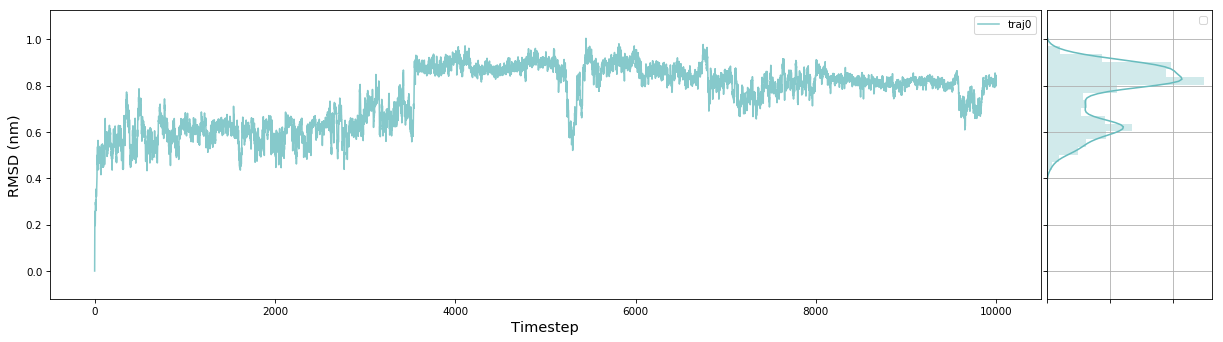
MSMExplorer is a Python visualization library for statistical models of biomolecular dynamics. It provides a high-level interface for drawing attractive statistical graphics with MSMBuilder.
Online documentation is available here. It includes IPython notebooks, detailed API documentation, and other useful info.
There are docs for the development version here. These should correspond with the github master branch.
from msmbuilder.example_datasets import FsPeptide
from msmbuilder.featurizer import RMSDFeaturizer
import msmexplorer as msme
# Load Fs Peptide Data
traj = FsPeptide().get().trajectories[0]
# Calculate RMSD
featurizer = RMSDFeaturizer(reference_traj=traj[0])
rmsd = featurizer.partial_transform(traj).flatten()
# Plot Trace
msme.plot_trace(rmsd, label='traj0', xlabel='Timestep', ylabel='RMSD (nm)')The documentation has an example gallery with short scripts showing how to use different parts of the package.
- Python 3.4+
The latest versions of the following packages are required:
The preferred installation mechanism for msmexplorer is with conda:
$ conda install -c omnia msmexplorerIf you don't have conda, or are new to scientific python, we recommend that you download the Anaconda scientific python distribution.
To install from PyPI, just do:
pip install msmexplorer
You may instead want to use the development version from Github, by running
pip install git+git://github.com/msmexplorer/msmexplorer.git#egg=msmexplorer
All development happens here, on Github.
If you're interested in contributing to MSMExplorer, please refer to our Contributing guide.
Please submit any bugs or questions to the Github issue tracker.
Released under a MIT license
@article{msmexplorer,
doi = {10.21105/joss.00188},
url = {https://doi.org/10.21105%2Fjoss.00188},
year = {2017},
month = {apr},
publisher = {The Open Journal},
volume = {2},
number = {12},
author = {Carlos X. Hern{\'{a}}ndez and Matthew P. Harrigan and Mohammad M. Sultan and Vijay S. Pande},
title = {{MSMExplorer}: Data Visualizations for Biomolecular Dynamics},
journal = {The Journal of Open Source Software}
}