-
Notifications
You must be signed in to change notification settings - Fork 8
Example and sample data
On this tutorial are shown two EvoMining examples.
- Utilizing default data, EvoMining web interface functionality is explained.
- Use of custom input databases is exemplified with sample data. To learn more about how to construct the databases consult the databases site.
start EvoMining on the prompt of the docker image with the default data by typing:
perl startEvoMining.pl
Once the script finished, start EvoMining web pipeline on firefox or any browser at the adress:
http://localhost/html/EvoMining/index.html.
When in doubt about how to launch EvoMining docker, consult Installation.
This should be the welcome screen, press the start button to initialize the web interface.
On a second screen EvoMining requires the e-value required to consider a sequence inside a family, default:0.001. Press submit button to launch the expansions script that for each enzymatic family identifies sequences belonging to the expanded family.

A heatmap table is presented to the user once the expanded family has been gathered. Organisms are distributed on the rows of the table. Each column represents an enzyme family. When an organism has a copy number above the average plus one standard deviation on one particular enzyme family, the correspondent square is filled in red, representing an expansion of that enzyme family on that particular organism. Once the heatmap is finished, continue to the search of genes of the family that has been recruited onto natural product biosynthetic gene clusters, press the submit button.

On this case, a table presents for each family its known recruitments into natural product (NPs) biosynthetic gene clusters. Metadata from MI-BiG such as type of natural product and taxonomy of the producer organism are shown. Select an enzyme family to continue into a phylogenetic reconstruction.

Once the phylogenetic history of an expanded family has been done, after alignment and curation, trees that were able to finish are shown. Select a tree and send it to the visualization interface.

Tree visualization integrates zooming and panning when trees are under mouse click. This characteristic allows to perform a detailed tree exploration. Trees are interactive in the sense that leaves can be clicked in and send it to context visualization to compare its genomic contexts. Context visualization displays on a tooltip metadata derived from RAST functional anotation. Trees are scalable vector graph (svg) files that remain available on you local directory.
This is an example of a tree coming from a family that has not been recruited. Phosphoserine aminotransferase the third enzyme from 3PGA aminoacid pathway is present mainly as a single copy on this set of organisms, this family has not been expanded on this example database. Recruitments from MI-BiG are not present on this tree, and there are not experimental evidence from MI-BiG that there are copies of this family devoted to secondary metabolism. Sequences used as seeds on the central-DB are orange colored, Best Bidirectional hits (BBH) of the seeds are red marked. The tree shows a leave marked on cyan, this color corresponds to the optional antiSMASH database, i.e. those copies that has been identified by antiSMASH software as candidate to belong to a BGC.
Metadata like organism name and gene id are displayed on tree leaves when mouseover.
This example corresponds to the Phosphoglycerate dehydrogenase, the first enzyme from the 3PGA aminoacid pathway. In contrast with the previous non expanded tree, Phosphoglycerate dehydrogenase tree posses an expanded family showing MI-BiG evidence that has been recruited multiple times into NPs metabolism. In addition to antismash (blue) and central colored copies: seeds (orange) and BBH (red), MI-BiG copies of this family are blue colored and all those leaves close to an MI-BiG mark that do not belong to the central family (red colored) and are not recognized by antiSMASH (cyan) are the EvoMining predictions (green).
On this tree there are also non colored leaves, this sequences are not BBH of the central database. Black branches could correspond to novel natural products non previously known by MI-BIG or may be diverging into another metabolic destiny.
Genomic context of selected genes are shown below. Metadata from RAST functional anotation are displayed on a tooltip when mouseover.
Trees are compatible with MicroReact svg and nwk EvoMining output can be uploaded to MicroReact to obtain visualization like this MicroReact EvoMining example
Results are stored at your local directory on the subdirectory
ALL_curado.fasta_MiBIG_DB.faa_los17
This part explains how to use databases introduced by the user, for that purpose this sample data will be used.
- Open a terminal and place your self on a local directory.
$ cd </path/to/mydirectory>
On the local prompt the starting character is $ on the docker prompt is #.
Get data:
curl https://zenodo.org/record/1219709/files/SampleData.tar.gz?download=1 > SampleData.tar.gz && tar -xzf SampleData.tar.gz
and go inside Sample data directory:
$ cd SampleData
Input sample data contains:
| File | Description |
|---|---|
| Genome-DB | Directory with the genome database |
| Rast.ids | Tab separated File with genome Names/Ids |
| central-DB.fasta | Central database file (fasta) |
the directory should look as follows:
The content of the Genome Database directory must be two file per genome, the aminoacids fasta file (faa) and the annotations file (txt), bot using the name of the RAST JobId:

Users data:
If you have genomes annotated in RAST that you whish to add to the database, first add your genome to the Rast.Ids file and then add the aminoacids (JobId.faa) and table_txt (JobId.txt) files to the Genome-DB directory.
-
Update Rast ids file:
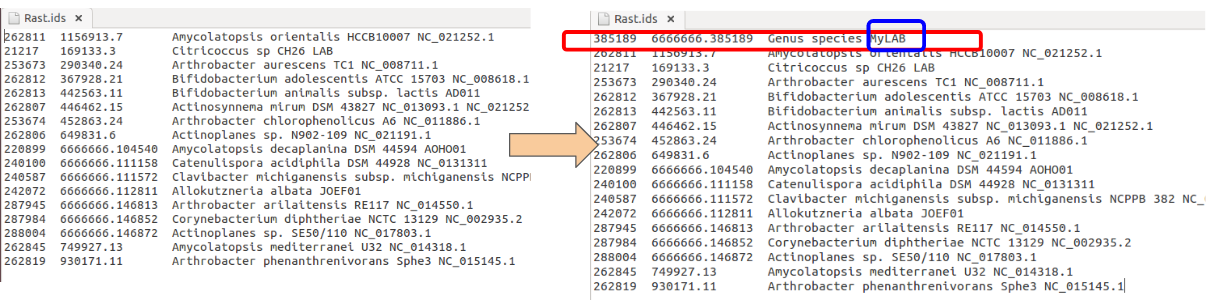
In the first column add the Job Id, in the second column the genome Id and in the third column your organisms name (red squared), at the end add an identification label (blue squared). The identification label is to prevent to organisms with the same name. Columns must be tab separated. -
Add your genome fasta and annotations files to the Genome Database directory:
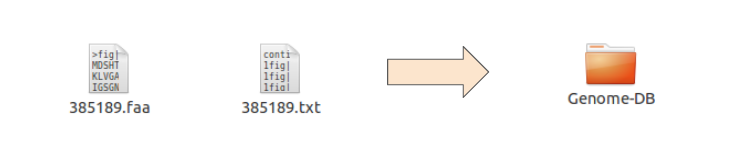
To download and upload Rast files in batch check my Rast docker image.
- Start EvoMining docker image.
$ docker run -i -t -v $(pwd):/var/www/html/EvoMining/exchange -p 80:80 nselem/evomining:latest /bin/bash
At this time the local directory /path/to/mydirectory/SampleData and docker directory /var/www/html/EvoMining/exchange are shared.
-
Run EvoMining
-
Start EvoMining with non default databases
# perl startEvoMining.pl
This command runs default EvoMining databases -
Start EvoMining with non default databases
# perl startEvoMining.pl -g Genome-DB -c central-DB.fasta -r Rast.ids
This command starts EvoMining using as central-DB the file central-DB.fasta, as natural-DB natural-DB.fasta and as genome DB the genomes contained on the directory Genome-DB. It also needs the file Rast.ids described previously. To know more about how to construct you own databases consult Databases Conformation
The natural products database may also be modified. To run EvoMining with a new natural products DB use:
# perl startEvoMining.pl -g Genome-DB -c central-DB.fasta -r Rast.ids -n natural-DB
-
EvoMining browser
Open EvoMining web interface and follow the steps as on the prior example.
In your browser got to
http://localhost/EvoMining/html/index.html -
Results
Results e.g. trees (nwk), alignments, and metadadta tables (cvs) will be stored in the directory:
<central-DB>/<natural-DB>/<genome-DB>in this case this information corresponds to:
central-DB.fasta_MiBIG_DB.faa_Genome-DB