-
Notifications
You must be signed in to change notification settings - Fork 17
Process Graph Building
As with the change of the openeo versions from 0.3.x to 0.4.x the process graph structure was completely overhauled. This mainly due to the fact that those graphs and much redundancy and it was not really possible to reuse intermediate results in different processes other than recalculating everything again. Also the change of the intentional data structure of multi-dimensional data cubes and the associated focus on reducing and aggregating of dimensions by using functions (callbacks) to control the reduction behavior pushed into this new leaner representation of process graphs introduced by the 0.4.x openeo API.
The R client supports this new process graph handling with versions >= 0.4.1 and the graph creation is only handled by the process graph builder which was introduced with version 0.2.1. The old functions process and collection are no longer available - collection was mainly removed since the openeo processes offer a load_collection function which would then be a process.
And version 0.6.0 brought some simplifications regarding process graph handling, "callback" creation and the redundant use of an openEO connection. In the prior versions the Graph object had all processes provided by the connected openEO service as functions attached. Starting with version 0.6.0 and future versions will divide this behavior, which means that the Graph or the now called user defined process will loose the attached functions, which will be available in the process collection (processes()). In terms of the callback sub graphs for reducer and aggregation processes, we added a new feature that transforms a function statement with the basic mathematical and logical operators into a process graph. The legacy way of creating callback graphs will be available until the final release at latest. Last, the connection to the latest connected openEO service will be stored in a package environment variable, so it is no longer necessary to pass the connection into each function which invokes a service interaction. However, it will be still possible to pass the connection explicitly, because one might want to work simultanously on multiple services.
During the development it became clear that we need an object-oriented programming style to realize the new process graph. The main reasons are that
- we have quite a lot of typed objects as argument values which relates to inheritance
- we do not want to lose overview (have private and public functions for objects)
- we want objects to behave similar, but in some cases differently (type checking, validation, serialization, etc.)
Those are some of the reasons why we modelled the process graph handling as R6 classes.
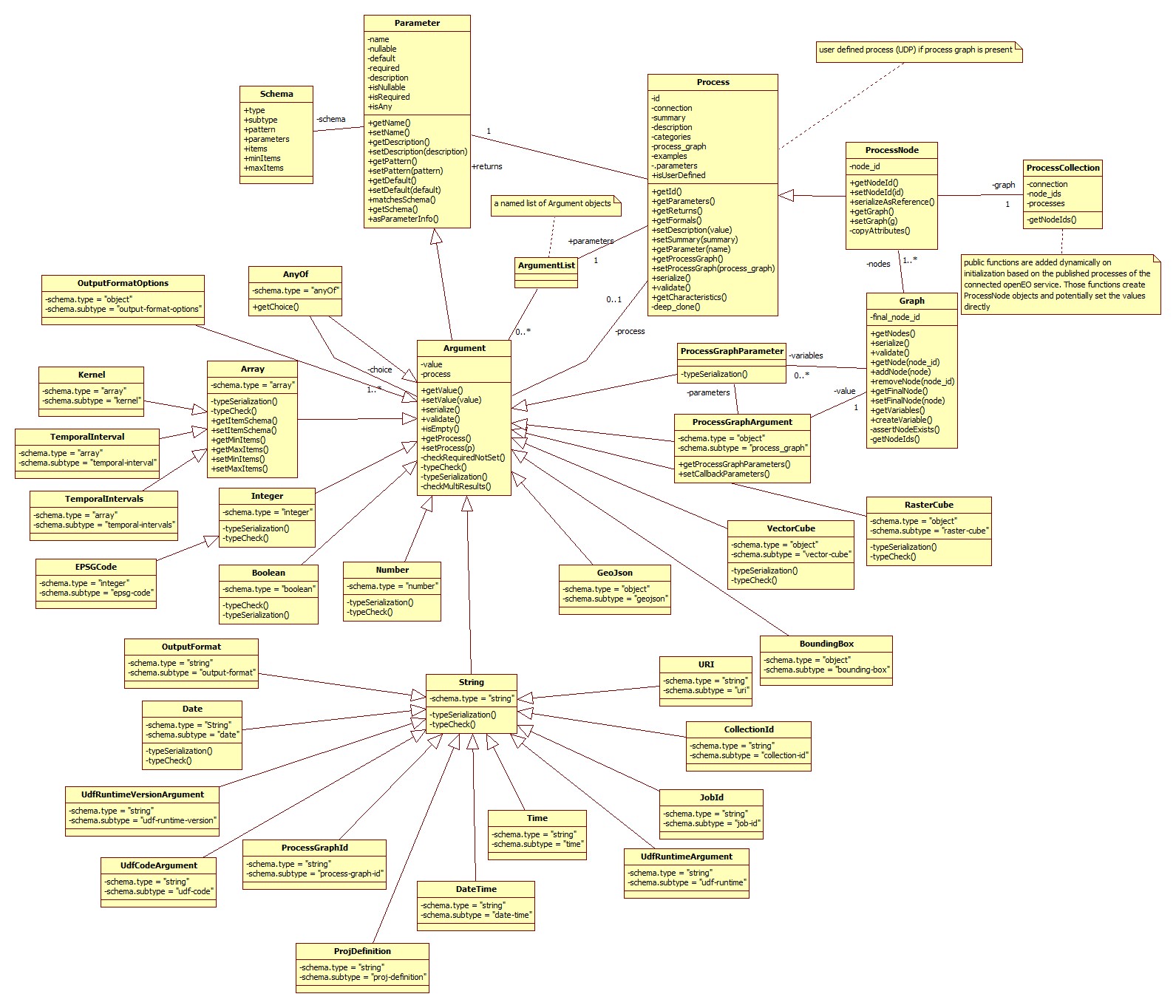
0.4.x process graph model (with selected argument classes)
The main take home message here is that a graph consists of process nodes which are derived from processes and extended with a node id and a special serialization. Parameters keep the information about the parameter of a process and the derived argument object keeps additionally the value which will be set by the user.
