-
Notifications
You must be signed in to change notification settings - Fork 3
04 RDF MassBank Resource Module
For an example of a MassBank record, please look at https://github.com/sneumann/SemanticMetabolomics/wiki/APPENDIX-02-MassBank-record-example
The current state of RDF MassBank:
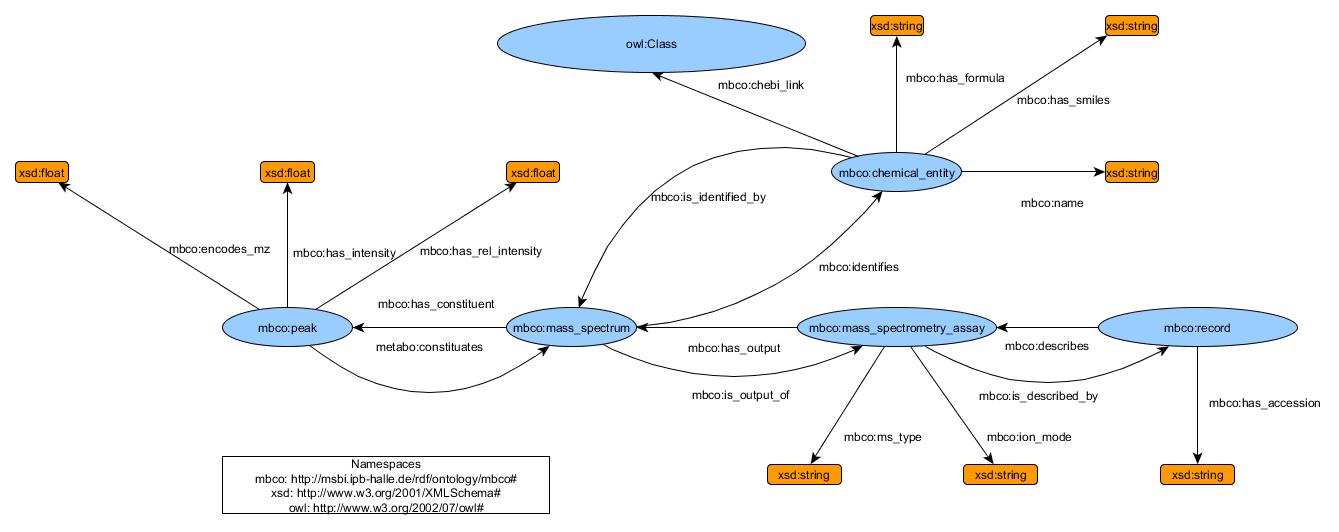
This model is described in RDF in our Git at https://github.com/sneumann/SemanticMetabolomics/blob/master/ontology/mbco.ttl (Terse Triple Language, the turtle RDF syntax). It does not contain all possible information described in Massbank records, but the model itself is set up so that predicates, classes and literals can easily be added in case the record specification changes.
In detail, this model consists of 5 classes and 17 predicates. Please note that the reference to "owl:class" is a reference to ChEBI. The classes are set up as a representation of the different segments that are contained in a massbank record according to the specification.
Every record itself is an instantiation of the class: mbco:record, as every record is a self-contained set of information. An example RDF-URL of a record is http://msbi.ipb-halle.de/rdf/ontologydata/record:EA029251, it is uniquely identifiable by its accession number.
The next connected class in the model is mbco:mass_spectrometry_assay, which is connected to mbco:record by the predicates mbco:describes and the intuitive inversion of it, mbco:is_described_by. The assay is intended to describe technical aspects of the experiment and by extension the concept of the experiment itself, that was conducted to produce the information for the record. This semantic granularity enabled us to follow formal ontological best practices along ontological realism. There currently are predicates (referring to literal strings) to note the ionization mode (mbco:ion_mode) and the ms type (mbco:ms_type). Since the mass spectrometry assay is unique to every experiment (and therefore record), it is identifiable by the record accession. Example:
http://msbi.ipb-halle.de/rdf/ontologydata/mass_spectrometry_assay:EA029251
A mass spectrometry assay (the process) results in a mass spectrum, so the next predicates mbco:has_output and mbco:is_output_of are logically a connection to the mbco:mass_spectrum class. This class is a necessary blank node (an abstract node that only exists to connect classes) to tie the experimental setup, the compound used for the experiment (mbco:chemical_entity) and the data resulting from the experiment (mbco:peak) together. Since these three things are clearly defined and separated in reality, they should (by OBO Foundry recommendation) also be defined as separate classes in the RDF model. By the real-world context in which these classes are used, an internal node is the only way to connect these classes while retaining this context in abstract form. A mass spectrum is unique to the experiment, which means that it is identifiable by the record accession.
Example:
http://msbi.ipb-halle.de/rdf/ontologydata/mass_spectrum:EA029251
The results of a mass spectrometry experiment are peaks, connected by the predicates mbco:constituates, mbco:has_constituent and represented by the class mbco:peak. Entities of this class
- encode the measured mass-to-charge (m/z) ratio (
mbco:encodes_mz) - contain the intensity that was measured for that ratio (
mbco:has_intensity) - contain the relative intensity of this peak compared to the highest peak in the mass spectrum (
mbco:has_relative_intensity) Peaks are identifiable by the record accession number concatenated with the m/z of the peak. Example: http://msbi.ipb-halle.de/rdf/ontologydata/peak:EA029251_185.9521
The chemical compound that was contained in the sample that generated the spectrum is represented by the class mbco:chemical_entity. Although storing information about the chemical compound in this model might seem superfluous (in light of this being a linked data project) it is necessary, because it is very likely in future cases that there is only limited information present for certain substances, i.e. not enough to identify a substance uniquely. There also already exist cases where very unusual substances were used in mass spectrometry experiments and these substances were not present in ChEBI at the time, so it is necessary to include this information in the MBCO to guarantee completeness of information.
In its current state, this class contains the possibility to note three literal types of information:
- the molecular formula of the substance (
mbco:has_formula) - the name of the substance (
mbco:name) - the smiles formula of the substance (
mbco:has_smiles) Substances are identifiable by the record accession number concatenated with the inchikey of the substance. Example: http://msbi.ipb-halle.de/rdf/ontologydata/chemical_entity:EA029251_XMTQQYYKAHVGBJ-UHFFFAOYSA-N
The final predicate in the model, mbco:chebi_link is the predicate that links the mbco database to the ChEBI database. In ChEBI, every substance constitutes its own class, henceforth the class node owl:class.