-
Notifications
You must be signed in to change notification settings - Fork 2
Stories
This is a 'growing' collection of day-to-day use cases where cwl-ica and ica-ica-lazy can help Some experience of ica-ica-lazy and cwl-ica is expected by the reader in this section.
- Story 1: Debugging a rogue sample by tinkering parameters
- Story 2: Uploading data from HPC
- Story 3: Using the CWL-ICA api to query run usages
- Story 4: Testing new versions of tools with engine parameter overrides
- Story 5: Sharing data to an external user with HPC access
- Story 6: Meeting notes
You've been given the task of inspecting an output vcf that just doesn't quite look right,
and have been tasked with re-running the variant calling workflow albeit with some alternative parameters.
The input and output data sit in the production project which you have read-only access to, and you would like to
re-run the workflow in the development project (a linked project to development_workflows).
The workflow name is dragen-somatic-pipeline and the version is 3.9.3.
The output data is in:
gds://production/analysis_data/SBJ01051/L2101235_L2101224/tumor_normal/2021-11-06__06-30-12/SBJ01051/
We can use the command gds-blame from ica-ica-lazy to determine the workflow id that created the output directory.
ica-context-switcher --project-name production --scope read-only
gds-blame --gds-path gds://production/analysis_data/SBJ01051/L2101235_L2101224/tumor_normal/2021-11-06__06-30-12/SBJ01051/From this we can confirm that workflow run id wfr.1a4523f298d746249187ff1b5ace7b8f was responsible for this output.
We can see the inputs to this workflow with the following command:
ica workflows runs get --output-format=json wfr.1a4523f298d746249187ff1b5ace7b8f | \
jq --raw-output '.input | fromjson'To replicate this workflow we need to copy over the fastq data into the development project.
Let's go ahead and do that now!
We can use the command gds-migrate from ica-ica-lazy to copy the data over to the development project.
Because we need to be in two-contexts at once here, gds-migrate will handle our tokens this time
(we don't need to be in any particular context to launch this command).
🚧
The command below will copy all fastqs in this directory over to the development project. Currently looking into regex support so that only certain files are copied over.
gds-migrate \
--src-project production \
--src-path gds://production/primary_data/211104_A00130_0181_AHWC25DSX2/WGS_TsqNano/ \
--dest-project development \
--dest-path gds://development/debugging/211104_A00130_0181_AHWC25DSX2/WGS_TsqNano/input-fastqs/You will see a task id (starting with trn.). You can use the command below to determine if your task has completed.
ica tasks runs get trn.... --output-format=json | \
jq --raw-output '.status'We can use the create-workflow-submission-template subcommand to mimic the inputs to the workflow run we found in step 1.
cwl-ica create-workflow-submission-template \
--workflow-path workflows/dragen-somatic-pipeline/3.9.3/dragen-somatic-pipeline__3.9.3.cwl \
--prefix debug-sample-SBJ01051 \
--project development_workflows \
--launch-project development \
--ica-workflow-run-instance-id wfr.1a4523f298d746249187ff1b5ace7b8f \
--ignore-workflow-id-mismatchThis will create the output files:
debug-sample-SBJ01051.template.yamldebug-sample-SBJ01051.launch.sh
Phoa, quite a lot to unpack there, let's break down the parameters a little:
-
workflow-path: Path to the cwl workflow used in original run -
prefix: The prefix for the output files, also used as the name of the workflow run. -
project: The project that the workflow we wish to launch is registered in -
launch-project: The linked-project to the 'project' parameter that we wish to launch the workflow from. -
ica-workflow-run-instance-id: The workflow run id that has the parameters we wish to copy -
ignore-workflow-id-mismatch: Ignore differences between workflow id in 'ica-workflow-run-instance-id' parameter and workflow id in 'project'.- Needed for when the project we launched from is different to the project the template run instance is from (in this case production_workflows).