The process graph builder is created by querying the back-end for all available collections and processes. The information is received as JSON objects. From the collection response the collection ids are extracted and stored in a named list at graph$data. The lists names and the values are the same - the collection id. The processes response is parsed and translated into Process objects with Argument objects instead of Parameter objects. Those Process objects are registrated within functions on the Graph object (the function process_graph_builder() returns a Graph object). The parameter names of this process creator functions are the same as those specified by the back-end and if the values are set, then they are also set in the corresponding Argument object of the Process.
As mentioned before, the process_graph_builder() function is deprecated and will be replaced with processes(), which returns a ProcessCollection object. This object behaves similarly as the Graph object in terms of process graph building. All provided openEO processes will be attached here and it has also the $data object, that contains the names of all the collections available at the openEO service.
con = connect(host = "http://demo.openeo.org", version="0.4.2",user = "some_user",password = "some_pwd",login_type = "basic")
# < 0.6.0
graph = process_graph_builder(con=con)
class(graph)## [1] "Graph" "R6"
# >= 0.6.0
p = processes()
class(p)## [1] "ProcessCollection" "R6"
head(p$data,5)## $`AAFC/ACI`
## [1] "AAFC/ACI"
##
## $`ASTER/AST_L1T_003`
## [1] "ASTER/AST_L1T_003"
##
## $`AU/GA/AUSTRALIA_5M_DEM`
## [1] "AU/GA/AUSTRALIA_5M_DEM"
##
## $`CAS/IGSNRR/PML/V2`
## [1] "CAS/IGSNRR/PML/V2"
##
## $`CIESIN/GPWv411/GPW_Basic_Demographic_Characteristics`
## [1] "CIESIN/GPWv411/GPW_Basic_Demographic_Characteristics"
str(p$reduce())## Classes 'ProcessNode', 'Process', 'R6' <ProcessNode>
## Inherits from: <Process>
## Public:
## clone: function (deep = FALSE)
## getCharacteristics: function ()
## getFormals: function ()
## getGraph: function ()
## getId: function ()
## getNodeId: function ()
## getParameter: function (name)
## getParameterOrder: function ()
## getParameters: function ()
## getReturns: function ()
## initialize: function (node_id = character(), process, graph = NULL)
## parameters: active binding
## serialize: function ()
## serializeAsReference: function ()
## setDescription: function (value)
## setGraph: function (g)
## setNodeId: function (id)
## setParameter: function (name, value)
## validate: function ()
## Private:
## .parameters: list
## categories:
## connection: NULL
## copyAttributes: function (process)
## deep_clone: function (name, value)
## description:
## examples: list
## graph: ProcessCollection, R6
## id: reduce
## node_id: reduce_FCUWR5349C
## parameter_order: list
## returns: list
## summary: Reduce dimensions
In this example we are going to calculate the minimum EVI and apply a linear scale into the byte value domain to store and view the results as PNG. The file format PNG works only with bytes - otherwise the image would be pitch black (NDVI value range is between -1 and 1).
