-
Notifications
You must be signed in to change notification settings - Fork 223
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
adding BetaBernoulli distribution with LogScore #132
Draft
guyko81
wants to merge
4
commits into
stanfordmlgroup:master
Choose a base branch
from
guyko81:beta-bernoulli
base: master
Could not load branches
Branch not found: {{ refName }}
Loading
Could not load tags
Nothing to show
Loading
Are you sure you want to change the base?
Some commits from the old base branch may be removed from the timeline,
and old review comments may become outdated.
Draft
Changes from all commits
Commits
Show all changes
4 commits
Select commit
Hold shift + click to select a range
File filter
Filter by extension
Conversations
Failed to load comments.
Loading
Jump to
Jump to file
Failed to load files.
Loading
Diff view
Diff view
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,180 @@ | ||
| from scipy.stats import betabinom as dist | ||
| import numpy as np | ||
| from ngboost.distns.distn import RegressionDistn | ||
| from ngboost.scores import LogScore | ||
| from scipy.special import digamma, polygamma | ||
| from array import array | ||
| import sys | ||
|
|
||
| class BetaBernoulliLogScore(LogScore): | ||
| def score(self, Y): | ||
| return -self.dist.logpmf(Y) | ||
|
|
||
| def d_alpha(self, Y): | ||
| return self.alpha * ( | ||
| digamma(self.alpha + self.beta) | ||
| + digamma(Y + self.alpha) | ||
| - digamma(self.alpha + self.beta + 1) | ||
| - digamma(self.alpha) | ||
| ) | ||
|
|
||
| def d_beta(self, Y): | ||
| return self.beta * ( | ||
| digamma(self.alpha + self.beta) | ||
| + digamma(-Y + self.beta + 1) | ||
| - digamma(self.alpha + self.beta + 1) | ||
| - digamma(self.beta) | ||
| ) | ||
|
|
||
| def d_alpha_alpha(self, Y): | ||
| return ( | ||
| (polygamma(1, Y + self.alpha) + | ||
| polygamma(1, self.alpha + self.beta) - | ||
| polygamma(1, self.alpha + self.beta + 1) - | ||
| polygamma(1, self.alpha) | ||
| ) * self.alpha**2 + self.d_alpha(Y) | ||
| ) | ||
|
|
||
| def d_alpha_beta(self, Y): | ||
| return ( | ||
| self.alpha * self.beta * ( | ||
| polygamma(1, self.alpha + self.beta) - | ||
| polygamma(1, self.alpha + self.beta + 1) | ||
| ) | ||
| ) | ||
|
|
||
| def d_beta_beta(self, Y): | ||
| return ( | ||
| (polygamma(1, -Y + self.beta + 1) + | ||
| polygamma(1, self.alpha + self.beta) - | ||
| polygamma(1, self.alpha + self.beta + 1) - | ||
| polygamma(1, self.beta) | ||
| ) * self.beta**2 + self.d_beta(Y) | ||
| ) | ||
| def d_score(self, Y): | ||
| D = np.zeros( | ||
| (len(Y), 2) | ||
| ) # first col is dS/d(log(α)), second col is dS/d(log(β)) | ||
| D[:, 0] = -self.d_alpha(Y) | ||
| D[:, 1] = -self.d_beta(Y) | ||
| return D | ||
|
|
||
| # Variance | ||
| def metric(self): | ||
| FI = np.zeros((self.alpha.shape[0], 2, 2)) | ||
| FI[:, 0, 0] = ( | ||
| self.d_alpha(0)*self.d_alpha(0)*self.dist.pmf(0) + | ||
| self.d_alpha(1)*self.d_alpha(1)*self.dist.pmf(1) | ||
| ) | ||
| # FI[:, 1, 0] = ( | ||
| # self.d_alpha(0)*self.d_beta(0)*self.dist.pmf(0) + | ||
| # self.d_alpha(1)*self.d_beta(1)*self.dist.pmf(1) | ||
| # ) | ||
| # FI[:, 0, 1] = ( | ||
| # self.d_alpha(0)*self.d_beta(0)*self.dist.pmf(0) + | ||
| # self.d_alpha(1)*self.d_beta(1)*self.dist.pmf(1) | ||
| # ) | ||
| FI[:, 1, 1] = ( | ||
| self.d_beta(0)*self.d_beta(0)*self.dist.pmf(0) + | ||
| self.d_beta(1)*self.d_beta(1)*self.dist.pmf(1) | ||
| ) | ||
| return FI | ||
|
|
||
| # Hessian | ||
| # def metric(self): | ||
| # FI = np.zeros((self.alpha.shape[0], 2, 2)) | ||
| # FI[:, 0, 0] = self.d_alpha_alpha(0) * self.dist.pmf(0) + self.d_alpha_alpha(1) * self.dist.pmf(1) | ||
| # # FI[:, 1, 0] = self.d_alpha_beta(0) * self.dist.pmf(0) + self.d_alpha_beta(1) * self.dist.pmf(1) | ||
| # # FI[:, 0, 1] = self.d_alpha_beta(0) * self.dist.pmf(0) + self.d_alpha_beta(1) * self.dist.pmf(1) | ||
| # FI[:, 1, 1] = self.d_beta_beta(0) * self.dist.pmf(0) + self.d_beta_beta(1) * self.dist.pmf(1) | ||
| # return FI | ||
|
|
||
| class BetaBernoulli(RegressionDistn): | ||
|
|
||
| n_params = 2 | ||
| scores = [BetaBernoulliLogScore] | ||
|
|
||
| def __init__(self, params): | ||
| # save the parameters | ||
| self._params = params | ||
|
|
||
| # create other objects that will be useful later | ||
| self.log_alpha = params[0] | ||
| self.log_beta = params[1] | ||
| self.alpha = np.exp(self.log_alpha) | ||
| self.beta = np.exp(self.log_beta) | ||
| self.dist = dist(n=1, a=self.alpha, b=self.beta) | ||