I would highly recommend using an IDE for this section
Let's go through and find-and-replace our gds-path inputs in debug-sample-SBJ01051.template.yaml:
- Replace
gds://production/primary_data/211104_A00130_0181_AHWC25DSX2/WGS_TsqNano/ - With
gds://development/debugging/211104_A00130_0181_AHWC25DSX2/WGS_TsqNano/input-fastqs/
Replace our reference tar ball:
- Replace:
gds://production/reference-data/dragen_hash_tables/v8/hg38/altaware-cnv-anchored/hg38-v8-altaware-cnv-anchored.tar.gz - With
gds://development/reference-data/dragen_hash_tables/v8/hg38/altaware-cnv-anchored/hg38-v8-altaware-cnv-anchored.tar.gz
Tinker with any additional parameters (you may need to 'uncomment' some lines to do this).
Let's set the workDirectory and outputDirectory engine parameters for this workflow.
Let's go ahead and use the ica-check-cwl-inputs from ica-ica-lazy to make sure all
of our input files exist and all input names are consistent with the workflow version registered on ICA.
You will need to enter the development context first
ica-context-switcher --project-name 'development' --scope 'admin'You can find this code snippet inside debug-sample-SBJ01051.launch.sh
# Convert yaml into json with yq
echo 'Converting debug-sample-SBJ01051.template.yaml to json' 1>&2
json_path=$(mktemp debug-sample-SBJ01051.template.XXX.json)
yq eval --output-format=json '.' debug-sample-SBJ01051.template.yaml > "$json_path"
# Validate workflow inputs against ICA workflow version
echo 'Validating workflow inputs against ICA workflow version definition (if ica-ica-lazy is installed)' 1>&2
if type "ica-check-cwl-inputs" >/dev/null 2>&1; then
ica-check-cwl-inputs \
--input-json "$json_path" \
--ica-workflow-id "wfl.32e346cdbb854f6487e7594ec17a81f9" \
--ica-workflow-version-name "3.9.3"
fiRun the following command to launch the workflow:
bash debug-sample-SBJ01051.launch.shThere have been some instances where using the ica binary has meant that files
larger than 80 Gb were not able to be uploaded to gds.
I would highly recommend using gds-sync-upload from ica-ica-lazy which
assumes the user has aws v2 installed over the ica binary for any upload or download of files to gds.
In this story we will go through two examples of uploading data to gds using gds-sync-upload.
One where the user has ica-ica-lazy installed on HPC and the latter where ica-ica-lazy is installed only on
the user's laptop.
gds-sync-upload and gds-sync-download allow the user to add in
additional aws s3 sync commands to customise the upload / download to contain only the files of interest.
This story exposes the user to the --include and --exclude aws s3 sync parameters.
These parameters work in the opposite order to rsync parameters.
Here, the parameters are prioritised from right to left.
For example, the following code will include all files ending in .fastq.gz but omit all else.
aws s3 sync ... --exclude '*' --include '*fastq.gz'
We can use the --dryrun parameter to ensure that our logic is correct before running the command.
This is by far the simplest way, we expect the user to have an admin token installed onto the project they wish to upload to.
ssh [email protected]
ica-context-switcher --scope admin --project developmentgds-sync-upload \
--src-path /g/data/gx8/projects/Mitchell_temp/DRAGEN_Debug/2021-11-15T0656_All_WGS_SBJ00040/data/ \
--gds-path gds://umccr-temp-data-dev/helen/wgs/SBJ00040/fastq/ \
--exclude='*' \
--include='190418_A00130_0101_BHKJT3DSXX_SBJ00040_MDX190025_L1900373_R*_001.fastq.gz' \
--dryrunWe should expect only the paired reads from the MDX190025 sample to be uploaded, observe the logs carefully.
