-
-
Notifications
You must be signed in to change notification settings - Fork 19
Manually distributed library example
This examples shows how to setup and run the ParMmg library on a manually defined distributed mesh (source file).
First, a description of the test case and the partitioning is provided. The essential code blocks for the library coupling are then commented.
In this example we want to setup and run the ParMmg library on a mesh of the unit cube shown below.
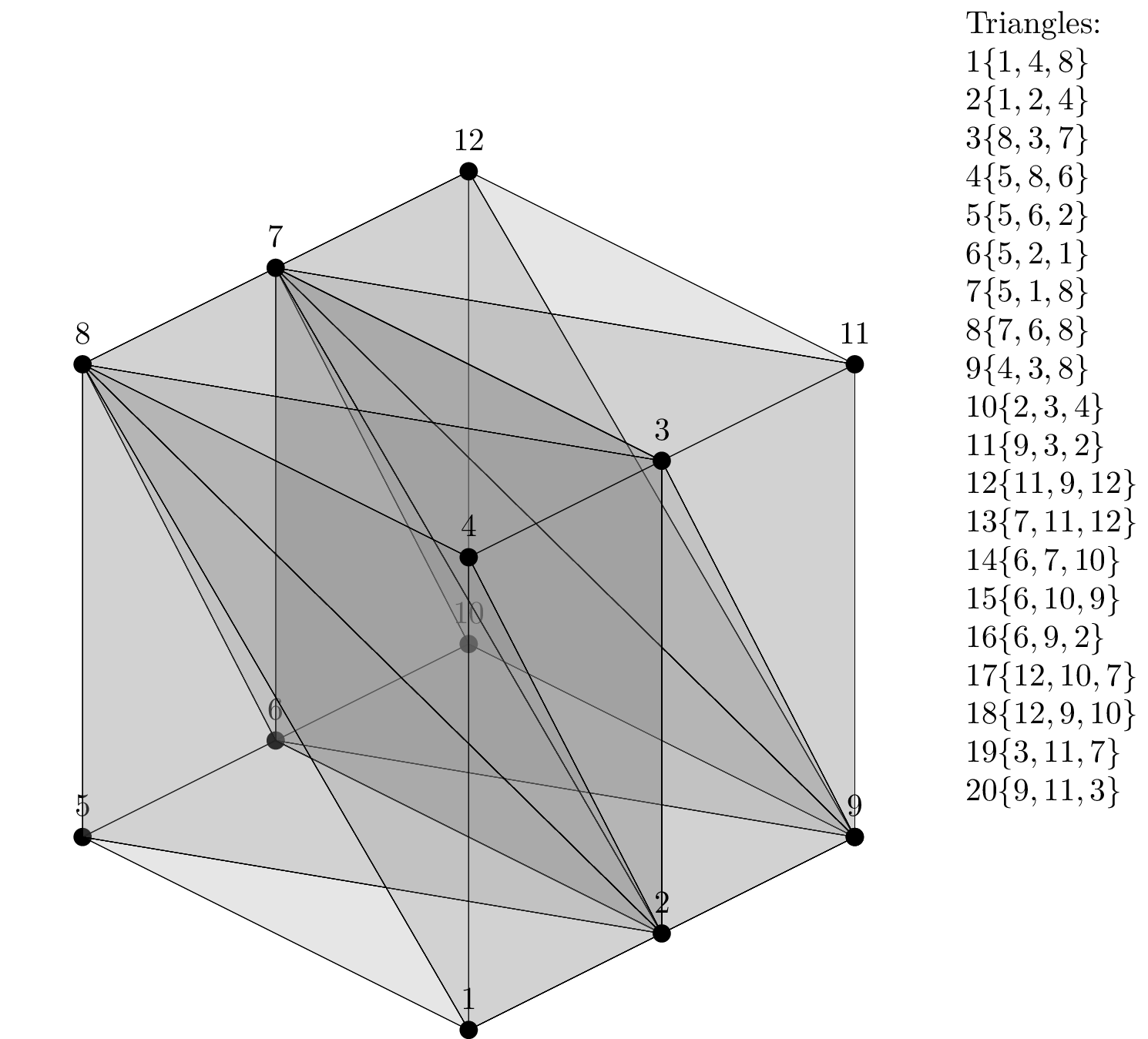
The figure above shows the global mesh of the unit cube and the list of boundary triangles, with a label and the list of nodes for each triangle. This mesh is not stored on any of the parallel processes, but it is visualized here as a reference model.
