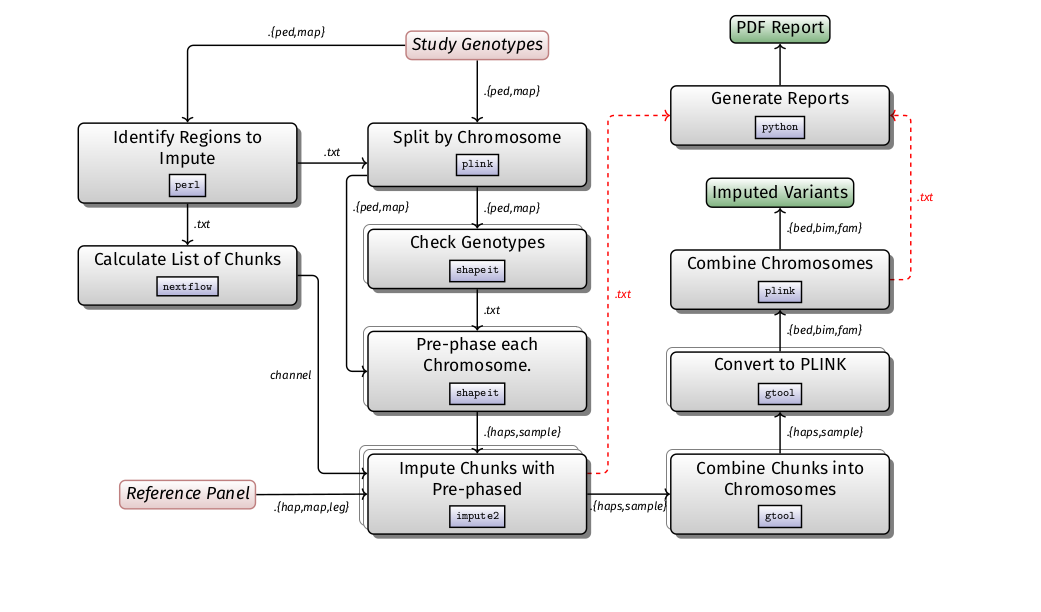
Imputation is likely to be run in the context of a GWAS, studying population structure, and admixture studies. It is computationally expensive in comparison to other GWAS steps. The basic steps of the pipeline is described in the diagram below:
The workflow is developed using 
This pipeline comes with docker/singularity containers making installation trivial and results highly reproducible.
This workflow which was developed as part of the H3ABioNet Hackathon held in Pretoria, SA in 2016. Should want to reference it, please use:
Baichoo S, Souilmi Y, Panji S, Botha G, Meintjes A, Hazelhurst S, Bendou H, Beste E, Mpangase PT, Souiai O, Alghali M, Yi L, O'Connor BD, Crusoe M, Armstrong D, Aron S, Joubert F, Ahmed AE, Mbiyavanga M, Heusden PV, Magosi LE, Zermeno J, Mainzer LS, Fadlelmola FM, Jongeneel CV, Mulder N. Developing reproducible bioinformatics analysis workflows for heterogeneous computing environments to support African genomics. BMC Bioinformatics. 2018 Nov 29;19(1):457. doi: 10.1186/s12859-018-2446-1. PubMed PMID: 30486782; PubMed Central PMCID: PMC6264621.
We track our open tasks using github's issues
The h3achipimputation pipeline comes with documentation about the pipeline, found in the docs/ directory:
- Installation
- Pipeline configuration
2.1. Configuration files
2.2. Software requirements
2.3. Other clusters - Running the pipeline
- Output and how to interpret the results
This pipeline itself needs no installation - NextFlow will automatically fetch it from GitHub. You can run the pipeline using test data hosted in github with singularity without have to install or change any parameters.
nextflow run h3abionet/chipimputation/main.nf -profile test,singularity
testprofile will download the testdata from https://github.com/h3abionet/chipimputation_test_data/tree/master/testdata_imputationsingularityprofile will download the singularity image from https://quay.io/h3abionet_org/imputation_tools
Check for results in ./output
wc -l output/impute_results/*
- Nextflow (can be installed as local user)
- NXF_HOME needs to be set, and must be in the PATH
- Note that we've experienced problems running Nextflow when NXF_HOME is on an NFS mount.
- The Nextflow script also needs to be invoked in a non-NFS folder
- Java 1.8+
- The compute nodes need access to shared storage for input, references, output.
- If you opt to use
singularityno software installation will be needed. - Otherwise, the following commands/softwares need to be available in PATH on the compute nodes