Now re-run the command but omit the the dryrun parameter
gds-sync-upload \
--src-path /g/data/gx8/projects/Mitchell_temp/DRAGEN_Debug/2021-11-15T0656_All_WGS_SBJ00040/data/ \
--gds-path gds://umccr-temp-data-dev/helen/wgs/SBJ00040/fastq/ \
--exclude='*' \
--include='190418_A00130_0101_BHKJT3DSXX_SBJ00040_MDX190025_L1900373_R*_001.fastq.gz'We use the --write-script-path to first write the script to a file and then execute elsewhere.
Rather than having to write the file locally and then upload to HPC, we can do this all in one step by
using /dev/stdout and the | parameter.
ica-context-switcher --scope admin --project developmentgds-sync-upload \
--src-path /g/data/gx8/projects/Mitchell_temp/DRAGEN_Debug/2021-11-15T0656_All_WGS_SBJ00040/data/ \
--gds-path gds://umccr-temp-data-dev/helen/wgs/SBJ00040/fastq/ \
--write-script-path /dev/stdout \
--exclude='*' \
--include='190418_A00130_0101_BHKJT3DSXX_SBJ00040_MDX190025_L1900373_R*_001.fastq.gz' \
--dryrun | \
ssh [email protected] 'cat > upload-files-to-gds.sh'We should expect only the paired reads from the MDX190025 sample to be uploaded, observe the logs carefully.
Observe the logs carefully, ensure output matches expected files to be uploaded
ssh [email protected]
bash upload-files-to-gds.shEdit the file upload-files-to-gds.sh on HPC and remove the --dryrun parameter from the aws s3 sync command.
Then re-run the script
bash upload-files-to-gds.shYour system may prevent you from running resource heavy commands on login nodes, therefore, please prefix the command
with the appropriate scheduling system command, i.e srun, qsub etc.
This uses a python object
You should have ica-ica-lazy installed and have installed cwl-ica through conda.
You may wish to install seaborn for step 5.
conda install seaborn \
--yes \
--channel conda-forge \
--name cwl-ica Create a token if it doesn't already exist
ica-add-access-token --project-name production --scope read-onlyEnter the project
ica-context-switcher --project-name production --scope read-onlyActivate the cwl-ica conda env
conda activate cwl-ica
Then open up an ipython console (every subsequent step is completed in this console)
-
Import the
ICAWorkflowRunclass fromcwl-ica, -
Import the environ module from
osso we can collect our token. -
And
attrgetterfrom theoperatorfor some list handling -
And
seabornfor working with some plot generation
from classes.ica_workflow_run import ICAWorkflowRun
from os import environ
from operator import attrgetter
import seaborn as sns
from matplotlib import pyplot as pltFor globals, we set our workflow run id we will be interacting with.
WORKFLOW_RUN_ID = "wfr.a448336414e14de0949812bbe135c4b1"cttso_run = ICAWorkflowRun(WORKFLOW_RUN_ID, project_token=environ["ICA_ACCESS_TOKEN"])Let's see what methods and attributes are available to us
[print(item)
for item in dir(cttso_run)
if not item.startswith("__")
];from_dict
get_ica_wes_configuration
get_run_instance
get_task_ids_and_step_names_from_workflow_run_history
get_task_run_objs
get_task_step_name_from_absolute_path_and_state_name
get_workflow
get_workflow_duration
get_workflow_run_history
ica_engine_parameters
ica_input
ica_output
ica_project_launch_context_id
ica_task_objs