For an easier graphical interpretation of the library setup, a partitioning of this mesh is manually defined for the case of two and four processes.
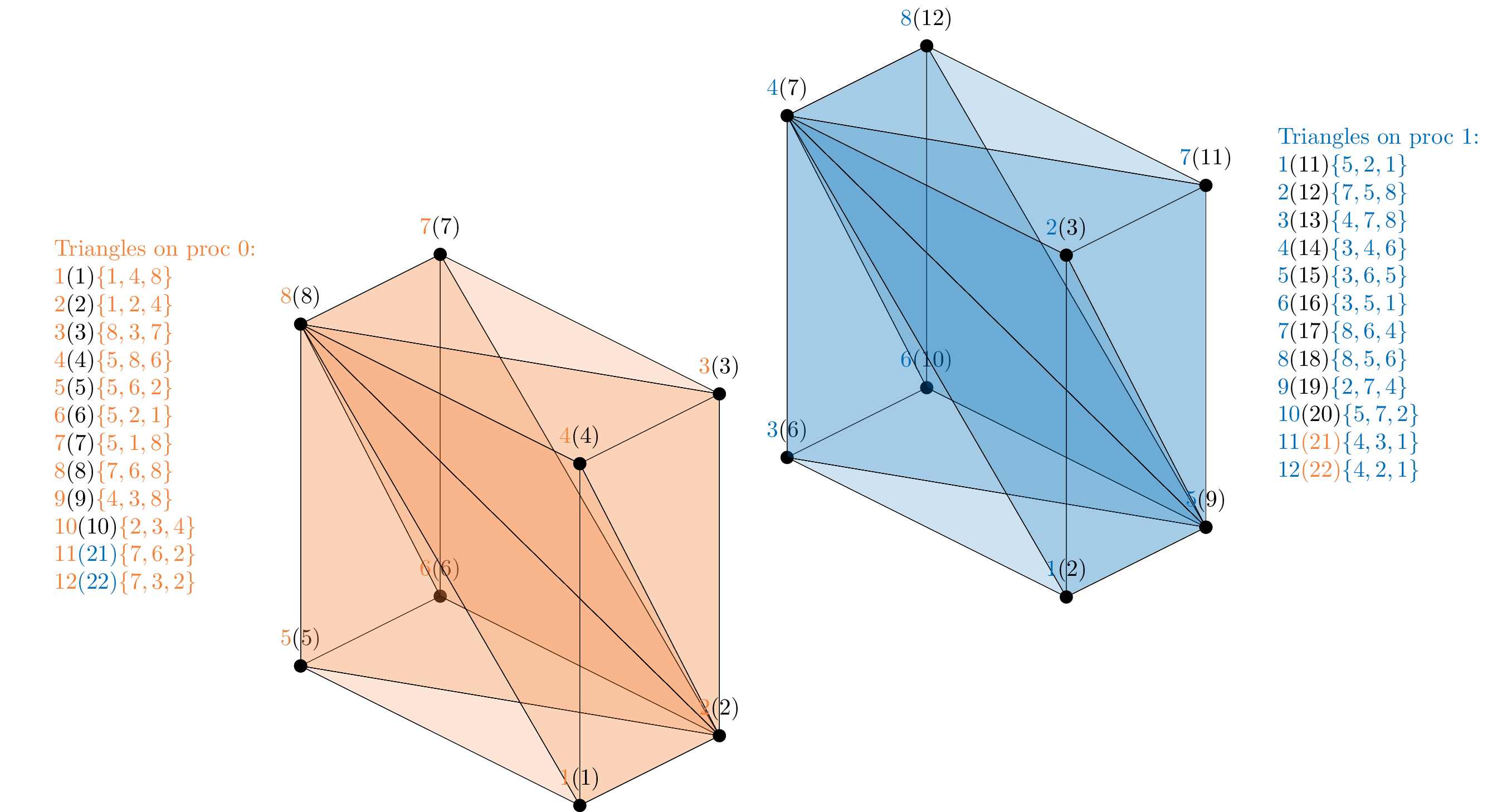
The figure above shows the mesh partitioning on two processes. Local labels are visualized together with their corresponding global labels in the reference model (black, in brackets). Note that on each process two new boundary triangles appear, that were not present in the global mesh. These are parallel interface triangles between processes 0 and 1, and the global labels 21 and 22 are arbitrarily assigned to make them recognizable (the brackets color correspond to the process they are shared with).
