip install -r requirements.txt
Configure the training parameters in the config.yaml file:
batch_size: Number of samples per batch.learning_rate: Initial learning rate for the optimizer.step_size: Epoch interval for learning rate adjustment.gamma: Factor for learning rate decay.epochs: Total number of training epochs.save_path: Directory to save model checkpoints.load_checkpoint: Path to a pre-trained checkpoint (orNoneto train from scratch).log_dir: Directory for TensorBoard logs.
To start the training process, execute:
python -m scripts.run_trainingThis command will initialize training based on the parameters specified in config.yaml and save the trained model checkpoints to the specified save_path.
Deploy the model using Gradio to create an interactive web interface that allows users to upload images and view segmentation results in real time.
-
Run the Gradio App:
python -m gradio_app.app
-
Access the Interface: Open your browser and go to the URL provided in the terminal to start interacting with the model.
- Mask2Former is a state-of-the-art model designed for instance and semantic segmentation tasks. It leverages transformer-based architecture to provide accurate and robust segmentation results.
- In this project, Mask2Former was fine-tuned on the FoodSeg103 dataset to adapt its capabilities for food-related segmentation tasks.
- FoodSeg103 is a comprehensive semantic segmentation dataset containing 103 food categories. It provides diverse and annotated food images to train and evaluate segmentation models.
- Mean Intersection over Union (mIoU): Achieved a mIoU score of 4.21 on the validation set. The model's performance could be further improved with enhanced computing resources and longer fine-tuning periods.
- Licensing: This project is licensed under the MIT License. See the LICENSE file for more details.
For questions, feedback, or contributions, please open an issue or reach out to me.