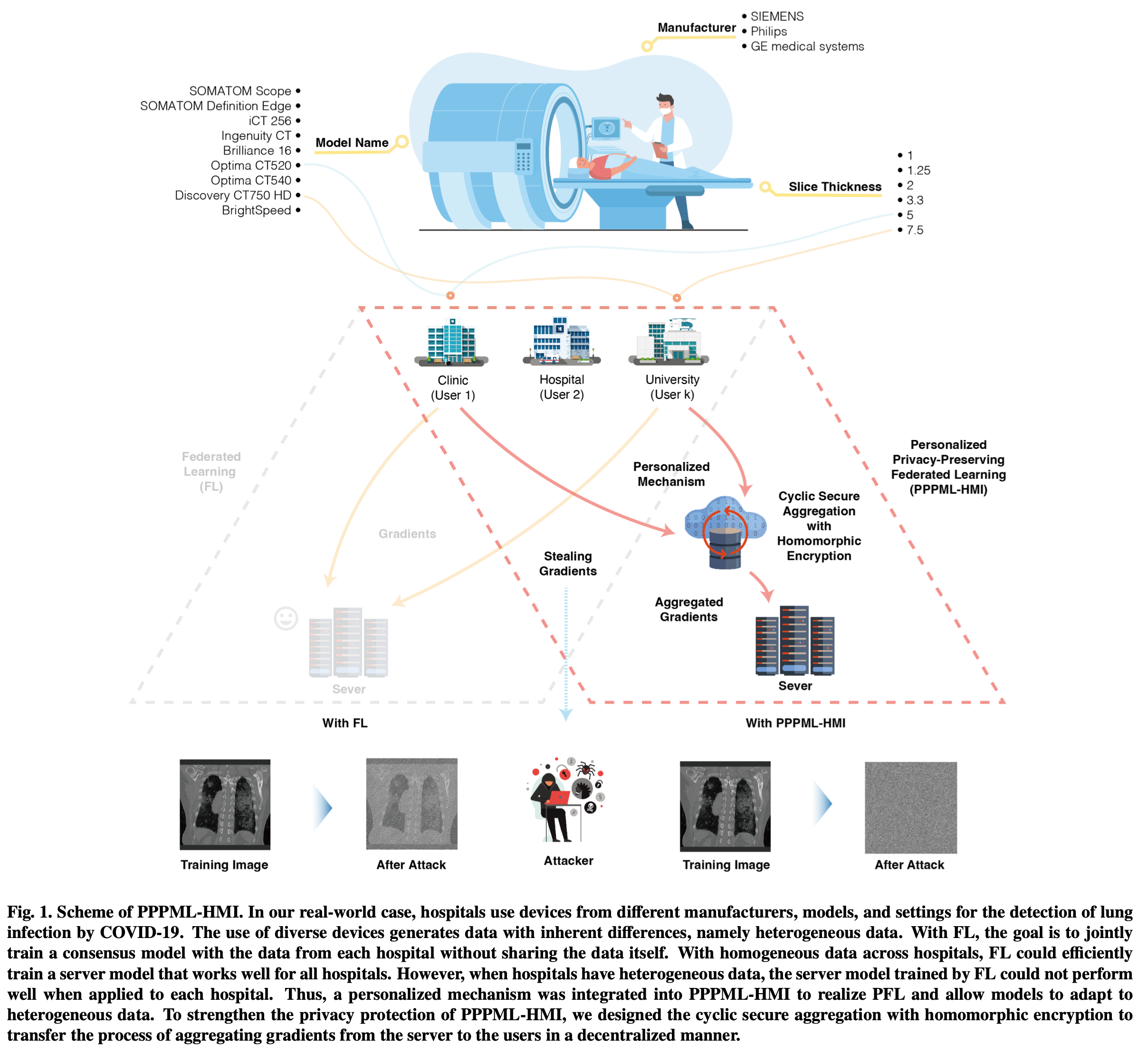
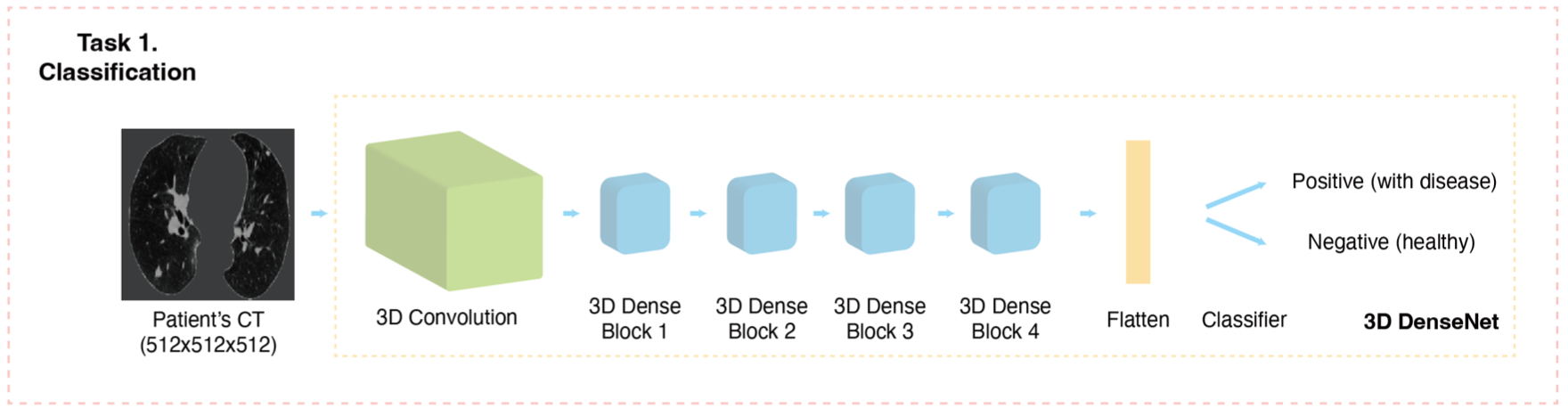
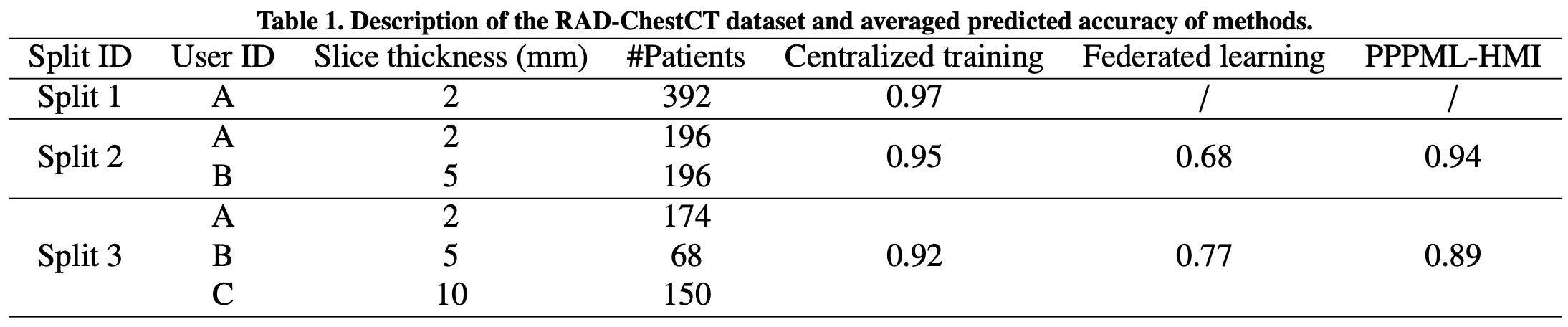
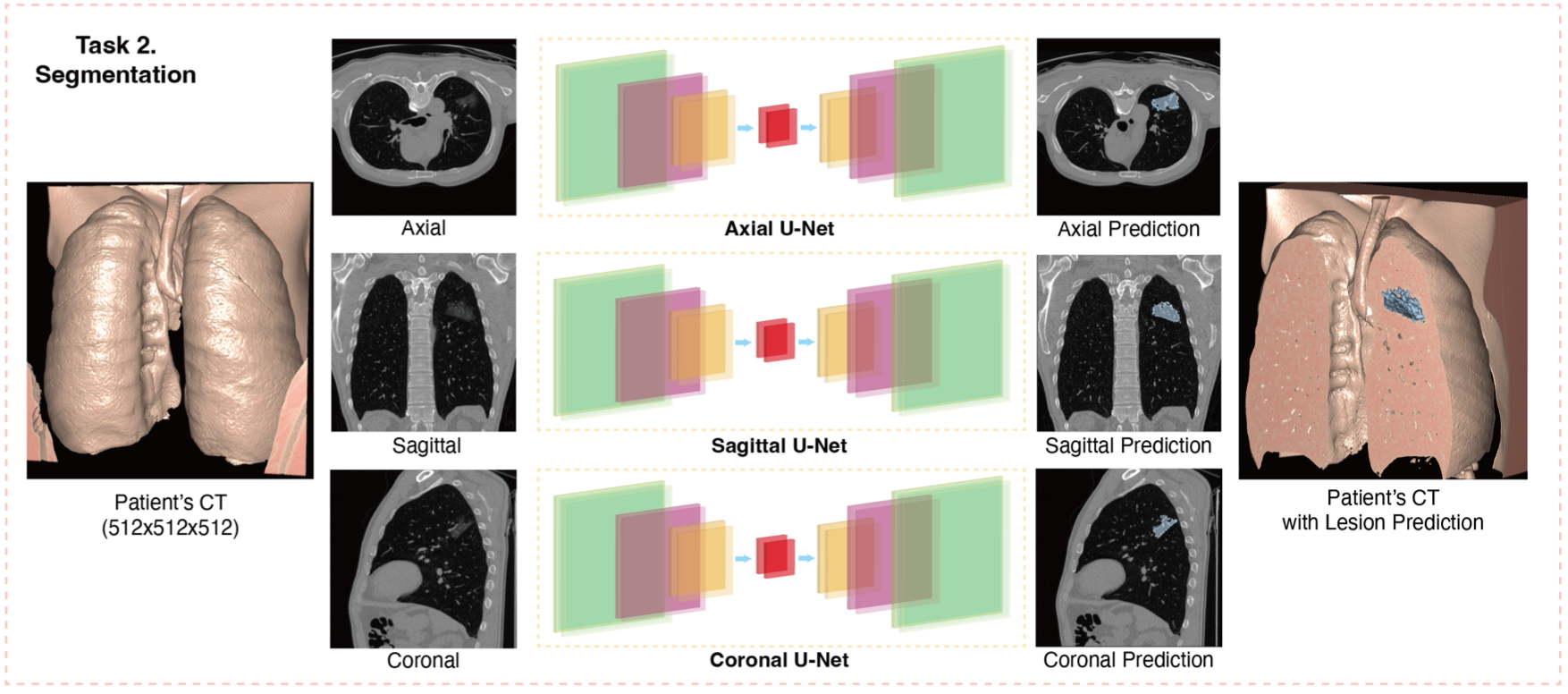
This repository contains the implementation of applications with PPPML-HMI from:
Juexiao Zhou, et al. "Personalized and privacy-preserving federated heterogeneous medical image analysis with PPPML-HMI"
conda env create -f environment.ymlFor the classification task, we used the publicly available RAD-ChestCT Dataset Draelos et al. (2021, 2020) at https://zenodo.org/record/6406114#.YxYo6S_qFf0. In order to access the data, users need to request access to files in this Zenodo. The decision whether to grant/deny access is solely under the responsibility of the record owner.
For the segmentation task, the data from partner hospitals are available upon request.