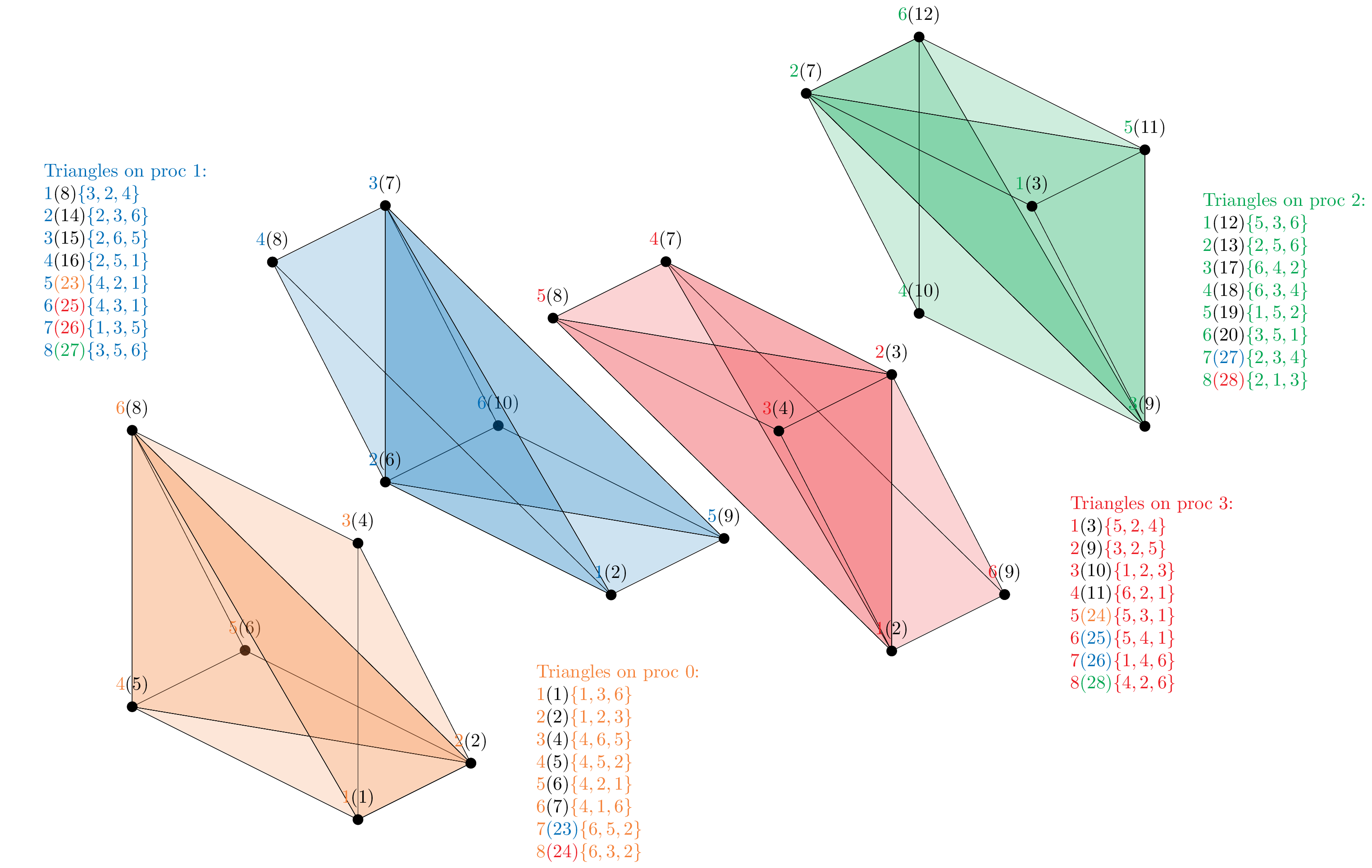
The figure above shows the mesh partitioning on four processes. Many more triangles arise from the parallel interfaces among pairs of processes, and we arbitrarily give them the global labels from 23 to 28 (note that the triangles that we previously labeled as 21 and 22 are no longer present neither as boundary nor as parallel triangles on any process).
