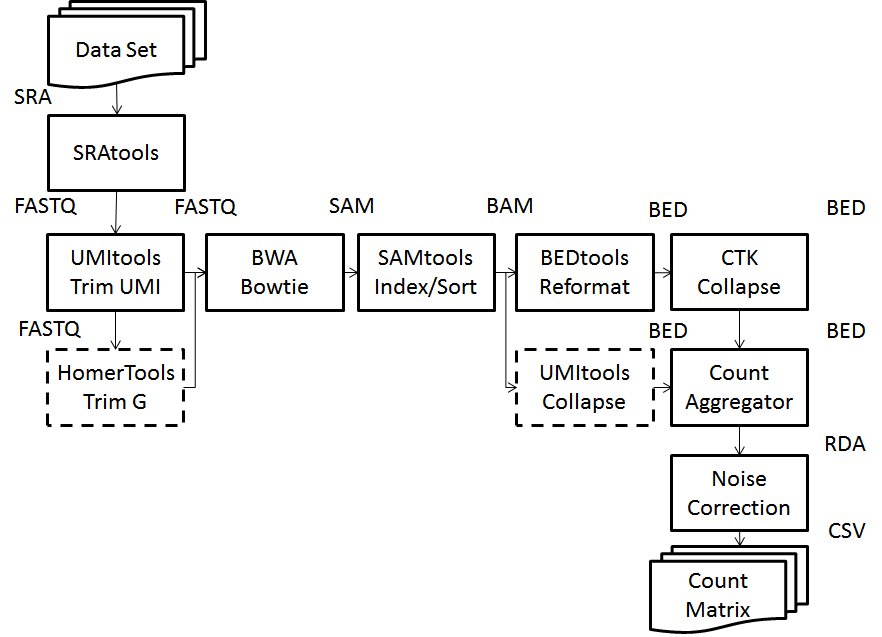
A streamlined pipeline for processing UMI-tagged single cell RNA-Seq data. The pipeline does the following workflow in three simple commands.
Read alignment and exact UMI collapsing: SRAToolkit, SAMtools, HomerTools ,BEDtools, UMItools, Bowtie and BWA
UMI sequencing error correction: CTK, CZPlib
Matrix imputation: impute
Multicore computing: doMC and parallel
Before running the pipeline, we need to prepare the index for the aligners.
For BWA, run the following command:
bwa index -a bwtsw Mus_musculus.GRCm38.cdna.all.faFor Bowtie:
bowtie-build Mus_musculus.GRCm38.cdna.all.fa GRCm38Install the prerequisite programs and set up the paths in SCUMI.sh
Then simply run
sh ./SCUMI.shA directory with resulting BED files will be created in the designated directory. Please run the following command to create the count matrix from the UMI-collapsed BED file in the directory. The output is an R object file (count_matrix.rda) and a CSV (count_matrix.csv) of the count matrix.
Rscript /path/to/BEDcounter_collapsed.RIf you want to collapse the file using multiple processors without CTK or UMItools. Please use the following command. The values after the R script is the number of cores.
Rscript /path/to/BEDcounter.R 12To correct the count matrix using the k-NN-based single cell expression noise model, please run the follows. The output is an R object file (count_matrix_imputed.rda).
Rscript /path/to/NoiseCorrection.R count_matrix.rdaThe reference transcriptomes can be downloaded from Ensembl.
Bose, S., Wan Z., Carr A., Rizvi, A.H. et al. Scalable microfluidics for single-cell RNA printing and sequencing. Genome Biol. 16:120.
Islam, S., Zeisel, A., Joost, S., La Manno G. et. al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 11:163-166.
Zhang, C., Darnell, R.B. 2011. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat. Biotech. 29:607-614.
We are grateful for the advice and help of Professor Chaolin Zhang, Professor Peter Sims, and Professor Yufeng Shen during the development of this toolkit as our final project of BINFG4017 DEEP SEQUENCING at Columbia University.