First, we prepare a graph with the process graph builder and then we define the data we want to load. In this case the load_collection function already offers parameter to subset the collection. If this back-end would not offer this, we would have to chain the individual functions afterwards - like filter_bbox, filter_time, etc.
p = processes()
data = p$load_collection(id = p$data$`COPERNICUS/S2`,
spatial_extent = list(west=-2.7634,south=43.0408,east=-1.121,north=43.8385),
temporal_extent = c("2018-04-30","2018-06-26"),
bands = c("B2","B4","B8"))Up next, perform the band arithmetics to calculate the EVI on Sentinel-2 data by reducing the spectral bands dimension with reduce.
spectral_reduce = p$reduce(data = data, dimension = "bands",reducer = function(x) {
B08 = x[3]
B04 = x[2]
B02 = x[1]
(2.5 * (B08 - B04)) / sum(B08, 6 * B04, -7.5 * B02, 1)
})In order to find the minimum value over time, we have to reduce the time dimension. This means we get rid of the time dimension by applying a callback function containing only the minimum function min(). It will work the way that the callback function is applied on every cell of the data cube with the shape of the array as before but without the temporal dimension.
Now, in the client we will just pass the openEO process min() as the reducer function.
temporal_reduce = p$reduce(data=spectral_reduce,dimension = "temporal", reducer = p$min)To apply the linear stretch we will litterally apply the function to each cell. This translates into modifying single values. Therefore we are going to create another callback with from an R function.
apply_linear_transform = p$apply(data=temporal_reduce,process = function(value) {
p$linear_scale_range(x = value,
inputMin = -1,
inputMax = 1,
outputMin = 0,
outputMax = 255)
})At the end we save the result as PNG file.
result = p$save_result(data=apply_linear_transform,format="PNG")Currently the ProcessNode objects form up the process graph. If you want to copy and paste the Process into another client you can use the coerce function for this.
graph = as(result,"Process")
graph## { "process_graph": {
## "load_collection_VPLSB6289N": {
## "process_id": "load_collection",
## "arguments": {
## "id": "COPERNICUS/S2",
## "spatial_extent": {
## "west": -2.7634,
## "south": 43.0408,
## "east": -1.121,
## "north": 43.8385
## },
## "temporal_extent": [
## "2018-04-30",
## "2018-06-26"
## ],
## "bands": [
## "B2",
## "B4",
## "B8"
## ]
## }
## },
## "reduce_VILVZ2625R": {
## "process_id": "reduce",
## "arguments": {
## "data": {
## "from_node": "load_collection_VPLSB6289N"
## },
## "reducer": {
## "callback": {
## "array_element_WQCIR9685L": {
## "process_id": "array_element",
## "arguments": {
## "data": {
## "from_argument": "data"
## },
## "index": 2,
## "return_nodata": false
## }
## },
## "array_element_GUBEJ3588S": {
## "process_id": "array_element",
## "arguments": {
## "data": {
## "from_argument": "data"
## },
## "index": 1,
## "return_nodata": false
## }
## },
## "subtract_DAPKL4482S": {
## "process_id": "subtract",
## "arguments": {
## "data": [
## {
## "from_node": "array_element_WQCIR9685L"
## },
## {
## "from_node": "array_element_GUBEJ3588S"
## }
## ],
## "ignore_nodata": true
## }
## },
## "multiply_EHBTP2482Q": {
## "process_id": "multiply",
## "arguments": {
## "data": [
## 2.5,
## {
