-
Notifications
You must be signed in to change notification settings - Fork 19
Counting isomiRs
Much miRNA expression profiling uses the read counts of all isoforms (isomiRs) of the canonical mature sequence summed together. While this may be appropriate for many cases it is also useful to consider the isoforms separately. This is true for several reasons:
- you only wish to count the exact canonical sequence.
- if the canonical mature sequence in miRBase is not even common in the tissue or samples you are studying and you wish to know this information.
- your samples contain two isoforms that differ such that one will not be detected in (for example) a PCR assay, giving an inconsistent result.
- one or more isoforms can better distinguish between sets of samples (i.e. control and disease) than the entire sum of counts.
Sequence variants include 5’ and 3’ trimming and extension, non-templated additions (enzymatically addition of a nucleotide to the 3’ end, i.e. adenylation, uridylation). A single read can have more than one modification so sRNAbench provided a useful non-redundant hierarchical method of classifying isomiRs. See reference below. A similar scheme has been used by Loher et al. The sRNAbench scheme is illustrated below:
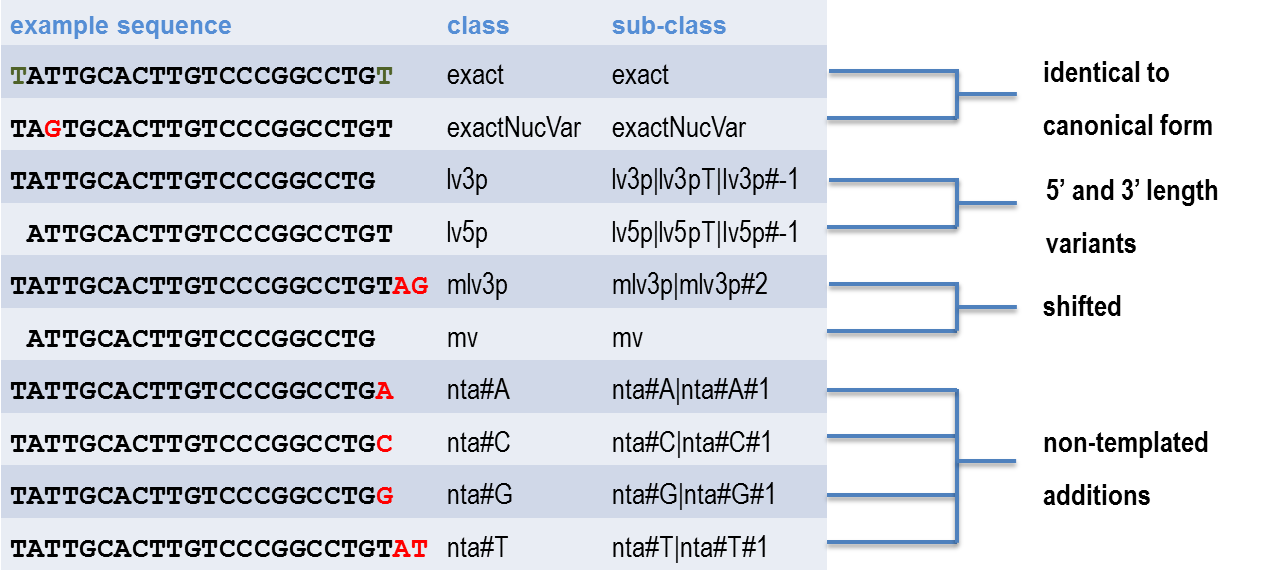
We have implemented the sRNAbench naming scheme. isomiR data is output by default when the map_mirbase method is called.
If required, you can explicitly call the routine to count isomirs, assuming you have already aligned to the mirbase sequences and have a sam file. You should also provide the original read counts loaded from the csv file which is always created when the reads are collapsed.
smrna.count_isomirs(samfile, truecounts, species='bta')
- Barturen, G., Rueda, A., … Hackenberg, M. (2014). sRNAbench: profiling of small RNAs and its sequence variants in single or multi-species high-throughput experiments. Methods in Next Generation Sequencing, 1(1), 21–31.
- Telonis, A.G. et al., 2015. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Research, 43(19), pp.9158–9175.
- Loher,P., Londin,E.R. and Rigoutsos,I. (2014) IsomiR Expression Profiles in Human Lymphoblastoid Cell Lines Exhibit Population and Gender Dependencies. Oncotarget, 5, 8790–8802.