-
Notifications
You must be signed in to change notification settings - Fork 5
MAPK
Saez-Rodriguez et al. (Saez-Rodriguez et al., 2009) showed in their work that large-scale protein signalling networks are useful to analyse complex biochemical pathways but these networks do not represent the cell-specific response to different stimuli. In this framework, we analyse the model converted from the Boolean Model proposed by Saez et al. during the DREAM4 challenge, which was a subset of a larger liver-specific model.
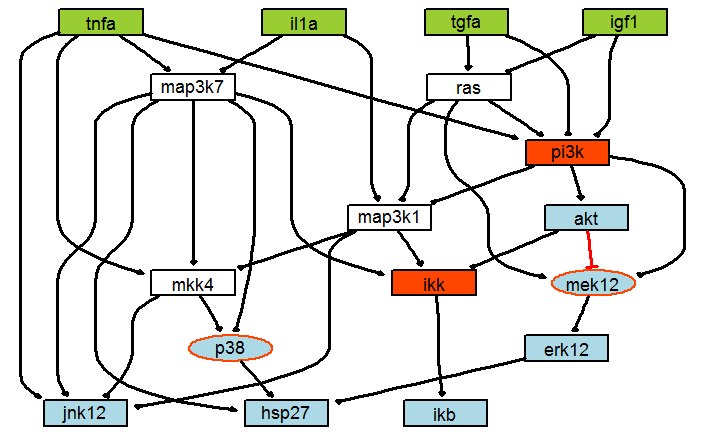
The model contains 22 nodes with 36 interactions (32 interactions are optimized), with one logical gates from MAP3K7 and MAP3K1 (OR gate) to IKK. The model contains 25 experimental conditions with 8 inputs and 7 outputs.
| Model Summary (Number of …) | MAPK |
|---|---|
| Nodes | 22 |
| (optimized) Parameters / Interactions | 32 |
| Logical gates | 1 |
| Inputs | 8 |
| Outputs | 7 |
| Experimental conditions | 25 |
| Optimisation time | 1.1 seconds |
The input files for this study are provided as Excel (.xlsx) files. Model fitting should be completed in a few seconds on a standard desktop computer. In our hands, the optimal fitting cost is 6.3025.