## "from_node": "subtract_DAPKL4482S"
## }
## ],
## "ignore_nodata": true
## }
## },
## "multiply_QFRIN1234O": {
## "process_id": "multiply",
## "arguments": {
## "data": [
## 6,
## {
## "from_node": "array_element_GUBEJ3588S"
## }
## ],
## "ignore_nodata": true
## }
## },
## "array_element_AEQLL7642M": {
## "process_id": "array_element",
## "arguments": {
## "data": {
## "from_argument": "data"
## },
## "index": 0,
## "return_nodata": false
## }
## },
## "multiply_OMXZF8184V": {
## "process_id": "multiply",
## "arguments": {
## "data": [
## -7.5,
## {
## "from_node": "array_element_AEQLL7642M"
## }
## ],
## "ignore_nodata": true
## }
## },
## "sum_FAZBT4933X": {
## "process_id": "sum",
## "arguments": {
## "data": [
## {
## "from_node": "array_element_WQCIR9685L"
## },
## {
## "from_node": "multiply_QFRIN1234O"
## },
## {
## "from_node": "multiply_OMXZF8184V"
## },
## 1
## ]
## }
## },
## "divide_NURWN8097L": {
## "process_id": "divide",
## "arguments": {
## "data": [
## {
## "from_node": "multiply_EHBTP2482Q"
## },
## {
## "from_node": "sum_FAZBT4933X"
## }
## ],
## "ignore_nodata": true
## },
## "result": true
## }
## }
## },
## "dimension": "bands"
## }
## },
## "reduce_LAIKH4472Y": {
## "process_id": "reduce",
## "arguments": {
## "data": {
## "from_node": "reduce_VILVZ2625R"
## },
## "reducer": {
## "callback": {
## "min_FBYYC9185K": {
## "process_id": "min",
## "arguments": {
## "data": {
## "from_argument": "data"
## }
## },
## "result": true
## }
## }
## },
## "dimension": "temporal"
## }
## },
## "apply_PHOBU4289T": {
## "process_id": "apply",
## "arguments": {
## "data": {
## "from_node": "reduce_LAIKH4472Y"
## },
## "process": {
## "callback": {
## "linear_scale_range_PSVPA3854R": {
## "process_id": "linear_scale_range",
## "arguments": {
## "x": {
## "from_argument": "x"
## },
## "inputMin": -1,
## "inputMax": 1,
## "outputMin": 0,
## "outputMax": 255
## },
## "result": true
## }
## }
## }
## }
## },
## "save_result_UVOTZ9626P": {
## "process_id": "save_result",
## "arguments": {
## "data": {
## "from_node": "apply_PHOBU4289T"
## },
## "format": "PNG",
## "options": {}
## },
## "result": true
## }
## }}
A callback is applied as a function within a reducer or an apply function. In case of a reducer function for a dimension, this means, that a particular dimension is reduced. The remaining dimensions are untouced but for the choosen dimension a resolve strategy has to be developed by the callback. Usually the input of a callback is an array of values. For example, consider a spatio-temporal data cube with some spectral bands. If we want to perform band arithmetics, then we need to reduce the spectral dimensions. This translate to the situation that we need to state a function that maps the band dimension values to a single value for each spatial and temporal dimension value.
In a technical sense callbacks are nothing else than sub process graphs. However, callbacks differ in terms of data injection. On the back-end data is pushed into the corresponding entry process that claims to receive a callback parameter (or in the R client terminology a callback value), whereas the usualprocess graph loads loads the data via an dedicated process like load_collection and the process results are passed on. Those callback parameter are usually vectors or two single values.