ica_workflow_id
ica_workflow_name
ica_workflow_run_instance_id
ica_workflow_run_name
ica_workflow_version_name
split_href_by_id_and_version
to_dict
workflow_duration
workflow_end_time
workflow_start_time
First let's see how many tasks there are
len(cttso_run.ica_task_objs)27
Wow! Let's see the methods for a given task run object
[print(item)
for item in dir(cttso_run.ica_task_objs[0])
if not item.startswith("__")
];compress_metrics
decompress_metrics
from_dict
get_gds_configuration
get_metrics
get_pod_metrics_file
get_pod_metrics_file_url
get_task_cpus
get_task_duration
get_task_memory
get_task_name
get_task_object
get_tes_configuration
get_usage_dict
ica_task_run_instance_id
pod_metrics_to_df
read_pod_metrics_file
task_cpus
task_duration
task_memory
task_metrics
task_name
task_start_time
task_step_name
task_stop_time
to_dict
Let's see which task took the longest
longest_task = max(cttso_run.ica_task_objs, key=lambda item: item.task_duration)
print(f"{longest_task.task_name} took {longest_task.task_duration} seconds to complete")tso500-ctdna-analysis-workflow__1.1.0--120.cwl took 16017 seconds to complete
Let's have a look at some of the metrics of this task
longest_task.task_metrics| timestamp | cpu | memory | |
|---|---|---|---|
| 0 | 1637747210 | 2.0 | 0.8 |
| 3 | 1637747269 | 0.5 | 3.7 |
| 4 | 1637747323 | 15.5 | 37.3 |
| 6 | 1637747387 | 3.3 | 13.1 |
| 8 | 1637747451 | 3.5 | 24.8 |
| ... | ... | ... | ... |
| 492 | 1637762094 | 1.0 | 30.1 |
| 494 | 1637762155 | 1.0 | 31.2 |
| 497 | 1637762207 | 1.0 | 32.5 |
| 498 | 1637762273 | 0.0 | 31.4 |
| 500 | 1637762329 | 0.0 | 31.4 |
253 rows × 3 columns
Let's plot the cpu and memory of this task over time
from matplotlib.ticker import FuncFormatter
from datetime import datetime
def epoch_to_human_readable(x, position):
# Convert time in seconds to hours or minutes
timestamp_obj = datetime.fromtimestamp(x)
if x == 0:
return 0
s = f"{timestamp_obj.hour:02d}:{timestamp_obj.minute:02d}"
return s# Set ax values
fig, ax = plt.subplots()
# Plot CPU
cpu_ax = sns.lineplot(x="timestamp", y="cpu", data=longest_task.task_metrics, ax=ax)
# Plot Mem
mem_ax = sns.lineplot(x="timestamp", y="memory",
data=longest_task.task_metrics,
color='orange', ax=cpu_ax.twinx())
# Set title
fig.suptitle(f"CPU / Mem overtime for {longest_task.task_name}")
# Set xlabel
ax.set_xlabel("Time (HH:MM)")
ax.set_xticklabels(cpu_ax.get_xticklabels(), rotation=45, ha='right');
ax.xaxis.set_major_formatter(FuncFormatter(epoch_to_human_readable));
# Set ylabel
cpu_ax.set_ylabel("CPUs");
mem_ax.set_ylabel("Memory (GB)");
# Set boundaries
cpu_ax.set_xlim(left=longest_task.task_metrics.timestamp.min(),
right=longest_task.task_metrics.timestamp.max())
plt.show()
plt.close()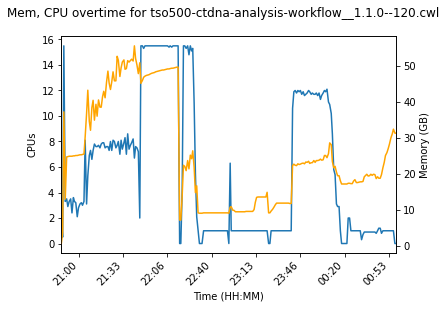
There are some CWL workflows we may wish to test frequently, with new versions of tools.
It may be useful to test with a plugin rather than updating the tool itself, which may be disruptive to other users in
that project.
An example of such a tool is umccrise.
You should have ica-ica-lazy installed and have installed cwl-ica through conda.
Activate the cwl-ica conda env and enter the context you wish to launch your project in.
