-
Notifications
You must be signed in to change notification settings - Fork 0
Background
Dengue virus (DENV) is a mosquito-borne virus that belongs to the Flavivirus genus of the Flaviviridae family. It is the causative agent of dengue fever, a disease that is endemic in tropical and subtropical regions around the world. Dengue fever can range from a mild illness to severe, life-threatening conditions such as dengue hemorrhagic fever (DHF) or dengue shock syndrome (DSS).
DENV represents the most significant mosquito-borne viral threat to global public health, causing over 100 million infections annually. Dengue's incidence continues to rise, with large outbreaks recorded across endemic tropical regions in 2023, alongside sustained local transmission in traditionally non-endemic areas such as Florida, USA, and Italy. As DENV continues to spread, it is critical to monitor its genetic evolution to better understand its transmission patterns on local, regional, and global scales.
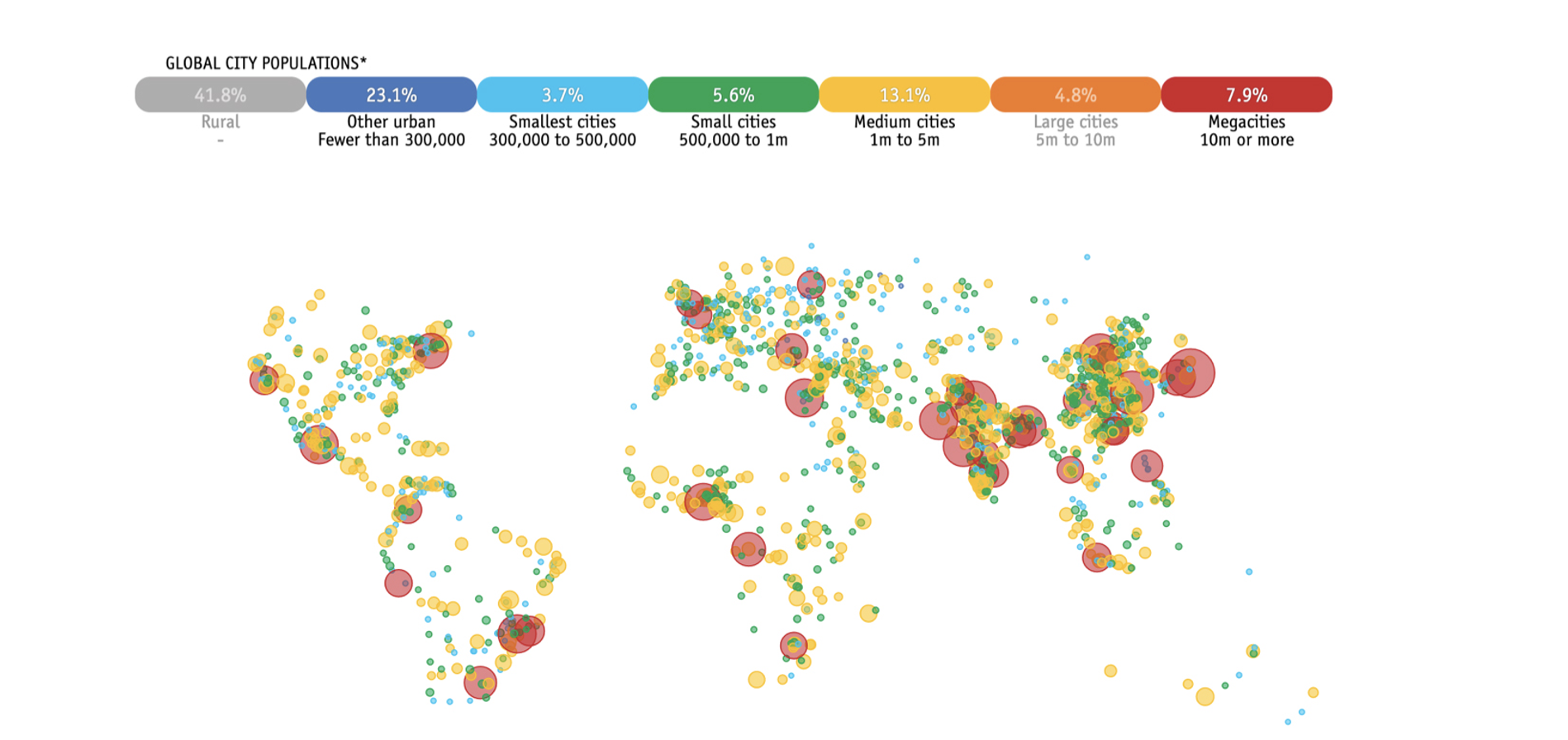
Projected urbanisation in 2027. Urbanisation can potentially impact dengue transmission, creating conditions that favour spread of mosquito-borne viruses.
Comparative genomics plays a crucial role in understanding the genetic diversity and evolution of Dengue virus (DENV). By analyzing the genomes of different DENV strains, researchers can identify genetic variations that influence important viral characteristics, such as transmissibility, virulence, and immune evasion. By integrating genomic data with epidemiological and clinical information, comparative genomics provides insights into the dynamics of DENV transmission, guiding strategies for vaccine development, vector control, and outbreak preparedness.
Dengue-GLUE is deigned to enable DENV researchers to carry out these large-scale analyses efficiently, facilitating the comparison of viral sequences and helping to standardize genomic data across different studies and regions.
DENV circulates in humans via four serotypes (DENV-1 to DENV-4), believed to have emerged from at least four distinct spillover events from a sylvatic (non-human primate) cycle centuries ago. These serotypes and genotypes have long formed the foundation of studies into DENV’s natural history, phenotypic diversity, and transmission dynamics. However, with advancements in global sequencing and its integration into public health, there is a growing need for greater granularity in DENV diversity classification, so that distinct evolutionary lineages within known genotypes can be identified.
Establishing a precise, standardized language for tracking DENV at various scales is essential for genomic epidemiology. This terminology should be accessible to both genomic researchers and public health practitioners, ensuring effective communication and response strategies.
Recently, a new dengue virus lineage system was developed that splits up the current genotypes into major and minor lineages to provide additional spatiotemporal resolution and a common language to discuss important genomic diversity. Dengue-GLUE implements a likelihood-based genotyping protocol that can be used to assign DENV sequences - ranging from full genomes to sub-genomic fragments - to the lineages defined in the newly established nomenclature system.
Sequences in the NCBI-Dengue-GLUE extension project, which incorporates all published DENV sequences, have been classified using this approach.