The following code snippet shows data injected to the main process graph, e.g. loading a data collection.
jsonlite::toJSON(data$serialize(),auto_unbox = TRUE,pretty=TRUE)## {
## "process_id": "load_collection",
## "arguments": {
## "id": "COPERNICUS/S2",
## "spatial_extent": {
## "west": -2.7634,
## "south": 43.0408,
## "east": -1.121,
## "north": 43.8385
## },
## "temporal_extent": [
## "2018-04-30",
## "2018-06-26"
## ],
## "bands": [
## "B2",
## "B4",
## "B8"
## ]
## }
## }
Since it is mostly of interest to distinguish binary operators and operators on arrays the number of variables in the signature is taken into account when chosing the correct callback if a particular parameter allows multiple callbacks with different callback values. The next two callback-value visualizations show examples of the two types of callback parameter - first the array input, second two values as input.
## $data
## {
## "from_argument": "data"
## }
and
## $x
## {
## "from_argument": "x"
## }
## $y
## {
## "from_argument": "y"
## }
The callback parameter can be thought of the signature of the callback function - meaning if we have a single data object as callback parameter we can assume in almost all cases that the data type is an array / vector.
The openeo package will do the transformation into a valid process graph internally. This is done since version v0.6.0 via reading the signature of a callback function.
The goal is that you as a user should not care about the callback parameter. For you it only depends on stating a function with eiter one or two parameters.
The p$data field contains the data available to a graph. In the case where the graph is build regularly with the processes() the data corresponds with the available collections on the back-end. In callback graphs data will be filled by the arguments callback parameter.
Processes are received from the back-end in a particular format. It contains general information about the process, but more important information about its parameter, e.g. name, order and type/format.
jsonlite::toJSON(describe_process(id = "reduce"),force=TRUE,auto_unbox = TRUE,pretty = TRUE)## {
## "id": "reduce",
## "summary": "Reduce dimensions",
## "description": "Applies a reducer to a data cube dimension by collapsing all the input values along the specified dimension into an output value computed by the reducer.\n\nThe reducer must be a callable process (or a set of processes as process graph) that accepts by default array as input. The reducer must compute a single or multiple return values of the same type as the input values were. Multiple values must be wrapped in an array. An example for a process returning a single value is ``median()``. In this case the specified dimension would be removed. If a callback such as ``extrema()`` returns multiple values, a new dimension with the specified name in `target_dimension` is created (see the description of the parameter for more information).\n\nA special case is that the reducer can be set to `null`, which is the default if no reducer is specified. It acts as a no-operation reducer so that the remaining value is treated like a reduction result and the dimension gets dropped. This only works on dimensions with a single dimension value left (e.g. after filtering for a single band), otherwise the process fails with a `TooManyDimensionValues` error.\n\nNominal values can be reduced too, but need to be mapped. For example date strings to numeric timestamps since 1970 etc.",