# Enter cwl-ica conda env
conda activate cwl-ica
# Add context switcher
ica-context-switcher --scope admin --project-name developmentSimilar to story 1, we will use a previous workflow run to initialise our workflow run yaml template.
cwl-ica create-workflow-submission-template \
--prefix umccrise-prod-test \
--workflow-path workflows/umccrise-with-dragen-germline-pipeline/2.0.0--3.9.3/umccrise-with-dragen-germline-pipeline__2.0.0--3.9.3.cwl \
--ica-workflow-run-instance-id wfr.191ae274ea4d4d29a5087f39306826c0 \
--project production_workflows \
--launch-project production \
--ignore-workflow-id-mismatchThis will create the files umccrise-prod-test.template.yaml and umccrise-prod-test.launch.sh.
In order to edit the umccrise tool, we will need to know the step ids of the umccrise pipeline.
cwl-ica get-workflow-step-ids \
--workflow-path workflows/umccrise-with-dragen-germline-pipeline/2.0.0--3.9.3/umccrise-with-dragen-germline-pipeline__2.0.0--3.9.3.cwlThis yields
[ 'get_tumor_bam_file_from_somatic_directory',
'get_tumor_name_from_bam_header',
'run_dragen_germline_pipeline_step/create_dummy_file_step',
'run_dragen_germline_pipeline_step/create_fastq_list_csv_step',
'run_dragen_germline_pipeline_step/dragen_qc_step',
'run_dragen_germline_pipeline_step/run_dragen_germline_step',
'run_umccrise_pipeline_step']Now we can update the docker tag through the engine parameter overrides in umccrise-prod-test.template.yaml like so:
engineParameters:
overrides:
"#run_umccrise_pipeline_step":
requirements:
DockerRequirement:
dockerPull: "<new_docker_image_path/tag>"Not all customers are going to have ICA installed or have an ICA account.
While we can provide them with a list of presigned urls, this isn't very practical for most users and does not provide
the correct directory structure.
In this tutorial we expect the external user to have:
- Some basic unix commandline knowledge
- Access to a unix system with
jq,python3andwgetinstalled - Access to a file system with adequate space to download data (such as HPC)
In this tutorial I will be providing an external client with a reference dataset from gds.
The client can then download the files to the NCI supercomputer cluster called gadi.
You should have ica-ica-lazy installed and have installed cwl-ica through conda.
Activate the cwl-ica conda env and enter the context you wish to launch your project in.
# Enter cwl-ica conda env
conda activate cwl-ica
# Add context switcher
ica-context-switcher --scope read-only --project-name developmentRun the following command to create a download script for a specific folder
gds-create-download-script \
--gds-path gds://development/qc_reference_data/ \
--output-prefix qc_reference_dataWe now have the file qc_reference_data.sh which has a suite of presigned urls inside a very long base64 string.
The external client does not need to have any knowledge of presigned urls or base64 in order to run the script.
In this example I will upload our script qc_reference_data.sh to my home directory on NCI through scp.
You may send through this script however means necessary (although securely) to your external client.
scp qc_reference_data.sh [email protected]:/home/563/al9223/This step is run by the external client
It is not recommended running anything on the login node on gadi so first the client runs an interactive qsub command.
qsub \
-I \
-P gx8 \
-q copyq -l walltime=10:00:00,ncpus=1,mem=10G,storage=gdata/gx8+scratch/gx8Once in the copy queue the client can run the following command
bash qc_reference_data.sh --help
Usage qc_reference_data.sh (--output-directory <output_directory>)
Description:
Download a suite of files from gds://development/qc_reference_data/
Options:
--output-directory: Local output path
Requirements:
* jq
* python3Specify the output directory for where we would like to store this data on our HPC
bash qc_reference_data.sh --output-directory /scratch/gx8/al9223/tmp/reference_data/And we're done!!