|
|
||
| def fit(Y): | ||
| def fit_alpha_beta_py(success, alpha0=1.5, beta0=5, niter=1000): | ||
| # based on https://github.com/lfiaschi/fastbetabino/blob/master/fastbetabino.pyx | ||
|
|
||
| # This optimisation works for Beta-Binomial distribution in general. | ||
| # For Beta-Bernoulli it's simplified by fixing the trials to 1. | ||
| trials = np.ones_like(Y) | ||
|
|
||
| alpha_old = alpha0 | ||
| beta_old = beta0 | ||
|
|
||
| for it in range(niter): | ||
|
|
||
| alpha = ( | ||
| alpha_old | ||
| * ( | ||
| sum( | ||
| digamma(c + alpha_old) - digamma(alpha_old) | ||
| for c, i in zip(success, trials) | ||
| ) | ||
| ) | ||
| / ( | ||
| sum( | ||
| digamma(i + alpha_old + beta_old) | ||
| - digamma(alpha_old + beta_old) | ||
| for c, i in zip(success, trials) | ||
| ) | ||
| ) | ||
| ) | ||
|
|
||
| beta = ( | ||
| beta_old | ||
| * ( | ||
| sum( | ||
| digamma(i - c + beta_old) - digamma(beta_old) | ||
| for c, i in zip(success, trials) | ||
| ) | ||
| ) | ||
| / ( | ||
| sum( | ||
| digamma(i + alpha_old + beta_old) | ||
| - digamma(alpha_old + beta_old) | ||
| for c, i in zip(success, trials) | ||
| ) | ||
| ) | ||
| ) | ||
|
|
||
| # print('alpha {} | {} beta {} | {}'.format(alpha,alpha_old,beta,beta_old)) | ||
| sys.stdout.flush() | ||
|
|
||
| if np.abs(alpha - alpha_old) and np.abs(beta - beta_old) < 1e-10: | ||
| # print('early stop') | ||
| break | ||
|
|
||
| alpha_old = alpha | ||
| beta_old = beta | ||
|
|
||
| return alpha, beta | ||
|
|
||
| alpha, beta = fit_alpha_beta_py(Y) | ||
| return np.array([np.log(alpha), np.log(beta)]) | ||
|
|
||
| def sample(self, m): | ||
| return np.array([self.dist.rvs() for i in range(m)]) | ||
|
|
||
| def __getattr__(self, name): | ||
| if name in dir(self.dist): | ||
| return getattr(self.dist, name) | ||
| return None | ||
|
|
||
| @property | ||
| def params(self): | ||
| return {"alpha": self.alpha, "beta": self.beta} | ||
Add this suggestion to a batch that can be applied as a single commit.
This suggestion is invalid because no changes were made to the code.
Suggestions cannot be applied while the pull request is closed.
Suggestions cannot be applied while viewing a subset of changes.
Only one suggestion per line can be applied in a batch.
Add this suggestion to a batch that can be applied as a single commit.
Applying suggestions on deleted lines is not supported.
You must change the existing code in this line in order to create a valid suggestion.
Outdated suggestions cannot be applied.
This suggestion has been applied or marked resolved.
Suggestions cannot be applied from pending reviews.
Suggestions cannot be applied on multi-line comments.
Suggestions cannot be applied while the pull request is queued to merge.
Suggestion cannot be applied right now. Please check back later.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
How did you derive this? In my derivation the Fisher Information is not diagonal.
Here's what I have; it'd be great if you could paste your independent derivation so that we can double check.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I made a mistake not including the other diagonal. My calculation was based on the definition of the FI matrix as the variance of the score. Therefore I just simply squared the gradient (but forgot that it's actually a vector and the square should be S*S.T).
Can we use the double derivative? Are we using this:
Claim: The negative expected Hessian of log likelihood is equal to the Fisher Information Matrix F.
https://wiseodd.github.io/techblog/2018/03/11/fisher-information/
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
As per my calculation the last row is different, it's
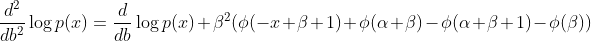
However when I use that formula the model doesn't work - it drops the singular matrix error again. And when I include the full FI matrix from the variance definition it also drops the singular matrix error. When I use the diagonal matrix from the Hessian definition it simply doesn't learn.
So the only working solution is the diagonal matrix from the variance definition. I have no clue why.
If you want to try the different approaches I have updated the code. All you need to do is to comment out the other diagonal or to comment out the other metric definition. For now I keep the working version as my pull request, but please check my code as I'm not sure I didn't miss something or didn't do a typo again.