In this sections, only the essential code blocks of libexamples/adaptation_example0/parallel_IO/manual_IO/main.c are detailed in order to explain the general methodology of the ParMmg library setup and call.
These are organized in five main steps which are detailed below and can be retrieved in the source file documentation. All the source file parts related to the definition of the mesh coordinates and partitioning are not detailed here, as the focus is on the library usage.
The library usage starts with the inclusion of ParMmg datatypes and parameters:
#include "libparmmg.h"This file (automatically copied from here when building) is normally present in the include/parmmg folder within the build directory or in your installation path, and contains all the available library functions and parameters together with their documentation.
A pointer to the ParMmg data structures needs then to be declared:
PMMG_pParMesh parmesh;The ParMmg data structures are initialized with:
parmesh = NULL;
PMMG_Init_parMesh(PMMG_ARG_start,
PMMG_ARG_ppParMesh,&parmesh,
PMMG_ARG_pMesh,PMMG_ARG_pMet,
PMMG_ARG_dim,3,PMMG_ARG_MPIComm,MPI_COMM_WORLD,
PMMG_ARG_end);
⚠️ This must be done after the initialization of the MPI library.
These variadic arguments, included between the parameters PMMG_ARG_start and PMMG_ARG_end, tell the library that we want to initialize a parmesh structure to perform adaptation using an input metrics, in three dimensions (the only supported case for now) using MPI_COMM_WORLD as MPI communicator.
The local mesh on each process is basically set up as an indipendent, self-contained Mmg mesh. The input mesh setup is thus reminiscent of the API functions used by the Mmg library. The notion of a parallel distributed mesh is provided by the additional definition of the parallel communicators, which are the set of mesh entities (triangular faces or nodes) that are shared between pairs of processes.
⚠️ Only element-based mesh partitioning is supported and no mesh overlapping is allowed, i.e. mesh elements are uniquely assigned to a process or another, and the only replicated mesh entities on more then one process are the element faces and nodes on a parallel interface.
First, the size of the mesh local to each process needs to be set:
if ( PMMG_Set_meshSize(parmesh,nVertices,nTetrahedra,nPrisms,nTriangles,
nQuadrilaterals,nEdges) != 1 ) {
MPI_Finalize();
exit(EXIT_FAILURE);
}
⚠️ Please note that only simplicial meshes are supported at the moment (i.e. zero prisms and quadrilaterals).
Mesh entities can then be passed either one-by-one or by arrays:
if ( !opt ) {
/* By array: give the array of the vertices coordinates such as the
* coordinates of the k^th point are stored in vert_coor[3*(k-1)],
* vert_coor[3*(k-1)+1] and vert_coor[3*(k-1)+2] */
if ( PMMG_Set_vertices(parmesh,vert_coor,vert_ref) != 1 ) {
MPI_Finalize();
exit(EXIT_FAILURE);
}
}
else {
/* Vertex by vertex: for each vertex, give the coordinates, the reference
and the position in mesh of the vertex */
for ( k=0; k<nVertices; ++k ) {
pos = 3*k;
if ( PMMG_Set_vertex(parmesh,vert_coor[pos],vert_coor[pos+1],vert_coor[pos+2],
vert_ref[k], k+1) != 1 ) {
MPI_Finalize();
exit(EXIT_FAILURE);
}
}
}
if ( !opt ) {
/* By array: give the array of the tetra vertices and the array of the tetra
* references. The array of the tetra vertices is such as the four
* vertices of the k^th tetra are respectively stored in
* tetra_vert[4*(k-1)],tetra_vert[4*(k-1)+1],tetra_vert[4*(k-1)+2] and
* tetra_vert[4*(k-1)+3]. */
if ( PMMG_Set_tetrahedra(parmesh,tetra_vert,tetra_ref) != 1 ) {
MPI_Finalize();
exit(EXIT_FAILURE);
}
}
else {
/* Vertex by vertex: for each tetrahedra,
give the vertices index, the reference and the position of the tetra */
for ( k=0; k<nTetrahedra; ++k ) {
pos = 4*k;
if ( PMMG_Set_tetrahedron(parmesh,tetra_vert[pos],tetra_vert[pos+1],
tetra_vert[pos+2],tetra_vert[pos+3],tetra_ref[k],k+1) != 1 ) {
MPI_Finalize();
exit(EXIT_FAILURE);
}
}
}
if ( !opt ) {
/* By array: give the array of the tria vertices and the array of the tria
* references. The array of the tria vertices is such as the three
* vertices of the k^th tria are stored in
* tria_vert[3*(k-1)], tria_vert[3*(k-1)+1] and tria_vert[4*(k-1)+2] */
if ( PMMG_Set_triangles(parmesh,tria_vert,tria_ref) != 1 ) {
MPI_Finalize();
exit(EXIT_FAILURE);
}
}
else {
/* Vertex by vertex: for each triangle, give the vertices index, the
* reference and the position of the triangle */
for ( k=0; k<nTriangles; ++k ) {
pos = 3*k;
if ( PMMG_Set_triangle(parmesh,
tria_vert[pos],tria_vert[pos+1],tria_vert[pos+2],
tria_ref[k],k+1) != 1 ) {
MPI_Finalize();
exit(EXIT_FAILURE);
}
}
}
⚠️ As in Mmg, mesh entities numbering starts from 1.