Following two cases can be used to reproduce the results in our paper only after obtaining data permissions as specified in Dataset.
for group in FedAvg PerAvg CSAPerAvg
do
for exp in new_c2_split1 new_c3_split1
do
python TrainPPPML.py \
--algorithm ${group} \
--num_global_iters 10 \
--local_epochs 2 \
--batch_size 4 \
--learning_rate 1e-2 \
--beta 0.001 \
--gpu 1 \
--dataset RAD \
--optimizer Adam \
--times 1 \
--exp ${exp} \
--onestepupdate 2 \
--HE 0 \
--RAD_checkpoint_root ./result/RAD_${group}
done
donefor group in FedAvg PerAvg CSAPerAvg
do
for direction in X Y Z
do
for test_id in {0..4}
do
python TrainPPPML.py \
--algorithm FedAvg \
--num_global_iters 10 \
--local_epochs 20 \
--batch_size 16 \
--learning_rate 1e-4 \
--beta 0.001 \
--gpu 0 \
--dataset COVID \
--test_ids ${test_id} \
--optimizer Adam \
--times 1 \
--directions ${direction} \
--covid_data_root data/raw_data/device_patients_oneletter \
--covid_checkpoint_root result/fedavg1 \
--covid_client_labels 'A B C D E'
done
done
doneYou may need to modify some functions in TrainPPPML.py to work on your own case. Required modifications includes:
main
data_loader, eg: read_user_data_COVID(index, args)
train, eg: User.train_COVID(self)
one-step finetune, eg: User.train_COVID_onestep(self)
test, eg: User.test_COVID(self)
evaluate, eg: evaluate_COVID(model, test_loader, params, device)
If you use our work in your research, please cite our paper:
Personalized and privacy-preserving federated heterogeneous medical image analysis with PPPML-HMI
Juexiao Zhou, Longxi Zhou, Di Wang, Xiaopeng Xu, Haoyang Li, Yuetan Chu, Wenkai Han and Xin Gao