- You're in the
cwl-icaconda env
This command updates our console's ${ICA_ACCESS_TOKEN} environment variable so we can use both ica-ica-lazy and ica commands.
ica-context-switcher --scope admin --project-name developmentica workflows runs list --max-items=0 | grep umccrise | lessWe then chose wfr.191ae274ea4d4d29a5087f39306826c0.
🚧 We then had an issue with running the following command
$ cwl-ica create-workflow-submission-template \
--access-token "${ICA_ACCESS_TOKEN}" \
--ica-workflow-run-instance-id "wfr.191ae274ea4d4d29a5087f39306826c0" \
--launch-project development \
--project development_workflows \
--prefix 'umccrise-testing' \
--workflow-path "${CWL_ICA_REPO_PATH}/workflows/umccrise-with-dragen-germline-pipeline/2.0.1--3.9.3/umccrise-with-dragen-germline-pipeline__2.0.1--3.9.3.cwl"Because it tries to get pod-metrics which have since been deleted - this error will be fixed at some point.
In future we can use the following logic to make sure that the workDirectory still exists for this workflow.
$ ica workflows runs get wfr.191ae274ea4d4d29a5087f39306826c0 --include engineParameters -o json | \
jq '.engineParameters | fromjson | .workDirectory'
$ ica folders get "gds://wfr.191ae274ea4d4d29a5087f39306826c0/umccrise-workflow-testing"
Folder not foundIf work directory does not exist we cannot use --ica-workflow-run-instance-id. Again, will be fixed in a coming release.
Same as above, just without the --ica-workflow-run-instance-id.
If we don't have the --ica-workflow-run-instance-id we also don't need the access token.
cwl-ica create-workflow-submission-template \
--launch-project development \
--project development_workflows \
--prefix 'umccrise-testing' \
--workflow-path "${CWL_ICA_REPO_PATH}/workflows/umccrise-with-dragen-germline-pipeline/2.0.1--3.9.3/umccrise-with-dragen-germline-pipeline__2.0.1--3.9.3.cwl"Now populate umccrise-testing.template.yaml replacing gds:// placeholders with actual gds file inputs.
Also comment out any parameters you don't need.
We can also use the following command to get the input attribute from the ica-workflow-run-instance-id.
If yq --prettyPrint doesn't work you may need to update your yq command
ica workflows runs get -o json wfr.191ae274ea4d4d29a5087f39306826c0 | \
jq --raw-output '.input | fromjson | {"input": .}' | \
yq --prettyPrintRemember to specify the engineParameters outputDirectory and workDirectory attributes.
You can also comment them out, just don't leave them blank
bash umccrise-testing.launch.yaml
Even though it's run through WES, WTS itself is a tool attribute. Therefor all of our cwl-ica commands will replace 'workflow' with 'tool'.
Generate the WTS tool template like so:
cwl-ica create-tool-submission-template \
--access-token "${ICA_ACCESS_TOKEN}" \
--launch-project development \
--project development_workflows \
--prefix 'wts-tool-testing' \
--tool-path "${CWL_ICA_REPO_PATH/tools/dragen-transcriptome/3.9.3/dragen-transcriptome__3.9.3.cwl"Again edit the template, comment out any parameters you don't need, and fix the engine parameters
Run bash wts-tool-testing.launch.sh to launch the tool.
For PON QC we have a workflow run instance id to copy that won't cause an error, yay!
cwl-ica create-workflow-submission-template \
--access-token "${ICA_ACCESS_TOKEN}" \
--ica-workflow-run-instance-id "wfr.e0cccb29cf44482182507bd33579270c" \
--launch-project development \
--project development_workflows \
--prefix 'pon-testing' \
--workflow-path "${CWL_ICA_REPO_PATH}/workflows/dragen-pon-qc/3.9.3/dragen-pon-qc__3.9.3.cwl"Again edit the template, comment out any parameters you don't need, and fix the engine parameters.
Most of this work will have been done for you with the template now mimicking the inputs of the workflow run instance id specified in the previous command
bash pon-testing.launch.sh