## "categories": [
## "cubes",
## "reducer"
## ],
## "gee:custom": true,
## "parameter_order": [
## "data",
## "reducer",
## "dimension"
## ],
## "parameters": {
## "data": {
## "description": "A data cube.",
## "schema": {
## "type": "object",
## "format": "raster-cube"
## },
## "required": true
## },
## "reducer": {
## "description": "A reducer to be applied on the specified dimension (see the process description for more details).",
## "schema": {
## "anyOf": [
## {
## "title": "Unary behaviour",
## "description": "Passes an array to the reducer.",
## "type": "object",
## "format": "callback",
## "parameters": {
## "data": {
## "description": "An array with elements of any data type.",
## "type": "array",
## "items": {
## "description": "Any data type."
## }
## }
## }
## },
## {
## "title": "No operation behaviour",
## "description": "Specifying `null` works only on dimensions with a single dimension value left. In this case the remaining value is treated like a reduction result and the dimension gets dropped.",
## "type": "null"
## }
## ],
## "default": {}
## }
## },
## "dimension": {
## "description": "The dimension over which to reduce. Fails with a `DimensionNotAvailable` error if the specified dimension does not exist.\n\n**Remarks:**\n\n* The default dimensions a data cube provides are described in the collection's metadata field `cube:dimensions`.\n* There could be multiple spatial dimensions such as `x`, `y` or `z`.\n* For multi-spectral imagery there is usually a separate dimension of type `bands` for the bands.",
## "schema": {
## "type": "string"
## },
## "required": true
## }
## },
## "returns": {
## "description": "A data cube with the newly computed values. The number of dimensions is reduced for callbacks returning a single value or doesn't change if the callback returns multiple values. The resolution and cardinality are the same as for the original data cube.",
## "schema": {
## "type": "object",
## "format": "raster-cube"
## }
## },
## "exceptions": {
## "TooManyDimensionValues": {
## "message": "The number of dimension values exceeds one, which requires a reducer."
## },
## "DimensionNotAvailable": {
## "message": "A dimension with the specified name does not exist."
## }
## }
## }
All those information are parsed and interpreted in order to create a suitable process representation in R. Those processes will be registered as constructor functions on the graph. This means by calling the function a parsed process will be copied, turned into a process node and added to the process graph.
p$reduce## function (data = NA, reducer = NA, dimension = NA)
## {
## exec_process = private$processes[[37]]$clone(deep = TRUE)
## node_id = .randomNodeId(exec_process$getId(), sep = "_")
## while (node_id %in% private$getNodeIds()) {
## node_id = .randomNodeId(exec_process$getId(), sep = "_")
## }
## private$node_ids = c(private$node_ids, node_id)
## arguments = exec_process$parameters
## this_param_names = names(formals())
## this_arguments = lapply(this_param_names, function(param) get(param))
## names(this_arguments) = this_param_names
## node = ProcessNode$new(node_id = node_id, process = exec_process,
## graph = self)
## lapply(names(this_arguments), function(param_name, arguments) {
## call_arg = this_arguments[[param_name]]
## arguments[[param_name]]$setValue(call_arg)
## }, arguments = arguments)
## return(node)
## }
## <bytecode: 0x0000000024d430f0>
## <environment: 0x0000000023ab4cc0>
Now if we want to apply the graph easier to different data sets, we can introduce variables and store this parametrized graph to the back-end. If the back-end offers the process to load process graphs we then can execute the parametrized graph by filling in the variables. If we don't fill in the variable values, the default value is assumed and if the default is not set and the graph shall be executed on the back-end it will result in an error at the back-end.
We can either use the variables directly during the graph creation as argument values or we can replace some of the values afterwards. Since the first part is quite obvious to do, we will give an example where we create four variables - for the collections and the three bands and replace them at the afore created process graph of the Example section.
collection_id = create_variable(id = "collection",description = "The collection id of a cube containing at least a red and a near infrared band",type = "string",default = "COPERNICUS/S2")
red_band = create_variable(id = "red_band",description="The band name of the red band",type="string",default="B4")
nir_band = create_variable(id = "nir_band",description="The band name of the NIR band",type="string",default="B8")
blue_band = create_variable(id = "blue_band",description="The band name of the blue band",type="string",default="B2")
data$parameters$id$setValue(collection_id)
data$parameters$bands$setValue(list(blue_band,red_band,nir_band))
sapply(variables(result),function(v)v$getName())data## {
## "process_id": "load_collection",
## "arguments": {
## "id": {
## "variable_id": "collection",
## "description": "The collection id of a cube containing at least a red and a near infrared band",
## "type": "string",
## "default": "COPERNICUS/S2"
## },
## "spatial_extent": {
## "west": -2.7634,
## "south": 43.0408,
## "east": -1.121,
## "north": 43.8385
## },
## "temporal_extent": [
## "2018-04-30",
## "2018-06-26"
## ],
## "bands": [
## {
## "variable_id": "blue_band",
## "description": "The band name of the blue band",
## "type": "string",
## "default": "B2"
## },
## {
## "variable_id": "red_band",
## "description": "The band name of the red band",
## "type": "string",
## "default": "B4"
## },
## {
## "variable_id": "nir_band",
## "description": "The band name of the NIR band",
## "type": "string",
## "default": "B8"
## }
## ]
## }
## }