The same procedure is followed to setup the metrics:
if ( PMMG_Set_metSize(parmesh,MMG5_Vertex,nVertices,MMG5_Scalar) != 1 ) {
MPI_Finalize();
exit(EXIT_FAILURE);
}
if ( !opt ) {
/* by array */
if ( PMMG_Set_scalarMets(parmesh,met) != 1 ) {
MPI_Finalize();
exit(EXIT_FAILURE);
}
}
else {
/* vertex by vertex */
for ( k=0; k<nVertices ; k++ ) {
if ( PMMG_Set_scalarMet(parmesh,met[k],k+1) != 1 ) {
MPI_Finalize();
exit(EXIT_FAILURE);
}
}
}The major difference with the Mmg library is the definition of the parallel communicators. First, the node or face communicator mode must be choosen, as only one of them can be used:
/* Set API mode */
if( !PMMG_Set_iparameter( parmesh, PMMG_IPARAM_APImode, API_mode ) ) {
MPI_Finalize();
exit(EXIT_FAILURE);
};As for the mesh, the size of the communicators and the entities on them need to be set. One communicator is needed for each pair of interacting processes (i.e. in the node API mode, multiple processes connected through one node communicate pairwise among all of them).
⚠️ For each communicator, the color of the neighbouring process (i.e. its MPI rank) needs to be passed, and a global label is passed for each entity in the communicators to make them recognizable on the two sides of the communicators in the case the entities are not passed in the same order on the two processes (last argument of thePMMG_Set_ithFaceCommunicator_faces()andPMMG_Set_ithNodeCommunicator_nodes()set to 1). If entities are listed in the same order on the two sides of the interface, ParMmg avoids additional computations to reorder them (in this case global labels for faces are ignored, while they are still needed for nodes).
Back to the example:
/* Select face or node interface API */
switch( API_mode ) {
case PMMG_APIDISTRIB_faces :
if( !rank ) printf("\n--- API mode: Setting face communicators\n");
/* Set the number of interfaces */
ier = PMMG_Set_numberOfFaceCommunicators(parmesh, ncomm);
/* Loop on each interface (proc pair) seen by the current rank) */
for( icomm=0; icomm<ncomm; icomm++ ) {
/* Set nb. of entities on interface and rank of the outward proc */
ier = PMMG_Set_ithFaceCommunicatorSize(parmesh, icomm,
color_face[icomm],
ntifc[icomm]);
/* Set local and global index for each entity on the interface */
ier = PMMG_Set_ithFaceCommunicator_faces(parmesh, icomm,
ifc_tria_loc[icomm],
ifc_tria_glob[icomm], 1 );
}
break;
case PMMG_APIDISTRIB_nodes :
if( !rank ) printf("\n--- API mode: Setting node communicators\n");
/* Set the number of interfaces */
ier = PMMG_Set_numberOfNodeCommunicators(parmesh, ncomm);
/* Loop on each interface (proc pair) seen by the current rank) */
for( icomm=0; icomm<ncomm; icomm++ ) {
/* Set nb. of entities on interface and rank of the outward proc */
ier = PMMG_Set_ithNodeCommunicatorSize(parmesh, icomm,
color_node[icomm],
npifc[icomm]);
/* Set local and global index for each entity on the interface */
ier = PMMG_Set_ithNodeCommunicator_nodes(parmesh, icomm,
ifc_nodes_loc[icomm],
ifc_nodes_glob[icomm], 1 );
}
break;
}The actual library call in the distributed case is performed by the PMMG_parmmglib_distributed()function and can be governed by several parameters (here showing two of them):
/* Set number of iterations */
if( !PMMG_Set_iparameter( parmesh, PMMG_IPARAM_niter, niter ) ) {
MPI_Finalize();
exit(EXIT_FAILURE);
};
/* Don't remesh the surface */
if( !PMMG_Set_iparameter( parmesh, PMMG_IPARAM_nosurf, 1 ) ) {
MPI_Finalize();
exit(EXIT_FAILURE);
};
/* remeshing function */
ierlib = PMMG_parmmglib_distributed( parmesh );At the end of the remeshing iterations, the entities on the parallel communicators will have changed due to parallel mesh redistribution. All node and face communicators entities can be recovered in arrays, in order for the user to be able to reconstruct its own parallel data structures:
int next_node_comm,next_face_comm,*nitem_node_comm,*nitem_face_comm;
int *color_node_out,*color_face_out;
int **out_tria_loc, **out_node_loc;
/* Get number of node interfaces */
ier = PMMG_Get_numberOfNodeCommunicators(parmesh,&next_node_comm);
/* Get outward proc rank and number of nodes on each interface */
color_node_out = (int *) malloc(next_node_comm*sizeof(int));
nitem_node_comm = (int *) malloc(next_node_comm*sizeof(int));
for( icomm=0; icomm<next_node_comm; icomm++ )
ier = PMMG_Get_ithNodeCommunicatorSize(parmesh, icomm,
&color_node_out[icomm],
&nitem_node_comm[icomm]);
/* Get IDs of nodes on each interface */
out_node_loc = (int **) malloc(next_node_comm*sizeof(int *));
for( icomm=0; icomm<next_node_comm; icomm++ )
out_node_loc[icomm] = (int *) malloc(nitem_node_comm[icomm]*sizeof(int));
ier = PMMG_Get_NodeCommunicator_nodes(parmesh, out_node_loc);
/* Get number of face interfaces */
ier = PMMG_Get_numberOfFaceCommunicators(parmesh,&next_face_comm);
/* Get outward proc rank and number of faces on each interface */
color_face_out = (int *) malloc(next_face_comm*sizeof(int));
nitem_face_comm = (int *) malloc(next_face_comm*sizeof(int));
for( icomm=0; icomm<next_face_comm; icomm++ )
ier = PMMG_Get_ithFaceCommunicatorSize(parmesh, icomm,
&color_face_out[icomm],
&nitem_face_comm[icomm]);
/* Get IDs of triangles on each interface */
out_tria_loc = (int **) malloc(next_face_comm*sizeof(int *));
for( icomm=0; icomm<next_face_comm; icomm++ )
out_tria_loc[icomm] = (int *) malloc(nitem_face_comm[icomm]*sizeof(int));
ier = PMMG_Get_FaceCommunicator_faces(parmesh, out_tria_loc);The output mesh can be recovered either one-by-one or by arrays through *_Get_* functions analogous to the *_Set_* ones. Taking as example just the mesh tetrahedra:
if ( !opt ) {
/* By array */
if ( PMMG_Get_tetrahedra(parmesh,tetra,ref,required) != 1 ) {
fprintf(inm,"Unable to get mesh tetra\n");
ier = PMMG_STRONGFAILURE;
}
for ( k=0; k<nTetrahedra; k++ ) {
if ( required && required[k] ) nreq++;
}
}
else {
/* Tetra by tetra */
for ( k=0; k<nTetrahedra; k++ ) {
pos = 4*k;
if ( PMMG_Get_tetrahedron(parmesh,
&(tetra[pos ]),&(tetra[pos+1]),
&(tetra[pos+2]),&(tetra[pos+3]),
&(ref[k]),&(required[k])) != 1 ) {
fprintf(inm,"Unable to get mesh tetra %d \n",k);
ier = PMMG_STRONGFAILURE;
}
if ( required && required[k] ) nreq++;
}
}At the end of the procedure, the ParMmg data structures can be freed:
PMMG_Free_all(PMMG_ARG_start,
PMMG_ARG_ppParMesh,&parmesh,
PMMG_ARG_end);